Abstract
TATA-binding protein (TBP)-related factor 2 (TRF2) is one of four closely related RNA polymerase II transcription factors. We compared the intracellular localizations of TBP and TRF2 during the cell cycle and mitosis in HeLa cells. We show that during interphase, endogenous or exogenously expressed TRF2 is located almost exclusively in the nucleolus in HeLa or Cos cells. TRF2 localization is not affected by stress or mitotic stimuli, but TRF2 is rapidly released from the nucleolus upon inhibition of pol I transcription or treatment by RNase. These results suggest that localization of HeLa TRF2 requires a nucleolar-associated RNA species. In contrast, in 3T3 fibroblast cells, exogenously expressed TRF2 localizes to the nucleoplasm. Constitutive expression of ectopic TRF2 in 3T3 cells leads to a prolonged S phase of the cell cycle and reduced proliferation. Together with previous data, our results highlight the cell-specific localization and functions of TRF2. Furthermore, we show that during cell division, HeLa TRF2 and TBP are localized in the mitotic cytoplasm and TRF2 relocalizes into the nascent nucleoli immediately after mitosis, whereas TBP reassociates with the chromatin. Although partially contradictory results have been reported, our data are consistent with a model where only small proportion of the cellular TBP remains associated with specific promoter loci during mitosis.
INTRODUCTION
RNA polymerase II (pol II) transcription initiation requires the formation of a multiprotein complex around the mRNA start site (Hampsey, 1998; Hampsey and Reinberg, 1999; Hochheimer and Tjian, 2003). A central factor in this process is the TATA binding protein (TBP). TBP associates with different sets of TBP-associated factors (TAFs) to form several distinct multiprotein complexes; SL1 required for pol I, TFIID and B-TFIID for pol II, and TFIIIB for pol III (Comai et al., 1992; Chiang et al., 1993; Hernandez, 1993).
TBP has a bipartite structure with a highly conserved C-terminal core domain that is 80% identical between yeast and mammals. The core folds as a molecular saddle responsible for binding DNA via the concave underside and interaction with general transcription factors as well as with a host of other regulatory proteins via the solvent exposed convex surface (Kim et al., 1993; Kim and Burley, 1994; Nikolov et al., 1995; Tan et al., 1996; Liu et al., 1998). To date, three proteins with strong sequence similarity to the core domain of TBP have been described. The first, Drosophila melanogaster TRF1, is expressed in a tissue-specific manner and functions as both an pol II and pol III transcription factor (Hansen et al., 1997; Holmes and Tjian, 2000; Takada et al., 2000). At present this gene has been described only in Drosophila.
A second protein with high homology to the TBP core domain found in most metazoans has received various names; TLF, TRF2, TLP, and TRP (Wieczorek et al., 1998; Dantonel et al., 1999; Maldonado, 1999; Moore et al., 1999; Ohbayashi et al., 1999; Rabenstein et al., 1999; Teichmann et al., 1999). More recently, a third protein TRF3 has been identified, but the function of this protein is unknown (Persengiev et al., 2003). In contrast TRF2/TLF has been well studied in vivo where it was first shown to be essential for expression of specific genes at the onset of zygotic pol II transcription in early Caenorhabditis elegans, Xenopus laevis, and zebrafish (Danio rerio) embryos (Dantonel et al., 2000; Kaltenbach et al., 2000; Veenstra et al., 2000; Muller et al., 2001; for review see Berk, 2000; Veenstra and Wolffe, 2001). In D. melanogaster, TRF2 is essential for embryogenesis and for cell cycle progression in SL2 cells (Hochheimer et al., 2002).
However, as it has been demonstrated recently, the situation is radically different in birds and in mammals. Targeted inactivation of mouse TRF2 shows that it is not required for embryogenesis, but that it is essential for spermatogenesis (Martianov et al., 2001; Zhang et al., 2001), where it plays a role in formation and/or maintenance of the chromocenter, a specialized chromatin structure present only in haploid spermatids (Martianov et al., 2002a). On the other hand, inactivation of TRF2 in chicken DT40 cells leads to a shortened G2 phase of the cell cycle and diminished apoptosis under stress conditions (Shimada et al., 2003). Aside from male sterility, trf2-/- mice otherwise appear normal, suggesting that there is no essential role of TRF2 in mammalian cell cycle in vivo.
Despite this wealth of data, the molecular function of TRF2 remains obscure. Although showing high sequence similarity to TBP, TRF2 does not bind classical TATA elements (Dantonel et al., 1999; Moore et al., 1999). In Drosophila, TRF2 is present in a large multiprotein complex containing proteins involved in chromatin remodeling and DREF (DNA replication-related element binding factor). DREF regulates a number of Drosophila genes involved in DNA replication and/or cell cycle progression whose promoters contain the cognate DRE element (Hochheimer et al., 2002). Hence in this case, TRF2 is recruited indirectly to the promoter of target genes and no cognate TRF2 binding sites have been identified in other organisms.
In HeLa cells, transfection, and overexpression studies in cells and in vitro experiments, have shown that TRF2 associates stably with TFIIA and can act as a repressor or activator of RNA polymerase II transcription (Moore et al., 1999; Teichmann et al., 1999; Ohbayashi et al., 2001). This observation suggests that TRF2 is a nuclear protein. However in DT40 and 3T3 cells, TRF2 has been suggested to be mainly present in the cytoplasm being imported into the nucleus only in G2 phase or in response to stress (Nakadai et al., 2004; Shimada et al., 2003).
Using specific antibodies, we have investigated the intracellular localizations of TRF2 and TBP. We show that during interphase, HeLa cell TRF2 is restricted to the nucleolus, whereas TBP localizes to the nucleoplasm. Nucleolar localization of TRF2 is not affected by stress or mitogenic signals, but it is rapidly translocated to the nucleoplasm upon inhibition of Pol I transcription or treatment with RNase A, suggesting that a nucleolar-associated RNA(s) directs intranuclear TRF2 localization. In contrast, in 3T3 cells TRF2 localizes to the nucleoplasm. Constitutive ectopic expression of TRF2 in these cells leads to reduced cell proliferation and a prolonged S phase. Furthermore, we also show that both TRF2 and TBP are largely excluded from the condensed chromatin during mitosis.
MATERIALS AND METHODS
Cell Culture
HeLa cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) at 1 g/L glucose supplemented with 2.5% fetal calf serum, 2.5% newborn calf serum. Cos-1 cells were grown in DMEM at 1 g/L glucose supplemented with 5% fetal calf serum. 3T3 cells were cultured in DMEM at 4.5 g/L glucose supplemented with 10% newborn calf serum.
UV Treatment
Irradiation was performed using a Stratagene UV Stratolinker 2400 (San Diego, CA) on confluent cells without medium. Immediately after irradiation fresh medium was added and the cells were incubated at 37°C in a humidified 5% CO2 incubator until they were fixed and stained at the indicated times.
Drug Treatments
Apoptosis was induced by treatment of cells with 10 or 20 ng · ml-1 of human tumor necrosis factor (Recombinant human TNF-α, R&D Systems Europe, Oxon, United Kingdom) directly added to the medium. Incubation was continued for the indicated times before fixing for different times. Tetradecanoyl phorbol 13-acetate (TPA; Sigma, St. Louis, MO) was added at a concentration of 100 ng · ml-1 as described in the results. Proteasome inhibitor MG132 (Calbiochem, La Jolla, CA) was used at a concentration of 50 μM. For treatment with epidermal growth factor (EGF; MC2, Sigma), cells were starved overnight in serum-free medium and rinsed with PBS before addition of fresh medium containing 50 μM EGF. For polymerase inhibition studies, cells were either treated by actinomycin D (Sigma) at 0.2 μg·ml-1 or α-amanitin (Boehringer, Indianapolis, IN) at 5 μg · ml-1 and incubated before fixing as described in Figure 4B.
Figure 4.
