Abstract
Reversible phosphorylation of myosin regulatory light chain (MRLC) is a key regulatory mechanism controlling myosin activity and thus regulating the actin/myosin cytoskeleton. We show that Drosophila PP1β, a specific isoform of serine/threonine protein phosphatase 1 (PP1), regulates nonmuscle myosin and that this is the essential role of PP1β. Loss of PP1β leads to increased levels of phosphorylated nonmuscle MRLC (Sqh) and actin disorganisation; these phenotypes can be suppressed by reducing the amount of active myosin. Drosophila has two nonmuscle myosin targeting subunits, one of which (MYPT-75D) resembles MYPT3, binds specifically to PP1β, and activates PP1β's Sqh phosphatase activity. Expression of a mutant form of MYPT-75D that is unable to bind PP1 results in elevation of Sqh phosphorylation in vivo and leads to phenotypes that can also be suppressed by reducing the amount of active myosin. The similarity between fly and human PP1β and MYPT genes suggests this role may be conserved.
INTRODUCTION
Nonmuscle myosin II, a molecular motor closely related to vertebrate smooth muscle myosin, powers the actomyosin cytoskeleton. It is required for the coordinated changes in the shape and position of individual cells during morphogenesis as well as for cytokinesis and other cell movements. Nonmuscle myosin II activity is also modulated by metastasis-related and tumor suppressor genes (reviewed Bresnick, 1999; Matsumura et al., 2001; Sellers, 2000; Somlyo and Somlyo, 2000).
The regulation of nonmuscle myosin is thought to be broadly similar to that of vertebrate smooth muscle myosin (Bresnick, 1999; Sellers, 2000). Contraction and relaxation of vertebrate smooth muscle are regulated by the reversible phosphorylation of myosin regulatory light chain (MRLC), principally on Ser-19. The motor activity of smooth muscle myosin is regulated by the balance of activatory phosphorylation, leading to muscle contraction, and inhibitory dephosphorylation, leading to relaxation. The spectrum of stimulating kinases includes myosin light-chain kinase (MLCK), Rho-associated protein kinase (ROK), p21-associated kinase (PAK), integrin-linked kinase (ILK) and leucine zipper–interacting protein kinase (Dlk/ZIP kinase; Somlyo and Somlyo, 2000; MacDonald et al., 2001; Winter et al., 2001; Kiss et al., 2002). The antagonistic protein phosphatase is the catalytic subunit of type 1 serine/threonine protein phosphatase (PP1c) in association with its myosin phosphatase targeting subunit MYPT1 or MYPT2, and a small subunit of unknown function (reviewed by Hartshorne, 1998). These kinases and phosphatases are themselves subject to regulation by reversible phosphorylation, for example ROK not only phosphorylates and activates MRLC, but also phosphorylates MYPT1 and inhibits MRLC dephosphorylation (reviewed Kaibuchi et al., 1999; Somlyo and Somlyo, 2000). The nonmuscle roles of these myosin-regulating kinases are less clear, though at least one (ROK) also regulates non-muscle myosin II in both mammals and Drosophila. Similarly, though PP1 is often assumed to be the major non-muscle MRLC phosphatase, PP2A has also been implicated (Holst et al., 2002). The various phosphorylation events have been investigated biochemically, but little is known about their physiological significance, particularly in nonmuscle cells.
Drosophila nonmuscle myosin II heavy chain zipper (zip) and regulatory light chain spaghetti squash (sqh) are essential for the normal development of a very wide range of cells and tissues (Karess et al., 1991; Young et al., 1993; Edwards and Kiehart, 1996; Hudson and Cooley, 2002). Drosophila Rho-kinase (Drok) phosphorylates both Sqh and DMBS (the single Drosophila homolog of MYPT1/2; Mizuno et al., 2002; Tan et al., 2003). By analogy to the vertebrate smooth muscle system it was proposed that this phosphorylation activates myosin and inhibits myosin phosphatase.
PP1 is involved in the regulation of many cellular functions including glycogen metabolism, muscle contraction, and mitosis (reviewed Bollen, 2001; Cohen, 2002). In Drosophila, the four genes encoding isoforms of PP1c are named by their chromosome location and subtype: PP1β9C, PP1α13C, PP1α87B, and PP1α96A (Dombrádi et al., 1990b, 1993). Of these, PP1α87B contributes 80% of the total PP1 activity, therefore the phenotypes of PP1α87B loss of function mutants (Axton et al., 1990; Dombrádi et al., 1990a; Baksa et al., 1993) may be due to a loss of overall PP1 activity, rather than identifying specific functions unique to the PP1α87B protein. Mice and humans have three PP1 genes: PP1α and PP1γ are related to the fly PP1α genes, although PP1δ (also known as PP1β) corresponds to fly PP1β. Of the mammalian genes, functional analysis by gene knockout in mice has so far only been performed for PP1γ (Varmuza et al., 1999). This knockout eliminated both the widely expressed PP1γ 1 and the testis-specific PP1γ 2. Homozygous mutant female mice were viable and fertile; homozygous mutant males were viable but sterile, with defects in spermatogenesis. Presumably the somatic and female germline functions of PP1γ are redundant with PP1α and/or PP1δ.
The in vitro biochemical activities of the PP1c isoforms are very similar. However, genetic analysis provides a powerful approach to analyze the specific, nonredundant functions of each isoform. We previously showed that the Drosophila PP1β catalytic subunit gene PP1β9C corresponds to flapwing (flw), weak alleles of which are viable but flightless (Raghavan et al., 2000). The semilethality of a strong allele, flw6, demonstrated that PP1β is essential in flies. flw6 larval body wall muscles appeared to form normally, but then detached and degenerated, leading to a semiparalyzed larva that could not feed properly (Raghavan et al., 2000). In addition to muscle defects, the occasional male flw6 survivors were sterile and had blistered wings, indicating a nonredundant role for PP1β9C in nonmuscle cells as well as in muscles.
Here we show that the essential role of PP1β in flies is to regulate nonmuscle actomyosin. The lethality of strong flw (PP1β) mutants was suppressed by reducing the level of phospho-Sqh (MRLC), either using nonphosphorylatable point mutants of sqh or by reducing the gene dosage of key regulators such as Rho1 or RhoGEF2. flw mutants were also suppressed by reducing the gene dosage of nonmuscle myosin heavy chain (zipper). Clones of ovarian follicle cells mutant for flw6 had increased levels of phospho-Sqh, leading to disorganized or absent F-actin and to increased levels of myosin. Therefore, although PP1 isoforms collectively have many known roles, the essential, nonredundant role for PP1β in Drosophila is in the regulation of nonmuscle myosin activity and actin organization.
Drosophila has been reported to have only one MYPT homolog, named DMBS (Mizuno et al., 2002; also known as DMYPT, Tan et al., 2003). We demonstrate that DMBS binds both α and β isoforms of PP1 and is therefore unlikely to mediate a PP1β-specific function. However we have also identified a Drosophila PP1β-specific regulatory subunit, MYPT-75D, which is similar to mammalian MYPT3, a prenylated MYPT1/2 paralog (Skinner and Saltiel, 2001). MYPT-75D binds specifically to PP1β in vitro and the two proteins coimmunoprecipitate from fly extracts. We show that MYPT-75D can stimulate PP1β's Sqh phosphatase activity in vitro and that MYPT-75D, PP1β and Sqh proteins coimmunoprecipitate. Expression of a nonPP1 binding form of MYPT-75D in flies results in elevation of phospho-Sqh and phenotypic consequences that can be suppressed by reducing the level of Sqh phosphorylation. We conclude that PP1β is targeted to Sqh by MYPT-75D, where it performs an essential role in the regulation of Sqh phosphorylation, and hence myosin activity, for which other PP1c isoforms cannot substitute. The conservation of all of these components, including the PP1α and β isoforms, suggests that regulation of nonmuscle myosin in mammals may also involve the activity of PP1β and an isoform-specific myosin targeting subunit.
MATERIALS AND METHODS
Fly Stocks
w67c23, P{lacW}l(1)GO172 (flw7) was kindly provided by Ulrich Schaefer and Tm2J8 by Andrew Dingwall. PP1α87B mutants were provided by Myles Axton. UAS MYPT-75DWT and UAS MYPT-75DF117A were generated by P element–mediated germline transformation of a y w strain. Other Drosophila stocks were obtained from the Bloomington Stock Center.
