Abstract
Large-scale gene expression analyses of microdissected primary tissue are still difficult because generally only a limited amount of mRNA can be obtained from microdissected cells. The introduction of the T7-based RNA amplification technique was an important step to reduce the amount of RNA needed for such analyses. This amplification technique produces amplified antisense RNA (aRNA), which so far has precluded its direct use for serial analysis of gene expression (SAGE) library production. We describe a method, termed ‘aRNA-longSAGE’, which is the first to allow the direct use of aRNA for standard longSAGE library production. The aRNA-longSAGE protocol was validated by comparing two aRNA-longSAGE libraries with two Micro-longSAGE libraries that were generated from the same RNA preparations of two different cell lines. Using a conservative validation approach, we were able to verify 68% of the differentially expressed genes identified by aRNA-longSAGE. Furthermore, the identification rate of differentially expressed genes was roughly twice as high in our aRNA-longSAGE libraries as in the standard Micro-longSAGE libraries. Using our validated aRNA-longSAGE protocol, we were able to successfully generate longSAGE libraries from as little as 40 ng of total RNA isolated from 2000–3000 microdissected pancreatic ductal epithelial cells or cells from pancreatic intraepithelial neoplasias.
INTRODUCTION
With the availability of histopathologically defined tumor progression models, it has become of key interest to identify important biological changes in the cell that are responsible for the development of the various tumor progression stages. Some insights came from the identification of activated oncogenes or inactivated tumor suppressor genes. To understand how these activated or inactivated tumor genes alter the complex cellular signaling and thus drive tumor progression, gene expression analyses of normal cells and their corresponding carcinoma precursor and carcinoma cells are crucial. Currently, gene expression analyses at the level of single candidate genes or broader expression analysis approaches, such as serial analysis of gene expression (SAGE) and microarray technology, are being used to reach this goal. It is expected that this knowledge will ultimately help not only to define new therapeutic target genes and prognostic gene expression patterns, but also to identify new (early) diagnostic markers.
To be able to analyze the expression profile of distinct histological cell types within a complex primary tissue, a method to isolate the cells of interest is needed. Microdissection using laser capture or manual techniques has been successfully used to produce such highly enriched cell preparations. Because the number of cells available through microdissection is limited in most instances, the amount of RNA that can be obtained from these samples is not sufficient for standard gene expression profiling protocols. In order to generate gene expression profiles from microdissected cells, it is necessary to amplify the amount of starting material, either by T7-based RNA amplification or by PCR amplification of the cDNA. The linear amplification of RNA by in vitro transcription, as introduced by Eberwine et al. (1), has been shown to result in less amplification bias than the PCR amplification of cDNA (2) and has been successfully applied for the gene expression profiling of microdissected cells using microarray technology (3–5).
An inherent limitation of microarrays is their ability to identify only predefined transcripts present on the array. SAGE, in turn, is a powerful alternative for performing expression analyses without prior knowledge of the genes to be identified (6). This technique creates gene expression profiles by generating libraries of short cDNA sequence tags, each tag representing an mRNA transcript. The gene expression profile is generated via concatenation, cloning and high-throughput sequencing of the tags (6). SAGE libraries have been widely used to study the genetic changes underlying the transformation from normal to cancer cells [for review see (7)]. A systematic analysis of publicly available SAGE tags has recently shown that a significant proportion of tags likely represent unknown genes or new isoforms of known genes, indicating that SAGE is truly complementary to current microarray technology (8). In addition, the recent introduction of longSAGE, a SAGE variant that produces 21 bp tags, instead of the 14 bp tags obtained from conventional SAGE libraries, further increases the reliability of the SAGE tag sequence to gene annotation. In addition, the 21 bp longSAGE tags can be used directly as primers for the isolation of novel transcripts using PCR technology (9).
Only few modifications of the current SAGE technology have so far been published that enable SAGE to be applied to <5 × 104 cells, a prerequisite for using SAGE for microdissected tissues. Two methods, PCR–SAGE (10) and SAGE-lite (11), rely on PCR amplification of the cDNA at the beginning of the SAGE procedure. A third method described by Schober et al. (12) requires an additional ditag PCR-amplification step. In all the cases, PCR amplification is likely to introduce a bias in the resulting expression profile (2,13).
The recently published small amplified RNA-SAGE approach (14) uses a modified protocol for T7-based RNA amplification that, in contrast to the Eberwine protocol, yields amplified sense RNA. The amplified RNA is then processed according to the standard SAGE protocol.
Here, we present a modification of the SAGE protocol which is the first to allow the direct use of amplified antisense RNA (aRNA) generated by means of the well-established and validated Eberwine protocol for SAGE library generation. Starting with as little as 40 ng of total RNA or 1.2 μg aRNA, we used a modified first and second strand cDNA synthesis strategy to successfully generate several longSAGE libraries from cultured cells and from 2000–3000 manually microdissected cells derived from normal pancreatic ducts, acinar cells and from PanIN lesions.
MATERIALS AND METHODS
Microdissection
Pancreatic resection specimens were immediately placed on ice, and tissue from the carcinoma, the peritumoral parenchyma and from the resection margin was removed, snap frozen and stored at −80°C. For the identification of acini, normal ducts and PanINs, 5 μm frozen sections prepared from tissue blocks of peritumoral pancreatic parenchyma, in particular tissue from the resection margins, were placed briefly in RNase-free ethanol (Merck, Darmstadt, Germany), stained with hematoxylin and eosin (H&E) and subsequently diagnosed by a pathologist (J. Lüttges and G. Klöppel). PanINs were classified according to published criteria (15). Tissue blocks containing the required cells were then serially sectioned. The slides were stained with H&E and immediately stored at −20°C. PanIN lesions from the stained 10 μm serial sections were manually microdissected under a microscope (BH2, Olympus, Wetzlar, Germany) using a sterile injection needle (size 0.65 × 25 mm, Fa. Braun, Melsungen, Germany). Manual microdissection was chosen because for this study the procedure was easier and faster than using the laser capture microdissection system (Arcturus PixCell II) available at our institute. Medium-sized interlobular ducts were preferably chosen in order to avoid contamination by the acinar tissue. Microdissected cells were sampled in a 100 μl reaction tube (Sarstedt, Nümbrecht, Germany) containing 50 μl extraction buffer (Arcturus, Moerfelden–Walldorf, Germany) and placed on ice. From step-sectioned lesions of the various cell types (PanIN-2, normal ductal epithelium and acinar cells), ∼2000–3000 cells were collected and pooled. Each cell pool contained cells from five to seven different cases.
