Abstract
Mitogen-activated protein kinases/extracellular signal regulated kinases (MAPKs/ERKs) are typically thought to be soluble cytoplasmic enzymes that translocate to the nucleus subsequent to their phosphorylation by their activating kinases or mitogen-activated protein/extracellular signal regulated kinase kinase. We report here the first example of nuclear translocation of a MAPK that occurs via temporally regulated exit from a membranous organelle. Confocal microscopy examining the subcellular localization of ERK3 in several cell lines indicated that this enzyme was targeted to the Golgi/endoplasmic reticulum Golgi intermediate compartment. Deletion analysis of green fluorescent protein (GFP)-ERK3 uncovered a nuclear form that was carboxy-terminally truncated and established a Golgi targeting motif at the carboxy terminus. Immunoblot analysis of cells treated with the proteasome inhibitor MG132 further revealed two cleavage products, suggesting that in vivo, carboxy-terminal cleavage of the full-length protein controls its subcellular localization. In support of this hypothesis, we found that deletion of a small region rich in acidic residues within the carboxy terminus eliminated both the cleavage and nuclear translocation of GFP-ERK3. Finally, cell cycle synchronization studies revealed that the subcellular localization of ERK3 is temporally regulated. These data suggest a novel mechanism for the localization of an MAPK family member, ERK3, in which cell cycle-regulated, site-specific proteolysis generates the nuclear form of the protein.
INTRODUCTION
Mitogen-activated protein kinases (MAPKs) or extracellular signal-regulated kinases (ERKs) mediate the intracellular response to a variety of extracellular signals, including mitogens, hormones, and stress (Kyriakis and Avruch, 2001). Activation of most members of the MAPK family by their activating kinases, or mitogen-activated protein/extracellular signal-regulated kinase kinase (MEKs) in the cytoplasm is typically followed by translocation to the nucleus. ERK3/MAPK6, in contrast, is the one member of the MAPK family for which this model of activation has not been established, despite the fact that ERK3 was first described over a decade ago (Boulton et al., 1991). To date, no physiological substrates have been described for ERK3, and conditions that result in the activation of this kinase have not been identified.
MAPKs do not have a transmembrane domain and have been classified as soluble cytoplasmic enzymes that translocate to the nucleus after a dual site-specific phosphorylation by MEKs. ERK2 is the only known example of an MAPK family member that stably associates with a membranous cytoplasmic compartment, the Golgi (Fiore et al., 1993). This localization has, however, only been described in neurons.
Presently, the subcellular localization of ERK3 is unresolved due to conflicting reports from laboratories studying the rat and human forms of the gene. The rat ERK3 has been reported to be a stable constitutively nuclear protein and to be specifically restricted to nuclear speckles (Cheng et al., 1996). In contrast, the human ERK3 protein is reported to be nuclear and cytoplasmic (Julien et al., 2003). The localization of ERK3 in nuclear speckles, was, in addition, not reported in this study. Interestingly, the 62 kDa rat ERK3 protein has been reported to be significantly smaller than the 100 kDa mouse and human proteins (Boulton et al., 1991). This rather unusual disparity in size between the human and rat ERK3 proteins could conceivably be responsible for the reported differences in the subcellular localization of this protein. We noticed that the human and mouse ERK3 coding sequences terminate with the residues KHLN, a sequence reminiscent of the carboxy-terminal dilysine (KKXX) motif, an endoplasmic reticulum retrieval/retention signal present in a number of transmembrane proteins found in the endoplasmic reticulum (ER), Golgi, as well as in the ER-Golgi intermediate compartment protein ERGIC-53 (Teasdale and Jackson, 1996). The KHLN motif is absent from the published sequence for the rat ERK3 protein (Boulton et al., 1991). The dilysine motif has not been found to be sufficient for membrane localization and is thought to function principally as retrieval motif for proteins cycling between the ER, ERGIC, and the Golgi (Teasdale and Jackson, 1996). Thus, it is conceivable that peripheral membrane proteins lacking a transmembrane domain and possessing a carboxy-terminal dilysine motif might recycle between these compartments by vesicular transport. Recently, site-directed mutagenesis of the KKXX motif of a subunit of the oligosaccharyltransferase enzyme complex (OST48), a type I membrane protein, revealed that histidine can functionally substitute for lysine in the 3 or 5 positions (Hardt and Bause, 2002). Neither ER localization nor COP I-mediated retrieval was impaired by this substitution (Hardt and Bause, 2002). Thus, the presence and location of the KHLN motif in ERK3 suggested that it might then be associated with the Golgi, ER, or the ERGIC. ERK3 has, however, not been reported to localize to any of these compartments.
Previous studies examining the subcellular localization of ERK3 have used either antipeptide antibodies against a region within the catalytic domain (Cheng et al., 1996) or antisera raised against amino acids 376–721, a region comprising the carboxy-terminal domain of ERK3 (Julien et al., 2003). Importantly, we noticed that the antigen used to raise the ERK3 antisera in the Julien study was a bacterial recombinant fusion protein whose amino terminus consisted of ERK3 sequence (amino acids 376–721) and whose carboxy terminus was the glutathione S-transferase protein (Coulombe et al., 2003). Thus, we could not exclude the possibility that this antisera may not have been optimized for the recognition of the carboxyl terminal KHLN motif in ERK3. We therefore decided to reexamine the subcellular localization of ERK3 by using an antisera raised against an antigen that was optimized to recognize the extreme end of the carboxy terminus. The antigen used in our study is therefore different from that used by Coulombe and colleagues. In our case, the bacterial recombinant fusion protein used to raise our ERK3 antisera contained ERK3 sequence (amino acids 376–720) at the carboxy-terminal end of the protein.
We have reexamined the subcellular localization of human ERK3 by using this affinity-purified antibody in several cell lines by confocal microscopy and have discovered that endogenous ERK3 localizes to the Golgi/ERGIC. Mutants of GFP-ERK3 were then used to map and characterize the Golgi-targeting domain. These experiments suggested that in vivo the full-length ERK3 protein is cleaved within the carboxy terminus thereby producing a truncated nuclear form. Immunoblot analysis of cells treated with the proteasome inhibitor MG132 subsequently revealed the presence of ERK3 cleavage products. We then used a mutagenesis strategy of green fluorescent protein (GFP)-ERK3 to test the hypothesis that the subcellular localization of ERK3 is controlled by a mechanism that requires in vivo cleavage of the protein. These mutants were characterized by immunoblot analysis and by immunofluorescent microcopy. Finally, to determine whether the subcellular localization of ERK3 was temporally regulated, cells expressing GFP-ERK3 were synchronized and analyzed by quantitative immunofluorescent microscopy. In this study, we also report an important revision to the published rat ERK3 cDNA sequence that reconciles it with those of the human and mouse genes. Thus, in all three species, the ERK3 protein is the same size and terminates with the sequence KHLN.
