Abstract
Archaeal cell division cycle protein 6 (Cdc6)/Origin Replication Complex subunit 1 (Orc1) proteins share sequence homology with eukaryotic DNA replication initiation factors but are also structurally similar to the bacterial initiator DnaA. To better understand whether Cdc6/Orc1 functions in an eukaryotic or bacterial-like manner, we have characterized the interaction of two Cdc6/Orc1 paralogs (mthCdc6-1 and mthCdc6-2) with the replication origin from Methanothermobacter thermoautotrophicus. We show that while both proteins display a low affinity for a small dsDNA of random sequence, mthCdc6-1 binds tightly to a short duplex containing a single copy of a 13 bp sequence that is repeated throughout the origin. Surprisingly, sequence comparisons show that this 13 bp sequence is a minimized version of the Origin Recognition Box element found in many euryarchaeotal origins. Analysis of mthCdc6-1 mutants demonstrates that the helix–turn–helix motif in the winged-helix domain mediates the interaction with this sequence. Association of both mthCdc6/Orc1 paralogs with the duplex containing the minimized Origin Recognition Box fits to an independent binding sites model, but their interaction with longer DNA ligands is cooperative. Together, our data provide the first detailed biophysical characterization of the association of an archaeal DNA replication initiator with its origin. Our observations also indicate that the origin-binding properties of Cdc6/Orc1 proteins closely resemble those of bacterial DnaA.
INTRODUCTION
The ability to faithfully duplicate and partition genetic information is essential to all cells. This process commences when specialized factors known as initiators bind to distinct origin regions in the chromosome (1–5). Proper recognition of replication origins is crucial for controlling the timing of origin unwinding and for recruiting and assembling a functional replisome. Regulation of these events helps to prevent over-initiation and maintains appropriate chromosomal copy number (3,6–10).
In eukaryotes, DNA replication occurs at multiple origin sites and is controlled by a six-subunit assembly called the Origin Recognition Complex (ORC) (2,3,11,12). In contrast, replication initiation in bacteria is typically controlled by a single protein, DnaA, which binds repeated sequence elements known as DnaA boxes that lie within a single chromosomal origin, oriC (13–16). Although a few ORC subunits share certain structural features with DnaA, most notably a series of ATP-binding motifs, extensive homology does not exist between the two initiators. As a result, it has not been clear to what extent the mechanisms of replication initiation in eukaryotes are related to those of bacteria.
Insights into this relationship have arisen from the studies of the archaea, which possess hallmarks of both bacterial and eukaryotic replication systems. For example, several replication proteins have been identified in archaea that share significant sequence homology with those found in eukaryotes. The total number and complexity of these factors are often reduced, however, suggesting that archaea may use a simplified eukaryotic-like replication system (17–23). One of these conserved proteins is Cdc6/Orc1 (cell division cycle protein 6/ORC subunit 1), which shares homology with both eukaryotic Cdc6 and the C-terminal half of Orc1 (17,19–21,24,25). Although the overall function of Cdc6/Orc1 is unclear, recent studies have shown that this protein can bind DNA and the Mcm (Mini-chromosome maintenance protein) helicase, activities also exhibited by eukaryotic ORC and Cdc6 (26–30). In addition, pioneering findings from several groups have shown that like ORC, Cdc6/Orc1 can associate with replication origins in vivo and in vitro (27,31).
Despite these similarities, data have emerged to suggest that the mechanisms of replication initiation might also be mechanistically similar between bacteria and archaea. For example, many euryarchaeotal organisms, like most bacteria, only have a single origin of replication composed of small sequence-repeat regions that flank A/T-rich elements (Figure 1) (21,32). In bacteria, the sequence repeats often mark binding sites for the DnaA initiator protein, while the A/T-rich segments are DNA unwinding elements (DUEs) (33–35). Structural studies and sequence analyses also have revealed congruencies between archaeal and bacterial initiators. The ATPase modules of Cdc6/Orc1 and DnaA are very similar structurally and belong to the ATPases Associated with various cellular Activities (AAA+) protein superfamily (36–40). A recent phylogenetic study has reinforced this relationship and has assigned the DnaA and Cdc6/Orc1 proteins to their own clade (41). Both proteins also contain helix–turn–helix (HTH) domains appended C-terminally to their AAA+ regions, although the folds of the two HTH modules are different, forming a winged-helix domain (WHD) in Cdc6/Orc1 and a NarL/FixJ/TrpR domain in DnaA. Originally, the presence of a WHD in Cdc6/Orc1 suggested that it might help localize the protein to replication origins as does the C-terminal HTH fold of DnaA (14,39). Recent data have shown that this region of one of the Sulfolobus solfataricus Cdc6/Orc1 proteins does mediate such associations (31).
Figure 1.
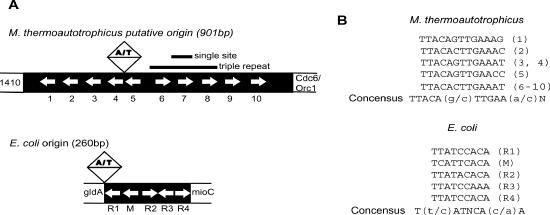
Comparison of M.thermoautotrophicus and E.coli origins of replication. (A) Schematic representation of the origins from M.thermoautotrophicus and E.coli. The M.thermoautotrophicus origin contains a central A/T-rich element flanked by 13 bp repeats spaced at approximately regular intervals along the origin (white arrows). In E.coli, the 260 bp origin contains five high-affinity A/T-rich DnaA boxes (white arrows) adjacent to an A/T-rich region. For both origins, repeats are present on the sense and antisense strand as indicated by the direction of the arrows. (B) The sequence of the repeats in both origins (32,64).
Taken together, these observations support the idea that Cdc6/Orc1 proteins may interact with and fire replication origins in a manner similar to DnaA. Nonetheless, there still exist several questions pertaining to this hypothesis. For example, it has not been determined whether Cdc6/Orc1 proteins interact with the short repeat sequences found in many archaeal origins in a manner analogous to the binding of DnaA to DnaA boxes nor has it been fully defined to what extent the HTH and ‘wing’ elements of the WHD respectively govern such associations. Similarly, it is not known whether Cdc6/Orc1 can associate with multiple binding sites in replication origins cooperatively, a property observed for DnaA (42–44). To date, in vitro studies have shown that Cdc6/Orc1 homologs from a variety of archaeons can bind non-specifically to DNA (28–30), and DNase I footprinting studies have revealed specific interactions between a S.solfataricus Cdc6/Orc1 homolog and a 36 bp sequence repeat in its origin of replication termed the Origin Recognition Box (ORB) (31). Interestingly, ORBs have not been identified in all archaea (31). For example, the predicted Methanothermobacter thermoautotrophicus origin (mth-ori) contains multiple 13 bp A/T-rich sequence repeats that are thought to be the specific binding sites for Cdc6/Orc1 (32).
In this study, we have examined the first step of archaeal replication initiation—the association of the initiator protein Cdc6/Orc1 with an origin of replication. Using filter binding assays we show that one of two M.thermoautotrophicus Cdc6/Orc1 paralogs, mthCdc6-1, specifically binds a short DNA duplex containing a 13 bp sequence element that is present in multiple copies throughout the M.thermoautotrophicus origin. In contrast, the second M.thermoautotrophicus Cdc6/Orc1 protein, mthCdc6-2, does not preferentially bind origin regions over other DNA sequences. Despite this difference, both Cdc6/Orc1 proteins associate cooperatively with DNA ligands longer than the duplex containing the 13 bp site. Mutagenesis studies further reveal that the WHD of mthCdc6-1, in particular a pair of conserved arginine residues in the HTH motif, is essential for the specific interaction between the protein and the origin repeat sequences. Moreover, we unexpectedly discovered that the mthCdc6-1 13 bp binding site constitutes a mini-ORB element as recognized by the Cdc6-1 ortholog of S.solfataricus (31). Together, these data provide the first biophysical characterization of an archaeal initiator with a replication origin, and show that some of the fundamental origin-binding properties of Cdc6/Orc1 proteins are conserved not only within several branches of archaea, but that they also reflect the types of interactions seen between bacterial origins and DnaA-class initiators.
