Abstract
Intraflagellar transport (IFT), the bidirectional movement of particles along flagella, is essential for flagellar assembly. The motor for retrograde IFT in Chlamydomonas is cytoplasmic dynein 1b, which contains the dynein heavy chain DHC1b and the light intermediate chain (LIC) D1bLIC. To investigate a possible role for the LIC in IFT, we identified a d1blic mutant. DHC1b is reduced in the mutant, indicating that D1bLIC is important for stabilizing dynein 1b. The mutant has variable length flagella that accumulate IFT-particle proteins, indicative of a defect in retrograde IFT. Interestingly, the remaining DHC1b is normally distributed in the mutant flagella, strongly suggesting that the defect is in binding of cargo to the retrograde motor rather than in motor activity per se. Cell growth and Golgi apparatus localization and morphology are normal in the mutant, indicating that D1bLIC is involved mainly in retrograde IFT. Like mammalian LICs, D1bLIC has a phosphate-binding domain (P-loop) at its N-terminus. To investigate the function of this conserved domain, d1blic mutant cells were transformed with constructs designed to express D1bLIC proteins with mutated P-loops. The constructs rescued the mutant cells to a wild-type phenotype, indicating that the function of D1bLIC in IFT is independent of its P-loop.
INTRODUCTION
Intraflagellar transport (IFT) is the rapid, bidirectional movement of granule-like particles along the length of eukaryotic cilia and flagella (Rosenbaum and Witman, 2002). IFT was first observed in the biflagellate green algae Chlamydomonas by differential interference contrast (DIC) microscopy (Kozminski et al., 1993). It also has been directly observed in the sensory cilia of Caenorhabditis elegans (Orozco et al., 1999; Signor et al., 1999; Qin et al., 2001) and in the primary cilia of mammalian kidney cells using green fluorescent protein–tagged IFT-particle proteins (Pazour, unpublished results). These IFT particles are moved in an anterograde direction from the cell body to the flagellar tip by kinesin-II (Kozminski et al., 1995) and then moved in a retrograde direction from the tip back to the cell body by cytoplasmic dynein 2/1b (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999). Studies in Chlamydomonas show that the IFT particles contain ∼16 different polypeptides with masses 20–172 kDa, which are organized into two complexes, A and B (Piperno and Mead, 1997; Cole et al., 1998; Cole, 2003). Homologues of these IFT-particle proteins are found in all ciliated organisms.
As a conserved process, IFT is important for assembly, maintenance, and normal function of flagella and cilia. Chlamydomonas cells with mutations in the genes encoding IFT motor subunits or IFT-particle proteins are nonmotile and either have no flagella or short flagella (Pazour et al., 2000; Brazelton et al., 2001). IFT transports flagellar precursors to the tip and returns turnover products to the cell body (Qin et al., 2004). IFT also is involved in mating (Pan and Snell, 2002) and flagella length control (Marshall and Rosenbaum, 2001) in Chlamydomonas. In the nematode C. elegans and the fly Drosophila melanogaster, mutations in genes encoding IFT-particle proteins and IFT motors affect the formation and function of the sensory cilia (Collet et al., 1998; Cole et al., 1998; Cole, 2003; Han et al., 2003; Haycraft et al., 2003; Sarpal et al., 2003). In the mouse, IFT defects cause a range of diseases including polycystic kidney disease, retinal degeneration, the laterality abnormality situs inversus (Pazour and Rosenbaum, 2002), and male sterility (San Agustin et al., unpublished results). Recent data on two mouse mutants, wimple (wim) and flexo (fxo), which are alleles of the mouse homologues of Chlamydomonas IFT172 and IFT88, show that the IFT machinery has an essential and vertebrate-specific role in Hedgehog signal transduction (Huangfu et al., 2003).
Although conserved among organisms and important in numerous cell types, the mechanism of IFT is not well understood. Studies from Chlamydomonas and C. elegans show that two dynein subunit genes are necessary for retrograde IFT. One encodes LC8, which is a light chain of several dynein isoforms and thus is presumably a subunit of the retrograde IFT motor (King et al., 1996; Pazour et al., 1998). The other encodes DHC1b, which is a cytoplasmic dynein heavy chain (DHC) isoform (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999). Its mammalian ortholog, DHC2, is associated with mammalian cilia (Mikami et al., 2002) and also was reported to play a role in the organization and/or function of the Golgi apparatus (Vaisberg et al., 1996).
Dyneins are typically large multisubunit complexes. For example, conventional cytoplasmic dynein 1 (a.k.a. dynein 1a in invertebrates) is composed of two heavy chains (DHC1/1a), 2 or 3 intermediate chains, several light intermediate chains (LICs), and numerous light chains (King, 2000; Reilein et al., 2001). It is likely that the retrograde IFT motor also contains subunits in addition to LC8 and DHC1b. Recently, a novel dynein LIC, D2LIC, was identified as a bona fide component of cytoplasmic dynein 2 in mammalian cells (Grissom et al., 2002) and was inferred to play a role in maintaining Golgi organization by binding cytoplasmic dynein 2 to its Golgi-associated cargo. Like dynein 1 LICs, mammalian D2LIC contains a phosphate-binding domain (P-loop) at its N-terminus; although P-loop domains are involved in binding the β,γ-phosphate group of ATP or GTP in many nucleotide-binding and -hydrolyzing proteins (Via et al., 2000), it is not known if this domain has any physiological role in dynein LICs.
Independently of the present study, the Chlamydomonas homologue of D2LIC was recently cloned and shown to be associated with DHC1b (Perrone et al., 2003), although its function was not examined. In C. elegans, the D2LIC homologue is XBX-1; deletion of XBX-1 resulted in stunted sensory cilia that accumulated IFT-particle proteins, suggesting that XBX-1 functions in retrograde IFT (Shafer et al., 2003). Surprisingly, however, XBX-1 accumulated, along with IFT-particle proteins, in the short sensory cilia of the C. elegans che-3 mutant (Shafer et al., 2003), which is defective in DHC1b/DHC2, but did not accumulate with the IFT-particle proteins in the short flagella of a Chlamydomonas DHC1b mutant (Perrone et al., 2003). Moreover, the P-loop that is conserved in mammalian and Chlamydomonas D2LIC homologues is absent in C. elegans XBX-1. These observations raise the possibility that the D2LIC homologues function differently in C. elegans, Chlamydomonas, and humans.
The current study, using Chlamydomonas reinhardtii as a model system, was undertaken to learn more about the role of this LIC in IFT and the possible function of its P-loop. We cloned the Chlamydomonas ortholog of mammalian D2LIC, which we term D1bLIC. Using D1bLIC cDNA as a probe, we identified a Chlamydomonas d1blic insertional mutant and characterized the mutant phenotype. We further analyzed the relationship between D1bLIC and DHC1b by immunostaining, flagellar fractionation, and coimmunoprecipitation. Finally, we used site-directed mutagenesis to investigate the function of the D1bLIC P-loop in IFT in vivo. Portions of this work were reported previously in abstract form (Hou et al., 2002).
MATERIALS AND METHODS
Strains and Antibodies
C. reinhardtii strains used in the work include: 137c (nit1, nit2, mt+; Chlamydomonas Genetics Center, Duke University, Durham, NC), CC124 (nit1, nit2, mt-; Chlamydomonas Genetics Center), A54-e18 (nit1–1, ac17, sr1, mt+; Schnell and Lefebvre, 1993; Nelson et al., 1994), and 3088.4 (dhc1b-1::NIT1, nit1, mt+; Pazour et al., 1999). TBD9–1 (d1blic::NIT1, nit1, mt+) was generated by transforming A54-e18 with the plasmid pMN56 linearized with EcoRI (Nelson et al., 1994) and was generously provided by T. Bui and C. Silflow (University of Minnesota at St. Paul) via W. Dentler (University of Kansas, Lawrence). YH43 (d1blic::NIT1, mt-) is a progeny of a cross between TBD9–1 and CC124. The mAb to Chlamydomonas β-tubulin was from Dr. G. Piperno (Mt. Sinai School of Medicine, NY). The monoclonal antibodies to Chlamydomonas IFT172 and IFT139 were from Dr. Douglas Cole (University of Idaho, Moscow, ID; Cole et al., 1998). The mAb to Chlamydomonas outer dynein arm IC IC1 and the polyclonal antibody to Chlamydomonas DHC1b were previously described (King et al., 1985, 1986; Pazour et al., 1999). The rat mAb (clone 3F10) to influenza hemagglutinin epitope (HA) tag was from Roche Molecular Biochemicals (Indianapolis, IN).
Growth Media
The following media were used: M (Sager and Granick [1953] medium I altered to have 0.0022 M KH2PO4 and 0.00171 M K2HPO4), R (M medium supplemented with 0.0075 M sodium acetate), M-N (M medium without nitrogen), and tris-acetate-phosphate (Gorman and Levine, 1965).
