Abstract
Sexual identity and mating are linked to virulence of the fungal pathogen Cryptococcus neoformans. Cells of the α mating type are more prevalent and can be more virulent than a cells, and basidiospores are thought to be the infectious propagule. Mating in C. neoformans involves cell-cell fusion and the generation of dikaryotic hyphae, processes that involve substantial changes in cell polarity. Two p21-activated kinase (PAK) kinases, Pak1 and Ste20, are required for both mating and virulence in C. neoformans. We show here that Ste20 and Pak1 play crucial roles in polarized morphogenesis at different steps during mating: Pak1 functions during cell fusion, whereas Ste20 fulfills a distinct morphogenic role and is required to maintain polarity in the heterokaryotic mating filament. In conclusion, our studies demonstrate that PAK kinases are necessary for polar growth during mating and that polarity establishment is necessary for mating and may contribute to virulence of C. neoformans.
INTRODUCTION
The ability to generate cell polarity, or subcellular asymmetry, is important for all eukaryotic cells. In multicellular eukaryotes, cell polarity is critical for embryogenesis, axon migration during neuronal development, and communication between lymphocytes and the immune system. In unicellular organisms, such as the budding yeast Saccharomyces cerevisiae, cell polarization is essential for mitotic cell division and mating (Johnson, 1999).
In S. cerevisiae, polarity is generated through reorganization of the actin cytoskeleton and is directed by the small GTPase Cdc42. Ste20 and Cla4, members of the highly conserved p21-activated kinase (PAK) family, act as downstream effectors of Cdc42 in a variety of morphogenic processes. Ste20 functions upstream of mitogen-activated protein (MAP) kinase signaling pathways mediating pheromone response, invasive growth, osmosensing, and cell wall integrity (Elion, 2000). In addition, Ste20 has been implicated in polarisome activation (Goehring et al., 2003). Cla4 functions in the organization and maintenance of septins at the mother-bud junction (Cvrckova et al., 1995; Weiss et al., 2000; Dobbelaere et al., 2003; Schmidt et al., 2003; Versele and Thorner, 2004). Ste20 and Cla4 also share overlapping essential functions in budding and cytokinesis and have recently been shown to function in mitotic exit (Cvrckova et al., 1995; Holly and Blumer, 1999; Weiss et al., 2000; Hofken and Schiebel, 2002; Jensen et al., 2002; Seshan et al., 2002).
We previously identified two PAK kinase homologues from the human fungal pathogen Cryptococcus neoformans, Ste20 and Pak1 (Wang et al., 2002). Ste20 and Pak1 each contain conserved N-terminal Cdc42-Rac interactive binding (CRIB) regulatory and C-terminal catalytic domains that characterize PAK kinases. In addition, Ste20 contains a pleckstrin homology domain that is found in a subgroup of PAK kinases that includes S. cerevisiae Cla4. C. neoformans is a dimorphic fungus that is the causative agent of cryptococcosis, a life-threatening fungal meningitis (Mitchell and Perfect, 1995). C. neoformans isolates are classified into four subtypes based on capsular antigens (serotype A var. grubii, serotype D var. neoformans, and serotype B and C var. gattii). Serotype A and D strains account for the majority of cryptococcal infections worldwide and occur most commonly in immunocompromised patients. In contrast, serotype B and C strains are primary pathogens and were recently found to be the causative agent of a cryptococcal outbreak on Vancouver Island, Canada (Speed and Dunt, 1995; Stephen et al., 2002; Fraser et al., 2003).
Several factors contribute to virulence of C. neoformans, including the ability to produce melanin and a polysaccharide capsule, growth at high temperature, and mating-type. C. neoformans is a budding yeast with a bipolar mating-type system. In response to external stimuli, including pheromones and nitrogen deprivation, a and α cells fuse and produce a dikaryotic hyphae. When filamentation ceases, a basidium forms at the filament tip where karyogamy, meiosis, and postmeiotic divisions occur. Multiple basidiospores, the meiotic progeny, bud from the surface of the basidium at four positions to result in four long chains of protruding basidiospores. The life cycle is completed when basidiospores germinate into budding yeast cells (Hull and Heitman, 2002; Wickes, 2002).
Because of their small size, basidiospores or desiccated yeast cells are thought to be the infectious propagules that enter the alveoli of the lung. In the clinic, strains of the α mating-type are more prevalent than strains of the a mating-type, and in serotype D α cells are more virulent than a cells in animal models of cyptococcosis (Kwon-Chung and Bennett, 1978; Kwon-Chung et al., 1992). The MAT locus of C. neoformans is unusually large, spans >100 kb, and encodes proteins involved in signal transduction (Ste20, Ste11), polar growth and organelle transport (Myo2), and transcription (Ste12 and Sxi1) (Lengeler et al., 2002). Because of the link between mating type and virulence, the genes contained within the MAT locus are of considerable interest.
Our initial analysis of the C. neoformans PAK kinases provided additional evidence supporting a link between mating-type and virulence in C. neoformans. The genes encoding Ste20α and Ste20a are located in the MAT locus, ste20 mutants are unable to mate, and the virulence of a clinical isolate deleted for STE20α was attenuated in animal models of cryptococcosis. However, we found that Pak1 is also required for mating and virulence in C. neoformans. To explore the links between mating and virulence, we have further analyzed the roles of Ste20 and Pak1 during mating in C. neoformans. We find that both Ste20 and Pak1 mediate cell polarity during mating. However, the roles of Ste20 and Pak1 are functionally and temporally distinct from each other; Pak1 is required during the initial fusion between α and a cells, whereas Ste20 maintains polarity during growth of the dikaryotic filament.
MATERIALS AND METHODS
Strains and Media
C. neoformans strains used in this study are listed in Table 1. Strains were grown on YPD medium, synthetic dextrose medium, and V8 medium (Sherman 1991; Kwon-Chung and Bennett 1992).
Table 1.
Strains and plasmids
| Strain | Genotype | Source/Reference |
|---|---|---|
| Serotype A | ||
| CSB1 | MATα pak1::URA5 ura5 | Wang et al., 2002 |
| CSB51 | MATα pak1::ura5 ura5 | Wang et al., 2002 |
| F99 | MATα ura5 | Wang et al., 2001 |
| H99 | MATα | Perfect et al., 1980 |
| JF99 | MATa ura5 | This study |
| JF219 | MATa ste20a::NAT ura5 | This study |
| JF265 | MATa pak1::NAT | This study |
| JF267 | MATa pak1::NAT ura5 | This study |
| KN99a | MATa | Nielsen et al., 2003 |
| PPW91 | MATα ste20α::URA5 ura5 | Wang et al., 2002 |
| PPW96 | MATα ste20α::ura5 ura5 | Wang et al., 2002 |
| Serotype D | ||
| CSB5 | MATa ste20a::ADE2 ade2 | Wang et al., 2002 |
| CSB8 | MATα pak1::URA5 ura5 ade2 | Wang et al., 2002 |
| CSB9 | MATα pak1::URA5 ura5 | Wang et al., 2002 |
| CSB10 | MATa pak1::URA5 ura5 | Wang et al., 2002 |
| CSB11 | MATα ste20α::URA5 ura5 ade2 | Wang et al., 2002 |
| KLB156-1 | MATa ste20a::ADE2 ura5 ade2 | Wang et al., 2002 |
| CSB15 | MATα ste20α::URA5 ura5 | Wang et al., 2002 |
| CSB21 | MATa ste20a::ADE2 ura5 ade2 lys1 | Wang et al., 2002 |
| CSB23 | MATα pak1::ura5 ura5 ade2 | Wang et al., 2002 |
| CSB25 | MATα pak1::ura5 ura5 | This study |
| CSB27 | MATa pak1::URA5 ura5 lys1 | This study |
| CSB48 | MATα ste20α::ura5 ura5 | Wang et al., 2002 |
| RDC5 | MATα cpk1::ADE2 ade2 | R. Davidson |
| JEC20 | MATa | J. Edman |
| JEC21 | MATα | J. Edman |
| JEC31 | MATa lys2 | J. Edman |
| JEC34 | MATa ura5 | J. Edman |
| JEC43 | MATα ura5 | J. Edman |
| JEC50 | MATα ade2 | J. Edman |
| JEC53 | MATa ura5 lys1 | J. Edman |
| JEC155 | MATα ura5 ade2 | J. Edman |
| Plasmids | ||
| pRCD83 | C. neoformans vector | R. Davidson |
| pCBN12 | serotype A PGPD1-STE20α | This study |
| pCBN19 | serotype D PGPD1-STE20α | This study |
| pCBN20 | serotype D PGPD1-STE20a | This study |
| pCBN23 | serotype D PGPD1-PAK1 | This study |
| pCBN34 | serotype A PGPD1-PAK1 | This study |
| pCBN47 | serotype A PGPD1-STE20a | This study |
Molecular Biology
Standard methods were performed as described by Sambrook et al., (1989). C. neoformans genomic DNA for Southern blot analysis was prepared as described previously (Pitkin et al., 1996). Biolistic transformations were performed as described previously (Toffaletti et al., 1993; Davidson et al., 2000a).