MATERIALS AND METHODS
Antibodies and Reagents
MG132 was obtained from Peptides International (Louisville, KY). Alexa-Fluor 488 goat anti-rabbit antibodies and cy3-conjugated donkey anti-mouse and anti-rabbit antibodies were obtained from Molecular Probes (Eugene, OR) and Jackson ImmunoResearch Laboratories (West Grove, PA), respectively. GM130, anti-calnexin, and anti-ERK3 (sc-156) antibodies were purchased from BD Biosciences (San Jose, CA), StressGen Biotechnologies (San Diego, CA), and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. An amino-terminal glutathione S-transferase (GST) fusion with amino acids 376–720 of the rat ERK3 protein was used to raise the rabbit carboxy-terminal ERK3 antisera. This antibody was precleared of GST antibodies and affinity purified with antigen coupled to Sepharose before use in experiments. The ERGIC 53 antibody G1/93 (Schweizer et al., 1988) was provided by Dr. Hans-Peter Hauri, University of Basel, Basel, Switzerland.
Cells and Cell Culture
HeLa, T98G, 293, and Rat 2 cells were obtained from American Type Culture Collection (Manassas, VA). All cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum (FBS).
Immunofluorescence Analysis
Cells were grown on coverslips, treated as indicated, and then fixed and stained. Briefly, coverslips were washed twice with phosphate-buffered saline (PBS) and then fixed and permeablized for 10 min with PBS/1% Triton X-100 and fixed for 10 min with 4% freshly prepared paraformaldehyde. Cells were blocked with PBS/0.1% Triton X-100/3% bovine serum albumin for 1 h at room temperature before immunostaining. Cells were incubated with the indicated primary and secondary antibodies in block for 1 h at room temperature.
The entire procedure was completed on the same day and performed at room temperature. Cells were visualized using a Zeiss LSM 510 confocal microscope by using the 100× objective and the fluorescein isothiocyanate/cy2, rhodamine/cy3, and cy5 filters.
Fluorescence-activated Cell Sorting (FACS) Analysis
Cell cycle distribution was evaluated with a BD Biosciences FACScan flow cytometer and CellQuest software.
Plasmids and Oligonucleotides
The plasmids described in this study were generated directly by subcloning (restriction fragments or polymerase chain reaction amplification) from the pBSK-rat ERK3 plasmid. The parental plasmid for all NGFP constructs was phrGFP-N1 (Stratagene, La Jolla, CA). All constructs were sequenced on both strands in the amplified regions to confirm that no mutations had been introduced during the subcloning.
RESULTS
Characterization of Rat ERK3
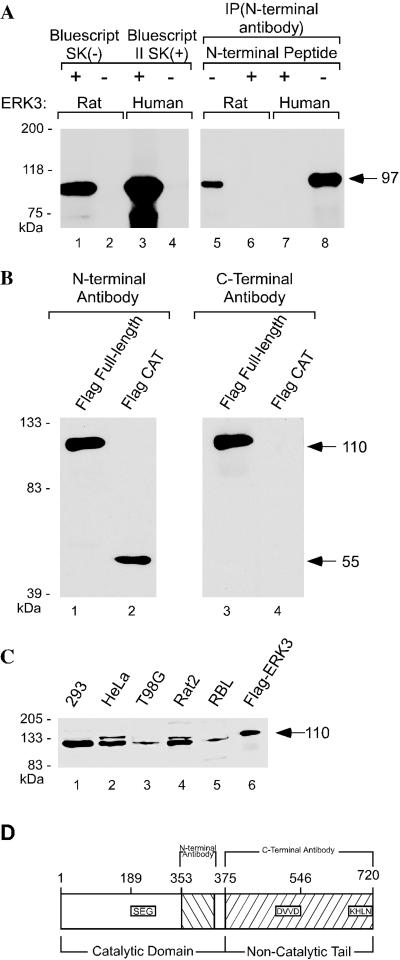
Although both the human and mouse ERK3 proteins have essentially identical amino acid sequence encoding a 721-amino acid protein of ∼100 kDa (Zhu et al., 1994, Meloche et al., 1996; Turgeon et al., 2000), the rat ERK3 gene is reported to encode a 62-kDa protein that is 543 amino acids in length (Boulton et al., 1991). We resequenced the entire open reading frame of rat ERK3 encoded by the original clone (GenBank accession no. M64301) and discovered that the GenBank sequence lacked one nucleotide (G) immediately after the codon specifying amino acid 503. A theoretical translation of the revised sequence predicted a polypeptide 720 amino acids long, essentially identical in sequence to the human and mouse proteins. In vitro translation of the rat and human ERK3 cDNAs revealed that both genes encoded proteins of the same molecular weight (Figure 1A).
Figure 1.
Rat ERK3 is the same size as human ERK3. (A) In vitro translation of rat and human ERK3 genes. rERK3 and huERK3 plasmids were translated in vitro from Bluescript SK(-) and II SK(+), respectively, by using the TNT-coupled reticulocyte system (Promega). In lanes 5–8, equal volumes of in vitro translation lysates were immunoprecipitated with the commercially available anti-ERK3 antibody sc-156 or “N-terminal antibody,” This antibody was preincubated with the antigenic peptide (sc-156P) before adding it to the immunoprecipitation reaction (lanes 6 and 7). (B) Immuno blots demonstrating the specificity of the polyclonal rabbit antisera for the ERK3 carboxy terminus. Human embryonic kidney 293 cells were transiently transfected with expression vectors carrying amino-terminally FLAG-tagged cDNAs encoding either full-length rat ERK3 (amino acids 1–720) or the catalytic domain (amino acids 1–375). Lanes 1 and 2 show an immunoblot of anti-FLAG immunoprecipitations from both transfections probed with the N-terminal antibody. This membrane was stripped and reprobed with the anti-carboxy-terminal ERK3 antibody (“C-terminal antibody,” lanes 3 and 4). (C) Immunoblot of whole cell extracts from human and rat cell lines probed with the anti-carboxy-terminal ERK3 antibody. Crude cell lysate (50 μg) was loaded per lane. An anti-FLAG immunoprecipitate of the full-length, epitope-tagged ERK3 serves as a positive control for the full-length protein and is shown in lane 6. (D) Schematic diagram of the full-length rat ERK3 protein.
Immunoprecipitation of in vitro-translated protein with a commercially purchased anti-ERK3 antibody raised against a peptide in the amino terminus within the catalytic domain yielded one major band in each of the reactions (Figure 1, A and D). Thus, the revised sequence reconciles the rat protein sequence with that of the mouse and human.
To examine the subcellular localization of the endogenous, full-length ERK3, we have raised a polyclonal antibody against a carboxy-terminal fragment spanning amino acids 375–720 of the rat ERK3 protein. To ensure that our antisera would be specific for the full-length form of the protein, the glutathione-S-ERK3 fusion protein used as our antigen was designed so that this protein terminated in ERK3 sequence (Figure 1D). This antibody recognizes the full-length form of the protein and is specific for the ERK3 carboxy terminus (Figure 1B). We then examined whether this antibody could detect endogenous ERK3 by immunoblot analysis in several human and rat cell lines (Figure 1C). A major band of ∼100 kDa was observed in crude cell lysates in all of the cell lines examined. The rat ERK3 gene therefore encodes a protein that is ∼100 kDa, similar to its human and mouse homologues.