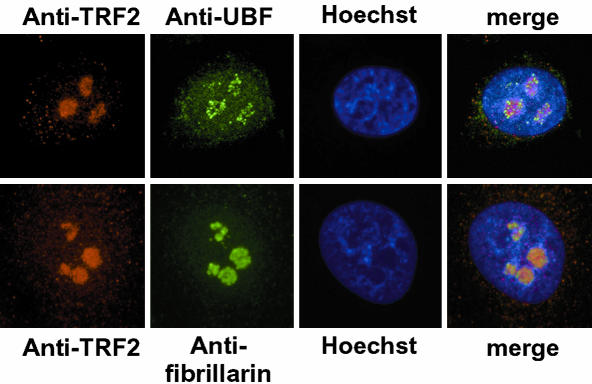
Localization of TRF2 within the nucleolus. HeLa cells were double-labeled with the indicated antibodies (TRF2-UBF, or TRF2-fibrillarin, red and green, respectively). A representative focal plane is shown to illustrate the localization of each protein within the nucleolus. Magnification, 400×.
RNase A Treatment
Cells were rinsed with PBS and then permeabilized with a low concentration of saponin (0.1 mg ·ml-1) at 4°C for 5 min. After washing with PBS, cells were incubated for 10 min at room temperature with 1 mg ·ml-1 RNase A (Sigma) in PBS or BPS alone. Then cells were rinsed twice in PBS before fixing.
Cell Synchronization
HeLa cells were incubated in the presence of 1 mM thymidine for 24 h. After three washes with PBS, fresh medium without thymidine was then added for 8 h. Cells were then blocked a second time in the presence of thymidine for 14 h before washing and release. Every hour for 8 h after release, an aliquot of cells was analyzed by fluorescence-activated cell sorting (FACS) and another, plated on glass slides, was fixed and stained for immunofluorescence. Nocodazole was purchased from Sigma and stored as a 10 mM stock solution in dimethylsulfoxide at -20°C. Immediately before use, stock solution was diluted to final concentration (10 μM) in complete culture medium. Cells cultured on coverslip were incubated at 37°C with 10 μM nocodazole for 9 h. For drug removal, cells were washed three times with PBS and then incubated at 37°C in a fresh complete culture medium for various time periods before fixing.
Transfection and Generation of Stable Cell Lines
HeLa cells were transfected by calcium phosphate precipitation as previously described (Mengus et al., 1995). 3T3 cells were transfected with using Fugene (Roche, Meylan, France). To generate stable cell lines, 3T3 cells were transfected with the Flag-TRF2 expression vector along with a vector encoding resistance to puromycin. Forty-eight hours after transfection, cell were selected in medium containing 0.3 μg ·ml-1 of puromycin for 10 d until well-isolated clones were observed. The clones were then picked and expanded. Flag-TRF2–expressing clones were identified by immunofluorescence and immunoblotting. A total of 10 Flag-TRF2–expressing clones were analyzed and the results of two representative clones are presented.
Immunofluorescence
Immunofluorescence was performed essentially as described (Martianov et al., 2002a). Cells were directly grown on glass slides inside 100-mm plastic tissue culture dishes. Before fixing, culture medium was removed and the slide was washed with PBS and then fixed by incubation for 15 min in PBS + 10% HCHO. Cells were then permeabilized by incubation for two times for 10 min in PBS + 0.1% Triton X-100 (PBST). Subsequently, the slide was washed three times with PBST + 5% normal goat serum (NGS) and incubated 9 h at 4°C with the primary antibody solubilized in PBS + NGS5%. The slide was washed three times with PBST and incubated again two times for 10 min in PBST. Afterward the slide was incubated with the second antibody solubilized in PBS + 0.1% Triton X-100 + NGS5% coupled with Cyanin3 or Alexa 488 for 1 h at room temperature. Next, the slide was washed three times with PBST and incubated again two times for 10 min in PBST, followed by the last wash for 5 min in PBS. Finally the slide was stained with Hoechst and covered with antifading medium (glycerol 80%, PBS 20%, propylgalate 5% p/v). The antibodies against TBP and TRF2 were as previously described (Lescure et al., 1994; Martianov et al., 2001). Anti-Flag antibody was purchased from Jackson ImmunoResearch (West Grove, PA) and antibodies against fibrillarin and UBF were gifts from D. Hernandez-Verdun (Fomproix et al., 1998), antibody against Pol I was a gift from C. Kedinger (Kedinger and Chambon, 1972), antibody against TFIIAαβ was a gift from L Tora, and rabbit polyclonal antibody against TBP was a gift from T. Oelgeschlager (Christova and Oelgeschlager, 2002). Immunoblots were performed by standard procedures and revealed by electrochemiluminescence.
Preparation of Cell Extracts
Extracts from HeLa cells were prepared by lysis in buffer A (10% glycerol, 50 mM Tris, pH 7.9, 1 mM EDTA, 0.1% NP40, 1 mM DTT) containing 0.05 M KCl by three cycles of freeze thaw. The lysate was centrifuged at 2500 rpm in a microfuge for 3 min to pellet the nuclei, and the supernatant was recovered as the cytoplasmic fraction. The pellet was resuspended and rinsed again with buffer A + 0.05 M KCl and recentrifuged as before. The nuclear pellet was then resuspended in buffer A + 0.5 M KCl and vortexed before incubation on ice for 30 min. The lysate was the centrifuged at 13,000 rpm in a microfuge for 10 min, and the supernatant containing the nuclear extract was recovered.
Enrichment of nucleoli was performed essentially as described by Andersen et al. (2002). HeLa cells were resuspended in buffer A without glycerol and 0.05 M KCl and then centrifuged at 4000 rpm for 10 min in a microfuge. The cell pellet was resuspended in 3 ml of 0.25 M sucrose with 0.01 M MgCl2 and layered on 3 ml of 0.35 M sucrose with 0.0005 M MgCl2 and centrifuged for 5 min at 1430 g at 4°C. The pellet was then resuspended in 350 μl of 0.35 M sucrose with 0.0005 M MgCl2 and pulse sonicated six times for 8 s. This was then layered on 1.5 ml of 0.88 M sucrose with 0.0005 M MgCl2 and centrifuged for 15 min at 5000 × g at 4°C. The pellet containing the enriched nucleoli fraction was then resuspended 50 μl of buffer A + 0.05 M KCl without glycerol.
Fluorescence-activated Cell Sorting
FACS analysis of the cell cycle by propidium iodide incorporation was performed by standard procedures on a FACSCalibur (Becton Dickinson, Erembodegem, Belgium). Data was analyzed using the ModFit LT v.3.1 program (Verity Software House, Topsham, ME).
RESULTS
TRF2 Localizes to the Nucleolus in HeLa and Cos Cells
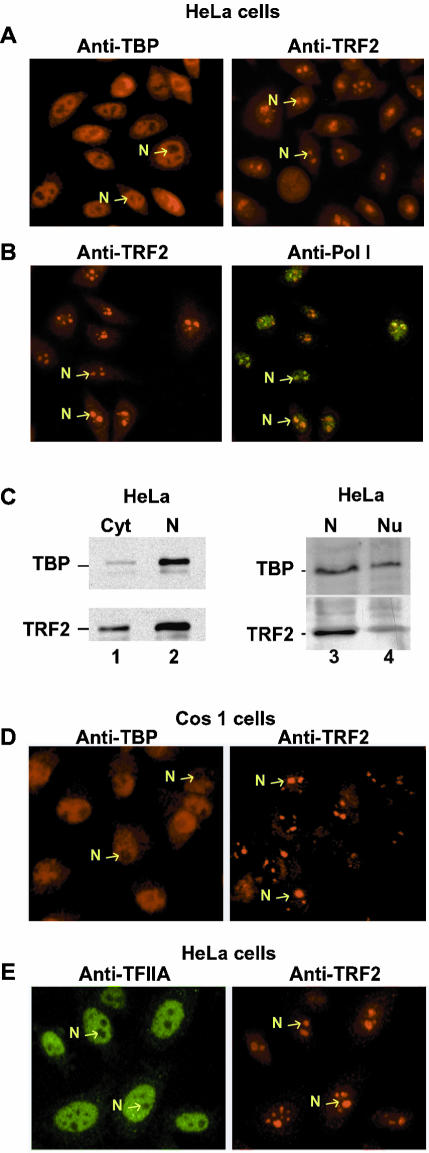
We have previously described monoclonal antibodies (mAb 3G3 and 2A1) that specifically recognize either TBP or TRF2 (Lescure et al., 1994; Martianov et al., 2001, 2002a). mAb 3G3 detects TBP in the nucleoplasm of interphase HeLa cells (Figure 1A, left panel). In contrast, antibody 2A1 reacts strongly with a nuclear structure(s), but not with the nucleoplasm (Figure 1A, right panel). Between 2 and 4 TRF2-reactive structures are observed per nucleus. Such large nuclear structures may correspond to the nucleoli. To test this, HeLa cells were double-stained with the 2A1 mAb (Figure 1B, left panel) and a polyclonal antibody against pol I (Figure 1B, right panel). Both of these antibodies clearly label the same structures, demonstrating that they correspond to the nucleoli. Similar results were obtained in Cos1 cells, where TRF2 was also located in the nucleoli, whereas as expected TBP was nucleoplasmic (Figure 1D). Although TBP is present in the nucleolar SL1 complex, it is not readily detected under these conditions because of its low abundance (Jordan et al., 1996 and see below).