RNA isolation and amplification
Total RNA from HeLa and Caco-2 cells was isolated by acid–phenol extraction. The same preparation of HeLa and Caco-2 RNA was used for Micro-longSAGE and aRNA-longSAGE library generation. Total RNA from microdissected cells (∼2000 cells from PanIN-2 lesions, 2500 pancreatic acinar cells and 3000 pancreatic duct cells) was isolated with a PicoPure RNA isolation kit (Arcturus, Moerfelden–Walldorf) following the manufacturer's instructions, including the optional DNase I digestion. An aliquot of 40 ng of total RNA (HeLa and Caco-2 RNA) or RNA corresponding to 1000–1250 cells (pancreatic acinar, ductal or PanIN-2 cells) was amplified in one round of linear RNA amplification using a MessageAmp™ aRNA Kit (Ambion, Huntingdon, UK). The quality of the RNA and aRNA was assessed and the amount of aRNA was estimated with an RNA 6000 Pico LabChip® on a Bioanalyzer platform (Agilent, Böblingen, Germany). For the samples derived from microdissected cells, amplifications were concentrated by using a YM-30 Microcon® centrifugal filter cartridge (Millipore, Schwalbach, Germany) according to the manufacturer's instructions and pooled to reach a minimum of 1.2 μg aRNA as starting material for SAGE library production.
Micro-longSAGE library production
To validate our aRNA-longSAGE protocol, standard Micro-longSAGE libraries with 100 μg total RNA as starting material were generated from two cell lines (HeLa and Caco-2).
Micro-longSAGE library generation was carried out according to the MicroSAGE protocol version 1.0e (which can be obtained from http://www.sagenet.org/protocol/index.htm) adapted to the longSAGE protocol published by Saha et al. (9), except for some modifications.
The RNA sample was adjusted to a final volume of 100 μl with DEPC–H2O and mixed with 200 μl Dynabeads Oligo(dT)25 (Dynal Biotech, Hamburg, Germany) that had been resuspended in 100 μl binding buffer (Dynal Biotech). Oligo(dT) beads and total RNA were incubated for 5 min at room temperature rotating on an overhead shaker. The beads were washed once with 200 μl of washing buffer A (Dynal Biotech), twice with 200 μl of washing buffer B (Dynal Biotech) and four times with 200 μl of first strand buffer (Invitrogen, Karlsruhe, Germany). First strand synthesis was carried out in a volume of 90 μl containing first strand buffer, 10 mM DTT (both Invitrogen), 0.5 mM of each dATP, dCTP, dGTP and dTTP (Promega, Mannheim, Germany), 1 μl RNAseOUT and 4.5 μl SuperScript™ III RNase H− reverse transcriptase (both Invitrogen). The reaction was incubated for 5 min at 37°C, 60 min at 50°C and 15 min at 70°C.
The volume of the second strand synthesis was downsized to 160 μl. After the second strand synthesis, the cDNA was digested with 50 U of the anchoring enzyme Hsp92II (an isoschizomer of NlaIII, Promega) in the buffer provided by the manufacturer in a reaction volume of 200 μl at 37°C for 1 h. The volume of the linker ligation was reduced to 10 μl. The cDNA tags were released by digestion with 2–8 U of the tagging enzyme MmeI (NEB, Frankfurt a.M., Germany) in 200 μl of the supplied buffer in the presence of 50 μM S-adenosylmethionine at 37°C for 1 h. The longSAGE tags were ligated as described by Saha et al. (9), diluted (1:175) and amplified with non-biotinylated primers for 26 cycles. All SAGE libraries were prepared from 96 pooled PCR reactions, purified by PC8 extraction and ethanol precipitation and digested with 200 U Hsp92II (Promega) in the buffer provided with the enzyme in a reaction volume of 200 μl for 1 h at 37°C. After the Hsp92II digestion, the PCR products were separated on a 12% polyacrylamide gel (19:1 acrylamide:bisacrylamide) and the ditag bands were excised and electroeluted at 4°C in 1× TAE for 2 h at 150 V (Elutrap system, Schleicher & Schuell, Dassel, Germany). Following the electroelution, the ditags were PC8 extracted, ethanol precipitated and resuspended in 7 μl of loTE (3 mM Tris–HCl, pH 7.5; 0.3 mM EDTA). Ditags were concatenated for 30 min at 16°C in a 10 μl reaction containing 5 U of T4 DNA ligase HC (Invitrogen) in the buffer supplied with the enzyme. Concatenated ditags were separated on an 8% polyacrylamide gel (19:1 acrylamide:bisacrylamide). Concatemers >300 bp were excised and electroeluted at 120 V for 30–60 min. After PC8 extraction and ethanol precipitation, the concatemers were cloned as described in the MicroSAGE protocol. The resulting longSAGE libraries consisted of more than 10 000 clones with an insert length of >600 bp. Sequencing of the libraries was carried out by MWG (MWG Biotech, Ebersberg, Germany).
LongSAGE tags were extracted from the sequence files with SAGE-PHRED 2003 software (can be obtained from je@bio.aau.dk) with the minimum quality of each base within a tag sequence set to PHRED20 and the maximum ditag length set to 36 (flanking CATGs not included), statistical analysis (Z-test) was carried out with the program SAGEstat (16) prior to normalization of the tag counts.
aRNA-longSAGE library production
For the validation of our aRNA-longSAGE protocol, aRNA-longSAGE libraries with ∼40 ng total RNA (amount of total RNA was estimated with an RNA 6000 Pico LabChip® on a Bioanalyzer platform) as starting material were generated from two cell lines (HeLa and Caco-2). For these two aRNA-longSAGE libraries, 1.2 μg of antisense amplified RNA (generated in one round of linear RNA amplification) was used for library production.
For aRNA-longSAGE library generation of the microdissected material, at least two RNA single-round amplifications were pooled for each library, which resulted in 1.3 μg (PanIN-2 library), 2.3 μg (pancreatic ductal cell library) and 8.9 μg (acinar cell library) aRNA as starting material for the respective libraries.
The aRNA was adjusted to a final volume of 10 μl with DEPC–H2O. Two microlitres of SAGE-random primer (5′-NNN NNN CAT G-3′, MWG Biotech), 125 ng/μl) and 1 μl dNTP-Mix [10 mM each of dATP, dCTP, dGTP and dTTP (Promega)] were added to the aRNA and the mixture was incubated at 65°C for 5 min and then frozen on dry ice. The reaction mixture was placed on ice and the following reagents (all from Invitrogen) were added to complete the reverse transcription reaction: 4 μl first strand buffer, 1 μl 0.1 M DTT, 1 μl RNaseOUT, 1 μl SuperScript™ III RNase H− reverse transcriptase. The reaction was incubated for 5 min at 37°C, 60 min at 50°C and 15 min at 70°C. After the addition of 1 μl of RNase H (2 U/μl, USB, Cleveland, OH), the sample was incubated at 37°C for 20 min. Afterwards, the sample was diluted with 79 μl DEPC–H2O and mixed with 200 μl Dynabeads Oligo(dT)25 (Dynal Biotech) that had been resuspended in 100 μl binding buffer (Dynal Biotech). Oligo(dT) beads and the first strand cDNA were annealed by rotating on an overhead shaker for 15 min at room temperature. The beads were washed twice with 200 μl of washing buffer B (Dynal Biotech), four times with 200 μl of second strand buffer (18.8 mM Tris–HCl, pH 6.9, 90.6 mM KCl, 4.6 mM MgCl2, 10 mM (NH4)2SO4, 0.15 mM β-NAD) and resuspended in 112.25 μl of ice-cold DEPC–H2O.