ERK3 Is Localized to the Golgi/ERGIC
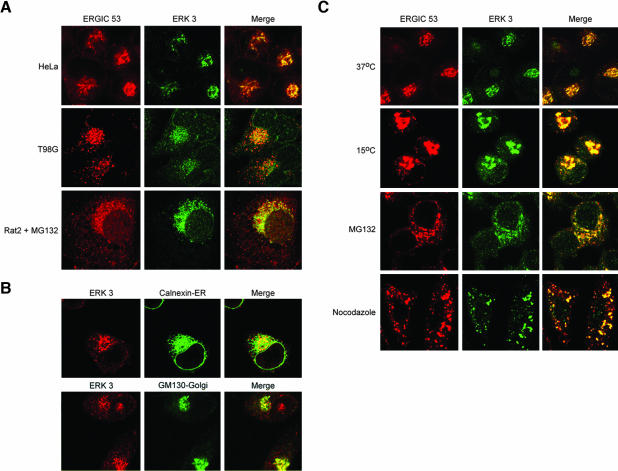
We then used this affinity-purified antibody to examine the subcellular localization of the endogenous ERK3 protein in Rat 2 cells as well as in two human cell lines, HeLa and T98G by confocal microscopy (Figure 2A). Because ERK3 protein levels are severalfold lower in Rat 2 cells than in the two human cell lines (Figure 1C), we took advantage of a previous observation in which a short treatment with proteasome inhibitors (MG132 and lactacystin) resulted in a threefold increase in ERK3 protein levels (Zimmermann et al., 2001). A 3-h incubation in 10 μM MG132 enabled us to visualize ERK3 localization in Rat 2 cells (Figure 2A). This short treatment had no visible effect on cell morphology or the cell cycle (unpublished data). Remarkably, ERK3 seemed to be extranuclear in all three cell lines and was principally restricted to a large perinuclear compartment suggestive of either Golgi, ERGIC, or the perinuclear portion of the ER (Figure 2A). To determine whether ERK3 was localized to the ERGIC or Golgi, we costained all three cell lines with our anti-ERK3 carboxy-terminal antibody and a monoclonal antibody (mAb) specific for the ERGIC-53 protein, a well-established marker for the ERGIC as well as the cis-Golgi (Schweizer et al., 1988). We observed prominent colocalization of ERGIC-53 and ERK3 in the perinuclear region of the cell (Figure 2A), a pattern consistent with cis-Golgi localization (Klumperman et al., 1998). We also observed colocalization of ERK3 with the Golgi matrix protein GM130 in the perinuclear Golgi ribbon (Figure 2B). ERK3 was not found to significantly colocalize with the endoplasmic reticulum protein calnexin (Figure 2B). Thus, our data in Figure 2, A and B, strongly suggested that ERK3 was present principally in the Golgi. The significant overlap of ERGIC-53 and ERK3 immunofluorescence, however, prompted us to test whether ERK3 could be detected in the ERGIC by examining ERK3 localization after a 3-h incubation at 15°C. This treatment promotes microtubule disassembly and results in the accumulation of ERGIC peripheral elements in the cytoplasm, because perinuclear localization is microtubule dependent (Hauri et al., 1992; Saraste and Kuismanen, 1992). Thus, if ERK3 is associated with the ERGIC, then it should accumulate and colocalize with ERGIC-53 in the peripheral elements at 15°C. We examined ERK3 localization at 15°C and observed significant overlap between ERGIC-53 and ERK3 (Figure 2C). Because a short incubation with the proteasome inhibitor MG132 can raise ERK3 protein levels, we next tested whether ERK3 levels in the ERGIC peripheral elements would be elevated in response to drug treatment. We found that indeed MG132 can raise the levels of ERK3 in the peripheral elements of the ERGIC (compare 15°C and MG132 panels in Figure 2C). ERK3 is therefore present in the ERGIC as well as the Golgi.
Figure 2.
ERK3 is localized to the Golgi/ERGIC. (A) HeLa, T98G, and Rat 2 cells were fixed and stained with the polyclonal anti-carboxy terminal ERK3 antibody and with the G1/93 (anti-ERGIC-53) antibody. The Rat 2 cells were incubated for 3 h in 10 μM MG132 before fixation. (B) HeLa cells were fixed and stained with the polyclonal anti-carboxy terminal ERK3 and mouse anti-GM-130 antibodies. To visualize the ER, the cells were costained with anti-carboxy terminal ERK3 ascites and with a polyclonal rabbit anti-calnexin antibody. (C) Four separate sets of coverslips were plated with HeLa cells. The control set of cells was incubated at 37°C. The second set was incubated at 15°C for 3 h before fixation. A third set of cells was incubated in 10 μM MG132-supplemented media for 3 h at 37°C. The final set was incubated 15 min on ice, and then immediately warmed to 37°C in the presence of 5 μg/ml nocodazole for an additional 2 h. All four sets of cells were costained with the ERK3 (“C-terminal”) antisera and with the anti-ERGIC-53 (G1/93 monoclonal) antibody.
Microtubule-depolymerizing agents such as nocodazole disrupt microtubule-dependent transport of Golgi proteins from the endoplasmic reticulum. A prolonged treatment (several hours) in this drug at 37°C leads to the disappearance of the Golgi perinuclear ribbon and the appearance of multiple Golgi structures of varying sizes scattered throughout the cytoplasm (Cole et al., 1996). The Golgi structures observed in nocodazole-treated cells are thought to result in part from the redistribution of Golgi proteins to ER exit sites (Hammond and Glick, 2000). Therefore, if ERK3 is a Golgi protein, then it should localize to Golgi structures in nocodazole-treated cells. We observed good overlap of ERGIC-53 and ERK3 immunofluorescence in Golgi structures in nocodazole-treated cells (Figure 2C). Together, the data strongly suggest that the bulk of cellular ERK3 is principally present in the Golgi/ERGIC.