Overexpression Analysis
The serotype A STE20α, STE20a, and PAK1 and serotype D STE20α, STE20a, and PAK1 open reading frames were amplified by polymerase chain reaction (PCR) with BamHI sites at each end. The PCR products were subcloned into pCRII-TOPO and sequenced. After digestion with BamHI, each open reading frame was subcloned into BamHI digested C. neoformans vector pRDC83 generating plasmids pCBN12 (serotype A STE20α), pCBN47 (serotype A STE20a), pCBN34 (serotype A PAK1), pCBN19 (serotype D STE20α), pCBN20 (serotype D STE20a), and pCBN23 (serotype D PAK1). Ura- strains PPW96, JF219, CSB51, CSB48, KLB156-1, and CSB25 were transformed with each plasmid, including pRDC83, which served as a control. Transformants were analyzed in bilateral crosses with the appropriate mating partner and for growth at 37 or 39°C (for ste20 transformants).
Northern Blot Analysis
Equal numbers (2 × 107 cells) of JEC21, α cpk1 (RDC5), ste20α (CSB5), and α pak1 (CSB9) cells were grown on separate V8 plates alone or in coculture with equal numbers (2 × 107 cells) of JEC20, ste20a (CSB5), or a pak1 (CSB10) cells for 24 h. Cells were harvested, lyophilized overnight, and RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA). Northern blots were performed according to standard protocols (Ausubel et al., 1992) by using 10 μg of total RNA per sample. The MFα, MFa, and ACT1 probes were amplified by PCR, radiolabeled with [α-32P]dCTP (Amersham Biosciences, Piscataway, NJ) by using a Rediprime II kit (Amersham Biosciences), and used in hybridization reactions as described previously (Church and Gilbert, 1984).
Fusion, Recombinant Spore, and Conjugation Assays
Fusion assays were performed as described previously (Alspaugh et al., 2000) with the following modifications. Bilateral crosses consisting of the wild-type (JEC50 × JEC34 and JEC50 × JEC31), ste20 (CSB11 × KLB156–1), and pak1 (CSB8 × CSB27) mutant strains containing equal numbers of cells were pipetted onto V8 agar medium. Each mating partner contained a complementing auxotrophic mutation (lys2 and ura5 or ade2 and ura5) to allow selection of prototrophic fusion products. After 48 h, the mating mixes were excised, resuspended in 1 ml of H2O, and plated in serial dilutions onto minimal medium. Colonies were counted after 3 d of incubation.
Recombinant spore production was assessed in a quantitative mating assay as described previously (Alspaugh et al., 2000) with the following modifications. Bilateral crosses consisting of the wild-type (JEC21 × JEC53 and JEC155 × JEC53), ste20 (CSB15 × CSB21), and pak1 (CSB23 × CSB10) mutant strains containing equal numbers of cells were pipetted onto V8 agar medium. After 7 d, the mating were excised, resuspended in 1 ml of H2O, and plated in serial dilutions onto 5-fluoroorotic acid agar medium lacking lysine or adenine. Colonies were counted after 3 d of incubation.
The number of conjugation products present in wild-type and pak1 cocultures was determined by mixing together equal numbers of wild-type (JEC21 and JEC30) and pak1 (CSB9 and CSB10) mutant cells on V8 agar medium. After 24 h, the cells were scraped off, resuspended in H2O, and examined microscopically for the presence of fused cells. Fifty cells were counted for each sample and a total of 300 cells were analyzed from each coculture.
Microscopy
Brightfield, differential interference microscopy (DIC) and fluorescent images were captured with a Zeiss Axioskop 2 Plus fluorescent microscope equipped with an AxioCam MRM digital camera. Additional brightfield images were captured with a Nikon Eclipse E400 microscope equipped with a Nikon CoolPix 990 digital camera.
For sterol staining, cells were harvested from V8 agar medium and resuspended in phosphate-buffered saline (PBS). Cells were incubated with 5 μg/ml filipin III (Cayman Chemical, Ann Arbor, MI) for 5 min, rinsed in PBS, and observed immediately. For concanavalin A (ConA)-filipin staining, overnight cultures were diluted and incubated for 1 h in either 50 μg/ml ConA Alexa Fluor 594 (α cells) or ConA Alexa Fluor 488 (a cells) (Molecular Probes, Eugene, OR). Cells were rinsed three times in PBS to remove unbound lectin, mixed, and pipetted onto V8 agar medium. After 4 h cells were harvested, counterstained with filipin, and visualized with the appropriate filters.
For filament staining, slide mounts were prepared as described previously (Fraser et al., 2003) with the following modifications. After incubation, each slide mating was transferred to a fresh petri dish, washed three times with 10 ml of PBS, fixed in 10 ml 10% formaldehyde for 20 min, washed three times with PBS, permeabilized with 1% Triton X-100 in PBS for 10 min, and washed three times with PBS before staining. To visualize septa and DNA, the slide matings were incubated in a 10-ml preparation of calcofluor (fluorescent brightener 28 F-3397; Sigma-Aldrich, St. Louis, MO) at 40 μg/ml and a 5000× dilution of a 5 mM solution of Sytox Green (Molecular Probes) in PBS for 20 min. Agar sections containing filaments were mounted on fresh microscope slides with 5 μl of antifade (1 mg/ml p-phenylenediamine in 50% glycerol). To visualize actin and DNA, similar mating preparations were fixed and incubated in a 10-ml preparation of rhodamine-conjugated phalloidin (P-195; Sigma-Aldrich) at 10 μg/ml. Agar sections containing filaments were mounted on fresh microscope slides with 5 μl of antifade mix containing 8 μg/ml 4,6-diamidino-2-phenylindole (DAPI) (D-21490; Molecular Probes).
Gene Disruption
The ura5-serotype A strain JF99 was isolated as a 5-fluoroorotic acid spontaneous mutants of KN99a. The construct for precise replacement of the serotype A STE20a open reading frame with the nourseothricin resistance cassette was generated as described previously (Davidson et al., 2002; Fraser et al., 2003) using primers to amplify STE20a 5′ (JOHE9252 AACGCTAAAGCCATGCCAACA; JOHE9253 AGCTCACATCCTCGCAGCGACGATGAAAGCCTGGTTCTAGGT) and STE20a 3′ (JOHE9254 CCGTGTTAATACAGATAAACCCCGCCTTATGATGGTATTTGTCCG; JOHE9255 CAGGTCGCTGGAATATCTTGT), with the overlap construct used to transform JF99 to create JF219 (MATa ste20a::NAT ura5). The construct for precise replacement of the serotype A PAK1 open reading frame with the nourseothricin resistance cassette was generated using primers to amplify PAK1 5′ (JOHE10447 AGGCCATCCATCACATTCCTT; JOHE10448 AGCTCACATCCTCGCAGCCCCCTTTTCTATCGCCGCGGTCGC) and PAK1 3′ (JOHE10449 CCGTGTTAATACAGATAAACCATTGGGTAGGAACGAAAACGTGGT; 110450 GCAAACTGGTCAATCGCATCT), with the overlap construct used to transform KN99a to create JF265 (MATa pak1::NAT) and JF99 to create JF267 (MATa pak1::NAT ura5). Putative deletion strains were confirmed by PCR and Southern blot analysis.