Chlamydomonas D1bLIC Gene Cloning and Sequence Analysis
A pair of the expressed sequence tag (EST) sequences (894070A09) was obtained by searching the Chlamydomonas EST database (http://www.biology.duke.edu/chlamy_genome/blast/blast_form.html) using the mammalian D2LIC sequence. Starting with the EST sequences, the intervening region of the Chlamydomonas D1bLIC cDNA was cloned from a Chlamydomonas cDNA library by PCR amplification using primer pairs D1bLIC5 (5′ACGTTGCTCAATCGCTTCTT 3′) and D1bLIC6 (5′ACCGTTCCTTCACACGAAAG 3′). The cDNA was used to screen a Chlamydomonas genomic library filter (http://www.genome.clemson.edu/orders/) and six positive bacterial artificial chromosome (BAC) clones (01O10, 18L01, 22E07, 31J16, 37P12, and 38N05) were obtained. When the BACs were digested with different restriction enzymes and the digests were analyzed by Southern blotting with the D1bLIC cDNA as a probe, each was confirmed to contain the same D1bLIC gene. The 01O10 BAC clone was subcloned into a 10-kb SmaI-HindIII cassette and then further subcloned into a 7.3-kb SmaI-SacI cassette that contained only the D1bLIC gene. The D1bLIC cDNA and gene were sequenced (GenBank accession number: AY616759). The predicted Chlamydomonas D1bLIC protein sequence was compared with the mammalian D2LIC using the Pairwise BLAST program at http://www.ncbi.nlm.nih.gov/BLAST/. The ProtParam tool at http://us.expasy.org/tools/protparam.html was used to calculate the predicted molecular weight and theoretical pI of D1bLIC. The ClustalW (1.82) Multiple Sequence Alignment program at http://www.ebi.ac.uk/clustalw/ was used for multiple sequence alignment and the TreeTop program with the cluster algorithm at http://www.genebee.msu.su/services/phtree_reduced.html was used to compute the phylogenetic tree for DLICs from different species. The ScanProsite program at http://us.expasy.org/prosite/ was used to search protein families and domains. The coils program (version 2.2) available at http://www.ch.embnet.org/software/COILS_form.html (Lupas et al., 1991; Lupas, 1996) was used for coiled-coil predictions. Protein motif fingerprints were identified using the PRINTS BLAST database at http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/.
D1bLIC Antibody Production, Protein Preparation, and Western Blotting
A 1.3-kb fragment of the Chlamydomonas D1bLIC cDNA was amplified by PCR from the cloned D1bLIC cDNA using primer pairs DLIC15 (5′GGAATTCACGTTGCTCAATCGCTTCTT 3′) and DLIC16 (5′ GGAATTCTTCCTTCACACGAAAGCGTA 3′). The PCR product was digested with EcoRI and inserted in the EcoRI site of pMAL-cR1 (New England Biolabs, Beverly, MA). Expression of this construct in Escherichia coli produced a protein in which the C-terminal 373 amino acids (starting at the sequence TLLNRFL) of D1bLIC were fused to the maltose-binding protein. The fusion protein was purified by amylose affinity chromatography, and antibodies were produced in rabbits (Research Genetics, Huntsville, AL). The same 1.3-kb PCR product was inserted into the EcoRI site of the pThioHisB vector (Invitrogen, Carlsbad, CA). The expression product of this construct was used to affinity purify anti-D1bLIC antibodies as described in Pazour et al. (1999). For Western blots, flagella were isolated and fractionated as described in Pazour et al. (1999). To obtain the flagellar matrix fraction, fresh flagella in HMDEK buffer (10 mM HEPES, pH 7.2, 5 mM MgSO4, 1 mM DTT, 0.5 M EDTA, and 25 mM KCl) were flash frozen in liquid nitrogen and thawed to effect disruption of the flagellar membrane as described by Zhang and Snell (1995). The sample was centrifuged at 16,000 rpm for 20 min at 4°C using an SS-34 rotor (Sorvall Products, Newtown, CT), and the supernatant was collected. Whole cell extracts were made as described in Pazour et al. (1999). Gel electrophoresis and Western blotting were performed as described in Pazour et al. (1999).
RNA and DNA Isolation and Analysis
DNA was isolated from Chlamydomonas by digesting the cells with proteinase K as described in Pazour et al. (1998). For determination of the patterns of gene expression, total RNA was obtained from wild-type (137c) cells or TBD9–1 cells before deflagellation and 30 min after deflagellation by pH shock as described in Witman et al. (1972) and Lefebvre and Rosenbaum (1986). Gel electrophoresis, Southern blotting, and Northern blotting were performed using standard procedures (Sambrook et al., 1987).
Genetic Analysis
TBD9–1 and CC124 cells were induced to mate by dibutyryl-cAMP and papaverine treatment and then were allowed to form mature zygotes on solid medium as described in Pazour et al. (1999). The plates were then kept at -20°C for 2 d to kill the vegetative cells. Plates were returned to light and 23°C for the tetrads to develop. The colonies that developed from each tetrad were streaked out and single colonies originating from each tetrad were transferred to 125 ml of M media. Cells were allowed to grow to midlog phase and then were scored for motility by microscopic observation of cells illuminated with dim red light. DNAs from the progenies were prepared for Southern analysis.
Transformation
Transformation was performed using the glass bead method of Kindle (1990) as described in Pazour et al. (1995). To rescue d1blic mutant cells, cloned D1bLIC genomic DNA and linearized plasmid pSP124S (from Dr. S. Purton, University College London) were cotransformed into cells. The zeocin-resistant cells were recovered as described in Lumbreras et al. (1998) except that zeocin (CAYLA, F-31405 Toulouse cedex 4, France) was used at 4 μg/ml in our experiments.
Electron and Immunofluorescence Microscopy
Cells were fixed in glutaraldehyde for EM (Hoops and Witman, 1983) and processed as described in Wilkerson et al. (1995). Cells were fixed and stained for immunofluorescence microscopy by the alternate protocol of Cole et al. (1998). Images were acquired using an AxioCam camera on an Axioskop 2 plus microscope (Carl Zeiss Microimaging, Thornwood, NY).
Sucrose Gradient Analysis
The flagellar matrix was fractionated through 12-ml 5–20% sucrose density gradients in a SWTi41 rotor at 36,000 rpm with the ultracentrifuge preset at 64,000 RAD2/SEC × 107. Gradients were fractionated into 0.5-ml aliquots and subsequently analyzed by SDS-PAGE and Western blotting. Thyroglobulin (19.4 S), catalase (11.3 S), and BSA (4.4 S) were used as sedimentation standards.
Immunoprecipitation
Protein A-Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ) were washed with HMDEK buffer + 3% BSA three times and then incubated with the anti-D1bLIC antibody in the same buffer for 1 h at 4°C. The beads were then washed twice with HMDEK buffer and incubated with freshly made flagellar matrix fraction for 1 h at 4°C. Beads with the immune complex were collected by centrifugation and were washed three times with HMDEK buffer. The supernatant and the final pellet were analyzed for D1bLIC and DHC1b protein levels using SDS-PAGE and Western blotting. In the control experiment, affinity-purified preimmune serum was used instead of affinity-purified anti-D1bLIC antibody.
HA-tagging of D1bLIC Genes and Site-directed Mutagenesis
A 2.3-kb SmaI-SacI fragment containing the 3′ end of the D1bLIC gene was removed from the 7.3-kb SmaI-SacI cassette containing the D1bLIC gene. It was cut by KasI and was blunted by T4 DNA polymerase. An ∼120-base pair fragment encoding three copies of the influenza hemagglutinin epitope (3HA) from the p3xHA plasmid (from Dr. C. Silflow, University of Minnesota; Silflow et al., 2001) was inserted into the blunted KasI site. The 3HA-tagged cassette was rejoined to the rest of the D1bLIC gene to yield a construct with the 3HA tag 63 nucleotides upstream from the stop codon.
A 2.8-kb SmaI-KpnI fragment containing the 5′ end of the D1bLIC gene was removed from the 7.3-kb SmaI-SacI D1bLIC gene cassette and was used for site-directed P-loop mutagenesis. Two sets of oppositely directed mutagenic primers were designed to amplify the whole plasmid containing the 2.8-kb SmaI-KpnI cassette. The ends of the PCR products were polished by T4 polymerase and T4 kinase. The modified PCR products were self-ligated to yield mutated circular plasmids. One set of primers (5′ gtcgacgttgctcaatcgcttcttgtatccc 3′ and 5′ gcgccggccgctcgcgagcc 3′) changed the wild-type P-loop from GSRAAGKS to GSRAAGAS and introduced a new SalI site at the DNA level. The second set of primers (5′ tatcgccacgttgctcaatcgcttc 3′ and 5′ ccggccgctcgcgagccc 3′) changed the wild-type P-loop from GSRAAGKS to GSRAAGIA and disrupted an existing NaeI site at the DNA level. Products were sequenced to verify that no additional mutation had been introduced by the PCR. The modified 2.8-kb fragments were inserted back into the 3HA-tagged D1bLIC gene to yield a K construct that has a K to A mutation in the P-loop and a KS construct that has a KS to IA mutation in the P-loop.
Detection of Exogenous D1bLIC Gene by PCR Amplification
Amplification by the PCR was used to check if a desired mutant D1bLIC gene was incorporated into the cells rescued with the K construct or the KS construct. The primer pair DLIC3 (5′CGGTTCTGCTAGGCGCAAAG 3′) and DLIC4 (5′TCCGAGTTCACGATCTCCTC 3′) were designed to amplify a ∼700-base pair fragment around the P-loop region from wild-type and rescued cells. No product should be amplified from YH43 (d1blic) cells. Products were then cut by SalI or NaeI, and analyzed in a 1.5% agarose gel.