ERK3 Has a Nuclear and a Golgi/ERGIC Form
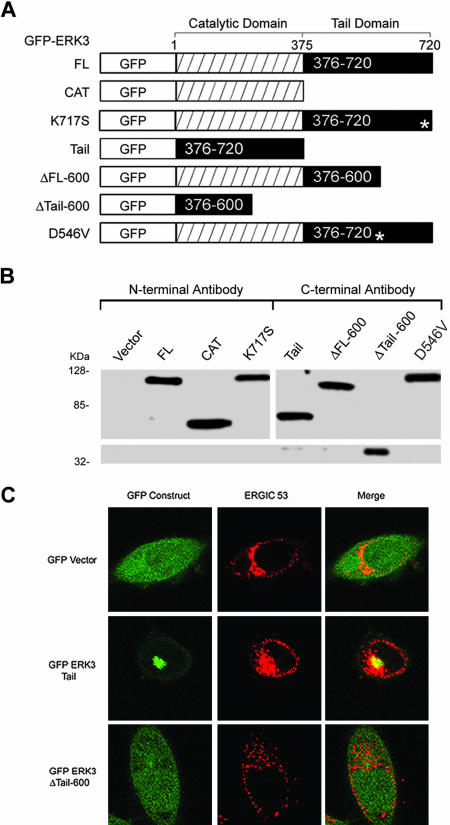
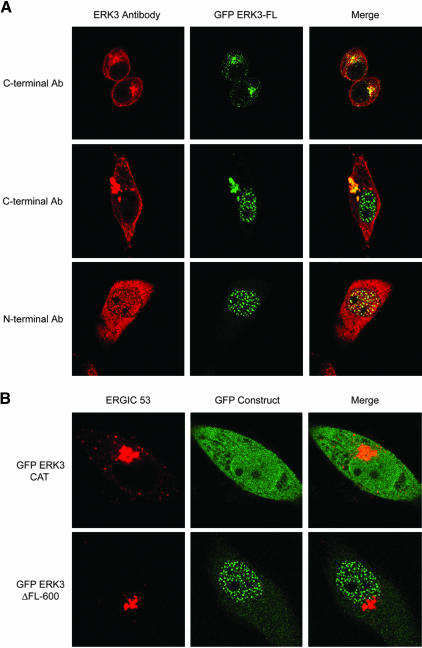
Our analysis of the 100-kDa endogenous rat and human ERK3 proteins suggested that ERK3 is localized to the Golgi/ERGIC. To our knowledge, this finding has not been reported in the literature. Importantly, our localization studies were performed with an affinity-purified carboxy-terminal–specific antibody that was raised with an antigen designed to detect the full-length protein (Figure 1D). We believe this to be the reason for the striking difference between our results and those of the recent report by Julien and colleagues. We next turned our attention toward determining whether we could detect ERK3 in the nucleus. Previous reports by Cheng and colleagues indicating that the 62-kDa rat endogenous ERK3 protein was constitutively nuclear had been performed using a peptide antibody against amino acids within the catalytic domain (Figure 1D). This antibody is available commercially, but in our hands has proved to be unsuitable for immunofluorescence studies. Indeed, we have not found any commercially available ERK3 antisera to be useful for in immunolocalization studies of the endogenous ERK3 protein. Importantly, we were unable to detect a nuclear form of ERK3 by confocal immunofluorescence microscopy by using our carboxy-terminal antisera. We, therefore, decided to examine whether a transiently expressed amino-terminally epitope-tagged ERK3 could be detected in the nucleus. To do this, we constructed a mammalian expression vector encoding full-length rat ERK3 with amino-terminal GFP and FLAG epitopes (GFP-ERK3 FL; Figure 4A). Remarkably, inspection of the subcellular localization of transiently expressed GFP-tagged full-length ERK3 by confocal microscopy revealed three distinct patterns (Figure 3A and Table 1), including an exclusively nuclear form of the protein. The nuclear form of GFP-ERK3 FL was recognized by a commercially purchased ERK3 antisera raised against an amino-terminal peptide but not by our carboxy-terminal antisera, suggesting that this protein was carboxy-terminally truncated (Figures 1D and 3A). Golgi localization of the GFP-ERK3 FL protein was confirmed by its colocalization with the ERGIC-53 protein. To test whether the nuclear form of ERK3 included part of the carboxy terminus (including residues 376–543 identified in the previous study), we examined the subcellular localization of two transiently expressed GFP-ERK3 fusion proteins lacking all (GFP-ERK3 CAT) or part of the carboxy terminus (GFP-ERK3 ΔFL-600). We found that all of the cells expressing the GFP-ERK3 ΔFL 600 protein exhibited a green fluorescent dot pattern in the nucleus (Figure 3B, bottom, and Table 1). None of the cells expressing this construct exhibited perinuclear green fluorescence representative of the Golgi perinuclear ribbon (compare with Figure 3A). In contrast, the GFP-ERK3 CAT protein was not localized to any particular compartment of the cell, and its overall distribution was indistinguishable from the GFP control (compare Figures 3B, top center, and 4C). We also examined the subcellular localization of transiently expressed, amino-terminally epitope-tagged ERK3 (M2 FLAG, GST, HA) full-length ERK3 and the catalytic domain alone in several cell lines and have obtained identical results (unpublished data). We concluded from these data that the nuclear form of ERK3 must be carboxy-terminally truncated, retaining the catalytic domain (amino acids 1–376) and perhaps as much as 224 amino acids carboxy-terminal to the catalytic domain. These data, together with our study of the endogenous ERK3 protein in human and rat cells, now provide a complete picture of the subcellular localization of this protein.
Figure 4.
The ERK3 carboxy terminus defines a Golgi/ERGIC targeting domain. (A) Amino-terminally GFP-tagged portions of the rat ERK3 open reading frame were created and expressed. All constructs that include the catalytic domain (GFP-ERK3 FL, ERK3 CAT, ERK3 ΔFL600, ERK3 K717S, and ERK3 D546V contain a FLAG epitope 3′ to the GFP open reading frame and 5′ to the ERK3 initiator methionine). Asterisks in the tail domains of the K717S and D546V mutants indicate positions of the mutations in the protein. The GFP-ERK3 tail construct (376–720) and the GFP-ERK3 Δtail600 (376–600) are direct fusions of the designated ERK3 sequence with GFP and contain no other intervening sequence. The GFP-ERK3 Δ383–398, GFP ERK3 Δ383–398/D546V, and GFP-ERK3 Δ KHLN have not been represented schematically here but were constructed in exactly the same way as the GFP-ERK FL construct. These expression vectors all contain a FLAG epitope 3′ to the GFP open reading frame and 5′ to the initiator methionine. (B) Immunoblots of GFP-ERK3 fusion proteins produced by transient expression in HEK 293 cells. Lanes 1–8 correspond to 50 μg of whole cell extracts prepared from cells transfected with the indicated vectors (A). Lanes 1–4 of the immunoblot were probed with the N-terminal anti-ERK3 antibody. Lanes 5–8 were probed with the C-terminal anti-ERK3 antibody. (C) HeLa cells were electroporated with the control GFP vector, GFP-ERK3 tail, or GFP-ERK3 Δ tail 600, plated onto coverslips, and then fixed after 24 h. The cells were then stained with anti-ERGIC-53 (G1/93) mAb.
Figure 3.
ERK3 has a nuclear and a Golgi/ERGIC form. (A) HeLa cells were electroporated with the GFP-ERK3 FL vector, plated onto coverslips, and then fixed after 24 h. The slides were stained either with the C-terminal antibody or with the N-terminal ERK3 antibody sc-156. The confocal microscopy images presented are representative of the three patterns observed over the entire field. (B) HeLa cells were electroporated with the GFP-ERK3 ΔFL600 or GFP-ERK3CAT vectors, plated onto coverslips, and then fixed after 24 h. The cells were stained with the ERGIC-53–specific antibody G1/93. The confocal microscopy images are representative cells over the entire field.