RESULTS
Pak1 Is Required for Cell Fusion during Mating in C. neoformans
Mating in C. neoformans is initiated when haploid α and a cells sense and respond to pheromone and fuse. The resulting dikaryon produces abundant dikaryotic filaments, basidia, and basidiospores (Kwon-Chung, 1976). Mating filament production is highly reduced in bilateral α pak1 × a pak1 and ste20α × ste20a mutant crosses (Wang et al., 2002). Decreased filamentation during mating could stem from a variety of reasons, including defects in cell fusion or filament morphogenesis. For example, cpk1, ste7, and ste11 MAP kinase cascade mutants all exhibit decreased filamentation during mating due to a defect in cell fusion (Davidson et al., 2003). Our previous epistasis analysis placed Ste20 upstream of the Ste11-Ste7-Cpk1 MAP kinase cascade (Wang et al., 2002). We therefore tested whether ste20 mutants exhibit a similar fusion defect.
To address this question, bilateral mutant crosses were conducted between serotype D ste20a, ste20α, a pak1, and α pak1 strains. Each strain was genetically marked to allow selection of prototrophic isolates formed by cell fusion after 48 h of incubation on mating medium. No prototrophic isolates were obtained from the bilateral pak1 mutant cross, whereas abundant prototrophic isolates were produced from both the wild-type control cross and the bilateral ste20 mutant cross (Figure 1A). Hence, Pak1 is required for cell fusion during mating but Ste20 is not. Ste20 likely functions at a later stage in mating, possibly during filament morphogenesis or hyphal elongation.
Figure 1.
Pak1 is required for fusion during mating. (A) Fusion assay of bilateral ste20 and pak1 crosses. Top, media used to select prototrophic fusion products from ste20α x ste20a (top left) and α pak1 x a pak1 (top right) crosses. Bottom, wild-type control crosses corresponding to the marker matched bilateral ste20 cross (bottom left) and the bilateral pak1 cross (bottom right). (B) MFα pheromone gene expression in wild-type α strain and α cpk1, α pak1, and ste20α strains alone, α and α cpk1 strains cocultured with wild-type a, α pak1 cocultured with a pak1, and ste20α cocultured with ste20a. Cells were incubated on V8 medium for 24 h at room temperature, RNA was prepared, and MFα gene expression was assessed by Northern blot analysis with an MFα1 gene probe. C. neoformans ACT1 was used as a loading control.
We also examined fusion in unilateral crosses between pak1 mutant and wild-type strains to determine whether one copy of PAK1 was sufficient for fusion. The number of prototrophic isolates produced from either pak1 unilateral cross was only modestly decreased compared with wild type, demonstrating that one copy of PAK1 suffices for fusion (unpublished data). This is in contrast to cpk1, ste7, and ste11 mitogen-activated protein kinase (MAPK) cascade mutants, which exhibit a fusion defect even in unilateral crosses with a wild-type mating partner (Davidson et al., 2003).
Pheromone Induction Is Normal in PAK Kinase Mutants
The MFα pheromone genes are induced in response to nutrient limitation and coculture with a cells via the Ste11-Ste7-Cpk1 MAPK cascade (Davidson et al., 2000b, 2003; Shen et al., 2002). If Pak1 were to function as the sole activator of the MAPK cascade then we would expect to observe a defect in MFα pheromone expression in α pak1 cells during coculture with a pak1 cells. MFα expression was examined by Northern blot analysis by using RNA extracted from α wild-type and α cpk1 cells cocultured with a wild-type cells and α pak1 cells cocultured with a pak1 cells. Consistent with previous observations, MFα expression was dramatically induced by 24 h when α cells were cocultured with a cells (Shen et al., 2002) (Figure 1B). MFα induction was absent in α cpk1 cells cocultured with wild-type a cells (Davidson et al., 2003), but it was induced when α pak1 cells were cocultured with a pak1 cells (Figure 1B). Wild-type levels of MFα also were observed in ste20α cells cocultured with ste20a cells (Figure 1B). In addition, MFa pheromone induction occurred normally in the bilateral pak1 and ste20 crosses (unpublished data). These results indicate that neither Pak1 nor Ste20 is required for activation of pheromone gene transcription during mating.
pak1 Mutants Fail to Polarize in Response to Pheromone
In S. cerevisiae, both α and a cells respond similarly to pheromone by arresting in G1, reorienting growth toward the pheromone gradient, and fusing with each other (Elion, 2000). In C. neoformans, α and a cells respond differently when confronted with pheromone; α cells produce conjugation tubes, whereas a cells become enlarged and refractile, and only occasionally produce conjugation tubes (Moore and Edman, 1993; Davidson et al., 2000b; Wang et al., 2000).
Previously, we demonstrated that α pak1 and a pak1 mutants failed to respond morphologically when confronted with mating pheromone, but both are able to produce and secrete pheromone (Wang et al., 2002). To address the role of the PAK kinases in pheromone-induced polarized growth, the initial morphological changes that α and a cells undergo before and during fusion were monitored using filipin, a fluorescent antibiotic that binds to 3-β-hydroxysterols (Wachtler et al., 2003). Sterols have recently been shown to be enriched in areas of polarized growth and mating projections in Schizosaccharamyces pombe and in mating projections in S. cerevisiae (Bagnat and Simons, 2002; Wachtler et al., 2003). Here, we examined the localization pattern of sterols in actively growing wild-type cells. In C. neoformans, filipin was localized to areas undergoing active polarized growth during vegetative growth: bud tips and septa (Figure 2A). Next, coculture experiments were performed with wild-type and pak1 mutant strains. Cells of opposite mating type were mixed and incubated for 4–24 h on V8 mating medium, harvested, and live cells were stained with filipin. By 4 h of incubation, morphological changes were discernable in wild-type cells (Figure 2, B–E). Whereas budding continued in some cells, the majority of cells (>50%) formed protrusions that stained intensely with filipin (Figure 2B). The protrusions elongated into tubes and grew until fusion occurred with a cell of the opposite mating type (Figure 2, C–E). Actin localized to the tips of the C. neoformans mating protrusions, as detected in samples that were fixed and stained with rhodamine-conjugated phalloidin (Figure 2G). In contrast to wild-type cells, only budding cells were observed in pak1 mutant cells at the 4-h time point (Figure 2F). Inspection of cocultured cells from later time points revealed that some fusion events did occur between α pak1 and a pak1 mutant cells, but these were less frequent and were delayed compared with those observed in cocultured wild-type cells.
Figure 2.
Morphogenic events precede fusion during mating in C. neoformans. Sterol distribution was analyzed in wild-type C. neoformans cells grown in YPD (A), or wild-type α cells cocultured with wild-type a cells (B–E), and α pak1 cells cocultured with a pak1 cells (F) on V8 medium for 4 h. Live cells were harvested, incubated for 1 min with filipin and observed immediately. By 4 h, a range of morphogenic changes were observed in the wild-type coculture, including shmoo-like projections (B), conjugation tubes (C and D), and conjugation (E). (G) C. neoformans shmoo-like projections contain polarized actin. Cells from the wild-type coculture were fixed and stained with rhodamine-conjugated phalloidin and DAPI to visualize F-actin and nuclei. (H) DIC image corresponding to G. Arrows denote shmoos and conjugation tubes. Bar, 10 μm.
To distinguish between mating types during fusion, wild-type α and a cells were differentially stained with fluorescent conjugates of ConA, a lectin that binds mannose and glucose residues. In C. neoformans, ConA localizes to septa and bud scars (Figure 3; Foster et al., 2004). Overnight cultures of α and a cells were incubated with Alexa 594 ConA (red) and Alexa Fluor 488 ConA (green) conjugates, respectively, for 1 h. Cells were washed to remove unbound lectin, and equal numbers of each mating partner were mixed and plated onto mating medium. After 4 h, the cells were harvested and counterstained with filipin. Fusion products consisting of ConA-labeled α and a cells were identified demonstrating that both mating types can produce conjugation tubes to fuse with a mating partner (Figure 3).
Figure 3.
C. neoformans conjugation tube formation. Wild-type α and a cells were pre-incubated in Alexa Fluor 594 ConA and Alexa Fluor 488 ConA, respectively, to distinguish between α and a cells during conjugation. Cells were mixed, incubated on V8 medium for 4 h, and counterstained with filipin to identify sites of conjugation between cells (arrows). Cells were observed with Texas Red (Alexa Fluor 594), fluorescein isothiocyanate (Alexa Fluor 488), and DAPI (filipin) filter sets and the images were merged. Bar, 5 μm.