Screens for Dominant Suppressors of flw6
For ethyl methanesulfonate (EMS) mutagenesis, w1118 males isogenic for all three autosomes were starved for 9–11 h and then fed a 1% sucrose solution containing 25 mM EMS for 20–24 h. Mutagenized males were crossed to y cho sn flw6/FM6; cn bw; e; ci eyR virgin females. Surviving flw6 males from the F1 progeny (y cho sn flw6/Y; cn bw/+*; e/+*; ci eyR/+*, where an asterisk indicates a mutagenized chromosome), were individually backcrossed to females of the parental flw6 strain as were their FM6 brothers (FM6/Y; cn bw/+*; e/+*; ci eyR/+*), once it became apparent that surviving flw6 F1 males were invariably sterile. FM6 brothers of surviving F2flw6males were used to generate a line for further analysis. Segregation of the suppressor activity relative to autosomal markers allowed us to determine the chromosomal linkage of the new mutations.
Our initial screens used y cho sn flw6 (Raghavan et al., 2000). However, when we realized that flw6 interacted with actomyosin genes, we removed singed (sn), which encodes an actin-bundling protein (Cant et al., 1994). Removing sn made no obvious difference to the flw phenotype, but in all later experiments we used w1118 flw6 or w67c23 flw7.
Mapping Analysis
Modifiers were recombination mapped by mating Su(flw)/Balancer males to females carrying a multiply marked second (aldp b pr cn c px sp/CyO) or third (ru h th st cu sr e ca/TM3, Sb) chromosome. In the case of a suppressor on the second chromosome, F1 Su(flw)/al dp b pr cn c px sp females were crossed to al dp b pr Bl cn c px sp/CyO males; equivalent crosses were used for the third chromosome, using ru h th st cu sr Pr e ca/TM6B. F2 recombinant males were individually crossed to y cho sn flw6/FM7c females to score suppression of flw6.
Isolation of MYPT-75D
The two-hybrid screen of 5 × 106 Drosophila 3rd instar larval cDNAs, using PP1β9C as bait, was described in Bennett and Alphey (2002) and Bennett et al. (1999). Database searches and sequencing revealed that BDGP clone LD46604 (Berkeley Drosophila Genome Project/HHMI EST Project, unpublished results) contained the entire MYPT-75D coding region.
MYPT-75D Expression Constructs
The start codon of MYPT-75D was modified to an NdeI site and the complete open reading frame inserted as an NdeI-NotI fragment into pET28a for expression as NH2-terminal His6-tagged protein in Escherichia coli and into pUAS-HM (Parker et al., 2001) for expression as His6Myc2-tagged protein in Drosophila. Substitution of Phe 117 to Ala in MYPT-75DF117A was by PCR-based site-directed mutagenesis.
Phosphatase Assays
Phosphatase assays of fly extracts were as in Bennett et al. (2003), from three independent extracts per genotype. Recombinant His6-Sqh was purified from E. coli as in Skinner and Saltiel (2001) and phosphorylated with MLCK as in Ichikawa et al. (1996). Phosphorylated Sqh was incubated with 0.5 μg of bacterially expressed PP1β9C or PP1α87B, prepared as in Bennett et al. (1999). Samples were analyzed by immunoblotting using anti-Sqh antibody and antiphospho-Sqh antibody. Assays using 32P-labeled Sqh and recombinant purified His6-MYPT-75D proteins were performed as Skinner and Saltiel (2001).
Anti-phospho-Sqh Antibody
The rabbit antiphospho-Sqh antibody was raised by Moravian-Biotechnology (Brno, Czech Republic) against a phosphopeptide (KKRAQRAT[phospho-S]NVFAM) corresponding to a fragment of Sqh phosphorylated at S21.
Immunoprecipitation from Adult Drosophila Extracts
Fly extracts from [UAS-HM-MYPT-75D, Sqh-FLAG, arm-GAL4 and UAS-HA-PP1 (PP1α87B or PP1β9C)] flies, which express FLAG-tagged Sqh, Myc-tagged MYPT-75D and either HA-tagged PP1α87B or HA-tagged PP1β9C, [arm-GAL4, UAS HA-PP1 (PP1α87B or PP1β9C)] flies, which express either HA-tagged PP1α87B or HA-tagged PP1β9C and [UAS-HM-MYPT-75D (wild-type or F117A); arm-GAL4, UAS-HA-PP1 (PP1α87B or PP1β9C)] flies, which express either Myc-tagged MYPT-75D wild-type or F117A and either HA-tagged PP1α87B or HA-tagged PP1β9C, were prepared as in Rudenko et al. (2003) and subjected to immunoprecipitation using anti-FLAG antibodies (M2, Sigma, St. Louis, MO), anti-DMBS antibodies (Mizuno et al., 2002), or anti-Myc antibodies (A14, Santa Cruz Biotechnology, Santa Cruz, CA). After absorption on protein G bound to GammaBind Plus Sepharose (Amersham Biosciences, Amersham, United Kingdom), we analyzed immunoprecipitates and total cell extracts by immunoblotting with antibodies against FLAG, Myc and/or HA (12CA5, Roche Diagnostics, Lewes, East Sussex, United Kngdom).
Immunostaining
Mosaic analysis of w flw6 clones: w1118 flw6 FRT18A/FM7c females were crossed to w1118 Ubi-GFP FRT18A;; MKRS, hs-FLP86E/+ males. Progeny from this cross were allowed to develop to second and third instar larvae, heat shocked at 37.5°C for 1.5h in a water bath, and then allowed to grow up to adulthood. Dissected ovaries from 3–5-d-old w1118 flw6 FRT18A/w1118 Ubi-GFP FRT18A;; MKRS, hs-FLP86 females were fixed for 30 min in 4% paraformaldehyde in PBS. F-actin was stained with 2.5 μg/ml TRITC phalloidin (Sigma) in PBS, 0.3% Tween-20. Rabbit antimyosin heavy chain (Zipper) antibody, provided by Christine Field (Foe et al., 2000), and antiphospho-Sqh were used at 1:600 and 1:400 in PBS, 0.3% Tween-20, respectively. Monoclonal mouse antiactin clone C4 (ICN Biomedicals, Costa Mesa, CA) was used at 1:5000. Secondary antibodies were Cy5-αRb, Cy5-αmouse, and Cy3-αRb (Jackson Labs, 1:1000). Homozygous w1118 flw6 follicle clones were visualized by the absence of Ubi-GFP.
Wing discs from MS1096-GAL4/+; UAS-MYPT-75D (wild-type or F117A)/+ larvae and ovaries from 3–5-d-old hs-FLP/+; UAS-MYPT-75D (wild-type or F117A)/AyGAL4, UAS GFP adult flies that had been heat-shocked as second and third instar larvae, were stained with mouse monoclonal antimyc antibody (9E10, Roche) at 1:5 and antiphospho-Sqh at 1:400.
Images were collected on a Bio-Rad Radiance Plus (Richmond, CA) scanning confocal microscope and processed with Adobe Photoshop 5.0 (San Jose, CA).
RESULTS
Isolation of Suppressors of flw
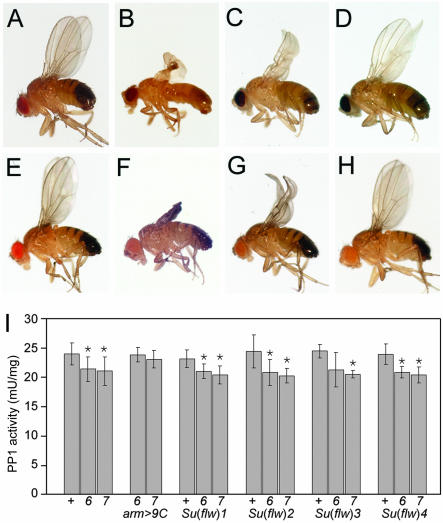
To analyze the essential, nonredundant function(s) of PP1β9C, we conducted an F2 EMS genetic screen for dominant extragenic suppressors of flw6, the design of which allowed us to screen all three autosomes (∼80% of the genome), but not the X chromosome. We recovered four suppressors from screening ∼1000 sets of mutant autosomes. Because of the way these suppressors were isolated, they must be due to independent mutagenic events. Each of these Su(flw) mutations dominantly suppressed the lethality, sterility, and wing defects of flw6, with variable penetrance (Table 1A and Figure 1).
Table 1.
