Abstract
Opposing fission and fusion events maintain the yeast mitochondrial network. Six proteins regulate these membrane dynamics during mitotic growth—Dnm1p, Mdv1p, and Fis1p mediate fission; Fzo1p, Mgm1p, and Ugo1p mediate fusion. Previous studies established that mitochondria fragment and rejoin at distinct stages during meiosis and sporulation, suggesting that mitochondrial fission and fusion are required during this process. Here we report that strains defective for mitochondrial fission alone, or both fission and fusion, complete meiosis and sporulation. However, visualization of mitochondria in sporulating cultures reveals morphological defects associated with the loss of fusion and/or fission proteins. Specifically, mitochondria collapse to one side of the cell and fail to fragment during presporulation. In addition, mitochondria are not inherited equally by newly formed spores, and mitochondrial DNA nucleoid segregation defects give rise to spores lacking nucleoids. This nucleoid inheritance defect is correlated with an increase in petite spore colonies. Unexpectedly, mitochondria fragment in mature tetrads lacking fission proteins. The latter finding suggests either that novel fission machinery operates during sporulation or that mechanical forces generate the mitochondrial fragments observed in mature spores. These results provide evidence of fitness defects caused by fission mutations and reveal new phenotypes associated with fission and fusion mutations.
INTRODUCTION
Mitochondria contribute to a variety of cellular processes, including carbon metabolism, fatty acid metabolism, amino acid and nucleic acid biosynthesis, and programmed cell death (Attardi and Schatz, 1988; Pon and Schatz, 1991; Green and Reed, 1998). However, these organelles are best known for their ability to generate ATP through oxidative phosphorylation reactions that occur on enzyme complexes embedded in the inner mitochondrial membrane. A few of the protein subunits of these respiratory complexes are encoded by mitochondrial DNA (mtDNA) and are synthesized on mitochondrial ribosomes. As a consequence, newly formed daughter cells must inherit both the mitochondrial compartment and (in some cases) mtDNA to survive. The mechanism of mitochondrial partitioning into daughter cells during mitotic division has been studied in a variety of organisms and cell types and is usually mediated by a molecular motor that transports the organelle along polarized cytoskeletal elements (Birky, 1983, 2001; Hollenbeck, 1996; Boldogh et al., 2001).
Mitochondrial partitioning during mitosis has been studied extensively in the budding yeast, Saccharomyces cerevisiae. Yeast mitochondria form a network of interconnected tubules that coalign with actin filaments and cables at the cell cortex (Stevens, 1981; Simon et al., 1995, 1997; Simon and Pon, 1996; Hermann et al., 1997; Hermann and Shaw, 1998). The morphology of this network is determined by opposing mitochondrial fission and fusion events that occur approximately every 2 min (Nunnari et al., 1997; Bleazard et al., 1999; Sesaki and Jensen, 1999; Shaw and Nunnari, 2002; Mozdy and Shaw, 2003). Mitochondrial fission occurs within a mitochondrial tubule or at a branch point in the network, whereas mitochondrial fusion occurs between two tubule tips or the tip and the side of a tubule. Approximately 10–50 structures called nucleoids are evenly distributed throughout the mitochondrial network (Williamson and Fennell, 1979). Each nucleoid consists of 5–10 copies of mtDNA packaged together with proteins (Williamson and Fennell, 1979). During mitotic division, a mitochondrial tubule with its resident nucleoids is transported along actin filaments and cables into the growing daughter cell (bud), where it appears to cluster at the bud tip (Yang et al., 1999). Subsequently, a new mitochondrial network is established in the bud before cytokinesis. Additional studies indicate that mtDNA nucleoids are also actively partitioned into daughter cells formed after mating and zygote formation (Azpiroz and Butow, 1993; Nunnari et al., 1997; Okamoto et al., 1997).
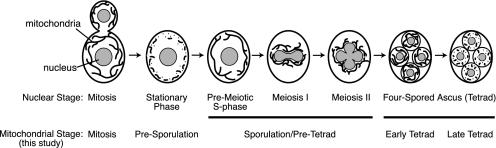
Mitochondrial partitioning during meiotic division appears to be more complex. During meiosis, the diploid yeast genome undergoes one round of replication and two rounds of segregation, yielding four haploid nuclei (Esposito and Klapholz, 1981; Kupiec et al., 1997) (Figure 1, nuclear stage). Because the nuclear envelope of S. cerevisiae does not break down during meiosis I or II, the result is a four-lobed, single nuclear compartment containing four haploid genomes (Figure 1, nuclear stage, meiosis II). Each haploid genome is ultimately enclosed by a prospore membrane and a cell wall, to form an individual spore (Figure 1, nuclear stage, fourspored ascus). Before completion of the prospore membrane and wall, additional cellular organelles fill the developing spore sac, and division of each nuclear lobe is completed (Esposito and Klapholz, 1981; Kupiec et al., 1997). The resulting four spores (a tetrad) are contained within the remains of the diploid mother cell, a protective structure called the ascus.
Figure 1.
Mitochondrial dynamics during meiosis and sporulation in budding yeast. Nuclei (gray) and mitochondria (black) are depicted during mitosis, meiosis, and spore formation in S. cerevisiae (Esposito and Klapholz, 1981). The corresponding nuclear stage or mitochondrial stage (Miyakawa et al., 1984; and this study) is indicated below each diagram. Mitotically dividing cells contain tubular mitochondrial networks (mitosis). Mitochondrial tubules fragment after cells arrest in G1 and prepare for meiosis and sporulation (stationary phase, presporulation). mtDNA and respiration are essential throughout this stage. During premeiotic S-phase (beginning of sporulation/pretetrad stage), mitochondrial fragments rejoin to form tubules. Throughout meiosis I and II (middle and end of sporulation/pretetrad stage), mitochondrial tubules associate with the lobes of the dividing nucleus. The organelles remain tubular through the end of meiosis II and during early tetrad formation (early tetrad). After tetrads mature, mitochondrial tubules fragment once again (late tetrad; day 12 of sporulation in this study).
A number of stage-specific changes in mitochondrial morphology and distribution occur during yeast meiosis and sporulation (Miyakawa et al., 1984) (Figure 1, mitochondrial stage). In the premeiotic stationary phase, mitochondria fragment and form puncta at the cell periphery (Figure 1, mitochondrial stage, presporulation). Each mitochondrial fragment contains one mtDNA nucleoid (Miyakawa et al., 1984). During this period, functional mitochondrial genomes, respiration, and protein synthesis are essential for expression of the meiosis regulatory gene IME1 (Treinin and Simchen, 1993) and progression through the meiotic program (Kuenzi et al., 1974; Jayachandran et al., 1977; Marmiroli et al., 1983). In contrast, functional mitochondrial genomes and mitochondrial protein synthesis are dispensable during the remainder of meiosis, sporulation, and germination, although respiration does continue (Kuenzi et al., 1974; Tingle et al., 1974; Jayachandran et al., 1977). Soon after transfer to sporulation medium, mitochondrial fragments generated during the stationary phase rejoin to form a tubular network (Miyakawa et al., 1984) (Figure 1, nuclear stage, premeiotic S-phase, and mitochondrial stage, beginning of sporulation/pretetrad). Within this network, the mtDNA nucleoids coalesce and form a continuous thread that resembles “beads-on-a-string” (Miyakawa et al., 1984). Before meiosis II, the mitochondrial network moves toward the cell center and becomes associated with the nuclear membrane, where it remains throughout meiosis II (Miyakawa et al., 1984) (Figure 1, mitochondrial stage, middle and end of sporulation/pretetrad). These changes in organelle distribution and morphology ensure that portions of the mitochondrial network and its resident mtDNA nucleoids are enclosed within the developing spores. The mitochondria remain tubular and well distributed throughout prospore membrane formation, prospore wall formation, and early tetrad development (Figure 1, mitochondrial stage, early tetrad). In mature or late tetrads, however, mitochondria again fragment and remain fragmented until spores begin to germinate (Miyakawa et al., 1984) (Figure 1, mitochondrial stage, late tetrad). Once tetrads form, discrete mtDNA nucleoids are visible within each spore, although some nucleoids are excluded from spores and remain in the residual cytoplasm of the ascus (Miyakawa et al., 1984).