Figure 1.
Distinct localizations of TBP and TRF2 in HeLa cells. (A) HeLa cells were labeled with antibodies against TBP or TRF2 as indicated. (B) HeLa cells were double-labeled with antibodies against TRF2 (red) or pol I (green) as indicated. (C) 20 μg of HeLa cell cytoplasmic (C), nuclear (N), or nucleolar-enriched fraction were separated by SDS-PAGE, and TBP and TRF2 were revealed by immunoblot using the 3G3 and 2A1 antibodies. (D) Cos1 cells labeled with antibodies against TBP or TRF2. (E) HeLa cells were double-labeled with antibodies against TFIIA (red) or TRF2 (green) as indicated. Representative nucleoli are indicated with arrows. Magnification, 40× in all panels.
Cytoplasmic and nuclear extracts were prepared from HeLa cells, and the presence of TBP and TRF2 was determined by immunoblotting. TRF2 was present in the cytoplasmic fraction, but was enriched in the nuclear fraction, whereas the majority of TBP was present in the nuclear extract (Figure 1C, lanes 1 and 2). TRF2 and TBP were also detected in extracts from fractions enriched in nucleoli (Figure 1C, lanes 3 and 4).
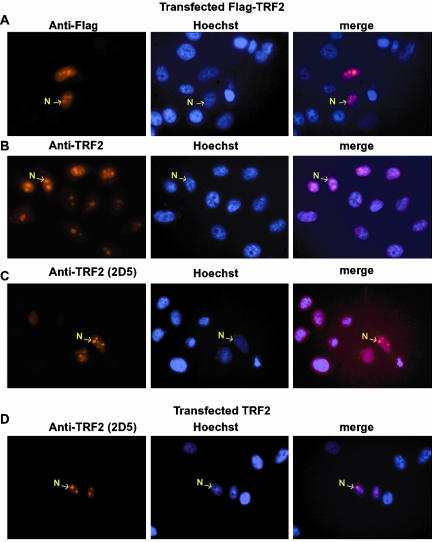
We have previously shown that mAb 2A1 recognizes TRF2 in Western blot and immunohistochemistry in testis from wild-type, but not trf2-/- mice (Martianov et al., 2002a). To further demonstrate the specificity of the HeLa cell nucleolar labeling, cells were transfected with a vector expressing a Flag-tagged TRF2 and the transfected cells labeled with either anti-TRF2 or anti-Flag antibodies. Using the anti-Flag antibody, the transfected Flag-TRF2 could be clearly seen to accumulate within the nucleoli of the transfected cells, while no labeling was seen in the adjacent untransfected cells (Figure 2A). Similarly, much stronger nucleolar labeling was seen with the anti-TRF2 antibody in the subpopulation of transfected cells (Figure 2B). We also used a second mAb 2D5 which is not sufficiently sensitive to detect endogenous TRF2, but clearly detects transfected Flag-TRF2 in the nucleoli of the transfected cells (Figure 2C). Analogous results were observed with a third independent antibody 2D7 (unpublished data). Transfected native TRF2 could also be detected within the nucleoli of transfected cells (Figure 2D). Together these results confirm that endogenous HeLa cell TRF2 is nucleolar and that the transfected protein also accumulates preferentially in the nucleoli.
Figure 2.
Nucleolar localization of TLF in transfected cells. (A–D) The antibodies used for labeling are indicated above each lane. Left panel, the immunostaining; center panel, the corresponding Hoechst-stained nuclei; and right panel, the merged images. Representative nucleoli are indicated with arrows. Magnification, 40× in all panels.
It has previously been suggested that TRF2 stably associates with TFIIA in HeLa cells (Teichmann et al., 1999). We therefore double-labeled HeLa cells with a polyclonal anti-TFIIA antibody and mAb 2A1. TFIIA is present in the nucleoplasm, while TRF2 is nucleolar and no double staining is observed (Figure 1E). Hence in intact HeLa cells, the majority of the TFIIA and TRF2 populations are not in the same cellular compartments and therefore cannot constitutively associate with each other.
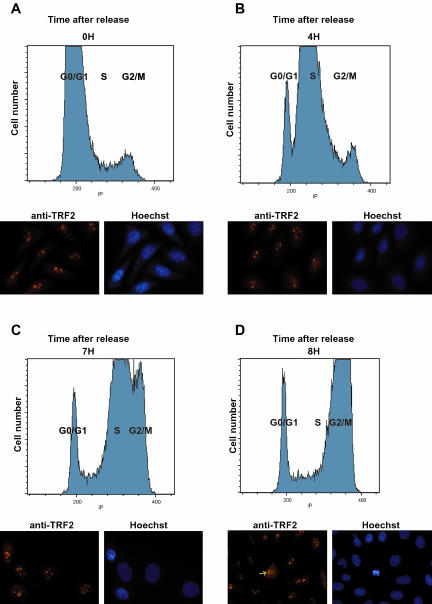
We next synchronized the HeLa cell population to follow TRF2 localization through the S and G2/M phases of the cell cycle. Cells were blocked at the entry to the S phase with thymidine and then analyzed from 0 to 8 h after release by FACS analysis and immunofluorescence. Three to 4 h after release the cells, predominantly in G1 at time 0, moved into the S phase (Figure 3, A and B). By 7 h after release, the majority of the cells were in late S phase (Figure 3C) and by 8 h in G2/M (Figure 3D). Immunofluorescence of these cell populations indicated that TRF2 remained in the nucleolus throughout the S phase and located only into the nucleoplasm upon disruption of the nucleoli during the prophase of mitosis (Figure 3, A–D, and see below). Thus, in HeLa cells TRF2 does not translocate to the nucleoplasm during the S and G2 phases of the cell cycle.
Figure 3.
(A–D) TRF2 is nucleolar during S and G2 phases of the cell cycle. Cells were synchronized with thymidine and aliquots of cells were analyzed every hour for 8 h after release. The FACS analysis at the indicated times after release is shown in the top half of each panel and the immunolabelling with anti-TRF2 antibody in the bottom half. In the 8-h panel a prophase cells is observed (arrow) where TRF2 is being released from the nucleolus.
To examine the localization of TRF2 within the nucleolus, confocal microscopy was performed with antibodies against the pol I transcriptional activator UBF or the structural component fibrillarin and TRF2. UBF is associated with the rDNA loci within the nucleolus, which can be seen as almost discrete speckles (Figure 4, top panel). In contrast, TRF2 labeling is much more homogenous and does not show the characteristic speckled labeling seen with UBF (Figures 4 and 1B). Labeling of TRF2 rather resembles that of fibrillarin (Figure 4, bottom panel). TRF2 is therefore not selectively associated with the rDNA loci, but more homogeneously localized within the nucleolus.