Suppression of flw6 by single-gene mutations.
| A | flw6 male survival [% (N)]
|
flw7 male survival [% (N)]
|
||
|---|---|---|---|---|
| Suppressor | flw6; Su(flw)/+ | flw6; Bal/+ | flw7; Su(flw)/+ | flw7; Bal/+ |
| Su(flw)1 | 56 (64) | 0 (40) | 84 (101) | 0 (42) |
| Su(flw)2 | 91 (61) | 0 (38) | 83 (54) | 3 (37) |
| Su(flw)3 | 90 (60) | 0 (39) | 49 (67) | 6 (52) |
| Su(flw)4 | 65 (71) | 0 (36) | 67 (60) | 0 (28) |
| B |
flw6 male survival [% (N)]
|
|
|---|---|---|
| Mutant | flw6; mut/+ | flw6; Bal/+ |
| arm > PP1β9C | 81 (107) | 0 (60) |
| zip1 | 71 (337) | 0 (167) |
| zip11×62 | 66 (328) | 0 (216) |
| zip02957 | 40 (81) | 0 (41) |
| Rho1E3,10 | 51 (262) | 0 (171) |
| RhoGEF204291 | 26 (113) | 0 (88) |
| Tm102299 | 85 (224) | 1 (107) |
| Tm2J8 | 0 (80) | 4 (72) |
| C |
flw1 male survival [% (N)]
|
flw6 male survival [% (N)]
|
||
|---|---|---|---|---|
| Mutant/construct | flw1; mut/+ | flw1; Bal/+ | flw6; mut/+ | flw6; Bal/+ |
| DMBSE1 | 90 (194) | 96 (139) | nd | nd |
| Hsp70-DMBSL | 99 (213) | — | 59 (177) | * |
| Hsp70 > MYPT-75D | 68 (67) | 127 (59) | 0 (22) | 0 (25) |
| arm > MYPT-75D | 57 (105) | 94 (101) | 0 (33) | 1 (68) |
| D |
flw/Y; P[sqhmut] male survival [% (N)]
|
||||
|---|---|---|---|---|---|
| flw mutant | sqhA20,A21 | sqhA21 | sqhA20 | sqhE21 | sqhE20,E21 |
| flw1 | nd | nd | 106 (185) | 90* (80) | 1 (96) |
| flw6 | 77 (274) | 92 (43) | 5 (112) | 0 (133) | 0 (63) |
| flw7 | 62 (52) | 52 (59) | 7 (65) | 0 (78) | 0 (56) |
(A) EMS-induced suppressors efficiently suppress the lethality of both flw6 and flw7.flw6/FM7c females were crossed to males heterozygous for the candidate suppressor and a balancer (Su(flw)/Bal); their adult progeny were scored. The percentage of expected progeny is the number of surviving male progeny divided by the number of the female progeny of the equivalent genotype ×100. N indicates total number of flies of these two classes.
(B) flw6/FM7c females were crossed to males heterozygous for the candidate suppressor and a balancer or dominant marker (mut/Bal); their adult progeny were scored. Expression of a wild type Flw/PP1β9C cDNA using the GAL4-UAS system, with the armadillo promoter (arm > PP1β9C) efficiently suppresses the lethality of flw6. Various zipper alleles with different molecular lesions also suppress, as do mutant alleles of Rho1, RhoGEF2, and Tm1, but not Tm2.
(C) flw1/flw1 or flw6/FM7 females were crossed to males carrying DMBS or MYPT-75D expression constructs. Overexpression of the DMBSL cDNA suppressed flw6, whereas overexpression of MYPT-75D did not. Overexpression of MYPT-75D weakly enhanced flw1. * The DMBS constructs are viable, no balancer was required.
(D) Genetic interaction between Sqh phosphorylation site mutants and flw. flw mutant females were crossed to males homozygous for various point mutants affecting the phosphorylation sites of Sqh (Jordan and Karess, 1997; Winter et al., 2001). These are autosomal insertions; all flies were wild type at the endogenous sqh locus. * Though viable, most flw1/Y; P[sqhE21]/+ males had crumpled wings, resembling the wing phenotype of strong flw mutants. Alanine (A) substitutions cannot be phosphorylated and thus reduce the requirement for flw function while glutamic acid (E) substitutions mimic phosphorylation and so increase the requirement for flw. sqhA20,A21 strongly suppresses both flw6 and flw7, whereas sqhE20,E21 is synthetic lethal with the weak, viable allele flw1.
nd, not done.
Figure 1.
Dominant suppression of flw by EMS-induced mutants. (A–H) Suppressors of flw [Su(flw)] can dominantly suppress flw wing phenotypes. (A) Wild-type (Oregon R) male. Rare flw 6/Y and flw 7/Y male survivors have severely crumpled wings (B and F). This is completely suppressed by expression of cDNA encoding wild-type PP1β9C (e.g., flw 7/Y; arm-GAL4, UAS-PP1β9C/+, E). The wing phenotype is partially rescued by the zipper alleles Su(flw)1 (C: flw 6/Y; Su(flw)1/+, G: flw 7/Y; Su(flw)1/+), Su(flw)2 and Su(flw)3 (unpublished data, indistinguishable from C and G) and completely rescued by the Tm1 allele Su(flw)4 (D: flw 6/Y; Su(flw)4/+, H: flw 7/Y; Su(flw)4/+). (I) Suppressor mutations do not restore PP1 activity of flw mutants. Extracts from wild-type (+) or flw 6 or flw 7 adult males with no suppressor or expressing a PP1β9C cDNA (arm >9C) or heterozygous for one of the suppressor mutations were assayed for PP1 activity (plotted as the mean ± SD; n = 3). Means that show significant difference from wild-type, as assessed by Student's t test at 95% confidence level, are indicated with an asterisk. Columns 1–3: Though PP1β represents only ∼10% of the total PP1 activity, extracts from flw mutants have a measurable reduction in PP1 activity (see also Raghavan et al., 2000). Columns 4–5: expression of a PP1β9C cDNA (arm-GAL4, UAS-PP1β9C) restored PP1 activity to wild-type levels (and suppressed the flw phenotype, see E and Table 2). Columns 6–17: flies heterozygous for any of the four EMS-induced suppressor mutations showed no increase in PP1 activity, even though these mutations suppressed the flw phenotype.
Suppressors Are Not Specific for flw6 and Do Not Increase Total PP1 Activity
flw6 is due to a point mutation in the PP1β coding region (Raghavan et al., 2000). Suppressors could perhaps be chaperones or binding proteins that stabilize or compensate for the changed structure of the protein. To test these possibilities we isolated a new flw allele, with a different molecular basis. flw7 has a P[lacW] element inserted in the 5′ untranslated region of PP1β9C, leading to a reduced level of expression of PP1β (Gross, 2001). flw7 mutant males show all the characteristic phenotypes of flw6, though slightly more (∼2%) of the mutant males survive. The semilethality and visible phenotypes of flw6 and flw7 can be rescued by ectopic expression of a PP1β9C cDNA (Table 1B and Figure 1). All four EMS-induced suppressors could also suppress flw7, indicating that they are not specifically compensating for the single amino acid change in flw6 (Table 1). The suppressors could potentially suppress flw6 and flw7 through a restoration of overall phosphatase activity, for example, by increased transcription of flw or by some posttranscriptional mechanism. We therefore measured the total PP1 activity of select mutant combinations. None of the suppressors led to a measurable increase in PP1 activity (Figure 1I).
PP1β9C Has a Single Essential Function
Having eliminated these mechanisms for global suppression of flw mutants, we concluded that the suppressors are components or targets of a pathway that requires flw. Because single gene mutations could suppress the lethality of flw mutants, regulation of this pathway must be the essential, nonredundant role of flw. The suppressors might represent an antagonistic kinase, a substrate, or other component of the pathway. These components might themselves be common to a small number of pathways, but still define a single nonredundant role for flw, e.g., a single key substrate.
Su(flw) Are Loss of Function Alleles of Nonmuscle Myosin Heavy Chain and Tropomyosin 1
We mapped the four Su(flw) using meiotic recombination. Su(flw)1, 2, and 3 mapped to the tip of 2R, distal to speck at 2-107.0 (60B13–60F). Su(flw)4 mapped to the third chromosome, between curled and stripe (3-55.1 ± 2.2 or 86D3-4; 90D6-E2). Complementation tests revealed that Su(flw)1, 2, and 3 are recessive lethal alleles of a single essential gene. All three also failed to complement existing alleles of zipper (zip), which encodes nonmuscle myosin heavy chain (Young et al., 1993), identifying the second chromosome Su(flw) as zipper. To test whether suppression is due to some allele-specific peculiarities, we tested a further two zip mutant alleles and a zip deficiency for their ability to suppress flw6 and flw7. All three suppressed the lethality and sterility of flw/Y (Table 1B), indicating that suppression is not allele specific and is most likely due to a simple reduction in the amount of myosin heavy chain.
