Abstract
Characteristic features of morphogenesis in filamentous fungi are sustained polar growth at tips of hyphae and frequent initiation of novel growth sites (branches) along the extending hyphae. We have begun to study regulation of this process on the molecular level by using the model fungus Ashbya gossypii. We found that the A. gossypii Ras-like GTPase Rsr1p/Bud1p localizes to the tip region and that it is involved in apical polarization of the actin cytoskeleton, a determinant of growth direction. In the absence of RSR1/BUD1, hyphal growth was severely slowed down due to frequent phases of pausing of growth at the hyphal tip. During pausing events a hyphal tip marker, encoded by the polarisome component AgSPA2, disappeared from the tip as was shown by in vivo time-lapse fluorescence microscopy of green fluorescent protein-labeled AgSpa2p. Reoccurrence of AgSpa2p was required for the resumption of hyphal growth. In the Agrsr1/bud1Δ deletion mutant, resumption of growth occurred at the hyphal tip in a frequently uncoordinated manner to the previous axis of polarity. Additionally, hyphal filaments in the mutant developed aberrant branching sites by mislocalizing AgSpa2p thus distorting hyphal morphology. These results define AgRsr1p/Bud1p as a key regulator of hyphal growth guidance.
INTRODUCTION
Polarized cell growth in both unicellular and multicellular organisms is a prerequisite for cellular and organ morphogenesis (Harold, 1995). Hyphal tip extensions of filamentous fungi, neuronal outgrowth in vertebrates, and extension of pollen tubes in plants are extreme examples of polarized cell growth. In the case of filamentous fungi, this leads to the production of long tubular cells, the hyphae. In general, polarized fungal growth occurs via the restriction of the delivery of new membrane and cell wall components to hyphal tips (Wendland, 2001; Momany, 2002). The actin cytoskeleton and microtubules play an important role in this targeted delivery of vesicles to sites of growth (Heath and Steinberg, 1999). The tendency of hyphae to grow in a stable direction for considerable distances is a property common to all filamentous fungi. This phenomenon requires growth guidance and the maintenance of growth direction similarly as found in neuronal cells (Luo, 2000; Dickson, 2001). As hyphae elongate at the tips, the growing tip is the prime place where growth guidance should be executed.
Although much research has been done to understand the ability of hyphae to change their growth direction in response to external stimuli such as electrical fields, calcium concentration, and nutrient gradients, the internal determinants of growth guidance largely remain unknown (Gow, 1994). An interesting class of mutants of the ascomycete Neurospora crassa, the ropy mutants, revealed distorted hyphal morphologies and frequent loss of growth directionality that were correlated with sustained misalignments of the Spitzenkorper (vesicle supply center) at the hyphal tip (Riquelme et al., 2000). N. crassa ropy mutants were found to encode components of the dynein/dynactin complex; ro-1, for example, is deficient in the dynein heavy chain (Plamann et al., 1994). However, ropy mutants show pleiotropic defects and most of the studies concentrated on nuclear distribution defects in these mutants (Plamann et al., 1994; Bruno et al., 1996; Tinsley et al., 1996). A recent study of morphogenesis mutants in Neurospora crassa has in addition identified signaling components as mediators of hyphal growth direction (Seiler and Plamann, 2003). Maintenance of polar growth also has been studied in the budding yeast Saccharomyces cerevisiae, however, these results may be difficult to apply to polar growth in filamentous fungi because one of the main differences between yeast-like growth and filamentous growth is in the maintenance of polarized growth (Wendland, 2001; Momany, 2002).
An evolutionarily conserved theme is the participation of the actin cytoskeleton in polarized growth from yeast and filamentous fungi to mammalian cells (Schmidt and Hall, 1998; Ayad-Durieux et al., 2000; Etienne-Manneville and Hall, 2002; Knechtle et al., 2003). Furthermore, Rho-GTPases have been involved in polarized morphogenesis during neuronal growth cone guidance as well as in hyphal cells (Luo, 2000; Dickson, 2001; Wendland and Philippsen, 2001; Nakamura et al., 2002; Zhang et al., 2003).
The Cdc42-Rho-GTPase plays a conserved role in the establishment of cell polarity and is required for the formation of hyphal tips in A. gossypii (Wendland and Philippsen, 2001). In other filamentous fungi, Cdc42p has been localized to the hyphal tips of growing hyphae, suggesting its continuous requirement during hyphal growth (Gorfer et al., 2001). In S. cerevisiae, the machinery regulating the process of bud-site selection acts upstream of CDC42 and acts in a cell type-specific way to impose either the axial budding pattern in MATa or MATα haploid cells or the bipolar budding pattern in diploid MATa/α cells (Pringle et al., 1995; Herskowitz et al., 1995; Casamayor and Snyder, 2002). We have previously studied the role of the Ashbya gossypii BUD3 homolog, which in S. cerevisiae plays a role in the axial budding pattern (Chant and Herskowitz, 1991). However, in our study, AgBUD3 was found not to play a role in growth guidance but rather was found to be involved in septum positioning (Wendland, 2003). Recently, a linkage between the Rho-GTPase Cdc42p and the Ras-GTPase Rsr1p/Bud1p has been described in S. cerevisiae via the direct interaction of activated Rsr1p/Bud1p (in its GTP-bound form) with Cdc24p, which represents the guanine-nucleotide exchange factor for Cdc42, or even a direct interaction with Cdc42p (Park et al., 1997; Kozminski et al., 2003).
To explore the potential role of a Ras-like GTPase in growth guidance of a filamentous fungus, we identified a RSR1/BUD1 homolog in A. gossypii and studied the growth dynamics of strains deleted for this gene by using in vivo time-lapse microscopy. We show that AgRsr1p/Bud1p localizes to the tip region, is essential for sustained polar growth, has an important function in maintaining the axis of polarity, and is involved in maintaining a strongly polarized distribution of the cortical actin cytoskeleton at the hyphal tip. In addition, monitoring the subcellular localization of a polarisome component via in vivo time-lapse fluorescence microscopy by using a AgSPA2-GFP fusion supports the conclusion that in A. gossypii Rsr1p/Bud1p exerts growth guidance control via polarisome components.
MATERIALS AND METHODS
A. gossypii Strains and Growth Conditions
The A. gossypii strains that were used in this study are listed in Table 1. A. gossypii strains were grown in full medium (AFM) or defined minimal medium (AMM) plus necessary amino acids (Ayad-Durieux et al., 2000). For selection of A. gossypii transformants that have integrated the GEN3 marker the aminoglycoside G418/Geneticin (Invitrogen, Carlsbad, CA) was used at a final concentration of 200 μg/ml. For selection of A. gossypii transformants that have integrated the ScLEU2 marker, transformants were plated on AMM lacking leucine. Escherichia coli XL1-blue was used as plasmid host. Bacteria were grown at 37°C in 2× YT medium (1.6% Tryptone, 1% yeast extract, and 0.5% NaCl) supplemented with 100 μg/ml ampicillin for selection.
Table 1.