To monitor nuclear migration with respect to cell fusion and postfusion events, we fixed and stained cells from 4-, 12-, and 24-h time points with DAPI. Staining revealed that by 12 h, α and a cells from the wild-type coculture had fused and formed mating filaments into which the α and a nuclei both migrated (Figure 4A); by 24 h, dikaryotic filaments were evident (Figure 4A). In contrast, mating filaments were not detected in the α pak1 × a pak1 coculture until 24 h (Figure 4A). To quantify the defect in pak1 conjugation, we counted the number of fusion products present in the wild-type and α pak1 X a pak1 mutant cocultures incubated for 24 h. On average, the wild-type coculture samples contained 3.8 ± 0.83 conjugation products per 50 cells, whereas the pak1 mutant coculture samples contained only 0.3 ± 0.26 conjugation products per 50 cells. Together, these results demonstrate that cells lacking Pak1 fail to polarize toward a mating partner in response to pheromone and, likely as a consequence, exhibit a relative fusion defect compared with wild-type cells.
Figure 4.
Fusion and spore production defect in pak1 mutants. (A) Wild-type and pak1 cocultures were incubated for 12 and 24 h on V8 medium. Cells were harvested, fixed, and stained with DAPI to visualize nuclei. DAPI and DIC images were merged to observe nuclear migration. At 12-h, nuclear migration (arrowhead) and mating filaments were detected in the wild-type cocultures. At 24 h, the initial mating filament cells had replicated to form dikaroytic filaments with clamp cells (asterisks), whereas in the pak1 coculture mating filaments were at the nuclear migration (arrowhead) stage of development. Bar, 10 μm. (B) Quantitative mating analysis. Bilateral crosses consisting of wild-type, pak1, and ste20 auxotrophic strains and prototrophic partners were prepared, incubated for 7 d on V8 medium, and serially diluted onto medium to select recombinants that were Ura- and either Lys+ (ste20 cross) or Ade+ (pak1 cross). Top, progeny from the pak1 bilateral cross (left) and the ste20 bilateral cross (right). Bottom, progeny from the wild-type control cross for pak1 (left) and ste20 (right).
pak1 Mutants Produce Few Meiotic Progeny
Our results suggest that despite a significant fusion defect, conjugation and filamentation might not be abolished in bilateral pak1 mutant crosses. To determine whether the pak1 fusion products produced functional mating filaments and completed the sexual cycle, we measured the number of viable recombinant meiotic progeny produced from wild-type and pak1 crosses. Cells from wild-type and bilateral pak1 crosses were genetically marked to permit selection of recombinant haploid progeny, mixed, incubated for 7 d on mating medium, and serially plated onto selective medium. A 200-fold reduction in the production of recombinant progeny was observed in the bilateral pak1 cross compared with the wild-type control cross (Figure 4B). Thus, α pak1 and a pak1 mutant cells exhibit a profound fusion defect and only rarely complete the sexual cycle to produce recombinant haploid meiotic progeny.
Filaments Lacking Pak1 Are Mononucleate
Although reduced compared with wild-type, bilateral pak1 crosses do produce filaments. The lack of recombinant progeny indicates that the majority of filaments produced in bilateral pak1 crosses are either dysfunctional, mononucleate, or both. Indeed, microscopic examination of the bilateral pak1 filaments revealed several abnormalities. Wild-type and bilateral pak1 crosses were incubated for 7 d on microscope slides covered in V8 agar medium. The slides were fixed and stained with calcofluor and Sytox Green to visualize clamp cells and nuclei, respectively.
Characteristic features of C. neoformans dikaryotic hypha include two paired nuclei per cell, nuclear migration into clamp cells, and fused clamp cell connections between filament cells (Figure 5A). However, DNA and cell wall staining of pak1 filaments revealed that the filament cells were mononucleate for the most part and contained many unfused clamp-cell-like or blastospore-like structures (Figure 5A). Two nuclei were occasionally observed in the leading filament cell as a result of replication of the single nucleus. Mononucleate filaments containing no or unfused clamp cells also are observed in several other physiological settings including conjugation tubes, asexual filaments (haploid fruiting), and diploid filamentous growth. Previously, we demonstrated that pak1 mutant cells are unable to undergo haploid fruiting, excluding this as a source of the mononucleate filaments observed (Wang et al., 2002). It is also unlikely that the mononucleate filaments are diploids because diploid fusion products were not recovered in our assay (Figure 1A). A more likely explanation is that the mononucleate filaments arise from pak1 cells that form mating projections but fail to fuse with a partner. In this model, Pak1 also is required for the physical fusion between α and a cells.
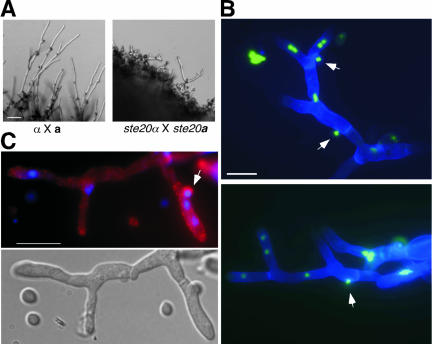
Figure 5.
α pak1 × a pak1 crosses produce mononucleate filaments. (A) Bilateral wild-type and pak1 crosses were prepared on glass slides coated with V8 agar medium and incubated for 5 d. The glass slide mounts were fixed and stained with calcofluor and Sytox Green to visualize septa, including clamp cell junctions, and DNA simultaneously. DIC images depict clamp cells in wild-type filaments and clamp cell-like protrusions in the pak1 filaments. The wild-type filaments contain two nuclei per filament cell (arrows), whereas the pak1 filaments contain one nuclei per filament cell with two exceptions: the terminal tip cell in which the single nucleus has divided before filament cell division and also multiple nuclei (asterisk) observed in the clamp cell-like protrusions. (B) Basidiospore formation is defective in pak1 mutants. Wild-type (top) and pak1 basidia and basidiospores (bottom) were stained with calcofluor to visualize septa (left). Left panels depict accompanying DIC images. Bar, 10 μm.
In C. neoformans, both dikaryotic and mononucleate (either haploid fruiting or diploid) hyphae produce basidia and basidiospores. Neither macroscopic nor microscopic examination of the bilateral pak1 filaments revealed the presence of normal basidia or basidiospores. Instead, the filaments terminated with deformed basidia that lacked basidiospores (Figures 5A and 8A) but did occasionally produce yeast-like cells or blastospores (Figure 5B). These observations suggest that Pak1 may play additional roles in basidia formation, meiosis, and basidiospore development.
Figure 8.
Ste20 and Pak1 are not interchangeable. (A) ste20α ura5 and α pak1 ura5 strains were transformed with plasmids expressing either STE20α or PAK1. Each strain also was transformed with a control plasmid. ste20 (left) and pak1 (right) bilateral crosses were prepared with each transformant and analyzed for filament production. Representative crosses were photographed at 100 and 400× (insets) magnification to visualize filamentation and basidiospore production. (B) ste20α transformants were assessed for growth at 37°C. Cells were grown overnight at 30 and 37°C and photographed at 1000× magnification with DIC optics. At 37°C ste20α mutants bearing vector alone or the PAK1 plasmid exhibited a cytokinesis defect, resulting in long chains of connected and elongated cells. Introduction of the STE20α gene complemented the cytokinesis defect, resulting in a wild-type budding yeast cell morphology.
Ste20 Is Required to Maintain Polarity
In contrast to the few filaments produced by bilateral pak1 crosses, bilateral ste20 mutant crosses produced abundant yet deformed filamentous structures that were rapidly over-grown by vegetative cells. Microscopic inspection revealed these filaments are much more branched than wild-type filaments (Figure 6A). Like other filamentous fungi, wild-type C. neoformans hyphae produce branches or secondary filaments that originate from subapical cells in the main filament (Figure 6A). In addition, branches also can form from the clamp cells that link adjacent filament cells (Kwon-Chung, 1976). To determine the origin of the branches in the ste20 mutant crosses, we stained the septa and cell walls of wild-type and bilateral ste20 filaments with calcofluor (Figures 5A and 6B). Staining revealed that the unusual branches in the ste20 mutant filaments did not originate from the clamp cells but were caused by the growing tip splitting in half (Figure 6B). Clamp cells were produced but were limited to one of the two branches, and nuclear costaining revealed that the filament cells contained unequal numbers of nuclei as a result. Whereas some filament cells contained two nuclei, others contained one, three, or zero nuclei (Figure 6B).