Maintenance of yeast mitochondrial morphology during mitosis is known to require conserved proteins that regulate either mitochondrial fission or fusion. Key components of the outer membrane fission machinery include the cytoplasmic GTPase Dnm1p (Otsuga et al., 1998; Bleazard et al., 1999; Fukushima et al., 2001), its binding partner Mdv1p/Fis2p/Gag3p/Net2p (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001), and the integral outer membrane protein Fis1p/Mdv2p (Mozdy et al., 2000; Tieu and Nunnari, 2000). These three proteins form a punctate complex on the outer mitochondrial membrane that mediates mitochondrial constriction and fission through an unknown mechanism (Tieu et al., 2002; Cerveny and Jensen, 2003). Proteins that regulate mitochondrial fusion include the outer membrane GTPase Fzo1p (Hermann et al., 1998; Rapaport et al., 1998; Fritz et al., 2001), the intermembrane space-localized GTPase Mgm1p (Shepard and Yaffe, 1999; Wong et al., 2000), and the outer membrane protein Ugo1p (Sesaki and Jensen, 2001). All three fusion proteins form a complex (Sesaki et al., 2003; Wong et al., 2003) that is required for mitochondrial fusion in mitotically growing cells and in zygotes formed during haploid yeast mating. In mitotic cells lacking one or more fission proteins, mitochondrial tip fusion continues, generating elaborate net-like structures (Bleazard et al., 1999; Sesaki and Jensen, 1999; Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001; Shaw and Nunnari, 2002). These fission mutants partition net-like mitochondria into buds as efficiently as wild-type cells but are reported to mutate or lose mtDNA at elevated rates under some growth conditions (Hanekamp et al., 2002). In contrast, mitochondria fragment in cells lacking one or more fusion proteins, because of unopposed fission (Hermann et al., 1998; Rapaport et al., 1998; Fekkes et al., 2000; Wong et al., 2000; Sesaki and Jensen, 2001; Mozdy and Shaw, 2003). As a consequence of this fragmentation, mtDNA is lost through an unknown mechanism, and fusion mutants become respiratory incompetent (Scott et al., 2003). Once again, these defects do not interfere with mitochondrial partitioning during division, inasmuch as fragments lacking mtDNA nucleoids are inherited normally by buds of yeast fusion mutants (Hermann et al., 1998; Wong et al., 2000; Sesaki and Jensen, 2001). Therefore, whereas fission and fusion are important for maintenance of network morphology and retention of mtDNA nucleoids, neither fission nor fusion is essential for organelle inheritance during mitosis. Moreover, when both fission and fusion are blocked in mitotically dividing yeast, tubular mitochondrial networks are restored, mtDNA loss is suppressed to some degree, and mitochondrial inheritance is unaffected (Bleazard et al., 1999; Sesaki and Jensen, 1999), leading some to wonder why fission and fusion have been conserved during evolution.
In this study, we tested the hypothesis that yeast mitochondrial fission and fusion are important for efficient sporulation and subsequent spore fitness. Here we report that mutant strains defective for mitochondrial fission alone or both fission and fusion are able to form spores. Meiosis and sporulation progress despite the fact that mitochondrial networks in mutant cells fail to fragment during presporulation and subsequently collapse to one side of the cell. These defects in mitochondrial dynamics and distribution ultimately have consequences for spore viability and fitness. In mutant sporulation/pretetrad cells (Figure 1) containing collapsed organelles, some spores fail to inherit mitochondria, resulting in decreased spore viability. Fluorescent labeling studies reveal additional mtDNA nucleoid distribution and segregation defects that give rise to spores lacking detectable nucleoids. These nucleoid-deficient spores are able to germinate but cannot respire, resulting in “petite” colonies that are unable to grow on glycerol. Unexpectedly, when fission proteins are absent during sporulation, mitochondria still fragment in spores of late tetrads. The latter finding suggests that 1) novel mitochondrial fission machinery operates during yeast sporulation or 2) mechanical forces generate the mitochondrial fragments observed in late spores. These combined results provide conclusive evidence of fitness defects caused by mitochondrial fission mutations and reveal new phenotypes associated with mitochondrial fission and fusion defects.
MATERIALS AND METHODS
Yeast Strains and Plasmids
All strains used in this study were derived from FY10 (Winston et al., 1995), including JSY4776, MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 TRP1/TRP1 mgm1::HIS3/mgm1::HIS3 dnm1::HIS3/dnm1::HIS3; JSY4806, MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 TRP1/TRP1 dnm1::HIS3/dnm1::HIS3; JSY4807, MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 TRP1/TRP1 fis1::HIS3/fis1::HIS3; JSY4812, MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 lys2Δ202/lys2Δ202 mdv1::HIS3/mdv1::HIS3; JSY4790, MATa/α leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ura3-52/ura3-52 trp1Δ63/trp1Δ63 fzo1::HIS3/fzo1::HIS3 dnm1::HIS3/dnm1::HIS3; JSY4609, MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 lys2Δ202/lys2Δ202. Standard methods were used to grow, transform, and manipulate yeast strains (Sherman et al., 1986; Guthrie and Fink, 1991). The strains were transformed with pVT100U-mtGFP (mito-green fluorescent protein [GFP]) (Westermann and Neupert, 2000; Fritz et al., 2001) or pRS424ADH-mtRFP (Mozdy and Shaw, unpublished data) to allow visualization of mitochondrial compartments during sporulation or pRS416ABF2-GFP (Abf2p is a mitochondrial nucleoid protein) (Zelenaya-Troitskaya et al., 1998) to allow visualization of mtDNA nucleoids during sporulation.
Sporulation and Quantification of Sporulation Phenotypes
Wild-type and mitochondrial fission and fusion mutant strains expressing mito-GFP or ABF2-GFP were grown for 12–18 h at 25°C in liquid synthetic dextrose lacking uracil or tryptophan. Cells were pelleted and resuspended at a final concentration of 0.5 OD600 U/ml in liquid YPG medium, to select for cells containing mtDNA. After 18–36 h at 30°C, <3% budded forms were present in the population. Cells (2 × 107) were harvested, washed twice in water, resuspended in 5 ml of sporulation medium (1% potassium acetate supplemented with 10 μg/ml adenine, arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, uracil, and valine) in a 50-ml glass flask, and shaken (225 rpm, gyratory G2 tabletop shaker; New Brunswick Scientific, Edison, NJ) at 25°C for 12 d. Aliquots of sporulating cells were examined daily with fluorescence and differential interference contrast (DIC) microscopy. Cultures were scored for sporulation efficiency, mitochondrial morphology and inheritance, and nucleoid distribution and inheritance as described below. Fluorescence microscopy and DIC microscopy were performed with an Axioplan Zeiss microscope, as described previously (Hermann et al., 1997). DIC and fluorescence microscopic images of sporulating cells were obtained with a Zeiss confocal microscope, as described previously (Mozdy et al., 2000).
Quantification of Spore Viability and Petite Spore Colony Formation
Wild-type and mutant tetrads were dissected onto YPD plates and incubated at 30°C for 5 d. Spores that formed visible colonies were designated viable. The viable spore colonies were replica-plated onto YPD and YPG media. Colonies that were able to grow on YPD medium but were unable to grow on YPG medium were designated petite.
