Abstract
The mouse vascular smooth muscle α-actin (SMA) gene enhancer is activated in fibroblasts by transforming growth factor β1 (TGFβ1), a potent mediator of myofibroblast differentiation and wound healing. The SMA enhancer contains tandem sites for the Sp1 transcriptional activator protein and Purα and β repressor proteins. We have examined dynamic interplay between these divergent proteins to identify checkpoints for possible control of myofibroblast differentiation during chronic inflammatory disease. A novel element in the SMA enhancer named SPUR was responsible for both basal and TGFβ1-dependent transcriptional activation in fibroblasts and capable of binding Sp1 and Pur proteins. A novel Sp1:Pur:SPUR complex was dissociated when SMA enhancer activity was increased by TGFβ1 or Smad protein overexpression. Physical association of Pur proteins with Smad2/3 was observed as was binding of Smads to an upstream enhancer region that undergoes DNA duplex unwinding in TGFβ1-activated myofibroblasts. Purβ repression of the SMA enhancer could not be relieved by TGFβ1, whereas repression mediated by Purα was partially rescued by TGFβ1 or overexpression of Smad proteins. Interplay between Pur repressor isoforms and Sp1 and Smad coactivators may regulate SMA enhancer output in TGFβ1-activated myofibroblasts during episodes of wound repair and tissue remodeling.
INTRODUCTION
Myofibroblasts arise as a consequence of tissue injury and acquire many of the contractile proteins normally expressed by smooth muscle cells such as vascular smooth muscle α-actin (SMA). These stromal cells typically contain numerous smooth muscle actin stress fibers that contribute to the generation of isometric mechanical force required for wound contraction and closure (Skalli and Gabbiani, 1988; Desmoulière and Gabbiani, 1996; Powell et al., 1999; Serini and Gabbiani, 1999; Hinz et al., 2001; Tomasek et al., 2002). Chronic myofibroblast activation has been associated with a number of pathological conditions associated with tissue remodeling, including hypertrophic scarring, interstitial fibrosis, fibromatosis, and stromal responses to certain neoplasias (Ronnov-Jessen et al., 1995; Weber, 1995; Shi et al., 1996; Zalewski and Shi, 1997; Nedelec et al., 2001; Cassiman et al., 2002). Studies on accepted heart grafts in the mouse showed that chronic allograft dysfunction (CAD), a leading cause of heart failure after transplantation, was associated with peritransplant SMA gene activation in cardiac myofibroblasts (Subramanian et al., 2002). Subsequent inappropriate elevation of SMA expression in cardiomyocytes, a sign of physiological stress (Black et al., 1991), and silencing of the SMA gene in coronary arterial smooth muscle cells was temporally linked to myofibroblast activation, fibrosis, and transplant vascular sclerosis that limits perfusion and compromises graft survival (Armstrong et al., 1997a,b,c; Subramanian et al., 1998).
Transforming growth factor β1 (TGFβ1) is an essential mediator of myofibroblast differentiation that contributes to the pathobiology of chronic fibrotic disease (Heldin et al., 1997; Derynck et al., 1998; Massague and Wotton, 2000). Reports not only demonstrate a key role for TGFβ1 in regulating extracellular matrix homeostasis (Fan et al., 1999; Branton and Kopp, 1999) but also point to its activating effect on the SMA gene required for stress fiber assembly and generation of tensile force in myofibroblasts (Ronnov-Jessen and Petersen, 1993; Serini et al., 1998). TGFβ1 binds a transmembrane receptor complex and activates Smad coregulatory proteins that are transported to the nucleus to form multisubunit transcriptional regulatory complexes (Massague and Wotton, 2000). Partner proteins that are cell type- and gene-specific interact with rate-limiting Smads to define the nature and duration of the transcriptional response. Such proteins may uniquely govern dynamic interplay between ubiquitous DNA-binding proteins that occupy closely positioned sites within the SMA enhancer. In this capacity, understanding the molecular mechanisms regulating the interactions of Smads and their partner proteins at the SMA enhancer in myofibroblasts will be crucial to understanding aberrant fibroproliferative processes associated with organ rejection after transplant.
Previous studies from our laboratory demonstrated that the SMA gene in stromal fibroblasts was regulated by an array of positive and negative cis-acting elements within a minimal enhancer required for basal and TGFβ1-inducible transcription (Kelm et al., 1996, 1999a,b, 2003; Carlini et al., 2002; Cogan et al., 2002). Whereas TGFβ1 amplified basal expression of the mouse SMA enhancer by concerted action at multiple positive regulatory elements, inducibility per se was confined to a site in the enhancer referred to as the THR that was previously shown to demonstrate TGFβ1-dependent hyperreactivity with single strand-specific DNA-modifying reagents (Becker et al., 2000; Cogan et al., 2002). Importantly, three single strand-specific DNA-binding factors were identified in fibroblasts and arterial smooth muscle cells that attenuated transcriptional output from the SMA enhancer (Cogan et al., 1995; Sun et al., 1995; Kelm et al., 1996, 1999a,b). These repressors bound opposite strands of a large purine/pyrimidine (Pur/Pyr)-rich region that encompassed the THR plus an adjacent MCAT element that binds the TEF1 protein required for SMA enhancer activation. Purα and Purβ bound to the purine-rich strand, whereas MSY1 was specific for the pyrimidine-rich strand. The level of all three proteins increased in accepted cardiac allografts in parallel with the development of peritransplant fibrosis and chronic rejection (Subramanian et al., 2002). Purα, Purβ, and MSY1 self-associate on single-stranded DNA as well as bind double strand-specific transcriptional activating proteins such as TEF1 and serum response factor (SRF) to form heteromeric complexes (Carlini et al., 2002). Because Purα, Purβ, and MSY1 prefer to bind single-strand DNA, they may block access of the TEF1-activating protein to its cognate double-stranded MCAT binding element within the Pur/Pyr-rich domain. Additionally, recent studies showed that the functional properties of the Purβ repressor can be influenced by SRF, a critical SMA gene activator in both myofibroblasts and arterial smooth muscle cells (Sun et al., 1995; Kelm et al., 2003).
We now know that the Sp1/3 DNA-binding proteins also function as transcriptional activators of the SMA enhancer in fibroblasts via interaction with a second TGFβ1 control element known as the TCE that was originally described in arterial smooth muscle cells (Cogan et al., 2002). In this report, we identify a novel, purine-rich domain that overlaps the core Sp1/3 consensus binding site within the TCE. Based on its proximity to the Sp1/3 site and sequence similarity to the Pur protein-binding element in the THR described above, we refer to this new regulatory component as the SPUR element. Although the DNA-binding activities of Purα, Purβ, and MSY1 are modulated after transplant chronic rejection (Subramanian et al., 2002), and Sp1 contributes to altered patterns of collagen gene expression typically associated with wound healing and fibrosis (Greenwel et al., 1997; Poncelet and Schnaper, 2001), we know nothing about interrelationships of these proteins in the context of TGFβ1-dependent SMA gene activation that represents an important feature of myofibroblast differentiation after tissue injury. Therefore, our goal was to determine whether single strand-specific, SMA repressor proteins interact with double strand-specific Sp1/3 DNA-binding proteins and investigate how these interactions change upon conversion of fibroblasts into myofibroblasts. In this report, we show that Pur and Sp1 proteins in fact share adjacent binding sites at the SPUR element of the SMA enhancer and physically interact in a TGFβ1-sensitive manner. The data suggest that fibroblast to myofibroblast conversion involves altered protein: DNA and protein:protein interactions localized at the SPUR and THR sites within the SMA enhancer. Dynamic interplay between Pur and Sp1 proteins may allow adjustment of SMA gene output in myofibroblasts to more efficiently repair damage inflicted by inflammatory cells in accepted organ grafts. Disruption of this interplay could cause excessive accumulation of hypercontractile myofibroblasts that may alter the structure of accepted cardiac grafts to the point of functional disruption. As such, these molecular interactions may be useful targets for novel forms of antirejection therapy after solid organ transplant.
MATERIALS AND METHODS
Cell Culture Methods
Mouse AKR-2B embryonic fibroblasts were maintained in McCoy's 5A medium (Cambrex Bio Science Walkersville, Walkersville, MD) supplemented with 5% heat-inactivated fetal bovine serum (hiFBS) and penicillin-streptomycin (Invitrogen, Carlsbad, CA). Nonhuman primate COS7 kidney fibroblasts were maintained in DMEM (4.5 g/l d-glucose) supplemented with penicillin-streptomycin and 10% hiFBS. All cell lines were cultivated in a humidified incubator at 37°C at 5% CO2. Fibroblasts were rendered quiescent by a 48-h exposure to HEPES-buffered DMEM (1.0 g/l d-glucose) containing 0.5% hiFBS, and penicillin-streptomycin-Fungizone. Recombinant human TGFβ1 (5 ng/ml, final concentration; R&D Systems, Minneapolis, MN) was added to cultures for varying periods before preparation of protein extracts.