RESULTS
Cloning and Sequence Analysis of D1bLIC
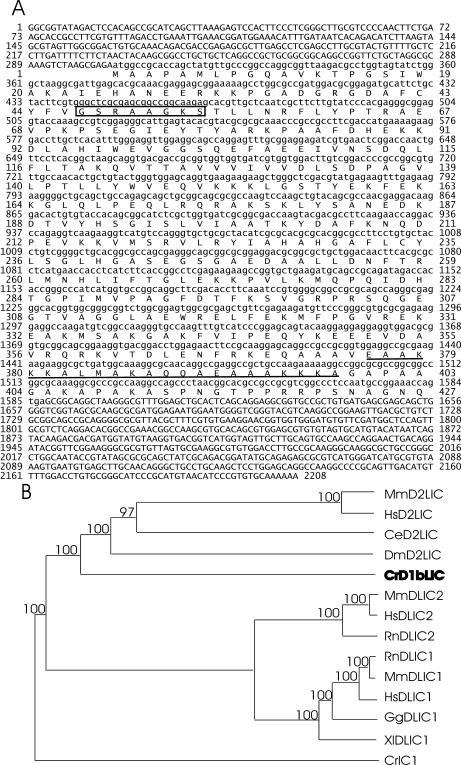
The D1bLIC gene, the Chlamydomonas ortholog of human D2LIC, was cloned as described in Materials and Methods, and 5692 base pairs of the D1bLIC gene region were sequenced. When the 5692 base pairs were blasted against the then newly released C. reinhardtti genome v1.0, an exact match on scaffold 319 was identified, which corresponded to the gene model genie.319.8 predicted by Green Genie.
The predicted D1bLIC protein is composed of 427 amino acids (Figure 1A), with a molecular weight of 46.6 kDa and a theoretical pI of 9.37. It is 28% identical to human D2LIC in the region of amino acids 28–372. Like all mammalian dynein LICs so far identified, D1bLIC has a P-loop near its N-terminus. Similar to D2LIC, it has a predicted coiled-coil domain near its C-terminus from amino acids 376–398. A BLAST search using D1bLIC as query yielded human D2LIC and the mouse homologue for D2LIC as the best matches as well as homologous proteins or ESTs from other species, such as rat, fly, nematode, chicken, Trypanosomiasis, etc., but not from nonciliated organisms such as Arabidopsis thaliana. A BLAST search of the Chlamydomonas genome (JGI v1.0 and v2.0) with human D2LIC did not reveal any genes that were more closely related to the query than D1bLIC. A phylogenetic analysis of D1bLIC, some of its probable homologues, and dynein 1 LICs shows that D1bLIC and human D2LIC group together, distinct from the dynein 1 LICs. (Figure 1B). These results indicate that the D1bLIC we cloned is the Chlamydomonas ortholog of human D2LIC.
Figure 1.
D1bLIC is the Chlamydomonas ortholog of mammalian D2LIC. (A) D1bLIC cDNA sequence and its predicted amino acid sequence. The P-loop from amino acids 47–54 is indicated by a box, and the predicted coiled-coil domain from amino acids 376–398 (predicted by COILS using the MTIDK matrix and a window of 21) is underlined. 5692 base pairs of genomic DNA, from 819 base pairs upstream of the D1bLIC start codon to 1354 base pairs downstream of the D1bLIC stop codon, were sequenced from a 7.3-kb SmaI-SacI cassette that was subcloned from the 01O10 BAC clone. The start of the 5′ untranslated region and the site of polyadenylation were determined by searching the Chlamydomonas EST database. The coding regions were determined by sequencing PCR products (containing sequences from nucleotides 58–1773) from a cDNA library. (B) Phylogenetic tree for dynein 1 and dynein 2 LICs from different species. The predicted D1bLIC sequence (in bold) was aligned with a subset of cytoplasmic dynein 1 and 1b/2 LIC sequences in GenBank using CLUSTAL W, and a phylogenetic tree was drawn by cluster algorithm. CrIC1 is Chlamydomonas outer arm dynein intermediate chain IC1. Branch lengths represent evolutionary relatedness. Numbers at branch points are bootstrap values. The Chlamydomonas D1bLIC groups closely with the mammalian and other presumptive D2LICs, but much less closely with cytoplasmic dynein 1 LICs. Cr, C. reinhardtii; Ce, C. elegans; Dm, D. melanogaster; Hs, Homo sapiens; Rn, Rattus novegicus; Mn, Mus musculus; Xl, Xenopus laevis; Gg, Gallus gallus. Sequences used for phylogenetic analysis are as follows: MnD2LIC (AAH39070); HsD2LIC (NP_057092); CeD2LIC (T20505); DmD2LIC (NP_609289); CrD1bLIC (AY616759); MnDLIC2 (XP_134573); HsDLIC2 (NP_006132); RnDLIC2 (I55514); RnDLIC1 (NP_665715); MmDLIC1 (NP_666341); HsDLIC1 (NP_057225); GgDLIC1 (I50637); XlDLIC1 (AAG42486); CrIC1 (Q39578).
Identification of a d1blic Mutant
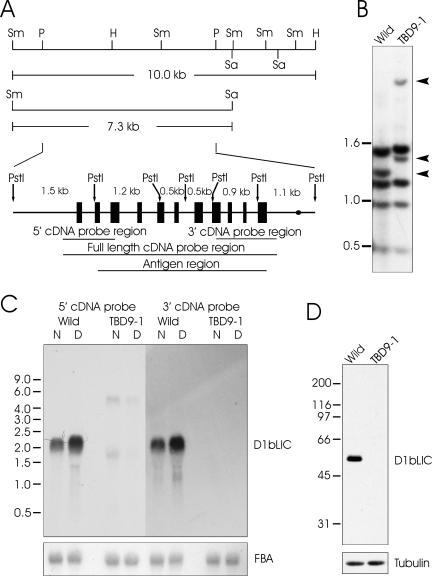
To investigate the function of D1bLIC, we used Southern blotting with a full-length D1bLIC cDNA probe (Figure 2A) to screen a collection of mutants created by insertional mutagenesis (Pazour and Witman, 2000). PstI cuts the wild-type D1bLIC gene into six fragments, of which two are about the same size and thus are not separated on the Southern blot (Figure 2A). For the wild-type cells, the full-length cDNA probe identified five bands on Southern blots digested with PstI (Figure 2B). From a screen of 95 mutants with swimming defects, one line (called TBD9-1) showed a restriction fragment length polymorphism with this probe. In this cell line, the 1.2-kb band, which corresponds to the 5′ end of the coding region, is missing, and two new bands of ∼1.4 and 3.5 kb are present (Figure 2B). Integration at this point is likely to produce a null allele. To test this, cells were analyzed by Northern and Western blot analysis. As reported (Perrone et al., 2003), the wild-type cells express a ∼2.2-kb D1bLIC message that is up-regulated by deflagellation, which is characteristic for messengers encoding flagellar proteins. However, this message is not detectable in the mutant cells using either 5′ or 3′ cDNA probes (Figure 2C). Consistent with the idea that this is a null allele, an affinity-purified polyclonal antibody to D1bLIC (see Materials and Methods) detected with high specificity a single band of ∼48.5 kDa in the wild-type whole-cell lysate, but detected no proteins in the mutant whole-cell lysate (Figure 2D).
Figure 2.
An insertional mutant, TBD9–1, is defective in D1bLIC. (A) Schematic diagram of D1bLIC gene structure. The 10.0-kb fragment from SmaI to HindIII and the 7.3-kb fragment from SmaIto SacI were used for later rescue experiments. The D1bLIC gene is composed of 11 exons (shown as black boxes). The site where the polyA signal is added is represented by a black dot. PstI cuts the gene into six fragments of the indicated sizes. The regions corresponding to the 5′ cDNA probe and 3′ cDNA probe used in the Northern blotting, the full-length cDNA probe used in the Southern blotting, and the antigen region for antibody preparation are indicated by lines underneath. Sm, SmaI; K, KpnI; P, PstI; H, HindIII; Sa, SacI. (B) Southern blotting indicates the D1bLIC gene is disrupted in TBD9–1. Genomic DNA from wild-type cells and TBD9–1 cells was cut by PstI and probed with the full-length D1bLIC cDNA probe. The TBD9–1 DNA is lacking one 1.2-kb PstI band present in wild-type DNA and has two additional bands of ∼1.4 and 3.5 kb as indicated by arrowheads. This implies that there is an insertion in the 1.2-kb PstI region of the D1bLIC gene in TBD9–1 cells, whereas the rest of the gene is intact. (C) Northern blotting indicates D1bLIC mRNA is disrupted in TBD9-1. Total RNA from TBD9-1 cells and wild-type cells before (N) and 30 min after deflagellation (D) were probed with 5′ or 3′ D1bLIC cDNA probes. The D1bLIC mRNA is ∼2.2 kb and is up-regulated by deflagellation in wild-type cells. No signal was detected by the 3′ cDNA probe in RNA from TBD9-1. Two faint bands of ∼5 and 1.8 kb were detected by the 5′ cDNA probe in RNA from TBD9-1 cells; they are slightly down-regulated by deflagellation. The bottom panel was probed by fructose-bisphosphate aldolase (FBA) cDNA as a loading control. (D) D1bLIC protein is missing in TBD9-1. A Western blot of whole cell lysates from TBD9-1 cells and wild-type cells probed with the anti-D1bLIC antibody shows that D1bLIC migrates at ∼48.5 kDa in wild-type cells and that no protein is present in TBD9-1 cells. The bottom panel was probed with an anti–β-tubulin antibody as a loading control.