RESULTS
Mitochondrial Fission and Fusion Proteins are Not Essential for Yeast Sporulation
The morphology of GFP-labeled mitochondrial compartments during sporulation was monitored with fluorescence microscopy. The presence/absence of prospore walls was simultaneously monitored with DIC microscopy. With this approach, mitochondria in sporulating diploid cells were observed to progress through the stages depicted in Figure 1, including mitosis, presporulation (equivalent to nuclear stationary phase), sporulation/pretetrad (includes nuclear premeiotic S-phase and meiosis I and II), early tetrad (earliest point at which spore walls are visible), and late tetrad (12 d in sporulation medium).
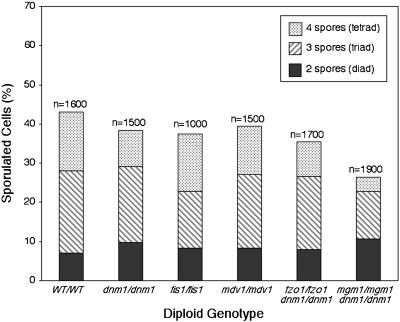
To determine whether mitochondrial fission was essential during sporulation, homozygous fission mutants (dnm1/dnm1, mdv1/mdv1, and fis1/fis1) were examined during a sporulation time course. We were unable to examine homozygous fusion mutants in isolation because mtDNA loss and respiratory defects in these mutants prevent progression beyond the respiration-dependent presporulation stage (Olempska-Beer, 1987; Treinin and Simchen, 1993; Hermann et al., 1998; Rapaport et al., 1998; Shepard and Yaffe, 1999; Wong et al., 2000). However, mitochondrial tubules and respiration can be maintained in fzo1 and mgm1 fusion mutants with deletion of the DNM1 gene, which abolishes mitochondrial fission (Bleazard et al., 1999; Sesaki and Jensen, 1999; Fekkes et al., 2000; Mozdy et al., 2000; Wong et al., 2000). For this reason, we monitored sporulation in fusion mutants that also contained dnm1 fission mutations (fzo1/fzo1 dnm1/dnm1 and mgm1/mgm1 dnm1/dnm1). Because the ugo1/ugo1 dnm1/dnm1 strain grew poorly and did not sporulate well, this strain was not analyzed in this study. As shown in Figure 2, both the homozygous fission mutant and dnm1/fusion mutant strains sporulated to some extent. Moreover, almost all of the strains formed similar percentages of two-spore diads, three-spored triads, and four-spored tetrads. Staining of nuclear DNA with the fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI) confirmed that meiosis I and II and formation of four haploid nuclei had occurred, suggesting that spore walls failed to enclose all four nuclei in diads and triads (Gorsich and Shaw, unpublished data). In the case of the mgm1/mgm1 dnm1/dnm1 strain, overall sporulation efficiency decreased to 26.4%, compared with 43% in wild-type diploids. These data indicate that the three fission proteins, Dnm1p, Mdv1p, and Fis1p, and two fusion proteins, Fzo1p and Mgm1p, are not essential for spore formation.
Figure 2.
Yeast diploids form spores in the absence of mitochondrial fusion and/or fission proteins. Diploid fission or dnm1/fusion mutant strains were constructed in the FY background. The strains were grown for 18–36 h in YPG medium to maintain mtDNA nucleoids, shifted to sporulation medium, and incubated at 30°C for 12 d. The percentage of sporulated cells (diads, triads, and tetrads) was scored daily for each strain. The bar graph shows combined peak sporulation day results for five to eight experiments (n = total number scored).
Fission and Fusion Defects Disrupt Mitochondrial Morphology, Distribution, and Inheritance During Sporulation
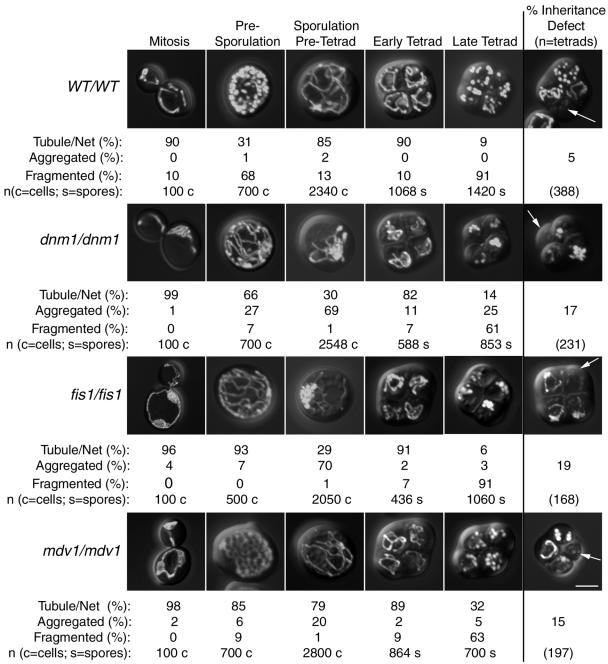
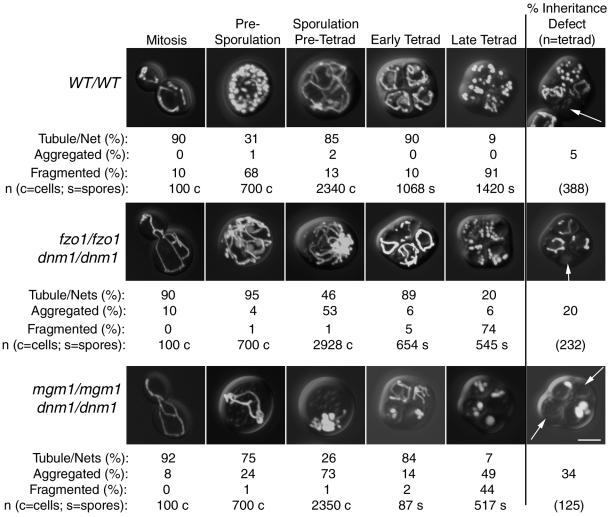
We next determined whether fission and fusion were important for mitochondrial morphology, distribution, and inheritance during the stages of sporulation depicted in Figure 1 (mitochondrial stages). All mutant and wild-type strains exhibited predominantly tubular mitochondrial morphology during mitotic growth (Figures 3 and 4, mitosis, images at far left in each row). Tubular mitochondria in fission mutant strains also contained nets, as expected. As described previously (Miyakawa et al., 1984), when wild-type cells were grown in presporulation medium (YPG), mitochondrial tubules in 68% of the cells were converted to fragments that remained well distributed (Figures 3 and 4, WT/WT, presporulation). In contrast, mitochondria remained predominantly tubular or net-like in the fission and dnm1/fusion mutants during presporulation (Figures 3 and 4, mutant strains, presporulation). These results suggest that Dnm1p, Mdv1p, and Fis1p fission proteins mediate the mitochondrial fragmentation that occurs during presporulation. After transfer to liquid sporulation medium, mitochondria again appeared tubular and well distributed in 85% of wild-type cells (Figures 3 and 4, WT/WT, sporulation/pretetrad). Although mitochondria in the fission and dnm1/fusion mutant cells remained tubular in the sporulation/pretetrad stage, the tubules began migrating toward one end of the cell, forming large aggregates (Figures 3 and 4, mutant strains, sporulation/pretetrad). Formation of these asymmetric mitochondrial aggregates was least severe in cells lacking Mdv1p (20% vs. 53–73% in other mutant strains) (Figures 3 and 4). Although mitochondrial tubules extended from the mitochondrial aggregates of most strains at this stage, mitochondria in cells lacking Mgm1p appeared globular or fragmented and contained fewer discernable tubules (Figure 4, dnm1/dnm1 mgm1/mgm1, sporulation/pretetrad).