Preparation of Protein Extracts and Electrophoretic Mobility Shift Assay (EMSA)
Cell monolayers were washed twice with Dulbecco's phosphate-buffered saline (PBS), scraped into fresh PBS, sedimented at 3000 rpm, washed once more in PBS, and resuspended in eight packed-cell volumes of hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM dithiothreitol [DTT]). Cells were allowed to swell for 10 min on ice before transfer to a Dounce homogenizer for processing with a type B pestle. Nuclei were collected from ruptured cells by centrifugation for 15 min at 4000 rpm and suspended in one-half packed-pellet volume of ice-cold, low salt buffer (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.02 M KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT). High salt buffer (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 1.2 M KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT) equal to one-half packed pellet volume was added and the nuclei further extracted with gentle rocking for 30 min at 4°C. Nuclei were collected by centrifugation for 30 min at 14,500 rpm and dialyzed against 50 volumes of dialysis buffer containing 20 mM HEPES, pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT. After dialysis, supernatants were collected by centrifugation at 14,500 rpm for 20 min for use in biochemical assays. Whole cell extracts were prepared from PBS-rinsed monolayers by using radioimmunoprecipitation assay (RIPA) buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail, 0.2 mM PMSF, 0.5 mM DTT) at 0.6 ml/100-mm culture plate followed by gentle rocking for 15 min at 4°C. Adherent lysed cell remnants were scraped into 0.3 ml of RIPA buffer and combined with the original lysate into a single microcentrifuge tube and the supernatant fraction was collected at 10,000 × g for 10 min at 4°C. Mouse ventricle tissue extracts were prepared from neonates (4, 6, 9, and 13 d old), 8- to 10-wk-old adults, and cardiac allografts as described previously (Subramanian et al., 2002). EMSA and super-shift EMSA was performed on cell and tissue extracts as described previously (Cogan et al., 2002; Subramanian et al., 2002).
DNA-binding Assay (DBA)
Synthetic oligonucleotide probes used in this study correspond to sequences present in the mouse SMA 5′-flanking region (Min et al., 1990). Reaction mixtures containing nuclear extract (100 μg of protein) and biotinylated oligonucleotides (100 pmol; Integrated DNA Technologies, Coralville, IA) were incubated in reaction mixtures containing poly(dI-dC), 10 mM Tris, pH 7.5, 50 mM NaCl, 0.5 mM DTT, 0.5 mM EDTA, 0.12 mM PMSF, 4% glycerol. Protein:biotin:DNA complexes were captured on streptavidin-immobilized paramagnetic particles (Promega, Madison, WI; 0.6 ml/reaction, 30-min incubation) as described previously (Kelm et al., 2003). After washing four times with buffer containing 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 100 mM NaCl, bound protein was eluted using 2× protein denaturing buffer and analyzed by SDS-PAGE and immunoblotting procedures.
Mammalian Protein Overexpression Plasmids, Cell Transfection, and Reporter Gene Assays
Fibroblasts at 40–50% confluence were transfected with optimized mixtures of the previously described SMA promoter:reporter fusion plasmids VSMP4 and VSMP8, and/or plasmids encoding various transcriptional regulatory proteins by using Mirus (Invitrogen, San Diego, CA) transfection reagent and a protocol provided by the manufacturer (Cogan et al., 2002). Plasmids encoding human Sp1 and the human Smad 2/3/4 proteins were kindly provided by Dr. J. Horowitz (North Carolina State University, Raleigh, NC) and Drs. L. Choy and R. Derynck (University of California San Francisco, San Francisco, CA), respectively. Plasmids were purified using QIAGEN preparative resin and a protocol provided by the manufacturer (QIAGEN, Valencia, CA). Forty-eight hours after transfection, cells were washed three times with cold PBS and then lysed using chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assay (ELISA) lysis buffer (Roche Applied Science, Indianapolis, IN). Whole cell extracts were clarified at 14,000 × g for 10 min at 4°C and stored at –20°C before assay. Total protein in extracts was determined by bicinchoninic acid colorimetric assay (Pierce Chemical, Rockford, IL). Equivalent amounts of lysed protein were evaluated by immunoblot to verify protein overexpression. CAT reporter gene activity was determined using a commercial ELISA kit (Roche Applied Science). Reporter gene expression was normalized with respect to total cell protein, and transfections were routinely performed in triplicate and repeated three to five times. Data sets were subjected to analysis of variance to assess statistical significance set at p < 0.05.
Protein Immunoprecipitation
Nuclear or whole cell protein extracts (100 μg) from fibroblasts were combined with commercial (Sigma-Aldrich, St. Louis, MO) H-tag (for Pur proteins) or DYKDDDDK-tag–specific (for Smads) antibodies (2 μg) in 200 μl of solution D (20 mM HEPES, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 1 mM DTT, 0.2 mM PMSF). After a 1-h incubation at 4°C on a rotating-platform mixer, a 20-μl aliquot of protein G-agarose (Sigma-Aldrich), that had been previously washed and suspended in solution D, was added and the mixture incubated 16 h at 4°C with rotation. Agarose beads with immobilized protein were collected by centrifugation at 2500 rpm for 5 min at 4°C, washed four times with PBS, suspended in 1× SDS-PAGE sample buffer, heated at 95°C for 5 min, and released proteins evaluated by SDS-PAGE and immunoblotting with selected antibodies.
Immunoblotting
Proteins (10-μg aliquots) were size fractionated by SDS-PAGE by using 10% polyacrylamide gels and then electrophoretically transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH). After overnight blocking at 4°C in Tris-buffered saline (TBS; 25 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 3% (wt/vol) nonfat dry milk and 0.5% bovine serum albumin, blots were incubated with selected rabbit polyclonal antibodies (2 μg/ml) for 90 min at room temperature with gentle rocking. Antibodies specific for Sp1 and Smad proteins were obtained commercially (Santa Cruz Biotechnology, Santa Cruz, CA) and two different Pur protein-specific antibodies (anti-Purα 291–313 and anti-Purβ 302–324) plus a pan-specific Purα/β antibody (42–69) were described previously (Kelm et al., 1999a; Subramanian et al., 2002). Blots were washed four times at room temperature over a 20-min period in TBS containing Tween 20 (0.05% vol/vol). Horseradish peroxidase-conjugated, goat anti-rabbit secondary antibody (1:1500) then was applied for 45 min after which time the blots were washed as described above and processed for antibody visualization by chemiluminescence (ECL; Amersham Biosciences, Piscataway, NJ) and imaged onto Biomax film (Eastman Kodak, Rochester, NY).
RESULTS
A Novel Double-Strand DNA-binding Site for Pur Proteins Is Located Proximal to the Sp1 Activation Site in the SMA Enhancer
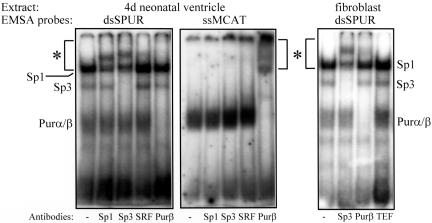
The double-strand TCE element in the mouse SMA enhancer represents a strong binding site for the Sp1/3-activating proteins in TGFβ1-activated myofibroblasts (Cogan et al., 2002). Inspection of the sequence flanking the TCE revealed three putative Pur protein binding sites (GGA) in both “forward” (also known as the “upper” or “sense” strand of a DNA duplex) and “reverse” (also known as the “lower” or “reverse” strand of a DNA duplex) orientations within and 3′ to the GC-rich core of the TCE required for Sp1/3 binding (Table 1). The original TCE probe was extended to include these sequence components, thus creating a new probe called SPUR that contains both Sp1/3- and Pur-binding sites. SPUR is located between –59 and –28 in the 5′-flanking region of the mouse SMA gene. To determine whether mouse Pur protein was able to bind the SPUR probe, EMSA was performed using protein extracts prepared from neonatal mouse ventricular tissue and fetal bovine serum-stimulated mouse AKR-2B fibroblasts. Immediately before birth, SMA is the dominant actin isoform expressed in mouse ventricular cardiomyocytes. Because SMA expression in these cells ceases by postnatal day 14, ventricular extracts from 4-d-old neonatal mice were expected to contain both activators (e.g., Sp1) and repressors (e.g., Pur α and β) of SMA gene transcription. Likewise, serum-stimulated fibroblasts transiently express SMA and contain both SMA gene activators and repressors to temper SMA transcriptional output. As shown in Figure 1, Sp1/Sp3 and Pur proteins were detected in heart tissue and fibroblast extracts by using the SPUR probe in double-strand (ds) context. Moreover, Sp1, Sp3, and Pur proteins were all independently size shifted on the dsSPUR probe by their respective polyclonal antibodies but not with antibodies specific for the TEF1 or SRF enhancer-activating proteins. For comparison, the forward, single-strand (ss) component of the MCAT probe was used in the heart extract EMSA as a positive control for Pur protein binding (Cogan et al., 1995; Sun et al., 1995; Subramanian et al., 2002) and to denote the position of supershifted Pur protein:DNA complexes. Antibody-supershifted complexes of Pur protein were evident in EMSAs by using the ssMCAT probe but less obvious in assays using the dsSPUR probe because the native Sp1: dsSPUR complex migrates to the same position as the supershifted Pur:dsSPUR complex (compare area marked with asterisk in the middle panel in Figure 1 with same areas of the left or right panels). Both Purα and Purβ DNA complexes were size shifted by the anti-Purβ 302–324 antibody owing to their known ability to form a heterodimer (Kelm et al., 1999a).
Table 1.
Sequences of DNA probes used to evaluate dynamic interplay of SMA enhancer-binding proteins
| GGA or AGG: potential Pur binding sites | |||||
| Sp1/Sp3 | |||||
| SPUR | GAAGCGAGTG | GGAGGGGA | TCAGAGCAAGGGGC | ||
| TCE | GAAGCGAGTG | GGAGGGGA | TCAGA | ||
| TCEm | GAAGCGAGTG |
TTAGGGGA
|
TCAGA | ||
| MCAT | GCAGAACAGAGGAATGCAGTGGAAGAGACC | ||||
| THR | GCAGTGGAAGAGACCCAGGCCTCTGGCCACCCAGA | ||||
| SPUR | GAAGCGAGTGGGAGGGGATCAGAGCAAGGGGC | ||||
| SPURm1 | GAAGCGAGTGTTAGGGGATCAGAGCAAGGGGC | ||||
| SPURm2 | GAAGCGAGTGGGAGGGGATCAGATACCTTTTA | ||||
| CAGA or AGAC: potential Smad binding site | |||||
| SBE (+ control) | CCCCAGACACCACCCACCCAGAGTGG | ||||
| SPUR | GAAGCGAGTGGGAGGGGATCAGAGCAAGGGGC | ||||
| MCAT | GCAGAACAGAGGAATGCAGTGGAAGAGACC | ||||
| THR | GCAGTGGAAGAGACCCAGGCCTCTGGCCACCCAGA | ||||
| NS (– control) | CAGGTGAATGGCAGCAGCTGTCGACTCAC | ||||
For simplicity, only the forward, upper strand sequence is shown. The salient features of Pur protein and Sp1/Sp3 binding motifs are bold lettered in the top group, whereas putative Smad-binding consensus sequences are bold lettered in the bottom group. Substitution mutations are underlined.