Figure 6.
Ste20 is required to maintain filament tip polarity. (A) Wild-type and bilateral ste20 crosses were incubated on V8 mating medium for 5 d at 24°C and photographed at 200× magnification with DIC optics. Wild-type filaments contain numerous lateral branches, whereas the ste20 filaments branch at the tip. (B) Glass slide mounts of bilateral ste20 mating filaments were costained with calcofluor and Sytox Green to visualize septa and nuclei. Each filament cell branches, leading to random segregation of nuclei. Whereas some filament cells maintain a dikaryotic state, other cells are mononucleate or anucleate. (C) Glass slide mounts of bilateral ste20 mating filaments were costained with rhodamine-conjugated phalloidin and DAPI to visualize actin and nuclei (top) and visualized with DIC optics (bottom). Arrows denote clamp cells containing nuclei. Bar, 10 μm.
Although not previously described in C. neoformans filaments, tip splitting has been described in Aspergillus nidulans polarity-maintenance mutants as dichotomous branching (Momany, 2002). Actin localizes as a cap at the tip of A. nidulans filaments and dichotomous branching occurs when the actin cap is perturbed or displaced, creating an additional axis of polarity. To determine whether actin localization was similarly perturbed in the filaments produced by a ste20α × ste20a bilateral cross, actin in wild-type and ste20 mutant filaments was stained with rhodamine-conjugated phalloidin. In wild-type filaments actin was localized in cortical patches and filaments along the length of the filament cell and was concentrated at the growing tip and in clamp cells, both areas of active polarized growth (Figure 7A). Transient actin rings also were observed at sites of septation between filament cells and at clamp connections (Figure 7, B and C). Comparison of wild-type and the ste20 mutant filaments revealed no overall difference in actin polarization (Figure 6C).
Figure 7.
Localization of actin in C. neoformans mating filaments. Glass slide mounts of wild-type mating filaments were costained with rhodamine-conjugated phalloidin and DAPI to visualize actin and nuclear dynamics during filamentous growth. (A) Actin patches are polarized to the tip of the growing dikaryotic filament and to the tip of the clamp cell (arrowheads). (B) Actin rings form at the site of septation during nuclear division. Nuclear migration has already occurred, separating one pair of duplicated nuclei (arrows) on either side of the actin ring, whereas one nucleus of the second duplicated pair prepares to migrate into the clamp cell (asterisk). (C) Actin rings also form at the clamp cell to separate the nucleus in the clamp cell from the terminal filament. Also depicted here is nuclear migration into the basidium (asterisk).
By microscopic examination, no normal basidia or basidiospores were identified in the bilateral ste20 cross. Instead, the basidia were deformed and either devoid of basidiospores or contained only a few (4 to 8) basidiospores (Figure 8A). Nuclear staining revealed that these basidiospores contain DNA, suggesting that they may be viable (unpublished data). To determine this, we measured the production of recombinant meiotic progeny in bilateral ste20 mutant crosses. Wild-type and ste20 bilateral mutant crosses were incubated for 7 d on mating medium. Each strain was genetically marked to allow for the selection of recombinant progeny. Whereas recombinant colonies were isolated from the bilateral ste20 cross, there was a fivefold reduction compared with a similarly marked auxotrophic wild-type cross (Figure 4B). Thus, bilateral ste20 crosses produce viable recombinant progeny although at a reduced rate. Together, these results indicate that Ste20 is required to maintain filament polarity and that loss of polarity leads to a reduction in the number of viable basidia and basidiospores.
C. neoformans PAK Kinases Are Not Interchangeable
In S. cerevisiae, Ste20 and Cla4 have overlapping functions and can substitute for each other in certain situations. For example, overexpression of Cla4 can suppress the mating and osmosensitivity defects associated with ste20 mutant strains (Sells et al., 1998; Raitt et al., 2000). A similar situation has been described in fission yeast Schizosaccharomyces pombe; overexpression of the Cla4 homolog Pak2/Shk2 suppresses the morphology and mating defects associated with deletion of the Ste20 homolog Pak1/Shk1 (Sells et al., 1998; Yang et al., 1998).
To determine whether overexpression of C. neoformans Pak1 can suppress the mating and morphology defects of the ste20 mutant or vice versa, we performed bilateral mating assays using α mutant strains overexpressing either the STE20α or the PAK1 gene. Filamentation equivalent to wild type was restored in control crosses (α pak1 + PAK1 × a pak1 and ste20α + STE20α × ste20a) (Figure 8A and Table 2). However, overexpression of PAK1 in the ste20α mutant strain did not restore filamentation in a bilateral ste20 cross, nor did STE20α restore filamentation in a bilateral pak1 cross when overexpressed in an α pak1 mutant strain (Figure 8A and Table 2). We also examined the ability of Pak1 to suppress the cytokinesis defects exhibited by ste20 mutant strains. Whereas overexpression of STE20α complemented the cytokinesis defect of the ste20α mutant strain, overexpression of PAK1 did not (Figure 8B and Table 2). Together, these results indicate that the C. neoformans PAK kinases play functionally specialized roles in mating and morphology.
Table 2.
Overexpression analysis
| Mutant | Gene | Restore filamentation? (bilateral cross)a | Suppress cytokinesis defect? |
|---|---|---|---|
| D ste20α | Vector | No | No |
| D STE20α | Yes | Yes | |
| D STE20a | Yes | Yes | |
| A STE20α | Yes | Yes | |
| A STE20a | Yes | Yes | |
| D PAK1 | No | No | |
| D ste20a | Vector | No | No |
| D STE20α | Yes | Yes | |
| D STE20a | Yes | Yes | |
| A STE20α | Yes | Yes | |
| A STE20a | Yes | Yes | |
| D PAK1 | No | No | |
| A ste20α | Vector | No | No |
| D STE20α | Yes | Yes | |
| D STE20a | Yes | Yes | |
| A STE20α | Yes | Yes | |
| A STE20a | Yes | Yes | |
| D PAK1 | No | No | |
| A ste20a | Vector | No | No |
| D STE20α | Yes | Yes | |
| D STE20a | Yes | Yes | |
| A STE20α | Yes | Yes | |
| A STE20a | Yes | Yes | |
| D PAK1 | No | No | |
| D α pak1 | Vector | No | NA |
| D STE20α | No | NA | |
| D STE20a | No | NA | |
| D PAK1 | Yes | NA | |
| A PAK1 | No | NA | |
| A α pak1 | Vector | No | NA |
| A STE20α | No | NA | |
| A STE20a | No | NA | |
| D PAK1 | No | NA | |
| A PAK1 | Yes | NA |
NA, not applicable.
With the exception of serotype A α pak1, which was crossed to wild-type a
Ste20 Function Is Mating-Type Independent
In contrast to a ste20 bilateral cross, unilateral ste20α × a and ste20a × α crosses produce abundant filaments with no morphology defects, indicating that one copy of either gene is sufficient. To determine whether there was any mating-type specificity associated with this Ste20 function, we overexpressed STE20α in a ste20a mutant and STE20a in a ste20α mutant. In bilateral ste20 crosses, Ste20α restored normal filament morphology when overexpressed in either a ste20α or a ste20a mutant strain (Table 2). Similarly, Ste20a restored filament morphology when overexpressed in either a ste20a or a ste20α mutant strain (Table 2). In addition, overexpression of Ste20α complemented the cytokinesis defect of the ste20a mutant strain and vice versa. These data indicate that, at least under these experimental conditions, Ste20α and Ste20a perform identical roles during vegetative growth and are redundant during mating. Thus, the functions of Ste20 seem independent of mating type even though the STE20 genes are located in the MAT locus and function in mating.