Nucleolar TRF2 Localization Is Not Affected by Stress or Mitogenic Response
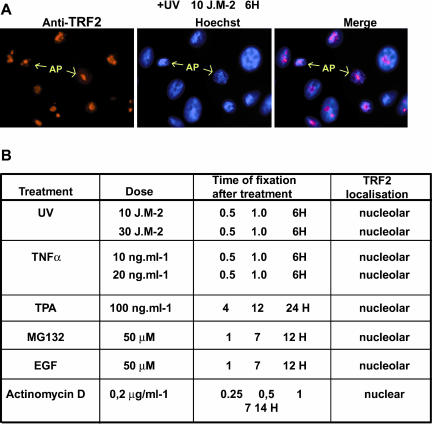
It has been shown that TRF2 translocates to the nucleoplasm in chicken DT40 after exposure to stress stimuli (Shimada et al., 2003). If TRF2 were to play a role in modulating stress response also in HeLa cells, we reasoned that such stimuli may induce a translocation of TRF2 from the nucleolus to the nucleoplasm where it could act to modulate gene expression. To test this idea, HeLa cells were treated with various stimuli, such as UV irradiation, and tumor necrosis factor alpha (TNF-α; see Figure 5A). Treatement with UV did not cause a change in TRF2 localization either shortly after exposure (unpublished data; see Figure 5A) or even 6 h after exposure (Figure 5B). At these late times, cells with condensed nuclei corresponding to apoptotic cells induced by the UV exposure could be detected. Even in these cells, TRF2 remained localized within the nucleolus (Figure 5A). Thus, TRF2 is not relocalized as part of the immediate early response to UV irradiation or during the subsequent apoptosis.
Figure 5.
TRF2 is not released from the nucleolus in response to stress signals. (A) HeLa cells were exposed to UV light as indicated and fixed 6 h after exposure. Representative apoptotic nuclei are indicated by the arrows. (B) A table summarizing TRF2 localization after the indicated treatments. The doses of each compound are indicated. Cells were fixed and labeled at the indicated times after treatments.
Treatment with TNF-α also induced apoptosis, but no relocalization of TRF2 (summarized in Figure 5B). Similarly, treatment with phorbol esters (TPA) to activate intracellular signaling pathways had no effect on TRF2 localization. It was also possible that TRF2 was localized in the nucleolus to protect it from targeted proteolysis. However in the presence of MG132, which blocks the proteosome and other targeted degradation pathways, no additional nucleoplasmic TRF2 was observed. The HeLa cells were also serum starved for 24 h and then treated with EGF. Neither serum starvation nor mitogenic stimuli had an effect on TRF2 localization. Together these results do not support the idea that TRF2 is involved in stress and growth response of HeLa cells.
Nucleolar Localization of TRF2 Requires pol I Transcription and an RNA
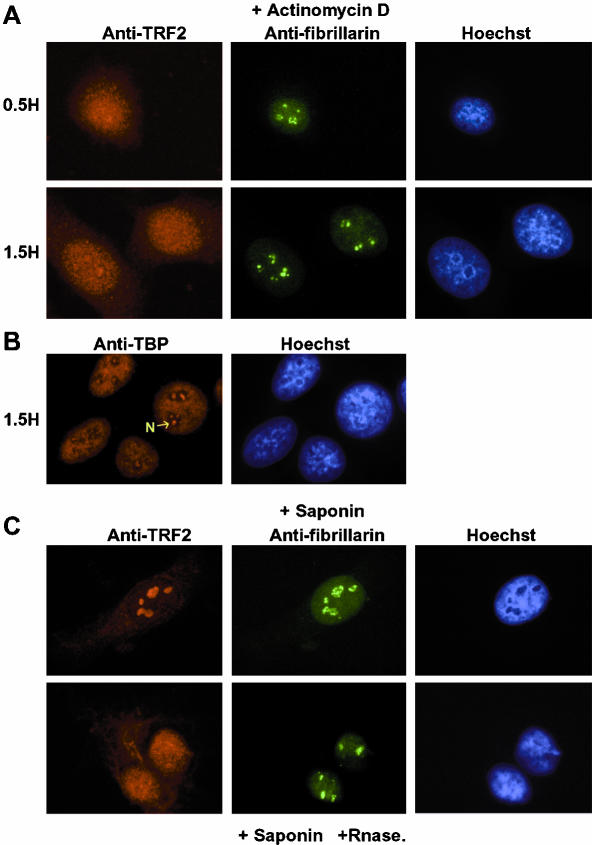
We next tested the effect or arresting pol I transcription using low doses of actinomycin D. In contrast to the above treatments, TRF2 rapidly within 15 min and stably relocalized to the nucleoplasm in the presence of actinomycin D (Figure 6A, summarized in Figure 5A). Moreover, a small amount of TRF2 was also detected in the cytoplasm. Treatment with actinomycin D for these short times does not lead to a total breakdown of the nucleoli as revealed by double-labeling with antifibrillarin (Figure 6A, top panel, and Andersen et al., 2002). After 1.5 h of actinomycin D treatment, residual nucleoli could still be observed by staining with fibrillarin, whereas TRF2 was now nucleoplasmic with detectable cytoplasmic staining (Figure 6A, bottom panel). Hence, nucleolar localization of TRF2 is rapidly abolished upon arrest of pol I transcription. In contrast, TRF2 localization was unaffected when pol II transcription was arrested by α-amanitin (unpublished data).
Figure 6.
Release of TRF2 from the nucleolus after treatment with actinomycin D and RNase. (A) Cells were treated with actinomycin D for the indicated times and double-labeled with antibody against TRF2 (red) and fibrillarin (green). (B) Labeling of actinomycin D–treated cells with antibody against TBP. (C) Cells were treated with saponin or saponin and RNase as indicated and double-labeled with antibody against TRF2 and fibrillarin.
Interestingly, after prolonged actinomycin D treatment, distinct foci of TBP within the nucleoli could now be seen. This likely corresponds to TBP bound to the rDNA loci which becomes visible due to a local concentration of the loci within the nucleolus (Figure 6B and Jordan et al., 1996).
Treatement of nuclei with RNase A has previously been reported to provoke disorganization of heterochromatin (Maison et al., 2002). To test whether RNA was involved in TRF2 nucleolar localization, we next permeabilized cells with saponin and treated the nuclei with RNase A. Treatment with saponin alone did not affect TRF2 localization (Figure 6C, top panel). In contrast, in the presence of saponin and RNase A, TRF2 was now distributed throughout the nucleoplasm with some also in the cytoplasm. RNase A treatment did not lead to dislocation of the nucleolus as revealed by double-labeling with antifibrillarin. This result indicates that an RNA(s) is required for nucleolar localization of TRF2.
Nucleolar Localization of TRF2 Is Cell Specific
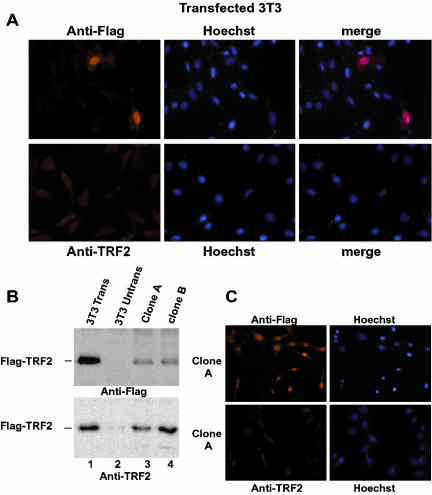
We also sought to localize TRF2 in 3T3 cells with our antibody. Surprisingly however, no significant signal could be observed despite the fact that mouse and human TRF2 have identical amino acid sequences. Labeling was equivalent to background seen with the anti-Flag antibody on untransfected cells (Figure 7A). We therefore transfected 3T3 cells with the vector expressing Flag-TRF2. Strikingly, TRF2 did not localize to the cytoplasm as may have been expected from previous reports, but accumulated mainly in the nucleoplasm as detected by the anti-Flag antibody (Figure 7A, top panel, where two transfected cells are surrounded by several untransfected cells). Thus in contrast to HeLa cells where transfected Flag-TRF2 localizes to the nucleoli, in 3T3 cells, it localizes to the nucleoplasm. Nevertheless, using the anti-TRF2 antibody, we did not detect the transfected Flag-TRF2 (Figure 7A, bottom panel). As a control we reprobed the slides of the transfected cells with the anti-Flag antibody, and although no transfected cells could be detected with the anti-TRF2 antibody, they were visible with the anti-Flag antibody (unpublished data).
Figure 7.