MATERIALS AND METHODS
Materials
All chemicals were of reagent grade and were purchased from Fisher. M.thermoautotrophicus genomic DNA was a gift from Dr J. Reeve (Ohio State University). Restriction and modification enzymes were from New England Biolabs. pET15b vectors containing the mthCdc6/Orc1 genes were a gift from Dr Z. Kelman, and pET21a was obtained from Novagen. Escherichia coli XL1-Blue and BL21 CodonPlus(DE3)-RIL cells were purchased from Stratagene and Novagen, respectively. Deoxyribonucleotides (dNTPs), [γ-32P]ATP and NAP-25 columns were bought from Amersham, and oligonucleotides were synthesized by MWG Biotech. All DNA purification kits and Ni-nitrilotriacetic acid (NTA) resin were obtained from Qiagen. Hexahistidine (His6)-tagged Tobacco Etch Virus (TEV) protease was expressed and purified in our laboratory from the plasmid pRK793 (45). Nytran paper and HAWP-25 filters used in the binding assay were from VWR International and Millipore, respectively.
Cloning of mthCdc6-1 and mthCdc6-2
Sense primers were designed to encode a His6-tag followed by a TEV protease cleavage site upstream of the mthCdc6-1 and mthCdc6-2 genes, respectively. Antisense primers included a double stop codon sequence after the terminal residues of the genes. mthCdc6-1 was amplified from the plasmid pET15bmt1412 using primer A; 5′-GATATCCCTACAACTGAGAACCTTTATTTTCAGGGCATGAACATTTTTGATGAGATAGG and primer B; 5′-GCACGATGACGGGGATCCTCATTAAACACCCCAGAGTGAGTC. The PCR product was gel purified and then used as the template for a second round of PCR using primer B and the forward primer C; 5′-GCGTAACCTGCATATGCACCATCACCATCACCATGATTATGATATCCCTACAACTGAGAAC to add the N-terminal His-tag. This PCR product was cloned into the expression vector pET21a between the NdeI and BamHI sites (pSCCdc6-1). mthCdc6-2 was amplified from pET15bmt1599 using the primer D; 5′-GATATCCCTACAACTGAGAACCTTTATTTTCAGGGCATGAAAGGCGATAAGAGGG and primer E; 5′-CGTGTAACTGCCGGATCCTTATCAGAAGCCAGGGCACCTT. This PCR product was then amplified using primers C and E, and cloned into the NdeI and BamHI sites in pET21a (pSCCdc6-2). All mthCdc6-1 mutants were created in pSCCdc6-1 using overlap PCR mutagenesis (46). All constructs were sequenced at the UC Berkeley DNA Sequencing Facility.
Expression and purification of mthCdc6/Orc1 homologs and mthCdc6-1 variants
E.coli BL21-CodonPlus(DE3)-RIL cells were transformed with the plasmid pSCCdc6-1 and grown at 37°C in 2× YT media containing 30 μg/ml of chloramphenicol and 100 μg/ml of ampicillin. When the cells reached an OD600 of 0.6, protein expression was induced using 1 mM isopropyl-1-thio-β-d-galactopyranoside and the cells were grown for five additional hours. Cells were harvested by centrifugation and resuspended in 2 ml lysis buffer [40 mM Tris–HCl (pH 8.0), 500 mM NaCl, 20% glycerol, 1 mM 2-mercaptoethanol and 10 mM imidazole] per gram of cells and supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A and 2 μM leupeptin). Cells were lysed by the addition of lysozyme to 0.05 mg/ml, followed by sonication using a Misonix Sonicator 3000. DNase I, RNase A and MgCl2 were added to the lysate to a final concentration of 10 μg/ml, 5 μg/ml and 6 mM, respectively, and incubated at 22°C for 10 min. The lysate was then centrifuged twice for 30 min at 10 000 g and the supernatant was loaded onto a superflow Ni-NTA column pre-equilibrated with lysis buffer at 22°C. The column was washed with 10 column volumes of wash buffer (lysis buffer containing 75 mM imidazole), and mthCdc6-1 was then eluted in elution buffer [40 mM Tris–HCl (pH 8.0), 400 mM KOAc, 20% glycerol and 125 mM imidazole plus protease inhibitors and 1 mM 2-mercaptoethanol]. Fractions containing mthCdc6-1 were pooled and exchanged into elution buffer without imidazole using a NAP-25 column. The N-terminal His6-tag was removed from mthCdc6-1 by incubating the protein with TEV protease (at a ratio of 10:1 mthCdc6-1 to protease) at 30°C for 2 h. Uncleaved mthCdc6-1 and the His6-tagged protease were subsequently removed by loading the cleaved protein solution onto a Ni-NTA column pre-equilibrated with imidazole-free elution buffer. Pure mthCdc6-1 was collected in the flow-through, flash frozen in liquid nitrogen, and stored at −80°C. mthCdc6-2 and all mthCdc6-1 mutants were expressed and purified as described for mthCdc6-1.
Circular dichroism (CD) studies of mthCdc6-1 wild-type and variant proteins
Purified wild-type and mutant mthCdc6-1 proteins were buffer exchanged into phosphate buffer (20 mM potassium phosphate, pH 7.0) for CD studies. Spectra were measured using an Aviv 62DS spectrometer in a cuvette with a 0.5 cm path length at 25°C. Data were recorded from 300 to 200 nm, at 1 nm intervals and each data point was averaged for 1 s. The molar ellipticity (Θ) was calculated using the following equation; Θ = ΨMW/104Lcn (47), where Ψ (degrees) is the observable signal, MW is the molecular weight, L is the path length (cm), c is the protein concentration (g/ml) and n is the number of amino acids. Θ was then plotted against wavelength for each of the mutants and compared to wild-type mthCdc6-1.
Sequences of DNA oligonucleotides used in the filter binding assay
The sequence of the sense strand for the single site, random site and triple repeat are listed below. The 13 bp repeats when present are underlined.
Single site: 5′-CATGGTCAGATTACACTTGAAATGGATGTCTCCC.
Random site: 5′-CAGGATCAGACAGTTGAGTTCTAGTATGTCTACC.
Triple repeat: 5′-cgagatcaattaatactaacTTACACTTGAAATgaatgtctcccttacaggtcatcagaaccatggtcagaTTACACTTGAAATggatgtctcccacatctagccatgaatcagagaactggataaggaacagcaggattttTTACACTTGAAATtcatccctcatgaattcccatcgag.
DNA substrates for filter binding assays
The short single- and random-site duplex oligonucleotides were prepared by annealing their respective sense and antisense strands. The sense strand was labeled with [γ-32P]ATP using T4 polynucleotide kinase (48), mixed with an equal concentration of its complementary strand in 10 mM HEPES (pH 7.5), 5 mM NaCl and 5 mM MgCl2, at 100°C for 5 min, and cooled overnight to 22°C. The M.thermoautotrophicus putative origin of replication was amplified from the genome and cloned into pUC19 (pUC19mth-ori), and then used as the template for amplification of the triple repeat. 32P-labeled forward primers were used to produce radiolabeled products, and these PCR products were gel-purified. The same procedure was used to generate radiolabeled random 282mer from the plasmid pJES38.