Figure 1.
EMSAs depicting interaction of native Sp1, Sp3, and Pur proteins in neonatal mouse ventricle (left and middle) and serum-treated AKR-2B fibroblast (right) extracts with either double-strand SPUR probe (dsSPUR) or a single-strand probe from a different region of the SMA enhancer (ssMCAT) that contains a site for Pur protein binding but not the other proteins. EMSA reactions were processed with antibodies specific for Sp1, Sp3, Purβ, SRF, and TEF1 to size shift DNA:protein complexes containing the cognate transcription factors (size-shifted complexes are within the bracketed area marked with asterisk). The Sp1, Sp3, and Pur proteins all were independently size shifted by their corresponding antibodies but not by unrelated SRF or TEF1 antibodies.
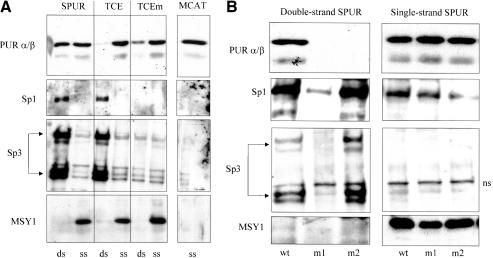
Pur proteins have always been regarded as single-strand–specific DNA-binding proteins in the context of SMA gene regulation. Thus, it was extremely interesting to observe a departure from this behavior with regard to their affinity for double-strand SPUR DNA. To further explore the significance of this new observation, DBA combined with Western blot analysis was performed using nuclear extracts prepared from mouse AKR-2B embryonic fibroblasts. Our intent was to examine sequence and strand specificity issues as well as clearly resolve binding of each Pur protein isoform. The SPUR and TCE probes, plus a TCE probe variant containing a mutated Sp1 site (TCEm; Table 1), in either double- or single (forward)-strand form were incubated with nuclear protein extracts. The forward strand of the MCAT element described above served as a positive control for single-strand binding because it is known to contain a high-affinity site for both Pur proteins (Cogan et al., 1995; Sun et al., 1995). Western blots of DNA-bound proteins were evaluated with antibodies specific for Pur α/β, Sp1, and Sp3. Whereas Pur proteins bound to the forward strands of all four probes, SPUR was the only double-stranded probe capable of binding Pur proteins (Figure 2A). As expected both Sp1 and Sp3 bound to double-strand forms of SPUR and TCE but failed to bind the double-strand TCEm nor did they bind any single-strand probes (Figure 2A, Sp1 and Sp3). Interestingly, the double-strand version of TCEm retained some Pur protein binding activity perhaps because of an intact GGA site that may be available for Pur occupancy in the absence of Sp1/3 binding to this mutant probe (refer to probe sequence data in Table 1). Because the single-strand–specific MSY1 transcriptional regulatory protein binds Pur proteins (Kelm et al., 1999b) and recently was shown to enhance loading of Purβ onto single-stranded Pur/Pyr-rich regions of the SMA enhancer (Kelm et al., 2003), we examined whether MSY1 was a component of the novel Pur-containing complexes identified on SPUR probes. Whereas MSY1 binding clearly was detected on single-strand versions of SPUR or TCE probes, this protein was not associated with the Pur complex formed on double-strand SPUR (Figure 2A). Moreover, the interaction of MSY1 with forward-strand DNA seemed specific for SPUR-based probes (SPUR, TCE, and TCEm) because the forward strand derived from the MCAT Pur/Pyrrich region was unable to bind MSY1 (Figure 2A). Thus, double-strand SPUR seemed to accomodate a novel, specific interaction with Pur repressor proteins that up to this point had only demonstrated clear preference for single-strand regions of the SMA enhancer.
Figure 2.
(A) DBA of fibroblast nuclear extracts using double- and single-strand SMA enhancer probes. DBA probes are indicated at the top margin. Antibodies used for Western blot analysis of affinity-isolated proteins are noted on the left. ds, double-strand DBA probes; ss, single-strand DBA probes. dsSPUR binds the Pur, Sp1, and Sp3 proteins. (B) DBA of fibroblast nuclear extracts by using mutant SPUR probes. SPUR mutations in double-strand context eliminated Pur binding but had no detrimental effect on binding to single-strand probes. wt, wild-type context; m1, m2, mutant context (Table 1); ns, nonspecific species. Antibodies for DBA Western blots are noted on the left.
The GC-rich and GGA components within double-strand SPUR were independently mutated to evaluate their importance in mediating Pur protein binding (Figure 2B, SPURm1 and SPURm2). Both the GC-rich and GGA sites were essential for Pur protein binding to double-strand SPUR. However, Sp1/3 binding was impaired only by mutation at the GC-rich site proper and not by mutation of the GGA motif. This observation demonstrated that Pur protein binding to double-strand probes was sequence specific. Qualitatively, single-strand mutant probes exhibited approximately the same ability to bind Pur and MSY1 proteins as probes in native context (Figure 2B, right), implying that intact GC-rich and GGA sites required for Pur binding to double-strand DNA were not important determinants for binding to single-strand DNA. These results are suggestive of relaxed single-strand sequence specificity compared with double-strand probes that may be more structurally constrained.
TGFβ1 and Its Receptor Coregulatory Smad Proteins Alter Pur Protein Interactions
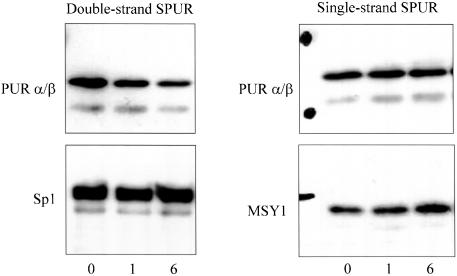
Because the SPUR region encompasses the TCE previously reported to be involved in TGFβ1 regulation of the SMA promoter in fibroblasts (Cogan et al., 2002), we examined Pur binding to SPUR in the presence and absence of this essential growth factor mediator of myofibroblast differentiation. Nuclear protein extracts prepared from myofibroblasts after either a 1- or 6-h exposure to 5 ng/ml TGFβ1 were evaluated by DBA. TGFβ1 treatment over a 6-h period caused a fourfold reduction in Pur binding that was specific for double-strand SPUR (Figure 3). Binding of the Sp1 transcriptional activator to double-strand SPUR did not change in the presence of TGFβ1 consistent with our previous observation that net Sp1/Sp3 binding to the SMA enhancer does not increase during myofibroblast differentiation (Cogan et al., 2002). Western blot analysis showed no reduction of Pur protein levels in fibroblast nuclear extracts after a 6-h exposure to TGFβ1 (unpublished data). This finding indicates that although Pur protein can be detected in TGFβ1-activated myofibroblasts, it seems less able to bind double-strand SPUR DNA. Interestingly, MSY1 binding to single-(forward) strand SPUR was modestly enhanced during TGFβ1 treatment (Figure 3). Similar trends were observed for Pur and MSY1 proteins in primary human pulmonary myofibroblasts after a 6-h exposure to TGFβ1 (unpublished data).
Figure 3.
DBA of nuclear extracts prepared from TGFβ1-activated myofibroblasts. Antibodies used for Western blots are shown at the left of each panel. Over a 6-h exposure to TGFβ1, there was a fourfold decrease in Pur protein binding to the dsSPUR probe.
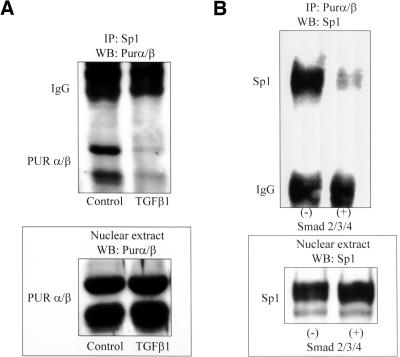
Protein immunoprecipitation experiments were performed to examine the possibility of a physical association between Sp1 and Pur proteins that might account for their joint affinity for double-strand SPUR as well as provide a control point for the modulating influence of TGFβ1 on Pur protein interaction with SPUR. Nuclear extracts prepared from myofibroblasts cultivated for 6 h in the presence or absence of TGFβ1 were incubated with a Sp1-specific antibody followed by Western blot evaluation of the precipitated material using a pan-specific Pur protein antibody. Whereas the Sp1 antibody immunoprecipitated Pur proteins from control extracts, significantly less Pur protein was collected from extracts prepared after TGFβ1 treatment (Figure 4A). Western blots of total nuclear protein revealed equivalent amounts of Pur proteins and Sp1 in control and treated extracts (Figure 4A, bottom), indicating that TGFβ1 most likely impaired protein:protein interaction rather than reduce net availability of these proteins.
Figure 4.