A. gossypii strains used in this study
| Name | Genotype | Source |
|---|---|---|
| Wild type | ATCC10895 | |
| Agleu2Δ | leu2Δthr4Δ | Altmann-Johl, 1996 |
| AYA27 | Agrsr1Δ::GEN3, leu2Δ thr4Δ | This study |
| AYA32 | Agrsr1Δ::LEU2, leu2Δ thr4Δ | This study |
| AYA38 | AgSPA2-GFP::GEN3, leu2Δ thr4Δ | Knechtle et al., 2003 |
| AYA40 | AgSPA2-GFP::GEN3, Agrsr1Δ::LEU2, leu2Δ thr4Δ | This study |
Sequence Analysis and Standard Procedures
Sequencing was performed on an ABI377A automated sequencer according to the manufacturer's instructions. The sequences of AgRSR1 plus flanking regions were obtained by partially sequencing genomic clones (pAG10683 obtained from F. Dietrich, University of Basel, Switzerland, and bAG1636, bAG1819 obtained from W. Choi and R. Wing, Clemson University, Clemson, SC). The protein sequence alignment was constructed using the MegAlign program that is part of the DNASTAR software package. Profile searches were done on the ISREC profile server. Standard DNA manipulations were performed as described by Sambrook et al., 1989.
Construction of AgRSR1 Deletion Mutants
Deletions of AgRSR1 were done using a polymerase chain reaction (PCR)-based method as described previously (Wendland et al., 2000). The oligonucleotides S1 and S2 used for the generation of the deletion cassettes are listed in Table 2. The S. cerevisiae LEU2 gene was used as alternative marker to delete AgRSR1 in the Agleu2Δ and AgSPA2-GFP strain strains. The pScLEU2 plasmid used as a template to generate the ScLEU2 deletion cassette was created by insertion of the entire ScLEU2 gene between the BglII site of pAF100 as described for the GEN3 marker (Wendland et al., 2000). Because pGEN3 and pScLEU2 share the same vector backbone, we were able to use the same S1 and S2 primer pair to generate a ScLEU2 disruption cassette with homology to the AgRSR1 gene. Transformation of A. gossypii strains was done by a modified electroporation method (Wendland et al., 2000). After electroporation, the mycelium was resuspended in 5 ml of AFM liquid medium and incubated for 6 h at 30°C to allow expression of the marker gene. Then mycelium was subsequently washed twice in sterilized water and spread on selective plates. Isolation of A. gossypii genomic DNA of transformants was performed as described (Wright and Philippsen, 1991). Correct genomic integration was verified by analytical PCR by using the primers listed in Table 2. The transformation of young multinucleate mycelia produces heterokaryotic transformants that carry in some nuclei the wild-type allele and in others the mutant allele. Isolation of spores containing a single haploid nucleus from heterokaryotic transformants on G418/Geneticin-containing medium or on minimal medium gives rise to homokaryotic transformants in which all nuclei carry the mutant allele (Table 1). After PCR verification of the resulting homokaryotic transformants, a phenotypic analysis was initiated. Homokaryotic spores of AgRSR1 deletion mutants were germinated on selective medium containing plates and incubated for 5 d at 16, 30, and 37°C to analyze radial growth speeds.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| S1-AgRSR1 | CTGATGGAGCTGCGGGAGCAGATCCTGCGGATCAAGGACTCGAAggctagggataacagggtaat |
| S2-AgRSR1 | GCACCAGGTCGATGAACACCTCGTCCACGTTGCTCTTCAGCAACgaggcatgcaagcttagatct |
| G1-AgRSR1 | GATAAGCTTGATCGCTTGCC |
| G4-AgRSR1 | AGCTCCGCGCGCATGATCTG |
| G2 | gtttagtctgaccatctcatctg |
| G3 | tcgcagaccgataccaggatc |
| L2 | tttaggaccagccacagcacc |
| L3 | aactggtgatttaggtggttcc |
| 5′-AgRSR1 | cggaattcATGAGGGACTACAAGTTGGTTGTGC |
| 3′-AgRSR1 | ggactagtCACAATAAGGAGACAACTACAGTGG |
Uppercase letters correspond to A. gossypii sequences; lowercase letters correspond to plasmid annealing region, internal marker regions, and restriction sites, respectively.
Microscopy Procedures and Green Fluorescent Protein (GFP) Tagging
Visualization of the actin cytoskeleton was done as described previously (Amberg, 1998; Knechtle et al., 2003). Briefly, 1.0 ml of an A. gossypii culture was fixed with 100 μl of 37% formaldehyde for 10 min, washed, and fixed a second time in phosphate-buffered saline (PBS) supplemented with 0.03% Triton X-100 (PBST) and 4% formaldehyde. Cells were washed twice with PBST, resuspended in PBST with 6.6 μm Alexa 488-phalloidin (Molecular Probes, Eugene, OR), and incubated for 1 h. Stained cells were washed five times and resuspended in mounting medium. This method produces substantial enhancements compared with previous staining methods for which the mycelia were centrifuged and resuspended in PBS before fixation. To stain chitin, calcofluor was added directly to the medium at a concentration of 1 mg/ml, incubated for 10 min, washed with PBS, and resuspended in mounting medium.
For in vivo time-lapse studies, strains were grown in liquid AFM medium until they had reached the required developmental phase. The preparation of the ground well microscopy slide was done according to Hoepfner et al. (2000). To this end, 1.0 ml of the culture containing pregerminating spores was briefly centrifuged and resuspended in 500 μl of fresh AFM medium. The spores were then placed on top of the ground well microscopy slide and covered by a coverslip that was partially sealed to avoid dehydration of the sample during the investigation time but that still allowed oxygenation. The temperature near the microscopy slide was 24°C as measured with a thermo-element.
To generate a GFP-tagged version of AgRSR1 the gene was amplified by PCR from genomic DNA with the oligonucleotides 5′-AgRSR1 and 3′-AgRSR1, resulting in a product with the complete AgRSR1 ORF (795 base pairs) followed by a terminator region (204 base pairs), a 5′-flanking EcoRI site, and a 3′-flanking SpeI site. This product was cloned into the EcoRI/SpeI site of pHPS250, a pYCP111 derivative for N-terminal GFP fusions that carries the AgRHO1 promoter from the ABR183W gene followed by the GFP (Heim and Tsien, 1996) and EcoRI/SpeI cloning sites for in frame integration of the gene of interest (Schmitz, unpublished data). pYCP111 is a CEN-ARS plasmid with the S. cerevisiae LEU2 gene (Gietz and Sugino, 1988), replicates freely in A. gossypii, and is staby maintained upon selection on leucine drop out medium (Wright and Philippsen, 1991). The newly generated plasmid pAG919 was transformed into strain AYA27, and transformants were selected on leucine drop out medium. The construct expressed a functional N-terminal GFP fusion of AgRsr1p.