Filter binding assay
Preliminary filter binding experiments between mthCdc6-1 and 1 nM of 32P single or random site were performed in the imidazole-free elution buffer plus 2 mM MgCl2, 3 mM DTT and 0.1 mg/ml BSA. Initial data from these assays showed that mthCdc6-1 binds tighter to the single site than to the random site (data not shown). However, under these conditions <45% of DNA was bound at saturating protein concentrations. We therefore screened buffers varying in the type and concentration of salt, buffering agent and pH to determine optimum binding conditions for the formation of the protein–DNA complex. From this analysis, we chose a solution containing 40 mM bis–Tris propane (pH 7.0), 70 mM KOAc, 2 mM MgCl2, 3 mM DTT, 20% glycerol and 0.1 mg/ml BSA (hereafter referred to as binding buffer) in which >65% of single site is bound to mthCdc6-1 at saturation but <20% interacts with the random site at the same protein concentration (data not shown). This buffer was then used for all subsequent experiments presented in this study. mthCdc6-1, mthCdc6-2 and all mthCdc6-1 mutants were buffer exchanged into binding buffer prior to carrying out binding assays.
To perform the assays, protein (0–4 μM) was mixed with 1 nM of 32P-labelled DNA in a total volume of 0.5 ml, incubated at 37°C for 1 h and then passed through nitrocellulose filters (pre-soaked in binding buffer) under vacuum controlled by a regulator at ∼300 mbar. Filters were then washed three times with 0.5 ml of binding buffer. The radioactivity retained on the filter was measured using a Beckmann LS6500 liquid scintillation counter.
Analysis of binding data
Binding curves were repeated in triplicate and the plots show the average and standard deviations from the three data sets. The data were analyzed using KaleidaGraph version 3.6 (Synergy Software), and where appropriate were fitted to a simple independent binding sites model (49):
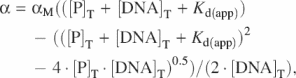
where α is the percentage of DNA bound, αM is the percentage of DNA bound at saturation, [P]T is the total protein concentration, [DNA]T is the total concentration of DNA and Kd(app) is the apparent dissociation constant. This equation describes the association of a protein with a ligand at equilibrium when each binding event is entirely independent and in such cases can be used to derive the dissociation constant for the interaction. Sigmoidal curves were fitted to the Hill equation and used to determine the Hill coefficient n; ![]() (49).
(49).
RESULTS
Selection of DNA targets for mthCdc6/Orc1 binding studies
To better understand the physical basis for the initiation of DNA replication in archaea, it is important to first clearly define and characterize molecular interactions that occur during the formation of a pre-replicative complex. Such studies have only recently become possible through the identification of replication proteins and origins from a variety of archaeal genomes (21,31,32,50–52). Here, we analyze the relationship between the archaeal Cdc6/Orc1 protein and the predicted origin of replication from M.thermoautotrophicus. This organism was selected for our studies since it has provided an excellent system for prior functional and structural studies of replication proteins (26,53–58), and contains at least one origin in its chromosome (32).
The candidate M.thermoautotrophicus origin (mth-ori) is bordered on one side by a gene coding for one of two Cdc6/Orc1 homologs (mthCdc6-1) (32). Curiously, this type of arrangement is also seen between DnaA and oriC of many bacteria, such as Streptomyces lividans (59) and Thermus thermophilus (34). The 901 bp mth-ori contains a central A/T-rich element flanked on either side by a series of symmetrically distributed 13 bp repeats of the consensus sequence TTACA(g/c)TTGAA(a/c)N (Figure 1). To characterize the association of mthCdc6-1 with these repeats, we used a radiolabeled filter binding assay to screen a number of different target DNAs, including a single 13 bp copy of the most commonly repeated sequence in the origin (single site), and a section of the origin containing three of these sequences (triple repeat; for sequences see Materials and Methods). As controls, we probed the binding of mthCdc6-1 to a random duplex of equal length to the single site (random site), and to a non-origin DNA that is slightly longer than the triple repeat (282mer). We also investigated the ability of the other mthCdc6/Orc1 paralog (mthCdc6-2) from M.thermoautotrophicus to bind these DNAs. Unlike mthCdc6-1, the mthCdc6-2 open reading frame is not located near the origin.
mthCdc6-1 specifically binds to the single site
We first examined whether mthCdc6-1 binds to the single site (Figure 2A, open circles). To quantitatively analyze the data for this interaction, the binding curve was fitted to an independent binding sites equation [see Methods and Materials, (49)]. Although the filter binding assay is a well-established method for quantitatively probing protein–ligand interactions, this technique does perturb the equilibrium during the washing steps. As a consequence, the dissociation constants (Kd) reported here are apparent Kds [Kd(app)]. Fitting the data for mthCdc6-1 in this way reveals that the affinity of this protein for the single site is 230 ± 30 nM (Table 1).
Figure 2.
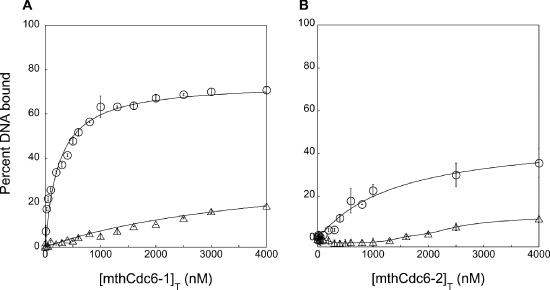
Binding of the mthCdc6/Orc1 homologs to the single and random sites. Binding data for single (open circles) and random (open triangles) site DNA oligonucleotide duplexes are shown for mthCdc6-1 (A) and mthCdc6-2 (B). The open points on the plot are the averaged values from three independent binding experiments with the standard deviation indicated by the error bars. For mthCdc6-1, the solid lines through the data are the best fit to the independent binding sites model (49). Data for mthCdc6-2 binding to the single site also fit to the independent binding sites model, while a smooth line is drawn through the points for the interaction between mthCdc6-2 and the random site.
Table 1. Apparent dissociation constants for the interaction between mtCdc6/Orc1 proteins and short oligonucleotides.
| mthCdc6/Orc1 protein | DNA | Kd(app) (nM) |
|---|---|---|
| mthCdc6-1 wt | Single site | 230 ± 30 |
| mthCdc6-1 wt | Random site | 9200 ± 3400 |
| mthCdc6-1 S330A/S332A | Single site | 2300 ± 180 |
| mthCdc6-1 N341A/E342A | Single site | 350 ± 70 |
| mthCdc6-1 R334A/R335A | Single site | No binding detected |
| mthCdc6-1 R334A | Single site | No binding detected |
| mthCdc6-1 R335A | Single site | 960 ± 190 |
| mthCdc6-1 S355A | Single site | 210 ± 30 |
| mthCdc6-1 K360A | Single site | 530 ± 40 |
| mthCdc6-1 R362A | Single site | 540 ± 30 |
| mthCdc6-1 R358A | Single site | 510 ± 40 |
| mthCdc6-2 wt | Single site | 1400 ± 230 |
| mthCdc6-2 wt | Random site | No binding detected |
To test whether the association between mthCdc6-1 and the single site is sequence-specific, we next assayed the binding of the protein to a random dsDNA of equal length to the single site. Under the same conditions, mthCdc6-1 binds the random site with much lower avidity [Kd(app) = 9200 ± 3400 nM; Figure 2A, open triangles]. The error on the fit to these data is larger than the single site, since the binding does not reach saturation in the protein concentration range that we have analyzed. Comparing these data to that for the single site shows that the affinity of mthCdc6-1 for the single site is 40-fold higher than for a DNA of random sequence. Overall, this observation is consistent with the hypothesis that the repeat sequences of the mth-ori are specific binding elements for the mthCdc6-1 protein.
mthCdc6-2 has a lower affinity for the single site than mthCdc6-1
To determine whether the sequence-specific binding we observed for the single site is unique to mthCdc6-1, we also probed the affinity of mthCdc6-2 for this sequence. These binding data, again fit to an independent binding sites model, and are shown in Figure 2B (open circles). The Kd(app) from this fit is 1400 ± 230 nM, an affinity that is moderately lower than seen for mthCdc6-1 (Table 1). The binding profiles for mthCdc6-2 also show that this protein does not bind the same total amount of DNA at saturation as mthCdc6-1.