Immunoprecipitation of a Sp1:Pur protein complex from fibroblasts. (A) Sp1 immunoprecipitate isolated from serum-free (control) AKR-2B fibroblast nuclear extracts contained prominent Purα and Purβ bands revealed by Western blot analysis by using a pan-specific anti-Pur antibody. Myofibroblast differentiation during a 6-h exposure to TGFβ1 was accompanied by diminished Sp1:Pur protein interaction. Exposure to TGFβ1 had no detectable effect on Pur protein isoform levels in nuclear extracts (bottom). (B) Pur protein immunoprecipitate isolated from COS7 fibroblasts transfected with empty expression plasmid (-) or equimolar mixtures of plasmids harboring cDNAs encoding Smad 2, 3, and 4 (+) were processed by Western blot by using a Sp1-specific antibody. Transfection with Smad expression plasmids did not effect net expression of Sp1 protein (bottom) but significantly impaired Sp1:Pur protein interaction (top).
Smad proteins 2, 3, and 4 mediate TGFβ1 receptor signaling in myofibroblasts during wound healing and strongly activate the native SMA gene (Cogan et al., 2002). To extend data presented in Figure 4A, we investigated the consequences of Smad protein overexpression on the physical association of Sp1 and Pur proteins in transfected COS7 fibroblasts. The Pur protein antibody was able to immunoprecipitate native Pur:Sp1 complexes from nontransfected COS7 fibroblasts but not cells transfected with an equimolar combination of expression vectors encoding Smad 2, 3, and 4 (Figure 4B). Western blot analysis demonstrated equal amounts of native Sp1 in both nontransfected control cells and cells overexpressing Smad proteins (Figure 4B, bottom). Together, the data presented in Figure 4, A and B, suggested that both TGFβ1 and its receptor-regulated Smad signaling agents can modify the physical interaction between Sp1 and Pur proteins.
Pur Proteins Are Potent SMA Transcriptional Repressors in Fibroblasts
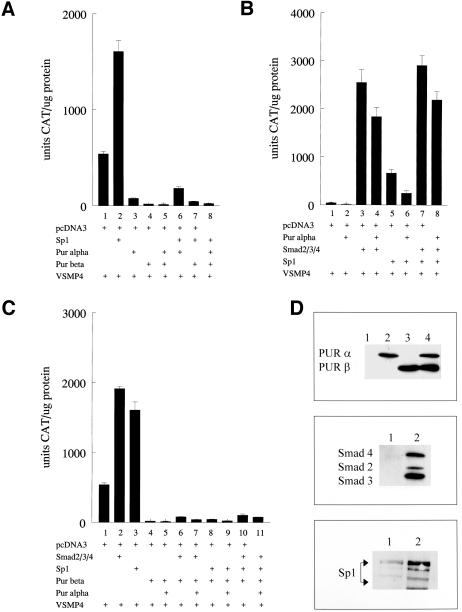
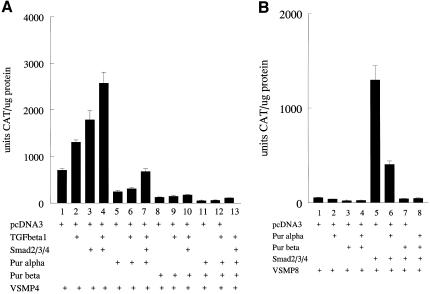
Previous results from our laboratories showed that Purβ, but not Purα, was a potent repressor of the mouse SMA promoter in aortic smooth muscle cells (Kelm et al., 2003). To examine the functional consequences of Sp1:Pur protein interactions on SMA enhancer activity in fibroblasts, we cotransfected combinations of Smads 2/3/4, Purα, Purβ, and Sp1 in COS7 kidney fibroblasts along with the SMA enhancer:CAT reporter fusion gene construct referred to as VSMP4. The 191-base pair enhancer fragment contained in VSMP4 includes the SPUR, TCE, MCAT, THR, and CArG B elements necessary for both basal and TGFβ1-inducible transcription of SMA in fibroblasts (Foster et al., 1992; Cogan et al., 2002). In contrast to their behavior in arterial smooth muscle cells, we were surprised to observe that both Pur proteins were equally effective in repressing basal and Sp1-augmented SMA enhancer output in transfected fibroblasts (Figure 5A). More interestingly, Smad 2/3/4 overexpression not only eliminated Purα repression of VSMP4 but also neutralized the negative influence of Purα on VSMP4 activation in the presence of Sp1 (Figure 5B). In contrast, Smad proteins were not effective in relieving SMA enhancer repression in fibroblasts that overexpressed Purβ either alone or in combination with Purα (Figure 5C). Apparently, Smad proteins were able to override repression of the SMA enhancer, either in a basal or Sp1-activated state, but only if repression was mediated by Purα and not Purβ. Smad 2/3/4 and Pur protein overexpression in transfected COS7 fibroblasts was verified by Western blot analysis (Figure 5D). To compare these results with TGFβ1 receptor-mediated myofibroblast activation, we examined the effect of Pur protein overexpression on SMA transcriptional activation in AKR-2B fibroblasts that were treated with TGFβ1 after transfection to activate endogenous Smads via the usual receptor-based pathway. Although both Pur proteins were capable of disrupting TGFβ1 activation of the SMA enhancer in myofibroblasts, Purβ showed a dominant repressor effect that was three to fourfold more potent than Purα (Figure 6A). When coexpressed with Purα, Smads were less potent derepressors of the SMA enhancer in AKR-2B fibroblasts compared with COS7 fibroblasts. This observation may be related to differences in the levels of native Pur proteins and/or availability of SMA enhancer activating factors such as Sp1, TEF1, or SRF in each fibroblast line. Finally, Smad activation of the 3.6-kb VSMP8 mouse SMA promoter construct, previously shown to drive high-level, smooth muscle tissue-specific overexpression in numerous transgenic mouse lines (Wang et al., 1997, 1998; Maeda et al., 1999; March et al., 1999; McGraw et al., 1999; Nobe et al., 2001; Gokturk et al., 2003) also was inhibited in COS7 fibroblasts by overexpression of either Pur protein (Figure 6B). Purβ was nearly 10-fold more potent than Purα as a repressor of Smad-activated VSMP8.
Figure 5.
Pur proteins behave as bimodal mediators of SMA enhancer activity in transfected COS7 fibroblasts. (A) Both Pur proteins were able to suppress Sp1-mediated activation of the SMA enhancer. (B) Smad proteins are robust activators of the SMA enhancer and can neutralize Purα-mediated repression with or without Sp1. (C) Purβ behaved as a dominant negative-type repressor and was unaffected by either Smad or Sp1 protein overexpression. For A to C, the expression plasmids included in individual transfections are noted by (+). (D) Protein overexpression in transfected COS7 fibroblasts was verified by Western blot analysis by using polyclonal antibodies specific for Pur proteins (top); Smad 2, 3, and 4 (middle); or Sp1 (bottom). Lane 1 (all panels), extracts from nontransfected COS7 fibroblasts; lanes 2–4, extracts from COS7 fibroblasts transfected with expression plasmids for Purα alone (lane 2, top), Smad2/3/4 (lane 2, middle), Sp1 (lane 2, bottom), Purβ alone (lane 3, top), or both Purα and Purβ (lane 4, top).
Figure 6.
Overexpression of Pur proteins in AKR-2B fibroblasts attenuates SMA enhancer and full-length promoter activation by TGFβ1 or Smad proteins. (A) VSMP4 enhancer activation in transfected fibroblasts by TGFβ1, Smad2/3/4, or a combination of both was substantially repressed by Purα overexpression and fully repressed by Purβ. Compare lanes 2, 3, and 4 with lanes 5, 6, and 7 (Purα-mediated repression) or lanes 8, 9, and 10 (Purβ-mediated repression). (B) Smad-mediated activation of the full-length VSMP8 promoter in AKR-2B fibroblasts was substantially repressed by Purα and fully so by Purβ. Compare lane 5 with either lane 6 (Purα-mediated repression) or lane 7 (Purβ-mediated repression). The VSMP8 promoter normally is transcriptionally silent in fibroblasts but activated by TGFβ1 or Smad signaling intermediary proteins (compare lanes 1 and 5). For A and B, the various treatments and/or combinations of expression plasmids used for each transfection are shown at the bottom.
The SMA-activating Proteins Smad2/3/4 and Sp1 Physically Engage Pur Repressor Proteins but Occupy Distinctly Different DNA-binding Sites within the SMA Enhancer
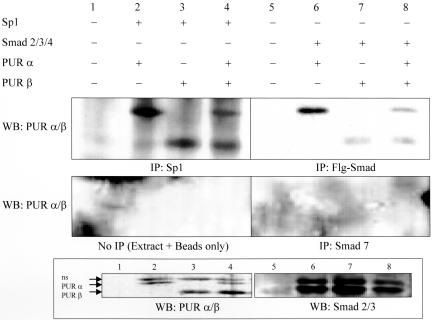
Results shown above point to a physical interaction between Sp1 and Pur proteins at double-strand SPUR and that Smad proteins seemingly counteract Purα repression of the SMA enhancer. We postulated that multiprotein complexes might exist in myofibroblasts that could, for example, facilitate Smad protein derepression of a Pur protein-blocked SMA enhancer. In support of this idea, Smad protein immunoprecipitates collected from COS7 fibroblasts that overexpressed H-tagged versions of Pur proteins in combination with DYKDDDDK-tagged versions of Smads 2/3/4 were observed to contain both Purα and Purβ (Figure 7, lanes 5–8). In addition, Sp1 immunoprecipitates were collected from cells coexpressing Sp1 and H-tagged Pur proteins in the absence of Smads to show that epitope tagging did not adversely affect Pur structure as assessed by its unimpaired ability to bind Sp1 (Figure 7, lanes 1–4). Moreover, negative control experiments to monitor nonspecific interaction with either the immunoprecipitation (IP) antibody (Figure 7, Smad7) or affinity beads used to collect the precipitated proteins (Figure 7, no IP) revealed no spurious binding interactions for the H-tagged Pur proteins.
Figure 7.
