Abstract
The germ cell lineage in Xenopus is specified by the inheritance of germ plasm, which originates within a distinct “mitochondrial cloud” (MC) in previtellogenic oocytes. Germ plasm contains localized RNAs implicated in germ cell development, including Xcat2 and Xdazl. To understand the mechanism of the early pathway through which RNAs localize to the MC, we applied live confocal imaging and photobleaching analysis to oocytes microinjected with fluorescent Xcat2 and Xdazl RNA constructs. These RNAs dispersed evenly throughout the cytoplasm through diffusion and then became progressively immobilized and formed aggregates in the MC. Entrapment in the MC was not prevented by microtubule disruption and did not require localization to germinal granules. Immobilized RNA constructs codistributed and showed coordinated movement with densely packed endoplasmic reticulum (ER) concentrated in the MC, as revealed with Dil16(3) labeling and immunofluorescence analysis. Vg1RBP/Vera protein, which has been implicated in linking late pathway RNAs to vegetal ER, was shown to bind specifically both wild-type Xcat2 3′ untranslated region and localization-defective constructs. We found endogenous Vg1RBP/Vera and Vg1RBP/Vera-green fluorescent protein to be largely excluded from the MC but subsequently to codistribute with Xcat2 and ER at the vegetal cortex. We conclude that germ line RNAs localize into the MC through a diffusion/entrapment mechanism involving Vg1RBP/Vera-independent association with ER.
INTRODUCTION
RNA localization is used by both somatic and germ cells to localize proteins to subcellular domains and to limit protein synthesis regionally (Bashirullah et al., 1998; Jansen, 2001; Kloc et al., 2002b). Oocytes provide particularly striking examples of the importance of RNA localization, with the protein products of maternal messages positioned before fertilization setting up local concentrations of regulatory proteins that establish regional fates in the embryo (St. Johnston, 1995; Bashirullah et al., 1998; King et al., 1999; Kloc et al., 2002b). Thus, in Drosophila oocytes, bicoid RNA tightly concentrated at one pole provides a local source of a transcription factor responsible for initiating programs of development to yield anterior fates. Similarly, nanos and oskar RNAs prepositioned at the opposite end of the oocyte direct posterior and germ line fates (for review, see Bashirullah et al., 1998).
In the amphibian Xenopus, more than a dozen maternal RNAs that are tightly localized to the vegetal cortex of the fully grown oocyte have been identified, as well as a small number of RNAs that are enriched in the animal cytoplasm (King et al., 1999). Many of the vegetal RNAs, including Xcat2, Xdazl, and Xpat, are associated with morphologically distinct islands of “germ plasm,” which contain clusters of mitochondria and electron-dense organelles called germinal granules (Heasman et al., 1984). Germ plasm segregates unequally during cleavage, into only a few blastomeres, and contains factors that specify these cells to become primordial germ cells (PGCs) (Houston and King, 2000a). Xcat2 encodes an RNA-binding protein (RBP) related to Nanos (Mosquera et al., 1993), the product of an essential gene in Drosophila and mouse that is required for the acquisition of germ line fate and for germ cell migration (Kobayashi et al., 1996; Forbes and Lehmann, 1998; Tsuda et al., 2003; Wang and Lin, 2004). In Xenopus, Xdazl is required for PGC differentiation, such that PGCs depleted of Xdazl fail to migrate (Houston and King, 2000a). Non-germ plasm–localized vegetal RNAs include the transforming growth factor-β family member Vg1 and the T-box transcription factor VegT, which are required for the development and patterning of the endoderm and mesoderm (Joseph and Melton, 1998; Zhang et al., 1998).
The vegetally localized RNAs in Xenopus oocytes follow two distinct localization pathways, arriving at the cortex during different periods of oogenesis (Forristall et al., 1995; Kloc and Etkin, 1995; Zhou and King, 1996b). The germ plasm RNAs use the early or METRO pathway and localize during very early stages of oogenesis (stage I) (Dumont, 1972) into a macroscopic structure called the mitochondrial cloud (MC), which lies close to the nucleus. Germ plasm RNAs become enriched in the region of the MC opposite the nucleus termed the METRO. Xcat2 RNA is incorporated into the germinal granules within this region (Heasman et al., 1984; Kloc et al., 1996, 2002a). Other germ line RNAs adopt distinct distributions within the germ plasm; Xpat RNA associates with the periphery of the granules, whereas Xdazl RNA is more generally distributed in the matrix (Kloc et al., 2002a). During stage II, the MC detaches from the nucleus and merges into a nearby cortical region, thereby bringing the germ plasm RNAs to the cortex. During subsequent stages of oogenesis, the MC fragments, giving rise to subcortical germ plasm islands (Heasman et al., 1984).
Late pathway RNAs, such as VegT and Vg1, are present at stage I but start to localize only after the MC has arrived at the cortex. These RNAs accumulate in a wedge-shaped zone surrounding the dispersing MC and then move to a thin subcortical layer during stages III and IV, through a process involving microtubules and kinesins I and II (Yisraeli et al., 1990; Kloc and Etkin, 1995; Betley et al., 2004; Yoon and Mowry, 2004). The early and late pathways show some mechanistic overlap. Certain RNAs, such as fatVg, show localization characteristics of both pathways (Kloc et al., 1996; Kloc and Etkin, 1998; Chan et al., 1999, 2001). In addition, RNAs that normally use the early pathway can localize by using the late pathway, although they may not enter the germ plasm. It has been proposed that components derived from the MC contribute to the establishment of a polarized microtubule network required for the late pathway (Kloc and Etkin, 1998).
Vegetal RNA localization in Xenopus oocytes requires specific sequences or localization signals (LSs) found in the 3′ untranslated region (UTR), similar to RNA localization in other cell types (King et al., 2004). The Xcat2 3′UTR has two distinct LSs. The 240-nucleotide MCLS is both required and sufficient to direct Xcat2 into the MC (Zhou and King, 1996a). In addition, the 160-nucleotide germinal granule localization element (GGLE) is required to direct Xcat2 into germinal granules, an event that requires the prior functioning of the MCLS (Kloc et al., 2000). Within the MCLS are short motifs that likely mediate protein binding to the signal in a redundant manner (Betley et al., 2002). A number of RBPs that interact with late pathway RNAs have been identified through direct interactions with the Vg1 LS, i.e., Vg1RBP (also known as Vera), VgRBP60/hnRNP1, VgRBP71, and Prrp (Schwartz et al., 1992; Mowry, 1996; Zhao et al., 2001; Kroll et al., 2002). Vg1RBP/Vera binds to both Vg1 and VegT RNAs and has been shown to be important for their localization (Deshler et al., 1998; Bubunenko et al., 2002; Kwon et al., 2002). Moreover, Vg1RBP/Vera can mediate the association of Vg1 RNA with microtubules in vitro (Elisha et al., 1995) and is found in a fraction containing endoplasmic reticulum (ER) membranes after centrifugation of undiluted oocyte extracts (Deshler et al., 1997). It has thus been proposed that Vg1RBP/Vera promotes late pathway localization by associating RNAs with distinctive ER in the wedge region, which becomes translocated to the vegetal cortex along polarized microtubules (Deshler et al., 1998). The Xcat2 MCLS was recently shown to bind Vg1RBP/Vera (Betley et al., 2002), consistent with its ability to use the late pathway (Zhou and King, 1996b).
The cellular and molecular mechanisms responsible for the localization of RNAs through the early pathway are not yet understood. Previous work suggested that neither microtubules nor microfilaments are required for the early pathway (Kloc et al., 1996; Zhou and King, 1996b), but the evidence was not conclusive. We examined the behavior of fluorescently labeled Xcat2 and Xdazl RNA constructs in living stage I and II oocytes by using time-lapse confocal microscopy and fluorescence recovery after photobleaching (FRAP) analysis. Our results confirmed that their localization does not require microtubules and provided evidence that these RNAs localize into the MC through a diffusion-entrapment mechanism. Unexpectedly, the RNAs were found to associate with a densely packed ER network in the MC. However, Vg1RBP/Vera, which is implicated in linking Vg1 RNA to the ER, appeared generally excluded from the MC, and no colocalization with ER or immobilized Xcat2 was detectable until the MC arrived at the cortex. These results indicate that, in contrast to the late/Vg1 pathway, localization of germ line RNAs to the MC occurs through a non–motor-driven mechanism involving diffusion in the cytoplasm and entrapment in the MC in association with ER. The ER association occurs independently of germinal granule formation and does not involve Vg1RBP/Vera. A similar mechanism was recently proposed for the posterior localization of the Xcat2-related RNA nanos in Drosophila oocytes (Forrest and Gavis, 2003), suggesting a conserved mechanism for the localization of RNAs involved in specification of the germ line.
MATERIALS AND METHODS
Oocytes and Microinjection
Adult or juvenile (3–7 cm from nose to anus) Xenopus laevis specimens were purchased from Xenopus Express (Plant City, FL) or CNRS, Rennes (Rennes, France). Stage I/II oocytes (staged according to the method described by Dumont, 1972) were released from dissected ovaries at various times after collagenase A treatment (0.8 mg/ml in 0.1 M NaH2PO4, pH 7.4) with gentle shaking into L-15 medium, i.e., 50% Leibowitz medium (Sigma, St. Louis, MO) supplemented with 1 mg/ml bovine serum albumin (BSA), 100 μg/ml gentamicin, 1 U penicillin, 1 μg/ml streptomycin, 1 mM l-glutamine, 1 μg/ml insulin, 15 mM HEPES (pH 7.8), and 50 U/ml nystatin. Freed oocytes were rinsed three times and transferred to fresh medium. Microinjection was performed with an Eppendorf Femtojet (Hamburg, Germany) apparatus, delivering 30–100 pl. After injection, oocytes were transferred to fresh L-15 medium containing 5–10% Xenopus serum with vitellogenin (Opresko, 1991) and were cultured in the dark at 18°C (Zhou and King, 1996b). To disturb microtubule organization, oocytes were incubated for 90 min on ice and/or for 24 h with 10 μM nocodazole in L-15 medium, diluted freshly from a 10 mM stock in dimethyl sulfoxide.