Image Acquisition and Computer Programs
The microscopy setup for inspection of mycelium stained for actin of chitin included a 75W/XBO epifluorescence illumination source, an axioplan2 microscope (Carl Zeiss, Feldbach, Switzerland) and a fluorescein isothiocyanate filter set (Chroma Technology, Brattelboro, VT). The microscope, the camera, and the fluorescence shutter were controlled by the MetaMorph 3.51 software (Universal Imaging, West Chester, PA). MetaMorph was used to collect z-axis and to process a series of pictures. The pictures were then converted to eight-bit files and finally contrast-enhanced using Photoshop 4.0 (Adobe Systems Europe, Edinburgh, Scotland). For time-lapse analysis, we used the same microscopy setup as described for the actin staining. Acquisition settings for the time-lapse studies were 2- or 5-min interval time, 1-s exposure time, and one z-axis plane. The phase contrast pictures files were assembled and movies were processed in QuickTime format (Apple Computer, Cupertino, CA) by using the program Adobe Premiere (Adobe systems Europe) as described previously (Hoepfner et al., 2000).
RESULTS
The A. gossypii Homolog of the S. cerevisiae RSR1/BUD1 Gene
We identified a Ras-related GTPase, AgRsr1p, based on amino acid sequence comparisons of A. gossypii sequences with other fungal GTP-binding proteins (Figure 1). This GTPase shares 64% identity on amino acid level with its S. cerevisiae homolog ScRsr1p/Bud1p, and the four GTP-binding domains are conserved up to 100%. We constructed a deletion inactivating two-thirds of the open reading frame (ORF) by using standard PCR-based gene targeting (see Materials and Methods). Analysis of radial growth of colonies at different temperatures revealed 40–50% decreased radial growth rates for the deletion mutant compared with wild type (Figure 7). To explore these differences in growth rate, we followed by brightfield microscopy the development of nine spores each from wild type and Agrsr1Δ on solid full medium at 30°C. For all spores, we observed the normal development of germlings as described previously (Ayad-Durieux et al., 2000; Knechtle et al., 2003). During 4–6 h after the first hyphal tube emerged from the germ bubble, the second hyphal tube started growing on the opposite site, followed by septum formation and branch initiation at the base of the first hyphal tube (Knechtle et al., 2003). During the next 4–6 h, young mycelia began to show differences in development. Tip extension speeds in all hyphae increased, but compared with wild type the elongation speed of mutant hyphae was on average 25% slower and hyphae frequently changed growth directions. In addition, only one-half as many lateral branches developed in the mutant. Hyphal growth speeds continued to increase in both strains and reached, 14–16 h after germination, 123±23 μm/h in wild-type but only 42±11 μm/h in mutant hyphae (unpublished data). The maximal speed determined from radial growth of fungal colonies is close to 200 μm/h for wild type and 100–120 μm/h for the AgRSR1 deletion strain (see below).
Figure 1.
Alignment of GTP-binding proteins of the Ras-superfamily. Identical amino acids residues are shaded. Conserved domains are highlighted. Sequence accession numbers are as follows: A. gossypii Rsr1 (AgRsr1), AE016819; S. cerevisiae ScRsr1-ScBud1, P13856; C. albicans CaRsr1, AAB81286; N. crassa Nckrev-1, BAA32410; S. cerevisiae Ras1 (ScRas1), P01119; and S. cerevisiae Ras2 (ScRas2), P01120.
Figure 7.
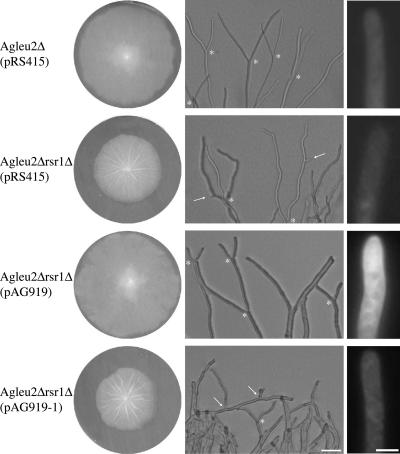
Functional complementation of Agleu2Δrsr1Δ and localization of AgRsr1p. Agleu2Δ and Agleu2Δrsr1Δ strains were transformed with the indicated plasmids: the control plasmid pRS415 carrying S. cerevisiae ARS and CEN sequences and the LEU2 gene, pAG919 carrying in addition an in-frame GFP-AgRSR1 fusion and pAG919-1 carrying an out-of-frame GFP-AgRSR1 fusion. Left column, fungal colonies grown for 5 d on leucine drop out agar. Middle column, images of colony edges of the respective transformants with tip branching indicated by asterisks and lateral branching by arrows. Right column, fluorescence microscopy images taken under identical conditions of hyphae grown for 16 h in liquid leucine drop out medium. Tip localization of GFP-fluorescence with a cap-like structure at the very tip can be observed for Agleu2Δrsr1Δ(pAG919). Bar, 50 μm (middle); 5 μm (right).
Dynamics of Young Mycelium Development in A. gossypii Wild Type and Agrsr1Δ
To explore in more detail differences in development of both A. gossypii strains, we used in vivo time-lapse microscopy. First, we monitored in 2-min intervals the development of A. gossypii wild type from germinating spores to multibranched young mycelia (Movie 1). Representative frames in Figure 2 show the typical developmental stages as described above. Under the same experimental conditions, mycelial development of Agrsr1Δ was characterized (Movie 2). The developmental pattern, as documented by representative frames in Figure 3, is similar to the wild type up to the stage of bipolar germlings (0–8 h). However, analysis of the video data revealed already short phases of pausing. After this time, development of the Agrsr1Δ mycelium was strikingly different from wild type (8–14 h). We observed 1) frequent pausing of hyphal tip elongation; 2) frequent deviations from the axis of polarity, resulting in zig-zag–shaped hyphae; and 3) frequent attempts of lateral branch formation that only lead to irregular thickening of originally evenly shaped hyphal segments but not to lateral branches. Furthermore, whereas in the wild-type lateral branching often occurred at sites of previous septation, the frequent attempts to initiate branch formation in the Agrsr1Δ strain occurred at random positions.
Figure 2.
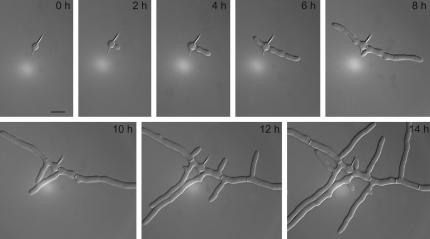
Development of an A. gossypii wild-type young mycelium monitored by in vivo time-lapse microscopy. Spores were pregrown for 4 h in complete liquid medium at 30°C before mounting for videomicroscopy (see Materials and Methods). Digital images were collected at 2-min intervals (see Movie 1). Representative frames taken at 2-h intervals show the development of wild-type mycelium. Typical landmarks are as follows: isotropic growth of the germ bubble (0 h), formation of the first (2 h) and second germ tube (6 h), and generation of septa and lateral branches (starting at 8–10 h). The time lapse was carried out at room temperature (25°C).
Figure 3.