Evidence for a Smad 2/3:Pur protein complex in transfected COS7 fibroblasts. Nuclear protein extracts prepared from COS7 fibroblasts overexpressing either Sp1 (lanes 1–4) or Smad 2/3/4 (lanes 5–8) were IPed with antibodies specific for Sp1 or Smads (as noted at the bottom of each panel) and processed for Western blot (WB) using antibodies shown on the left. Both Sp1 (IP:Sp1) and Smad (IP:Flg-Smad) immunoprecipitates contained Purα and Purβ but samples processed with the Smad 7 negative control antibody did not (IP:Smad 7). None of the overexpressed proteins exhibited nonspecific binding to the affinity isolation beads (no IP). Protein overexpression was verified by Western blot analysis of nuclear protein extracts as shown in the bottom two panels. The band denoted “ns” was not studied in detail but may represent a covalently modified Pur protein isoform (unpublished data).
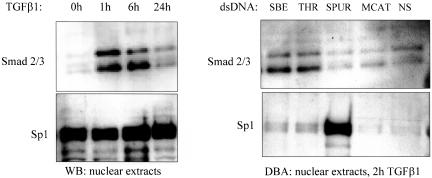
We reasoned that identification of DNA-binding sites for Smad proteins would help clarify spatial relationships between interacting Pur proteins, Smads, and Sp1 within the 191-base pair SMA enhancer. As shown by Western blot analysis in Figure 8, Smads 2 and 3 accumulated in AKR-2B fibroblast nuclei within 1 h after exposure to 5 ng/ml TGFβ1. However, Sp1 levels were not influenced by growth factor treatment and were constitutively high in the nucleus. Interestingly, DBA using fibroblast nuclear extracts prepared 2 h after exposure to TGFβ1 revealed preferential Smad 2/3 binding to the double-strand THR probe, whereas Sp1, confirming previous observations, was most strongly associated with a probe containing the more 3′, double-strand SPUR site (Figure 8). Smad protein binding to SPUR was not noticeably higher than that observed with the double-strand MCAT probe or a double-strand, nonspecific sequence control but comparable with a double-strand, positive-control probe containing a known Smad-binding element (SBE). In summary, although the Pur repressors physically interact with the Sp1 and Smad activators in myofibroblasts, each activator binds a different region of the SMA enhancer. The preferential interaction of Smad 2/3 with the THR component of the SMA enhancer is interesting in view of a previous report showing that TGFβ1 activates SMA transcription in myofibroblasts by facilitating DNA conformational changes within the THR region (Becker et al., 2000).
Figure 8.
SMA enhancer-activating proteins in myofibroblasts exhibit different subcellular localization and enhancer-binding specificity. Left, Smad and Sp1 western blots of nuclear protein extracts prepared from AKR-2B fibroblasts after exposure to TGFβ1 over a 24-h period. Smad 2 and 3 entered the nucleus within 1 h after TGFβ1 treatment, whereas nuclear Sp1 levels were uniformly high. Right, DBA using nuclear extracts from TGFβ1-activated myofibroblasts and various dsDNA probes derived from the SMA enhancer (noted at the top). The Smad and Sp1 enhancer activating proteins bind to different regions of the SMA enhancer.
Cardiac Remodeling Is Associated with Altered Pur Protein Isoform Ratio
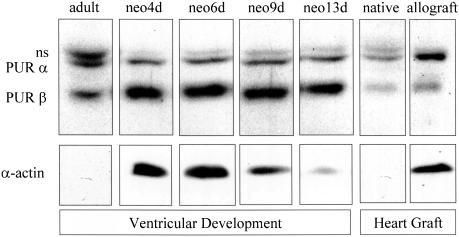
The ability of both Pur proteins to bind the Sp1 and Smad2/3 SMA gene-activating proteins and the different functional responses of Purα and Purβ to TGFβ1 signaling prompted us to speculate that SMA gene output may be governed by Pur protein isoform ratio within remodeling cells and tissue beds. For example, accumulation of the Purβ isoform may silence the SMA gene owing to the dominant negative effect of this protein on basal transcription regardless of whether activating Smad proteins coexist in the nucleus. In contrast, stoichiometric dominance of the Purα isoform may be associated with more dynamic states of SMA gene transcription because it seems to operate bimodally depending on whether or not Sp1 and Smad activators are present. To further examine these ideas, we evaluated two mouse models of SMA gene reprogramming and cardiac remodeling to discern relationships between SMA gene output and Pur protein isoform ratio. During postnatal cardiac development in the mouse, the fetal SMA isoform expressed between 8 and 13 d of embryonic development is gradually replaced by the cardiac isoform of α-actin (Parker et al., 1990; Black et al., 1991; Subramanian et al., 2002). Notably, ventricular expression of Purβ was dominant during the first postnatal week as SMA protein levels decreased (Figure 9). Nine days after birth, Purα, Purβ, and SMA levels in the maturing mouse ventricle all decreased and were barely detectable in the 8 wk-old adult mouse heart. In contrast, reactivation of the fetal SMA gene in accepted, chronically rejected, adult mouse heart grafts was accompanied by elevation of both Pur proteins, especially the Purα isoform that exhibits TGFβ1 sensitivity (Figure 9).
Figure 9.
Cardiac remodeling is accompanied by changes in Pur protein isoform content and SMA expression. Protein extracts prepared from neonatal (postpartum day 4, 6, 9, and 13) and adult mouse ventricles as well as ventricles from explanted allogeneic (allograft) heart grafts (30 d posttransplant) were evaluated by Western blot by using Pur- and SMA-specific antibodies. The postnatal developmental period was characterized by dominance of the Purβ isoform and diminishing SMA expression, whereas chronically rejected allografts expressed more Purα and SMA protein compared with nontransplanted native hearts from graft recipients. The band denoted “ns” was not studied in detail but may represent a covalently modified Pur protein isoform (unpublished data).
DISCUSSION
Faulty SMA Gene Expression and Cardiac Remodeling
SMA is expressed constitutively in vascular smooth muscle cells, transiently in stromal myofibroblasts, and under strict developmental control in cardiomyocytes to impart functional characteristics as required by each specialized cell type. Aberrant expression of the SMA isoform can lead to impaired vascular compliance and contractility (Schlidmeyer et al., 2000; Hao et al., 2003) and has been linked to abnormal accumulation of myofibroblasts and hypertrophic scarring of healing wounds (Powell et al., 1999; Tomasek et al., 2002) as well as inappropriate assembly of stress fibers and sarcomere disruption in cardiomyocytes (Kumar et al., 1997). The long-term consequences of SMA gene misregulation are clearly exemplified in accepted heart grafts via a chronic pathogenic process referred to as CAD. CAD is a cardiac remodeling abnormality associated with loss of SMA from the coronary arterial bed and gain in the myocardial parenchyma and stroma (Shi et al., 1994; Suzuki et al., 1996; Subramanian et al., 1998). Using a model of murine heterotopic cardiac transplant, we recently demonstrated that cardiac fibroblasts were more susceptible than cardiomyocytes to SMA gene reprogramming as evidenced by their early accumulation of the SMA transcriptional reprogramming proteins, TEF1, MSY1, Purα, and Purβ (Subramanian et al., 2002). Cardiac myofibroblasts may instigate ventricular remodeling in accepted heart grafts owing to their ability to synthesize extracellular matrix protein and form scars that theoretically could biomechanically stress adult cardiomyocytes and thus activate fetal SMA gene expression in these cells in a manner analogous to the situation observed in some cardiac hypertrophic disease states (Black et al., 1991; Sussman et al., 2002). In this report, we have attempted to characterize interrelationships between transcriptional activators and repressors of SMA expression first identified in TGFβ1-activated myofibroblasts and transplanted hearts. We hypothesize that disruption of the balance between SMA gene activation and repression may be responsible for certain aspects of chronic rejection histopathology observed in accepted heart grafts that may be precipitated by excess myofibroblasts, particularly interstitial and perivascular fibrosis and coronary vascular sclerosis.
DNA Structure and SMA Transcriptional Control in Myofibroblasts
In myofibroblasts, the TEF1, Sp1/3, Smad 2/3, and SRF transcriptional regulatory proteins all bind double-strand DNA and function as activators of the SMA enhancer, whereas Purα, Purβ, and MSY1 show a clear preference for single-strand DNA and act as repressors (Cogan et al., 1995; Sun et al., 1995; Kelm et al., 1999a; Cogan et al., 2002). Kelm and coworkers have shown that competitive interplay between the TEF1 activator and Purα/Purβ/MSY1 repressors can regulate promoter output at a Pur/Pyr-rich region of the SMA enhancer containing an inverted repeat with high potential to form exposed single-stranded loops within a thermodynamically stable cruciform structure (Kelm et al., 1999a; Becker et al., 2000; Carlini et al., 2002). We now extend these findings by showing that Pur repressor proteins also compete with the Sp1 activator but at a distinctly different, double-strand region of SMA enhancer referred to as the SPUR element. SPUR, unlike the Pur/Pyr-rich region, does not contain an obvious inverted repeat sequence that would allow unfolded DNA structure. Functional interplay between Pur and Sp1 proteins previously has been observed in mouse brain where the complex is believed to control transcription from the myelin basic protein (MBP) gene (Tretiakova et al., 1999). In this system, physical interaction with Sp1 enhanced the ability of Pur α to interact with its target single-strand sequence to drive MBP promoter activation. Similarly, during differentiation of the U937 monocytic cell line, Purα and Sp1 collaborated to induce CD11c β-integrin gene transcription required for promoting monocyte:endothelial cell adhesion during inflammation (Shelley et al., 2002). In the CD11c promoter, Sp1 bound to the double-strand sequence, whereas Purα binding was strictly confined to a single-strand DNA target. In contrast, our results indicate that Pur proteins interact with both double-strand as well as single-strand elements in the region of the SMA enhancer that contains a high-affinity Sp1 binding site. Moreover, Pur binding to double-strand SPUR DNA was significantly disrupted in the presence of TGFβ1, suggesting that the association of Pur protein with double-strand DNA at SPUR was associated with a state of gene repression, not activation as in the MBP and CD11c promoters. Pur binding to double-strand SPUR required a novel 3′ GAA-containing motif plus an intact GC-rich Sp1 site consistent with the idea that prior Sp1 occupancy was essential, although not the sole deciding factor, for Pur interaction with double-strand SPUR. Binding of Pur to double-strand SPUR could require physical interaction with Sp1 and/or DNA conformational changes mediated by the protein complex that make the GAA region of SPUR more receptive to Pur binding. Sp1:Pur protein complexes demonstrated a strict requirement for double-strand DNA. Single-strand SPUR probes did not bind Sp1:Pur protein complexes despite the fact that these probes were fully capable of Pur protein binding alone in either native or mutant sequence context. Single-strand SPUR probes capable of binding Pur protein may demonstrate conformational flexibility not afforded by double-strand SPUR perhaps due to constraints imposed by bound Sp1/3 proteins. These observations are important because they show that the Sp1:Pur:dsSPUR ternary complex was not formed by opportunistic interaction of Pur protein with flexible, single-strand DNA. Rather, the data suggest that a novel Sp1:Pur protein complex specifically binds to a conformationally constrained, double-strand SPUR element.