Localization of TRF2 in 3T3 cells. (A) Transfected 3T3 cells were labeled with antibody against TRF2 or Flag as indicated. Transfected cells expressing TRF2 could be observed with the anti-Flag antibody, whereas only background staining was seen with the anti-TRF2 antibody. (B) Immunoblots of extracts cell lines stably expressing Flag-TRF2. Ten micrograms of cell extract were used in each lane. Lane 1, extracts from transiently transfected cells; lane 2, un transfected cells; and lanes 3 and 4, extracts from stably cell lines. Flag-TRF2 was detected by either anti-TRF2 or anti-Flag as indicated. (C and D) Immunofluorescence of cell line 1 with anti-Flag or anti-TRF2 as indicated. Strong nuclear expression of TRF2 is observed using the Flag antibody, whereas only background staining was seen with the anti-TRF2 antibody.
To confirm these results, we isolated 3T3 cell lines stably expressing Flag-TRF2. TRF2 could be detected in extracts from these cells using both the anti-Flag and anti-TRF2 antibodies (Figure 7B). In these cells, Flag-TRF2 also locates mainly to the nucleoplasm with a small amount present in the cytoplasm (Figure 7C). Therefore, there is no specific nucleolar localization of TRF2 in 3T3 cells it is in fact excluded from the nucleoli in these cells (Figure 7C). As observed in the transiently transfected cells, no specific nucleoplasmic signal could be seen using the anti-TRF2 antibody. Thus although these cells express Flag-TRF2, it is not detected by immunofluorescence using the anti-TRF2 antibody, suggesting that the epitope is masked or modified in this cell type.
Ectopic TRF2 Nuclear Expression in 3T3 Cells Affects the S phase of the Cell Cycle
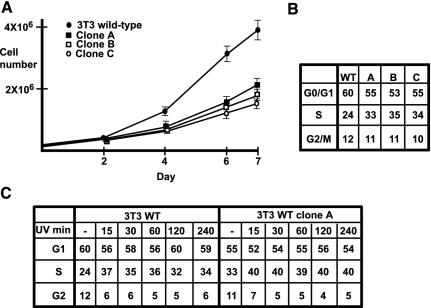
We analyzed the effect of constitutive TRF2 expression on the growth properties of 3T3 cells. We first compared the growth rate of the wild-type parental cells with those of three independent TRF2-expressing clones. Compared with wild-type, the TRF2-expressing clones showed a significantly lower proliferation rate (Figure 8A). To determine whether TRF2 expression affects a particular phase of the cell cycle, we subjected the cells to FACS analysis. In each clone, a significant increase in the number of cells in S phase was observed compared with the wild-type cells (Figure 8B). In wild-type cells, an average of 24% was found, whereas in each clone this value was 33–35%. In contrast, each TRF2-expressing clone showed a slightly reduced proportion of G1 phase cells, but no significant difference in the G2 population. On exposure to UV, the proportion of cells in S phase increased and the proportion in G2 decreased in a similar manner in wild-type and TRF2-expressing cells (Figure 8C, analogous results were obtained for clones B and C, unpublished data). The overall proportion of S phase cells is higher in the TRF2-expressing cells, but this simply reflects the higher proportion in S phase in the absence of UV treatment. Hence, constitutive ectopic TRF2 expression in 3T3 cells affects the S phase of the cell cycle, but does not affect response to UV-induced stress.
Figure 8.
Properties of TRF2-expressing 3T3 cells. (A) Growth curves of each cell line; 2.5 × 104 cells of each line were plated at day 0. Viable cells were counted 2, 4, 6, and 7 d after seeding. The results show the average of three different experiments. (B) FACS analysis of the cell cycle. The values represent the proportion of cells at each stage of the cycle. The results are averages of three experiments rounded to the nearest whole number. (C) Cells were exposed to UV irradiation (150 J · m-2) and the cell cycle analyzed by FACS after the indicated times. The values are averages of two experiments rounded to the nearest whole number.
Localization of TRF2 and TBP during Mitosis
We next examined localization of HeLa cell TRF2 during mitosis. In the thymidine-blocked cells TRF2 remained in the nucleoli throughout the S and G2 phases, but appeared to be released into the nucleoplasm upon disruption of the nucleoli during prophase. To better investigate this, cells were blocked in G2/M for 9–12 h using nocodazole, which was then washed away. Cells were then fixed at various times after release from the block and immunolocalization of TRF2 was performed.
Immediately after release from blocking with nocodazole, in late G2/M, TRF2 could be seen in the nucleoplasm (Figure 9A). This is due to disruption of the nucleoli after prolonged exposure to nocodazole. At prophase, TRF2 remained in the nucleoplasm, but was progressively excluded from the condensing chromatin (Figure 9A). Exclusion of TRF2 from the condensed mitotic chromatin was observed during metaphase and anaphase (Figure 9, A and B). These results were confirmed by confocal microscopy (unpublished data and see below). However, TRF2 was seen concentrating in the nascent nucleoli immediately after cell division (Figure 9B). Thus during mitosis, TRF2 is not associated with the residual nucleolar organizing regions (NORs), but like other pol II transcription factors, is excluded from the mitotic chromatin.
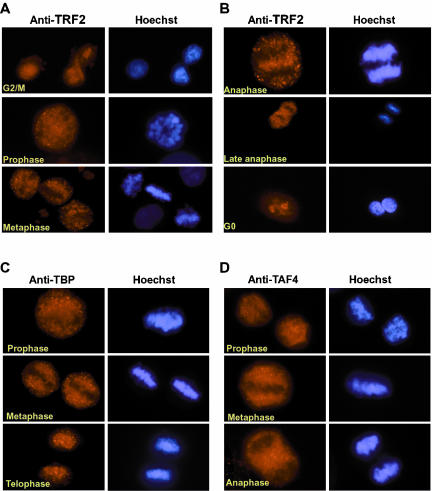
Figure 9.
Localization of TRF2, TBP, and TAF4 during mitosis. (A and B) Cells were labeled with antibody against TRF2 and counterstained with Hoechst. The stage of mitosis is indicated. (C and D) A similar analysis of mitotic stages with monoclonal antibodies against TBP and TAF4.
As controls in these experiments, we also labeled cells with antibodies against TBP. Somewhat conflicting results concerning TBP localization during mitosis have been reported. TBP was originally observed in the mitotic cytoplasm (Segil et al., 1996), whereas a more recent report rather suggested that TBP associates with the condensed mitotic chromatin (Chen et al., 2002). Using our monoclonal anti-TBP antibody, we observe TBP to be excluded from the condensing mitotic chromatin during prophase (Figures 9C and 10A). At later stages, TBP remains located in the mitotic cytoplasm and relocalizes with the chromatin only at late anaphase/telophase (Figures 9C and 10B).
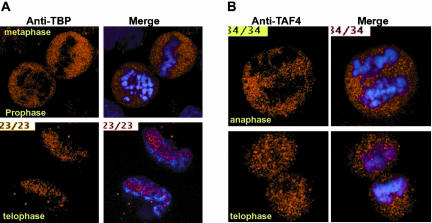
Figure 10.
Exclusion of TBP and TAF4 from condensed mitotic chromatin. (A and B) Representative focal planes from confocal sections are shown. The left panel shows the antibody staining with the polyclonal rabbit antisera against TBP or mAb against TAF4 and the right panel the merge with the Hoeschst stained DNA. The mitotic stages are indicated.
To confirm these results, we performed similar experiments with a rabbit polyclonal sera that has previously been used to perform chromatin immunoprecipitations on mitotic cell chromatin (Christova and Oelgeschlager, 2002). Using this antisera, confocal microscopy confirms that the vast majority of TBP is excluded form the condensed mitotic chromatin until telophase, where it reassociates with the decondensing chromatin (Figure 10A). However even at this stage, confocal microscopy shows that TBP remains excluded from the most condensed chromatin regions. At each stage, the exclusion can be seen much better by confocal analysis than by conventional microscopy, where the rounding of the cell and the superposition of the different focal planes gives a less clear image. Similar experiments were performed with antibodies against the TFIID/TFTC subunit TAF4 (Mengus et al., 1997). Like TBP, TAF4 remained excluded from the condensed chromatin until telophase, where reassociation with the chromatin was observed (Figures 9D and 10B and unpublished data). These results indicate that the vast majority of TBP and TAF4 are not associated with the chromatin during mitosis.