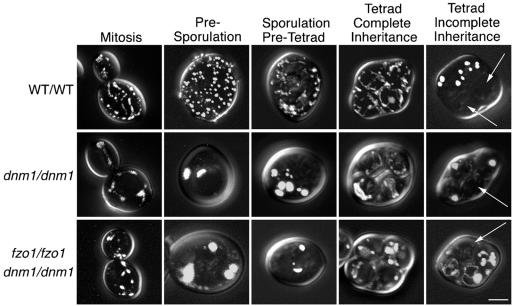
Figure 3.
Mitochondrial distribution and morphology in sporulating wild-type and fission mutant yeast cells. Wild-type and homozygous mutant diploids expressing mito-GFP were sporulated. Mitochondrial morphology was scored daily for 12 d after transfer to sporulation medium. Representative images of cells in mitosis and presporulation, sporulation/pretetrad, early tetrad, and late tetrad stages are shown. The percentages of cells containing mitochondria with different morphological features (tubule/net, aggregated, or fragmented) are indicated below each image. Examples of tetrads that failed to distribute GFP-labeled mitochondria into one or more spores are shown at the far right (% inheritance defect, white arrows). Bar, 5 μm.
Figure 4.
Mitochondrial distribution and morphology in sporulating wild-type and dnm1/fusion mutant yeast cells. Wild-type and dnm1/fusion mutant diploids expressing mito-GFP were sporulated. Mitochondrial morphology was scored daily for 12 d after transfer to sporulation medium. Representative images of cells in mitosis and presporulation, sporulation/pretetrad, early tetrad, and late tetrad stages are shown. The percentages of cells containing mitochondria with different morphological features (tubule/net, aggregated, or fragmented) are indicated below each image. Examples of tetrads that failed to distribute GFP-labeled mitochondria into one or more spores are shown at the far right (% inheritance defect, white arrows). Bar, 5 μm.
Mitochondria were largely tubular in the early tetrads of both wild-type cells and cells lacking fission and fusion proteins. However, unlike spores formed from wild-type diploids, all mutant strains generated some spores with aggregated mitochondria. As wild-type tetrads matured (Figures 3 and 4, WT/WT, late tetrad), mitochondrial tubules fragmented in the majority (91%) of spores. Unexpectedly, mitochondrial compartments also fragmented in 44–91% of spores formed by fission and dnm1/fusion mutant strains (Figure 3, mutant strains, late tetrad). This result was surprising, inasmuch as mitochondria cannot fragment in haploid strains lacking Dnm1p, Mdv1p, and Fis1p, because of the defect in fission.
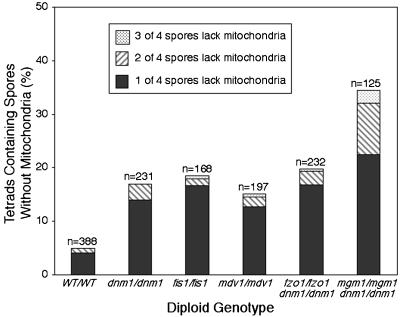
We reasoned that sporulation/pretetrad-stage cells with defects in mitochondrial distribution might fail to incorporate mitochondrial compartments into newly formed spores. As shown in Figures 3 and 4, one or more spores of a tetrad often failed to inherit detectable levels of mitochondria when fission or dnm1/fusion proteins were absent (Figures 3 and 4, images at far right). Although an inheritance defect was observed in a small percentage of wild-type tetrads (5%), the number of tetrads lacking mitochondria in one or more spores increased when fission proteins were absent (dnm1/dnm1, 17%; fis1/fis1, 19%; mdv1/mdv1, 15%) or when both fusion and Dnm1p fission proteins were absent (fzo1/fzo1 dnm1/dnm1, 20%; mgm1/mgm1 dnm1/dnm1, 34%).
In Figure 5, the results indicate the percentages of tetrads lacking mitochondria in one, two, or three of the four spores. Among the wild-type and mutant tetrads with an inheritance defect, usually only one of the four spores in a tetrad lacked detectable mitochondria. However, a small percentage of tetrads lacked mitochondria in two or three of the spores. Cells lacking the Mgm1p fusion protein produced a larger percentage of defective tetrads in all three categories, consistent with our observation that mgm1 mutant strains exhibited the most severe defects in mitochondrial aggregation and distribution (Figure 4, mgm1/mgm1 dnm1/dnm1).
Figure 5.
Mitochondrial partitioning to spores is defective in diploid fission and dnm1/fusion mutants. Wild-type and homozygous mutant diploids expressing mito-GFP were sporulated for 12 d. Wild-type and fission or dnm1/fusion mutant tetrads were scored for the presence or absence of GFP-labeled mitochondria in one (solid bars), two (hatched bars), or three (stippled bars) spores of a tetrad. Tetrads lacking GFP-labeled mitochondria in all four spores were not scored, because these cells might have lost the mito-GFP-expressing plasmid (n = total number scored).
Spore Viability and Respiratory Competence are Reduced When Mitochondrial Fission or dnm1/Fusion Proteins are Absent
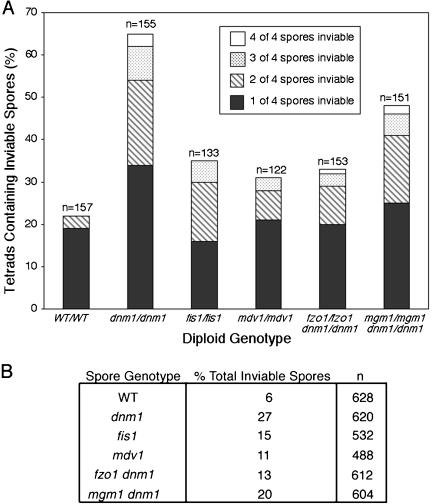
To determine whether reduced mitochondrial inheritance affected spore viability, wild-type (n = 157), dnm1/dnm1 (n = 155), fis1/fis1 (n = 133), mdv1/mdv1 (n = 122), fzo1/fzo1 dnm1/dnm1 (n = 153), and mgm1/mgm1 dnm1/dnm1 (n = 151) tetrads were dissected onto YPD plates, and colony formation was monitored. Spores that were unable to form a visible colony were scored as inviable. Although 22% of wild-type tetrads contained at least one inviable spore, the percentage increased in all strains lacking fission and dnm1/fusion proteins (Figure 6A). The number of tetrads with one, two, three, or four inviable spores also increased in the mutant strains. When total spore inviability was calculated for each strain (no. of inviable spores/no. of spores dissected × 100), the differences between strains were similar, with the most severe defects occurring in dnm1 and mgm1 dnm1 spores (Figure 6B). In fact, the percentage of dnm1 mutant tetrads containing inviable spores (Figure 6A) was larger than expected if this inviability was attributable solely to mitochondrial inheritance defects in these tetrads (Figure 5). This discrepancy suggests that Dnm1p plays a role in spore germination and/or viability independent of its role in mitochondrial division and inheritance.
Figure 6.
Viability (colony formation) is reduced in diploid fission and dnm1/fusion mutants. (A) Wild-type, fission mutant, and dnm1/fusion mutant tetrads were dissected onto YPD medium, and the ability of each spore to form a colony was assessed. For each strain, the number of tetrads with one (solid bars), two (hatched bars), three (stippled bars), or four (white bars) inviable spores is shown (n = total number scored). (B) The percentages of total inviable spores produced by wild-type and mutant tetrads dissected in A are shown (n = total number scored).