Having shown that several zip alleles could suppress at least two alleles of flw, we investigated whether zip mutants could suppress alleles of PP1α87B.zip did not suppress PP1α87B1/PP1α87B 87Bg-3, a semilethal allelic combination, indicating that zip specifically suppresses PP1β (unpublished data). This does not preclude a role for PP1α87B in myosin regulation, but implies that this is not the only essential role of PP1α87B, consistent with the mutant phenotype of PP1α87B (Axton et al., 1990).
Previous genetic analysis identified several zip interacting loci (Halsell and Kiehart, 1998), one of which, Tropomyosin 1 (Tm1), maps to the same region as Su(flw)4. Su(flw)4 failed to complement Tm102299, identifying this suppressor as an allele of Tm1. Tm102299 suppressed both flw6 and flw7, again indicating that this genetic interaction is not allele-specific (Table 1B and unpublished data). Drosophila has two tropomyosin genes, the other being the muscle-specific Tm2 (Kreuz et al., 1996). We therefore tested whether Tm2J8 could also suppress flw6, but found no such suppression (Table 1B), indicating that the essential role of PP1β specifically relates to regulation of nonmuscle myosin. We also tested the known upstream regulators of MRLC activity, Rho1 and RhoGEF2, for interaction with flw. Mutations in both genes strongly suppressed flw6 (Table 1B).
Phosphorylation Mutants of Sqh
In view of the known role of PP1 as a MRLC phosphatase in mammals, we speculated that mutants of the phosphorylation site(s) in Sqh might interact genetically with flw. We obtained a set of such phosphorylation site mutants in which the critical residues Thr-20 and Ser-21 (equivalent to Thr-18 and Ser-19 in vertebrate MRLC) had been changed to Ala (sqhA20A21), to prevent phosphorylation, or to Glu (sqhE20E21), to mimic constitutive phosphorylation (Jordan and Karess, 1997; Winter et al., 2001). Phosphorylation of these sites leads to myosin activation; dephosphorylation reduces myosin motor activity. These sqh mutant transgenes are under the control of the sqh promoter and are expressed at levels similar to the native protein. All these experiments were carried out in a background containing the wild-type sqh gene. The nonphosphorylatable sqhA20A21 strongly suppressed both flw6 and flw7 mutant phenotypes (Table 1). In contrast, increasing the level of “phospho”-Sqh with the phosphorylation mimic sqhE20E21 enhanced the weak flw mutant, flw1 to lethality. By using single mutants in either Thr-20 or Ser-21, we found that Ser-21 had the greater effect (Table 1). The phosphorylation state of Sqh is therefore closely related to the viability of flw mutants and hence to the essential role of PP1β.
Phospho-Sqh Levels Are Elevated in flw6 Mutant Clones
To examine further the role of PP1β in actomyosin regulation, we investigated the effect of loss of PP1β9C on myosin distribution and phosphorylation, and on actin organization. Clones of ovarian follicle cells homozygous for flw6 were stained for actin, phosphorylated Sqh and myosin heavy chain (Zip, Figure 2). The level of phospho-Sqh was dramatically increased in the majority of these clones (Figure 2G), as was the level of Zip (Figure 2K). A nonphosphospecific anti-Sqh antibody showed no increase in staining in the clones (unpublished data). In most clones with elevated phospho-Sqh or Zip, the amount of filamentous actin (F-actin) was decreased, and the apical F-actin network was clearly disorganized (e.g., Figure 2B). Disruption of F-actin, visualized with TRITC-phalloidin, correlated with accumulation of cytoplasmic actin, visualized by immunofluorescence with antiactin antibody (Figure 2, A–D). Because this antibody does not appear to stain F-actin efficiently under our fixation conditions (note the absence of overlap between the actin antibody signal and the phalloidin staining in Figure 2D), we propose that there is an increase in the cytoplasmic pool of G-actin in the mutant cells. These data clearly show that the correct structure of the actin cytoskeleton, and Sqh dephosphorylation, are both dependent on PP1β9C.
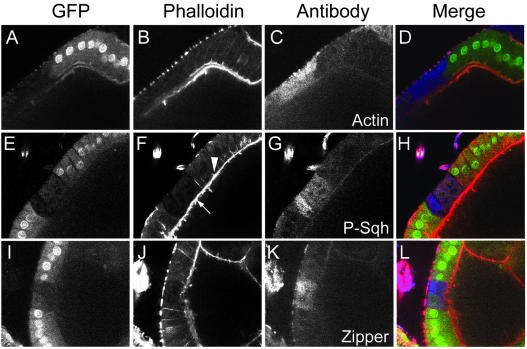
Figure 2.
Loss of PP1β leads to hyper-phosphorylation of Sqh and disruption of the actin cytoskeleton. (A–L) Egg chambers including clones of follicle cells homozygous for flw 6 stained with TRITC-phalloidin (column 2, red in merge), and antibodies against either actin, phospho-Sqh or Zip (column 3, blue in merge). flw 6 homozygous cells are marked by absence of GFP (column 1, green in merge). (A–D) Actin organization is disrupted in cells homozygous for flw 6. Lateral F-actin was undetectable, whereas cortical F-actin is absent or disrupted (B; also F and J). Staining with an antiactin antibody, which failed to detect filamentous actin, showed that G-actin levels are elevated in the flw 6 clone relative to adjacent normal cells (C). (E–H) Cells homozygous for flw 6 have elevated phospho-Sqh and disrupted F-actin. Phospho-Sqh in normal cells is barely detectable. Within the mutant clones, both apical and lateral F-actin is missing or disrupted (arrowhead indicates normal actin in cells adjacent to the clone). The cortical actin in the oocyte is unaffected (arrow). (I–L) Cells homozygous for flw 6 have elevated levels of Zip.
The phenotypes described above were not fully penetrant. On careful examination we noticed that some clones had increased levels of phospho-Sqh and myosin but normallooking F-actin. This suggests that the F-actin disruption is a secondary consequence of increased phospho-Sqh. The increase in phospho-Sqh and Zip in these clones appeared to be concentrated toward the basal region of the cell (see Supplementary Information).
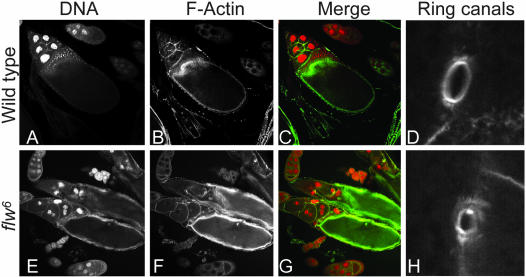
In addition to somatic clones, we analyzed germline clones mutant for flw6, and also generated mutant egg chambers by somatic rescue of the flw6 phenotype with arm-GAL4, UAS-PP1β9C, which expresses PP1β9C in somatic cells but not in the female germline (Rorth, 1998). Like follicle cells, mutant egg chambers had elevated levels of phospho-Sqh and Zip and defects in their actin cytoskeleton organization. In particular, they had aggregates of myosin, resembling those seen in Sqh mutants (Jordan and Karess, 1997). Later, mutant egg chambers showed a clear “dumping” defect, a failure of the normally rapid transfer of the contents of the nurse cells into the oocyte at stages 10B and 11 (Figure 3, E–G). This process depends on actomyosin (Jordan and Karess, 1997; Hudson and Cooley, 2002), and so may be a consequence of the defective actin and myosin organization, but we also observed that the ring canals, which join the nurse cells to each other and to the oocyte, were defective in flw6 mutant egg chambers. The ring canals appear normal initially, but then fail to grow properly, leading to ring canals that are much smaller than wild-type by stage 10 (Figure 3H). Small ring canals may not permit fast cytoplasmic transport from nurse cell to cytoplasm. These germline phenotypes—dumping failure, disorganized actin and myosin, and small ring canals—are all dependent on the germline genotype: they are present in mutant germline clones with wild-type follicle cells, but not in egg chambers which have wild-type germ line but predominantly mutant follicle cells.
Figure 3.
Loss of PP1β in egg chambers leads to abnormal ring canals and dumping defects. Cytoplasmic dumping and ring canal growth are defective in flw 6 homozygous egg chambers. Egg chambers were stained with propidium iodide to visualize DNA (A and E, red in merge) and with phalloidin to visualize F-actin (B and F, green in merge) and ring canals (D and H). Relative to wild-type (A–D) flw 6 homozygous egg chambers (E–H) have abnormal actin and nuclear structures characteristic of dumping defects. Two ring canals are shown from the same individual: (D) a normal ring canal from a stage 10 egg chamber heterozygous for flw 6 and (H) a small, abnormal ring canal from a stage 10 egg chamber homozygous for flw 6.