DISCUSSION
Regulation of TRF2 Activity through Compartmentalization
In this study we show that TRF2 localizes to the nucleolus in cultured HeLa and Cos cells. Experiments using synchronized cell populations show that TRF2 is nucleolar throughout the interphase. Our conclusions are based on experiments involving the use of three different monoclonal antibodies directed against endogenous and exogenously expressed TRF2 and the use of anti-Flag antibody to detect exogenously expressed Flag-TRF2. In all cases, TRF2 is observed in the nucleolus. Biochemical fractionation of HeLa cells shows the presence of TRF2 in nuclear extracts and in fractions enriched in nucleoli. We do however detect significant amounts of TRF2 in the cytoplasmic fraction. In this respect, we have also observed significant amounts of TRF2 in cytoplasmic extracts from mouse testis (our unpublished data) despite the fact that TRF2 is clearly nuclear in male germ cells (Martianov et al., 2002a). It is unclear why TRF2 leaks out from the nucleus at low ionic strength, whereas in both male germ cells and HeLa cells immunofluorescence clearly shows the protein to be nuclear. This tendency to leak from the nucleus may help to explain a previous suggestion (Shimada et al., 2003) that HeLa TRF2 was localized to the cytoplasm.
TRF2 does not display the characteristic intranucleolar localization seen with pol I or UBF, which are associated with the rDNA loci (Zatsepina et al., 1993). It is therefore unlikely that TRF2 is required for pol I transcription. This conclusion is supported by the observation that TRF2 is not ubiquitously expressed and that trf2-/- mice and DT40 cells are viable. Instead, the localization of TRF2 within the nucleolus resembles that of a structural protein like fibrillarin. This rather suggests that TRF2 is perhaps stored within the nucleolus in an inactive form. Indeed, regulation of transcription factors by confinement within the nucleolus has previously been described, for example, in the case of Mdm2, where its translocation to the nucleolus physically separates it from p53, allowing p53 to escape proteolytic degradation and accumulate in the nucleoplasm (for review see Horn and Vousden, 2004). Similarly, several other proteins involved in growth control such as basic and acidic fibroblast growth factors are also localized in the nucleolus. These observations highlight the idea of a multifunctional nucleolus as more than just a specialized site of rRNA transcription and ribosome biosynthesis (see Pederson, 1998; Andersen et al., 2002). TRF2 was not, however, detected in proteomic screens of the HeLa cell nucleolus (Leung et al., 2003), perhaps because it is not a particularly abundant protein. Indeed, other less abundant nucleolar proteins have not been observed in this screen.
Why would HeLa cells store TRF2 in the nucleolus? It has been shown that TRF2 stably associates with TFIIA in HeLa cell extracts (Teichmann et al., 1999). This interaction appears to be stronger than that with TBP (Nakadai et al., 2004). Nevertheless, little or no TFIIA was detected in the nucleolus and consequently the majority of TRF2 and TFIIA do not colocalize within the nucleus. It is possible that the observed complex is formed only in the extracts after cell lysis, although we cannot exclude that possibility that a small fraction of each protein constitutively associate with one another. Moreover, addition of TRF2 to in vitro transcription systems can sequester TFIIA and repress transcription (Moore et al., 1999). The ability of TRF2 to sequester TFIIA and interfere with transcription may explain why its presence in the nucleoplasm is regulated. The compartmentalization of TRF2 through localization in the nucleolus may correspond to a strategy for preventing an inappropriate interaction with TFIIA and limiting its transcriptional activity. Why this may be necessary in HeLa and Cos cells, but not in other cell types (see below) is unknown.
Distinct Intracellular Localization of TRF2 in Different Cell Types
Although TRF2 is nucleolar in HeLa and Cos cells, this is not observed in all cell types. In our experiments, transiently transfected TRF2 localizes to the nucleoplasm of 3T3 cells and is also mainly nucleoplasmic in 3T3 cell lines stably expressing TRF2. We obtained analogous results using transfected mouse embryonic fibroblasts (unpublished data). Most importantly however, these results show that exogenous TRF2 does not concentrate in the nucleoli of 3T3 cells, as it does in HeLa cells. Moreover, in vivo in mice, TRF2 is imported rapidly into the nucleus in early stage pachytene cells during spermatogenesis and accumulates in both the hetero- and euchromatin compartments of the nucleoplasm. At no stage does TRF2 accumulate in the nucleolus of pre- or postmeiotic male germ cells (Martianov et al., 2002a). Similarly, TRF2 is also nucleoplasmic in early C. elegans and zebrafish embryos (Dantonel et al., 2000; Muller et al., 2001). The ability of TRF2 to localize in the nucleolus is therefore cell specific and not a ubiquitous feature in cultured cells in vitro or in tissues in vivo.
We also found in these experiments that the TRF2 expressed in 3T3 and embryonic fibroblast cells could not be detected by immunofluorescence using our anti-TRF2 antibody, but it could be readily detected on immunoblots. One possibility is that, in 3T3 cells, the epitope for our antibody in TRF2 is masked possibly because TRF2 forms a complex with other proteins that cover the epitope. This possibility is consistent with previous reports showing that Drosophila TRF2 can form a macromolecular complex (Hochheimer et al., 2002). It is striking that we can detect TRF2 in HeLa cells and in male germ cells, but not in 3T3 cells. This suggest that TRF2 may associate with distinct partners in different cell types. The potential TRF2 partner proteins in 3T3 cells remain to be identified.
Nucleolar TRF2 Localization Requires an RNA
In DT40 and 3T3 cells TRF2 is proposed to translocate to the nucleus in response to stress signals (Shimada et al., 2003) and regulate genes involved in cell cycle and stress response. The stress signals that induce import of TRF2 into the nucleus in 3T3 cells do not release TRF2 from the nucleolus in HeLa cells. This observation strongly suggests that TRF2 is not required for gene expression during the stress response of HeLa cells. Interestingly, UV and other stress inducers also disrupt nucleolar structure (for review see Rubbi and Milner, 2003), but the changes in nucleolar structure induced by stress do not lead to release of TRF2.
In contrast, our results show that TRF2 can be rapidly released from the nucleolus upon inhibition of pol I transcription. This treatment does not release TBP or other known pol I transcription factors, which remain associated with the rDNA loci (Zatsepina et al., 1993; Jordan et al., 1996; Gebrane-Younes et al., 1997). Similarly, treatment with RNase also releases TRF2 from the nucleolus. Neither treatment leads a total dislocation of the nucleolus, but they do release TRF2 perhaps as the result of more minor alterations in nucleolar structure. One interpretation of these observations is that TRF2 nucleolar localization requires rRNA. However, rRNA is long lived compared with the rapid dissociation of TRF2 from the nucleoli in actinomycin D–treated cells. Moreover, as rRNA is obviously ubiquitous, one would expect rRNA to direct TRF2 to the nucleolus in all cell types, but this is not what is observed.
Alternatively, because TRF2 nucleolar localization is cell specific, it is possible that another RNA species may be involved. This RNA may be cell specific or at least its ability to localize TRF2 to the nucleolus would be cell specifically regulated. Because actinomycin D treatment leads to defined changes in nucleolar structure (Andersen et al., 2002), it is possible that this RNA would be released from the nucleolus along with TRF2 under these conditions. One possible candidate would be a member of the small nucleolar (sno)RNA family. These RNAs associate with proteins to form a large number of small nucleolar ribonucleoprotein particles (snoRNPs) found associated with fibrillarin. SnoRNAs are involved in modifications of rRNA and also of U6 snRNA, mainly 2′-O-ribose methylation and conversion of uridine to pseudouridine (for review see Bachellerie et al., 2002; Kiss-Laszlo et al., 1996; Pederson, 1998). Several hundred snoRNAs are known, and some are cell specific, and for many their functions remain undefined (Bachellerie et al., 2002; Filipowicz and Pogacic, 2002). One or several snoRNAs would therefore be good candidates for localizing TRF2 to the nucleolus in a cell-specific manner.