Table 1.
Subcellular distribution of GFP-ERK3 constructs
| A. GFP DNA | Cytoplasmica | Nuclear | Compartmentb | Compartment and nuclearc |
|---|---|---|---|---|
| Vector | 100 | |||
| Full length | 26 | 62 | 12 | |
| CATd | 84 | |||
| Tail | 100 | |||
| ΔTail600 | 100 | |||
| ΔFL600 | 100 | |||
| D546V | 17 | 64 | 19 | |
| Δ383–98e | 4 | 82 | 14 | |
| Δ/D546Ve | 6 | 83 | 12 | |
| K717S | 50 | 45 | 5 | |
| ΔKHLNf | 44 | 36 | 20 |
Subcellular localization is expressed as percentage of the population. Asynchronously growing HeLa cells were transfected by the calcium phosphate method with the designated constructs, allowed to express for 24 or 48 h, then fixed and stained as necessary. The subcellular localization of protein expressed by each construct was then quantified using immunofluorescence microscopy. The GFP protein was also costained with a FLAG cy3-conjugated antibody since all GFP ERK3 constructs contain a M2 FLAG epitope located 3′ to the GFP and immediately 5′ to the ERK3 initiator methionine. The data are the mean value representative of at least three independent experiments. The standard error for the data is listed below in the key to the legend. At least 200 cells were scored per construct per experiment. In an independent experiment, the Golgi colocalization of these constructs was confirmed or negated by costaining with rabbit Giantin antibody and a cy3-conjugated anti-rabbit secondary antibody.
“Cytoplasmic” localization indicates that the protein is excluded from the nucleus and is not associated with the Golgi (does not colocalize with Giantin)
“Compartment” localization refers to perinuclear colocalization with a Golgi marker (Giantin)
These cells contained nuclear and compartmental (Golgi) fluorescence
16% of the protein expressed by this construct localized to the nucleus and the cytoplasm
Δ383–98 represents full-length GFP-ERK3 lacking amino acids 383–398 in the ERK3 coding region. Δ/D546V represents the Δ383–98 construct together with the D546V mutation
ΔKHLN represents full-length GFP-ERK3 lacking the carboxyterminal KHLN residues
Table 2.
| B. | Nuclear | Compartment | Compartment and nuclear |
|---|---|---|---|
| Full-length (wild type): | 1.0 | 1.5 | 1.5 |
| D546V: | 3.8 | 7.0 | 10 |
| Δ383–98: | 1.5 | 1.2 | 2.5 |
| Δ/D546V: | 1.3 | 0.6 | 1.1 |
| K717S: | 4.0 | 2.0 | 7.0 |
| ΔKHLN: | 2.5 | 4.0 | 2.5 |
The standard deviation for the subcellular localization for cells expressing the GFP vector, GFP-tail, ΔTail600, and ΔFL600 constructs was zero. The standard deviation for the subcellular localization for cells expressing the following constructs are listed above.
Lysine 717 Is a Critical Part of a Carboxy-terminal Dilysine-like Motif in ERK3
We have proposed that the ERK3 KHLN motif might function in a manner similar to the KKXX retrieval/retention motif present in some Golgi proteins, such as ERGIC-53 (Hauri et al., 1992). Importantly, although this motif might be expected to stabilize the association of ERK3 with the Golgi, it alone should not be sufficient. We therefore explored next the contribution of the KHLN motif in the context of the full-length protein and in the absence of the catalytic domain (amino acids 1–376) to the Golgi localization of these proteins. We began these studies by first transiently expressing GFP fusions of amino acids 376–720 (GFP-ERK3 tail) and amino acids 376–600 (GFP-ERK3 ΔTail-600) in HeLa cells and examined whether either protein stably associated with the Golgi (Figure 4, A and C, and Table 1). Expression of the GFP-ERK3 tail construct (376–720) revealed prominent perinuclear localization that overlapped significantly with the Golgi perinuclear ribbon (Figure 4C) in all transfected cells examined (Table 1). In contrast, the GFP-ERK3 ΔTail-600 protein failed to localize to the Golgi, and cells expressing this construct resembled those expressing the control GFP vector (Figure 4C and Table 1). Together, these data strongly indicate that at least part of the Golgi targeting domain in ERK3 lies within amino acids 600–720. To directly test the contribution of the KHLN motif to Golgi targeting in the context of the full-length form of the protein, we performed site-directed mutagenesis on GFP-ERK3 FL, altering lysine 717 to a serine (Figure 1D). Remarkably, this single point mutation resulted in a doubling in the fraction of the population expressing only the nuclear form of ERK3 compared with cells expressing the wild-type construct (50 vs. 26%; Table 1). Deletion of the entire carboxy-terminal KHLN motif from GFP-ERK3 produced a distribution similar to that of the K717S mutant (Table 1). Thus, lysine 717 is a critical part of a carboxy-terminal dilysine-like motif that stabilizes the association of ERK3 with the Golgi/ERGIC.
Nuclear Translocation of ERK3 Is Cell Cycle Dependent
In asynchronously growing cells, we had observed that cells containing nuclear ERK3 could be classified into two categories, namely, those that contained only the nuclear form of the protein and those that contained ERK3 in both the Golgi and the nucleus (Table 1). The remainder of the population consisted of cells in which ERK3 was restricted to the Golgi. Remarkably, the percentage of cells in each of these three categories was essentially constant over multiple experiments, suggesting that the subcellular localization of ERK3 could be temporally regulated (Table 1). These data, together with our deletion analysis of GFP-ERK3, led us to postulate that the full-length, Golgi-bound protein might be subject to cell cycle-regulated cleavage within the carboxy-terminal domain. Such a mechanism would result in the nuclear translocation of the amino-terminal portion of ERK3, which includes the catalytic domain. Accordingly, we examined the subcellular localization of transiently expressed GFP-ERK3 in HeLa cells synchronized by a thymidine protocol and then released from the block over a period of 9 h. To quantify the extent of exit of ERK3 from the Golgi, we collectively refer to all cells containing nuclear ERK3 under one category designated “nuclear entry.” Importantly, this category pools the number of cells containing exclusively nuclear ERK3 with those in which the protein is found in both the Golgi and the nucleus. Remarkably, we found that a thymidine block resulted in a population that was significantly enriched in cells containing the nuclear form of ERK3 compared with the control population (62 vs. 38%). Resumption of cell cycle progression 4 h after release from the block; also resulted in a population which was enriched for the nuclear form of ERK3 (59 vs. 38%). Importantly, both of these synchronized populations were enriched in cells that had exited the G1 phase of the cell cycle but had not yet completed mitosis. In contrast, 9 h after release from the thymidine block the fraction of cells containing the nuclear form of ERK3 plummeted to levels found in the control population (30 vs. 38%). The percentage of cells with exclusive Golgi localization also returned the levels observed in the control population (Figure 5). Importantly, 68% of this population had completed mitosis and was in the G1 phase of the subsequent cell cycle. Thus, the subcellular localization of ERK3 is cell cycle dependent, shifting from the Golgi to the nucleus as the cells proceed toward M phase and then returning to the Golgi in the next cycle. Together, these findings strongly support the premise that nuclear translocation of ERK3 is temporally regulated in cycling cells. To our knowledge, this has not been described for any other member of the MAPK family.