To determine whether the interaction of mthCdc6-2 with the single site is sequence-specific, we investigated the ability of mthCdc6-2 to bind the random site. Unlike the curve produced for the binding of this protein to the single site, the data for this interaction do not fit a simple hyperbolic function and thus cannot be described by the independent binding sites model (Figure 2B, open triangles). Moreover, binding of mthCdc6-2 to the random DNA is only detected when the protein concentration is 1000-fold higher than the DNA concentration; even here, however, mthCdc6-2 does not bind more than 10% of the total DNA. Taken together, these observations suggest that while mthCdc6-2 may possess some affinity for the 13 bp repeats within the mth-ori, these regions serve as more robust recognition elements for the mthCdc6-1 protein.
The WHD of mthCdc6-1 is responsible for DNA binding
We next wanted to identify the region of mthCdc6-1 responsible for binding to the single site, and to define the relative contributions of key amino acid residues to DNA binding. Structural studies of Pyrobaculum aerophilum Cdc6/Orc1 revealed that the protein contains a WHD, which was proposed to act as an origin binding element that contacts DNA through the HTH and wing motifs (39). Using homology modeling as a guide, we made double and single alanine substitutions at polar and charged amino acids in the second, or ‘recognition’, helix of the HTH and in the wing of the mthCdc6-1 WHD (Figure 3). All mthCdc6-1 variants were purified to near homogeneity as for wild-type (Figure 4A). To determine whether any of the mutations had caused substantial structural changes that might alter their binding properties, we analyzed the purified variants by far ultraviolet CD. In all instances, the spectra for the mutants were found to be the same as wild-type mthCdc6-1; a comparison of the most severe mutant R334A/R335A and mthCdc6-1 is shown in Figure 4B. The WHD mutants were then assayed for binding to the single site, and their affinities compared to wild-type mthCdc6-1.
Figure 3.
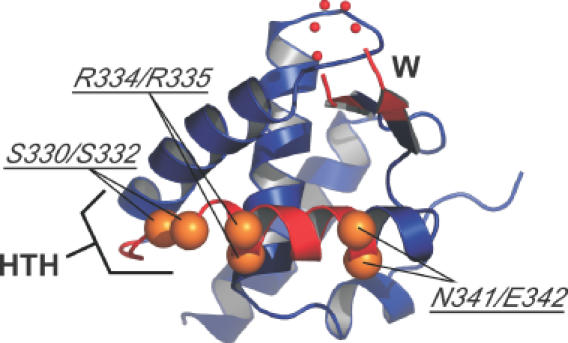
Position of mutations in the WHD. Ribbon representation of P.aerophilum Cdc6/Orc1 WHD, with the HTH and wing (W) regions labeled and shaded red (39). The position of the M.thermoautotrophicus recognition helix mutations in the WHD based on homology modeling are shown in orange. The location of wing mutations are not shown since this region of the domain is disordered in the P.aerophilum structure (red dots).
Figure 4.
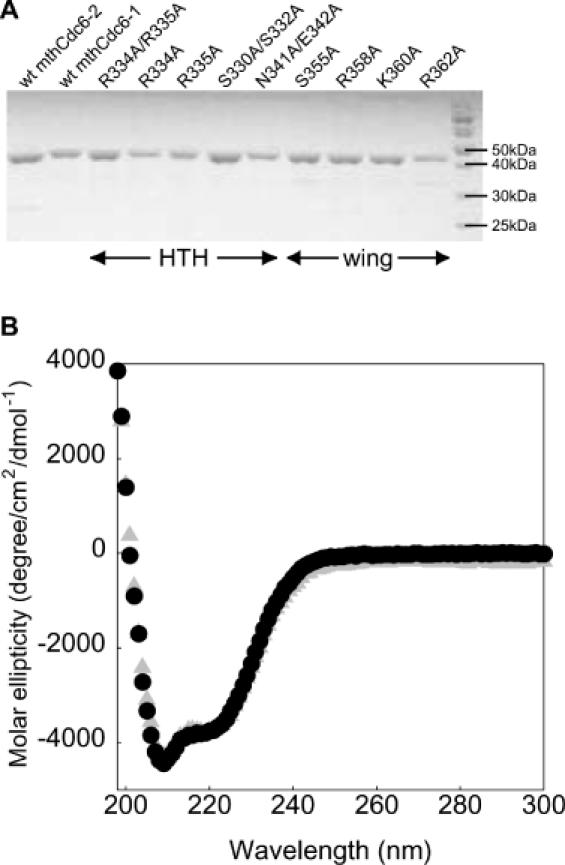
SDS–PAGE analysis and CD spectra of mthCdc6/Orc1 variants. (A) SDS polyacrylamide gel (12%) of mthCdc6-1 variants and wild-type mthCd6-1 and mthCd6-2 used in filter binding studies. (B) CD spectra wild-type mthCdc6-1 (gray triangles) compared to R334A/R335A (black circles).
Mutated residues in the recognition helix had a range of effects on binding to the single site (Figure 5A). For example, the N341A/E342A double mutant binds to the single site with an affinity only 1.5 times weaker than wild-type, although the total amount of DNA bound is ∼15% less than wild-type at the end of the titration. In contrast, mutating both Ser330 and Ser332 to alanine leads to a more drastic decrease in affinity for the single site but binds close to the same total amount of DNA as wild-type [Kd(app) = 2300 ± 180 nM; Table 1]. The most dramatic effect was observed for the R334A/R335A double mutant, which displayed no significant binding to the single site even at a 4000-fold molar excess of protein (Figure 5A). To further map the relative contributions of Arg334 and Arg335, we made single alanine substitutions of these residues and tested each for binding to the single site (data not shown). R334A showed no significant affinity for the single site, indicating that this residue is key for mthCdc6-1 binding to DNA, while R335A exhibited a moderate affinity for this sequence that was approximately four times weaker than wild-type protein (Table 1).
Figure 5.
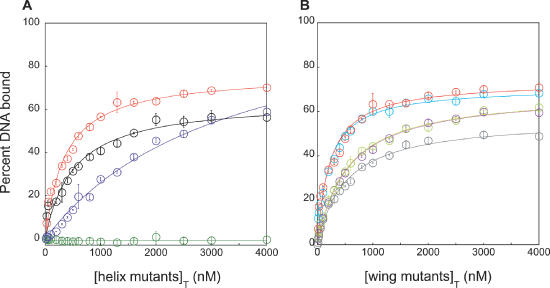
Binding of mthCdc6-1 WHD mutants to the single site. (A) Binding curves for the recognition helix mutants N341A/E342A (black), S330A/S332A (dark blue), R334A/R335A (dark green) and for comparison wild-type protein (red). All curves have been fitted to the independent binding sites model. (B) Binding curves for the wing mutants S355A (light blue), K360A (light green), R362A (purple) and R358A (gray) have also been fitted to the independent binding sites model.