Other potential Pur protein partners include TEF1 previously shown to bind double-strand DNA encompassing a muscle-specific MCAT element in the SMA enhancer (Sun et al., 1995). Whereas MSY1 also was identified as a Pur protein partner at the MCAT site, this DNA binding protein was not a component of the newly identified double-strand SPUR protein complex although, like Pur, MSY1 binds well to the forward strand of SPUR. Curiously, the forward strand of SPUR is purine rich and substantially different from the reverse, pyrimidine-rich strand of the MCAT that exhibits high affinity for MSY1 (Cogan et al., 1995). One explanation for this discrepancy is that MSY1 binding to the forward, purine-rich strand of SPUR may be indirect and mediated by direct physical interaction with Pur protein, the primary occupant of purine-rich single-strand elements in the SMA enhancer (Kelm et al., 1999b; Carlini et al., 2002). Regardless of the actual basis for MSY1 interaction with forward-strand SPUR, our data clearly show that formation of a Sp1:Pur complex at double-strand SPUR renders the element unavailable for MSY1 binding. Given the remarkable ability of MSY1 to potentiate Pur protein-mediated repression of the SMA gene in arterial smooth muscle cells (Kelm et al., 2003), its exclusion from double-strand SPUR may provide a specific functional advantage in myofibroblast differentiation by allowing for rapid and robust activation of the SMA enhancer by TGFβ1. In accord with its disabling effect on SMA transcriptional activity, MSY1 is well known in other systems for its ability to promote single-strand DNA conformational states that may further reduce the incidence of transcriptional activation at the SPUR element in the absence of TGFβ1-based signaling (Matsumoto and Wolffe, 1998).
The Influence of TGFβ1 Signaling on Transcriptional Regulatory Protein Interactions at the SMA Enhancer
One important concept emerging from our studies is that Pur protein is a natural inhibitor of SMA gene transcription unless microenvironmental conditions prevail that either alter DNA conformation in the enhancer or provide ratelimiting activation proteins capable of neutralizing Pur protein repression. We have found that TGFβ1 is able to alter the physical interaction of Sp1 and Pur proteins with the net result that affinity for Pur binding at double-strand SPUR DNA is reduced during SMA enhancer activation. Smad proteins become available in the fibroblast nucleus soon after TGFβ1 receptor engagement and seem to play an important, rate-limiting role in governing the magnitude and/or duration of SMA gene expression during myofibroblast differentiation (Cogan et al., 2002). Our present studies do not address whether Smad proteins directly alter the physical interaction of Pur proteins with dsSPUR DNA. However, Smad proteins do seem to influence Pur:Sp1 protein:protein interaction, which is especially interesting in view of evidence in the literature supporting the notion that Smad proteins may activate genes by physically sequestering transcriptional repressors. Members of the Hox family of homeodomain proteins repress transcription when bound to their cognate DNA sites. One member of the TGFβ superfamily, bone morphogenetic protein, uses a Smad protein intermediary to relieve repression of the osteopontin promoter (Shi et al., 1999). Derepression seems to occur by Smad1 protein binding and displacement of Hoxc-8 from DNA. This group also has shown more recently that Smad4 physically displaces the Hoxc-9 repressor from the same promoter in response to TGFβ1-mediated activation of osteopontin gene transcription (Shi et al., 2001). Whereas the details of Smad-based derepression of the SMA gene are incomplete, the Pur/Pyr-rich THR region does contain a functionally important DNA binding site for Pur protein that could indirectly recruit Smad to the THR via the demonstrated physical interaction between Pur and Smad2/3. Moreover, the THR region contains consensus CAGA motifs that may stabilize Smad protein binding after recruitment by Pur. However, protein:DNA interactions at the THR are likely to be very complex. In particular, a recent report showed that interferon-γ, a potent antifibrosis cytokine, blocked TGFβ1 activation of the α2 procollagen gene by enhancing YB-1 interaction with Smad3 (Higashi et al., 2003). The THR contains a binding site for MSY1, the mouse homologue of human YB-1, raising the interesting possibility that dynamic interplay between Pur:Smad3 and MSY1: Smad3 complexes could in some unknown way govern transcriptional output from this region of the SMA enhancer. Additionally, nucleotide sequences flanking the THR may be required in view of reports indicating that CAGA motifs by themselves are neither necessary nor sufficient for Smad binding (Massague and Wotton, 2000).
After TGFβ1 activation, we speculate that nuclear Smads may bind and displace Pur proteins from double-strand SPUR but not alter binding of the Sp1 transcriptional activator (Cogan et al., 2002). Concurrently, TGFβ1 signaling could alter Pur protein binding at the THR to uncover a cryptic double-strand, Smad-binding site among the several CAGA motifs located there. THR and SPUR are separated by 105 base pairs and theoretically could lie within two adjacent 80-base pair DNA loops in a single nucleosome (Wolffe, 1998). Thus, physical and functional links between SPUR and THR in TGFβ1-activated myofibroblasts may be accomplished in the context of nucleosome packing and/or chromatin folding. One working model for SMA gene repression places Pur protein at the double-strand SPUR as well as THR where a purine-rich, single-strand site may be exposed due to cooccupancy by MSY1 on the opposite, pyrimidine-rich strand. Transient activation, a key feature of the known physiological response of stromal fibroblasts to TGFβ1, could be accomplished in this model by 1) deployment of nuclear Smads, 2) Smad-assisted removal of Pur protein from Sp1 at SPUR, 3) subsequent, rapid turnover of nuclear Smads, and finally 4) reengagement of the Sp1:Pur repressor complex at SPUR within 24 h after TGFβ1 receptor occupancy. Turnover of nuclear Smads may enable Pur and MSY1 proteins to quickly reconfigure a repressed, single-strand DNA conformational state at the THR. Unlike the situation at SPUR, TGFβ1 signaling does not seem to physically displace Pur from the THR site, so it may be readily available to collaborate with MSY1 and efficiently reform the repressed cruciform configuration at THR. These hypothetical events at THR also may terminate transcriptional activity at the nearby MCAT element via impairment of its cognate TEF1 binding protein (Carlini et al., 2002), thus reestablishing a low, basal level of SMA gene output. Later association of MSY1 with SPUR may further stabilize transcriptional repression by enhancing the inhibitory grip of Pur protein (Kelm et al., 2003) and/or disrupting duplex DNA structure at this region of the SMA enhancer to prevent Sp1 binding.
Differential Roles of Pur Protein Isoforms in Myofibroblast Activation and Its Significance in the Pathobiology of Chronic Rejection
Of the two Pur proteins associated with mouse SMA gene transcriptional control, only Purα inhibition of the SMA enhancer was neutralized by TGFβ1 or Smads. Purβ seemingly lacks the ability to participate in TGFβ1-based SMA gene activation in myofibroblasts and functions as a dominant negative type regulatory protein. However, our recent work on SMA promoter regulation in aortic smooth muscle cells clearly showed that Purβ-based repression can be alleviated by serum response factor, an important lineage determinant in smooth muscle cells that interacts with myocardin to direct cardiovascular development and maintenance of blood vessels in the mouse (Kelm et al., 2003). These observations suggest that the negative effect of Purβ on SMA transcription might be countered by developmental signals mediated by SRF designed to allow for long-term accumulation of essential muscle contractile proteins such as SMA in vascular smooth muscle cells. In contrast, activators such as Smad proteins that accumulate temporarily in response to inflammatory injury seem to lack the ability to override Purβ repression and thus may be more relevant in pathways that require SMA gene activation to be strategically controlled in a spatially or temporally restricted manner such as myofibroblast-based wound healing. Our data clearly shows that Purα is released from SPUR in parallel with SMA transcriptional derepression and additional studies will be required to determine whether this DNA binding behavior is unique to the Purα isoform. If functional differences exist, misregulation of Pur protein gene expression or their interactions with enhancer DNA and/or transcriptional activators such as Sp1 or Smads could explain some pathobiological features of chronic fibrotic diseases, including CAD. We found that Pur protein levels were very low in the nontransplanted, adult mouse heart but became significantly elevated in accepted cardiac allografts showing signs of chronic rejection. Immunohistological analysis previously showed that accumulation of interstitial and perivascular myofibroblasts were largely responsible for the observed increase in nuclear Pur protein levels in accepted cardiac allografts 30 d after transplant when chronic rejection and transplant vascular sclerosis first becomes evident (Subramanian et al., 2002).