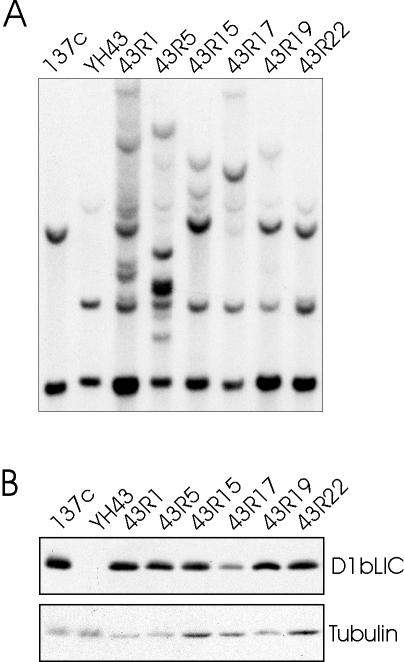
To make sure that the mutant was defective only in D1bLIC, we crossed the original mutant with wild-type cells (CC124), and selected one progeny (called YH43) that is defective in D1bLIC as determined by Southern and Western blots (Figure 3, A and B). When aerated in 125 ml M media, most of the mutant cells are nonmotile with short, stumpy flagella, whereas a few have longer flagella of variable length, which sometimes can even be near normal length, and swim abnormally. The mutant cells were cotransformed with plasmid pSP124S that contains the ble gene and with either a 10-kb SmaI-HindIII genomic fragment or a 7.3-kb SmaI-SacI genomic fragment, both of which contain the D1bLIC gene (Figure 2A). Both genomic fragments rescued the phenotype to wild-type swimming behavior (7 of 43 ble transformants for the 10-kb fragment and 10 of 216 ble transformants for the 7.3-kb fragment). Figure 3, A and B, show Southern and Western blot analyses of some of the cells rescued by the 10-kb fragment, confirming that the exogenous D1bLIC gene was incorporated into the genome to express D1bLIC protein. The 10-kb fragment contains the D1bLIC gene, most of a predicted gene downstream of the D1bLIC gene, and the 3′ end of a predicted gene upstream of D1bLIC. The 7.3-kb fragment contains only the D1bLIC gene and the 3′ end of the upstream gene, which is of unknown function (JGI v2.0). Thus, the rescue of the phenotype via these fragments is strong evidence that the flagellar defect observed in this cell line is caused by the defect in the D1bLIC gene.
Figure 3.
The phenotype of YH43 can be rescued by the cloned D1bLIC gene. (A) D1bLIC DNA is restored in cells rescued for the motility defect by transformation with the D1bLIC gene. Genomic DNA from wild-type cells (137c), YH43, and some of the cells rescued by the 10-kb D1bLIC genomic fragment (43R1, 43R5, 43R15, 43R17, 43R19, and 43R22) was cut by SmaI and analyzed on Southern blots probed with the full-length D1bLIC cDNA. In addition to the mutated D1bLIC gene, different bands were detected in the rescued cells, indicating the incorporation of the wild-type D1bLIC transgene at different sites. (B) D1bLIC protein is detected in rescued cells. Western blots of wild-type cells (137c), YH43, and some of the rescued cells (43R1, 43R5, 43R15, 43R17, 43R19, and 43R22) probed with the anti-D1bLIC antibody showed that the rescued cells express D1bLIC protein. β-tubulin was probed in the bottom panel as a loading control.
Retrograde IFT Is Defective in the d1blic Mutant
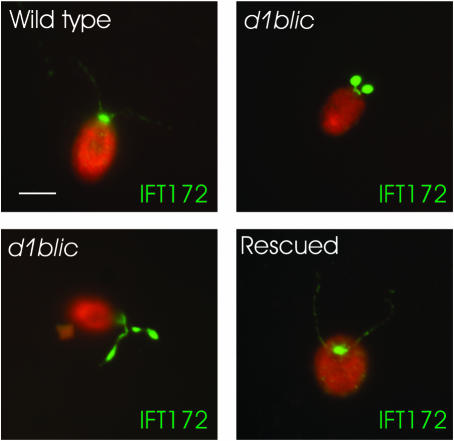
To investigate if D1bLIC is involved in retrograde IFT, we examined the localization of IFT particles in d1blic mutant cells by immunofluorescence and electron microscopy. In wild-type cells, the majority of IFT-particle proteins localize in the peri-basal body region, while a few are distributed along the length of the flagella (Cole et al., 1998; Pazour et al., 1999; Deane et al., 2001) as illustrated in Figure 4 with an antibody to IFT172. In the d1blic mutant, IFT172 accumulates at the tip of the short stumpy flagella or in large inclusions randomly located along the flagella of cells with longer flagella. Accumulation of IFT-particle proteins in short flagella is characteristic of mutants with defects in retrograde IFT (Pazour et al., 1998, 1999; Porter et al., 1999). In contrast, cells rescued for the motility phenotype by transformation with the cloned gene showed a normal distribution of IFT172.
Figure 4.
Immunofluorescence microscopy shows that the YH43 cell accumulates IFT-particle proteins in its short flagella. Wild-type cells, YH43 cells (d1blic), and rescued cells were stained with an anti-IFT172 antibody (green). Cell bodies are red due to auto-fluorescence. Most of the IFT172 protein is in the peri-basal body region with a lesser amount spread along the flagella in the wild-type cells. In the d1blic mutant cells, IFT172 is redistributed from the peri-basal body region to the flagella, which are usually short and stumpy but sometimes longer. In mutant cells rescued with the wild-type D1bLIC gene, IFT172 is distributed as in wild-type cells. Scale bar, 5 μm.
EM analysis showed that flagella from d1blic mutants are filled with an electron-dense substance not observed in flagella from wild-type cells or in rescued cells (Figure 5). This material is identical in appearance to the IFT particles that accumulate in fla14 mutant flagella (Pazour et al., 1998) and dhc1b mutant flagella (Pazour et al., 1999). These results confirm that IFT particles accumulate in the short flagella of the d1blic mutant. Therefore, the primary defect in the d1blic mutant flagella is a loss of retrograde IFT. The IFT particles are moved into the flagellum by anterograde IFT, but become trapped there because some aspect of retrograde IFT is abnormal.
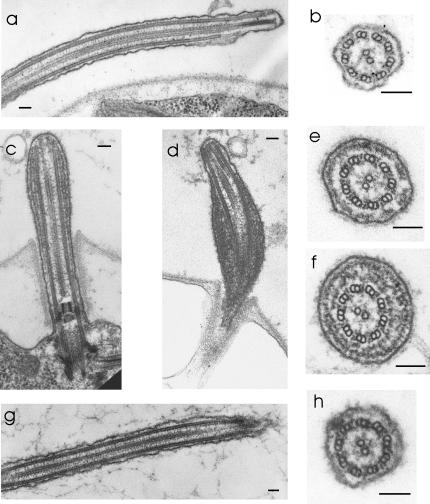
Figure 5.
Electron microscopy shows that d1blic flagella accumulate IFT particles. In wild-type cells (a and b) and the rescued cells (g and h) the space between the flagellar membrane and doublet microtubules is usually devoid of material. In contrast, in the d1blic mutant cells (c, d, e, and f) the space between the doublet microtubules and the flagellar membrane is filled with electron-dense material identical in appearance to IFT particles (Kozminski et al., 1993, 1995; Pazour et al., 1998). Note that the d1blic mutant cells have an apparently normal axoneme. Scale bars, 100 nm.
In contrast to what was observed in fla14 cells, which are defective in LC8, the d1blic mutation does not appear to affect components of the axoneme, as inner and outer dynein arms and radial spokes are present along with a normalappearing central pair of microtubules that are properly positioned in the axoneme. The flagellar defects in d1blic mutant cells are less severe than those in dhc1b mutant cells, whose flagella are always short and stumpy and may lack a normal axoneme.
The d1blic Mutant Is Not Defective in Growth Rate or in Golgi Localization or Morphology
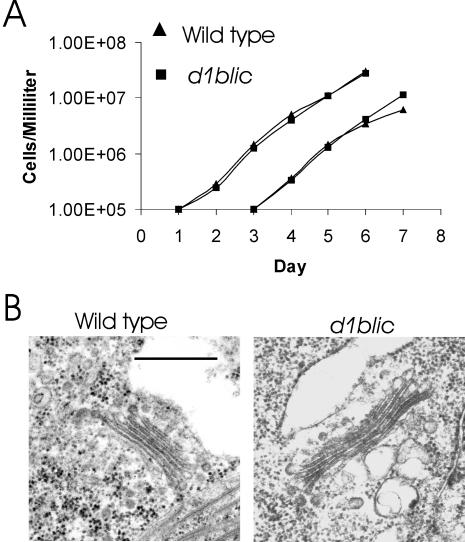
Cytoplasmic dynein is involved in many cell activities. To see if D1bLIC is involved in cell division or cell cycle control, we checked the growth rate of the d1blic mutant (Figure 6A). There was no significant difference in growth rate between wild-type cells and d1blic mutant cells, indicating that D1bLIC is not involved in any essential cell process in Chlamydomonas.