Binding curves for mutants with substitutions in the wing region are shown in Figure 5B. As with the HTH mutants, each of the wing variants shows hyperbolic binding profiles and fits well to an independent binding-sites model. The effects of the wing mutations, however, appear to be much more subtle (Table 1). For example, the binding curve for S355A is virtually superimposable with wild-type (Figure 5B, light blue versus red) and similarly shows a nearly identical Kd(app) for the single site (210 ± 30 nM; Table 1). Similarly, the Kd(app)s for R358A (gray), R362A (purple) and K360A (light green), are only 2.5-fold weaker than wild-type (Table 1). As seen with the N341A/E342A and S330A/S332A variants, the R358A, K360A and R362A mutants all bind less DNA than wild-type mthCdc6-1 at the highest ratio of protein to DNA tested here. The fits to the binding data, however, suggest that if the protein concentration was raised significantly above 4 μM most of these mutants would eventually become saturated with the same amount of DNA as wild-type mthCdc6-1.
Taken together, these data indicate that the HTH amino acids Ser330, Ser332, Arg334 and Arg335 appear to be directly involved in contacting the single site. Changes in the wing do not appear to have a substantial impact on DNA binding, although we cannot rule out a more significant role for other residues in the wing that we were unable to examine here (e.g. the wing mutation R356A as well as a deletion of the wing from residues 356 to 362 both failed to express soluble protein). Regardless, given the striking effect of the R334A/R335A mutation, our results clearly show that the mthCdc6-1 WHD is the primary region responsible for binding to the 13 bp repeats in the mth-ori.
mthCdc6-1 binds cooperatively to DNAs longer than the single site
The origins of E.coli and S.lividans contain a number of closely spaced, repeating elements that are bound cooperatively by DnaA monomers (16,42–44). The origin of M.thermoautotrophicus also contains closely spaced multiple repeats, and gel-filtration analysis of mthCdc6-1 shows that like DnaA this protein is a monomer (E. Cunnigham and J. Berger, unpublished data). Therefore, it is possible that mthCdc6-1 binds to multiple sites in a replication origin in a manner similar to DnaA. To test this hypothesis, we analyzed the interaction between mthCdc6-1 and a 181 bp segment of the origin containing three identical copies of the most commonly repeated sequence [hereafter referred to as the triple repeat (Figure 6A)]. Interestingly, the binding curve for this substrate when fitted to the simple binding-sites model shows a higher affinity [Kd(app) of 5.5 ± 0.6 nM (fit not shown)] than would be expected if the proteins were binding the sites independently. Indeed, the affinity of mthCdc6-1 for the triple repeat is ∼40-fold greater than that observed for the single site, and 10-fold higher than that expected for binding DNA containing three independent sites. These findings indicate that the association of mthCdc6-1 with the triple repeat is more complex when compared to the single site. Since the three sites in the repeat are spaced relatively close to each other (37–58 bp apart), we hypothesized that mthCdc6-1 might be binding cooperatively to the triple repeat. To examine this further, we compared fits of the data to both the Hill equation and the independent binding sites model. The data fit best to the Hill equation [Figure 6A, (49)] which becomes more apparent when plotted on a semi-log scale (Figure 6A, inset). The Kd(app(0.5)) for the interaction is ∼5 nM (corresponding to the protein concentration at which 50% half saturation is reached) and the Hill coefficient from the fit is 1.6 ± 0.10, consistent with the idea that the interaction is positively cooperative (n > 1).
Figure 6.
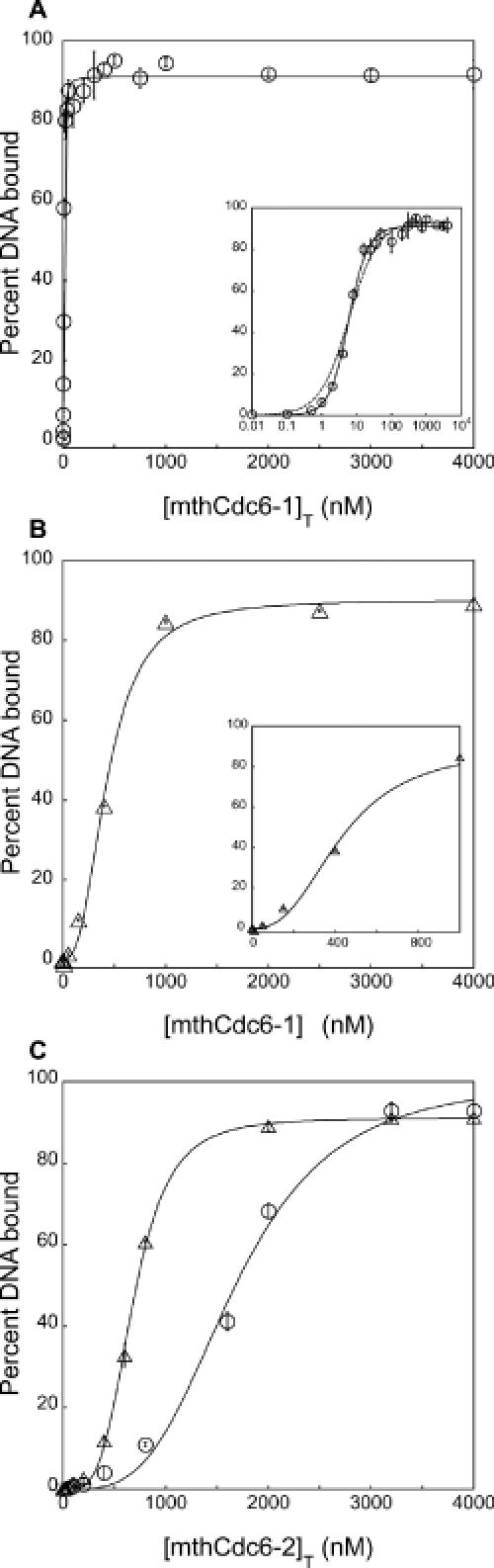
Binding of mthCdc6-1 and mthCdc6-2 to the triple repeat and 282mer. Binding of mthCdc6-1 to the triple repeat (A) and random 282mer (B) fitted to the Hill equation (49). In (A) the inset shows the binding of mthCdc6-1 to the triple repeat on a semi-log plot fitted to the Hill equation (solid line) and independent binding sites model (dashed line). The inset in (B) is an enlarged view of the data for the binding of mthCdc6-1 to the random 282mer at protein concentrations <1000 nM. (C) Binding of mthCdc6-2 to the triple repeat (open circles) and 282mer (open triangles) have also been fitted to the Hill equation.
Because the physiological temperature of M.thermoautotrophicus is 65–70°C, we wanted to examine whether this elevated temperature could either alter the Kd(app) and increase/decrease the cooperative binding we observe for mthCdc6-1 to the triple repeat. We therefore carried out binding assays between mthCdc6-1 and the triple repeat at 65°C. This experiment showed that the binding affinity of the protein is almost the same as we observed at 37°C and the plots produced at the two temperatures are essentially superimposable (data not shown). Thus, temperature does not appear to significantly affect the binding affinity or cooperative nature of the association between mthCdc6-1 and its target origin sites.
As a second control for this study, we also probed the binding of mthCdc6-1 to a random 282 bp DNA sequence (random 282mer). At first glance, the shape of the binding curve produced for this interaction is hyperbolic (Figure 6B). However, upon closer inspection we found that <1% of this DNA is bound at 50 nM protein or less (Figure 6B, inset), whereas ∼80% of the triple repeat is bound by mthCdc6-1 at this concentration (Figure 6A). As the protein concentration is raised above 50 nM, the percentage of random 282mer bound to protein dramatically increases, producing a sigmoidal binding curve that is diagnostic of positive cooperativity. Accordingly, the binding data obtained for the random 282mer fit best to the Hill equation, producing a Hill coefficient with a value of 2.6 ± 0.31 and Kd(app(0.5)) of 470 nM.