We also demonstrated in this report that postnatal α-actin gene reprogramming in the mouse ventricle occurred during a developmental period characterized by high level expression of Purβ. Although circumstantial, this finding nonetheless is consistent with the potent suppressive effect of Purβ on SMA gene transcription in fibroblasts, shown in this report, and our recent study on aortic smooth muscle cells (Kelm et al., 2003). Late embryonic mouse heart development is characterized by complete down-regulation of the fetal SMA gene in cardiomyocytes and coordinate up-regulation of the gene encoding the cardiac muscle-specific isoform of α-actin (McHugh et al., 1991; Kumar et al., 1997). We speculate that Purβ contributes to this α-actin gene reprogramming process and that high level expression of Purβ in the newborn mouse ventricle reflects both legacy and ongoing SMA gene repression as the cardiac α-actin protein accumulates in the maturing heart. Interestingly, high levels of Purβ were associated with an animal model of heart failure and similarly observed in biopsies removed from human patients afflicted with chronic heart failure (Gupta et al., 2003). These investigators showed that Purβ can suppress expression of the α-myosin heavy chain gene promoter in transfected cardiomyocytes and thus could represent a new marker for gene silencing and ventricular remodeling in human heart disease.
In summary, we have shown that the SPUR region of the SMA promoter, which shows high affinity binding to Sp1, also binds to Pur proteins. The Sp1:Pur complex was dependent on DNA conformation and displayed sensitivity to TGFβ1. The Smad 2/3 proteins, key mediators of TGFβ1 receptor signaling, disrupted physical interaction between Sp1 and Pur proteins yet themselves bound to Pur protein as well as DNA sequence elements in the THR, a region known to undergo chromatin conformational changes in response to TGFβ1 (Becker et al., 2000). The Pur repressor protein may therefore occupy a strategic position in the SMA enhancer that takes advantage of its unusual ability to bind both single- and double-strand DNA as well as transiently coordinate the action of two newly identified protein partners, Smad 2/3 and Sp1, that drive activation of the SMA gene in TGFβ1-activated myofibroblasts. The emerging and highly interesting functional properties of Pur proteins provide new insight for developing therapeutic strategies for treating and preventing chronic diseases such as arteriosclerosis, idiopathic fibrosis of the liver, lung, kidney and heart, and cardiac allograft dysfunction where SMA gene expression is known to be poorly regulated.
Acknowledgments
This work supported by National Institutes of Health grants HL-70294 and HL-60876 to A.R.S., HL-54281 to R.J.K., and American Heart Association Predoctoral Fellowship grant 0215142B to J.A.P.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–04–0348. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–04–0348.
References
- Armstrong, A.T., Strauch, A.R., Starling, R.C., Sedmak, D.D., and Orosz, C.G. (1997a). Morphometric analysis of neointimal formation in murine cardiac allografts. Transplantation 63, 941-947. [DOI] [PubMed] [Google Scholar]
- Armstrong, A.T., Strauch, A.R., Starling, R.C., Sedmak, D.D., and Orosz, C.G. (1997b). Morphometric analysis of neointimal formation in murine cardiac allografts. II. Rate and location of lesion formation. Transplantation 64, 322-328. [DOI] [PubMed] [Google Scholar]
- Armstrong, A.T., Strauch, A.R., Starling, R.C., Sedmak, D.D., and Orosz, C.G. (1997c). Morphometric analysis of neointimal formation in murine cardiac grafts. III. Dissociation of interstitial fibrosis from neointimal formation. Transplantation 64, 1198-1202. [DOI] [PubMed] [Google Scholar]
- Becker, N.A., Kelm, R.J., Jr., Vrana, J.A., Getz, M.J., and Maher, L.J.I. (2000). Altered sensitivity to single-strand-specific reagents associated with the genomic vascular smooth muscle alpha-actin promoter during myofibroblast differentiation. J. Biol. Chem. 275, 15384-15391. [DOI] [PubMed] [Google Scholar]
- Black, F.M., Packer, S.E., Parker, T.G., Michael, L.H., Roberts, R., Schwartz, R.J., and Schneider, M.D. (1991). The vascular smooth muscle α-actin gene is reactivated during cardiac hypertrophy provoked by load. J. Clin. Investig. 88, 1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branton, M.H., and Kopp, J.B. (1999). TGF-β and fibrosis. Microbe Infect. 1, 1349-1365. [DOI] [PubMed] [Google Scholar]
- Carlini, L.E., Getz, M.J., Strauch, A.R., and Kelm, R.J., Jr. (2002). Cryptic MCAT enhancer regulation in fibroblasts and smooth muscle cells. Suppression of TEF-1 mediated activation by the single-stranded DNA-binding proteins, Purα, Purβ, and MSY1. J. Biol. Chem. 277, 8682-8692. [DOI] [PubMed] [Google Scholar]
- Cassiman, D., Libbrecht, L., Desmet, V., Denef, C., and Roskams, T. (2002). Hepatic stellate cell/myofibroblast subpopulations in fibrotic human and rat livers. J. Hepatol. 36, 200-209. [DOI] [PubMed] [Google Scholar]
- Cogan, J.G., Subramanian, S.V., Polikandriotis, J.A., Kelm, R.J., Jr., and Strauch, A.R. (2002). Vascular smooth muscle α-actin gene transcription during myofibroblast differentiation requires Sp1/3 protein binding proximal to the MCAT enhancer. J. Biol. Chem. 277, 36433-36442. [DOI] [PubMed] [Google Scholar]
- Cogan, J.G., Sun, S., Stoflet, E.S., Schmidt, L.J., Getz, M.J., and Strauch, A.R. (1995). Plasticity of vascular smooth muscle α-actin gene transcription. Characterization of multiple, single-, and double-strand specific DNA-binding proteins in myoblasts and fibroblasts. J. Biol. Chem. 270, 11310-11321. [DOI] [PubMed] [Google Scholar]
- Derynck, R., Zhang, Y., and Feng, X.H. (1998). Smads: transcriptional activators of TGF-β responses. Cell 95, 737-740. [DOI] [PubMed] [Google Scholar]
- Desmoulière, A., and Gabbiani, G. (1996). The role of myofibroblasts in wound healing and fibrocontractile diseases. In: The Molecular and Cellular Biology of Wound Repair, ed. R.A.F. Clark, New York: Plenum Press, 391-423.
- Fan, J.M., Ng, Y.Y., Hill, P.A., Nikolic-Paterson, D.J., Mu, W., Atkins, R.C., and Lan, H.Y. (1999). Transforming growth factor-β regulates tubular epithelialmyofibroblast transdifferentiation in vitro. Kidney Int. 56, 1455-1467. [DOI] [PubMed] [Google Scholar]
- Foster, D.N., Min, B., Foster, L.K., Stoflet, E.S., Sun, S., Getz, M.J., and Strauch, A.R. (1992). Positive and negative cis-acting regulatory elements mediate expression of the mouse vascular smooth muscle α-actin gene. J. Biol. Chem. 267, 11995-12003. [PubMed] [Google Scholar]
- Gokturk, C., et al. (2003). Overexpression of semicarbazide-sensitive amine oxidase in smooth muscle cells leads to an abnormal structure of the aortic elastic laminus. Am. J. Pathol. 163, 1921-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwel, P., Imagaki, Y., Hu, W., Walsh, M., and Ramirez, F. (1997). Sp1 is required for the early response of α2(I)collagen to transforming growth factor-β1. J. Biol. Chem. 272, 19738-19745. [DOI] [PubMed] [Google Scholar]
- Gupta, M., Sueblingvong, V., Raman, J., Jeevanadam, V., and Gupta, M.P. (2003). Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. Implications in the repression of alpha-myosin heavy chain during heart failure. J. Biol. Chem. 278, 44935-44948. [DOI] [PubMed] [Google Scholar]
- Hao, H., Gabbiani, G., and Bochaton-Piallat, M.-L. (2003). Arterial smooth muscle cell heterogeneity. Implications for atherosclerosis and restenosis development. Circ. Res. 23, 1510-1520. [DOI] [PubMed] [Google Scholar]
- Heldin, C.-H., Miyazono, K., and Ten Dijke, P. (1997). TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465-471. [DOI] [PubMed] [Google Scholar]
- Higashi, K., Inagaki, Y., Fujimori, K., Nakao, A., Kaneko, H., and Nakatsuka, I. (2003). Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J. Biol. Chem. 278, 43470-43479. [DOI] [PubMed] [Google Scholar]
- Hinz, B., Celetta, G., Tomasek, J.J., Gabbiani, G., and Chaponnier, C. (2001). Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol. Biol. Cell 12, 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm, R.J., Jr., Cogan, J.G., Elder, P.K., Strauch, A.R., and Getz, M.J. (1999a). Molecular interactions between single-stranded DNA-binding proteins associated with all essential MCAT element in the mouse smooth muscle α-actin promoter. J. Biol. Chem. 274, 14238-14245. [DOI] [PubMed] [Google Scholar]
- Kelm, R.J., Jr., Elder, P.K., and Getz, M.J. (1999b). The single-stranded DNA-binding proteins, Purα, Purβ, and MSY1 specifically interact with an exon 3-derived mouse vascular smooth muscle α-actin messenger RNA sequence. J. Biol. Chem. 274, 38268-38275. [DOI] [PubMed] [Google Scholar]