Mutations and Cloning
The Xcat2 (accession no. X72340), Vg1 (accession no. M18055), and Xdazl (accession no. AF017778) wild-type and mutant clones used in this study were prepared by PCR. Two external primers containing a BamHI site (on forward primers) and a XhoI site (on reverse primers) were designed for the cloning of the 3′UTRs from Xcat2 and Vg1. Primers for the Xcat2 3′UTR were GCATGGGATCCTGAGACACTGAACTGGAAGG (forward) and ATAGACTCGAGTTTTTTTTTTTTTTTTTTCCGG (reverse). The alternative reverse primer to generate the 240-nucleotide Xcat2 MCLS was ATAGACTCGAGCCCCGGGGACAGAAACAA (reverse). Two overlapping internal primers containing the desired mutation were prepared. Two rounds of PCR were required for each mutation. PCR conditions were essentially as described previously (MacArthur et al., 1999). The resulting PCR inserts were purified with the QIAquick PCR purification kit (Qiagen, Chatsworth, CA) and then digested with BamHI and XhoI. All inserts were gel purified, extracted with phenol:chloroform, precipitated with isopropanol, and resuspended in water. All inserts were cloned into pCS2+ plasmid (Turner and Weintraub, 1994) and sequenced to verify the mutations. The plasmid pTZ18.XdazlLE, containing the minimal LS for Xdazl (nucleotides 1335–1780) in a modified version of the pTZ18U vector (Bio-Rad Laboratories, Hercules, CA), was a gift from Jim Deshler (Boston University, Boston, MA) (Betley et al., 2002). A construct containing the Xcat2 open reading frame and the 3′UTR of Xenopus β-globin (600 nucleotides) served as a control.
RNA Synthesis
Sense RNAs were transcribed from NotI-linearized plasmids with SP6 RNA polymerase and the mMessage Machine kit (Ambion, Austin, TX). For fluorescent probes, 600 μM Alexa 488-UTP or tetramethylrhodamine-UTP (Molecular Probes, Eugene, OR) was used with 3 mM unlabeled NTP mixture (final concentrations), giving a ratio of 1:5. Unincorporated nucleotides were removed and transcripts were purified with Sephadex G-50 Quick Spin columns (Roche Diagnostics, Indianapolis, IN). The pSP73XβG5′ plasmid (Mowry and Melton, 1992) was linearized and transcribed with T7 RNA polymerase to produce sense RNA.
Construction of Vg1RBP/Vera-EGFP (Enhanced Green Fluorescent Protein)
Vg1RBP/Vera (full-length coding region minus the stop codon, allelic variant D, accession no. AF064634) was cloned into pSP64TSN. This vector is a derivative of pSP64TS (www.xenbase.org/WWW/Marker_pages/PlasMaps/pSP64TS.html) in which the EcoRV site is replaced with XmaI and NcoI sites. The coding sequence from pEGFP-N1 (Clontech, Palo Alto, PA) was generated with primers that included the SpeI (reverse primer: CCCAATTGGACTAGTTTACTTGTACAGCTCGTCCATG) and NcoI (forward primer: CATCATGCCATGGCAATGGTGAGCAAGGGCGAGGAG) sites. The resulting 731-nucleotide fragment was cloned into the NcoI/SpeI site in-frame with Vg1RBP/Vera, followed by the stop codons and the 3′UTR of β-globin. The sequence was verified with sequencing and tested with in vitro translation.
In Vitro Binding Assay
Radiolabeled probes for UV cross-linking were transcribed from pSP73-vg340 (Mowry, 1996), pCS2+Xcat2FLWT, pCS2+Xcat2SR1/E2, and pCS2+Vg1E2/R1 as described by Lewis et al. (2004). Sequence-specific competitor RNAs were synthesized from linearized pSP73-vg340 and pCS2+Xcat2FLWT with the ME-GAscript kit (Ambion). Escherichia coli rRNA, which was used as a nonspecific competitor, was a generous gift from Albert Dahlberg (Brown University, Providence, RI). Oocyte S10 lysate was prepared by homogenizing defolliculated Xenopus stage I and II oocytes in an equal volume of S10 buffer (50 mM Tris, pH 8, 50 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 25% glycerol). After centrifugation at 10,000 × g at 4°C for 15 min, the supernatant was aliquoted and stored at –80°C. UV cross-linking experiments and partial purification of Vg1RBP/Vera and VgRBP60/hnRNPI were performed as described by Lewis et al. (2004).
Confocal Imaging of Living Oocytes
For visualization of ER, live oocytes were injected with 30-pl drops of saturated DilC16(3) (Dil) in Wesson vegetable oil (Terasaki and Jaffe, 1991). Images were acquired 3–30 h after injection. Mitochondria were visualized with brief (∼30 s) incubation of oocytes with 2.5 μg/ml DiOC6(3) (DiO) in OR2, diluted from a 2.5 mg/ml stock in ethanol just before use (Savage and Danilchik, 1993). Fluorescent RNAs were injected at 1 mg/ml. Oocytes in which the MC localization machinery had been saturated, as indicated by cortical accumulation of injected RNAs, were not used. Vg1RBP/Vera-EGFP RNA (5–30 pl at 1 mg/ml) was injected and allowed to translate for at least 16 h before imaging. Oocytes were mounted in L-15 medium, with supplements as described above, in sealed chambers consisting of two glass coverslips separated by a spacer of humidified filter paper.
Images were acquired with an inverted Leica SP2 confocal microscope (Leica, Wetzlar, Germany) equipped with argon ion, helium-neon, and green neon lasers and were processed with Leica and NIH Image/ImageJ software. Photobleaching of GFP and Alexa 488 was achieved by increasing the laser power of the 488-nm line of the 25-mW argon laser and scanning a reduced region of the field defined with the zoom function and a 16-pass line average. Photobleaching of DiI was similarly achieved by increasing the power of the 488-nm line to allow indirect excitation via coinjected Alexa 488- or GFP-labeled probes and the power of the 543-nm line of the weaker helium-neon laser. Quantification was achieved with Leica SP2 or Metamorph software (Universal Imaging, Downingtown, PA). For estimation of two-dimensional diffusion coefficients from FRAP data (after correction for acquisition photobleaching), curves were fitted to the equation F = A(1 - ekt) + B, with F being fluorescence intensity and t time and A + B giving the plateau level after recovery. The half-time of recovery is then given by thalf = ln0.5/-k and the effective diffusion constant by DE = w2/4thalf, where w is one-half the width of the bleached zone.
Immunofluorescence Assays
Oocytes were fixed in methanol/1% formaldehyde at –20°C for a minimum of 2 h. After stepwise rehydration to phosphate-buffered saline (PBS) containing 0.05% Triton X-100, oocytes were extracted for 10 or 30 min in PBS/0.25% Triton X-100, rinsed in PBS, and blocked for at least 1 h in 1% BSA/PBS at 4°C. Samples were incubated with primary antibodies overnight at 4°C and with secondary antibodies for 2 h at room temperature, washed with PBS/0.1% Tween-20, and mounted with Citifluor (Citifluor, London, United Kingdom). The antibodies were rabbit antisera raised against full-length recombinant Vg1RBP/Vera protein from Nancy Standart (University of Cambridge, Oxford, United Kingdom) (Git and Standart, 2002), used at 1:1000, and from William L. Taylor (Vanderbilt University, Memphis, TN) (Pfaff and Taylor, 1992; Zhang et al., 1999), used at 1:500, a rat mAb against the ER marker GRP94 (StressGen, Victoria, Canada), used at 1:500, anti-α-tubulin mouse mAb DMIA (Sigma), used at 1:1000, and preabsorbed rhodamine- or fluorescein isothiocyanate-labeled anti-Ig antibodies (Jackson Immunoresearch Laboratories, West Grove, PA), used at 1:75 to 1:150. Control experiments in which primary antibodies were omitted confirmed that no cross-reaction between antibodies occurred in double-staining experiments. To confirm the specificity of anti-Vg1RBP/Vera staining, antisera diluted 1:100 or 1:200 were preincubated with 50–60 μg/ml Vg1RBP/Vera-his fusion protein or BSA overnight at 4°C and then centrifuged for 15 min at 4°C in an Eppendorf centrifuge before the final dilution and staining. Images were acquired with the Leica SP2 confocal microscope, as described above.