In tetrads from all sporulation experiments, there were more inviable spores than spores that failed to inherit mitochondria. Therefore, some spores scored as containing mitochondria must not have been able to germinate and/or form colonies. This phenomenon is likely attributable to the fact that mitochondrial inheritance by spores was scored very conservatively. A spore was characterized as containing mitochondria regardless of how little mito-GFP fluorescence was detected. Many of the inviable spores might have inherited insufficient mitochondria to sustain subsequent germination and/or colony growth. To determine whether inviable wild-type and mutant spores were able to germinate, spores that did not form visible colonies were examined microscopically. For each strain, the percentage of inviable spores that germinated and divided at least once was as follows: wild-type, 93%; dnm1, 85%; fis1, 86%; mdv1, 92%; fzo1 dnm1, 64%; mgm1 dnm1, 93%. Although large percentages of mgm1 dnm1 (20%) and dnm1 (27%) spores were inviable, the ability of spores from these two strains to germinate was identical or only slightly decreased, relative to wild-type spores. These findings suggest that some spores inherit sufficient mitochondria to support germination and one or more cell divisions but not growth of an entire colony.
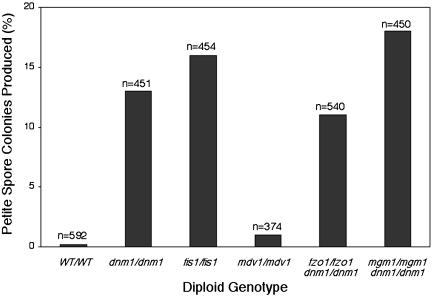
Viable spore colonies indicated in Figure 6 were analyzed for their ability to respire and grow on the nonfermentable carbon source glycerol. As shown in Figure 7, viable spores derived from cells lacking mitochondrial fission or fusion proteins gave rise to glycerol growth-defective (petite) colonies more often than did spores derived from wild-type cells (wild-type, 0.2%; dnm1, 13%; fis1, 16%; mdv1, 1%; fzo1 dnm1, 11%; mgm1 dnm1, 18%). DAPI staining of spore colonies from a dnm1/dnm1 sporulation experiment revealed that 64% of the petite spore colonies contained rho0 cells (lacking DAPI-stained mtDNA nucleoids), whereas 36% of the petite spore colonies contained rho- cells (containing DAPI-stained nucleoids; we presume that these cells are rho- because nucleoids are present but respiration is compromised). These data suggest that petite spore colonies arising in these experiments result both from a failure to inherit nucleoids and from increased occurrence of nonfunctional (rho-) mtDNA in nucleoids. Very few viable spores derived from the mdv1/mdv1 strain were petite, suggesting that Mdv1p is less critical for formation of respiration-competent spores during meiotic division.
Figure 7.
Fission and dnm1/fusion mutant strains produce more petite spore colonies than do wild-type cells. Wild-type, fission mutant, and dnm1/fusion mutant tetrads were dissected onto YPD medium. Respiration-deficient (petite) spore colonies were identified by their inability to grow after replica plating onto YPG medium (n = total number scored).
To determine whether the results reported above were specific for the FY strain background, we repeated these studies with WT/WT and dnm1/dnm1 diploids generated in the W303 strain background. Compared with WT/WT and dnm1/dnm1 diploids in the FY background, W303 strains produced a slightly higher percentage of inviable spores and a similar percentage of respiration-incompetent spores. On the basis of this comparison, it seems unlikely that the FY strain contains background mutations responsible for the defects observed in our fission and fusion mutant studies.
Inheritance of mtDNA Nucleoids is Disrupted When Mitochondrial Fission or dnm1/Fusion Proteins are Absent
The increase in petite colony formation described above suggested that partitioning of mtDNA nucleoids might be disrupted in sporulating cells lacking fission and dnm1/fusion proteins. To test this idea, mitochondrial nucleoids were visualized in wild-type and mutant diploid cells with a GFP-tagged form of the Abf2 nucleoid-binding protein (Zelenaya-Troitskaya et al., 1998). Although costaining of cells with DAPI confirmed that Abf2-GFP puncta contained mtDNA (data not show), Abf2-GFP proved to be a more reliable marker of mtDNA nucleoid distribution during sporulation, because DAPI-labeled mtDNA was often obscured by brightly labeled nuclear DNA in mutant cells containing mitochondrial aggregates.
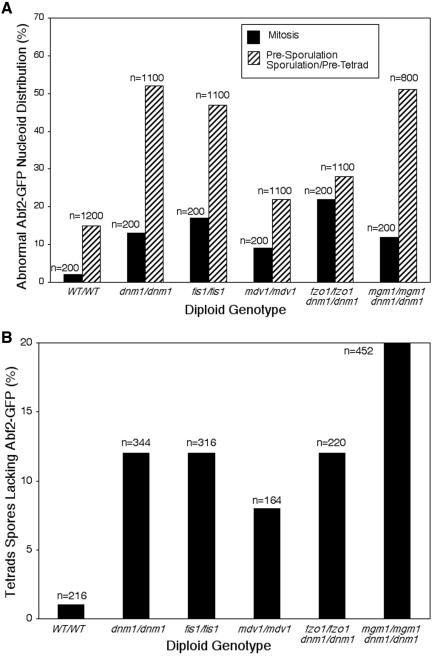
Abf2-GFP distribution was quantified in wild-type and mutant cells in mitosis or in presporulation and sporulation/pretetrad stages (Figure 8A). Abf2-GFP distribution was considered wild-type if five or more green fluorescent puncta were present and evenly distributed on mitochondria or if nucleoid staining resembled the beads-on-a-string pattern reported previously (Miyakawa et al., 1984). Representative images of Abf2-GFP-labeled nucleoids in wild-type (WT/WT) cells during mitosis and presporulation and sporulation/pretetrad stages are shown in Figure 9. When wild-type cells were in mitosis or in presporulation and sporulation/pretetrad stages, only 2% or 15% of the population, respectively, exhibited abnormal Abf2-GFP distribution (Figure 8A, WT/WT). The percentage of cells with abnormal Abf2-GFP distribution during mitosis increased in the fission and dnm1/fusion mutants, although the defects were moderate (9–22%) (Figure 8A). More severe mtDNA nucleoid distribution defects occurred in many mutant strains in presporulation and sporulation/pretetrad stages (22–52%) (Figure 8A). In cells with abnormal distribution, Abf2-GFP-labeled nucleoids appeared as two or more large aggregates that were often asymmetrically localized. Representative images of abnormal Abf2-GFP-labeled nucleoid distribution in dnm1/dnm1 and dnm1/dnm1 fzo1/fzo1 mutant strains during mitosis and presporulation and sporulation/pretetrad stages are shown in Figure 9.
Figure 8.
mtDNA nucleoid distribution and inheritance are disrupted in sporulating fission and dnm1/fusion mutants. (A) The distribution of Abf2-GFP-labeled mtDNA nucleoids was examined in wild-type and mutant strains during mitosis (black bars) and presporulation and sporulation/pretetrad stages (gray bars). Mitochondrial nucleoid appearance and distribution in single cells were scored as wild-type (five or more dispersed nucleoids or beads-on-a-string appearance) or abnormal (less than four nucleoids or aggregates). Abnormal Abf2-GFP nucleoid distribution is shown (n = total number scored). (B) Wild-type and mutant tetrad spores were scored for the presence or absence of Abf2-GFP-labeled mitochondrial nucleoids in each spore (n = total number scored).
Figure 9.
Distribution of Abf2-GFP-labeled mtDNA nucleoids is abnormal in sporulating fission and dnm1/fusion mutants. Wild-type (WT/WT), fission mutant (dnm1/dnm1), and dnm1/dnm1 fzo1/fzo1 mutant strains expressing Abf2-GFP were examined during a sporulation time course. Representative confocal microscopic images of cells in mitosis and presporulation, sporulation/pretetrad, and tetrad stages are shown. In each row, the fourth panel from the left shows an image of a tetrad containing Abf2-GFP labeling in all four spores. The far right panels show images of tetrads in which one or more spores lack detectable Abf2-GFP labeling (white arrows). Bar, 5 μm.