Second strand synthesis and all the other steps of longSAGE library generation were performed according to the Micro-longSAGE protocol described above. A detailed aRNA-longSAGE protocol can be obtained from the authors.
Northern blotting
Northern blots and hybridization were performed as described previously (17). Equal loading of the northern blots was assured by assessing the signal intensities of the 28S and 18S rRNA bands. Autoradiographic figures were prepared using a Typhoon 9400 (Amersham Biosciences, Freiburg, Germany) PhosphorImager system. Signal intensities were quantified using Image Quant 5.2 software (Amersham Biosciences). Probes for the individual genes used for hybridization were derived from IMAGE clones obtained from RZPD (Berlin, Germany) which were sequence-verified prior to probe labeling.
Quantitative real-time PCR
cDNA was synthesized using 1 μg of total RNA, oligo(dT)18 primers and SuperScript™ III RNase H− reverse transcriptase (Invitrogen) following the manufacturer's protocol. Four parallel cDNA syntheses were carried out, diluted to a final volume of 50 μl with 1× first strand buffer and pooled. Two such pools were prepared on different days (‘cDNAs-1’ and ‘cDNAs-2’). Primer sets for quantitative real-time PCR (qRT–PCR) were generally designed using Primer Express 2.0 software (Applied Biosystems, Foster City, CA) and can be obtained from the authors. For several housekeeping genes (GAPD, HMBS, HPRT1, UBC, YWHAZ), published primer sequences were used (18). qRT–PCR was performed using a SYBR Green I reaction mixture containing 75 mM Tris–HCl, pH 8.8, 20 mM ammonium sulfate, 0.01% (v/v) Tween-20, 2 mM MgCl2 (all Sigma–Aldrich, Munich, Germany), 1 μl of a 600-fold dilution of SYBR Green I (BioWhittaker, Rockland, ME), 2.5 U Taq polymerase (NEB) and 0.2 μM of forward and reverse primer (QIAgen, Hilden, Germany) in a final reaction volume of 20 μl. Reactions were run on a DNA Engine Opticon®2 cycler (MJ Research, Waltham, MA). The cycling conditions consisted of 3 min initial denaturation at 94°C and 40 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 30 s and 80°C for 3 s. Fluorescence was measured at the last step of each cycle. Melting curves were obtained after each PCR run and showed single PCR products. cDNAs were run in triplicate, non-RT (without reverse transcriptase) and no-template controls were run in duplicates. C(T) values of non-RT controls were at least eight cycles higher than C(T) values of the corresponding cDNAs.
PCR efficiencies were determined using serial dilutions of a 1:1 mixture of HeLa and Caco-2 cDNA. Expression levels for the genes of interest and for housekeeping genes were measured for cDNAs-1 and cDNAs-2 in independent PCR runs. Expression ratios were calculated as described by Pfaffl (19) using the geometric mean expression of the housekeeping genes GAPD, HMBS, HPRT1, PPIA, UBC and YWHAZ to normalize the expression data for the genes of interest.
RESULTS
For the analysis of microdissected pancreatic cells via SAGE, we attempted to adapt the cDNA synthesis step for SAGE library production in such a manner that amplified aRNA produced via the well-established and validated linear RNA amplification method introduced by Eberwine et al. could be used directly as the starting material.
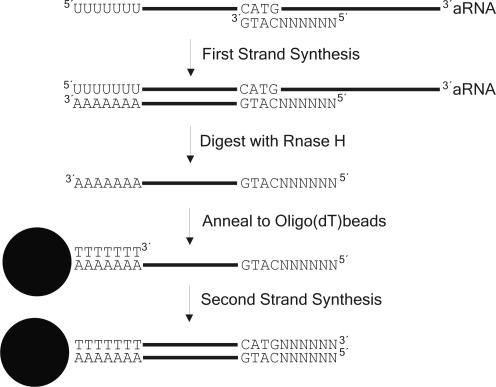
To be able to use linearly amplified aRNA as the starting material for the generation of SAGE libraries, we modified the cDNA synthesis steps in the SAGE protocol as follows. First of all, the aRNA was reverse transcribed with a random primer that included the recognition site of the SAGE anchoring enzyme NlaIII (SAGE-random primer 5′-NNN NNN CAT G-3′, Figure 1). The recognition site for NlaIII was introduced, because we wanted to specifically enrich for target sequences that are needed for subsequent steps in the SAGE protocol. The RNA was then removed from the resulting DNA–RNA hybrid by digestion with RNase H. At this point, the cDNA first strand corresponds to the mRNA before amplification and has a 3′ poly(A) tail which can be annealed to magnetic oligo(dT) beads. After coupling of the first strand synthesis product to the magnetic beads, the (dT)25 oligonucleotide linked to the beads served as primer for the second strand cDNA synthesis step. The resulting cDNA can be used directly in either the conventional MicroSAGE protocol or the MicroSAGE protocol modified for longSAGE (9).
Figure 1.
Scheme of the modified cDNA synthesis protocol within the aRNA-longSAGE procedure.
The cDNA that is generated with our protocol carries at its 3′ end the recognition site for the anchoring enzyme Hsp92II (CATG) followed by six additional base pairs. After completion of the cDNA synthesis, the next step in the SAGE protocol requires digestion with the anchoring enzyme. In order to assess whether the 6 bp next to the recognition site was sufficient to ensure complete Hsp92II digestion of the cDNA, we used the PCR technique to generate a 344 bp fragment of the elongation factor 1 alpha 1 transcript, which contained a terminal 6 bp overhang 3′ next to the CATG site. Digestion of 1 μg of this PCR product with Hsp92II under the conditions we used for the digestion of the cDNA during longSAGE library generation showed the required complete digestion of the PCR product (Figure 2).
Figure 2.