It is interesting to note that the interaction between mthCdc6-1 and the triple repeat does not produce a binding curve with as clear a sigmoidal shape or as great a Hill coefficient as seen for the 282mer (Figure 6B). Binding assays using the full-length mth-ori region and a random 900mer gave similar results (data not shown). It is possible that the high affinity of mthCdc6-1 for DNAs containing three or more 13 bp repeats may obscure the pre-ascent phase of the curve over the lowest protein concentrations assayed here, whereas the weak affinity of mthCdc6-1 for the random 282mer and 900mer at low-protein concentrations enables the more dramatic sigmoidal profiles to be observed. Regardless, all mthCdc6-1 binding data for long DNA ligands were fit best by the Hill equation and produce Hill coefficients >1, consistent with a positively cooperative binding mechanism for the interaction between this protein and DNA.
mthCdc6-2 also exhibits cooperative binding behavior
To determine whether the cooperative DNA-binding interactions observed for mthCdc6-1 are specific to this protein, we investigated the association of mthCdc6-2 with the triple repeat and random 282mer. Remarkably, binding of mthCdc6-2 to both substrates also revealed sigmoidal behavior (Figure 6C). These data, when fit to the Hill equation produced Hill coefficients >3.5. Inspection of the curves shows that mthCdc6-2 has an even lower affinity for the triple repeat and random 282mer than mthCdc6-1 (1680 and 720 nM, respectively). Moreover, even though mthCdc6-2 binds the single site tighter than the random site (Figure 2B), it does not preferentially bind the triple repeat over the random 282mer (Figure 6C, open circles versus open triangles). Similar binding behavior was also observed for the interaction between mthCdc6-2 and both the entire mth-ori region and a random 900mer (data not shown). Together, these data indicate that both Cdc6/Orc1 proteins of M.thermoautotrophicus cooperatively associate with extended DNA sequences, but that the repeat sequences in the mth-ori are principally recognized by mthCdc6-1.
DISCUSSION
All cellular organisms use a defined set of proteins to initiate the replication of chromosomal DNA. In bacteria and eukaryotes, this role is fulfilled by DnaA and ORC, respectively (3–5,12,13,16,60). For archaea, genomic sequence data show that these organisms possess proteins homologous to those of eukaryotes, suggesting that they may initiate replication in a similar manner (17,19–21,24,25). However, biochemical and structural data indicate that there may also be significant functional, and perhaps evolutionary overlap between the replication initiation mechanisms of bacteria and archaea (31,39,40). In this study, we have attempted to better explore these issues by characterizing the associations of two Cdc6/Orc1 paralogs with the predicted origin of replication from M.thermoautotrophicus.
It is known that certain archaeons contain replication origins bearing short repeat sequences whose general organization is similar to that of bacterial origins (21,32). A prediction from this finding is that Cdc6/Orc1 may preferentially interact with these sequences much like DnaA binds DnaA boxes. This hypothesis was recently validated by studies of Cdc6/Orc1 homologs from S.solfataricus, which were shown to bind 36 bp sequence elements known as ORBs in the predicted origin of replication. On the basis of sequence analyses, ORBs are predicted to exist in a number of archaea including Pyrococcus abyssi, Aeropyrum pernix, Archaeoglobus fulgidus and the Halobacterium species NRC-1 (31). For M.thermoautotrophicus, Forterre and colleagues have proposed that multiple copies of a 13 bp sequence in its predicted replication origin mark binding sites for Cdc6/Orc1 (32). Although no ORB sequences were initially identified in the M.thermoautotrophicus genome (31), we found upon inspection of the single site that the 13 bp sequence repeat is actually a mini-ORB of the consensus sequence (t/c)TNCANNNGAA(a/c) (Table 2).
Table 2. Consensus mini-ORB elements (t/c)TNCANNNGAA(a/c) in archaeal genomes.

Many archaeal genomes contain more than one Cdc6/Orc1 open reading frame (50,51,61). Because the gene encoding mthCdc6-1 is located near the putative origin, an arrangement seen for the initiators and origins of many archaea and bacteria, we felt it likely that the mthCdc6-1 protein is an origin binding protein. Consistent with this hypothesis, we show that mthCdc6-1 specifically binds to these 13 bp mini-ORB sequences over other DNAs. However, as a control we also studied the binding affinity of mthCdc6-2. These experiments show that the association of mthCdc6-2 with a single 13 bp binding site in the origin is 6-fold weaker than mthCdc6-1. Moreover mthCdc6-2 binds multi-site origin segments with an affinity comparable to that observed for random DNA sequences of similar length, while mthCdc6-1 binds multi-site origin regions extremely tightly (Figure 6A). These data indicate that there are defined roles for the two Cdc6/Orc1 proteins in M.thermoautotrophicus. For mthCdc6-1, that function appears to be the specific recognition of replication origins. In contrast, the role of mthCdc6-2 is less clear. Recent two-hybrid studies by Kelman and colleagues suggest that mthCdc6-2 uses the wing in the WHD to interact with the M.thermoautotrophicus Mcm, recruiting the helicase to mthCdc6-1 coated origins (30). However, at this stage it is not apparent whether mthCdc6-2 may have other roles in replication as well.
During origin recognition, archaeal Cdc6/Orc1 proteins have been proposed to contact DNA using a WHD located at the protein's C-terminus (26,28,39). Consistent with this idea, Bell and colleagues recently showed that a double mutation in the WHD of a S.solfataricus Cdc6/Orc1 homolog renders the protein unable to contact the ORB-1 binding site (31). Studies have not been forthcoming however, on whether the WHD of Cdc6/Orc1 proteins are used for binding to archaeal origins that lack full-length ORB elements, or to what extent individual amino acids in the wing and HTH motifs contribute to DNA-binding affinity. To address these questions, we made a number of single and double point mutations in the wing and HTH motifs of the mthCdc6-1 WHD. Our binding data show clearly that residues in the recognition helix, in particular a pair of arginines (Arg334 and Arg335) located near the turn of the HTH, are extremely important for stabilizing mthCdc6-1•DNA interactions (Figure 5A). In contrast, mutations in the wing have little or no effect on the affinity of mthCdc6-1 for the single site (Figure 5B). Thus, for mthCdc6-1 it appears that the HTH is more significant than the wing for regulating DNA affinity. This observation is consistent with the behavior of certain types of WHD proteins that use the HTH to bind sequence-specifically to the major groove of DNA, while the wing makes relatively non-specific contacts with the phosphodiester backbone (62).
These observations are also congruent with sequence comparisons, which have revealed that there are at least two major classes of archaeal Cdc6/Orc1 proteins (63,64). One of these classes, which includes mthCdc6-1, S.solfataricus Cdc6-1 and A. fulgidus Cdc6-1, contains the DNA binding motif φT/SXRRφD (where φ is any hydrophobic residue) in the turn and recognition helix of the WHD. In contrast, mthCdc6-2 belongs to a second class of WHDs that do not posses this sequence. Given that our results show that serine and arginine residues in the φT/SXRRφD motif are essential for DNA binding, this distinction may account for the different binding affinities of mthCdc6-1 and mthCdc6-2 for the single site. It is also interesting to note that S.solfataricus Cdc6-1 binds to ORB and mini-ORB elements (31). Based on these findings, it seems plausible that the φT/SXRRφD motif identified in all archaeal Cdc6-1 orthologs is required for the recognition of all mini-ORBs (t/c)TNCANNNGAA(a/c) present in replication origins.