To determine whether this abnormal nucleoid appearance and distribution affected nucleoid inheritance by spores, wild-type and mutant tetrads were examined for spores lacking Abf2-GFP labeling (only asci containing four spores were used for this calculation) (Figure 8B). Although very few wild-type spores lacked detectable Abf2-GFP (1%), the nucleoid inheritance defect increased in spores derived from all fission and dnm1/fusion mutant strains (dnm1, 12%; fis1, 12%; mdv1, 8%; fzo1 dnm1, 12%; mgm1 dnm1, 20%). The observation that mtDNA nucleoid inheritance by spores is reduced in fission and dnm1/fusion mutants is consistent with our finding that these mutant strains give rise to glycerol growth-defective (petite) colonies more often than the wild-type strain (Figure 7). Moreover, this observation is consistent with our DAPI staining studies showing that 64% of petite colonies contain cells with no visible mtDNA nucleoids. Together, these results indicate that mitochondrial fission and fusion proteins are important for proper mtDNA nucleoid distribution and inheritance during meiosis and sporulation.
DISCUSSION
In the wild, yeast cells are found as mitotically dividing diploids (Mortimer et al., 1994; Mortimer, 2000). However, their ultimate survival depends on their ability to undergo meiosis and sporulation when exposed to adverse environmental conditions. In this study, we demonstrate that loss of fission proteins alone or both fission and fusion proteins causes significant defects in mitochondrial distribution and inheritance during meiosis and sporulation. These defects also result in decreased mtDNA nucleoid inheritance, germination, and viability, all of which are important for yeast survival and fitness in the wild.
As reported by Miyakawa et al. (1984) and observed in this study, mitochondria undergo what appear to be temporally regulated fission and fusion events during meiosis and sporulation. Although fission and fusion are balanced to maintain mitochondrial tubules during mitotic growth, they are shifted toward mitochondrial fragmentation during the presporulation stage of the meiosis and sporulation program. This shift toward fragmentation could be achieved by increasing the rate of fission or decreasing the rate of fusion. Although we were unable to evaluate mitochondrial morphology in strains lacking fusion proteins alone, we were able to monitor mitochondrial fragmentation during presporulation in cells lacking fission proteins. Our finding that mitochondria remained tubular during presporulation in cells lacking Dnm1p, Mdv1p, or Fis1p indicates that these proteins contribute to mitochondrial fragmentation at this stage of the meiotic program.
When wild-type cells are sporulated, tubular mitochondria also fragment during the late tetrad stage. We anticipated that this fragmentation would be blocked in cells lacking the fission proteins Dnm1p, Fis1p, or Mdv1p. Unexpectedly, mitochondria were still able to fragment in late tetrads formed by cells lacking any one of these fission proteins or all three of these proteins (Gorsich and Shaw, unpublished data). We further explored the role of Dnm1p at this stage by examining the localization of Dnm1p-GFP during a wild-type sporulation time course. Studies of mitotic wild-type cells showed that Dnm1p-GFP localized in many discrete puncta (fission complexes) along mitochondrial tubules (Otsuga et al., 1998; Bleazard et al., 1999). In our sporulating wild-type culture, we conservatively scored cells with five or more evenly distributed Dnm1p-GFP puncta as wild-type and cells with zero to four puncta as mutant. As shown in Table 1, we observed a wild-type distribution of Dnm1p-GFP puncta (>90% of cells) when cells were in mitosis or presporulation or pretetrad stages. However, visible Dnm1p-GFP puncta redistributed into one or a few large aggregates in ∼50% of the cells in early and late tetrad stages. With the assumption that such aggregates are not functional, these results suggest that Dnm1p may be less active during late stages of sporulation. Alternatively, active Dnm1p-GFP puncta may be less visible at these late stages. If Dnm1p activity is reduced or absent during late stages of sporulation, then two alternative explanations could account for the mitochondrial fragmentation observed in late tetrads. First, there may be additional genes specifically required for mitochondrial morphology maintenance during the later stages of sporulation, particularly during tetrad maturation. If mutations in such genes failed to produce mitotic phenotypes, then they would have escaped detection in mitochondrial morphology mutant screens of the viable gene disruption collection (Dimmer et al., 2002). Second, some type of mechanical force could cause mitochondrial fragmentation during the late tetrad stage of sporulation, independent of Dnm1p, Fis1p, or Mdv1p. This type of mechanical fragmentation could result from changes in actin-mitochondria interactions. In support of this model, mitochondria are reported to aggregate, fragment, and become immobile during sporulation in various actin (act1) mutants (Smith et al., 1995) and in haploid cells treated with the actin filament- and cable-disrupting drug latrunculin A (Boldogh et al., 1998). Two additional observations argue against this model. First, dnm1Δ net-like mitochondria do not fragment when actin cables are disrupted by a mutation in a gene called MDM20 (Bleazard et al., 1999). Second, mitochondrial nets in dnm1Δ or mdv1Δ cells are not disrupted when these cells are treated with latrunculin A (Cerveny et al., 2001; Jensen et al., 2001). Additional studies will be necessary to determine the basis of mitochondrial fragmentation in mature spores of fission and dnm1/fusion mutants.
Table 1.
Dnm1p-GFP localization during sporulation
| Stage | 0 Dnm1p-GFP puncta (%) | 1–4 Dnm1p-GFP puncta (%) | >5 Dnm1p-GFP puncta (%) | n |
|---|---|---|---|---|
| Mitosis | 0.0 | 8.0 | 92.0 | 100(c) |
| Presporulation | 0.0 | 6.7 | 93.3 | 150(c) |
| Pretetrad | 0.0 | 6.0 | 94.0 | 400(c) |
| Early tetrad | 4.6 | 50.4 | 45.0 | 480(s) |
| Late tetrad | 2.6 | 48.7 | 48.7 | 880(s) |
Data from sporulation time course of a wild-type diploid in the FY background. (c), cells; (s), spores.
We observed that homozygous fission mutants (dnm1/dnm1, fis1/fis1, and mdv1/mdv1) and dnm1/fusion mutants (fzo1/fzo1 dnm1/dnm1 and mgm1/mgm1 dnm1/dnm1) were able to form spores. Clearly, mitochondrial fission and fusion are not essential for prospore membrane and cell wall formation. Moreover, DAPI staining of sporulated cultures revealed no binucleate or multinucleate spores, suggesting that nuclear DNA replication and meiotic divisions were occurring normally (Gorsich and Shaw, unpublished data). Instead, strains lacking mitochondrial fission and fusion proteins exhibited defects in mitochondrial morphology, the most striking of which was the aggregation and collapse of mitochondrial membranes during the sporulation/pretetrad stage. The fission and fusion mutants with asymmetrically distributed mitochondria produced spores lacking this organelle, providing direct evidence that mitochondria must be evenly distributed at or near the nuclear lobes to be included in all four spores. Finally, we showed that cells lacking fission proteins alone or both fission and fusion proteins produced more inviable spores than did wild-type cells. This viability defect may be a direct consequence of the mitochondrial inheritance defect, because mitochondria cannot be generated de novo. In contrast, spores that fail to inherit wild-type amounts of vacuoles/lysosomes do not exhibit viability defects (Roeder and Shaw, 1996). Instead, such spores regenerate new vacuoles before germination, either de novo or from an undetected pool of inherited membranes. Two additional observations suggest that production of colony-forming spores may require the inheritance of a critical amount of mitochondria. First, for most mutant strains, the number of tetrads with inviable spores was greater than the number of tetrads with mitochondria-deficient spores (lacking detectable mitochondria). Therefore, some spores that receive detectable amounts of mitochondria may receive too little for effective germination. Second, in many strains, a significant fraction of spores that did not form colonies were able to germinate and divide at least once. Therefore, it is possible that some spores inherit insufficient mitochondria to support germination and/or colony formation.