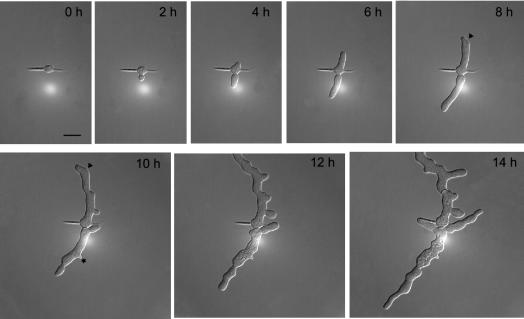
Development of a Agrsr1Δ young mycelium monitored by in vivo time-lapse microscopy (see Movie 2). Spores were prepared and images taken as described in Figure 2. Arrowheads point to changes in growth direction and the asterisk marks a complete growth arrest of a lateral branch.
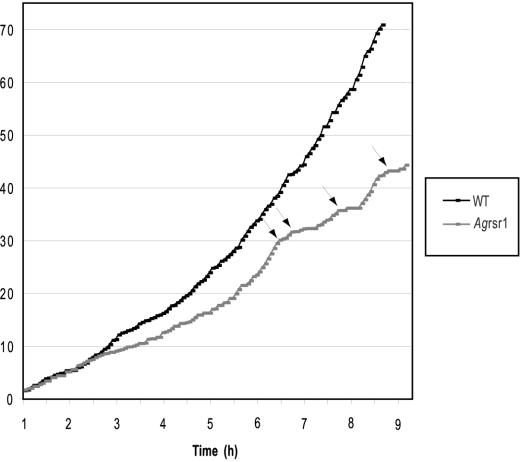
Quantitative data for tip extension rates and stability of growth axes, both representing essential parameters of growth guidance, and, in addition, hyphal diameters were determined using Movies 1 and 2 and two additional movies (unpublished data). Both sets of data were very similar. Figure 4 documents tip extensions of the main hypha in steps of 4 min for wild type (black dots) and the Agrsr1Δ (gray dots) based on Movies 1 and 2. In both strains, elongation rate was 3.4–3.8 μm/h during 2 h after formation of the first germ tube. During the next 6 h, this rate progressively increased to 15 μm/h in wild type and 9 μm/h in Agrsr1Δ. The lower number for the deletion is due to 24 growth pauses from 4 to 16 min. After pausing, growth resumed at the hyphal tip. In wild type, we saw only five short pausing events of up to 2 min during the same time. Thus, acceleration of tip extension speed, one hallmark of hyphal maturation, still functions in Agrsr1Δ. The other hallmark, hyphal splitting at all fast-growing tips into two fast-growing branches (Ayad-Durieux et al., 2000) also occurs in the deletion mutant but much delayed and less efficiently (Figure 7).
Figure 4.
Analysis of hyphal elongation rate in young mycelium of A. gossypii wild type and in Agrsr1Δ. The source of these measurements is Movie 1 and Movie 2, and the elongation of the first germ tube was measured over the entire length of the movies. Time zero corresponds to the last steps of isotropic growth before germination of the first germ tube (see Figures 3 and 4). Arrows mark prolonged periods of pausing in Agrsr1Δ.
To evaluate the stability of the axes of polarity, we measured the length of straight hyphal segments in both strains. Within the limits of our microscopic setup, we observed that the wild-type hyphae grew straight for up to 60 μm. Sometimes minor changes in growth direction were observed, the maximum angle of deviation from the original axis was 20°. In contrast, Agrsr1Δ hyphae grew straight only for a maximum of 28 μm followed by changes in growth direction up to 55°. Diameters at the tip region ranged from 2.0 to 4.7 μm for wild type and from 1.7 to 5.3 μm for the Agrsr1Δ mutant. In subapical regions, wild-type hyphae exhibited constant hyphal diameters corresponding to tube-like hyphae. The Agrsr1Δ strain, however, showed variable hyphal diameters indicated by a swollen appearance of the hyphae. Our time-lapse analysis revealed that Agrsr1Δ hyphae initially elongated mainly as regular hyphal tubes but that at later times bulges (up to 9 μm) formed in subapical regions indicative of additional irregular growth events (Figure 3, 10–14 h).
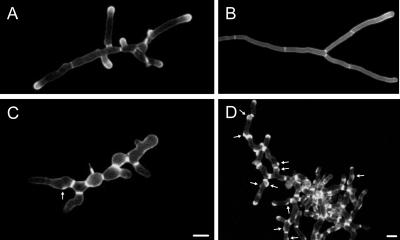
To find out whether chitin deposition in Agrsr1 was influenced by the frequent pausing and changes in growth direction, we stained young and mature mycelium of both strains with calcofluor (Figure 5). In wild-type hyphae, calcofluor marks sites of septation and areas of intense polarized growth such as tips and branching sites. Similarly to the wild type, Agrsr1Δ hyphae showed an accumulation of chitin at sites of septation and at tip regions. This indicates that the processes resulting in correct septation were not defective. However, in Agrsr1Δ the distances between chitin rings was much shorter in comparison to wild type, particularly in mature mycelium (Figure 5, B and D). In addition, the deletion mutant showed frequent deposition of chitin at cortical sites close to septin rings (Figure 5D, white arrows). The majority of these sites developed into small bulges, indicating that new axes of polarity at selected branch sites had been established but were not maintained.
Figure 5.
Chitin distribution in wild type (A and B) and in Agrsr1Δ (C and D). Septa and growth regions of hyphal tips are stained with calcofluor. Septa are thicker in Agrsr1Δ and develop at shorter distances compared with wild type, very likely due to frequent pausing events. Arrows mark sites of abortive branch initiations. Bar, 10 μm.
During A. gossypii wild-type development, the formation of bipolar germlings results from the initiation of a second germ tube at an angle of 180° from the first one (91/100) or in less frequent cases at an angle of 90° (9/100). Germ cells of the Agrsr1Δ mutant were still able to correctly produce the bipolar germination pattern (75/100) with an increase in perpendicular branching (25/100). The Agrsr1Δ mutant did not produce a random germination pattern. This indicates that for the very early phases of development AgRsr1p is not required for the establishment of correct axes of polarity.
AgRsr1p Influences the Polarity of the Actin Cytoskeleton
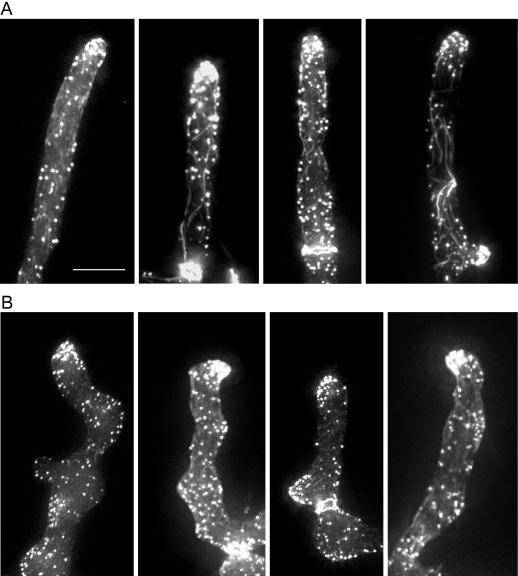
Because polarization of the cortical actin cytoskeleton is indicative of the axis of cell polarity, we examined the organization of the actin cytoskeleton in the A. gossypii wild type and the Agrsr1Δ mutant. Young and mature mycelia were fixed, and the actin cytoskeleton was stained with Alexa 488 phalloidin. In wild-type hyphae, we could observe clusters of cortical actin patches in >95% of tips, indicative of active growth, as well as actin rings at sites of developing septa, actin cables, and single patches at subapical cortical sites, confirming previous observations (Figure 6A; Wendland and Philippsen, 2000; Knechtle et al., 2003). In Agrsr1Δ, only 77% of tips in young mycelium and 66% of tips in mature mycelium showed clusters of actin patches, often in an asymmetric distribution (Figure 6B and Table 3). These data are in agreement with the observed frequent pausing of tip extension in Agrsr1Δ. Interestingly, in Agrsr1Δ strains the asymmetric distribution of the actin patches at the hyphal tip was often associated with a bent hyphal tip and therefore indicated a change in growth direction. These findings suggested that AgRsr1p plays a role in stabilizing the polarity axis at the tip.