Ste20 Function Is Serotype Independent
Serotype is correlated with the ability of C. neoformans to cause disease. In the clinic, cryptococcosis caused by serotype A strains predominates over cases involving serotype B, C, or D strains. Recent analyses have shown that certain gene products also have a serotype-dependent impact on C. neoformans virulence. For example, the Ste12 transcription factor is important for virulence in a serotype D strain but not in a serotype A strain (Yue et al., 1999). Conversely, Ste20 is required for virulence in serotype A but not in serotype D. In addition, whereas both serotype A and D ste20 mutants exhibit defects in cytokinesis and mating filament polarity, only serotype A ste20α mutants are temperature sensitive for growth (Wang et al., 2002).
To address the role of Ste20 serotype specificity, we generated a serotype A ste20a mutant by disrupting STE20a in the serotype A MATa strain KN99a (Nielsen et al., 2003). Our previous analysis of Ste20 in serotype A was limited to ste20α due to the unavailability of a serotype A MATa strain. The serotype A ste20a mutant behaved exactly as the serotype A ste20α mutant with respect to cytokinesis and high-temperature growth (unpublished data) (Wang et al., 2002). In addition, the few filaments produced in a bilateral serotype A ste20 cross exhibited defects similar to the filaments produced in a bilateral serotype D ste20 cross (Figure 9). Next, we overexpressed the serotype A STE20α gene in the serotype D ste20α mutant and the serotype D STE20α gene in the serotype A ste20α mutant. When overexpressed, serotype D Ste20α complemented the cytokinesis defect, high-temperature growth defect, and mating filamentation defect of the serotype A ste20α mutant (Table 2). Similarly, serotype A Ste20α overexpression complemented the cytokinesis and mating filamentation defects of the serotype D ste20α mutant (Table 2). Identical results were obtained using serotype A and D STE20a genes (Table 2). Additionally, overexpression of serotype A STE20α complemented the serotype D ste20a mutant (summarized in Table 2). In summary, Ste20 function is independent of both serotype and mating type.
Figure 9.
Serotype A bilateral ste20 and pak1 crosses exhibit filamentation defects. Serotype A ste20a and a pak1 strains were each crossed to wild-type α, ste20α, and α pak1 strains. Few or no filaments were observed in the mutant bilateral crosses and in crosses between ste20 and pak1 mutants. Filamentation also was reduced in the pak1 unilateral crosses but the few basidia and basidiospores that were produced were normal morphologically (insets). Representative crosses were photographed at 100 and 400× (insets) magnification.
Pak1 Function Is Serotype Dependent
We also examined whether Pak1 could function between the divergent serotypes. To this end, we first attempted to generate a serotype A a pak1 mutant by crossing the serotype A α pak1 mutant with the congenic mating partner KN99a. However, in contrast to serotype D pak1 unilateral crosses, very few filaments were produced (Figure 9). In an alternative approach, we generated a serotype A a pak1 mutant strain by targeted gene disruption. Similar to a serotype A α pak1 unilateral cross, few filaments were produced when the serotype A a pak1 mutant was crossed to a wild-type α strain (Figure 9). No filamentation was observed in the serotype A bilateral pak1 cross (Figure 9). Interestingly, no filamentation was observed in crosses between ste20α and a pak1 mutants or between α pak1 and ste20a mutants (Figure 9).
Overexpression of serotype A PAK1 in either α or a A pak1 mutants restored filamentation in unilateral crosses (Table 2). However, overexpression of the serotype A PAK1 gene in the serotype D pak1 mutant did not restore filamentation to a serotype D pak1 bilateral cross (Table 2). Similar results were obtained with serotype D PAK1; overexpression of serotype D PAK1 complemented the serotype D pak1 mutant but not the serotype A pak1 mutant (Table 2). Thus, in contrast to Ste20, Pak1 function is serotype specific.
DISCUSSION
In this study, we find that Ste20 and Pak1 have polarity specific roles in C. neoformans mating. Our studies have revealed that the C. neoformans PAK kinases have both conserved and unique roles. Whereas Pak1 is required for fusion, a role similar to that performed by S. cerevisiae Ste20, C. neoformans Ste20 performs a novel role in maintaining polarity in the filament. In addition, the morphogenic roles of each kinase seem to be nonoverlapping and temporally distinct from one another.
A PAK Kinase for Cell Fusion
We find that in response to pheromone, C. neoformans α and a cells generate mating projections and conjugation tubes and that Pak1 is required for this response. Mating projections and conjugation tubes occur by 4 h, the same time that pheromone transcript levels increase (Shen et al., 2002; Chang et al., 2003). In the basidiomycete Ustilago maydis, pheromone gene induction and conjugation tube formation also coincide and are regulated by a pheromone-responsive MAP kinase cascade (Muller et al., 2003). The C. neoformans pheromone-responsive MAP kinase pathway, composed of Ste11, Ste7, and Cpk1, is required for pheromone gene induction and cell fusion (Davidson et al., 2000b, 2003). In S. cerevisiae the PAK kinase Ste20 activates the pheromone-responsive MAP kinase cascade which in turn induces the processes required for mating, including shmoo formation and cell fusion (Elion, 2000). In U. maydis, the Ste20 homolog Smu1 has been recently characterized and smu1 mutant strains exhibit mating defects and decreased pheromone expression (Smith et al., 2004). C. neoformans Pak1 is most closely related to S. cerevisiae Ste20. However, our data show that pheromone induction is not compromised in pak1 mutants, indicating that the fusion defect is either independent of MAP kinase cascade activation or that Pak1 functions downstream of the MAP kinase cascade, or plays a redundant role with other elements that activate MAP kinase signaling.
Why then do pak1 mutants fail to fuse? In S. cerevisiae, Ste20 has additional roles in bud and shmoo morphogenesis. Polarization at the bud tip is maintained by a complex of proteins called the polarisome. This complex consists of Spa1, Pea2, Bud6, and Bni1. Ste20 is thought to activate the polarisome via phosphorylation of Bni1, a formin homology protein and a Cdc42 effector (Goehring et al., 2003). Polarisome components, including Bni1, are required for shmoo formation and cell fusion (Gehrung and Snyder, 1990; Chenevert et al., 1994; Dorer et al., 1997; Evangelista et al., 1997; Gammie et al., 1998). Our findings are consistent with a model that C. neoformans Pak1 induces mating projection formation by activating polarisome components.
Origin of pak1 Filaments
The filaments produced in bilateral pak1 crosses seem similar to the conjugation tubes produced by haploid or diploid cells (unpublished data). Typically, fusion occurs only between α and a cells, leading to the production of a dikaroytic filament, basidia, and basidiospores. However, filamentation is thermally repressed and fusion products incubated at 37°C undergo nuclear fusion and become diploid. When grown at 24°C these diploids reenter the sexual cycle and form monokaroytic filaments, basidia, and basidiospores. Also, haploid cells can reproduce asexually (haploid fruiting) under nutrient-limiting conditions by forming monokaryotic filaments. Common to all three processes is the ability of haploid cells to form conjugation tubes during nutrient-limiting conditions, suggesting that this may be a default mechanism. A possible explanation for this model is given by the appearance of mononucleate filaments in pak1 crosses. The pak1 mutant cells are unable to respond to pheromone and identify a mating partner, and as a consequence form long conjugation tubes that mature into mononulceate filaments. Our data support this interpretation, but other possibilities also can be considered. For example, pak1 filaments may originate from cell fusion but are unable to maintain both nuclei, either as a diploid or as a dikaryon, thus resulting in mononucleate filaments.
In contrast to mononucleate filaments produced by haploid fruiting or diploid filaments, pak1 mutant filaments do not produce normal basidia or basidiospores; either no basidiospores are formed on the basidia or budding cells are produced instead. Little is known about C. neoformans basidia and basidiospore development. In other filamentous fungi, asexual spores form from a specialized structure called a conidiophore. Polarity mutants defective in spore and conidiophore formation have been identified and include a Cla4 homolog from the rice blast fungus Magnaporthe grisea, a septin homolog from Aspergillus nidulans, a Rac homolog from the human pathogen Penicillium marneffei, and a Ras homolog from the alfalfa pathogen Colletotrichum trifolii (Truesdell et al., 1999; Westfall and Momany, 2002; Boyce et al., 2003; Li et al., 2004). Interestingly, both Rac and Ras are signaling components known to function in mediating PAK kinase activation (Daniels and Bokoch, 1999; Pruyne and Bretscher, 2000).