MYPT-75D Is a PP1β-specific Binding Protein
We have shown that loss of PP1β9C in clones leads to hyperphosphorylation of Sqh in vivo, and the lethality of strong mutants of flw can be suppressed by a nonphosphorylatable mutant version of Sqh. This demonstrates that flw regulates the phosphorylation of Sqh and that this is the essential role of PP1β. We also analyzed phospho-Sqh level by immunoblotting. We detected phospho-Sqh in extracts from rare hemizygous flw6 survivors, whereas we found no detectable phospho-Sqh in extracts from wild-type flies or flies hemizygous for flw1 (Figure 4A), presumably reflecting a low wild-type abundance relative to the sensitivity of our antibody, consistent with our immunofluorescence data. Phosphatase assays revealed that both PP1β9C and PP1α87B efficiently dephosphorylate phospho-Sqh in vitro as immunoreactivity to our phospho-Sqh–specific antibody decreased over time on incubation with purified PP1β or PP1α (Figure 4A). However, as purified PP1 catalytic subunits have broad substrate specificity in vitro, this was not surprising. The greater substrate specificity of PP1c in vivo is due to association of the catalytic subunit with regulatory subunits (Bollen, 2001; Cohen, 2002). Although PP1c need interact only transiently with its substrates to dephosphorylate them, it is nonetheless sometimes found in stable complexes with specific substrates, mediated by the relevant regulatory subunit.
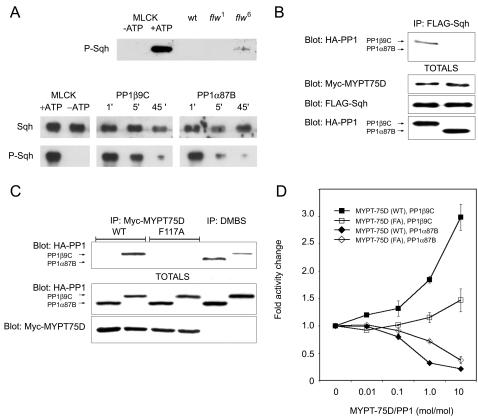
Figure 4.
MYPT-75D specifically binds PP1β and stimulates its activity toward Sqh in vitro. (A) Top, levels of phosphorylated-Sqh are elevated in flw 6 mutant larvae. Equal amounts of total protein extracted from wild-type, flw 1, and flw 6 males was subjected to immunoblotting with an antiphospho-Sqh specific antibody. For comparison, samples were run alongside unphosphorylated and phosphorylated recombinant Sqh. Phospho-Sqh was detected in flw 6 but not wild-type or flw 1 extracts. (A) Bottom, Sqh is dephosphorylated in vitro by both PP1β9C and PP1α87B. Recombinant Sqh was phosphorylated by myosin light chain kinase (MLCK) and then incubated with either PP1β9C or PP1α87B for 1, 5, or 45 min. The reaction products were separated by SDS-PAGE, blotted, and probed with either an Sqh-specific antibody (top panels) or a phospho-Sqh-specific antibody (bottom panels). Left-most panels, MLCK efficiently phosphorylated Sqh. A control reaction without (-) ATP demonstrated the specificity of the phospho-Sqh antibody for the phosphorylated form. Both PP1β9C (center panels) and PP1α87B (right panels) efficiently dephosphorylated phospho-Sqh, as determined by loss of immunoreactivity against the phospho-Sqh antibody (bottom panels) relative to reactivity against the Sqh antibody (top panels). (B) Sqh coimmunoprecipitates with PP1β9C. FLAG-tagged Sqh was immunoprecipitated from flies expressing FLAG-Sqh, HA-PP1β9C and myc-MYPT-75D. HA-PP1β9C coprecipitates with FLAG-Sqh. (C) Isoform specificity of MYPT-75D and DMBS. MYPT-75D was immunoprecipitated from flies expressing myc-tagged MYPT-75D WT or myc-tagged MYPT-75D F117A and either HA-tagged PP1β9C or HA-tagged PP1α87B. Top panel, first four lanes: PP1β9C and not PP1α87B coprecipitated with MYPT-75D WT but not MYPT-75D F117A. Middle and bottom panels, blots of total extracts showing that there were equivalent levels of MYPT-75D and HA-tagged PP1 in the various flies. The third and fourth lanes show an equivalent experiment, but precipitating DMBS with an anti-DMBS antibody, rather than MYPT-75D. Both PP1β9C and PP1α87B coprecipitate with DMBS. (D) MYPT-75D stimulates PP1β9C's Sqh-directed phosphatase activity in vitro. Wild-type and mutant MYPT-75D were preincubated for 10 min at 30°C at various concentrations with purified recombinant PP1β9C or PP1α87B, and then the reaction was started by the addition of 32P-labeled Sqh. Sqh phosphatase activity without the addition of MYPT-75D was taken as 100%. PP1 activity is plotted as the mean ± SD (n = 3).
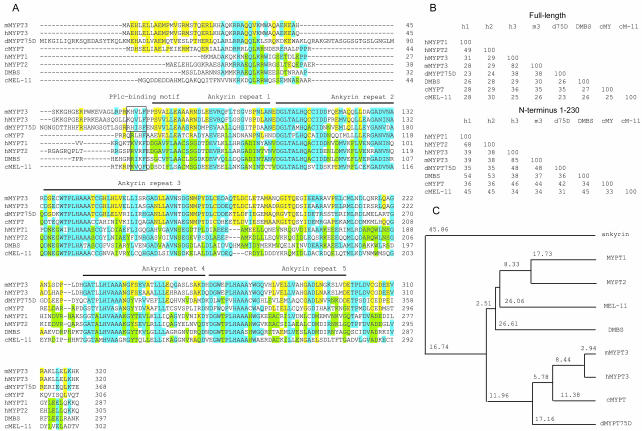
Drosophila has two genes that might encode regulatory subunits capable of targeting PP1c to Sqh. These are DMBS, homologous to mammalian MYPT1/2 (Mizuno et al., 2002; Tan et al., 2003) and MYPT-75D, homologous to mammalian MYPT3 (Figure 5). We isolated MYPT-75D in a two-hybrid screen for proteins capable of binding PP1β9C (Bennett et al., 1999; Bennett and Alphey, 2002). Of 36 genes identified in this screen, including homologues of known PP1 regulatory subunits such as Inhibitor-2 and NIPP1 (Bennett et al., 1999; Parker et al., 2002), only MYPT-75D discriminated between the PP1 isoforms. Unfortunately, sequence comparison of the different MYPT proteins has not revealed any sequences that might be responsible for the observed binding specificity of MYPT-75D. MYPT-75D contains a canonical PP1c-binding motif (R/K,[x], V/I, x, F, here RHISF, residues 113–117) followed by five ankyrin repeats (residues 119–148, 151–180, 182–216, 280–310, 312–346, Figure 5) and a potential CaaX prenylation motif (CCVLM, residues 737–741). Unlike DMBS, MYPT-75D does not contain a regulatory Rho-kinase phosphorylation site.
Figure 5.
MYPT-75D is a second myosin targeting subunit molecule in Drosophila that is most closely related to mammalian MYPT3. (A) Multiple sequence alignment of the predicted sequence of the NH2-terminal ankyrin-repeat region of MYPT proteins: hMYPT1, human MYPT1 (NP_002471); hMYPT2, human MYPT2 (NP_002472); hMYPT3, human MYPT3 (AAH07854); mMYPT3, mouse MYPT3 (AAG40949); dMYPT-75D, Drosophila MYPT-75D (AY075426); DMBS, Drosophila DMBS (AAF49547); cMYPT, C. elegans MYPT (yk603e4); cMEL-11, C. elegans MEL-11 (AAB47273). MYPT1/2 specific residues are in green; MYPT3 specific sequences are in yellow and common residues are in blue. The putative PP1c-binding motifs are boxed and the position of the ankyrin repeats is indicated with lines. The PP1c-binding motif in cMYPT does not conform to the consensus sequence as it has L in place of V or I. Although the first ankyrin repeat shows only weak homology to the consensus sequence, it is generally accepted to be an ankyrin repeat motif in MYPT (Skinner and Saltiel, 2001). (B) Pair-wise percentage identities between MYPT proteins over the whole length or over the NH2 terminal ankyrin-repeat region. (C) Phylogenetic tree analysis of MYPT proteins (over their whole length) with Drosophila ankyrin (T13940) as outgroup, where branch length is proportional to sequence difference. Alignment was performed using CLUSTALW at http://www.ebi.ac.uk/clustalw/. The phylogenetic tree was reconstructed using the Neighbor-Joining algorithm, as implemented in the PHYLIP sequence analysis package (Phylogeny Inference Package, 3.5c Dept of Genetics, University of Washington, Seattle, WA).