Beyond the realm of the archaea, it is notable that several aspects of binding we observe for mthCdc6-1 to segments of the mth-ori are also reminiscent of the interaction between DnaA and bacterial origins. For example, the affinities of DnaA proteins for DnaA boxes range from 1 nM [E.coli (65)] to 2.7 μM [T.thermophilus (34)]. Our filter binding studies show that the association of mthCdc6-1 with a single mini-ORB site lies within this range of values [Kd(app) = 230 nM, Table 1]. Similarly, as seen in many bacterial origins, the mth-ori contains multiple copies of short sequence repeats that serve as initiator binding sites. We found that mthCdc6-1 binds cooperatively to these repeats, as observed for the association of DnaA with oriC (15,44,66). It is interesting to note that the cooperativity seen in DnaA arises from the self-association of AAA+ modules between initiator molecules bound to DNA (34,66–68). Given the high degree of structural similarity between the DnaA and Cdc6/Orc1 AAA+ domains, it seems plausible that this region might be likewise responsible for the cooperative binding of the archaeal proteins observed here. Future studies will be needed to further investigate the mechanism of cooperative binding by mthCdc6/Orc1.
In summary, our results bear on several facets of archaeal Cdc6/Orc1 function. We have shown that while mthCdc6-1 of M.thermoautrophicus can associate weakly with random DNA sequences, it specifically recognizes conserved, 13 bp mini-ORB sequence repeats that reside in the predicted origin of replication. This function contrasts with that of the mthCdc6-2 paralog, which we show can also bind duplex DNA, but displays little selectivity for the origin regions. In addition, we have demonstrated that mthCdc6-1 binds to mini-ORBs through the WHD, and that the principle determinants of this association map to a signature sequence motif in the HTH region that is conserved across numerous archaeal Cdc6/Orc1 orthologs; such properties are reminiscent of the association between the conserved HTH signature sequence element of bacterial DnaA and its target 9 bp DnaA boxes in oriC (43,69). Finally, our observations that mthCdc6/Orc1 paralogs display cooperative behavior when binding to extended DNA segments indicates that these proteins may likewise mimic DnaA by possessing an ability to assemble into a higher-order complex on the mth-ori. Overall, our data provide strong biochemical evidence that certain fundamental mechanisms by which archaeal and bacterial initiator proteins act to bind replication origins and control the onset of DNA replication are likely to be shared across these two domains of life.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful for the gift of the pET15bmtCdc6-1 and pET15bmtCdc6-2 plasmids from Dr Z. Kelman and for M.thermoautotrophicus genomic DNA from Dr J. Reeve. We thank Dr J. Liu for constructing pUC19Mtori, Dr R. Schekman for using his Beckman LS 6500 liquid scintillation counter and Dr S. Marqusee for using her Aviv 62DS spectrometer. Finally, we thank all members of the Berger laboratory for helpful discussions and assistance in the preparation of this manuscript. This research was generously funded by a grant from the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Seki T. and Diffley,J.F. (2000) Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl Acad. Sci. USA, 97, 14115–14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielinsky A.K., Blitzblau,H., Beall,E.L., Ezrokhi,M., Smith,H.S., Botchan,M.R. and Gerbi,S.A. (2001) Origin recognition complex binding to a metazoan replication origin. Curr. Biol., 11, 1427–1431. [DOI] [PubMed] [Google Scholar]
- 3.Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 4.Gerbi S.A., Strezoska,Z. and Waggener,J.M. (2002) Initiation of DNA replication in multicellular eukaryotes. J. Struct. Biol., 140, 17–30. [DOI] [PubMed] [Google Scholar]
- 5.Messer W. (2002) The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev., 26, 355–374. [DOI] [PubMed] [Google Scholar]
- 6.Skarstad K., Boye,E. and Steen,H.B. (1986) Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J., 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boye E., Lobner-Olesen,A. and Skarstad,K. (1988) Timing of chromosomal replication in Escherichia coli. Biochim. Biophys. Acta, 951, 359–364. [DOI] [PubMed] [Google Scholar]
- 8.Boye E., Lobner-Olesen,A. and Skarstad,K. (2000) Limiting DNA replication to once and only once. EMBO Rep., 1, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diffley J.F. (2001) DNA replication: building the perfect switch. Curr. Biol., 11, R367–R370. [DOI] [PubMed] [Google Scholar]
- 10.Ogura Y., Imai,Y., Ogasawara,N. and Moriya,S. (2001) Autoregulation of the dnaA–dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol., 183, 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowley A., Cocker,J.H., Harwood,J. and Diffley,J.F. (1995) Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J., 14, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePamphilis M.L. (2003) The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene, 310, 1–15. [DOI] [PubMed] [Google Scholar]
- 13.Kaguni J.M. (1997) Escherichia coli DnaA protein: the replication initiator. Mol. Cells, 7, 145–157. [PubMed] [Google Scholar]
- 14.Speck C., Weigel,C. and Messer,W. (1997) From footprint to toeprint: a close-up of the DnaA box, the binding site for the bacterial initiator protein DnaA. Nucleic Acids Res., 25, 3242–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigel C., Schmidt,A., Ruckert,B., Lurz,R. and Messer,W. (1997) DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J., 16, 6574–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messer W., Blaesing,F., Jakimowicz,D., Krause,M., Majka,J., Nardmann,J., Schaper,S., Seitz,H., Speck,C., Weigel,C. et al. (2001) Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading. Biochimie, 83, 5–12. [DOI] [PubMed] [Google Scholar]
- 17.Bernander R. (1998) Archaea and the cell cycle. Mol. Microbiol., 29, 955–961. [DOI] [PubMed] [Google Scholar]
- 18.Bernander R. (2000) Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol., 8, 278–283. [DOI] [PubMed] [Google Scholar]
- 19.Kelman Z. (2000) DNA replication in the third domain (of life). Curr. Protein Pept. Sci., 1, 139–154. [DOI] [PubMed] [Google Scholar]
- 20.Kelman L.M. and Kelman,Z. (2003) Archaea: an archetype for replication initiation studies? Mol. Microbiol., 48, 605–615. [DOI] [PubMed] [Google Scholar]
- 21.Myllykallio H., Lopez,P., Lopez-Garcia,P., Heilig,R., Saurin,W., Zivanovic,Y., Philippe,H. and Forterre,P. (2000) Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science, 288, 2212–2215. [DOI] [PubMed] [Google Scholar]
- 22.Giraldo R. (2003) Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev., 26, 533–554. [DOI] [PubMed] [Google Scholar]
- 23.Grabowski B. and Kelman,Z. (2003) Archeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol., 57, 487–516. [DOI] [PubMed] [Google Scholar]
- 24.Brown J.R. and Doolittle,W.F. (1997) Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev., 61, 456–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgell D.R. and Doolittle,W.F. (1997) Archaea and the origin(s) of DNA replication proteins. Cell, 89, 995–998. [DOI] [PubMed] [Google Scholar]
- 26.Grabowski B. and Kelman,Z. (2001) Autophosphorylation of archaeal Cdc6 homologues is regulated by DNA. J. Bacteriol., 183, 5459–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsunaga F., Forterre,P., Ishino,Y. and Myllykallio,H. (2001) In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl Acad. Sci. USA, 98, 11152–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Felice M., Esposito,L., Pucci,B., Carpentieri,F., De Falco,M., Rossi,M. and Pisani,F.M. (2003) Biochemical characterization of a CDC6-like protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem., 278, 46424–46431. [DOI] [PubMed] [Google Scholar]