A PAK Kinase for Polarity Maintenance
The tip-branching defect exhibited by ste20 mutants during filamentous growth implicates Ste20 in hyphal tip maintenance. PAK kinase homologues from filamentous fungi have been characterized and play roles in filament establishment and hyphal maturation; however, none has been implicated in hyphal tip maintenance (Leberer et al., 1996; Leberer et al., 1997; Ayad-Durieux et al., 2000; Szabo, 2001; Smith et al., 2004). Proteins involved in tip maintenance have been identified in P. marneffei, A. nidulans, and Neurospora crassa. Not surprisingly, some of the genes corresponding to the tip-branching mutants encode cytoskeletal-related proteins, including the Rac homolog CflA (P. marneffei), actin (N. crassa), and a Bni1 homolog SepA (A. nidulans) (Sharpless and Harris, 2002; Seiler and Plamann, 2003; Virag and Griffiths, 2004). Because Bni1 homologues are involved in both tip splitting and shmoo formation, albeit in different organisms, it raises the intriguing possibility that a C. neoformans formin protein may be a common target for both Pak1 and Ste20 during morphogenesis. Although no formin proteins have been identified in C. neoformans, our BLAST searches of C. neoformans sequence databases have revealed that formin domain sequences are present.
In C. neoformans, α and a nuclei must be segregated to maintain the dikaryotic filament. In contrast, P. marneffei, A. nidulans, and N. crassa filaments contain multiple nuclei per filament cell (Coppin et al., 1997). Thus, in C. neoformans, tip-splitting has a unique and detrimental impact on nuclear segregation during filamentous growth that affects subsequent basidiospore formation and viability, and Ste20 has a pivotal role in this process.
Dosage Dependency and Serotype Specificity of PAK Kinases
During our analysis we found that a single copy of PAK1 was sufficient for function, and this also held true for STE20, where a single copy of either MAT variant would suffice for mating to occur. This redundancy of STE20 function, specifically the lack of discrimination between the mating-type–specific alleles of the gene, was highly unexpected. It is thought that the products of the MAT locus contribute to α- and a-specific signaling pathways controlling reproduction and virulence (Hull and Heitman, 2002; Fraser and Heitman, 2003). The finding that the allelic components of MAT may be interchangeable between mating types without alteration of phenotype implies that despite the large cohort of genes present in this structure, the different MAT alleles may be functionally equivalent with the exception of those components considered the most ancient; the homeodomain protein Sxi1α (Hull et al., 2002) and the pheromones/pheromone receptors (Chang et al., 2003).
Also puzzling is the serotype specificity of Pak1 compared with Ste20. Serotype A and D Ste20α and Pak1 are 94 and 90% identical at the amino acid level, respectively. However, serotype A Pak1 contains several short insertions not shared by serotype D Pak1. Population genetic studies show that C. neoformans A and D serotypes diverged ∼18 million years thus it is likely that the serotype A and D Pak1 homologues have acquired unique functions during mating (Fan et al., 1994; Xu et al., 2000). However, the two different STE20 alleles of each serotype have clearly been diverging for a much longer time due to their presence in the nonrecombining MAT locus. In contrast to the divergence between the serotype alleles of each gene, at the nucleotide level the coding sequences of the MATa and MATα alleles of STE20 are only 66% identical. The divergence of function of the PAK1 genes between serotype A and D is therefore all the more exceptional; the function has changed dramatically over the past ∼20 million years, where the STE20 genes (which are much more divergent) each seem to encode identical functions.
Cell Polarity and Virulence
One goal of our study was to gain insight into the connections between mating and virulence in C. neoformans. In U. maydis, the hyphal form is pathogenic and the sexual cycle occurs in the host thus there is a clear connection between mating and virulence. For example, mutations in components of the pheromone responsive MAP kinase cascade or the Ste20 homolog Smu1 exhibit reduced virulence (Mayorga and Gold, 1999; Muller et al., 1999, 2003; Smith et al., 2004). In contrast, there is no evidence that mating or filamentation occurs during cryptococcal infections. However, there is a link between mating and virulence and many of the gene products found to impact virulence also are required for mating. In the specific case of Ste20, the link between mating and virulence may result from the temperature-sensitive growth defect exhibited by serotype A ste20 mutants. Serotype D ste20 mutants exhibit a cytokinesis defect, but they are not temperature sensitive for growth and exhibit no virulence defect (Wang et al., 2002).
In contrast, pak1 mutants are not temperature sensitive nor do they exhibit defects in any known virulence factor. Why then are pak1 mutants avirulant in animal models of cyptococcosis? Within the host, C. neoformans cells encounter a combination of stresses, including high temperature, limiting nutrients, and high osmolarity. Our results indicate that Pak1 plays a positive role establishing polarity during mating. During stress, Pak1 also may be required for bud morphogenesis or some other essential function. However, pak1 mutant strains do not seem to be stress sensitive and are slightly more resistant to salt stress than wild-type strains incubated at 37°C (unpublished data). In conclusion, our studies reveal an intriguing association between cell polarity mechanisms and virulence in the human pathogen C. neoformans. Further characterization of Pak1 and Ste20 and the identification of their downstream targets will be necessary to elucidate the connections between mating, cell polarity, and virulence.
Acknowledgments
We thank Andrew Alspaugh, Toshiaki Harashima, and Peter Kraus for critical reading of the manuscript and Marie-Josee Boily for technical assistance. These studies were supported in part by R01 grants AI39115 and AI50113 from the National Institute of Allergy and Infectious Diseases and by P01 grant AI44975 from the National Institute of Allergy and Infectious Diseases to the Duke University Mycology Research Unit. J.H. is an associate investigator of the Howard Hughes Medical Institute and a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–05–0370. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–05–0370.
References
- Alspaugh, J.A., Cavallo, L.M., Perfect, J.R., and Heitman, J. (2000). RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36, 352-365. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1992). Current Protocols in Molecular Biology, New York: Greene Publishing Associates and Wiley-Interscience.