In extracts from flies moderately overexpressing myc-MYPT-75D along with HA epitope-tagged PP1β9C or PP1α87B and FLAG-tagged Sqh (using arm-GAL4), PP1β9C coimmunoprecipitated with Sqh, but PP1α87B did not (Figure 4B), suggesting that PP1c is in a stable complex with MYPT-75D and Sqh in vivo. We further examined the PP1 binding specificities of MYPT-75D and DMBS by coimmunoprecipitation from flies. We found that DMBS bound both PP1β9C and PP1α87B, whereas MYPT-75D bound only PP1β9C (Figure 4C). Therefore Drosophila contains at least three distinct PP1-MYPT complexes: PP1α +DMBS, PP1β +DMBS and PP1β +MYPT-75D. To test directly the role of the “RVXF” site in MYPT-75D for binding to PP1c, we disrupted the motif by changing the critical Phe residue to Ala. This mutant, MYPT-75DF117A, failed to bind to PP1β9C (Figure 4C).
flw Interacts Differentially with Two Myosin-targeting Subunits
We examined whether DMBS or MYPT-75D interact genetically with flw mutants (Table 1). We found that reduction in the gene dosage of DMBS (DMBSE1/+) did not enhance flw1, but that overexpression of a DMBS cDNA did suppress flw6. Unfortunately, no mutants for MYPT-75D are available. High-level expression of the MYPT-75D cDNA (hsp70-GAL4, UAS-MYPT-75D with heat shock) is lethal to flw1 mutants but also to wild-type (unpublished data). Moderate overexpression of a MYPT-75D cDNA (arm-GAL4, UAS-MYPT-75D or hsp70-GAL4, UAS-MYPT-75D without heat shock) did not suppress flw6 and indeed somewhat enhanced flw1.
MYPT-75D:PP1β Regulates Phosphorylation of Sqh In Vitro and In Vivo
We measured the effect of MYPT-75D on PP1c activity in vitro using 32P-Sqh as substrate. Recombinant MYPT-75D stimulated the Sqh phosphatase activity of PP1β9C, but inhibited that of PP1α87B (Figure 4D). Disruption of the PP1c-binding motif abolished the ability of MYPT-75D to activate PP1β9C, showing that binding to MYPT-75D is necessary for the stimulation of PP1β9C's Sqh phosphatase activity.
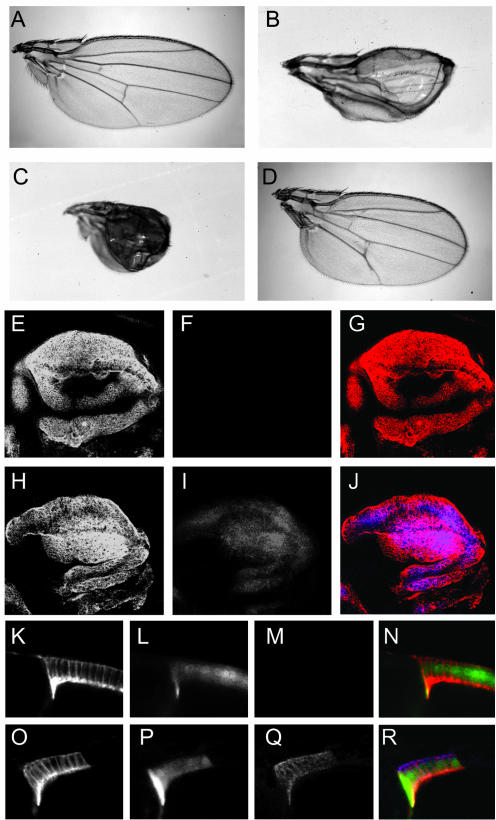
We then used this nonbinding mutant to test the in vivo role of MYPT-75D-PP1β. Ectopic expression of wild-type MYPT-75D (MYPT-75DWT) in the wing had no phenotypic effect, but expression of MYPT-75DF117A resulted in crumpled, blistered wings (Figure 6, A and B). The wing phenotypes induced by expression of MYPT-75DF117A were suppressed by zip1, Tm102299, and sqhA20, A21 and enhanced by sqhE20, E21 (Figure 6, C and D), indicating that the effect of loss of binding of PP1β to MYPT-75D corresponds to changes in nonmuscle myosin activity.
Figure 6.
An MYPT-75D:PP1β9C regulates Sqh phosphorylation in vivo. (A–D). Effect of expression of wild-type MYPT- 75D and MYPT-75D F117A in wild-type and mutant backgrounds. (A) Wings from flies ectopically expressing wild-type MYPT-75D using the wing specific driver MS1096-GAL4 resemble wild-type. (B) Ectopic expression of MYPT-75D F117A using MS1096-GAL4 leads to blistered and crumpled wings. The effect of MYPT-75D F117A is enhanced by sqh E20,E21 (C) and suppressed by sqh A20,A21 (D). (E–J) Levels of antiphospho-Sqh staining in wing imaginal discs expressing wild-type MYPT-75D or MYPT-75D F117A under the control of MS1096-GAL4. Two wing discs are shown, one (E–G) expressing wild-type MYPT-75D, and the other (H–J) MYPT-75D F117A, stained with antibodies against myc (column 1, red in merge) to detect ectopic myc-tagged MYPT-75D, and phospho-Sqh (column 2, blue in merge). (K–R) Levels of antiphospho-Sqh staining in clones of follicle cells expressing wild-type MYPT-75D (K–N) or MYPT-75D F117A (O–R). Egg chambers were stained with antibodies against myc (column 1, red in merge) and phospho-Sqh (column 3, blue in merge). Clones of cells expressing myc-MYPT-75D are marked by GFP (column 2, green in merge).
When we examined levels of phospho-Sqh in wing discs from larvae of these genotypes, we saw elevated phospho-Sqh staining in cells expressing MYPT-75DF117A but not those expressing MYPT-75DWT (compare Figure 6, F and I). The myc-tagged MYPT-75D localized to the cell periphery. We also examined the effect of MYPT-75D overexpression on phospho-Sqh in follicle cell clones in the ovary. Just as we observed in wing discs, phospho-Sqh was elevated in clones expressing MYPT-75DF117A but not MYPT-75DWT (Figure 6, M and Q). Therefore, MYPT-75D-bound PP1β can stimulate dephosphorylation of nonmuscle MRLC both in vitro and in vivo.
DISCUSSION
We have previously shown that PP1β mutants are semilethal, therefore PP1β is an essential gene in Drosophila (Raghavan et al., 2000). Here we have shown that two semilethal mutant alleles of PP1β can be dominantly suppressed by loss-of-function extragenic mutations. The existence of single-gene extragenic suppressors indicates that PP1β has a single essential role, the identity of the suppressors indicates that this role is in the regulation of actin and/or myosin. Though the main defect observed in flw mutants is muscle detachment and degeneration, it is clear from our data that it is nonmuscle myosin, rather than muscle myosin, that is affected. Zipper and Sqh are components of nonmuscle myosin; the muscle version of Sqh, Mlc2, does not interact with flw (Raghavan et al., 2000). Similarly, Tm1, but not the muscle-specific Tm2, suppresses flw. Disruption of nonmuscle myosin in flw mutants may lead to disruption of the actin cytoskeleton and affect cell adhesion in many cell types, but seems to be most readily apparent in contractile muscle, particularly the highly specialized indirect flight muscles (Raghavan et al., 2000). Though not directly involved in generation of contractile force, nonmuscle myosin seems to be necessary for the correct development of striated myofibrils (Bloor and Kiehart, 2001).