Figure 6.
The d1blic mutant is normal in cell growth rate and localization and morphology of the Golgi apparatus. (A) Growth curves for wild-type cells and d1blic mutant cells. Wild-type cells and d1blic cells were grown in liquid M medium with aeration with 5% CO2. Every 24 h a sample was removed and the cells counted with a hemocytometer. On day 3, a second set of cultures was inoculated by diluting cells from the first series to 105 cells/ml. (B) The Golgi apparatus of d1blic mutant cells is morphologically normal. Like the wild-type cells, the d1blic cells have a pair of Golgi apparatuses on the opposite side of the nucleus from the basal bodies. The Golgi apparatus is composed of 4–8 cisternae and is adjacent to a large membrane-bound vesicle on the trans side.
It has been suggested that human D2LIC is involved in maintenance of the Golgi apparatus (Grissom et al., 2002). To investigate a possible role for D1bLIC in Golgi maintenance in Chlamydomonas, we examined Golgi localization and morphology in the d1blic mutant by EM. As seen in Figure 6B, the Golgi had normal morphology in the d1blic mutant cells. Golgi apparatuses in both wild-type and mutant cells are always observed on the opposite side of the nucleus from the flagella and are adjacent to a large membrane-bound vacuole on the side toward the cell surface.
DHC1b Is Reduced in d1blic Mutant Cells and Vice Versa
Previous studies showed that DHC1b is the heavy chain of the retrograde IFT motor. To investigate the relationship between DHC1b and D1bLIC, we looked at the level of each protein in a mutant defective in the other protein and found that in the absence of one protein, the level of the other protein in the whole cell lysate is greatly decreased (Figure 7). This result implies that DHC1b and D1bLIC exist in the same complex. Because the majority of DHC1b (Pazour et al., 1999) and D1bLIC (see below and Figure 8) are in the peri-basal body region in wild-type cells, this result also implies that DHC1b and D1bLIC are associated with one another in the cell body before they move into the flagella. Lack of either one of the proteins probably affects stability of the complex and causes the other protein to be degraded.
Figure 7.
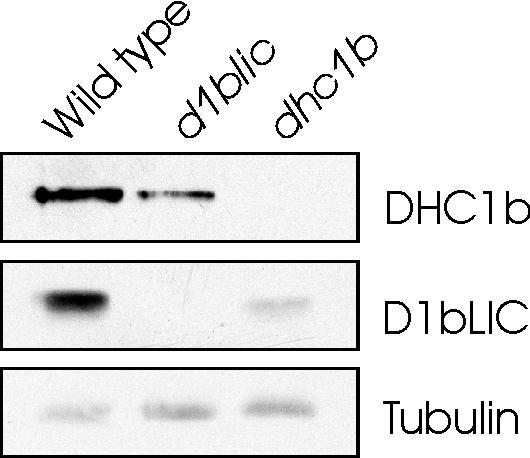
The level of DHC1b is reduced in d1blic mutant cells and vice versa. Western blots of whole-cell lysates from wild-type cells, d1blic mutant cells, and dhc1b mutant cells were probed with anti-DHC1b or anti-D1bLIC antibodies. The DHC1b level is reduced in the d1blic mutant; the D1bLIC level is greatly reduced in the dhc1b mutant. β-tubulin was probed with an anti–β-tubulin antibody as a loading control.
Figure 8.
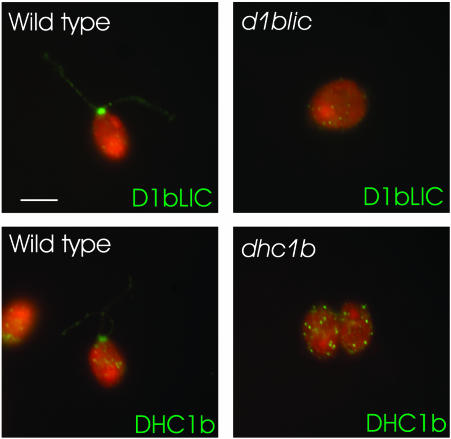
D1bLIC and DHC1b have a similar cellular localization. Wild-type cells (left panels) were stained with anti-D1bLIC antibody or anti-DHC1b antibody. The null mutants (right panels) served as controls and were stained in the same way. The antigens are in green and the cell bodies are in red. Both DHC1b and D1bLIC are located primarily in the peri-basal body region and sparsely distributed along the flagella.
D1bLIC Is in the Same Complex as DHC1b
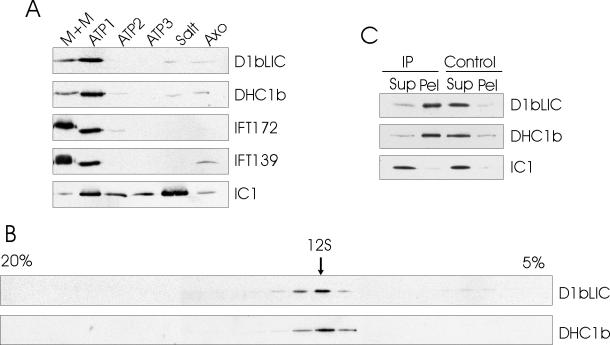
To further investigate if DHC1b and D1bLIC exist in the same complex, we first looked at the cellular localization of both DHC1b and D1bLIC by immunofluorescence microscopy (Figure 8). As reported (Perrone et al., 2003), DHC1b and D1bLIC have a similar cellular localization: they both localize in the peri-basal body region and along the flagella in the wild-type cell. To investigate if they distribute together in various flagellar fractions, we fractionated flagella into the membrane plus matrix fraction, different ATP-wash fractions, a high-salt extract fraction and an extracted axoneme fraction. Western blotting by antibodies to IFT172, IFT139, DHC1b, D1bLIC, and outer dynein arm IC IC1 showed that DHC1b and D1bLIC had a similar distribution in the different flagella fractions, which was different from that of IFT-particle proteins or the outer dynein arm (Figure 9A). These results indicate that DHC1b and D1bLIC are in the same subcompartment in the flagella. We then separated the proteins of the flagellar matrix fraction by sucrose density gradient centrifugation and found that DHC1b and D1bLIC comigrate at ∼12S (Figure 9B). To further confirm that D1bLIC is in the same complex as DHC1b, we immunoprecipitated D1bLIC from the flagella matrix fraction using the anti-D1bLIC antibody and found that DHC1b, but not outer dynein arm component IC1, was coimmunoprecipitated (Figure 9C). These results confirm that D1bLIC and DHC1b are subunits of the same dynein complex.
Figure 9.
D1bLIC is in the same complex as DHC1b. (A) D1bLIC and DHC1b have a similar distribution in flagellar fractions. Wild-type flagella were fractionated into detergent-soluble membrane plus matrix (M+M), ATP wash 1 (ATP1), ATP wash 2 (ATP2), ATP wash 3 (ATP3), salt extract (Salt), and salt-extracted axonemes (Axo). Extracts from an equal number of flagella were probed with anti-DHC1b, anti-D1bLIC, anti-IFT172, anti-IFT139, and anti-IC1 antibodies. As reported by Pazour et al. (1999), DHC1b is contained primarily in the first ATP wash, with a lesser amount in the membrane plus matrix fraction and very little in the other fractions. In contrast, IFT172 and IFT139 are concentrated primarily in the membrane plus matrix fraction, whereas outer dynein arm intermediate chain IC1 is located primarily in the salt extract. Like DHC1b, D1bLIC also is contained primarily in the first ATP wash. (B) D1bLIC and DHC1b cosediment with each other. The matrix fraction from wild-type flagella was fractionated by centrifugation in a 5–20% sucrose gradient. Fractions were probed with anti-D1bLIC and anti-DHC1b antibodies. D1bLIC and DHC1b comigrate in a sharp peak at ∼12 S. (C) DHC1b is coimmunoprecipitated by anti-D1bLIC antibody. Anti-D1bLIC antibody (IP) or preimmune serum (Control) was used for the immunoprecipitation experiments. The resulting supernatants (Sup) and pellets (Pel) were probed with anti-D1bLIC, anti-DHC1b, and anti-IC1 antibodies. Both D1bLIC and DHC1b were immunoprecipitated by the anti-D1bLIC antibody but not by preimmune serum. IC1 remained in the supernatant from both the anti-D1bLIC antibody and preimmune serum immunoprecipitations.