Digestion of the PCR product EF alpha with the anchoring enzyme Hsp92II. The PCR product EF alpha was generated with the primers 5′-CAAGCCCATGTGTGTTGAG-3′and 5′-GAAAACCAAAGTGGTCCAC-3′ using cDNA from the cell line BxPC3 as template. One microgram of the EF alpha PCR product was digested with 50 U Hsp92II in 200 μl reaction volume. Fifty nanograms of PCR products before (‘EF alpha’) and after digestion with Hsp92II (‘EF alphaΔHsp92II’) were separated on an 8% polyacrylamide gel (80:1 acrylamide:bisacrylamide) and silver stained. The stained gel was scanned with a UMAX Mirage II Scanner and edited using the program Corel Photo-Paint (values set to brightness: 33; contrast: 33; intensity: 14).
To validate our modified protocol and to keep our validation experiments as close as possible to the application we were aiming at (differential expression analyses of microdissected cells), we chose to compare the differences in gene expression between two cell lines (HeLa and Caco-2) under ‘non-amplified’ (starting with total RNA) versus ‘amplified’ (starting with aRNA) conditions. Thus, we generated the Micro-longSAGE libraries ‘HeLa’ (14 766 tags sequenced) and ‘Caco-2’ (11 222 tags sequenced) as well as the aRNA-longSAGE libraries ‘HeLa-amp’ (8967 tags sequenced) and ‘Caco-2-amp’ (13 502 tags sequenced). The percentage of unique tags identified was consistently higher in the Micro-longSAGE libraries (67% for both libraries) than in the aRNA-longSAGE libraries (53% in Caco-2-amp and 54% in HeLa-amp), whereas the percentage of tags found more than once was consistently higher in the aRNA-longSAGE libraries (20% in both Caco-2-amp and HeLa-amp) than in the Micro-longSAGE libraries (15% in both Caco-2 and HeLa; see also Table 1).
Table 1. Tag abundances for the Micro-longSAGE and aRNA-longSAGE libraries of HeLa and Caco-2 cells.
| Micro-longSAGE | aRNA-longSAGE | |||
|---|---|---|---|---|
| Caco-2 | HeLa | Caco-2-amp | HeLa-amp | |
| Sequenced tagsa | 11 222 | 14 766 | 13 502 | 8967 |
| Unique tags | 7488 (66.7%b) | 9858 (66.8%b) | 7161 (53.0%b) | 4840 (54.0%b) |
| Tag count = 1 | 6367 (85.0%c) | 8348 (84.7%c) | 5703 (79.6%c) | 3885 (80.3%c) |
| Tag count > 1 | 1121 (15.0%c) | 1510 (15.3%c) | 1458 (20.4%c) | 955 (19.7%c) |
| Tag count > 2 | 508 (6.8%c) | 693 (7.0%c) | 724 (10.1%c) | 499 (10.3%c) |
| Tag count > 5 | 153 (2.0%c) | 237 (2.4%c) | 263 (3.7%c) | 196 (4.0%c) |
| Tag count > 10 | 68 (0.91%c) | 94 (0.95%c) | 118 (1.65%c) | 90 (1.86%c) |
aWithout linker tags.
bPercentage of unique tags among sequenced tags.
cPercentage of tags with a given tag count among the number of unique tags.
Subsequently, tag counts were compared and analyzed for statistically significant differences among the ‘reference libraries’ (Micro-longSAGE) derived from HeLa and Caco-2 total RNA as well as among the ‘test libraries’ (aRNA-longSAGE) derived from the same total RNAs. For the reference libraries, the comparison of expression profiles between HeLa and Caco-2 cells yielded 71 differentially expressed genes with a P-value below 0.01. Of these 71 genes, 52 (73%) were also found in the test libraries (HeLa-amp versus Caco-2-amp) with a high (26 genes with P < 0.01; Table 2) to moderate (26 genes with 0.01 < P < 0.2) probability of being differentially expressed. For 50 of the 52 (96%) genes found in both the reference and test libraries, the direction of the gene regulation was concordant. For the remaining 19 genes that were differentially expressed according to the Micro-longSAGE analysis, either no tags were found in the aRNA-longSAGE analysis (five genes) or the difference in tag counts was not significant (14 genes with P > 0.2).
Table 2. List of genes which were differentially expressed with a high statistical significance (P < 0.01) in the Micro-longSAGE and aRNA-longSAGE experiments.
| Tag sequencea | Caco-2-amp | HeLa-amp | Caco-2 | HeLa | RefSeqb | Gene |
|---|---|---|---|---|---|---|
| AGCACCTCCAGCTGTAC | 8.1 | 25.6 | 2.7 | 14.2 | NM_001961 | EEF2 |
| AGGGCTTCCAATGTGCT | 19.3 | 39.0 | 18.7 | 6.1 | XM_209178 XM_371781 XM_209500 | – |
| ATGTAAAAAATACAAAC | 43.7 | 0.0 | 15.1 | 0.0 | NM_000239 | LYZ |
| ATTATTTTTCTAAGCTG | 26.7 | 10.0 | 13.4 | 2.7 | NM_000971 | RPL7 |
| CAATAAATGTTCTGGTT | 151.1 | 72.5 | 40.1 | 22.3 | NM_000997 | RPL37 |
| CACAAACGGTAGTTTTG | 64.4 | 34.6 | 31.2 | 11.5 | NM_001030 | RPS27 |
| CACCTAATTGGAAGCGC | 0.0 | 10.0 | 0.0 | 14.9 | NM_173702 | MTATP6 |
| CGCCGCCGGCTCAACAA | 15.6 | 127.1 | 11.6 | 48.8 | NM_007209 | RPL35 |
| CTTGTAATCCTACTTGG | 10.4 | 0.0 | 9.8 | 0.0 | n.a. | n.a. |
| GAGGGAGTTTCATTAAA | 56.3 | 31.2 | 27.6 | 12.2 | NM_000990 | RPL27A |
| GCATAATAGGTGTTAAA | 25.2 | 3.3 | 30.3 | 6.1 | NM_000982 | RPL21 |
| GCCCCCAATAAAGGCAG | 0.0 | 14.5 | 0.0 | 10.2 | NM_002305 | LGALS1 |
| GCGTGGTGTGTCCCTGG | 0.0 | 31.2 | 0.0 | 6.8 | n.a. | n.a. |
| GGCCCTCCCGGGACATC | 0.0 | 8.9 | 0.0 | 31.8 | NM_001631 | ALPI |
| GGCTCCCACTGCAGCCT | 0.0 | 22.3 | 0.0 | 6.1 | NM_007355 | HSPCB |
| GGGCTGGGGTCCTCCTG | 3.7 | 60.2 | 5.3 | 33.9 | NM_000992 | RPL29 |
| GGGGGCCCCGTCCCCCG | 0.0 | 6.7 | 0.9 | 9.5 | NM_020470 | YIF1 |
| GTTGTGGAGGGTTGTGG | 0.0 | 6.7 | 0.9 | 11.5 | NM_001200 | BMP2 |
| GTTGTGGTTAATCTGGT | 99.2 | 13.4 | 42.8 | 10.8 | NM_004048 | B2M |
| TAAATTGCAAATAAACC | 8.1 | 0.0 | 8.0 | 0.0 | NM_019010 | KRT20 |
| TAATAAAGGTGTTTATT | 48.1 | 13.4 | 57.9 | 20.3 | NM_001012 | RPS8 |
| TAGGTTGTCTAAAAATA | 50.4 | 3.3 | 28.5 | 4.1 | NM_003295 | TPT1 |
| TAGTTTGAAGGCGGTCA | 11.1 | 0.0 | 13.4 | 0.0 | NM_012413 | QPCT |
| TCCGTGGTTGGGTGCAC | 0.0 | 19.0 | 0.0 | 31.8 | NM_006317 | BASP1 |
| TTCAATAAAAAGCTGAA | 102.2 | 32.3 | 41.0 | 18.3 | NM_001003 | RPLP1 |
| TTCATACACCTATCCCC | 7.4 | 23.4 | 6.2 | 21.0 | NM_173712 | MTND4L |
n.a., No annotation available. All tag counts given in the table are normalized to 10 000 tags per library.