Figure 5.
Nuclear translocation of ERK3 is cell cycle dependent. HeLa cells growing on coverslips were transiently transfected with GFP-ERK3 FL and then incubated in 2 mM thymidine containing media for 18 h. Six slides were collected and fixed after this first treatment with thymidine. Two time points were collected after release from the thymidine block; 4 and 9 h. Six slides/condition and time point were subsequently processed for immunofluorescence analysis. Percentage of Golgi refers to the percentage of transfected cells with GFP-ERK3 restricted to the Golgi. Percentage of nuclear entry includes all transfected cells containing nuclear ERK3. The slides were fixed and immunostained with the cy3FLAG antibody. Only cells exhibiting green and red fluorescence were scored. The cell cycle profiles of asynchronously growing and synchronized cells (as determined by FACS analysis) are presented below the quantitation of the localization data.
Deletion of a Carboxy-terminal Acidic-rich Region Hinders ERK3 Exit from the Golgi/ERGIC
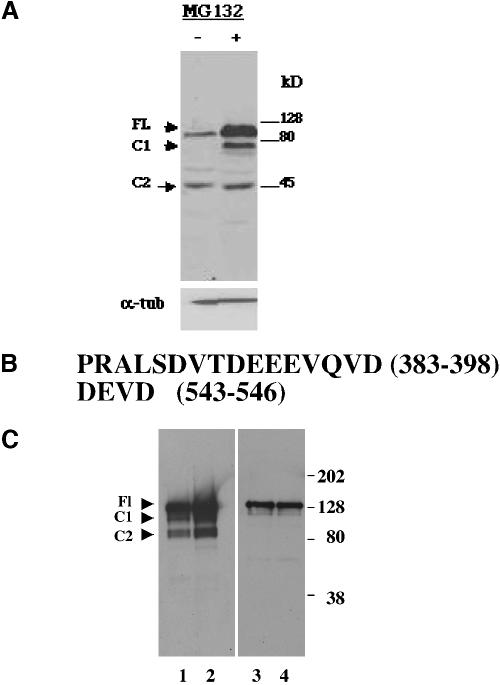
Our analysis of the subcellular localization of carboxy-terminal deletions of GFP-ERK3 FL suggested that that the full-length form of the protein was subject to site-specific proteolytic cleavage in a carboxy-terminal region before nuclear entry. These experiments suggested that the size of the nuclear form of ERK3 was larger than the catalytic domain (amino acids 1–376) and could encompass as much as the amino-terminal 600 amino acids. Thus, the nuclear form of ERK3 is likely to be between 45 and 68 kDa. Our cell cycle analysis of ERK3 experiments further suggested that the nuclear form of the protein was labile because it was not present throughout the entire cell cycle. We therefore reasoned that ERK3 cleavage products might be detectable by immunoblot analysis of cells cultured in the presence of proteasome inhibitors. It has been reported that highly specific inhibitors of the proteasome, such as MG132 and lactacystin, elevated the levels of full-length ERK3 protein severalfold in asynchronously growing cell lines (Zimmermann et al., 2001). We confirmed this result by immunoblot analysis by using our carboxy-terminal antisera and further found that treatment of an asynchronously growing cell line with MG132 stabilized the levels of a highly labile truncated ERK3 protein (henceforth termed C1) migrating below the 80-kDa marker (Figure 6A). The levels of a second truncated ERK3 protein (termed C2) migrating ∼45 kDa was not observed to change in this experiment. Because these proteins were detected by our carboxy-terminal antisera, we concluded that the electrophoretic migration of these cleaved forms of ERK3 together with our deletion analysis supported the notion that site-specific cleavage of the full-length form of the protein might be occurring at two sites within the carboxy-terminal domain of the protein, somewhere between amino acids 377 and 600.
Figure 6.
Deletion of a carboxy-terminal acidic-rich region hinders ERK3 exit from the Golgi/ERGIC. (A) Immunoblot of whole cell extracts prepared from exponentially growing HeLa cells incubated in 10 μM MG132 for 3 h (5-min exposure of the film). Protein (50 μg) was loaded in each lane. The upper blot was probed with C-terminal anti-ERK3 antibody. Protein normalization of all the samples (50 μg/lane; bottom) was confirmed by probing a Western blot of total cell lysates with an anti-α tubulin antibody (lower blot). (B) Amino acid sequences of the two mutant regions tested as putative cleavage sites. A 15-nucleotide sequence immediately 5′ to the nucleotides encoded for the region of interest (amino acids 388–98) also was deleted because it contained a unique enzymatic restriction site that aided in the identification of bacterial colonies expressing this recombinant plasmid. Thus, all constructs containing the Δ383–398 deletion lack the sequence PRALS immediately amino-terminal to DVTDEEEVQVD (amino acids). (C) Immunoblot of FLAG immunoprecipitations of extracts of HeLa cells transiently expressing various forms of GFP-ERK3. To normalize for differences in level of protein expression, 1 mg of protein was used for the GFP-ERK3 Δ383–398 and GFP ERK3 Δ383–398/D546V immunoprecipitations and 200 μg of protein was used for the GFP-ERK3 FL and GFP-ERK3 D546V immunoprecipitations. The blots were probed with the C-terminal anti-ERK3 antibody. The expression constructs used for transfections in lanes 1–4 are, respectively, GFP-ERK3 FL, GFP-ERK3 D546V, GFP-ERK3 Δ383–398, and GFP ERK3 Δ383–398/D546V.