Localization of DHC1b in the d1blic Mutant Flagella Is Normal
As noted above, the phenotype of the d1blic mutant is less severe than that of the dhc1b mutant. This suggests that the DHC1b remaining in the d1blic mutant retains some function even in the absence of the LIC subunit. To investigate the specific role of D1bLIC in cytoplasmic dynein 1b function and IFT, we examined the distribution of DHC1b in d1blic mutant cells with near full-length flagella, because these cells are likely to have the highest levels of DHC1b protein. We envisioned three main possibilities: 1) DHC1b is abnormally concentrated in the peri-basal body region with very little in the flagella, suggesting that D1bLIC has a role in attaching cytoplasmic dynein 1b as cargo to IFT particles moving in an anterograde direction; 2) DHC1b accumulates along with the IFT particles in the mutant flagella, suggesting that DHC1b motor function is defective in the absence of D1bLIC; or 3) DHC1b is normally distributed in the flagella, suggesting that motor activity is unimpaired in the absence of the LIC. When d1blic mutant cells were stained with the antibody to DHC1b, the distribution of DHC1b was not noticeably different than that observed in wild-type cells (Figure 10). Significantly, DHC1b did not accumulate in patches throughout the flagella, as was observed for the IFT particles. These observations suggest that DHC1b retains motor activity and can move out of flagella normally in the absence of D1bLIC. The fact that the mutant flagella accumulate IFT particles but not DHC1b protein further suggests that D1bLIC has a role in binding the IFT particle to the cytoplasmic dynein 1b motor for retrograde transport.
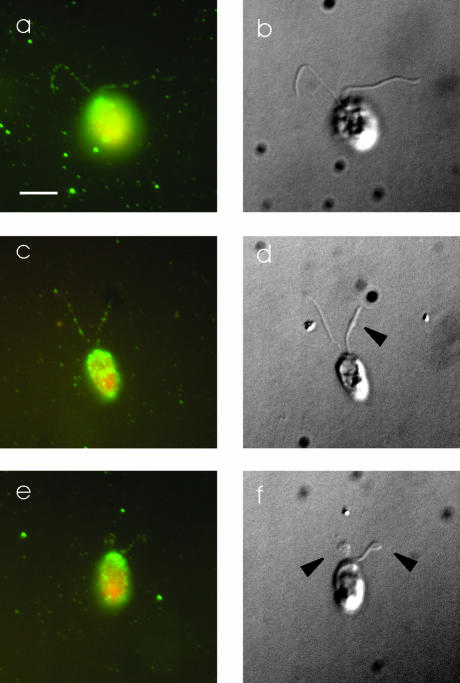
Figure 10.
DHC1b has a normal localization in d1blic mutant cells. Wild-type (a and b) and d1blic mutant cells with near full-length flagella (c and d) or shorter flagella (e and f) were stained with the anti-DHC1b antibody and were imaged by fluorescence microscopy (a, c, and e) or DIC microscopy (b, d, and f). Arrowheads indicate the bulges, shown in Figure 4 to be filled with IFT particles, in d1blic mutant flagella. DHC1b protein is in green and cell bodies in red. The DHC1b protein was present as punctae along the length of both wild-type and d1blic mutant flagella. No obvious accumulation of DHC1b protein was observed in the bulges on the d1blic flagella. Scale bar, 5 μm.
D1bLIC's P-loop Is Not Required for the Protein's Function in Retrograde IFT
Sequence analyses show that both D1bLIC and D2LIC have a P-loop at their N-termini. The conservation of the P-loop throughout evolution suggests that it is involved in D1bLIC function. To investigate this, we first fused the 3′ end of the wild-type D1bLIC gene to DNA encoding a 3HA tag. The 3HA-tagged gene rescued the mutant cells to wild-type swimmers, which expressed 3HA-tagged D1bLIC (43R, Figure 11B). Site-directed mutagenesis was then applied to the 3HA-tagged D1bLIC gene to create two constructs with a mutated P-loop. The K construct has a K to A mutation. The KS construct has a KS to IA mutation. These mutations have been shown to disrupt the P-loop function in other proteins (Saraste et al., 1990; Silvanovich et al., 2003). PCR amplification showed that the constructs were incorporated into the genomes (Figure 11A) and Western blots with anti-HA antibody and anti-D1bLIC antibody showed that these transformed cells express HA-tagged D1bLIC protein (Figure 11B). These transformants have normal length flagella (Figure 11C) and swim like wild-type cells. Immunostaining of these cells with anti-IFT172 antibody shows that IFT172 protein is normally distributed in these cells (Figure 11D). This result implies that the P-loop is not required for the normal function of D1bLIC in retrograde IFT. Because IFT is required for the mating process (Pan and Snell, 2002), we checked the mating ability of these cells by mixing the gametes of the rescued cells with wild-type gametes and observed that they can form normal mating clusters and subsequently quadriflagellate cells. This result shows that the P-loop also is not important for the mating process.
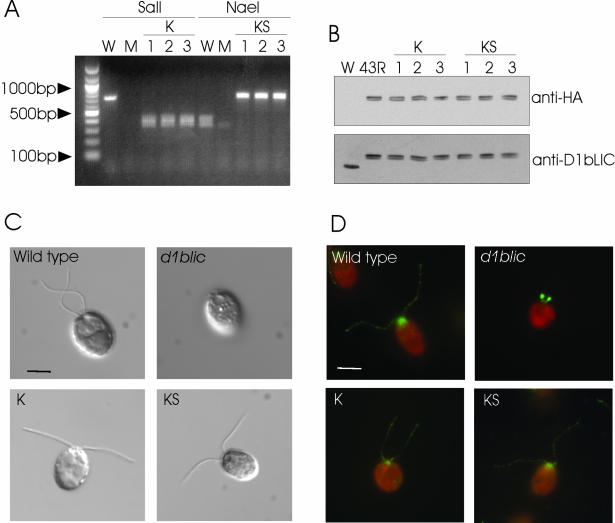
Figure 11.
The P-loop is not required for D1bLIC's function in IFT. (A) PCR amplification shows that the mutated exogenous D1bLIC genes were incorporated into the cell lines transformed with mutated P-loop constructs K and KS. The P-loop region was amplified using genomic DNA from wild-type cells (W), d1blic mutant cells (M), and cells transformed with mutated P-loop construct K (1, 2, and 3) or construct KS (1, 2, and 3). As expected, a ∼700-base pair product obtained from wild-type cells was cut by NaeI into two ∼350-base pair products, but was not cut by SalI. No specific product was obtained from mutant cells. The ∼700-base pair products obtained from cells transformed with the K construct contained a SalI site and thus could be cut by SalI into two ∼350-base pair fragments. The ∼700-base pair products obtained from cells transformed with the KS construct lack the NaeI site and thus remained ∼700 base pairs in length after NaeI treatment. (B) Western blotting shows that the transformed cells express the 3HA-tagged D1bLIC protein. Whole-cell lysate from wild-type cells (W), cells transformed with the 3HA-tagged wild-type D1bLIC gene (43R), cells transformed with the K construct (1, 2, and 3), and cells rescued with the KS construct (1, 2, and 3) were probed with the anti-HA antibody or the anti-D1bLIC antibody. The anti-HA antibody did not recognize any band in the wild-type cell lysate, but recognized a doublet of ∼ 56–57 kDa in the transformed cell lysates. Western blotting using the anti-D1bLIC antibody on an equivalent blot confirmed that these doublets represented HA-tagged D1bLIC protein, which migrated at a higher position than the endogenous D1bLIC protein in the wild-type cells. (C) DIC images of wild-type cells, d1blic cells, and d1blic cells transformed with the K (K) and KS (KS) constructs. Flagellar length is completely restored in cells transformed with the K or KS constructs. Scale bar, 5 μm. (D) IFT172 is normally distributed in d1blic cells transformed with the K and KS constructs. Wild-type cells, d1blic cells, and d1blic cells transformed with the K (K) and KS (KS) constructs were stained with an anti-IFT172 antibody (green). Cell bodies are red due to auto-fluorescence. Scale bar, 5 μm.
DISCUSSION
D1bLIC Is a Subunit of the Retrograde IFT Motor
Previously, two genes were identified as being required for retrograde IFT in Chlamydomonas. One, FLA14, encodes LC8, whereas the other, DHC1b, encodes the heavy chain of cytoplasmic dynein 1b. Both fla14 and dhc1b mutants accumulate IFT-particle proteins in their short flagella, indicating that retrograde IFT is defective in these mutants. Here, we show that a d1blic mutant also accumulates IFT-particle proteins in its flagella, which is consistent with a defect in retrograde IFT. These results are also consistent with the recent report that XBX-1, the C. elegans homologue of D1bLIC, is required for retrograde IFT and the assembly of sensory cilia in the nematode (Schafer et al., 2003).
Our findings that the level of DHC1b is decreased in the d1blic mutant and vice versa provide strong genetic evidence for an interaction between DHC1b and D1bLIC. Such an interaction is supported by our observations that D1bLIC and DHC1b 1) colocalize in the peri-basal body region and along the flagella, 2) have a similar distribution among different flagella fractions, 3) comigrate with each other when the flagella matrix is fractionated by sucrose density gradient centrifugation, and 4) are coimmunoprecipitated from the flagellar matrix fraction by an anti-D1bLIC antibody. Similar biochemical observations were made in a recent parallel study on D1bLIC (Perrone et al., 2003). Taken together, these observations conclusively demonstrate that D1bLIC is a subunit of the retrograde IFT motor, cytoplasmic dynein 1b, in the flagella. The results support that D1bLIC also is associated with DHC1b in the cell body, where a large pool of cytoplasmic dynein 1b is maintained in the peri-basal body region.