The dominant suppression of the lethality of flw6 and flw7 mutants by SqhA20A21, coupled with the enhancement of flw1 by SqhE20E21, implies that the essential role of PP1β 9C is related to the regulation of the phosphorylation state of Sqh. To address whether this interaction is direct or indirect we have shown that PP1β can directly dephosphorylate phospho-Sqh in vitro and that the two proteins coimmunoprecipitate from Drosophila extracts. Furthermore we have identified a new PP1β-specific MYPT, and shown that binding of MYPT-75D to PP1β stimulates dephosphorylation of nonmuscle MRLC both in vitro and in vivo. We therefore conclude that the major or only essential role of PP1β in Drosophila is to dephosphorylate Sqh and that this role is mediated, at least in part, by association with a β-specific MYPT protein. Although flw6 behaves as a null allele by genetic tests (Raghavan et al., 2000), we cannot rule out the possibility that it has some residual activity and that this is sufficient to perform one or more additional essential functions of PP1β, which for some reason require only a very low level of PP1β activity. PP1β, MRLC and MYPT proteins are highly conserved between flies and mammals, so it seems likely that dephosphorylation of MRLC is also an essential role of PP1β in humans.
Though PP1β can dephosphorylate Sqh directly and manipulating the phosphorylation state of Sqh is sufficient to suppress strong mutants of flw, we cannot exclude the possibility that flw has other substrates in the same pathway. For example, nonmuscle myosin heavy chain, which in mammals can be phosphorylated by PKC and CKII (Murakami et al., 1998; Bresnick, 1999), could also be a substrate for PP1β.
What is the molecular basis of the suppression of flw? We believe that the key defect, both in flw mutants and in flies expressing MYPT-75DF117A, is the hyperphosphorylation of Sqh, particularly on Ser-21; this is directly suppressed by the nonphosphorylatable Sqh mutants. In these experiments a pool of normal Sqh remains, so we are essentially manipulating the ratio of phosphorylated and nonphosphorylated Sqh. Phosphorylation of Sqh leads to activation of the myosin motor; reduction in the amount of myosin heavy chain in zipper+/- presumably reduces the amount of active motor. Sqh is known to be a substrate for Rho-kinase, itself activated by a pathway that includes two more suppressors: Rho1 and RhoGEF2. Rho-kinase itself is located on the X chromosome and was therefore not accessible to our genetic screen.
Tm1, a strong suppressor of flw6, is not a member of Rho-kinase pathway but a cytoskeletal actin-binding protein (Tetzlaff et al., 1996). Several functions have been ascribed to nonmuscle tropomyosin in mammals: modulation of myosin function (Strand et al., 2001), actin polymerization (Wen et al., 2000), regulating microfilament branching (Blanchoin et al., 2001), and suppression of neoplastic transformation (Mahadev et al., 2002). Reduction in the amount of Tm1 appears to mitigate the consequences of hyper-phosphorylated Sqh; the obvious mechanism is by reducing the binding of active myosin to actin, though Tm1 could have its effect through regulation of actin structure and polymerization.
The phenotypes we have described for flw somewhat resemble those of DMBS, particularly in the female germ line (Tan et al., 2003) and in that they both lead to the accumulation of phospho-Sqh (Mizuno et al., 2002), though DMBS mutants do not show the accumulation of myosin aggregates (Tan et al., 2003). The differences in lethal phase (embryonic for DMBS, predominantly larval for flw) might be accounted for by maternal contribution and differences in protein stability; we were unable to investigate this further as both DMBS and flw are required for oogenesis. Furthermore, the flw suppressors sqhA20A21, Rho1 and zipper have been shown or deduced to modify at least some of the DMBS phenotypes. This might indicate that the critical role of flw is mediated by DMBS. However, we have shown that DMBS is not specific for PP1β. PP1α87B is much more abundant than PP1β, so flw mutants should have little effect on the DMBS: PP1c complex. It is possible that DMBS:PP1β has a unique role not shared by DMBS:PP1α; it is also possible that DMBS, which is phosphorylated by Rho-kinase, is itself directly or indirectly activated by a PP1β-specific phosphatase complex. However, because we have identified an additional, PP1β-specific MYPT, it seems much more likely that this is the key targeting subunit that mediates the essential role of PP1β in vivo and that the suppression of flw by Rho and RhoGEF is through a decrease in phosphorylation of nonmuscle MRLC by Rho-kinase.
Why do flies have two MYPTs apparently doing the same job, one PP1β-specific and the other not? Clearly DMBS is not completely redundant with MYPT-75D, as DMBS mutants are lethal; mutants for MYPT-75D are not available to test the converse. One possible explanation for the presence of multiple myosin targeting subunits in mammals, flies and nematodes lies at the C-termini: MYPT-75D/MYPT3 have a CaaX prenylation motif, whereas DMBS/MYPT1/2 do not. MYPT-75D localizes to the cell periphery; this implies the existence of two different nonmuscle myosin phosphatases in different compartments of the cell: DMBS:PP1c (PP1α or PP1β) in the cytoplasm and MYPT-75D:PP1β at the plasma membrane. These myosin phosphatases have different roles and may be subject to different regulation. However, gross perturbation, such as complete removal of one complex in either DMBS or flw mutants, may lead to hyperphosphorylation of Sqh throughout the cell and hence to similar phenotypic consequences. Similarly, overexpression of the cytoplasmic form at a sufficiently high level may compensate for loss of the membrane-associated form: we found that overexpression of a DMBS cDNA can suppress flw6, indicating that greatly increased levels of DMBS:PP1α87B can partially compensate for loss of functional MYPT-75D: PP1β9C complexes. A reduction in DMBS gene dose did not enhance flw1, indicating that DMBS is not itself the key targeting subunit for PP1β. Overexpression of MYPT-75D did not suppress flw6, presumably because MYPT-75D is not limiting or because increased levels of a defective MYPT-75D:PP1β9C complex are not helpful. High-level overexpression of MYPT-75D was lethal to wild-type flies, and modest overexpression somewhat reduced the viability of flw1 flies. We interpret both of these as being due to excess MYPT-75D diverting some PP1β from its normal role or location. flw1 flies, in which the MYPT-75D:PP1β9C myosin phosphatase is already somewhat defective, would be predicted to be more sensitive to this effect, as we observed.
In conclusion, we have shown that PP1β has an essential role, which is in the regulation of nonmuscle myosin, and this can be entirely explained by its role as an MRLC phosphatase. It associates with two different myosin-targeting subunits, one of which is specific for PP1β. These two myosin phosphatases have different roles, though sufficiently high-level expression of the putative cytoplasmic form can partially compensate for loss of the putative membrane-associated form. Loss of PP1β, and hence the PP1β-specific myosin phosphatase, leads to cytoskeletal defects and death, as does loss of the other myosin phosphatase, indicating that each has an important, nonredundant role. All of the components of the system we have analyzed are well conserved between flies and humans, suggesting that the PP1β-specific myosin phosphatase may also be conserved.
Supplementary Material
Acknowledgments
We thank Roger Karess, Yasuyoshi Nishida, Ulrich Schaefer, Myles Axton, Andrew Dingwall, and the Bloomington Drosophila Stock Center for flies; Roger Karess, Christine Field, and Yasuyoshi Nishida for antibodies; David Hartshorne for calmodulin and MLCK. We thank other members of the MRC Drosophila Cooperative Group for their help and advice, particularly to Karen Clifton, Attila Tasnadi, and Graham Knight for technical assistance. This work was supported by grants to L.A. from the Medical Research Council and BBSRC. D.B. is a Todd-Bird Research Fellow at New College, Oxford.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–02–0139. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–02–0139.
The online version of this article contains supplemental material accessible through http://www.molbiolcell.org.