RESULTS
Progressive Accumulation and Immobilization of Germ Plasm RNAs in the MC
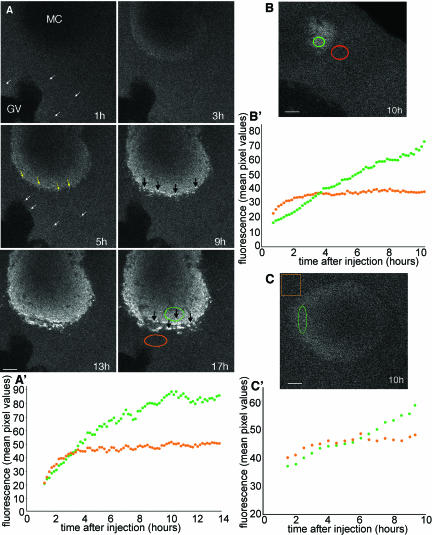
To investigate the localization mechanism of germ plasm RNAs, we followed the accumulation of Alexa 488-labeled Xcat2 3′UTR RNA into the MC after microinjection into previtellogenic (stage I and II) oocytes with time-lapse confocal microscopy (Figure 1A and video 1). The microinjection of Alexa 488-labeled RNAs has been established as a valuable method for tracking RNA localization in Drosophila oocytes and embryos (Bullock and Ish-Horowicz, 2001; Cha et al., 2001; Wilkie and Davis, 2001). Previous work showed that exogenous injected Xcat2 RNA forms particles in the cytoplasm independent of the site of injection (Kloc et al., 1996) but did not characterize the process of localization. Accumulation of fluorescent particles (<0.4 μm in diameter) in the MC was typically first detectable 3 h after injection at a distant site (Figure 1A). No directed translocation of particles was detected, nor was any accumulation observed around the MC before localization. Accumulation of the Xcat2 3′UTR within the MC was accompanied by progressive enlargement of the detectable aggregates, which attained 0.7 μm by 5 h after injection, up to 1 μm by 9 h after injection, and up to 1.9 μm by 17 h after injection (Figure 1A). These fluorescent Xcat2 RNA aggregates in the MC presumably include germinal granules, which range from 50 to 200 nm in diameter in stage I oocytes (Kloc et al., 2002a). Quantification of fluorescence accumulation revealed that accumulation of the Xcat2 3′UTR occurred in a linear manner (Figure 1, A′ and B′), proceeding from the periphery of the MC and often favoring the cortical side, where the METRO region is located (Kloc and Etkin, 1995; Kloc et al., 2002a). Fluorescent Xdazl LE, which is not incorporated into germinal granules, was similarly found to show linear accumulation in the MC (Figure 1, C and C′).
Figure 1.
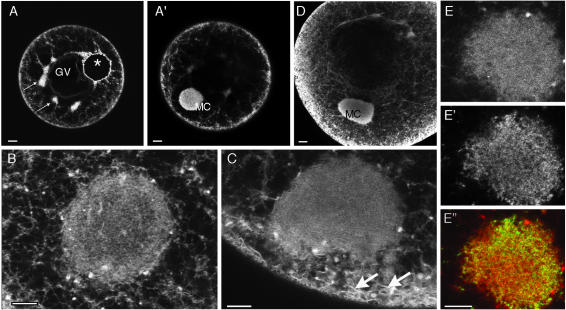
Progressive accumulation of Xcat2 3′UTR in the MC. (A) Confocal images from series acquired every 10 min during a 17-h period after microinjection of Alexa 488-Xcat2 RNA into a stage I oocyte (video 1). The particulate appearance of the fluorescent probe in the cytoplasm (white arrows) and the subsequent concentration of small particles in peripheral regions of the MC (yellow arrows), before the accumulation of progressively larger aggregates (black arrows) in peripheral regions of the MC, should be noted. (A′) Quantification of Xcat2 fluorescence in a peripheral region of the MC (green) and a nearby cytoplasmic region (orange). In the cytoplasmic region, fluorescence initially increased for a period of 2–3 h as it spread from the injection site (on the far side of the nucleus) and then remained at a constant level. In the MC, accumulation of Xcat2 3′UTR was linear for the first 5 h after injection. (B and B′) Quantification of fluorescent Xcat2 3′UTR accumulation in a peripheral region of the MC (green) (MC seen in tangential section) and a nearby cytoplasmic region (orange) in another stage I oocyte. (C and C′) Quantification of fluorescent Xdazl LE accumulation in a peripheral region of the MC (green) and a nearby cytoplasmic region (orange) in another stage I oocyte. All scale bars, 10 μm.
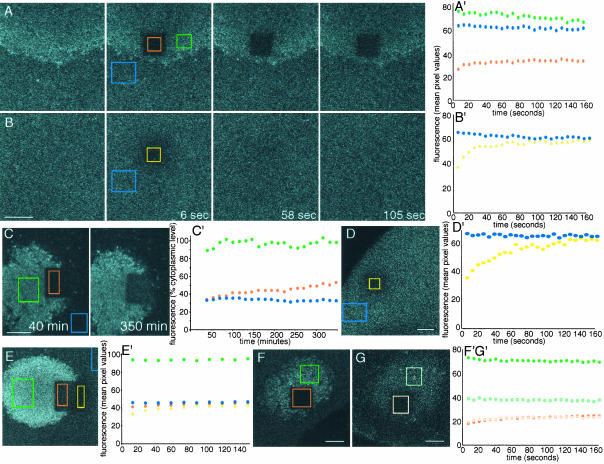
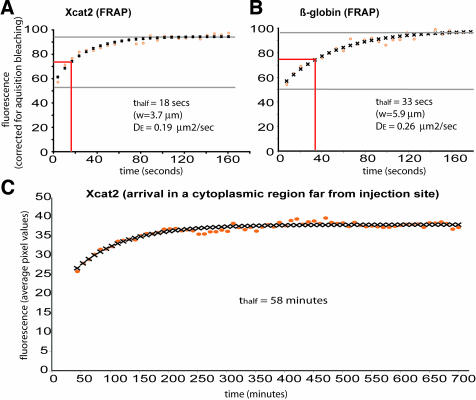
To compare the mobility of the Xcat2 3′UTR within different regions of the oocyte, FRAP analysis was performed after overnight localization of Alexa 488-labeled RNA (Figure 2). Fluorescence was bleached with intensive laser scanning of central square regions (7–12-μm sides) within the observed field, and images were acquired in the subsequent few minutes (see Materials and Methods). Fluorescence recovery in cytoplasmic regions was symmetrical and rapid, increasing from 40–60% of adjacent cytoplasmic values at ∼10 s after bleaching to 80-90% within 2 min (Figure 2B). The recovery exhibited the typical characteristics of diffusion and closely fit a theoretical diffusion curve (Figure 3A) (see Materials and Methods). It should be noted that the cytoplasmic fluorescence was entirely attributable to the injected probe; no autofluorescence was detected in uninjected oocytes under the imaging conditions used. In cases in which the bleached zone overlapped the MC, fluorescence recovery occurred both inside the MC and outside the MC within 2–3 min, indicating that RNA can enter the MC from the cytoplasmic pool (Figure 2E, orange region, and video 2). Fluorescence recovery within the MC region did not always reach cytoplasmic levels and varied among different regions of the MC and among oocytes, presumably because the volume of cytoplasm within this dense structure is small and its distribution is inhomogeneous. The small volume of cytoplasm within the MC precluded quantification of diffusion within this region. The fluorescence recovery in the cytoplasm of the Xcat2 3′UTR and the Xdazl LE was very similar to that of a construct in which the 3′UTR of Xcat2 was replaced with that of β-globin, which does not localize to the MC (Figures 2, B′ and D′, and 3, A and B), with mean effective diffusion constants being calculated as ∼0.2 μm2/s in each case (Table 1). Such slow diffusion rates for RNAs in the oocyte cytoplasm, compared with those for smaller molecules such as monomeric proteins, are consistent with measurements made in cultured mammalian cells (Luby-Phelps et al., 1986). These diffusion rates account for the time required for the injected fluorescent RNAs to reach steady-state levels in the cytoplasm (3–4 h; see orange lines in Figure 1, A′, B′, and C′), through a process that also followed diffusion kinetics (Figure 3C). It should be noted, however, that the observed diffusion of injected fluorescent RNAs in neighboring cytoplasm was initially faster than its linear accumulation in the MC (indicated by green lines in Figure 1, A′, B′, and C′), even at the very periphery of the MC (Figure 1B), indicating that diffusion is not rate-limiting for the localization process.
Figure 2.
Immobilization of Xcat2 RNA in the MC. (A) Confocal images from a FRAP experiment performed on a stage I oocyte 20 h after injection of Alexa 488-Xcat2 3′UTR. Throughout the figure, unbleached regions of the MC are shown in green, unbleached regions of the cytoplasm in blue, bleached regions of the MC in orange, and bleached regions of the cytoplasm in yellow. Times after photobleaching are indicated. (A′) Quantification of fluorescence recovery in the bleached zone (orange), compared with nearby regions of the MC (green) and cytoplasm (blue). No transfer of fluorescence to the photobleached zone from surrounding regions was detected, indicating that Xcat2 3′UTR had become immobilized in the MC. (B and B′) In a cytoplasmic region (yellow) of the same oocyte, fluorescence recovered to the level of an unbleached region (blue) within 2 min after photobleaching. (C and C′) Long-term FRAP analysis of another oocyte injected with fluorescent Xcat2 3′UTR revealed ongoing linear RNA accumulation in the bleached zone of the MC (orange), indicating that photobleaching did not damage this region. The frontier of bleached material with the MC remained largely unaltered after several hours, as did unbleached MC (green) and cytoplasm (blue). (D and D′) FRAP analysis of a fluorescent β-globin RNA 3′UTR construct, which served as a nonlocalized control RNA, in a cytoplasmic region (yellow) of a stage I oocyte. Fluorescence recovery to the level of an unbleached cytoplasmic region (blue) was complete within 3 min. (E and E′) FRAP experiment with another oocyte injected with fluorescent Xcat2 3′UTR, showing fluorescence recovery to cytoplasmic levels both inside (orange) and outside (yellow) the MC during the 3 min after bleaching but no redistribution of localized fluorescent RNA within the MC (see video 2). (F, F′, G, and G′) Localized fluorescent Xdazl RNA (F and F′) (dark orange/dark green) behaved similarly to Xcat2 3′UTR (A and A′) after photobleaching, as did Xcat2 3′UTR lacking the GGLE (G and G′) (light orange/light green) in another oocyte, showing that RNA immobilization does not require incorporation into germinal granules (see video 3). In each case, the y axis is given in arbitrary units of fluorescence as mean pixel values within the region shown. All scale bars, 10 μm.