Figure 6.
Organization of the actin cytoskeleton in wild type (A) and in Agrsr1Δ (B). Germinated spores of both strains were grown in liquid complete medium for 12 h at 30°C, fixed with formaldehyde, stained with Alexa 488-phalloidin, and analyzed by fluorescence microscopy. Four representative images of hyphal tips (n = 100 for each strain) are presented. Bar, 10 μm.
Table 3.
Actin polarization frequency and symmetry at hyphal tips
| Wild type (%)
|
Agrsr1Δ (%)
|
|||
|---|---|---|---|---|
| Polarizeda | Asymmetricb | Polarizeda | Asymmetricb | |
| Young mycelia (12 h) | 95 | 0 | 75 | 33 |
| Mature mycelia (20 h) | 97 | 0 | 66 | 12 |
Spores of each strain were inoculated in full liquid medium and incubated for the indicated period of time at 30°C. Samples were removed and stained with Alexa 488-phalloidin to visualize actin polymers. Polarization of actin patches at hyphal tips and the respective symmetry was scored.
n = 500
n = 100
A Functional GFP Fusion to AgRsr1p Locates to the Tip Region
AgRsr1p seems to play a major role in controlling sustained surface growth along a predetermined polarity axis. To locate this GTPase in growing hyphae, we constructed amino-terminal GFP fusions under control of promoters of three different A. gossypii GTPase genes. The vector plasmid is able to replicate in A. gossypii by using an S. cerevisiae ARS element, and it can be selected in an Agleu2 delta strain due to the S. cerevisiae LEU2 gene (see Materials and Methods). As control of functional expression, we tested complementation of the deletion phenotype in mature mycelium: decreased hyphal tip extension, instability of hyphal growth axis, suppressed tip branching, and unscheduled lateral branching in apical compartments. Transformants of Agleu2Δrsr1Δ with pAG919 fully complemented all four phenotypes as shown in Figure 7. When this plasmid carried a frame-shift in the first codon of the AgRSR1 ORF (pAG919-1), all transformants showed the complete deletion phenotype. Also, transformants expressing the GFP fusion from a weak promoter did not complement. GFP fluorescence in hyphae of pAG919 transformants was highly enriched in tip regions with a sharp zone at the very tip (Figure 7). We inspected up to 100 μm in apical compartments but could not observe enriched zones of GFP fluorescence at presumptive septa.
AgRsr1 as Stability Factor for the Polarisome Complex
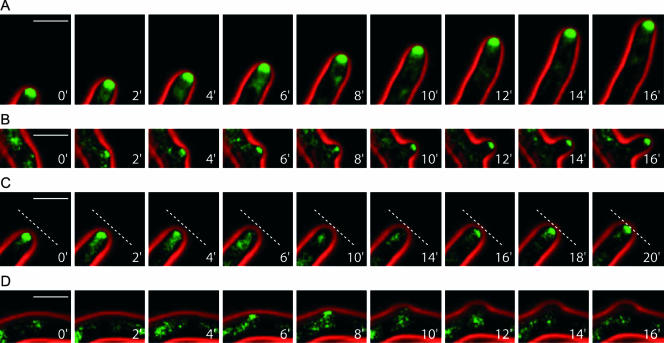
To investigate the changes in morphogenesis of Agrsr1Δ in more detail, we used in vivo time-lapse microscopy and monitored the distribution of a polarisome component in A. gossypii. Recently, AgSpa2p, a homolog of the S. cerevisiae polarisome component Spa2p, was shown by fusion to GFP to permanently localize to the tips of growing hyphae in A. gossypii and to occur as small patches at initiation sites for lateral branches to establish new axes of polarity (Knechtle et al., 2003). Localization of AgSpa2p-GFP could therefore be used to analyze events that occurred during pausing of hyphal growth in Agrsr1Δ strains. To this end, the genomic copy of AgSPA2 was tagged with GFP at its 3′ end in the Agrsr1Δ strain (see Materials and Methods). This allowed the expression of AgSPA2-GFP from its endogenous SPA2-promoter. Proper function of Spa2p-GFP was demonstrated previously (Knechtle et al., 2003). By using in vivo time-lapse fluorescence microscopy, accumulation of Spa2p-GFP was observed in growing hyphal tips of the wild type and also the Agrsr1Δ mutant strain (Movies 3 and 4). In A. gossypii wild type, AgSpa2p-GFP permanently localized to the growing hyphal tips and polarized at sites of branch initiation (Figure 8, A and B; Knechtle et al., 2003). Pausing of growth at tips, typical for Agrsr1Δ strains (Movie 2), however, was associated with the disappearance of AgSpa2-GFP patches at hyphal tips (Movie 4 and Figure 8), whereas reinitiation of tip growth coincided with the reappearance of AgSpa2-GFP at the corresponding hyphal tips (Figure 8C). Furthermore, we observed accumulation of AgSpa2-GFP at subapical cortical sites corresponding to lateral branch initials (Figure 8D). At these sites, however, cortical AgSpa2-GFP patches frequently disappeared in Agrsr1Δ shortly after lateral growth was induced which resulted in the abandonment of growth at these sites (Figure 8D). Movie 4 monitors tip elongation of three Agrsr1Δ hyphae (numbered 1–3 starting from the left) and growth of one lateral branch in the upper part of the frames during a period of 4 h. Brightfield (red) and fluorescence images (green) were taken every 2 min (see Materials and Methods). During the observation period, the GFP label at hyphal tips transiently disappeared seven times for 10, 4, and 12 min in hypha 1, for 105 min in hypha 2, and for 80, 10, and 4 min in hypha 3. Hyphal tips did not or did only marginally extend in the absence of AgSpa2p-GFP fluorescence at these tips. The longest noninterrupted growth periods were 80 min (hypha 1) and 130 min (hypha 2). Resumption of hyphal growth was detected almost immediately after the reappearance of Spa2p-GFP at the hyphal tips (with a delay of up to 2 frames, which corresponds to ∼2–4 min). Growth of cortical protrusions immediately after the appearance of a GFP signal at these sites was observed six times. These events were captured in hypha 2 (frames 56, 70/71 and 94/95) and in hypha 3 (frames 47/49, 99/100 and 105–107). Of particular interest is a second growth phase of the first cortical protrusion in hypha 1, the tip of which shows a transient GFP signal during frames 113 and 114. In addition to these observations, Movie 4 also captured three clearly asymmetric tip localization patterns of AgSpa2 in hypha 3, frames 10/11 and 28 and in the branch, frames 80/81. We assume that these asymmetric positions coincided with changes in growth direction as well as with asymmetrically located actin patch in hyphal tips (Figure 7). These time-lapse studies show that Agrsr1Δ hyphae do not achieve a state of permanent polarity at their tips and that they initiate but do not maintain branches at apparently random cortical sites.