In this study, sporulating wild-type cells produced respiration-competent spores with high efficiency (1 of 589 dissected spores was petite), indicating that a high-fidelity mechanism exists to faithfully partition mtDNA during sporulation. This partitioning mechanism is disrupted in cells lacking fission proteins alone or both fission and fusion proteins, inasmuch as spores from these strains sometimes failed to inherit mtDNA nucleoids and formed respiration-incompetent spore colonies more frequently than did wild-type spores. Therefore, mitochondrial fission and fusion indirectly contribute to yeast survival and fitness by enhancing the fidelity of mtDNA nucleoid inheritance during meiotic division and sporulation. Together, our observations suggest that mitochondrial fission and fusion have been conserved during evolution in part because of their importance during meiosis and sporulation. These processes are also important for survival and fitness in mitotically dividing yeast, inasmuch as mitochondrial fragmentation in fusion mutants causes mtDNA loss and respiratory defects (Hermann et al., 1998; Rapaport et al., 1998; Shepard and Yaffe, 1999; Sesaki and Jensen, 2001) and dnm1 fission mutants exhibit defects in mtDNA stability (Hanekamp et al., 2002).
Because studies in mitotically growing cells demonstrated that the fission proteins Dnm1p, Fis1p, and Mdv1p function in the same pathway, we expected that diploid strains lacking these proteins would display similar sporulation phenotypes. However, the sporulation phenotypes of cells lacking individual fission proteins differed in severity. Specifically, dnm1/dnm1 sporulation defects were the most severe, whereas mdv1/mdv1 defects were the least severe. These different phenotypes suggest that the need for each protein during fission may vary. During mitotic growth, the Dnm1p GTPase assembles to form punctate structures on the sides of mitochondrial tubules (Otsuga et al., 1998). These structures (mitochondrial fission complexes) are associated with sites of inner and outer membrane constriction in immunogold labeling studies (Bleazard et al., 1999) and often occur at sites of mitochondrial fission in time-lapse imaging studies (Sesaki and Jensen, 1999; Legesse-Miller et al., 2003). The Mdv1p protein binds to Dnm1p and is also found in these mitochondrial fission complexes (Fekkes et al., 2000; Tieu and Nunnari, 2000; Tieu et al., 2002; Cerveny et al., 2001; Cerveny and Jensen, 2003). Previous studies showed that, when Mdv1p is absent during mitotic growth, Dnm1p is able to form punctate complexes on the outer mitochondrial membrane (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001). However, fission is still reduced or blocked, indicating that Mdv1p plays an important role in fission. Because less severe mitochondrial morphology defects are observed in mdv1/mdv1 sporulating cells, the function of Mdv1p may be less important during sporulation.
Mitochondria also undergo dramatic morphological changes during human spermatogenesis (Seitz et al., 1995; Meinhardt et al., 1999) and oogenesis (Pozo et al., 1990; Motta et al., 2000; Sathananthan and Trounson, 2000), and a number of human fertility disorders have been linked to defects in mitochondrial distribution and function (Mundy et al., 1995; Piasecka and Kawiak, 2003). Moreover, defects in mitochondrial function appear to contribute to chromosomal nondisjunction and age-related trisomies such as Down's syndrome (Schon et al., 2000; Arbuzova et al., 2001, 2002). Other studies have demonstrated that postmeiotic fertility and developmental defects can be caused by abnormal mitochondrial morphology in mammals (Van Blerkom et al., 2000; Chen et al., 2003). The studies presented here raise the possibility that some inherited human diseases, fertility disorders, and developmental defects result from or are compounded by meiotic defects in mitochondrial fission and fusion. Now that mammalian homologues of some mitochondrial fission and fusion proteins have been identified, the importance of these processes during gametogenesis, fertilization, and early development in mammals can be investigated.
Acknowledgments
We thank members of the Shaw laboratory for stimulating discussions and comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM-53466), awarded to J.M.S.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–12–0875. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–12–0875.
Abbreviations used: DIC, differential interference contrast; GFP, green fluorescent protein; mtDNA, mitochondrial DNA.
References
- Arbuzova, S., Cuckle, H., Mueller, R., and Sehmi, I. (2001). Familial Down syndrome: evidence supporting cytoplasmic inheritance. Clin. Genet. 60, 456-462. [DOI] [PubMed] [Google Scholar]
- Arbuzova, S., Hutchin, T., and Cuckle, H. (2002). Mitochondrial dysfunction and Down's syndrome. Bioessays 24, 681-684. [DOI] [PubMed] [Google Scholar]
- Attardi, G., and Schatz, G. (1988). Biogenesis of mitochondria. Annu. Rev. Cell Biol. 4, 289-333. [DOI] [PubMed] [Google Scholar]
- Azpiroz, R., and Butow, R.A. (1993). Patterns of mitochondrial sorting in yeast zygotes. Mol. Biol. Cell 4, 21-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky, C.W. (1983). The partitioning of cytoplasmic organelles at cell division. Int. Rev. Cytol. 15, 49-89. [DOI] [PubMed] [Google Scholar]
- Birky, C.W., Jr. (2001). The inheritance of genes in mitochondria and chloro-plasts: laws, mechanisms, and models. Annu. Rev. Genet. 35, 125-148. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I., Vojtov, N., Karmon, S., and Pon, L.A. (1998). Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J. Cell Biol. 141, 1371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I.R., Yang, H.C., and Pon, L.A. (2001). Mitochondrial inheritance in budding yeast. Traffic 2, 368-374. [DOI] [PubMed] [Google Scholar]
- Cerveny, K.L., and Jensen, R.E. (2003). The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 14, 4126-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DNM1-interacting protein, Net2p. Mol. Biol. Cell 12, 309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Detmer, S.A., Ewald, A.J., Griffin, E.E., Fraser, S.E., and Chan, D.C. (2003). Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer, K.S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W., and Westermann, B. (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, R.E., and Klapholz, S. (1981). Meiosis and axcospore development. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, ed. J.N. Strathern, E.W. Jones, and J.R. Broach, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 211-287.
- Fekkes, P., Shepard, K.A., and Yaffe, M.P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151, 333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, S., Rapaport, D., Klanner, E., Neupert, W., and Westermann, B. (2001). Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J. Cell Biol. 152, 683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, N.H., Brisch, E., Keegan, B.R., Bleazard, W., and Shaw, J.M. (2001). The AH/GED sequence of the dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol. Biol. Cell 12, 2756-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281, 1309-1312. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (eds.) (1991). Guide to Yeast Genetics and Molecular Biology, San Diego, CA: Academic Press.
- Hanekamp, T., Thorsness, M.K., Rebbapragada, I., Fisher, E.M., Seebart, C., Darland, M.R., Coxbill, J.A., Updike, D.L., and Thorsness, P.E. (2002). Maintenance of mitochondrial morphology is linked to maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Genetics 162, 1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G.J., King, E.J., and Shaw, J.M. (1997). The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J. Cell Biol. 137, 141-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G.J., and Shaw, J.M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265-303. [DOI] [PubMed] [Google Scholar]
- Hermann, G.J., Thatcher, J.W., Mills, J.P., Hales, K.G., Fuller, M.T., Nunnari, J., and Shaw, J.M. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck, P.J. (1996). The pattern and mechanism of mitochondrial transport in axons. Front. Biosci. 1, 91-102. [DOI] [PubMed] [Google Scholar]
- Jayachandran, S., Sidhu, R.S., Ethiraj, S., and Tauro, P. (1977). Dependence of yeast sporulation on mitochondrial protein synthesis. Folia Microbiol. (Praha) 22, 74-78. [DOI] [PubMed] [Google Scholar]
- Jensen, R.E., Aiken-Hobbs, A.E., Cerveny, K.L., and Sesaki, H. (2001). Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc. Res. Tech. 51, 573-583. [DOI] [PubMed] [Google Scholar]
- Kuenzi, M.T., Tingle, M.A., and Halvorson, H.O. (1974). Sporulation of Saccharomyces cerevisiae in the absence of a functional mitochondrial genome. J. Bacteriol. 117, 80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec, M., Byers, B., Esposito, R.E., and Mitchell, A.P. (1997). Meiosis and sporulation in Saccharomyces cerevisiae. In: The Molecular Biology of the Yeast Saccharomyces, ed. J.R. Pringle, J.R. Broach, and E.W. Jones, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 889-1036.