Figure 3.
Diffusion of injected fluorescent RNAs in oocyte cytoplasm. Diffusion of fluorescent Xcat2 3′UTR (A and C) and the β-globin RNA 3′UTR construct (B) injected into stage I oocytes and incubated overnight, as in Figures 1 and 2, was analyzed. FRAP was monitored in A and B, and the arrival of fluorescence in a cytoplasmic region far from the injection site was monitored in C. Theoretical diffusion curves (black crosses) were found to fit closely to the experimental data (orange circles), plotted as mean pixel values in a cytoplasmic region within the bleached zone, normalized relative to an unbleached zone at each time point in the FRAP experiments. Effective diffusion constants (DE) were calculated from the FRAP data (see Materials and Methods).
Table 1.
Cytoplasmic diffusion constants calculated from FRAP data
| RNA construct | Size (nucleotides) | n | Dε (μm2/s), mean (minimum, maximum) |
|---|---|---|---|
| Xcat2 3′UTR | 400 | 5 | 0.23 (0.19, 0.30) |
| Xdazl (LE) | 450 | 7 | 0.19 (0.15, 0.25) |
| β-Globin 3′UTR+Xcat2 open reading frame | 600 | 2 | 0.23 (0.19, 0.26) |
In contrast to the diffusion of RNA in the cytoplasm, fluorescence within the MC did not redistribute after photobleaching (Figure 2A). The frontier of bleached material in the MC remained essentially unaltered, even after several hours. This was not attributable to structural damage to the bleached region, because long-term reaccumulation of Xcat2 3′UTR from the cytoplasmic pool could occur (Figure 2C′), proceeding from the periphery of the MC and showing linear kinetics similar to those of the initial RNA accumulation (Figure 1A′). Similar FRAP experiments were performed with oocytes injected with fluorescent RNA constructs that localize to the MC but not to germinal granules, i.e., Xdazl RNA (Figure 2F and video 3), which localizes mainly to granular fibrillar material, and a truncated version of the Xcat2 3′UTR lacking the GGLE region (Figure 2G) responsible for germinal granule localization (Kloc et al., 2002a). In both cases, immobilization in the MC was equivalent to that of Xcat2. We conclude from these experiments that localization of RNAs through the early pathway does not depend on germinal granule formation and proceeds through immobilization within the MC of previously diffusible cytoplasmic particles.
RNA Accumulation in the MC Is Microtubule Independent
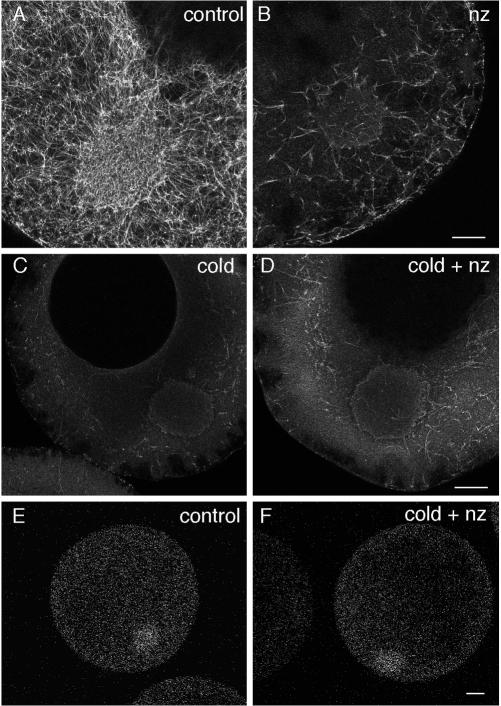
Immobilization of Xcat2 3′UTR in the MC and its free movement in the cytoplasm suggest a non–motor-driven mechanism for Xcat2 localization. This is consistent with previous work showing that Xcat2 was concentrated within the MC after 24 h in the presence of the microtubule-depolymerizing drug nocodazole or the microfilament-disrupting drug cytochalasin (Kloc et al., 1996). We found, however, that a significant population of the microtubules present in stage I and II oocytes (Figure 4A) was resistant to nocodazole treatment equivalent to that used in previous studies (Figure 4B). These microtubules might represent a stable population observed at this stage (Gard, 1991, 1993) and could potentially mediate MC accumulation of RNAs in the presence of nocodazole. We were able to depolymerize almost the entire microtubule network with a 90-min cold treatment (Figure 4C) (Gard, 1991) before oocytes were cultured at 18°C with nocodazole, to prevent repolymerization (Figure 4D). Under these conditions, fluorescent Xcat2 3′UTR localization in the MC after 24 h (Figure 4F) was indistinguishable from that in control experiments (Figure 4E). Microtubule networks could reform in such oocytes, indicating that the oocytes were still viable (our unpublished results). We conclude that Xcat2 3′UTR localizes to the MC through a diffusion/entrapment mechanism that does not require microtubules. We confirmed that treatment with cytochalasin B, which caused dramatic disruption of the cortex, indicating that microfilaments had been extensively disrupted, did not prevent accumulation or trapping of Xcat2 3′UTR in the MC (our unpublished results).
Figure 4.
Microtubule-independent Xcat2 localization. (A–D) Confocal images of oocytes fixed for antitubulin immunofluorescence after treatments to disrupt microtubules. In oocytes cultured in control medium for 24 h, microtubules were detected as a dense network both inside and outside the MC (A). After 24 h of culture at 18°C with 10 μM nocodazole (∼30 μg/ml), a significant population of microtubules remained (B). In contrast, a 90-min cold treatment destroyed almost all detectable microtubules (C), and regrowth was prevented with culture of such oocytes at 18°C for 24 h with 10 μM nocodazole (D). (E and F) Fluorescent Xcat2 3′UTR in oocytes cultured at 18°C in control medium (E) and with nocodazole after 90-min cold treatment (F) localized to the MC to equivalent degrees (groups of 6 or 7 were examined). Scale bars, 25 μm.
A Dense ER Network in the MC
The ER has been implicated in the late pathway localization of Vg1 RNA to the vegetal cortex (Deshler et al., 1997; Alarcon and Elinson, 2001) and as an anchoring site for localized RNAs in ascidian eggs (Sardet et al., 2003). The ER has also been observed in ultrastructural studies as a component of the germ plasm in a variety of organisms, including Xenopus (Heasman et al., 1984; Houston and King, 2000b). To examine possible involvement of ER in trapping germ line RNAs in the MC, we visualized ER distribution in live oocytes after microinjection of oil droplets loaded with the long-chain lipophilic dye DiI. This in vivo visualization technique relies on transfer of the dye out of the oil drop upon contact with internal cell membranes and rapid spreading throughout the continuous lipid membranes of the ER network (Terasaki and Jaffe, 1991). One to 2 h after injection of DiI-loaded oil droplets, the dye decorated a network extending into most regions of the oocyte, with characteristic concentrations beneath the oocyte surface and around the germinal vesicle (GV) (Figure 5, A–C). A striking accumulation of fluorescence was also observed in the MC in all stage I and II oocytes examined. In most cases, the accumulation was too dense to allow structure to be resolved; however, in some smaller oocytes, a fine network of tubules could be distinguished (Figure 5B). The dense DiI labeling in the MC was not attributable to transfer of DiI to the abundant mitochondria in the MC, as shown by incubation of DiI-injected oocytes with the cell-permeable dye DiO to label mitochondria (Savage and Danilchik, 1993). DiO-stained structures occupied positions in the cell similar to those of the DiI-labeled network but were clearly distinct from the DiI-labeled network and were punctate rather than tubular in appearance (Figure 5, E and E″). FRAP analysis revealed rapid spreading of DiI but not DiO fluorescence in the MC (our unpublished results), supporting the idea that the DiI in the MC was inserted in a continuous membrane network.
Figure 5.
The MC contains a dense ER network. (A—C) Confocal images of stage I (A and B) and stage II (C) oocytes injected with oil droplets (*) loaded with the lipophilic dye DiI. The dye spread into a network extending throughout the cell (A), was enriched in cortical regions as well as in small clumps around the GV (small arrows), and was highly concentrated in the MC, lying in a different focal plane (A′). At higher magnification, a dense network could be discriminated within the MC in some oocytes (B). In stage II oocytes (C), ER sheets in the subcortical region (arrows) appeared to merge with ER in the MC. (D) Immunofluorescence assays with an antibody to the ER luminal protein GRP94 revealed a distribution very similar to that of DiI staining, as seen in this stage II oocyte. (E, E′, and E″) Comparison of labeling with DiI (E) (at high magnification, in an optical plane through the periphery of a MC) after oil droplet injection the previous day with that with the short-chain permeable dye DiO (E′). As seen in the overlay (E″, DiI in red and DiO in green), the two dyes labeled clearly distinct membranous organelles, probably mainly ER (DiI) and mitochondria (DiO). Scale bars, 20 μm in A and D, 10 μm in B, C, and E.
In a previous study, immunodetection with an antibody recognizing an ER luminal protein suggested that both the MC and the cortex of stage I oocytes are relatively lacking in ER (Kloc and Etkin, 1998). To resolve the discrepancy with our data, we performed immunofluorescence assays with an antibody recognizing another ER luminal protein, GRP94 (Argon and Simen, 1999; Brunati et al., 2000). The distribution of GRP94 mirrored that of the DiI staining seen in live oocytes, including dense labeling of the MC as well as strands running through the cytoplasm and a cortical layer (Figure 5D). The difference between these results and those of the previous study is likely attributable to the fixation and detection methods used. We conclude that the dense membranous network revealed in the MC with DiI labeling is comprised of ER.