aOnly the variable part of the 21 bp longSAGE tag is shown. The corresponding 21 bp tag can be obtained by adding the NlaIII recognition site CATG 5′ to the variable part of the tag.
bOnly tags that are the 3′ most tag of a RefSeq (release 4) sequence are annotated.
The comparison between the test libraries HeLa-amp and Caco-2-amp yielded a total of 134 statistically significant (P < 0.01), differentially expressed genes (Supplementary Table 1). Of these 134 genes, 68 were also found to be differentially expressed in the reference libraries with a high (26 genes with P < 0.01) or moderate (42 genes with 0.01 < P < 0.2) statistical probability. The direction of regulation between the reference and test libraries was concordant for 63 of these 68 (93%) genes. The remaining 66 genes were either found not to be regulated or were not found at all in the reference libraries and were therefore considered to be potential candidates for newly identified, differentially expressed genes.
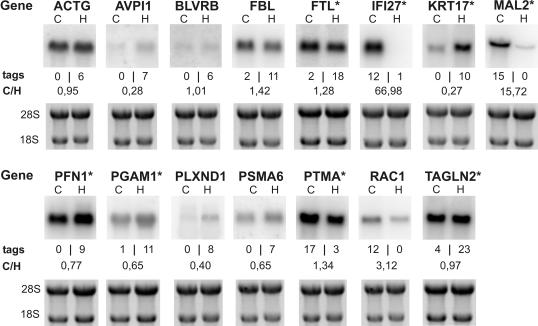
Next, we sought to confirm a representative number of genes found to be differentially regulated in the aRNA-longSAGE libraries (P < 0.01) using independent technologies. We hypothesized that the confirmation rate would likely reach an optimum for highly significant, differentially expressed genes (P < 0.01) found in both the amplified and non-amplified libraries. Therefore, to get a conservative estimation, we chose to validate via standard northern blots (n = 15; Figure 3) or qRT–PCRs (n = 10; Table 3) a representative number of genes which either were also differentially expressed in the reference libraries with a moderate P-value (n = 12) or belonged to the genes not identified as being differentially expressed in Micro-longSAGE (n = 13). The confirmation rate was higher for the first group of genes (9/12; 75%) than it was for the second group (8/13; 62%). Taken together, with our conservative strategy for 17 of 25 genes (68%), the direction of regulation could be confirmed without regard to the magnitude of the expression ratio. Confirmed genes reached a minimum expression ratio of 1.3 in either direction in the qRT–PCR or the northern blot experiments.
Figure 3.
Northern blots of genes found to be differentially expressed between Caco-2 and HeLa cells by the aRNA-longSAGE protocol. Asterisk, genes that were also found to be differentially expressed in Micro-longSAGE with a moderate P-value (0.01 < P < 0.2); C, Caco-2; H, HeLa; tags, tag counts for aRNA-longSAGE; C/H, expression ratio Caco-2/HeLa; 18S, 18S rRNA; 28S, 28S rRNA. Electropherograms of total RNA are given as a loading control.
Table 3. qRT–PCR analysis for validation of the aRNA-longSAGE data.
| Tag sequencea | aRNA-longSAGE | qPCR: Caco-2/HeLa | Gene | |||
|---|---|---|---|---|---|---|
| Caco-2-amp | HeLa-amp | P-value | cDNAs-1 | cDNAs-2 | ||
| CTTCCAGCTAACAGGTC | 1.5 | 11.2 | 0.002 | 1.14 | 1.19 | ANXA2b |
| GCTGGCTGGCTGCTGGG | 0.7 | 16.7 | 0.000 | 0.47 | 0.47 | CCT7 |
| GTGACCTCCTTGGGGGT | 2.2 | 26.8 | <0.001 | 0.68 | 0.89 | COX8A |
| TCTTAATGAAGTTTGAA | 8.1 | 0.0 | 0.007 | 2.03 | 2.04 | EIF4A2 |
| GTGTAATAAGACATAAC | 17.8 | 3.3 | 0.002 | 1.04 | 1.18 | HNRPA2B1 |
| GACGACACGAGCCGATC | 3.0 | 13.4 | 0.004 | 0.50 | 0.59 | RPS28 |
| CCCCAGCCAGTCCCCAC | 34.8 | 143.9 | <0.001 | 0.75 | 0.73 | RPS3b |
| TGAGCCCGGCCGGCCCA | 0.7 | 8.9 | 0.003 | 0.75 | 0.68 | SCYL1b |
| TGCGGAGGCCCTGCGCA | 1.5 | 10.0 | 0.005 | 0.47 | 0.41 | SSSCA1 |
| TAGACTAGCAAAATAGT | 13.3 | 0.0 | 0.001 | 2.99 | 2.79 | TM4SF8b |
All tag counts given in the table are normalized to 10 000 tags per library.
aOnly the variable part of the 21 bp longSAGE tag is shown. The corresponding 21 bp tag can be obtained by adding the NlaIII recognition site CATG 5′ to the variable part of the tag.
bGenes also found to be differentially expressed in Micro-longSAGE with a moderate P-value (0.01 < P < 0.2).
The expression ratios from qRT–PCRs and northern blots were generally lower than the corresponding expression ratios from aRNA-longSAGE experiments.