We observed that, after their release from the flagellum by freeze-thaw, DHC1b and D1bLIC comigrated in a sharp peak at ∼12S in sucrose density gradients. Sedimentation at 12S implies that the particle is a single-headed dynein containing one heavy chain, as in the case of the 12S particle consisting of the γ DHC and two light chains (Pfister et al., 1982, Witman et al., 1983) or the 11.9S particle consisting of the α DHC and one light chain (Pfister and Witman, 1984), both obtained by dissociation of outer arm dynein. In contrast, Perrone et al. (2003) observed that cytoplasmic dynein 1b sedimented in a broad peak at ∼19S after its release from demembranated axonemes by treatment with 10 mM MgATP. A particle sedimenting at 19S would likely represent a two-headed dynein containing two DHCs, as in the case of the 18S particle consisting of the α and β DHCs and their associated subunits from the outer arm dynein (Pfister et al., 1982), or the ∼21S particle containing inner arm DHCs 1α and 1β plus an intermediate chain (Piperno et al., 1990). It is possible the freeze-thaw step that we used to release cytoplasmic dynein 1b from the flagellum caused it to dissociate into the smaller 12S complex. However, an alternative possibility is that dynein 1b is in a two-headed form when actively transporting IFT particles, but is in a one-headed form when being passively transported to the tip of the flagellum as cargo. The former would be expected to remain associated with the axoneme by rigor bonds after demembranation and then be released by ATP, whereas the latter would be released with the IFT particles by demembranation or freeze-thaw. If dynein 1b is transported in an anterograde direction as a single-headed species, this would severely limit its ability to interact with the doublet microtubules to produce competing force in the retrograde direction.
Role of D1bLIC in Cytoplasmic Dynein 1b
The phenotype of the d1blic mutant is not as severe as that of the dhc1b mutant. The dhc1b mutants are nonmotile with short, stumpy flagella (Pazour et al., 1999; Porter et al., 1999). Although most d1blic mutant cells are nonmotile with short, stumpy flagella, some have variable length flagella of up to normal length. Moreover, EM showed that some dhc1b mutant cells have an apparently normal axoneme and some do not (Pazour et al., 1999), whereas all d1blic mutant cells, including those with short stumpy flagella, have an apparently normal axoneme. A likely explanation for the difference between the d1blic and dhc1b mutants is that D1bLIC serves as an intimate partner for DHC1b to help stabilize the dynein 1b complex, but that DHC1b can function to some extent even in the absence of D1bLIC. A role for D1bLIC in stabilizing dynein 1b is strongly supported by the observation that the level of DHC1b is reduced in the mutant cells lacking D1bLIC. Moreover, even though the cellular level of DHC1b is reduced in the d1blic mutant, what remains must be at least partially functional to support sufficient IFT to assemble flagella of up to normal length and maintain them at that length. It is probable that in the d1blic mutant cells, DHC1b, reduced in amount and probably impaired in cargo binding (see below), is unable to transport IFT particles out of the flagellum as effectively as the fully functional anterograde IFT motor can move them into the flagellum, so that the particles gradually accumulate in the flagellum. The variability in the length of the flagella in the d1blic mutant probably reflects cell-to-cell differences in the remaining amount of DHC1b. The large accumulation of IFT particles undoubtedly impairs the movement of the flagella, so that even cells with normal length flagella swim in an abnormal manner.
Studies on isoforms of DLIC1, the LIC for cytoplasmic dynein 1/1a, imply that LICs may be involved in cargo binding (Purohit et al., 1999; Tynan et al., 2000) and motor regulation (Niclas et al., 1996; Dell et al., 2000; Bielli et al., 2001). The phenotype of the d1blic mutant suggests a similar role for D1bLIC. Indeed, the observation that the distribution of DHC1b in d1blic mutant flagella is not noticeably different than that observed in wild-type flagella, whereas IFT-particle proteins accumulate dramatically in d1blic mutant flagella, strongly implies that D1bLIC stabilizes the interaction of cytoplasmic dynein 1b with its cargo, or increases the affinity of the motor for its cargo. However, because the stability of DHC1b is dependent on the presence of D1bLIC, it will be necessary to test the cargo binding and motor regulatory functions of D1bLIC by means of site-directed mutations that do not prevent expression of the subunit and its incorporation into the dynein 1b complex. The d1blic null mutant described here should be very useful as a recipient of such transgenes in future studies.
Role of the Conserved P-loop
The P-loop is commonly found in many adenine and guanine nucleotide-binding proteins, where it coordinates the β, γ-phosphate group of the nucleotide and is necessary for nucleotide hydrolysis (Via et al., 2000). Although LICs are not known to bind ATP or GTP, Chlamydomonas D1bLIC and mammalian D2LIC have a conserved P-loop motif, suggesting that this P-loop has a function in D1bLIC activity. To test this hypothesis in vivo, we transformed the d1blic null mutant with constructs designed to express D1bLIC subunits carrying mutations known to disrupt P-loop function in other proteins (Saraste et al., 1990; Silvanovich et al., 2003). The mutated constructs fully rescued the flagella assembly defect of d1blic mutant cells, implying that D1bLIC's P-loop is not critical for the protein's function in retrograde IFT. The cells rescued with the mutated constructs also mate normally, implying that the P-loop is not essential for the mating process. This result is consistent with previous results on the P-loops of C. elegans and mammalian cytoplasmic dynein 1 LICs (Tynan et al., 2000; Yoder and Han, 2001) and with the fact that the C. elegans dynein 2 LIC does not have a P-loop (Schafer et al., 2003).
Other Functions of Cytoplasmic Dynein 1b
Our results together with previous studies show that in Chlamydomonas, D1bLIC/DHC1b is involved in flagella formation, maintenance and function by acting as the retrograde IFT motor. Our observation that the d1blic null mutant grows normally indicates that D1bLIC is not important for any essential cell function. However, it might have other functions, in addition to being the retrograde IFT motor, related to flagellar assembly. One possibility is that, as a microtubule minus-end directed motor, cytoplasmic dynein 1b also is involved in transporting flagellar proteins from the cell body to the basal body region. However, because the dhc1b null mutant has short, stumpy flagella filled with IFT particles and sometimes containing an apparently normal axoneme (Pazour et al., 1999), these IFT particle-proteins and many axonemal structural proteins apparently are not dependent on DHC1b for their transport from the cell body to the basal body region. Moreover, mastigoneme protein, which normally is an extracellular component of the flagellar membrane, localizes at the apex of dhc1b mutant cells (M. Chapman and G. Witman, unpublished results), and the major flagellar membrane glycoprotein is highly localized to the short stumpy flagella of dhc1b mutant cells (unpublished results). Therefore, it is unlikely that the transport of these proteins from their sites of synthesis to their site of incorporation into the cell or flagellar plasma membrane is disrupted by the DHC1b mutation.
A second possibility is that the DHC1b complex is involved in maintaining the Golgi apparatus. DHC2, the mammalian homologue of DHC1b, has been localized to the Golgi apparatus, and microinjection of DHC2 antibodies caused the Golgi complex to disperse (Vaisberg et al., 1996). Moreover, D2LIC colocalized with DHC2 at the Golgi apparatus throughout the cell cycle, and the two proteins colocalized with Golgi fragments induced by the depolymerization of microtubules with nocodazole (Grissom et al., 2002). After brefeldin A treatment, some D2LIC lost its association with the Golgi and remained localized in the vicinity of the centrosome (Grissom et al., 2002). However, in Chlamydomonas, it appears that DHC1b/D1bLIC is not important for Golgi maintenance, because both d1blic (this study) and dhc1b (Pazour et al., 1999) mutants have normal Golgi.
Although our observations in Chlamydomonas do not address the role of DHC2/D2LIC in mammalian cells, it has been reported that antibodies to DHC2, D2LIC, and an IFT-particle protein strongly stained the ependymal layer lining the lateral ventricles (Mikami et al., 2002). Both DHC2 and D2LIC staining also were observed associated with connecting cilia in the retina and within primary cilia of nonneuronal cultured cells, supporting a specific role for dynein 2 in the generation and maintenance of cilia (Mikami et al., 2002). Perrone et al. (2003) also found that DHC2 and D2LIC colocalized in the apical cytoplasm and axonemes of ciliated epithelia in the lung, brain, and efferent duct. Therefore, it is very likely that cytoplasmic dynein 2 is involved in cilia formation in mammals. It remains controversial whether it also has a role in Golgi maintenance in mammals.
Acknowledgments
We thank John A. Follit for help with the D1bLIC antibody production. We are grateful to Dr. Carolyn Silflow, Dr. William Dentler, and Mr. Trung Bui for graciously providing the TBD9–1 mutant. We thank Dr. Douglas Cole for the generous gift of antibodies to Chlamydomonas IFT172 and IFT139 and Dr. Gianni Piperno for the antibody to Chlamydomonas β-tubulin. We thank Dr. Carolyn Silflow for providing the p3xHA plasmid, and Dr. Saul Purton for the pSP124S plasmid. This work was supported by National Institutes of Health grants GM60992 (G.P.) and GM30626 (G.W.) and by the Robert W. Booth fund at the Greater Worcester Community Foundation (G.W.). We also gratefully acknowledge grant P30 DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases for support of core facilities used in this research.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–05–0377. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–05–0377.
Abbreviations used: BAC, bacterial artificial chromosome; DHC, dynein heavy chain; DIC, differential interference contrast; EM, electron microscopy; EST, expressed sequence tag; HA, influenza hemagglutinin epitope; IFT, intraflagellar transport; LIC, light intermediate chain; P-loop, phosphate-binding domain.