Localized RNAs Associate with ER in the MC
Parallel imaging of oocytes injected with DiI-loaded oil droplets and Alexa 488-labeled Xcat2 or Xdazl RNA revealed a general correlation between the distribution of localized RNA and ER in the MC. We also observed accumulations of RNA in ER clumps around the MC (Figure 6, A and B, arrows), but no association with the general ER network was detectable. In time-lapse recordings, ER clumps were sometimes seen to join the MC from surrounding regions but did not appear to carry fluorescent RNAs with them. The RNA accumulated progressively within regions already highly enriched in ER (video 4). Within the MC, DiI labeling was too dense to allow the precise relationship between the RNAs and the ER to be discriminated; however, in peripheral regions of the MC and in nearby ER clumps, RNA particles could be shown to be intimately associated with the ER. Quantification of the extent of colocalization between Xcat2 and ER, through counting of 14–20 particles in five distinct fields around the MC in three separate experiments, showed that an average of 68% of well-resolved RNA particles colocalized with ER clumps (compared with 37% for points randomly distributed in the same fields; significantly different by t test, with 99% confidence). Furthermore, examination of sequences of images acquired at 6-s intervals in peripheral regions of the MC revealed that the local jostling movements of ER and Xcat2 particles occurred in precise coordination (Figure 6B and video 5). Both Xcat2 RNA constructs lacking the GGLE and Xdazl RNAs similarly “danced” with ER around the MC (unpublished data). Localization of germ plasm RNAs is thus correlated with their association with dense ER in the MC region, independent of germinal granule formation.
Figure 6.
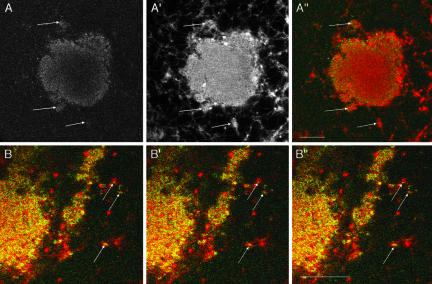
Localized RNAs associate with ER. (A) Confocal images of a live stage I oocyte injected the previous day with Alexa 488-Xcat2 3′UTR (A) and DiI-loaded oil drop (A′). RNA aggregates (green in the overlay A″) are observed to coincide in positions in and around the MC (e.g., at arrows) and with accumulations of ER (red in A″). (B) Higher magnification, consecutive, overlaid images (colors as in A″) from a sequence acquired every 6.5 s in a confocal plane tangential to the edge of the MC, showing that Xcat2 particles (green) undergo jostling movements together with the ER (red) (e.g., at arrows; see also video 4).
Vg1RBP/Vera Binding to Early Pathway RNAs
Vg1RBP/Vera, which was previously implicated in linking late pathway RNAs with ER (Deshler et al., 1997), is a candidate protein for mediating an interaction between Xcat2 and the ER. To examine this possibility, we analyzed the binding of Xcat2 RNA to proteins present in stage I oocyte lysates, including Vg1RBP/Vera. The sets of proteins that bound to Xcat2 and Vg1 RNAs were strikingly similar (Figure 7A). A set of six VgRBPs recognized the Xcat2 3′UTR in a sequence-specific manner, including Vg1RBP/Vera and VgRBP60/hnRNPI (Schwartz et al., 1992; Mowry, 1996; Deshler et al., 1997; Cote et al., 1999). Binding of these proteins to the Xcat2 3′UTR (Figure 7A, lane 4) was competed by a molar excess of either unlabeled Vg1 LS (Figure 7A, lane 5) or Xcat2 3′UTR (Figure 7A, lane 6) but was not affected by nonspecific competitor RNA. The interaction of Xcat2 RNA with Vg1RBP/Vera and VgRBP60/hnRNP I was confirmed using partially purified preparations of Vg1RBP/Vera and VgRBP60/hnRNP I (Figure 7B).
Figure 7.
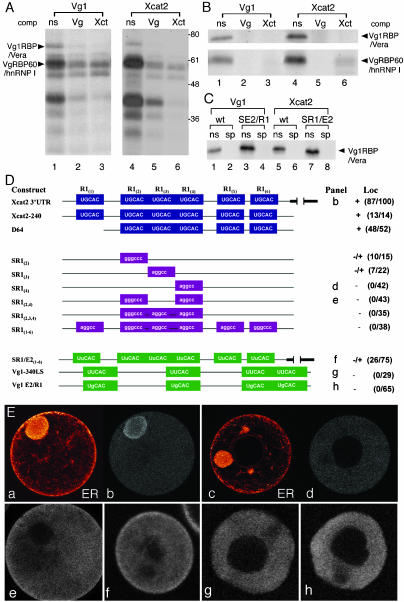
Vg1RBP/Vera binding to early pathway RNA. (A) The Vg1 LS and Xcat2 3′UTR bind to similar oocyte factors. Radiolabeled Vg1 LS (lanes 1–3) and Xcat2 3′UTR (lanes 4–6) transcripts were tested with UV cross-linking for the ability to be bound in vitro by proteins from an S10 oocyte lysate. Specificity of binding was assessed by competition with unlabeled non-specific competitor RNA (ns, lanes 1 and 4), Vg1 LS (Vg, lanes 2 and 5), or Xcat2 3′UTR (Xct, lanes 3 and 6). Vg1RBP/Vera and VgRBP60/hnRNP I are indicated on the left; molecular weight markers are indicated on the right. (B) Both Vg1RBP/Vera and VgRBP60/hnRNP I specifically bind to the Xcat2 3′UTR. Radiolabeled Vg1 LS (lanes 1–3) and Xcat2 3′UTR (lanes 4–6) transcripts were tested by UV cross-linking for the ability to be bound in vitro by partially purified Vg1RBP/Vera (top) and VgRBP60/hnRNP I (bottom). Specificity of binding was assessed by competition with unlabeled nonspecific competitor RNA (ns, lanes 1 and 4), Vg1 LS (Vg, lanes 2 and 5), or Xcat2 3′UTR (Xct, lanes 3 and 6). (C) Mutations within E2 or R1 motifs do not affect binding of Vg1RBP/Vera to the Vg1 LS and Xcat2 3′UTR. Radiolabeled Vg1 LS (wt, lanes 1 and 2), Vg1 E2/R1 (SE2/R1, lanes 3 and 4), Xcat2 3′UTR (wt, lanes 5 and 6), or SR1/E2(1–6) (SR1/E2, lanes 7 and 8) transcripts were tested with UV cross-linking for the ability to be bound in vitro by partially purified Vg1RBP/Vera. Specificity of binding was assessed by competition with unlabeled nonspecific competitor RNA (ns, odd-numbered lanes) or Vg1 LS (sp, even-numbered lanes). (D) UGCAC motifs are required for Xcat2 MC entrapment. The schematic representation of the 3′UTR of Xcat2 shows the UGCAC (R1) motifs and deletion (D) or noncognate substitution (S) mutants tested by microinjection into stage I oocytes. Numbers identify which R1 repeat is changed in the Xcat2 3′UTR. Point mutations changed R1 to E2 repeats in Xcat2 [SR1/E2(1–6)] or E2 to R1 repeats in the Vg1–340 LS (Vg1 E2/R1). Scores for localization (Loc) (right) indicate the number of oocytes showing localization per the total number of injected oocytes in which a MC was detected. Data were compiled from three independent experiments. Letters preceding the scores refer to panels in E. (E) Confocal images showing localization of representative oocytes injected with selected Alexa 488-labeled transcripts diagramed in D. Panels a and c show the ER network labeled with DiI of the oocytes shown in b and d, respectively. Oocyte diameters 150–300 μm.
Vg1RBP/Vera binds the late pathway RNAs Vg1 and VegT in a manner dependent on the presence of multiple E2 motifs (UUCAC) in their 3′UTRs (Deshler et al., 1997, 1998; Bubunenko et al., 2002; Kwon et al., 2002). The 240-nucleotide Xcat2 MCLS contains very similar repeats (UGCAC) termed R1, and deletion of all six R1 motifs was previously shown to abolish MC localization (Betley et al., 2002). Our analysis of the R1 motifs in the Xcat2 RNA 3′UTR (Figure 7D) showed that substitution of single R1 motifs [R1(2), R1(3), or R1(4) but not R1(1)] was sufficient to severely reduce MC localization (Figure 7, D and Ed), whereas substitution of both R1(2) and R1(4) resulted in more severe defects in localization (Figure 7, D and Ee). Conversion of all R1 motifs to E2 motifs caused a 65% reduction in the number of oocytes showing MC localization, compared with control values (three independent experiments) (Figure 7, D and Ef). In contrast, conversion of E2 motifs to R1 repeats in Vg1 RNA was not sufficient to drive ectopic localization to the MC (Figure 7, D and Eg). Therefore, R1 motifs are required but not sufficient for entrapment within the MC, and E2 repeats can partially substitute for R1 repeats. Vg1RBP/Vera binding to Xcat2 RNA was not dependent on the presence of R1 repeats (Figure 7C) (Betley et al., 2002), and we showed that changing R1 repeats in Xcat2 to E2 repeats also had no effect on Vg1RBP/Vera binding (Figure 7C, lanes 3, 5, and 7). More surprisingly, given that binding of partially purified Vg1RBP/Vera to VegT and Vg1 RNAs requires intact E2 repeats (Deshler et al., 1997; Bubunenko et al., 2002; Kwon et al., 2002), we found that R1 motifs were able to substitute for E2 repeats in Vg1 RNA without affecting Vg1RBP/Vera binding or its ability to compete with wild-type Vg1 RNA for binding (Figure 7C, lanes 3 and 4). This could reflect the ability of E2 and RI motifs placed in equivalent positions to mediate the formation of similar secondary structural features.