Figure 8.
Loss of localization of the polarisome component AgSpa2-GFP during tip growth in Agrsr1Δ. AgSpa2p-GFP distribution in growing hyphal tips as well as during lateral branch initiation was monitored by in vivo time-lapse microscopy in A. gossypii wild type and in Agrsr1Δ. The series of images represent sections of selected frames of Movies 3 and 4. (A) Permanent localization of AgSpa2p-GFP in a growing tip. (B) Polarized AgSpa2p-GFP at a branching site in A. gossypii wild type. (C) Transient disappearance and reappearance of AgSpa2-GFP in Agrsr1Δ at a hyphal tip. (D) Transient assembly of AgSpa2p-GFP at a cortical site. The dashed lines are at constant positions in each image to allow monitoring of growth arrest and resumption of tip growth. The time interval represents minutes in real time. Bar, 10 μm.
DISCUSSION
Fungal hyphae in general are capable of maintaining a growth direction for a substantial period. From time to time, hyphae establish a new axis of growth, for example, to produce a lateral branch. Hyphal growth guidance defined as the ability of hyphae to maintain continuous polarized growth (in contrast to yeast cells) and the direction of growth has not been addressed at the molecular level in detail. In this study, we have used A. gossypii to investigate the molecular requirements for growth guidance of fungal hyphae and to provide evidence that the Ras-like GTPase, encoded by AgRSR1, is required for growth guidance by regulating the actin cytoskeleton and the localization of the polarisome component Spa2p.
BUD-Genes in A. gossypii versus BUD-Genes in S. cerevisiae
In S. cerevisiae, BUD-genes take part in the selection of a new bud site (Herskowitz et al., 1995; Pringle et al., 1995; Madden and Snyder, 1998). This process is cell type specific. Haploid cells (either a or α) exhibit an axial budding pattern in which cells use the proximal cell pole (the pole with which the daughter cell was connected to the mother cell) for budding. Diploid cells use both poles for budding, thus displaying a bipolar budding pattern. Genes were specifically assigned to one of these budding patterns. For example, ScBUD3 and ScBUD4 are required for the axial budding pattern. Deletion of either of these genes in haploid cells results in bipolar bud site selection, whereas their deletion in diploid cells does not affect the bipolar budding pattern. On the other hand, ScBUD8 and ScBUD9 are specific for the diploid budding pattern and do not affect axial budding of haploid cells (Casamayor and Snyder, 2002). These genes are thought to encode landmark proteins that act upstream of a GTPase module consisting of the Ras-like GTPase ScRsr1p/Bud1p, its guanine-nucleotide exchange factor ScBud5p, and its GT-Pase-activating protein ScBud2p. Deletion of any of these three module proteins leads to random budding regardless of cell type.
Evidence that BUD-gene homologs are present in A. gossypii came from the characterization of the A. gossypii AgBUD3 gene (Wendland, 2003). AgBud3p shows landmark protein features because it is found at sites of future septation and also may play a role in lateral branch formation, which in A. gossypii often takes place adjacent to sites of previous septation (resembling axial budding in yeast). More importantly, however, AgBud3p is involved in septum construction by recruiting AgCyk1p (the homolog of an S. cerevisiae protein controlling cytokinesis) to sites of septation. Loss of the AgBud3p ring at septal sites results in the formation of “linear actin rings” and of delocalized septa.
Here, we analyzed the A. gossypii homolog of ScRsr1p/Bud1p that is essential for cell type-specific selection of new bud sites but has no known function in vegetative growth (Pringle et al., 1995). In the Agrsr1Δ mutant, lateral branching was still initiated but often abandoned after a short period of polarized growth resulting in bulges along the hyphae. This indicates a role of AgRsr1p in preventing false branch site selection and in maintaining polarized hyphal growth of lateral branches.
AgRsr1p Is Required for Maintenance of Polarized Hyphal Growth and Growth Guidance
Hyphal maturation in A. gossypii as well as in other filamentous fungi is a two-phase process. Hyphae of young mycelia grow with a relatively slow elongation rate, ∼5–10 μm/h in A. gossypii. In contrast, mature hyphae reach a growth speed of ∼170–200 μm/h and characteristically show tip branching (Ayad-Durieux et al., 2000; Knechtle et al., 2003). Maturation also was found to occur in the Agrsr1Δ mutant, but hyphal growth speed reached only a maximum of ∼100 μm/h. With the use of videomicroscopy, we could demonstrate that Agrsr1Δ hyphae show frequent pausing and regain of growth. Pausing of growth was not accompanied by swelling of the hyphal tips as was observed in the Agbem2Δ and Agrho3Δ mutants (Wendland and Philippsen, 2000, 2001). Growth direction of Agrsr1Δ hyphae, however, was affected because reinitiation of growth after pausing occurred in an unpredictable manner. This resulted in zig-zag–shaped hyphae instead of the straight wild-type hyphal tubes. Some colony growth mutants in other filamentous fungi, for example, A. nidulans, N. crassa, and Schizophyllum commune are known to exhibit distorted hyphal growth, indicating defects in growth guidance. In N. crassa, these mutants of the ropy class were shown to carry defects in the dynein/dynactin complex that destabilize the apical vesicle supply center (Spitzenkörper) and result in deviations from an axial growth trajectory (Riquelme et al., 2002). Some hyphal morphology mutants isolated in N. crassa have defects in genes coding for dynein/dynactin and signaling proteins such as Cdc42p and Lrg1p (Seiler and Plamann, 2003). Disruption of the thn1 gene of S. commune, which encodes a putative regulator of G protein signaling, led to defects of vegetative growth and a corkscrew-like hyphal morphology (Fowler and Mitton, 2000).
Role of AgRsr1p in Polarisome Stability and Actin Cortical Patch Distribution
Loss of AgRSR1 affects the localization of the polarisome component AgSpa2p. The homolog in S. cerevisiae belongs to a group of proteins that localize to sites of growth in a cell cycle-dependent manner (Sheu et al., 2000). AgSpa2p-GFP signals were observed as a stable marker of growing hyphal tips of A. gossypii corresponding to the fact that in filamentous fungi hyphal tip growth is uncoupled from cell cycle events (Knechtle et al., 2003). During pausing in Agrsr1Δ hyphae, however, AgSpa2p-GFP signals were completely lost from the hyphal tips. Only upon the reoccurrence of AgSpa2p-GFP in the hyphal apex did growth resume, indicating that the maintenance of polarized hyphal growth depends on polarisome functions that are coordinated via AgRsr1p. AgSPA2 in A. gossypii was found to control the area of surface extension but not polarization of the actin cytoskeleton at the hyphal tips (Knechtle et al., 2003). However, in Agrsr1Δ defects in distribution of actin cortical patches at the hyphal tips were observed. An uneven distribution of patches in the growth zone presumably triggers the zig-zagging of hyphal growth in Agrsr1Δ. Mutant strains of Candida albicans in which the CaRSR1 homolog is deleted show characteristics of both the S. cerevisiae and A. gossypii mutants in that the yeast-phase cells show random budding and that upon hyphal induction germ tube emergence and hyphal elongation are affected (Yaar et al., 1997). These studies, however, did not provide any insight into the growth guidance of Candida hyphae.