References
- Axton, J.M., Dombradi, V., Cohen, P.T., and Glover, D.M. (1990). One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell 63, 33-46. [DOI] [PubMed] [Google Scholar]
- Baksa, K. et al.. (1993). Mutations in the protein phosphatase 1 gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. Genetics 135, 117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, D., and Alphey, L. (2002). PP1 binds dSARA to antagonise Dpp signalling. Nat. Genet. 31, 419-423. [DOI] [PubMed] [Google Scholar]
- Bennett, D., Szoor, B., and Alphey, L. (1999). The chaperone-like properties of mammalian inhibitor-2.are conserved in a Drosophila homologue. Biochemistry 38, 16276-16282. [DOI] [PubMed] [Google Scholar]
- Bennett, D., Szoor, B., Gross, S., Vereshchagina, N., and Alphey, L. (2003). Ectopic expression of inhibitors of protein phosphatase type 1 (PP1) can be used to analyze roles of PP1 in Drosophila development. Genetics 164, 235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin, L., Pollard, T.D., and Hitchcock-DeGregori, S.E. (2001). Inhibition of the Arp2/3 complex-nucleated actin polymerisation and branch formation by tropomyosin. Curr. Biol. 11, 1300-1304. [DOI] [PubMed] [Google Scholar]
- Bloor, J., and Kiehart, D. (2001). zipper nonmuscle myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev. Biol. 239, 215-228. [DOI] [PubMed] [Google Scholar]
- Bollen, M. (2001). Combinatorial control of protein phosphatase-1.Trends Biochem. Sci. 26, 426-431. [DOI] [PubMed] [Google Scholar]
- Bresnick, A. (1999). Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 11, 26-33. [DOI] [PubMed] [Google Scholar]
- Cant, K., Knowles, B., Mooseker, M., and Cooley, L. (1994). Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 125, 369-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, P. (2002). Protein phosphatase 1—targeted in many directions. J. Cell Sci. 115, 241-256. [DOI] [PubMed] [Google Scholar]
- Dombrádi, V., Axton, J.M., Barker, H.M., and Cohen, P.T. (1990a). Protein phosphatase 1 activity in Drosophila mutants with abnormalities in mitosis and chromosome condensation. FEBS Lett. 275, 39-43. [DOI] [PubMed] [Google Scholar]
- Dombrádi, V., Axton, J.M., Brewis, N.D., da Cruz e Silva, E.F., Alphey, L., and Cohen, P.T.W. (1990b). Drosophila contains three genes that encode distinct isoforms of protein phosphatase 1. Eur. J. Biochem. 194, 739-745. [DOI] [PubMed] [Google Scholar]
- Dombrádi, V., Mann, D.J., Saunders, R.D.C., and Cohen, P.T.W. (1993). Cloning of the fourth functional gene for protein phosphatase 1 in Drosophila melanogaster from its chromosomal location. Eur. J. Biochem. 212, 177-183. [DOI] [PubMed] [Google Scholar]
- Edwards, K., and Kiehart, D. (1996). Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development 122, 1499-1511. [DOI] [PubMed] [Google Scholar]
- Foe, V.E., Field, C.M., and Odell, G.M. (2000). Microtubules and mitotic cycle phase modulate spatiotemporal distributions of F-actin and myosin II in Drosophila syncytial blastoderm embryos. Development 127, 1767-1787. [DOI] [PubMed] [Google Scholar]
- Gross, S. (2001). Genetic and biochemical analysis of Protein Phosphatase type 1 in Drosophila melanogaster, DPhil, University of Oxford, Oxford.
- Halsell, S., and Kiehart, D. (1998). Second-site noncomplementation identifies genomic regions required for Drosophila nonmuscle myosin function during morphogenesis. Genetics 148, 1845-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne, D. (1998). Myosin phosphatase: subunits and interactions. Acta Physiol. Scand. 164, 483-493. [DOI] [PubMed] [Google Scholar]
- Holst, J., Sim, A., and Ludowyke, R. (2002). Protein phosphatases 1 and 2A transiently associate with myosin during the peak rate of secretion from mast cells. Mol. Biol. Cell 13, 1083-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A., and Cooley, L. (2002). Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annu. Rev. Genet 36, 455-488. [DOI] [PubMed] [Google Scholar]
- Ichikawa, K., Hirano, K., Ito, M., Tanaka, J., Nakano, T., and Hartshorne, D.J. (1996). Interactions and properties of smooth muscle myosin phosphatase. Biochemistry 35, 6313-6320. [DOI] [PubMed] [Google Scholar]
- Jordan, P., and Karess, R. (1997). Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J. Cell Biol. 139, 1805-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaibuchi, K., Kuroda, S., and Amano, M. (1999). Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 68, 459-486. [DOI] [PubMed] [Google Scholar]
- Karess, R.E., Chang, X.J., Edwards, K.A., Kulkarni, S.J., Aguilera, I., and Kiehart, D.P. (1991). The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65, 1177-1189. [DOI] [PubMed] [Google Scholar]
- Kiss, E., Muranyi, A., Csortos, C., Gergely, P., Ito, M., Hartshorne, D., and Erdodi, F. (2002). Integrin-linked kinase phosphorylates the myosin phosphatase target subunit at the inhibitory site in platelet cytoskeleton. Biochem. J. 365, 79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuz, A.J., Simcox, A., and Maughan, D. (1996). Alterations in flight muscle ultrastructure and function in Drosophila tropomyosin mutants. J. Cell Biol. 135, 673-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, J., Borman, M., Muranyi, A., Somlyo, A., Hartshorne, D., and Haystead, T. (2001). Identification of the endogenous smooth muscle myosin phosphatase-associated kinase. Proc. Natl. Acad. Sci. USA 98. [DOI] [PMC free article] [PubMed]
- Mahadev, K., Raval, G., Shantaram, B., Willingham, M.C., Lange, E.M., Vonderhaar, B., Salomon, D., and Prasad, G.L. (2002). Suppression of the transformed phenotype of breast cancer by Tropomyosin-1.Exp. Cell Res. 279, 40-51. [DOI] [PubMed] [Google Scholar]
- Matsumura, F., Totsukawa, G., Yamakita, Y., and Yamashiro, S. (2001). Role of myosin light chain phosphorylation in the regulation of cytokinesis. Cell Struct. Funct. 26, 639-644. [DOI] [PubMed] [Google Scholar]
- Mizuno, T., Tsutsui, K., and Nishida, Y. (2002). Drosophila myosin phosphatase and its role in dorsal closure. Development 129, 1215-1223. [DOI] [PubMed] [Google Scholar]
- Murakami, N., Chauhan, V.P.S., and Elzinga, M. (1998). Two nonmuscle isoforms expressed in rabbit brains: filament forming properties, the effects of phosphorylation by protein kinase C and casein kinase II, and location of the phosphorylation sites. Biochemistry 37, 1989-2003. [DOI] [PubMed] [Google Scholar]
- Parker, L., Gross, S., and Alphey, L. (2001). Vectors for the expression of tagged proteins in Drosophila. BioTechniques 31, 1280-1286. [DOI] [PubMed] [Google Scholar]
- Parker, L., Gross, S., Beullens, M., Bollen, M., Bennett, D., and Alphey, L. (2002). Functional interaction between NIPP1 and PP1 in Drosophila: consequences of over-expression of NIPP1 in flies and suppression by co-expression of PP1. Biochem. J. 368, 789-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan, S. et al. (2000). Protein phosphatase 1beta is required for the maintenance of muscle attachments. Curr. Biol. 10, 269-272. [DOI] [PubMed] [Google Scholar]
- Rorth, P. (1998). Gal4 in the Drosophila female germline. Mech. Dev. 78, 113-118. [DOI] [PubMed] [Google Scholar]
- Rudenko, A., Bennett, D., and Alphey, L. (2003). Trithorax interacts with type 1 serine/threonine protein phosphatase Drosophila. EMBO Rep. 4, 59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers, J. (2000). Myosins: a diverse superfamily. Biochim. Biophys. Acta 1496, 3-22. [DOI] [PubMed] [Google Scholar]
- Skinner, J., and Saltiel, A. (2001). Cloning and identification of MYPT 3, a prenylatable myosin targetting subunit of protein phosphatase 1. Biochem. J. 356, 257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlyo, A., and Somlyo, A. (2000). Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J. Physiol. 522, 177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, J., Nili, M., Homsher, E., and Tobacman, L.S. (2001). Modulation of myosin function by isoform-specific properties of Saccharomyces cerevisiae and muscle tropomyosin. J. Biol. Chem. 276, 34832-34839. [DOI] [PubMed] [Google Scholar]
- Tan, C., Stronach, B., and Perrimon, N. (2003). Roles of myosin phosphatase during Drosophila development. Development 130, 671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff, M.T., Jackle, H., and Pankratz, M.J. (1996). Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oscar mRNA required for germ cell formation. EMBO J. 15, 1247-1254. [PMC free article] [PubMed] [Google Scholar]
- Varmuza, S., Jurisicova, A., Okano, K., Hudson, J., Boekelheide, K., and Shipp, E.B. (1999). Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1cγ gene. Dev. Biol. 205, 98-110. [DOI] [PubMed] [Google Scholar]
- Wen, K.K., Kuang, B., and Rubenstein, P.A. (2000). Tropomyosin-dependent filament formation by a polymerisation-defective mutant yeast actin (V266G, L267G). J. Biol. Chem. 275, 40594-40600. [DOI] [PubMed] [Google Scholar]
- Winter, C., Wang, B., Ballew, A., Royou, A., Karess, R., Axelrod, J., and Luo, L. (2001). Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105, 81-91. [DOI] [PubMed] [Google Scholar]
- Young, P., Richman, A., Ketchum, A., and Kiehart, D. (1993). Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 7, 29-41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.