Vg1RBP/Vera Does Not Codistribute with Xcat2 RNA or ER
Having shown that Vg1RBP/Vera was capable of binding Xcat2 RNA in vitro, we next investigated whether Vg1RBP/Vera codistributed with Xcat2 RNA in vivo. We localized Vg1RBP/Vera in stage I–III oocytes in immunofluorescence assays with two distinct anti-Vg1RBP/Vera antisera. Similar staining patterns were observed, and in both cases staining could be abolished with preabsorption with bacterially expressed Vg1RBP/Vera protein (our unpublished results). In stage I–III oocytes, Vg1RBP/Vera staining was strong throughout the cytoplasm, being enriched in a cortical/subcortical layer, and was detectable but weaker in the GV (Figure 8). Staining was generally excluded from the MC (Figure 8, A, D, and E). No obvious codistribution with fluorescent Xcat2 3′UTR was detected either after or during localization (Figure 8, A′ and B′). In stage II oocytes, a slight accumulation of anti-Vg1RBP/Vera was detected around the dispersing MC, with strong enhancement in the cortical region adjacent to it (Figure 8C). In this region, although not elsewhere, there was overlap of strong Vg1RBP/Vera staining with localized fluorescent RNAs (Figure 8, C″, arrows).
Figure 8.
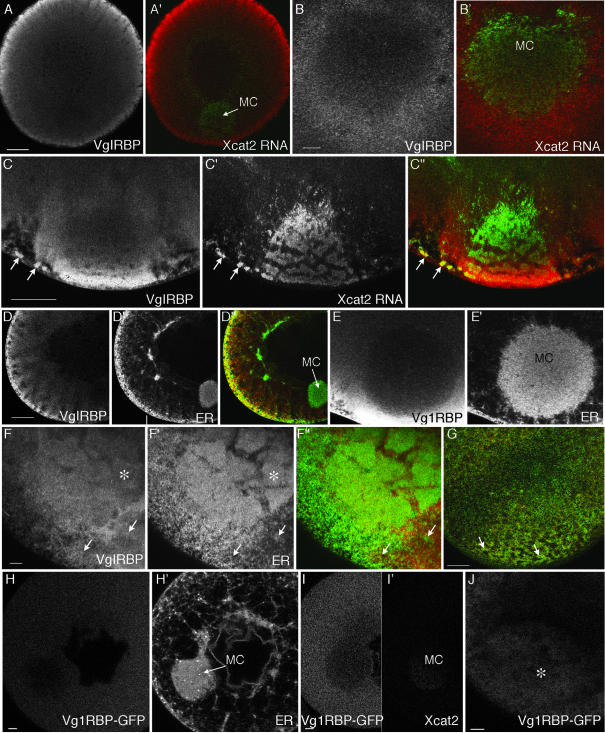
Vg1RBP/Vera is excluded from the MC but enriched in the vegetal cortex. (A–G) Confocal images showing immunofluorescence of stage I (A, B, D, and E), early stage II (C and F), and stage III (G) oocytes with an anti-Vg1RBP/Vera antibody. At all stages, staining was observed throughout the cytoplasm, being enriched in cortical regions and relatively weak in the MC. (A′, B′, and C″) Superimposed images with anti-Vg1RBP/Vera (red) and Alexa 488-labeled Xcat2 3′UTR injected 20 h before fixation (green; also shown in C′). Anti-Vg1RBP/Vera and localized fluorescent RNAs (arrows in C″) are found together in stage II oocytes around the MC and in the region of MC-cortex contact but not elsewhere. (D′, E′, and F′) Costaining with anti-GRP94 to visualize ER. (D″, F″, and G) Superimposed images, with anti-Vg1RBP/Vera in red and anti-GRP94 in green. Again, localization in the same region is observed only once the MC meets the vegetal cortex (F and F″), remaining colocalized in patches at the vegetal cortex in stage III (G). (H–J) Confocal images of live oocytes, 24 h after injection of RNA coding for Vg1RBP/Vera-GFP (H, I, and J) and either DiI (H′) or rhodamine-Xcat2 3′UTR (I′). Vg1RBP/Vera-EGFP protein was distributed homogeneously in stage I oocytes (H and I), with relatively little in the MC. It did not codistribute with either ER or exogenous localized Xcat2 3′UTR. In stage II (J), like exogenous Vg1RBP/Vera, it became concentrated at the vegetal pole (*). Scale bars, 10 μm in B, E, F, H, I, and J, 50 μm in A, C, D, and G.
The relative lack of Vg1RBP/Vera staining in the ER-rich MC suggested that, in early stages, Vg1RBP/Vera does not associate with ER. Double-immunofluorescence assays with anti-Vg1RBP/Vera and the ER marker-specific anti-GRP94 confirmed that there was no colocalization in the cytoplasm, MC, or cortex in stage I; indeed, the distributions were largely mutually exclusive (Figure 8, D and E′). The strong anti-GRP94 staining in the MC (Figure 8, D′ and E′) also ruled out the possibility that antibodies were generally excluded from this region in our experiments. In stage II, the only region where both antibodies were concentrated was the region where the MC was in contact with the cortex (Figure 8F). In stage III oocytes, the distinct vegetal ER subpopulation described previously (Deshler et al., 1997; Kloc and Etkin, 1998) could be distinguished in the vegetal region, and double-immunofluorescence assays demonstrated colocalization of Vg1RBP/Vera with ER (Figure 8G). This colocalization was exclusive to this patch and was not observed elsewhere. It should be emphasized that the observed staining overlap does not prove that there is any functional interaction between Vg1RBP/Vera and ER in this region. We conclude that Vg1RBP/Vera does not function to link RNAs to the ER during localization through the early pathway. It may play a role in the cortical RNA-anchoring step of the late pathway.
The lack of association of endogenous Vg1RBP/Vera with Xcat2 RNA suggested by our immunofluorescence studies was confirmed with expression of a Vg1RBP/Vera-EGFP fusion protein. Vg1RBP/Vera-EGFP was distributed throughout the cytoplasm of stage I/II oocytes, with relatively little being detectable within the MC (Figure 8, H and I). As in the case of endogenous Vg1RBP/Vera, we could not detect significant codistribution of Vg1RBP/Vera-EGFP with DiI-labeled ER in stage I oocytes (Figure 8H) or with rhodamine-labeled Xcat2 3′UTR (Figure 8I). FRAP analysis revealed that Vg1RBP/Vera-EGFP, like Xcat2 3′UTR, was freely diffusible in the cytoplasm of both stage I and stage II/III oocytes (our unpublished results). In contrast, from late stage II, Vg1RBP/Vera-GFP was observed to be concentrated in the vegetal cortex (Figure 8J) and could be shown by FRAP to be immobilized there (our unpublished results). These results are consistent with a role for Vg1RBP/Vera in vegetal anchoring during the late pathway but not the early pathway.
DISCUSSION
We have presented evidence that the localization of the germ plasm RNAs Xcat2 and Xdazl to the MC is driven by an entrapment mechanism, rather than by directed transport. This conclusion is supported by FRAP analyses showing free movement of germ line RNAs in the cytoplasm, equivalent to that of nonlocalizing RNA, and by the demonstration that localization does not require microtubules or microfilaments. Xcat2 3′UTR became immobilized upon reaching the MC and accumulated there in a linear manner, whereas cytoplasmic levels of exogenous RNA remained constant, indicating progressive entrapment rather than selective cytoplasmic degradation as the mechanism for localization. Fluorescent Xcat2 3′UTR progressively condensed into large aggregates within the MC, including the METRO region where germinal granules are concentrated. Immobilization of Xcat2 3′UTR in the MC was not, however, dependent on germinal granule formation.
Equivalent diffusion/entrapment mechanisms have been shown to underlie RNA localization in other cell types, with the most notable case being the Xcat2-related nanos RNA in late-stage Drosophila oocytes (Forrest and Gavis, 2003). As with Xcat2 RNA, nanos RNA does not require microtubules to localize, although the dramatic microtubule-mediated cytoplasmic streaming that occurs at that time facilitates localization. Cytoplasmic streaming also appears to play a role in localizing RNAs to the blastoderm in the yolky zebrafish embryo (Maegawa et al., 1999). Entrapment of RNAs in the MC in Xenopus oocytes occurs before yolk uptake begins, and the RNA-protein particles are able to spread readily throughout the oocyte in the absence of observable cytoplasmic flow. These small particles probably contain single RNA types with associated proteins (this study; Kloc and Etkin, 1998), with large aggregates forming only after their arrival in the MC. Similarly, nanos RNA forms small ribonucleoprotein particles in the cytoplasm, forming larger granules only in association with germ (pole) plasm. Therefore, diffusion/entrapment mechanisms may be characterized by the formation of small ribonucleoprotein particles, whereas motordriven transport involves larger complexes (Ferrandon et al., 1994; Ainger et al., 1997; Kohrmann et al., 1999; Mallardo et al., 2003).