Function of TRF2 in Mammalian Cells
We established 3T3 cells that constitutively express TRF2 in the nucleoplasm. TRF2 expression leads to reduced cell proliferation and a prolonged S phase of the cell cycle, but no change in the G2/M phase is observed. Nuclear expression of TRF2 does not significantly affect the stress response of the cells to UV. However, it is worth noting that the levels of TRF2 expression in our wild-type 3T3 cells are below the levels of detection, whereas the ectopically expressed protein is readily detectable. Despite this overexpression, the effects on the cell cycle remain rather modest. Moreover, in HeLa cells TRF2 is sequestered in the nucleolus throughout the S phase and hence is unlikely to have a significant function at this stage. Together with the observation that, aside from male sterility, trf2-null mice appear normal, these results do not support a general role of TRF2 in the mammalian cell cycle.
The effects of TRF2 expression in 3T3 cells differ from the role attributed to TRF2 in chicken DT40 cells. In DT40 cells, inactivation of TRF2 leads to a shortened G2/M checkpoint, whereas its constitutive expression in the nucleus prolongs the cell cycle because of accumulation of cells at the G2/M transition. These results show that TRF2 expression has distinct effects on the cell cycle in 3T3 and DT40 cells. These observations are reminiscent of those obtained with TBP. DT40 cells heterozygous for TBP have an altered cell cycle (Um et al., 2001), whereas TBP heterozygous mouse ES cells and mice are normal (Martianov et al., 2002b). Thus, results obtained in DT40 cells may not be always directly transposable to mammalian cells.
Release of TRF2 and TBP from Chromatin during Mitosis
Unlike pol I factors that remain associated with the residual NORs (Sirri et al., 1999), TRF2 is released from the nucleolus as it breaks down in the prophase of mitosis and rapidly reassociates with nascent nucleoli immediately after cell division. Otherwise, during mitosis, TRF2 localizes in the cytoplasm and is excluded from the condensed DNA. In this respect, it behaves similarly to TBP and TAF4, which are also localized in the cytoplasm during most of mitosis.
Our observation of TBP in the mitotic cytoplasm is in agreement with previous observations of Segil et al. (1996). More recently, however, it has been suggested that a large proportion of the TBP pool remains stably associated with the condensed mitotic chromatin (Chen et al., 2002). These experiments were performed with transfected GFP-TBP fusion proteins. Fluorescence from GFP-TBP revealed colocalization with mitotic chromatin, whereas antibody staining showed the endogenous TBP in the mitotic cytoplasm. This difference was interpreted as the result of masking of the antibody epitope in endogenous TBP in the condensed chromatin, a limitation not encountered with GFP fluorescence. Immunofluorescence is not, however, an inherent limitation, as we have shown previously that almost all the cellular pool of TAF7L, a testis-specific TAF, remains associated with condensed meiotic chromatin in spermatocytes, whereas TBP and several other TAFs are excluded (Pointud et al., 2003).
Here we have used a polyclonal antibody that has previously been shown to immunoprecipitate TBP from mitotic chromatin in ChIP experiments (Christova and Oelgeschlager, 2002). The epitopes for this antisera are therefore not masked by chromatin condensation. Using both this and the mAb, we observe that the vast majority of the TBP is present in the cytoplasm through metaphase and early anaphase, but by late anaphase/telophase TBP and TAF4 are reconcentrated around the chromatin. Confocal microscopy shows that at this stage, these proteins remain excluded from the most condensed chromatin, but may well associate with the decondensing chromatin.
Our results are consistent with the idea that only a very small proportion of the cellular TBP remains associated with specific promoter loci during mitosis. This TBP can be detected by ChIP experiments in which PCR allows an amplification of the genomic DNA associated with the TBP and is therefore more sensitive than immunofluorescence (Christova and Oelgeschlager, 2002). Our data are not however consistent with the idea that a large proportion of the cellular TBP remains associated with the mitotic chromatin as suggested by Chen et al. (2002). The difference may result from the overexpression of the GFP-TBP fusion in the transfection experiments of Chen et al., whereas we detect endogenous cellular TBP. Consequently, the overexpressed GFP-fusion proteins may not exhibit exactly the same behaviors as the endogenous proteins. In addition, Chen et al. (2002) do not observe GFP-TFIIB associated with mitotic chromatin, whereas the endogenous protein is detected associated with promoter loci by ChIP (Christova and Oelgeschlager, 2002). Thus, even using GFP fluorescence, the small amounts of promoter bound TFIIB cannot be readily detected. Together our results would rather indicate that the majority of the cellular TBP and TAF4 is excluded from the condensed mitotic chromatin, only a small proportion remaining associated with the inactive promoters. Nevertheless, this promoter-bound TBP likely plays a central role in restarting gene transcription after mitosis.
Supplementary Material
Acknowledgments
We thank I. Michel for technical assistance, C. Ebel, and J. Barths for invaluable help in the FACS analysis, Dr. Thomas Oelgeschlager for the polyclonal TBP antisera, Dr. Danièle Hernandez-Verdun for the anti-UBF and fibrillarin antibodies, Dr. Laszlo Tora for the anti-TFIIA antibody, M. Oulad-Abdelghani and the mAb facility, and J. L. Vonesch and staff of the confocal microscopy facility, the DNA sequencing, and peptide synthesis facilities. I.M. was supported by a fellowship from the Fondation pour la Recherche Médicale. This work was supported by grants from the CNRS, the INSERM, the Hôpital Universitaire de Strasbourg, the Ministère de la Recherche et de la Technologie, the European Union research grant RTN(2002) 00026, the Association pour la Recherche contre le Cancer, and the Ligue Nationale contre le Cancer.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–02–0138. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–02–0138.
The online version of this article contains supplemental material accessible through http://www.molbiolcell.org.