An examination of the amino acid sequence in this region revealed two acidic, caspase-like sites, located at amino acids 388–398 (DVTDEEEVQVD) and amino acids 543–546 (DVVD), that might result in cleavage products of the sizes observed in our experiments (Figure 6B). We then mutagenized GFP-ERK3 FL to determine whether either or both of these regions defined in vivo cleavage sites within the ERK3 carboxy terminus. As a first approach toward mutagenesis of this region, we made use of what is known about the proteolytic preferences of caspases based on studies with peptide substrates. Caspases hydrolyze their substrates on the carboxyl side of an aspartic acid residue (the so-called P1 residue), which is part of a tetrapeptide defining the consensus cleavage motif (Earnshaw et al., 1999). Interestingly, although this motif often begins and terminates with aspartic acid (D/P4–XX-D/P1), cleavage strictly occurs at the P1 aspartate residue. To determine whether amino acids 543–546 define an in vivo cleavage site for ERK3, we tested whether glutamic acid 546 was a potential cleavage substrate by mutating it to a valine. We created a second mutant by deleting the DVTDEEEVQVD sequence from the full-length protein, thereby ruling out the possibility that cleavage could occur at any one of several potential sites within this region (Figure 6B). Our model predicts that elimination of all cleavage sites within the ERK3 carboxy terminus should prevent cleavage of ERK3 and therefore exit from the Golgi. Such mutants should exhibit little or no nuclear translocation. To directly test whether our mutations removed ERK3 cleavage sites, we examined both cleavage and subcellular localization of all three GFP-ERK3 mutants (Δ 383–98, D546V, and Δ 383–98/D546V) by immunoblot analysis of whole cell extracts and by immunofluorescence microscopy. The predicted molecular weight of GFP-FL ERK3 is 116 kDa. Cleavage of this protein at or near aspartic acid 546 would be expected to produce a protein migrating with a mobility of ∼93 kDa, whereas cleavage somewhere within amino acids 388–398 would produce a protein of ∼75 kDa. Immunoprecipitation of extracts of asynchronously growing cells expressing the wild-type or D546V proteins and subsequent immunoblotting revealed a carboxy-terminally truncated protein (C1) migrating between the 128-and 80-kDa molecular weight markers (Figure 6C). A smaller ERK3 cleavage product (C2) is also generated, and it migrated near the 80-kDa molecular weight standard. Remarkably, we were not able to detect significant levels of either cleaved form of GFP-ERK3 (C1 or C2) when we performed analogous experiments with cells expressing the 383–98 deletion individually or in combination with the D546V mutation. These data suggest that amino acids 383–398 are required for cleavage of the full-length protein. In contrast, in cells expressing the D546V point mutation, ERK3 underwent cleavage at both sites, suggesting that either this single mutation was not sufficient to abrogate cleavage in this region or that there are alternative cleavage sites nearby. Next, we examined whether either of these mutations affected the translocation of ERK3 to the nucleus (Table 1). Nuclear translocation of each mutant was compared with that of the full-length, wild-type protein and ΔFL600, which we designated as 100%, by using conventional immunofluorescence microscopy. We found that the subcellular localization of all of the mutant ERK3 proteins (Δ 383–98, D546V, Δ 383–98/D546V) strongly supported our interpretation of the data obtained by immunoblot analysis: cells expressing the deletion mutant exhibited greater than sixfold reduction in nuclear localization (Table 1). In contrast, the D546V mutant was able to translocate to the nucleus nearly as efficiently as the wild-type protein. The region spanned by amino acids 383–398 therefore contains a region critical to both the cleavage and nuclear translocation of ERK3. These experiments provide compelling evidence that the nuclear translocation of ERK3 is achieved by a mechanism directed through an acidic patch and requiring site-specific cleavage within the carboxy-terminal region of the protein.
DISCUSSION
Our finding that ERK3 resides in the ERGIC and the Golgi was unexpected because MAPKs are thought to be soluble cytoplasmic enzymes. Because ERK3 lacks a trans-membrane domain, its association with these membranous organelles must rely on relatively stable interactions with other molecules in these compartments. We have explored the contribution of a carboxy-terminal dilysine motif (KHLN) and find that it stabilizes but is not sufficient for the association of the full-length form of ERK3 with the Golgi. Importantly, this motif is not present in any other member of the MAPK family, including the highly related p63 MAPK, which lacks the carboxy-terminal KHLN motif that is unique to ERK3.
Remarkably, an examination of the subcellular localization of GFP-ERK3 in synchronized cells reveals that nuclear entry increases as cells progress through S phase, concomitant with diminished Golgi partitioning. Reentry into the subsequent cell cycle was accompanied by a dramatic reversal as ERK3 localization shifts from the nucleus back to the Golgi. These studies further revealed a nuclear form of the protein that was carboxy-terminally truncated, suggesting that proteolytic cleavage releases ERK3 from the Golgi. Thus, ERK3, like other members of the MAPK family, translocates to the nucleus, but it does so through a wholly unique mechanism not shared by other ERKs. This interesting mechanism of regulating the nuclear translocation of ERK3 via a temporal sequestration in the Golgi raises the question as to the purpose for this precise localization. It is conceivable that ERK3 could have two different sets of substrates, one located in the cytoplasm and the other in the nucleus. In support of this hypothesis, we have found that both forms of this enzyme are produced and are active in cells stably expressing an epitope-tagged version of the protein in in vitro kinase assays in which myelin basic protein, histone H1, or H3 (unpublished data) was used as substrates. Residency in the Golgi may therefore be part of a mechanism that ensures that ERK3 does not have access to nuclear substrates at an improper time.
The presence of ERK3 in the Golgi further raises the question as to whether this enzyme has any role in the biology of this organelle. Malholtra and colleagues have implicated an unidentified MAPK in the fragmentation of the Golgi before mitosis (Acharya et al, 1998; Colanzi et al, 2003). Moreover, we find that both the PLK and cdc2/B cyclin, two enzymes also reported to participate in this process, robustly phosphorylate ERK3 in vitro (data not shown). Thus, although the role of ERK3 in Golgi biology has not been the focus of this report, the answer to this question will be of great interest to cell biologists and is the subject of an ongoing investigation. Finally, our study of ERK3 corroborates previous reports that there is a nuclear form of ERK3 and establishes for the first time that it is generated via proteolysis of a region within the carboxy-terminal part of the protein somewhere between amino acids 380–600. Thus, we are left with the very interesting question of the identity of the ERK3 specific proteolytic activity. In this report, we have uncovered several important clues as to the nature of the ERK3 protease. Our experiments indicate that the exit of ERK3 from the Golgi is principally mediated by an acidic patch spanned by amino acids 383–398. Second, ERK3 cleavage occurs in cycling cells. Thus, the ERK3 protease must be active in growing cells and may demonstrate some preference for short stretches of acidic residues. These requirements have led us to begin to consider caspases and/or the proteasome. Caspases prefer to cleave carboxy-terminal to acidic residues, and have been best studied in cells undergoing apoptosis. However, cell permeable caspase inhibitors have been shown to inhibit the growth of proliferating T cells (Fischer et al., 2003) Moreover, negative regulators of the cell cycle such as Wee1 and CDC27 are cleaved by caspases (Fischer et al., 2003). Finally, caspase 2 has been found to associate with the Golgi and to promote cleavage of golgin 160 during apoptosis thereby promoting disintegration of the Golgi (Mancini et al., 2000). Thus, it is conceivable that a nonapoptotic caspase activity could be the ERK3 protease.