Anchoring or entrapment is a vital step in both motor protein–driven and diffusion-based RNA localization processes; however, the mechanisms involved remain little understood. Not surprisingly, microfilaments are involved in anchoring germ plasm to the actin-rich cortex both in Drosophila (Forrest and Gavis, 2003) and in Xenopus (Savage and Danilchik, 1993) but they do not appear to be required for the direct association of the RNAs with the germ plasm in either organism. Actin filaments do participate in anchoring Vg1 RNA to the cortex (Yisraeli et al., 1990) but probably do so indirectly, by maintaining the organization of cytokeratin filaments (Gard et al., 1997), which have been implicated in anchoring both Vg1 RNA and the germ plasm component Xwnt-11 RNA to the vegetal cortex in fully grown oocytes (Alarcon and Elinson, 2001). RNAs themselves may also play a structural role in anchoring other RNAs (Kloc and Etkin, 1994; Heasman et al., 2001).
We have uncovered a distinct ER domain in the MC of Xenopus oocytes that is well placed to provide support for mRNA entrapment. This subdomain of ER is continuous with cytoplasmic and cortical networks. Its boundaries may be maintained by the surrounding basket of intermediate filaments and stable microtubules (Gard, 1991). Our timelapse observations do not support a role for ER in moving Xcat2 to the MC, but they suggest that it contributes to immobilizing RNAs to the MC region and maintains a close association with accumulated RNA particles. This association is not attributable to translation-related ribosome recruitment to ER, because Xcat2 and Xdazl RNAs are not translated within germ plasm but are stored untranslated until the blastula stage (Houston and King, 2000b). A distinct ER population was previously implicated in the late RNA localization pathway in Xenopus, occupying a characteristic “wedge” region between the GV and the vegetal cortex from late stage II onward (Deshler et al., 1997). In this case too, the ER may be fulfilling a trapping function, because RNA association with the vegetal wedge region (although not its subsequent displacement to the cortex) is microtubule independent (Deshler et al., 1997; Kloc and Etkin, 1998). The wedge domain of ER at stage II arises around the MC when it contacts the cortex and could be derived partially from the dense ER of the MC. Resolution of this question awaits the development of oocyte culture systems that support the stage I-stage II transition.
How might RNAs be linked to ER during the early and late localization pathways? One candidate for mediating this association is Vg1RBP/Vera. This protein was found to cofractionate with ER from oocyte extracts, although not if the extracts were diluted, which suggests a weak association (Deshler et al., 1997). More recently, it has been shown that Vg1RBP/Vera, as well as all other Vg1 LS-binding proteins, can specifically bind early pathway RNAs as well as late pathway RNAs in vitro (Betley et al., 2002). The ability of these proteins, which are present in stage I oocytes, to bind LEs in vitro does not necessarily indicate that they function in vivo for localization. We think it unlikely that Vg1RBP/Vera functions in the early pathway, because Vg1RBP/Vera codistributed with the wedge region ER used in the late pathway but not with localized RNAs or ER in the MC. One possible explanation for this probable lack of involvement of Vg1RBP/Vera in the early pathway is that, in stage I of oogenesis, Vg1RBP/Vera associates with Xcat2 RNA in the cytoplasm but cannot associate with ER. Alternatively, Vg1RBP/Vera may exhibit insignificant binding to Xcat2 RNA in vivo in stage I, perhaps because of competition with other Xcat2 RBPs. It is clear that Vg1RBP/Vera associates with late and early pathway RNAs in distinct ways. Binding to late pathway RNAs is dependent on E2 motifs (Deshler et al., 1997; Bubunenko et al., 2002; Kwon et al., 2002) or equivalently placed R1 motifs (this study). For Xcat2, R1 repeats are not required for Vg1RBP/Vera binding and, although the canonical E2 motif UUCAC is not present within the Xcat2 LS, four WYCAY motifs capable of binding Vg1RBP/Vera in vitro are present (Deshler et al., 1998). Failure of Xcat2 localization to the MC caused by loss of the R1 motifs can be partially, but not fully, compensated for with E2 motifs. These results may reflect differences in protein binding affinities for early and late pathway RNAs leading to competition between RNAs for protein binding and/or the presence of cofactors specific to one or the other pathway. In this regard, it is interesting to note that several of the WYCAY motifs are close to or overlap with the R1 motifs. Binding of an unknown protein to the R1 repeats in stage I might exclude Vg1RBP/Vera from binding Xcat2 RNA in vivo. This interpretation would account for the ability of Xcat2 to localize through the late pathway (Zhou and King, 1996b). Vg1RBP/Vera binding to germ plasm RNAs may function to localize residual populations through association with the wedge region ER through the late pathway in stage II oocytes.
There are other examples in animals and plants of localized RNA associating with ER subdomains. In rice seeds, prolamin mRNA is targeted to a specific subdomain of ER (Choi et al., 2000); in ascidians, specific RNAs including HrPEM and macho-1, associate with the rough ER beneath the vegetal cortex (Sardet et al., 2003). Cortical ER in fully grown Xenopus oocytes was also implicated in the anchoring of Vg1 and Xwnt-11 RNAs through detergent treatment of isolated cortices (Alarcon and Elinson, 2001). Xcat2 RNA anchoring was not affected in those experiments, perhaps because the RNA is found within dense germinal granules at that stage. Within the Drosophila germ line, a distinct structure, the sponge body, that contains ER-like vesicles, electron-dense masses associated with mitochondria, and RNA has been described (Wilsch-Brauninger et al., 1997). The sponge body may act as the MC does in Xenopus and facilitate the localization of germ plasm components during oogenesis. In neuronal dendrites, the double-stranded RBP Staufen forms large RNA-containing granules that are associated with the ER, as well as smaller particles containing dendritically localized RNAs (Kohrmann et al., 1999; Mallardo et al., 2003). Staufen has also recently been shown to colocalize with dense ER patches beneath the vegetal cortex of Xenopus stage VI oocytes (Allison et al., 2004).
The ER is being found to be a key factor in RNA localization in many systems, and this study has revealed how different ER domains within a single cell may assume distinct localization functions. The challenge now is to understand how such specialized ER domains come into being and how their association with specific RNAs and proteins is mediated.
Supplementary Material
Acknowledgments
We are grateful to William Taylor, Nancy Standart, and Anna Git for providing Vg1RBP/Vera antibodies and recombinant protein and to Jim Deshler for providing the Xdazl LE construct. We thank Christian Rouviere for developing and applying Metamorph macros for image realignment, Antoine Sciandra for helping with curve fitting, and Mark Terasaki for encouragement with FRAP experiments. This research was financed by the Centre National pour la Recherche Scientifique and by French Government ACI 167 (E.H.), by the National Institutes of Health Grant GM-33932 and National Science Foundation Grant BN-9601209 (M.L.K.), and by National Institutes of Health Grant HD-30699 (K.L.M.). E. H. and M.L.K. were also supported by a Human Frontiers Science Project network.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0265. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0265.
Abbreviations used: PGC, primordial germ cell; FRAP, fluorescence recovery after photobleaching; MC, mitochondrial cloud; UTR, untranslated region; RBP, RNA-binding protein; LS, localization signal; GGLE, germinal granule localization element; ER, endoplasmic reticulum; BSA, bovine serum albumin; GFP, green fluorescent protein; PBS, phosphate-buffered saline; GV, germinal vesicle; LE, localization element; Dil, DilC16(3); DiO, DiOC(6)(3).