- Ayad-Durieux, Y., Knechtle, P., Goff, S., Dietrich, F., and Philippsen, P. (2000). A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113, 4563-4575. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., and Simons, K. (2002). Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99, 14183-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce, K.J., Hynes, M.J., and Andrianopoulos, A. (2003). Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J. Cell Sci. 116, 1249-1260. [DOI] [PubMed] [Google Scholar]
- Chang, Y.C., Miller, G.F., and Kwon-Chung, K.J. (2003). Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect. Immun. 71, 4953-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert, J., Valtz, N., and Herskowitz, I. (1994). Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics 136, 1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin, E., Debuchy, R., Arnaise, S., and Picard, M. (1997). Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61, 411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F., De Virgilio, C., Manser, E., Pringle, J.R., and Nasmyth, K. (1995). Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9, 1817-1830. [DOI] [PubMed] [Google Scholar]
- Daniels, R.H., and Bokoch, G.M. (1999). p21-Activated protein kinase: a crucial component of morphological signaling? Trends Biochem. Sci. 24, 350-355. [DOI] [PubMed] [Google Scholar]
- Davidson, R.C., Blankenship, J.R., Kraus, P.R., de Jesus Berrios, M., Hull, C.M., D'Souza, C., Wang, P., and Heitman, J. (2002). A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148, 2607-2615. [DOI] [PubMed] [Google Scholar]
- Davidson, R.C., Cruz, M.C., Sia, R.A., Allen, B., Alspaugh, J.A., and Heitman, J. (2000a). Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29, 38-48. [DOI] [PubMed] [Google Scholar]
- Davidson, R.C., Moore, T.D., Odom, A.R., and Heitman, J. (2000b). Characterization of the MFalpha pheromone of the human fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 38, 1017-1026. [DOI] [PubMed] [Google Scholar]
- Davidson, R.C., Nichols, C.B., Cox, G.M., Perfect, J.R., and Heitman, J. (2003). A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49, 469-485. [DOI] [PubMed] [Google Scholar]
- Dobbelaere, J., Gentry, M.S., Hallberg, R.L., and Barral, Y. (2003). Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345-357. [DOI] [PubMed] [Google Scholar]
- Dorer, R., Boone, C., Kimbrough, T., Kim, J., and Hartwell, L.H. (1997). Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics 146, 39-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion, E.A. (2000). Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3, 573-581. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M.S., Chow, C.J., Adames, N., Pringle, J.R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118-122. [DOI] [PubMed] [Google Scholar]
- Foster, A.J., Bird, R.A., Kelly, S.L., Nishimura, K., Poyner, D., Taylor, S., and Smith, S.N. (2004). FITC-lectin avidity of Cryptococcus neoformans cell wall and capsular components. Mycologia 96, 1-8. [PubMed] [Google Scholar]
- Fraser, J.A., and Heitman, J. (2003). Fungal mating-type loci. Curr. Biol. 13, R792-795. [DOI] [PubMed] [Google Scholar]
- Fraser, J.A., Subaran, R.L., Nichols, C.B., and Heitman, J. (2003). Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2, 1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A.E., Brizzio, V., and Rose, M.D. (1998). Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9, 1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung, S., and Snyder, M. (1990). The SPA2gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 111, 1451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring, A.S., Mitchell, D.A., Tong, A.H., Keniry, M.E., Boone, C., and Sprague, G.F., Jr. (2003). Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell 14, 1501-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofken, T., and Schiebel, E. (2002). A role for cell polarity proteins in mitotic exit. EMBO J. 21, 4851-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly, S.P., and Blumer, K.J. (1999). PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147, 845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, C.M., and Heitman, J. (2002). Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36, 557-615. [DOI] [PubMed] [Google Scholar]
- Hull, C.M., Davidson, R.C., and Heitman, J. (2002). Cell Identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1a. Genes Dev. 16, 3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., Geymonat, M., Johnson, A.L., Segal, M., and Johnston, L.H. (2002). Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 115, 4977-4991. [DOI] [PubMed] [Google Scholar]
- Johnson, D.I. (1999). Cdc 42, an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63, 54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung, K.J. (1976). Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68, 821-833. [PubMed] [Google Scholar]
- Kwon-Chung, K.J., and Bennett, J.E. (1978). Distribution of alpha and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108, 337-340. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung, K.J., and Bennett, J.E. (1992). Cryptococcosis. In: Medical Mycology, Malvern, PA: Lea & Febiger, 397-446.
- Kwon-Chung, K.J., Edman, J.C., and Wickes, B.L. (1992). Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60, 602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer, E., Harcus, D., Broadbent, I.D., Clark, K.L., Dignard, D., Ziegelbauer, K., Schmidt, A., Gow, N.A., Brown, A.J., and Thomas, D.Y. (1996). Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93, 13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer, E., Ziegelbauer, K., Schmidt, A., Harcus, D., Dignard, D., Ash, J., Johnson, L., and Thomas, D.Y. (1997). Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7, 539-546. [DOI] [PubMed] [Google Scholar]
- Lengeler, K.B., Fox, D.S., Fraser, J.A., Allen, A., Forrester, K., Dietrich, F.S., and Heitman, J. (2002). Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1, 704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Xue, C., Bruno, K., Nishimura, M., and Xu, J. (2004). Two PAK kinase genes, CHM1 and MST1 have distinct functions in Magnaporthe grisea. Mol. Plant-Microbe Interact. 17, 547-556. [DOI] [PubMed] [Google Scholar]
- Mayorga, M.E., and Gold, S.E. (1999). A MAP kinase encoded by the ubc3gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34, 485-497. [DOI] [PubMed] [Google Scholar]
- Mitchell, T.G., and Perfect, J.R. (1995). Cryptococcosis in the era of AIDS–100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8, 515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5, 580-585. [DOI] [PubMed] [Google Scholar]
- Moore, T.D., and Edman, J.C. (1993). The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13, 1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P., Aichinger, M., Feldbrugge, and Kahmann, R. (1999). The M.A.P. kinase kpp2regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34, 1007-1017. [DOI] [PubMed] [Google Scholar]
- Muller, P., Weinzierl, G., Brachmann, A., Feldbrugge, M., and Kahmann, R. (2003). Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2, 1187-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K., Cox, G.M., Wang, P., Toffaletti, D.L., Perfect, J.R., and Heitman, J. (2003). Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 71, 4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect, J.R., Lang, S.D.R., and Durack, D.T. (1980). Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101, 177-194. [PMC free article] [PubMed] [Google Scholar]
- Pitkin, J.W., Panaccione, D.G., and Walton, J.D. (1996). A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142, 1557-1565. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000). Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113, 365-375. [DOI] [PubMed] [Google Scholar]
- Raitt, D.C., Posas, F., and Saito, H. (2000). Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19, 4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Schafer, W. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schmidt, M., Varma, A., Drgon, T., Bowers, B., and Cabib, E. (2003). Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell 14, 2128-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., and Plamann, M. (2003). The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells, M.A., Barratt, J.T., Caviston, J., Ottilie, S., Leberer, E., and Chernoff, J. (1998). Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J. Biol. Chem. 273, 18490-18498. [DOI] [PubMed] [Google Scholar]
- Seshan, A., Bardin, A.J., and Amon, A. (2002). Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12, 2098-2110. [DOI] [PubMed] [Google Scholar]
- Sharpless, K.E., and Harris, S.D. (2002). Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13, 469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.C., Davidson, R.C., Cox, G.M., and Heitman, J. (2002). Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell 1, 366-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Smith, D.G., Garcia-Pedrajas, M.D., Hong, W., Yu, Z., Gold, S.E., and Perlin, M.H. (2004). An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot. Cell 3, 180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed, B., and Dunt, D. (1995). Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21, 28-34; discussion 35-26. [DOI] [PubMed] [Google Scholar]
- Stephen, C., Lester, S., Black, W., Fyfe, M., and Raverty, S. (2002). Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 43, 792-794. [PMC free article] [PubMed] [Google Scholar]
- Szabo, R. (2001). Cla4 protein kinase is essential for filament formation and invasive growth of Yarrowia lipolytica. Mol. Genet. Genomics 265, 172-179. [DOI] [PubMed] [Google Scholar]
- Toffaletti, D.L., Rude, T.H., Johnston, S.A., Durack, D.T., and Perfect, J.R. (1993). Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175, 1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell, G.M., Jones, C., Holt, T., Henderson, G., and Dickman, M.B. (1999). A Ras protein from a phytopathogenic fungus causes defects in hyphal growth polarity, and induces tumors in mice. Mol. Gen. Genet. 262, 46-54. [DOI] [PubMed] [Google Scholar]
- Versele, M., and Thorner, J. (2004). Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164, 701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag, A., and Griffiths, A.J. (2004). A mutation in the Neurospora crassa actin gene results in multiple defects in tip growth and branching. Fungal. Genet. Biol. 41, 213-225. [DOI] [PubMed] [Google Scholar]
- Wachtler, V., Rajagopalan, S., and Balasubramanian, M.K. (2003). Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 116, 867-874. [DOI] [PubMed] [Google Scholar]
- Wang, P., Nichols, C.B., Lengeler, K.B., Cardenas, M.E., Cox, G.M., Perfect, J.R., and Heitman, J. (2002). Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot. Cell 1, 257-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Perfect, J.R., and Heitman, J. (2000). The G-protein beta subunit GPB1is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20, 352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E.L., Bishop, A.C., Shokat, K.M., and Drubin, D.G. (2000). Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2, 677-685. [DOI] [PubMed] [Google Scholar]
- Westfall, P.J., and Momany, M. (2002). Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and condioiophore development. Mol. Biol. Cell 13, 110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes, B.L. (2002). The role of mating type and morphology in Cryptococcus neoformans pathogenesis. Int. J. Med. Microbiol. 292, 313-329. [DOI] [PubMed] [Google Scholar]
- Yang, P., Kansra, S., Pimental, R.A., Gilbreth, M., and Marcus, S. (1998). Cloning and characterization of shk2, a gene encoding a novel p21-activated protein kinase from fission yeast. J. Biol. Chem. 273, 18481-18489. [DOI] [PubMed] [Google Scholar]
- Yue, C., Cavallo, L.M., Alspaugh, J.A., Want, P., Cox, G.M., Perfect, J.R., and Heitman, J. (1999). The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153, 1601-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]