- 29.Grainge I., Scaife,S. and Wigley,D.B. (2003) Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res., 31, 4888–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin J.H., Grabowski,B., Kasiviswanathan,R., Bell,S.D. and Kelman,Z. (2003) Regulation of minichromosome maintenance helicase activity by Cdc6. J. Biol. Chem., 278, 38059–38067. [DOI] [PubMed] [Google Scholar]
- 31.Robinson N.P., Dionne,I., Lundgren,M., Marsh,V.L., Bernander,R. and Bell,S.D. (2004) Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell, 116, 25–38. [DOI] [PubMed] [Google Scholar]
- 32.Lopez P., Philippe,H., Myllykallio,H. and Forterre,P. (1999) Identification of putative chromosomal origins of replication in Archaea. Mol. Microbiol., 32, 883–886. [DOI] [PubMed] [Google Scholar]
- 33.Hwang D.S. and Kornberg,A. (1992) Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem., 267, 23083–23086. [PubMed] [Google Scholar]
- 34.Schaper S., Nardmann,J., Luder,G., Lurz,R., Speck,C. and Messer,W. (2000) Identification of the chromosomal replication origin from Thermus thermophilus and its interaction with the replication initiator DnaA. J. Mol. Biol., 299, 655–665. [DOI] [PubMed] [Google Scholar]
- 35.Speck C. and Messer,W. (2001) Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J., 20, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- 37.Ogura T. and Wilkinson,A.J. (2001) AAA+ superfamily ATPases: common structure-diverse function. Genes Cells, 6, 575–597. [DOI] [PubMed] [Google Scholar]
- 38.Davey M.J., Jeruzalmi,D., Kuriyan,J. and O'Donnell,M. (2002) Motors and Switches: AAA+ machines within the replisome. Nature Rev., 3, 1–10. [DOI] [PubMed] [Google Scholar]
- 39.Liu J., Smith,C.L., DeRyckere,D., DeAngelis,K., Martin,G.S. and Berger,J.M. (2000) Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell, 6, 637–648. [DOI] [PubMed] [Google Scholar]
- 40.Erzberger J.P., Pirruccello,M.M. and Berger,J.M. (2002) The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J., 21, 4763–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.lyer M.L., Leipe,D.D., Koonin,E.V. and Aravind,L. (2004) Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol., 146, 13–31. [DOI] [PubMed] [Google Scholar]
- 42.Doran K.S., Helinski,D.R. and Konieczny,I. (1999) A critical DnaA box directs the cooperative binding of the Escherichia coli DnaA protein to the plasmid RK2 replication origin. J. Biol. Chem., 274, 17918–17923. [DOI] [PubMed] [Google Scholar]
- 43.Messer W., Blaesing,F., Majka,J., Nardmann,J., Schaper,S., Schmidt,A., Seitz,H., Speck,C., Tungler,D., Wegrzyn,G. et al. (1999) Functional domains of DnaA proteins. Biochimie, 81, 819–825. [DOI] [PubMed] [Google Scholar]
- 44.Majka J., Zakrzewska-Czerwinska,J. and Messer,W. (2001) Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J. Biol. Chem., 276, 6243–6252. [DOI] [PubMed] [Google Scholar]
- 45.Kapust R.B., Tozer,J., Anderson,D.E., Cherry,S., Copelend,T.D. and Waugh,D.S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng., 12, 993–1000. [DOI] [PubMed] [Google Scholar]
- 46.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 47.Campbell I.D. and Dwek,R.A. (1984) Biological Spectroscopy. The Benjamin/Cummings Publ. Co., Menlo Park, CA. [Google Scholar]
- 48.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Clarke A.R. (1996) In Engel,P.C. (ed.), Enzymology, Labfax. BIOS Scientific Publishers and Academic Press, Chapter 6, pp. 203–204. [Google Scholar]
- 50.Ng W.V., Kennedy,S.P., Mahairas,G.G., Berquist,B., Pan,M., Shukla,H.D., Lasky,S.R., Baliga,N.S., Thorsson,V., Sbrogna,J. et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. USA, 97, 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.She Q., Singh,R.K., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C.Y., Clausen,I.G., Curtis,B.A. and De Moors,A. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus. Proc. Natl Acad. Sci. USA, 98, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang R. and Zhang,C.T. (2003) Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun., 302, 728–734. [DOI] [PubMed] [Google Scholar]
- 53.Kelman Z., Lee,J.K. and Hurwitz,J. (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl Acad. Sci. USA, 96, 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelman Z., Pietrokovski,S. and Hurwitz,J. (1999) Isolation and characterization of a split B-type DNA polymerase from the archaeon Methanobacterium thermoautotrophicum deltaH. J. Biol. Chem., 274, 28751–28761. [DOI] [PubMed] [Google Scholar]
- 55.Kelman Z. and Hurwitz,J. (2000) A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum deltaH. J. Biol. Chem., 275, 7327–7336. [DOI] [PubMed] [Google Scholar]
- 56.Sriskanda V., Kelman,Z., Hurwitz,J. and Shuman,S. (2000) Characterization of an ATP-dependent DNA ligase from the thermophilic archaeon Methanobacterium thermoautotrophicum. Nucleic Acids Res., 28, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poplawski A., Grabowski,B., Long,S.E. and Kelman,Z. (2001) The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J. Biol. Chem., 276, 49371–49377. [DOI] [PubMed] [Google Scholar]
- 58.Yu X., VanLoock,M.S., Poplawski,A., Kelman,Z., Xiang,T., Tye,B.K. and Egelman,E.H. (2002) The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep., 3, 792–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jakimowicz D., Majka,J., Messer,W., Speck,C., Fernandez,M., Martin,M.C., Sanchez,J., Schauwecker,F., Keller,U., Schrempf,H. et al. (1998) Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology, 144, 1281–1290. [DOI] [PubMed] [Google Scholar]
- 60.Mendez J. and Stillman,B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol., 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith D.R., Doucette-Stamm,L.A., Deloughery,C., Lee,H., Dubois,J., Aldredge,T., Bashirzadeh,R., Blakely,D., Cook,R., Gilbert,K. et al. (1997) Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol., 179, 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gajiwala K.S. and Burley,S.K. (2000) Winged helix proteins. Curr. Opin. Struct. Biol., 10, 110–116. [DOI] [PubMed] [Google Scholar]
- 63.Singleton M.R., Morales,R., Grainge,I., Cook,N., Isupov,M.N. and Wigley,D.B. (2004) Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 64.Berquist B.R. and DasSarma,S. (2003) Archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bact. 185, 5959–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaper S. and Messer,W. (1995) Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem., 270, 17622–17626. [DOI] [PubMed] [Google Scholar]
- 66.Jakimowicz D., Majkadagger,J., Konopa,G., Wegrzyn,G., Messer,W., Schrempf,H. and Zakrzewska-Czerwinska,J. (2000) Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol., 298, 351–364. [DOI] [PubMed] [Google Scholar]
- 67.Weigel C., Schmidt,A., Seitz,H., Tungler,D., Welzeck,M. and Messer,W. (1999) The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol. Microbiol., 34, 53–66. [DOI] [PubMed] [Google Scholar]
- 68.Simmons L.A., Felczak,M. and Kaguni,J.M. (2003) DnaA protein of Escherichia coli: oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol., 49, 849–858. [DOI] [PubMed] [Google Scholar]
- 69.Fujikawa N., Kurumizaka,H., Nureki,O., Terada,T., Shirouzu,M., Katayama,T. and Yokoyama,S. (2003) Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res., 31, 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]