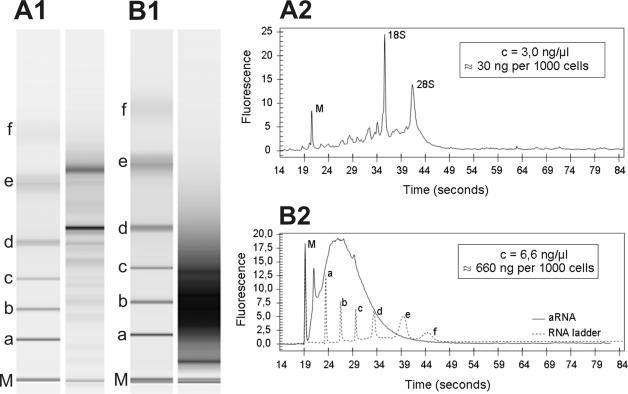
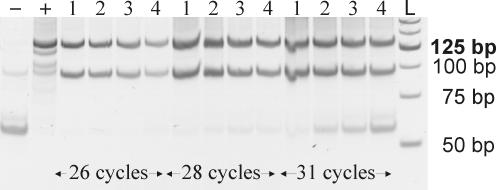
Having thus established that our protocol can be used to generate reliable longSAGE libraries from as little as 40 ng total RNA, we went on to use microdissected cells as the starting material. RNA was isolated from 2000 microdissected cells from PanIN-2 lesions, 2500 microdissected acinar cells and 3000 microdissected normal pancreatic ductal cells yielding 40–200 ng total RNA (for an elecropherogram of total RNA see Figure 4). One round of T7-based amplification yielded 1.3 μg (PanIN-2 cells, Figure 4), 8.9 μg (acinar cells) and 2.3 μg (pancreatic ductal cells) aRNA, which was subsequently used to produce aRNA-longSAGE libraries (for an example of ditag PCR see Figure 5).
Figure 4.
Analysis of RNA quality and length for RNA derived from microdissected cells. Representative RNA gel images generated with an RNA 6000 Pico LabChip®on a Bioanalyzer platform are shown for total RNA (A1) and aRNA (B1). Corresponding electropherograms are shown for total RNA (A2) and aRNA (B2). M, marker for sample synchronization; 18S, 18S rRNA; 28S, 28S rRNA; a–f, RNA ladder (0.2, 0.5, 1.0, 2.0, 4.0, 6.0 kb).
Figure 5.
PCR amplification of ditags. The ditag ligation derived from 3000 pancreatic ductal cells was diluted to various end concentrations and amplified by PCR. An aliquot of 5 of 50 μl from each PCR product were analyzed on a 12% polyacrylamide gel. −, negative control; +, positive control; 1–4, 1:175, 1:350, 1:700, 1:1400 dilution factor of the ditag ligation, respectively; L, 25 bp ladder.
From the PanIN-2, ductal and acinar cell longSAGE libraries, we sequenced 42 808, 31 767 and 44 137 tags (without linker tags), respectively. The validity of the gene expression profiles was judged by comparing the normalized tag counts of typical acinar and ductal marker genes in the acinar cell library with their respective tag counts in the ductal cell library (Table 4). The cytokeratin profile typical for both acinar and ductal pancreatic cells [KRT8 and KRT18 for acinar cells, KRT7, KRT8, KRT18 and KRT19 for ductal cells (20)] was detected in the aRNA-longSAGE libraries. Typical acinar marker genes were found with high tag counts in the acinar cell aRNA-longSAGE library but with very low tag counts in the ductal aRNA-longSAGE library indicating a high quality of the manual microdissection procedure. Similar results were also obtained with the PanIN-2 library.
Table 4. Expression level of acinar and ductal cell marker genes identified by aRNA-longSAGE.
| Tag sequencea | PanAc | PanDuc | PanIN-2 | Gene |
|---|---|---|---|---|
| GCGTGACCAGCTTTGTT | 1645 | 7 | 5 | Elastase 3B (A) |
| GAGCACACCCTGAATCA | 926 | 8 | 6 | Carboxypeptidase A1 (A) |
| TGCGAGACCACCCCTAT | 654 | 5 | 7 | Carboxypeptidase A2 (A) |
| TCAGGGTGATTCTGGTG | 1541 | 20 | 15 | Protease, serine 1 (trypsin 1) (A) |
| ACGCTGGACGCTCCAAG | 411 | 1 | 0 | Colipase, pancreatic (A) |
| CCTCCAGCTACAAAACA | 3 | 49 | 26 | Keratin 8 (A, D) |
| CAAACCATCCAAAAGAC | 10 | 79 | 30 | Keratin 18 (A, D) |
| CCTGGTCCCAAGACAGT | 0 | 3 | 5 | Keratin 7 (D) |
| GACATCAAGTCGCGGCT | 0 | 12 | 16 | Keratin 19 (D) |
A, acinar marker gene; D, ductal marker gene; PanAc, pancreatic acinar library; PanDuc, pancreatic ductal library; PanIN-2, PanIN-2 library.
aOnly the variable part of the 21 bp longSAGE tag is shown. The corresponding 21 bp tag can be obtained by adding the NlaIII recognition site CATG 5′ to the variable part of the tag. All tag counts given in the table were normalized to 45 000 tags per library.
An initial search for genes differentially expressed in our PanIN-2 lesions in comparison with the ductal cell aRNA-longSAGE library identified S100 P and mucin 5AC as the two genes that were most highly upregulated in our PanIN-2 library. Both genes are known to be upregulated in PanIN-2 cells [J. Lüttges, personal communication; (21,22)]. In addition, several other genes found to be differentially expressed in our study had previously been shown to be either upregulated or downregulated in pancreatic ductal adenocarcinoma (Supplementary Table 2).
DISCUSSION
Differential gene expression analysis of microdissected tissue has become an important approach enabling scientists to define gene expression patterns of specific cell types from normal and pathological tissues. The minimum amount of starting RNA required to perform expression analysis using standard protocols for the two most commonly used technology platforms, microarrays and SAGE, generally cannot be isolated from microdissected tissues.
This limitation was overcome for microarray experiments with the introduction of the linear amplification of RNA using T7 RNA polymerase (1). Importantly, a number of research groups have shown that the correlation between expression profiles generated through amplified aRNA versus total RNA as starting material is sufficiently good to allow reliable profiling of gene expression with limited starting material (23–26). For SAGE, it was previously not possible to use the aRNA produced by the available standard protocols directly as starting material. Here, we present a method that overcomes this limitation. Our method, termed aRNA-longSAGE, is based on an alternative cDNA synthesis approach using aRNA in combination with an NlaIII-anchored random primer for first strand cDNA synthesis, followed by on-bead second strand synthesis (Figure 1).