Our detection of ERK3 cleavage in growing cells with active proteasomes, the stabilization of ERK3 protein levels by proteasome inhibitors, and our analysis of the 383–98 deletion mutant also have led us to consider whether the ERK3 protease might be the postacidic cleavage activity specific to the β1 subunits of the 20S proteasome (Kisselev and Goldberg, 2001). This preference of the proteasome for cleavage after short stretches of acidic residues has, however, been studied exclusively with peptide substrates and has not been demonstrated with a protein. The proteasome is a particularly attractive candidate for the ERK3 protease because a number of proteins, including the transcription factor nuclear factor-κB (NF-κB) have been shown to be undergo limited proteolytic processing by this protease. Indeed, the nuclear translocation of NF-κB is dependent on the proteasomal generation of the 55-kDa form of the IκB subunit from a 110-kDa precursor (Palombella et al., 1994). A direct test of whether the proteasome can generate a carboxy-terminally truncated protein similar in size to the nuclear form of ERK3 from the full-length protein will require the reconstitution of ERK3 site-specific proteolysis in vitro by using purified proteasomes. This very interesting experiment is, however, well beyond the scope of this report.
We propose here that our biochemical data, together with our microscopic studies, are consistent with site-specific cleavage of the full-length form of ERK3, resulting in the nuclear translocation of the cleaved protein. In addition we find that the subcellular localization of ERK3 also is temporally regulated in cycling cells. Many of the details of this rather elaborate mechanism governing the intracellular location of ERK3 are not currently understood, but given that ERK3 is an MAPK, it is likely that phosphorylation may play a role. In conclusion, our study of ERK3 provides the first example of a mechanism linking a proteolytic activity to the nuclear translocation of a member of the MAPK family.
Supplementary Material
Acknowledgments
I.S. is grateful to Dr. R. Erikson for support during the duration of this project. We thank Dr. Z. Chen for help with confocal microscopy. We thank Dr. H. P. Hauri for the G1/93 mAb and Drs. J. Flier and G. Yancopoulos for human and rat ERK3 plasmids, respectively. This project was funded by a Mentored Career Development Award (5KO1CA77103) awarded to I.S. by the National Cancer Institute and by a Pew Scholar Award from the Pew Charitable Trusts awarded to B.D.D.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0234. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0234.
References
- Acharya, U., Mallabiabarrena, A., Acharya, J.K., and Malhotra, V. (1998). Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell 92, 183-192. [DOI] [PubMed] [Google Scholar]
- Boulton, T.G., Nye, S.H., Robbins, D.J., Ip, N.Y., Radziejewska, E., Morganbesser, S.D., Depinho, R.A., Panayotatos, N., Cobb, M.H., and Yancopoulos, G.D. (1991). ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65, 663-675. [DOI] [PubMed] [Google Scholar]
- Cheng, M., Boulton, T.G., and Cobb, M.H. (1996). Erk3 is a constitutively nuclear kinase. J. Biol. Chem. 271, 8951-8958. [DOI] [PubMed] [Google Scholar]
- Colanzi, A., Suetterlin, I., and Malholtra, V. (2003). Cell cycle specific Golgi fragmentation: how and why? Curr. Opin. Cell Biol. 15, 462-467. [DOI] [PubMed] [Google Scholar]
- Cole, N.B., Sciaky, N., Marotta, A., Song, J., and Lippincott-Schwartz, J. (1996). Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell 7, 631-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe, P., Rodier, G., Pelletier, S., Pellerin, J., and Meloche, S. (2003). Rapid Turnover of extracellular signal regulated kinase 3 by the ubiquitin proteasome pathway defines a novel paradigm of mitogen activated protein kinase regulation during cellular differentiation. Mol. Cell. Biol. 13, 4542-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., Martins, L.M., and Kauffmann, S.H. (1999). Mammalian caspase structure, activation, substrates and functions during apoptosis. Annu. Rev. Biochem. 68, 383-424. [DOI] [PubMed] [Google Scholar]
- Fiore, R.S., Bayer, V.E., Pelech, S.I., Posada, J., Cooper, J.A., and DeCamilli, P. (1993). p42 mitogen activated protein kinase in brain: prominent colocalization in neuronal cell bodies and dendrites. Neuroscience 55, 463-472. [DOI] [PubMed] [Google Scholar]
- Fischer, U., Janicke, R.U., and Schulze-Osthoff, K. (2003). Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10, 76-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, A.T., and Glick, B.J. (2000). Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11, 3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt, B., and Bause, E. (2002). Lysine can be replaced by histidine and not arginine as the ER retrieval motif for type I membrane proteins. Biochem. Biophys. Res. Commun. 291, 751-757. [DOI] [PubMed] [Google Scholar]
- Julien, C., Coulombe, P., and Meloche, S. (2003). Nuclear export of ERK3 by a CRM1 dependant mechanism regulates its inhibitory action on cell cycle progression. J. Biol. Chem. 278, 42615-42624. [DOI] [PubMed] [Google Scholar]
- Kisselev, A.F., and Goldberg, A.L. (2001). Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 121, 1-20. [DOI] [PubMed] [Google Scholar]
- Klumperman, J. Schweizer, A., Clausen, H., Tang, B.L., Hong, W., Oorschot, V., and Hauri, H.P. (1998) The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J. Cell Sci. 111, 3411-3425. [DOI] [PubMed] [Google Scholar]
- Kyriakis, J.M., and Avruch, J. (2001). Mammalian mitogen activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81, 807-869. [DOI] [PubMed] [Google Scholar]
- Mancini, M., Machamer, C.E., Roy, S., Nicholson, D.W., Thornberry, N.A., Casciola-Rosen, L.A., and Rosen, A. (2000). Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J. Cell Biol. 149, 603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche, S., Beatty, B.G., and Pellerin, J. (1996). Primary structure, expression and chromosomal locus of a human homolog of rat ERK3. Oncogene 13, 1575-1579. [PubMed] [Google Scholar]
- Palombella, V.J., Rando, O.J., Goldberg, A.L., and Maniatis, T. (1994). The ubiquitin proteasome proteasome pathway is required for processing the NF-Kappa B1 precursor protein and the activation of NF-Kappa B. Cell 78, 773-785. [DOI] [PubMed] [Google Scholar]
- Saraste, J., and Kuismanen, E. (1992). Pathways of protein sorting and membrane traffic between the rough endoplasmic reticulum and the Golgi complex. Semin. Cell Biol. 3, 343-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, A., Fransen, J., Bachi, T., Ginsel, L., and Hauri, H.P. (1988). Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 107, 1643-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale, R.D. and Jackson, M.R. (1996). Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Biol. 12, 27-54. [DOI] [PubMed] [Google Scholar]
- Turgeon, B., Saba-Eleil, M.K., and Meloche, S. (2000). Cloning and characterization of mouse ERK3 as a unique gene product of 100 kD. Biochem. J. 346, 169-175. [PMC free article] [PubMed] [Google Scholar]
- Zhu, A.X., Zhao, Y., Moller, D.E., and Flier, J.S. (1994). Cloning and characterization of p97 MAPK, a novel human homolog of rat ERK3. Mol. Cell. Biol. 12, 8202-8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, J., Lamerant, N., Grossenbacher, R., and Furst, P.A. (2001). Proteasome- and p38 dependent regulation of ERK3 expression. J. Biol. Chem. 276, 10759-10766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.