- Legesse-Miller, A., Massol, R.H., and Kirchhausen, T. (2003). Constriction and dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 14, 1953-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmiroli, N., Ferri, M., and Puglisi, P.P. (1983). Involvement of mitochondrial protein synthesis in sporulation: effects of erythromycin on macromolecular synthesis, meiosis, and ascospore formation in Saccharomyces cerevisiae. J. Bacteriol. 154, 118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt, A., Wilhelm, B., and Seitz, J. (1999). Expression of mitochondrial marker proteins during spermatogenesis. Hum. Reprod. Update 5, 108-119. [DOI] [PubMed] [Google Scholar]
- Miyakawa, I., Aoi, H., Sando, N., and Kuroiwa, T. (1984). Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 66, 21-38. [DOI] [PubMed] [Google Scholar]
- Mortimer, R.K. (2000). Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10, 403-409. [DOI] [PubMed] [Google Scholar]
- Mortimer, R.K., Romano, P., Suzzi, G., and Polsinelli, M. (1994). Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10, 1543-1552. [DOI] [PubMed] [Google Scholar]
- Motta, P.M., Nottola, S.A., Makabe, S., and Heyn, R. (2000). Mitochondrial morphology in human fetal and adult female germ cells. Hum. Reprod. 15(suppl 2), 129-147. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy, A.D., and Shaw, J.M. (2003). A fuzzy mitochondrial fusion apparatus comes into focus. Nat. Rev. Mol. Cell. Biol. 4, 468-478. [DOI] [PubMed] [Google Scholar]
- Mundy, A.J., Ryder, T.A., and Edmonds, D.K. (1995). Asthenozoospermia and the human sperm mid-piece. Hum. Reprod. 10, 116-119. [DOI] [PubMed] [Google Scholar]
- Nunnari, J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K., Newman, S.M., Perlman, P.S., and Butow, R.A. (1997). Sorting patterns of mitochondrial proteins in yeast zygotes. Mol. Biol. Cell 8, 240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olempska-Beer, Z. (1987). Current methods for Saccharomyces cerevisiae. II. Sporulation. Anal. Biochem. 164, 278-286. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B.R., Brisch, E., Thatcher, J.W., Hermann, G.J., Bleazard, W., and Shaw, J.M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka, M., and Kawiak, J. (2003). Sperm mitochondria of patients with normal sperm motility and with asthenozoospermia: morphological and functional study. Folia Histochem. Cytobiol. 41, 125-139. [PubMed] [Google Scholar]
- Pon, L., and Schatz, G. (1991). Biogenesis of yeast mitochondria. In: The Molecular Biology of the Yeast Saccharomyces, ed. J.R. Broach, J.R. Pringle, and E.W. Jones, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 334-406.
- Pozo, J., Corral, E., and Pereda, J. (1990). Subcellular structure of prenatal human ovary: mitochondrial distribution during meiotic prophase. J. Submicrosc. Cytol. Pathol. 22, 601-607. [PubMed] [Google Scholar]
- Rapaport, D., Brunner, M., Neupert, W., and Westermann, B. (1998). Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150-20155. [DOI] [PubMed] [Google Scholar]
- Roeder, A.D., and Shaw, J.M. (1996). Vacuole partitioning during meiotic division in yeast. Genetics 144, 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan, A.H., and Trounson, A.O. (2000). Mitochondrial morphology during preimplantational human embryogenesis. Hum. Reprod. 15(suppl 2), 148-159. [DOI] [PubMed] [Google Scholar]
- Schon, E.A., Kim, S.H., Ferreira, J.C., Magalhaes, P., Grace, M., Warburton, D., and Gross, S.J. (2000). Chromosomal non-disjunction in human oocytes: is there a mitochondrial connection? Hum. Reprod. 15(suppl 2), 160-172. [DOI] [PubMed] [Google Scholar]
- Scott, S.V., Cassidy-Stone, A., Meeusen, S.L., and Nunnari, J. (2003). Staying in aerobic shape: how the structural integrity of mitochondria and mitochondrial DNA is maintained. Curr. Opin. Cell Biol. 15, 482-488. [DOI] [PubMed] [Google Scholar]
- Seitz, J., Mobius, J., Bergmann, M., and Meinhardt, A. (1995). Mitochondrial differentiation during meiosis of male germ cells. Int. J. Androl. 18(suppl 2), 7-11. [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (2001). UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., Southard, S.M., Yaffe, M.P., and Jensen, R.E. (2003). Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell 14, 2342-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J.M., and Nunnari, J. (2002). Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12, 178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard, K.A., and Yaffe, M.P. (1999). The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 144, 711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J.B. (1986). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Simon, V.R., Karmon, S.L., and Pon, L.A. (1997). Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton 37, 199-210. [DOI] [PubMed] [Google Scholar]
- Simon, V.R., and Pon, L.A. (1996). Actin-based organelle movement. Experientia 52, 1117-1122. [DOI] [PubMed] [Google Scholar]
- Simon, V.R., Swayne, T.C., and Pon, L.A. (1995). Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J. Cell Biol. 130, 345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.G., Simon, V.R., O'Sullivan, H., and Pon, L.A. (1995). Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 6, 1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, B. (1981). Mitochondrial structure. In: The Molecular Biology of the Yeast Saccharomyces, ed. J.M. Strathern, E.W. Jones, and J.R. Broach, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 471-504.
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingle, M.A., Kuenzi, M.T., and Halvorson, H.O. (1974). Germination of yeast spores lacking mitochondrial deoxyribonucleic acid. J. Bacteriol. 117, 89-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treinin, M., and Simchen, G. (1993). Mitochondrial activity is required for the expression of IME1, a regulator of meiosis in yeast. Curr. Genet. 23, 223-227. [DOI] [PubMed] [Google Scholar]
- Van Blerkom, J., Davis, P., and Alexander, S. (2000). Differential mitochondrial distribution in human pronuclear embryos leads to disproportionate inheritance between blastomeres: relationship to microtubular organization, ATP content and competence. Hum. Reprod. 15, 2621-2633. [DOI] [PubMed] [Google Scholar]
- Westermann, B., and Neupert, W. (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421-1427. [DOI] [PubMed] [Google Scholar]
- Williamson, D.H., and Fennell, D.J. (1979). Visualization of yeast mitochondrial DNA with the fluorescent stain “DAPI.” Methods Enzymol. 56, 728-733. [DOI] [PubMed] [Google Scholar]
- Winston, F., Dollard, C., and Ricupero-Hovasse, S.L. (1995). Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S228C. Yeast 11, 53-55. [DOI] [PubMed] [Google Scholar]
- Wong, E.D., Wagner, J.A., Gorsich, S.W., McCaffery, J.M., Shaw, J.M., and Nunnari, J. (2000). The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E.D., Wagner, J.A., Scott, S.V., Okreglak, V., Holewinske, T.J., Cassidy-Stone, A., and Nunnari, J. (2003). The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160, 303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.C., Palazzo, A., Swayne, T.C., and Pon, L.A. (1999). A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr. Biol. 9, 1111-1114. [DOI] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya, O., Newman, S.M., Okamoto, K., Perlman, P.S., and Butow, R.A. (1998). Functions of the HMG box protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics 148, 1763-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]