To validate our aRNA-longSAGE protocol, we prepared two Micro-longSAGE ‘reference’ libraries and two aRNA-longSAGE ‘test’ libraries from two different cell lines (HeLa and Caco-2) and compared the expression profiles obtained from reference and test libraries. Of the 71 differentially expressed genes (P < 0.01) identified with the reference libraries (Micro-longSAGE), 52 (73%) were also identified in the test libraries (aRNA-longSAGE) with a high to moderate probability of differential expression. Importantly, nearly all of these genes (50 of 52; 96%) were found with the same direction of regulation. Taken together, our data indicate that aRNA-longSAGE is able to maintain 70% (50/71) of the differences identified in the reference libraries. This identification rate is somewhat lower than what has been reported for similar experiments using microarrays, which show an identification rate of 81–94% (24,25). A possible explanation for this discrepancy is the limited number of tags we collected for each of the test and reference libraries.
To get a conservative estimate for the confirmation rate of the 134 genes identified as being differentially expressed by aRNA-longSAGE, we validated a representative number of genes which either were also differentially expressed in the reference libraries with a moderate P-value or belonged to the candidates for newly identified genes (Supplementary Table 1). qRT–PCR or northern blot analyses of 25 genes using the original total RNA confirmed the direction of differential expression for 17 (68%) of these genes. Our data are comparable to the data from a study by Polacek et al. (25), who reported a concordance rate of 67% between microarray results generated with aRNA and qRT–PCR using total RNA. Our confirmation rate is likely to be an underestimation because we did not analyze genes which were found to be differentially expressed with a high statistical significance in both the test and reference libraries.
The number of differentially expressed genes identified by aRNA-longSAGE was almost twice as high as for the analysis using the conventional Micro-longSAGE protocol (134 genes with P < 0.01 versus 71 genes with P < 0.01). The higher overall discovery rate for differentially expressed genes using aRNA-longSAGE is in good agreement with previous reports of microarray analyses with aRNA as starting material (24,25). A possible explanation for this improved discovery rate is either the preferential amplification of certain sequences during the T7-based amplification of RNA or the preferential reverse transcription of certain sequences during the random-primed reverse transcription of aRNA, which is part of our aRNA-longSAGE protocol. Therefore, both enzymatic steps potentially lead to some reduction of the complexity of the amplified library. Our observation that the overall number of identified unique tag sequences was reduced in the amplified library supports this notion (Table 1). Furthermore, this reduction led to a higher overall tag count per individual tag sequence and thus a greater number of genes reached the threshold required to be statistically significant (Table 1). The identified tendency toward an overestimation of the expression ratios by aRNA-longSAGE has also been reported previously for standard SAGE experiments (27).
The current literature contains one SAGE protocol that also includes T7-RNA polymerase amplification in order to augment the starting material prior to SAGE library production. This method, called small amplified RNA-SAGE by Vilain et al. (14), is a modified RNA amplification procedure combined with the standard SAGE protocol. The RNA amplification by in vitro transcription takes place after the on-bead cDNA synthesis, digestion of cDNA with NlaIII and ligation of a linker that contains the T7 RNA promoter. This protocol, therefore, has the drawback of introducing two additional enzymatic steps (an additional digestion of cDNA with NlaIII and subsequently an additional linker ligation both analogous to the steps carried out subsequently during the standard SAGE protocol) to the RNA amplification procedure introduced by Eberwine et al. (1), which is likely to reduce the overall yield of amplified material. Although the amount of amplified RNA generated by small amplified RNA-SAGE has not been reported by Vilain et al., the amplification yield is likely to be reflected in the amplification conditions used for the subsequent ditag PCR step. The aRNA-longSAGE protocol involved pooling of 96 ditag PCR reactions run at 26 cycles compared to the pooling of 192 reactions run at 30 cycles required by the small amplified RNA-SAGE protocol (14). As both protocols started with roughly the same amount of total RNA (40 ng for aRNA-longSAGE and 50 ng for small amplified RNA-SAGE), the RNA amplification step in the aRNA-longSAGE procedure is likely to be more efficient than the RNA amplification step in the procedure described by Vilain et al.
The standard RNA amplification procedure used herein is generally able to produce at least 1.2 μg of aRNA from 40 ng of total RNA. This aRNA input for the aRNA-longSAGE protocol corresponds to ∼100 μg of total RNA. Therefore, it could be speculated that the aRNA-longSAGE protocol also works with much less input total RNA, but most likely at the expense of higher PCR cycle numbers which may have the drawback of increasing the PCR amplification bias during the ditag amplification step.
Another potential disadvantage of the protocol published by Vilain et al. is that up to 5% of T7-linker tags are identified in their SAGE libraries, increasing the sequencing effort for SAGE data generation.
Finally, our aRNA-longSAGE protocol in contrast to the small amplified RNA-SAGE procedure allows the direct use of aRNA produced by standard protocols for the creation of SAGE libraries, thus offering the opportunity to use the same pool of amplified RNA for microarray experiments and for the generation of a complementary SAGE library.
Comparing the pancreatic ductal and acinar cell aRNA-longSAGE profiles with each other, we readily identified a number of known acinar and ductal cell-specific genes (Table 4). Furthermore, we found only few acinar cell specific gene tags in the ductal cell preparation, confirming the high purity of our cell pools generated through manual microdissection.
Comparing the PanIN-2 and the ductal library, the two most highly upregulated genes identified, S100 P and MUC5AC, have previously been shown to be upregulated in PanIN-2 lesions [J. Lüttges, personal communication; (21,22)]. In an ongoing study, we are aiming at the confirmation of additional candidate genes by immunohistochemistry (IHC) and qRT–PCR.
In summary, the validated aRNA-longSAGE protocol presented here successfully combines microdissection and standard T7 amplification with longSAGE library production for the analysis of microdissected primary tissue samples. This method should be readily translatable to similar experiments in other tissue types.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Britta Redeker, Matthias Becker and Susanne Braun for excellent technical assistance as well as Katherine Dege for critical reading of the manuscript. This work was supported by grants from the Deutsche Krebshilfe (J.L., G.K., W.S., T.G. and S.A.H, 70-2988-Schm3) and the Bundesministerium für Bildung und Forschung (S.A.H., 0311878). J.E. and K.L.N. were supported by the Danish Technical Research Council (26-00-0141) and the Danish Veterinarian and Agricultural Research Council (23-02-0034). Coordination was funded by EU-Grant BMH4-QLG1-CT-2002-01196 to S.A.H., T.G., J.L. and G.K.