References
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1-11. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. (2002). The expanding snoRNA world. Biochimie 84, 775-790. [DOI] [PubMed] [Google Scholar]
- Berk, A.J. (2000). TBP-like factors come into focus. Cell 103, 5-8. [DOI] [PubMed] [Google Scholar]
- Chen, D., Hinkley, C.S., Henry, R.W., and Huang, S. (2002). TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13, 276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, C.M., Ge, H., Wang, Z., Hoffmann, A., and Roeder, R.G. (1993). Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 12, 2749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova, R., and Oelgeschlager, T. (2002). Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4, 79-82. [DOI] [PubMed] [Google Scholar]
- Comai, L., Tanese, N., and Tjian, R. (1992). The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68, 965-976. [DOI] [PubMed] [Google Scholar]
- Dantonel, J.C., Quintin, S., Lakatos, L., Labouesse, M., and Tora, L. (2000). TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol. Cell 6, 715-722. [DOI] [PubMed] [Google Scholar]
- Dantonel, J.C., Wurtz, J.M., Poch, O., Moras, D., and Tora, L. (1999). The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem. Sci. 24, 335-339. [DOI] [PubMed] [Google Scholar]
- Filipowicz, W., and Pogacic, V. (2002). Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14, 319-327. [DOI] [PubMed] [Google Scholar]
- Fomproix, N., Gebrane-Younes, J., and Hernandez-Verdun, D. (1998). Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J. Cell Sci. 111, 359-372. [DOI] [PubMed] [Google Scholar]
- Gebrane-Younes, J., Fomproix, N., and Hernandez-Verdun, D. (1997). When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J. Cell Sci. 110, 2429-2440. [DOI] [PubMed] [Google Scholar]
- Hampsey, M. (1998). Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62, 465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey, M., and Reinberg, D. (1999). RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev. 9, 132-139. [DOI] [PubMed] [Google Scholar]
- Hansen, S.K., Takada, S., Jacobson, R.H., Lis, J.T., and Tjian, R. (1997). Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 91, 71-83. [DOI] [PubMed] [Google Scholar]
- Hernandez, N. (1993). TBP, a universal eukaryotic transcription factor? Genes Dev. 7, 1291-1308. [DOI] [PubMed] [Google Scholar]
- Hochheimer, A., and Tjian, R. (2003). Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17, 1309-1320. [DOI] [PubMed] [Google Scholar]
- Hochheimer, A., Zhou, S., Zheng, S., Holmes, M.C., and Tjian, R. (2002). TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420, 439-445. [DOI] [PubMed] [Google Scholar]
- Holmes, M.C., and Tjian, R. (2000). Promoter-selective properties of the TBP-related factor TRF1. Science 288, 867-870. [DOI] [PubMed] [Google Scholar]
- Horn, H.F., and Vousden, K.H. (2004). Cancer: guarding the guardian? Nature 427, 110-111. [DOI] [PubMed] [Google Scholar]
- Jordan, P., Mannervik, M., Tora, L., and Carmo-Fonseca, M. (1996). In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133, 225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach, L., Horner, M.A., Rothman, J.H., and Mango, S.E. (2000). The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell 6, 705-713. [DOI] [PubMed] [Google Scholar]
- Kedinger, C., and Chambon, P. (1972). Animal DNA-dependent RNA polymerases. 3. Purification of calf-thymus BI and BII enzymes. Eur. J. Biochem. 28, 283-290. [DOI] [PubMed] [Google Scholar]
- Kim, J.L., and Burley, S.K. (1994). 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat. Struct. Biol. 1, 638-653. [DOI] [PubMed] [Google Scholar]
- Kim, J.L., Nikolov, D.B., and Burley, S.K. (1993). Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365, 520-527. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., and Kiss, T. (1996). Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85, 1077-1088. [DOI] [PubMed] [Google Scholar]
- Lescure, A., Lutz, Y., Eberhard, D., Jacq, X., Krol, A., Grummt, I., Davidson, I., Chambon, P., and Tora, L. (1994). The N-terminal domain of the human TATA-binding protein plays a role in transcription from TATA-containing RNA polymerase II and III promoters. EMBO J. 13, 1166-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A.K., Andersen, J.S., Mann, M., and Lamond, A.I. (2003). Bioinformatic analysis of the nucleolus. Biochem. J. 376, 553-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Ishima, R., Tong, K.I., Bagby, S., Kokubo, T., Muhandiram, D.R., Kay, L.E., Nakatani, Y., and Ikura, M. (1998). Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94, 573-583. [DOI] [PubMed] [Google Scholar]
- Maison, C., Bailly, D., Peters, A.H., Quivy, J.P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. (2002). Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329-334. [DOI] [PubMed] [Google Scholar]
- Maldonado, E. (1999). Transcriptional functions of a new mammalian TATA-binding protein-related factor. J. Biol. Chem. 274, 12963-12966. [DOI] [PubMed] [Google Scholar]
- Martianov, I., Brancorsini, S., Gansmuller, A., Parvinen, M., Davidson, I., and Sassone-Corsi, P. (2002a). Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development 129, 945-955. [DOI] [PubMed] [Google Scholar]
- Martianov, I., Fimia, G.M., Dierich, A., Parvinen, M., Sassone-Corsi, P., and Davidson, I. (2001). Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7, 509-515. [DOI] [PubMed] [Google Scholar]
- Martianov, I., Viville, S., and Davidson, I. (2002b). RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 298, 1036-1109. [DOI] [PubMed] [Google Scholar]
- Mengus, G., May, M., Carre, L., Chambon, P., and Davidson, I. (1997). Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 11, 1381-1395. [DOI] [PubMed] [Google Scholar]
- Mengus, G., May, M., Jacq, X., Staub, A., Tora, L., Chambon, P., and Davidson, I. (1995). Cloning and characterization of hTAFII18, hTAFII20 and hTAFII 28, three subunits of the human transcription factor TFIID. EMBO J. 14, 1520-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, P.A., Ozer, J., Salunek, M., Jan, G., Zerby, D., Campbell, S., and Lieberman, P.M. (1999). A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol. Cell Biol. 19, 7610-7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, F., Lakatos, L., Dantonel, J., Strahle, U., and Tora, L. (2001). TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 11, 282-287. [DOI] [PubMed] [Google Scholar]
- Nakadai, T., Shimada, M., Shima, D., Handa, H., and Tamura, T.A. (2004). Specific interaction with transcription factor IIA and localization of the mammalian TATA-binding protein-like protein (TLP/TRF2/TLF). J. Biol. Chem. 279, 7447-7455. [DOI] [PubMed] [Google Scholar]
- Nikolov, D.B., Chen, H., Halay, E.D., Usheva, A.A., Hisatake, K., Lee, D.K., Roeder, R.G., and Burley, S.K. (1995). Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377, 119-128. [DOI] [PubMed] [Google Scholar]
- Ohbayashi, T., Makino, Y., and Tamura, Ta. (1999). Identification of a mouse TBP-like protein (TLP) distantly related to the drosophila TBP-related factor. Nucleic Acids Res. 27, 750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi, T., Shimada, M., Nakadai, T., and Tamura, T.A. (2001). TBP-like protein (TLP/TLF/TRF2) artificially recruited to a promoter stimulates basal transcription in vivo. Biochem. Biophys. Res. Commun. 285, 616-622. [DOI] [PubMed] [Google Scholar]
- Pederson, T. (1998). Growth factors in the nucleolus? J. Cell Biol. 143, 279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev, S.P., Zhu, X., Dixit, B.L., Maston, G.A., Kittler, E.L., and Green, M.R. (2003). TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc. Natl. Acad. Sci. USA 100, 14887-14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud, J.C., Mengus, G., Brancorsini, S., Monaco, L., Parvinen, M., Sassone-Corsi, P., and Davidson, I. (2003). The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116, 1847-1858. [DOI] [PubMed] [Google Scholar]
- Rabenstein, M.D., Zhou, S., Lis, J.T., and Tjian, R. (1999). TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. USA 96, 4791-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi, C.P., and Milner, J. (2003). Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22, 6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil, N., Guermah, M., Hoffmann, A., Roeder, R.G., and Heintz, N. (1996). Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10, 2389-2400. [DOI] [PubMed] [Google Scholar]
- Shimada, M., Nakadai, T., and Tamura, T.A. (2003). TATA-binding protein-like protein (TLP/TRF2/TLF) negatively regulates cell cycle progression and is required for the stress-mediated G(2) checkpoint. Mol. Cell. Biol. 23, 4107-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri, V., Roussel, P., and Hernandez-Verdun, D. (1999). The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J. Cell Sci. 112, 3259-3268. [DOI] [PubMed] [Google Scholar]
- Takada, S., Lis, J.T., Zhou, S., and Tjian, R. (2000). A TRF 1, BRF complex directs Drosophila RNA polymerase III transcription. Cell 101, 459-469. [DOI] [PubMed] [Google Scholar]
- Tan, S., Hunziker, Y., Sargent, D.F., and Richmond, T.J. (1996). Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381, 127-151. [DOI] [PubMed] [Google Scholar]
- Teichmann, M., Wang, Z., Martinez, E., Tjernberg, A., Zhang, D., Vollmer, F., Chait, B.T., and Roeder, R.G. (1999). Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc. Natl. Acad. Sci. USA 96, 13720-13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um, M., Yamauchi, J., Kato, S., and Manley, J.L. (2001). Heterozygous disruption of the TATA-binding protein gene in DT40 cells causes reduced cdc25B phosphatase expression and delayed mitosis. Mol. Cell. Biol. 21, 2435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra, G.J., Weeks, D.L., and Wolffe, A.P. (2000). Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in xenopus. Science 290, 2312-2315. [DOI] [PubMed] [Google Scholar]
- Veenstra, G.J., and Wolffe, A.P. (2001). Gene-selective developmental roles of general transcription factors. Trends Biochem. Sci. 26, 665-671. [DOI] [PubMed] [Google Scholar]
- Wieczorek, E., Brand, M., Jacq, X., and Tora, L. (1998). Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393, 187-191. [DOI] [PubMed] [Google Scholar]
- Zatsepina, O.V., Voit, R., Grummt, I., Spring, H., Semenov, M.V., and Trendelenburg, M.F. (1993). The RNA polymerase I-specific transcription initiation factor UBF is associated with transcriptionally active and inactive ribosomal genes. Chromosoma 102, 599-611. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Penttila, T.L., Morris, P.L., Teichmann, M., and Roeder, R.G. (2001). Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292, 1153-1155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.