Mechanistic Implications Regarding Growth Control Executed by Rsr1p
In S. cerevisiae, Rsr1p/Bud1p localizes to sites of growth in a cell cycle-dependent manner (Park et al., 2002). This is of peculiar interest because it points to a more important role than simple bud-site selection at the G1/S transition. However, S. cerevisiae rsr1/bud1 mutants do not show drastic growth defects, besides their random budding pattern. In contrast, loss of AgRSR1 profoundly decreased hyphal growth rate. Because lack of AgRSR1 did not eliminate hyphal growth in A. gossypii, downstream events seem to be able to initiate growth independently of AgRSR1. In S. cerevisiae, GTP-bound Rsr1p/Bud1p recruits Cdc24p to the membrane. This local enrichment triggers activation of Cdc42p at sites of bud formation. Downstream S. cerevisiae targets of Cdc42p-GTP, for example, Gic1p, Gic2p, and the formin Bni1p, are then activated (Brown et al., 1997; Dong et al., 2003; Kawasaki et al., 2003). The formin Bni1p interacts in S. cerevisiae with the polarisome component Spa2p, which forms a complex with Pea2p and Bud6p (Sheu et al., 1998). This network is central for the targeted delivery of secretory vesicles and the polarized organization of the actin cytoskeleton S. cerevisiae. Therefore, activation of Cdc24p in an Rsr1p/Bud1p-independent manner may be sufficient to trigger the polarity establishment machinery into a new cycle of polarized growth, resulting in bud formation in S. cerevisiae. Similarly, AgCdc24p also may be able to reinitiate polar growth in A. gossypii via AgCdc42p independently of AgRsr1p. This is in line with a recent report that suggested that stochastic activation of Cdc42p may occur in S. cerevisiae, which was shown to require the ScBem1 protein to result in actin polymerization (Irazoqui et al., 2003; Wedlich-Söldner et al., 2003). A similar stochastic activation of AgCdc42p in Agrsr1Δ hyphae may be able to explain the defect in growth guidance in these hyphae. After pausing, the reinitiation of growth may occur at a random position, although still at the hyphal apex, and thus result in the zig-zag phenotype of hyphal growth in these mutants. Furthermore, activation of AgCdc42p along the hyphae may be responsible for the emergence of bulges that distort hyphal morphology in subapical parts. Additionally, in S. cerevisiae regulation of polarized growth during mating projection formation was found to depend on polarisome components, Rsr1p/Bud1p and Cdc42p regulators (Nern and Arkowitz, 2000; Bidlingmaier and Snyder, 2004). One observation that is in line with this hypothesis is the different pausing intervals that occurred. Loss of AgRsr1p in A. gossypii led to lengthy intervals of pausing reaching up to 2 h. This roughly equals the duration of a cell cycle in A. gossypii. Therefore, we suggest that pausing is put to an end via the cell cycle machinery at the beginning of each new cycle. Short pauses may be ended by other proteins (e.g., AgCdc24p) that can activate AgCdc42p once they reach a threshold activity at a certain site.
Implications for Growth Guidance in Other Cell Systems
Recently, the mammalian Rin-GTPase and its Drosophila Ric homolog were shown to induce neurite outgrowth (Hoshino and Nakamura, 2002; Spencer et al., 2002). Both proteins were classified as members of the Ras-superfamiliy of GT-Pases (Lee et al., 1996; Wes et al., 1996). Interestingly, the Rin GTPase is expressed specifically in neurons (Lee et al., 1996). Rin binds to calmodulin and functions via Rac/Cdc42 signaling pathways (Hoshino and Nakamura, 2003). In addition to the Ras-like GTPase Rin another GTPase of the Rho-subfamily, RhoG, activates neurite outgrowth upon nerve growth factor (NGF) induction (Katoh et al., 2000). In these cell systems, NGF treatment also activates Ras and results in neurite outgrowth (Goi et al., 1999). This indicates that growth guidance in neurons is a process that can be influenced via extracellular stimuli. In filamentous fungi, growth guidance also may be influenced by the environment similarly to mating projection formation in S. cerevisiae. But using in vitro assays with a most uniform environment, intrinsic cues could be used as a default. These and other results let emerge a common theme for the genetic regulation of growth guidance in eukaryotic cells and make filamentous fungi powerful model systems to elucidate the molecular mechanisms that regulate these processes.
Supplementary Material
Acknowledgments
We thank Flora Banuett and Ira Herskowitz for encouraging discussions and Fred Dietrich, Sangdun Choi, and Rod Wing for A. gossypii clones and sequence information. We especially thank Hans-Peter Schmitz for providing GFP-fusion vectors before publication and Sylvia Voegeli for verification sequencing. This work was supported by grants from the Swiss National Science Foundation grant 31-55941.98 (to P.P. and J.W.) and the Deutsche Forschungs Gemeinschaft (WE2634/2-1 to J.W.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–02–0104. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–02–0104.
The online version of this article contains supplemental material accessible through http://www.molbiolcell.org.