REFERENCES
- 1.Eberwine J., Yeh,H., Miyashiro,K., Cao,Y., Nair,S., Finnell,R., Zettel,M. and Coleman,P. (1992) Analysis of gene expression in single live neurons. Proc. Natl Acad. Sci. SA, 89, 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puskas L.G., Zvara,A., Hackler,L.,Jr and Van Hummelen,P. (2002) RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques, 32, 1330–1334, 1336,, 1338,, 1340. [DOI] [PubMed] [Google Scholar]
- 3.Kitahara O., Furukawa,Y., Tanaka,T., Kihara,C., Ono,K., Yanagawa,R., Nita,M.E., Takagi,T., Nakamura,Y. and Tsunoda,T. (2001) Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res., 61, 3544–3549. [PubMed] [Google Scholar]
- 4.Grutzmann R., Foerder,M., Alldinger,I., Staub,E., Brummendorf,T., Ropcke,S., Li,X., Kristiansen,G., Jesnowski,R., Sipos,B. et al. (2003) Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch., 443, 508–517. [DOI] [PubMed] [Google Scholar]
- 5.Ohyama H., Zhang,X., Kohno,Y., Alevizos,I., Posner,M., Wong,D.T. and Todd,R. (2000) Laser capture microdissection-generated target sample for high-density oligonucleotide array hybridization. Biotechniques, 29, 530–536. [DOI] [PubMed] [Google Scholar]
- 6.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 7.Polyak K. and Riggins,G.J. (2001) Gene discovery using the serial analysis of gene expression technique: implications for cancer research. J. Clin. Oncol., 19, 2948–2958. [DOI] [PubMed] [Google Scholar]
- 8.Chen J., Sun,M., Lee,S., Zhou,G., Rowley,J.D. and Wang,S.M. (2002) Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc. Natl Acad. Sci. USA, 99, 12257–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha S., Sparks,A.B., Rago,C., Akmaev,V., Wang,C.J., Vogelstein,B., Kinzler,K.W. and Velculescu,V.E. (2002) Using the transcriptome to annotate the genome. Nat. Biotechnol., 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 10.Neilson L., Andalibi,A., Kang,D., Coutifaris,C., Strauss,J.F.,III, Stanton,J.A. and Green,D.P. (2000) Molecular phenotype of the human oocyte by PCR-SAGE. Genomics, 63, 13–24. [DOI] [PubMed] [Google Scholar]
- 11.Peters D.G., Kassam,A.B., Yonas,H., O'Hare,E.H., Ferrell,R.E. and Brufsky,A.M. (1999) Comprehensive transcript analysis in small quantities of mRNA by SAGE-lite. Nucleic Acids Res., 27, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schober M.S., Min,Y.N. and Chen,Y.Q. (2001) Serial analysis of gene expression in a single cell. Biotechniques, 31, 1240–1242. [PubMed] [Google Scholar]
- 13.Ye S.Q., Zhang,L.Q., Zheng,F., Virgil,D. and Kwiterovich,P.O. (2000) miniSAGE: gene expression profiling using serial analysis of gene expression from 1 microg total RNA. Anal. Biochem., 287, 144–152. [DOI] [PubMed] [Google Scholar]
- 14.Vilain C., Libert,F., Venet,D., Costagliola,S. and Vassart,G. (2003) Small amplified RNA-SAGE: an alternative approach to study transcriptome from limiting amount of mRNA. Nucleic Acids Res., 31, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hruban R.H., Adsay,N.V., Albores-Saavedra,J., Compton,C., Garrett,E.S., Goodman,S.N., Kern,S.E., Klimstra,D.S., Klöppel,G., Longnecker,D.S. et al. (2001) Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol., 25, 579–586. [DOI] [PubMed] [Google Scholar]
- 16.Ruijter J.M., Van Kampen,A.H. and Baas,F. (2002) Statistical evaluation of SAGE libraries: consequences for experimental design. Physiol. Genomics, 11, 37–44. [DOI] [PubMed] [Google Scholar]
- 17.Schwarte-Waldhoff I., Klein,S., Blass-Kampmann,S., Hintelmann,A., Eilert,C., Dreschers,S., Kalthoff,H., Hahn,S.A. and Schmiegel,W. (1999) DPC4/SMAD4 mediated tumor suppression of colon carcinoma cells is associated with reduced urokinase expression. Oncogene, 18, 3152–3158. [DOI] [PubMed] [Google Scholar]
- 18.Vandesompele J., De Preter,K., Pattyn,F., Poppe,B., Van Roy,N., De Paepe,A. and Speleman,F. (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol., 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res., 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouwens L. (1998) Cytokeratins and cell differentiation in the pancreas. J. Pathol., 184, 234–239. [DOI] [PubMed] [Google Scholar]
- 21.Luttges J., Zamboni,G., Longnecker,D. and Kloppel,G. (2001) The immunohistochemical mucin expression pattern distinguishes different types of intraductal papillary mucinous neoplasms of the pancreas and determines their relationship to mucinous noncystic carcinoma and ductal adenocarcinoma. Am. J. Surg. Pathol., 25, 942–948. [DOI] [PubMed] [Google Scholar]
- 22.Kim G.E., Bae,H.I., Park,H.U., Kuan,S.F., Crawley,S.C., Ho,J.J. and Kim,Y.S. (2002) Aberrant expression of MUC5AC and MUC6 gastric mucins and sialyl Tn antigen in intraepithelial neoplasms of the pancreas. Gastroenterology, 123, 1052–1060. [DOI] [PubMed] [Google Scholar]
- 23.Attia M.A., Welsh,J.P., Laing,K., Butcher,P.D., Gibson,F.M. and Rutherford,T.R. (2003) Fidelity and reproducibility of antisense RNA amplification for the study of gene expression in human CD34+ haemopoietic stem and progenitor cells. Br. J. Haematol., 122, 498–505. [DOI] [PubMed] [Google Scholar]
- 24.Feldman A.L., Costouros,N.G., Wang,E., Qian,M., Marincola,F.M., Alexander,H.R. and Libutti,S.K. (2002) Advantages of mRNA amplification for microarray analysis. Biotechniques, 33, 906–912, 914. [DOI] [PubMed] [Google Scholar]
- 25.Polacek D.C., Passerini,A.G., Shi,C., Francesco,N.M., Manduchi,E., Grant,G.R., Powell,S., Bischof,H., Winkler,H., Stoeckert,C.J.,Jr et al. (2003) Fidelity and enhanced sensitivity of differential transcription profiles following linear amplification of nanogram amounts of endothelial mRNA. Physiol. Genomics, 13, 147–156. [DOI] [PubMed] [Google Scholar]
- 26.Jenson S.D., Robetorye,R.S., Bohling,S.D., Schumacher,J.A., Morgan,J.W., Lim,M.S. and Elenitoba-Johnson,K.S. (2003) Validation of cDNA microarray gene expression data obtained from linearly amplified RNA. Mol. Pathol., 56, 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menssen A. and Hermeking,H. (2002) Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl Acad. Sci. USA, 99, 6274–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.