References
- Bielli, A., Thornqvist, P.O., Hendrick, A.G., Finn, R., Fitzgerald, K., and McCaffrey, M.W. (2001). The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1.Biochem. Biophys. Res. Commun. 281, 1141-1153. [DOI] [PubMed] [Google Scholar]
- Brazelton, W.J., Amundsen, C.D., Silflow, C.D., and Lefebvre, P.A. (2001). The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11, 1591-1594. [DOI] [PubMed] [Google Scholar]
- Cole, D.G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4, 435-442. [DOI] [PubMed] [Google Scholar]
- Cole, D.G., Diener, D.R., Himelblau, A.L., Beech, P.L., Fuster, J.C., and Rosenbaum, J.L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet, J., Spike, C.A., Lundquist, E.A., Shaw, J.E., and Herman, R.K. (1998). Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148, 187-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane, J.A., Cole, D.G., Seeley, E.S., Diener, D.R., and Rosenbaum, J.L. (2001). Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11, 1586-1590. [DOI] [PubMed] [Google Scholar]
- Dell, K.R., Turck, C.W., and Vale, R.D. (2000). Mitotic phosphorylation of the dynein light intermediate chain is mediated by cdc2 kinase. Traffic 1, 38-44. [DOI] [PubMed] [Google Scholar]
- Gorman, D.S., and Levine, R.P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54, 1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom, P.M., Vaisberg, E.A., and McIntosh, J.R. (2002). Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol. Biol. Cell 13, 817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y.G., Kwok, B.H., and Kernan, M.J. (2003). Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr. Biol. 13(19), 1679-1686. [DOI] [PubMed] [Google Scholar]
- Haycraft, C.J., Schafer, J.C., Zhang, Q., Taulman, P.D., and Yoder, B.K. (2003). Identification of CHE-13,a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 284, 251-263. [DOI] [PubMed] [Google Scholar]
- Hoops, H.J., and Witman, G.B. (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97, 902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Cole, D.G., Dentler, W., Pazour, G., and Witman, G. (2002). A dynein light intermediate chain is required for retrograde intraflagellar transport (IFT). Mol. Biol. Cell 13, 41a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., Liu, A., Rakeman, A.S., Murcia, N.S., Niswander, L., and Anderson, K.V. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83-87. [DOI] [PubMed] [Google Scholar]
- Kindle, K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87, 1228-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M. (2000). The dynein microtubule motor. Biochim. Biophys. Acta 1496, 60-75. [DOI] [PubMed] [Google Scholar]
- King, S.M., Dillman, J.F., 3rd, Benashski, S.E., Lye, R.J., Patel-King, R.S., and Pfister, K.K. (1996). The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J. Biol. Chem. 271, 32281-32287. [DOI] [PubMed] [Google Scholar]
- King, S.M., Otter, T., and Witman, G.B. (1985). Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc. Natl. Acad. Sci. USA 82, 4717-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., Otter, T., and Witman, G.B. (1986). Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 134, 291-306. [DOI] [PubMed] [Google Scholar]
- Kozminski, K.G., Beech, P.L., and Rosenbaum, J.L. (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131, 1517-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K.G., Johnson, K.A., Forscher, P., and Rosenbaum, J.L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90, 5519-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, P.A., and Rosenbaum, J.L. (1986). Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu. Rev. Cell Biol. 2, 517-546. [DOI] [PubMed] [Google Scholar]
- Lumbreras, V., Steven, D.R., and Purton, S. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14, 441-447. [Google Scholar]
- Lupas, A. (1996). Prediction and analysis of coiled-coil structures. Methods Enzymol. 266, 513-525. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Marshall, W.F., and Rosenbaum, J.L. (2001). Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155, 405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami, A., Tynan, S.H., Hama, T., Luby-Phelps, K., Saito, T., Crandall, J.E., Besharse, J.C., and Vallee, R.B. (2002). Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 115, 4801-4808. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Savereide, P.B., and Lefebvre, P.A. (1994). The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14, 4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclas, J., Allan, V.J., and Vale, R.D. (1996). Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J. Cell Biol. 133, 585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, J.T., Wedaman, K.P., Signor, D., Brown, H., Rose, L., and Scholey, J.M. (1999). Movement of motor and cargo along cilia. Nature 398, 674. [DOI] [PubMed] [Google Scholar]
- Pan, J., and Snell, W.J. (2002). Kinesin-II is required for flagellar sensory transduction during fertilization in Chlamydomonas. Mol. Biol. Cell 13, 1417-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., Vucica, Y., Seeley, E.S., Rosenbaum, J.L., Witman, G.B., and Cole, D.G. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., and Witman, G.B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., and Rosenbaum, J.L. (2002). Intraflagellar transport and ciliadependent diseases. Trends Cell Biol. 12, 551-555. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., Sineschekov, O.A., and Witman, G.B. (1995). Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131, 427-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Wilkerson, C.G., and Witman, G.B. (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141, 979-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., and Witman, G.B. (2000). Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods 22, 285-298. [DOI] [PubMed] [Google Scholar]
- Perrone, C.A., Tritschler, D., Taulman, P., Bower, R., Yoder, B.K., and Porter, M.E. (2003). A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell 14, 2041-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, K.K., Fay, R.B., and Witman, G.B. (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. 2, 525-547. [DOI] [PubMed] [Google Scholar]
- Pfister, K.K., and Witman, G.B. (1984). Subfractionation of Chlamydomonas 18 S dynein into two unique subunits containing ATPase activity. J. Biol. Chem. 259, 12072-12080. [PubMed] [Google Scholar]
- Piperno, G., and Mead, K. (1997). Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 94, 4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Ramanis, Z., Smith, E.F., and Sale, W.S. (1990). Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J. Cell Biol. 110, 379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E., Bower, R., Knott, J.A., Byrd, P., and Dentler, W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit, A., Tynan, S.H., Vallee, R., and Doxsey, S.J. (1999). Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147, 481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Diener, D.R., Geimer, S., Cole, D.G., and Rosenbaum, J.L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Rosenbaum, J.L., and Barr, M.M. (2001). An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11, 457-461. [DOI] [PubMed] [Google Scholar]
- Reilein, A.R., Rogers, S.L., Tuma, M.C., and Gelfand, V.I. (2001). Regulation of molecular motor proteins. Int. Rev. Cytol. 204, 179-238. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, J.L., and Witman, G.B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3, 813-825. [DOI] [PubMed] [Google Scholar]
- Sager, R., and Granick, S. (1953). Nutritional studies with Chlamydomonas reinhardtii. Ann. NY Acad. Sci. 56, 831-838. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1987). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 545.
- Saraste, M., Sibbald, P.R., and Wittinghofer, A. (1990). The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430-434. [DOI] [PubMed] [Google Scholar]
- Sarpal, R., Todi, S.V., Sivan-Loukianova, E., Shirolikar, S., Subramanian, N., Raff, E.C., Erickson, J.W., Ray, K., and Eberl, D.F. (2003). Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr. Biol. 13, 1687-1696. [DOI] [PubMed] [Google Scholar]
- Schafer, J.C., Haycraft, C.J., Thomas, J.H., Yoder, B.K., and Swoboda, P. (2003). XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell 14, 2057-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell, R.A., and Lefebvre, P.A. (1993). Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134, 737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor, D., Wedaman, K.P., Orozco, J.T., Dwyer, N.D., Bargmann, C.I., Rose, L.S., and Scholey, J.M. (1999). Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 147, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow, C.D., LaVoie, M., Tam, L.W., Tousey, S., Sanders, M., Wu, W., Borodovsky, M., and Lefebvre, P.A. (2001). The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153, 63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvanovich, A., Li, M.G., Serr, M., Mische, S., and Hays, T.S. (2003). The third P-loop domain in cytoplasmic dynein heavy chain is essential for dynein motor function and ATP-sensitive microtubule binding. Mol. Biol. Cell 14, 1355-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan, S.H., Purohit, A., Doxsey, S.J., and Vallee, R.B. (2000). Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 275, 32763-32768. [DOI] [PubMed] [Google Scholar]
- Vaisberg, E.A., Grissom, P.M., and McIntosh, J.R. (1996). Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J. Cell Biol. 133, 831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via, A., Ferre, F., Brannetti, B., Valencia, A., and Helmer-Citterich, M. (2000). Three-dimensional view of the surface motif associated with the P-loop structure: cis and trans cases of convergent evolution. J. Mol. Biol. 303, 455-465. [DOI] [PubMed] [Google Scholar]
- Wilkerson, C.G., King, S.M., Koutoulis, A., Pazour, G.J., and Witman, G.B. (1995). The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129, 169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B., Carlson, K., Berliner, J., and Rosenbaum, J.L. (1972). Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54, 507-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B., Johnson, K.A., Pfister, K.K., and Wall, J.S. (1983). Fine structure and molecular weight of the outer arm dyneins of Chlamydomonas. J. Submicrosc. Cytol. 15, 193-197. [Google Scholar]
- Yoder, J.H., and Han, M. (2001). Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell 12, 2921-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Snell, W.J. (1995). Flagellar adenylyl cyclases in Chlamydomonas. Methods Cell Biol. 47, 459-465. [DOI] [PubMed] [Google Scholar]