References
- Amberg, D.C. (1998). Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol. Biol. Cell 9, 3259-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayad-Durieux, Y., Knechtle, P., Goff, S., Dietrich, F., and Philippsen, P. (2000). A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113, 4563-4575. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier, S., and Snyder, M. (2004). Regulation of polarized growth initiation and termination cycles by the polarisome and Cdc42 regulators. J. Cell Biol. 164, 207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.L., Jaquenoud, M., Gulli, M.P., Chant, J., and Peter, M. (1997). Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11, 2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K.S., Tinsley, J.H., Minke, P.F., and Plamann, M. (1996). Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc. Natl. Acad. Sci. USA 93, 4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor, A., and Snyder, M. (2002). Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5, 179-186. [DOI] [PubMed] [Google Scholar]
- Chant, J., and Herskowitz, I. (1991). Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203-1212. [DOI] [PubMed] [Google Scholar]
- Dickson, B.J. (2001). Rho GTPases in growth cone guidance. Curr. Opin. Neurobiol. 11, 103-110. [DOI] [PubMed] [Google Scholar]
- Dong, Y., Pruyne, D., and Bretscher, A. (2003). Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161, 1081-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- Fowler, T.J., and Mitton, M.F. (2000). Scooter, a new active transposon in Schizophyllum commune, has disrupted two genes regulating signal transduction. Genetics 156, 1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527-534. [DOI] [PubMed] [Google Scholar]
- Goi, T., Rusanescu, G., Urano, T., and Feig, L.A. (1999). Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol. Cell. Biol. 19, 1731-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfer, M., Tarkka, M.T., Hanif, M., Pardo, A.G., Laitiainen, E., and Raudaskoski, M. (2001). Characterization of small GTPases Cdc42 and Rac and the relationship between Cdc42 and actin cytoskeleton in vegetative and ectomy-corrhizal hyphae of Suillus bovinus. Mol. Plant Microbe Interact. 14, 135-144. [DOI] [PubMed] [Google Scholar]
- Gow, N.A. (1994). Growth and guidance of the fungal hypha. Microbiology 140, 3193-3205. [DOI] [PubMed] [Google Scholar]
- Harold, F.M. (1995). From morphogenes to morphogenesis. Microbiology 141, 2765-2778. [DOI] [PubMed] [Google Scholar]
- Heath, I.B., and Steinberg, G. (1999). Mechanisms of hyphal tip growth: tube dwelling amebae revisited. Fungal Genet. Biol. 28, 79-93. [DOI] [PubMed] [Google Scholar]
- Heim, R., and Tsien, R.Y. (1996). Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178-182. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I., Park, H.O., Sanders, S., Valtz, N., and Peter, M. (1995). Programming of Cell Polarity in Budding Yeast by Endogenous Signals. In: Cold Spring Harbor Symposia on Quantitative Biology, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [DOI] [PubMed]
- Hoepfner, D., Brachat, A., and Philippsen, P. (2000). Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol. Biol. Cell 11, 1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, M., and Nakamura, S. (2002). The Ras-like small GTP-binding protein Rin is activated by growth factor stimulation. Biochem. Biophys. Res. Commun. 295, 651-656. [DOI] [PubMed] [Google Scholar]
- Hoshino, M., and Nakamura, S. (2003). Small GTPase Rin induces neurite outgrowth through Rac/Cdc42 and calmodulin in PC12 cells. J. Cell Biol. 163, 1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui, J.E., Gladfelter, A.S., and Lew, D.J. (2003). Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5, 1062-1070. [DOI] [PubMed] [Google Scholar]
- Katoh, H., Yasui, H., Yamaguchi, Y., Aoki, J., Fujita, H., Mori, K., and Negishi, M. (2000). Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol. Cell. Biol. 20, 7378-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, R., Fujimura-Kamada, K., Toi, H., Kato, H., and Tanaka, K. (2003). The upstream regulator, Rsr1p, and downstream effectors, Gic1p and Gic2p, of the Cdc42p small GTPase coordinately regulate initiation of budding in Saccharomyces cerevisiae. Genes Cells 8, 235-250. [DOI] [PubMed] [Google Scholar]
- Knechtle, P., Dietrich, F., and Philippsen, P. (2003). Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14, 4140-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K.G., Beven, L., Angerman, E., Tong, A.H., Boone, C., and Park, H.O. (2003). Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell 14, 4958-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.H., Della, N.G., Chew, C.E., and Zack, D.J. (1996). Rin, a neuron-specific and calmodulin-binding small G-protein, and Rit define a novel subfamily of ras proteins. J. Neurosci. 16, 6784-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. (2000). Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173-180. [DOI] [PubMed] [Google Scholar]
- Madden, K., and Snyder, M. (1998). Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52, 687-744. [DOI] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance and mew axes. Curr. Opin. Microbiol. 12, 580-585. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Komiya, M., Sone, K., Hirose, E., Gotoh, N., Morii, H., Ohta, Y., and Mori, N. (2002). Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 22, 8721-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern, A., and Arkowitz, R.A. (2000). G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell. 5, 853-864. [DOI] [PubMed] [Google Scholar]
- Park, H.O., Bi, E., Pringle, J.R., and Herskowitz, I. (1997). Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94, 4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.O., Kang, P.J., and Rachfal, A.W. (2002). Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J. Biol. Chem. 277, 26721-26724. [DOI] [PubMed] [Google Scholar]
- Plamann, M., Minke, P.F., Tinsley, J.H., and Bruno, K.S. (1994). Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 127, 139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R., Br, E., Harkins, H.A., Zahner, J.E., De Virgilio, C., Chant, J., Corrado, K., and Fares, H. (1995). Establishment of Cell Polarity in Yeast. In: Cold Spring Harbor Symposia on Quantitative Biology, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [DOI] [PubMed]
- Riquelme, M., Gierz, G., and Bartnicki-Garcia, S. (2000). Dynein and dynactin deficiencies affect the formation and function of the Spitzenkorper and distort hyphal morphogenesis of Neurospora crassa. Microbiology 146, 1743-1752. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Roberson, R.W., McDaniel, D.P., and Bartnicki-Garcia, S. (2002). The effects of ropy-1 mutation on cytoplasmic organization and intracellular motility in mature hyphae of Neurospora crassa. Fungal Genet. Biol. 37, 171-179. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schmidt, A., and Hall, M.N. (1998). Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14, 305-338. [DOI] [PubMed] [Google Scholar]
- Sheu, Y.J., Barral, Y., and Snyder, M. (2000). Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y.J., Santos, B., Fortin, N., Costigan, C., and Snyder, M. (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., and Plamann, M. (2003). The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, M.L., Shao, H., and Andres, D.A. (2002). Induction of neutrite extension and survival in phenochromocytoma cells by the Rit GTPase. J. Biol. Chem. 277, 20160-20168. [DOI] [PubMed] [Google Scholar]
- Tinsley, J.H., Minke, P.F., Bruno, K.S., and Plamann, M. (1996). p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol. Biol. Cell 7, 731-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner, R., Altschuler, S., Wu, L., and Li, R. (2003). Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231-1235. [DOI] [PubMed] [Google Scholar]
- Wendland, J. (2001). Comparison of morphogenetic networks of filamentous fungi and yeast. Fungal Genet. Biol. 34, 63-82. [DOI] [PubMed] [Google Scholar]
- Wendland, J. (2003). Analysis of the landmark protein Bud3 of Ashbya gossypii reveals a novel role in septum construction. EMBO Rep. 4, 200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, J., Ayad-Durieux, Y., Knechtle, P., Rebischung, C., and Philippsen, P. (2000). PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242, 381-391. [DOI] [PubMed] [Google Scholar]
- Wendland, J., and Philippsen, P. (2000). Determination of cell polarity in germinated spores and hyphal tips of the filamentous ascomycete Ashbya gossypii requires a rhoGAP homolog. J. Cell Sci. 113, 1611-1621. [DOI] [PubMed] [Google Scholar]
- Wendland, J., and Philippsen, P. (2001). Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 157, 601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes, P.D., Yu, M., and Montell, C. (1996). RIC, a calmodulin-binding Ras-like GTPase. EMBO J. 15, 5839-5848. [PMC free article] [PubMed] [Google Scholar]
- Wright, M.C., and Philippsen, P. (1991). Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene 109, 99-105. [DOI] [PubMed] [Google Scholar]
- Yaar, L., Mevarech, M., and Koltin, Y. (1997). A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology 143, 3033-3044. [DOI] [PubMed] [Google Scholar]
- Zhang, X.F., Schaefer, A.W., Burnette, D.T., Schoonderwoert, V.T., and Forscher, P. (2003). Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron 40, 931-944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.