- Kelm, R.J., Jr., Sun, S., Strauch, A.R., and Getz, M.J. (1996). Repression of transcriptional enhancer factor-1 and activator protein-1-dependent enhancer activity by vascular actin single-stranded DNA binding factor 2. J. Biol. Chem. 271, 24278-24285. [DOI] [PubMed] [Google Scholar]
- Kelm, R.J., Jr., Wang, S.X., Polikandriotis, J.A., and Strauch, A.R. (2003). Structure/function analysis of mouse purβ, a single-stranded DNA-binding repressor of vascular smooth muscle α-actin gene transcription. J. Biol. Chem. 278, 38749-38757. [DOI] [PubMed] [Google Scholar]
- Kumar, A., et al. (1997). Rescue of cardiac-actin-deficient mice by enteric smooth muscle gamma-actin. Proc. Natl. Acad. Sci. USA 94, 4406-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S., et al. (1999). Targeted overexpression of parathyroid hormone-related protein (PTHrP) to vascular smooth muscle in transgenic mice lowers blood pressure and alters vascular contractility. Endocrinology 140, 1815-1825. [DOI] [PubMed] [Google Scholar]
- March, K.L., Sandusky, G., and Fan, L. (1999). Hyperplasia in multiple smooth muscle tissues in transgenic mice expressing a temperature-sensitive SV40 T-antigen under the control of smooth muscle alpha-actin regulatory sequences. Oncogene 18, 3773-3782. [DOI] [PubMed] [Google Scholar]
- Massague, J., and Wotton, D. (2000). Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19, 1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, K., and Wolffe, A.P. (1998). Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 8, 318-323. [DOI] [PubMed] [Google Scholar]
- McGraw, D.W., Forbes, S.L., Kramer, L.A., Witte, D.P., Fortner, C.N., Paul, R.J., and Liggett, S.B. (1999). Transgenic overexpression of β2-adrenergic receptors in airway smooth muscle alters myocyte function and ablates bronchial hyperreactivity. J. Biol. Chem. 274, 32241-32247. [DOI] [PubMed] [Google Scholar]
- McHugh, K.M., Crawford, K., and Lessard, J.L. (1991). A comprehensive analysis of developmental and tissue-specific expression of the isoactin multigene family in the rat. Dev. Biol. 148, 442-458. [DOI] [PubMed] [Google Scholar]
- Min, B., Foster, D.N., and Strauch, A.R. (1990). The 5′-flanking region of the mouse vascular smooth muscle α-actin gene contains evolutionarily conserved sequence motifs within a functional promoter. J. Biol. Chem. 265, 16667-16675. [PubMed] [Google Scholar]
- Nedelec, B., Shankowsky, H., Scott, P.G., Ghahary, A., and Tredget, E.E. (2001). Myofibroblasts and apoptosis in human hypertrophic scars: the effect of interferon-α2b. Surgery 130, 798-808. [DOI] [PubMed] [Google Scholar]
- Nobe, K., Sutliff, R.L., Kranias, E.G., and Paul, R.J. (2001). Phospholamban regulation of bladder contractility: evidence from gene-altered mouse models. J. Physiol. 535, 867-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, T.G., Packer, S.E., and Schneider, M.D. (1990). Peptide growth factors can provoke “fetal” contractile protein gene expression in rat cardiac myocytes. J. Clin. Investig. 85, 507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncelet, A.-C., and Schnaper, H.W. (2001). Sp1 and Smad proteins cooperate to mediate transforming growth factor-β1-induced α2(I) collagen expression in human glomerular mesangial cells. J. Biol. Chem. 276, 6983-6992. [DOI] [PubMed] [Google Scholar]
- Powell, D.W., Mifflin, R.C., Valentich, J.D., Crowe, S.E., Saada, J.I., and West, A.B. (1999). Myofibroblasts. I. Paracrine cells important in health and disease. Am. J. Physiol. 277, C1-C19. [DOI] [PubMed] [Google Scholar]
- Ronnov-Jessen, L., and Petersen, O.W. (1993). Induction of α-smooth muscle actin by transforming growth factor-β1 in quiescent human breast gland fibroblasts. Implications for myofibroblast generation in breast neoplasia. Lab. Investig. 68, 696-707. [PubMed] [Google Scholar]
- Ronnov-Jessen, L., Petersen, O.W., Koteliansky, V.E., and Bissell, M.J. (1995). The origin of the myofibroblasts in breast cancer. Recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J. Clin. Investig. 95, 859-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlidmeyer, L.A., Braun, R., Taffet, G.E., Debiasi, M., Burns, A.E., Bradley, A., and Schwartz, R.J. (2000). Impaired vascular contractility and blood pressure homeostasis in the smooth muscle alpha-actin null mouse. FASEB J. 14, 2213-2220. [DOI] [PubMed] [Google Scholar]
- Serini, G., Bochaton-Piallat, M.L., Ropraz, P., Geinoz, A., Borsi, L., Zardi, L., and Gabbiani, G. (1998). The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-β1. J. Cell Biol. 142, 873-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serini, G., and Gabbiani, G. (1999). Mechanisms of myofibroblast activity and phenotypic modulation. Exp. Cell Res. 250, 273-283. [DOI] [PubMed] [Google Scholar]
- Shelley, C.S., Teodoridis, J.M., Park, H., Farokhzad, O.C., Bottinger, E.P., and Arnaout, M.A. (2002). During differentiation of the monocytic cell line U937, Pur alpha mediates induction of the CD11c beta 2 integrin promoter. J. Immunol. 168, 3887-3893. [DOI] [PubMed] [Google Scholar]
- Shi, C.W., Russell, M.E., Bianchi, C., Newell, J.B., and Haber, E. (1994). Murine model of accelerated transplant arteriosclerosis. Circ. Res. 75, 199-207. [DOI] [PubMed] [Google Scholar]
- Shi, X., Bai, S.T., Li, L., and Cao, X. (2001). Hoxa-9 represses transforming growth factor-beta-induced osteopontin gene transcription. J. Biol. Chem. 276, 850-855. [DOI] [PubMed] [Google Scholar]
- Shi, X., Yang, X., Chen, D., Chang, Z., and Cao, X. (1999). Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem. 274, 13711-13717. [DOI] [PubMed] [Google Scholar]
- Shi, Y., O'Brien, J.E., Fard, A., Mannion, J.D., Wang, D., and Zalewski, A. (1996). Adventitial myofibroblasts contribute to neointimal formation in injured porcine coronary arteries. Circulation 94, 1655-1664. [DOI] [PubMed] [Google Scholar]
- Skalli, O., and Gabbiani, G. (1988). The biology of the myofibroblast. Relation to wound contraction and fibrocontractive diseases. In: The Molecular and Cellular Biology of Wound Repair, ed. R.A.F. Clark and P.M. Henson, New York: Plenum Press, 373-401.
- Subramanian, S.V., Kelm, R.J., Jr., Polikandriotis, J.A., Orosz, C.G., and Strauch, A.R. (2002). Reprogramming of vascular smooth muscle alpha-actin gene expression as an early indicator of dysfunctional remodeling following heart transplant. Cardiovasc. Res. 54, 539-548. [DOI] [PubMed] [Google Scholar]
- Subramanian, S.V., Orosz, C.G., and Strauch, A.R. (1998). Vascular smooth muscle α-actin expression as an indicator of parenchymal cell reprogramming in cardiac allografts. Transplantation 65, 1652-1656. [DOI] [PubMed] [Google Scholar]
- Sun, S., Stoflet, E.S., Cogan, J.G., Strauch, A.R., and Getz, M.J. (1995). Negative regulation of the vascular smooth muscle α-actin gene in fibroblasts and myoblasts: disruption of enhancer function by sequence-specific single-stranded-DNA-binding proteins. Mol. Cell. Biol. 15, 2429-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, M.A., McCulloch, A., and Borg, T.K. (2002). Dance band on the titanic. Biomechanical signaling in cardiac hypertrophy. Circ. Res. 91, 888-898. [DOI] [PubMed] [Google Scholar]
- Suzuki, J.I., et al. (1996). Nonmuscle and smooth muscle myosin heavy chain expression in rejected cardiac allografts - a study in rat and monkey models. Circulation 94, 1118-1124. [DOI] [PubMed] [Google Scholar]
- Tomasek, J.J., Gabbiani, G., Hinz, B., Chaponnier, C., and Brown, R.A. (2002). Myofibroblasts and mechano-regulation of connective tissue remodeling. Nat. Rev. 3, 349-363. [DOI] [PubMed] [Google Scholar]
- Tretiakova, A., Steplewski, A., Johnson, E.M., Khalili, K., and Amini, S. (1999). Regulation of myelin basic protein gene transcription by Sp1 and Purα: evidence for association of Sp1 and Purα in brain. J. Cell Physiol. 181, 160-168. [DOI] [PubMed] [Google Scholar]
- Wang, J.W., Niu, W., Nikiforov, Y., Naito, S., Chernausek, S., Witte, D., LeRoith, D., Strauch, A., and Fagin, J.A. (1997). Targeted overexpression of IGF-I evokes distinct patterns of organ remodeling in smooth muscle cell tissue beds of transgenic mice. J. Clin. Investig. 100, 1425-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.W., Niu, W., Witte, D.P., Chernausek, S.D., Nikiforov, Y.E., Clemens, T.L., Sharifi, B., Strauch, A.R., and Fagin, J.A. (1998). Overexpression of insulin-like growth factor-binding protein-4 (IGFBP-4) in smooth muscle cells of transgenic mice through a smooth muscle α-actin-IGFBP-4 fusion gene induces smooth muscle hypoplasia. Endocrinology 139, 2605-2614. [DOI] [PubMed] [Google Scholar]
- Weber, K.T. (1995). Wound Healing in Cardiovascular Disease, Armonk, NY: Futura Publishing Co., Inc.
- Wolffe, A.P. (1998). Chromatin. Structure and Function, 3rd ed., San Diego: Academic Press.
- Zalewski, A., and Shi, Y. (1997). Vascular myofibroblasts - lessons from coronary repair and remodeling. Arterioscler. Thromb. Vasc. Biol. 17, 417-422. [DOI] [PubMed] [Google Scholar]