The online version of this article contains supplemental material accessible through http://www.molbiolcell.org
References
- Allison, R, Czaplinski, K., Git, A., Adegbenro, E., Stennard, F., Houliston E., and Standart, N. (2004). Xenopus Staufen 1 and 2 were vegetally localized during oogenesis. RNA (in press). [DOI] [PMC free article] [PubMed]
- Ainger, K., Avossa, D., Diana, A.S., Barry, C., Barbarese, E., and Carson, J.H. (1997). Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 138, 1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon, V.B., and Elinson, R.P. (2001). RNA anchoring in the vegetal cortex of the Xenopus oocyte. J. Cell Sci. 114, 1731-1741. [DOI] [PubMed] [Google Scholar]
- Argon, Y., and Simen, B.B. (1999). GRP94, an ER chaperone with protein and peptide binding properties. Semin. Cell Dev. Biol. 10, 495-505. [DOI] [PubMed] [Google Scholar]
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. (1998). RNA localization in development. Annu. Rev. Biochem. 67, 335-394. [DOI] [PubMed] [Google Scholar]
- Betley, J.N., Frith, M.C., Graber, J.H., Choo, S., and Deshler, J.O. (2002). A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 12, 1756-1761. [DOI] [PubMed] [Google Scholar]
- Betley, J.N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F., and Deshler, J.O. (2004). Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14, 219-224. [DOI] [PubMed] [Google Scholar]
- Brunati, A.M., Contri, A., Muenchbach, M., James, P., Marin, O., and Pinna, L.A. (2000). GRP94 (endoplasmin) co-purifies with and is phosphorylated by Golgi apparatus casein kinase. FEBS Lett. 471, 151-155. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M., Kress, T.L., Vempati, U.D., Mowry, K.L., and King, M.L. (2002). A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 248, 82-92. [DOI] [PubMed] [Google Scholar]
- Bullock, S.L., and Ish-Horowicz, D. (2001). Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414, 611-616. [DOI] [PubMed] [Google Scholar]
- Cha, B.J., Koppetsch, B.S., and Theurkauf, W.E. (2001). In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 106, 35-46. [DOI] [PubMed] [Google Scholar]
- Chan, A.P., Kloc, M., Bilinski, S., and Etkin, L.D. (2001). The vegetally localized mRNA fatvg is associated with the germ plasm in the early embryo and is later expressed in the fat body. Mech. Dev. 100, 137-140. [DOI] [PubMed] [Google Scholar]
- Chan, A.P., Kloc, M., and Etkin, L.D. (1999). fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development 126, 4943-4953. [DOI] [PubMed] [Google Scholar]
- Choi, S.B., Wang, C., Muench, D.G., Ozawa, K., Franceschi, V.R., Wu, Y., and Okita, T.W. (2000). Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407, 765-767. [DOI] [PubMed] [Google Scholar]
- Cote, C.A., Gautreau, D., Denegre, J.M., Kress, T.L., Terry, N.A., and Mowry, K.L. (1999). A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell 4, 431-437. [DOI] [PubMed] [Google Scholar]
- Deshler, J., Highett, M., and Schnapp, B. (1997). Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276, 1128-1131. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., Abramson, T., and Schnapp, B.J. (1998). A highly conserved RNA-binding protein for cytoplasmic mRNA localisation in vertebrates. Curr. Biol. 8, 489-496. [DOI] [PubMed] [Google Scholar]
- Dumont, J.N. (1972). Oogenesis in Xenopus laevis (Daudin). 1. Stages of oocyte development in laboratory maintained animals. J. Morphol. 136, 153-180. [DOI] [PubMed] [Google Scholar]
- Elisha, Z., Havin, L., Ringel, I., and Yisraeli, J.K. (1995). Vg1 RNA binding protein mediates the association of Vg1 RNA with microtubules in Xenopus oocytes. EMBO J. 14, 5109-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., Elphick, L., Nusslein-Volhard, C., and St. Johnson, D. (1994). Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221-1232. [DOI] [PubMed] [Google Scholar]
- Forbes, A., and Lehmann, R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679-690. [DOI] [PubMed] [Google Scholar]
- Forrest, K.M., and Gavis, E.R. (2003). Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13, 1159-1168. [DOI] [PubMed] [Google Scholar]
- Forristall, C., Pondel, M., and King, M.L. (1995). Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development 121, 201-208. [DOI] [PubMed] [Google Scholar]
- Gard, D.L. (1991). Organization, nucleation, and acetylation of microtubules in Xenopus laevis oocytes: a study by confocal immunofluorescence microscopy. Dev. Biol. 143, 346-362. [DOI] [PubMed] [Google Scholar]
- Gard, D.L. (1993). Confocal immunofluorescence microscopy of microtubules in amphibian oocytes and eggs. Methods Cell Biol. 38, 241-264. [DOI] [PubMed] [Google Scholar]
- Gard, D.L., Cha, B.J., and King, E. (1997). Organisation and animal-vegetal asymmetry of cytokeratin filaments in stage VI Xenopus oocytes is dependent upon F-actin and microtubules. Dev. Biol. 184, 94-114. [DOI] [PubMed] [Google Scholar]
- Git, A., and Standart, N. (2002). The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA 8, 1319-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman, J., Quarmby, J., and Wylie, C.C. (1984). The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev. Biol. 105, 458-469. [DOI] [PubMed] [Google Scholar]
- Heasman, J., Wessely, O., Langland, R., Craig, E.J., and Kessler, D.S. (2001). Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Dev. Biol. 240, 377-386. [DOI] [PubMed] [Google Scholar]
- Houston, D.W., and King, M.L. (2000a). A critical role for Xdazl, a germ plasm-localised RNA, in the differentiation of primordial germ cells in Xenopus. Development 127, 447-456. [DOI] [PubMed] [Google Scholar]
- Houston, D.W., and King, M.L. (2000b). Germ plasm and molecular determinants of germ cell fate. Curr. Top. Dev. Biol. 50, 155-181. [DOI] [PubMed] [Google Scholar]
- Jansen, R.P. (2001). mRNA localization: message on the move. Nat. Rev. Mol. Cell. Biol. 2, 247-256. [DOI] [PubMed] [Google Scholar]
- Joseph, E.M., and Melton, D.A. (1998). Mutant Vg1 ligands disrupt endoderm and mesoderm formation in Xenopus embryos. Development 125, 2677-2685. [DOI] [PubMed] [Google Scholar]
- King, M., Messitt, L., and Mowry, K. (2004). Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell. (in press). [DOI] [PubMed]
- King, M.L., Zhou, Y., and Bubunenko, M. (1999). Polarizing genetic information in the egg: RNA localization in the frog oocyte. Bioessays 21, 546-557. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Bilinski, S., Pui-Yee Chan, A., and Etkin, L.D. (2000). The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3′UTR. Dev. Biol. 217, 221-229. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Dougherty, M.T., Bilinski, S., Chan, A.P., Brey, E., King, M.L., Patrick, C.W., Jr., and Etkin, L.D. (2002a). Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Dev. Biol. 241, 79-93. [DOI] [PubMed] [Google Scholar]
- Kloc, M., and Etkin, L.D. (1994). Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science 265, 1101-1103. [DOI] [PubMed] [Google Scholar]
- Kloc, M., and Etkin, L.D. (1995). Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development 121, 287-297. [DOI] [PubMed] [Google Scholar]
- Kloc, M., and Etkin, L.D. (1998). Apparent continuity between the messenger transport organiser and late RNA localisation pathways during oogenesis in Xenopus. Mech. Dev. 73, 95-106. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Larabell, C., and Etkin, L.D. (1996). Elaboration of the messenger transport organizer pathway for localization of RNA to the vegetal cortex of Xenopus oocytes. Dev. Biol. 180, 119-130. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Zearfoss, N.R., and Etkin, L.D. (2002b). Mechanisms of subcellular mRNA localization. Cell 108, 533-544. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., Yamada, M., Asaoka, M., and Kitamura, T. (1996). Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380, 708-711. [DOI] [PubMed] [Google Scholar]
- Kohrmann, M., Luo, M., Kaether, C., DesGroseillers, L., Dotti, C.G., and Kiebler, M.A. (1999). Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10, 2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, T.T., Zhao, W.M., Jiang, C., and Huber, P.W. (2002). A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development 129, 5609-5619. [DOI] [PubMed] [Google Scholar]
- Kwon, S., Abramson, T., Munro, T.P., John, C.M., Kohrmann, M., and Schnapp, B.J. (2002). UUCAC- and Vera-dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 12, 558-564. [DOI] [PubMed] [Google Scholar]
- Lewis, R.A., Kress, T.L., Cote, C.A., Gautreau, D., Rokop, M.E., and Mowry, K.L. (2004). Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech. Dev. 121, 101-109. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps, K., Taylor, D.L., and Lanni, F. (1986). Probing the structure of cytoplasm. J. Cell Biol. 102, 2015-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur, H., Bubunenko, M., Houston, D.W., and King, M.L. (1999). Xcat2 RNA is a translationally sequestered germ plasm component in Xenopus. Mech. Dev. 84, 75-88. [DOI] [PubMed] [Google Scholar]
- Maegawa, S., Yasuda, K., and Inoue, K. (1999). Maternal mRNA localization of zebrafish DAZ-like gene. Mech. Dev. 81, 223-226. [DOI] [PubMed] [Google Scholar]
- Mallardo, M., Deitinghoff, A., Muller, J., Goetze, B., Macchi, P., Peters, C., and Kiebler, M.A. (2003). Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. USA 100, 2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera, L., Forristall, C., Zhou, Y., and King, M.L. (1993). A mRNA localised to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 117, 377-386. [DOI] [PubMed] [Google Scholar]
- Mowry, K. (1996). Complex formation between stage-specific oocyte factors and a Xenopus mRNA localization element. Proc. Natl. Acad. Sci. USA 93, 14608-14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry, K., and Melton, D. (1992). Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science 255, 991-994. [DOI] [PubMed] [Google Scholar]
- Opresko, L.K. (1991). Vitellogenin uptake and in vitro culture of oocytes. Methods Cell Biol. 36, 117-132. [DOI] [PubMed] [Google Scholar]
- Pfaff, S.L., and Taylor, W.L. (1992). Characterization of a Xenopus oocyte factor that binds to a developmentally regulated cis-element in the TFIIIA gene. Dev. Biol. 151, 306-316. [DOI] [PubMed] [Google Scholar]
- Sardet, C., Nishida, H., Prodon, F., and Sawada, K. (2003). Maternal mRNAs of PEM and macho 1, the ascidian muscle determinant, associate and move with a rough endoplasmic reticulum network in the egg cortex. Development 130, 5839-5849. [DOI] [PubMed] [Google Scholar]
- Savage, R., and Danilchik, M. (1993). Dynamics of germ plasm localization and its inhibition by ultraviolet irradiation in early cleavage Xenopus embryos. Dev. Biol. 157, 371-382. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.P., Aisenthal, L., Elisha, Z., Oberman, F., and Yisraeli, J.K. (1992). A 69-kDa RNA-binding protein from Xenopus oocytes recognizes a common motif in two vegetally localized maternal mRNAs. Proc. Natl. Acad. Sci. USA 89, 11895-11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Johnston, D. (1995). The intracellular localisation of messenger RNAs. Cell 81, 161-170. [DOI] [PubMed] [Google Scholar]
- Terasaki, M., and Jaffe, L.A. (1991). Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J. Cell Biol. 114, 929-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, M., Sasaoka, Y., Kiso, M., Abe, K., Haraguchi, S., Kobayashi, S., and Saga, Y. (2003). Conserved role of nanos proteins in germ cell development. Science 301, 1239-1241. [DOI] [PubMed] [Google Scholar]
- Turner, D.L., and Weintraub, H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434-1447. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and Lin, H. (2004). Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303, 2016-2019. [DOI] [PubMed] [Google Scholar]
- Wilkie, G.S., and Davis, I. (2001). Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 105, 209-219. [DOI] [PubMed] [Google Scholar]
- Wilsch-Brauninger, H., Schwarz, H., and Nusslein-Volhard, C. (1997). A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J. Cell Biol. 139, 817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli, J.K., Sokol, S., and Melton, D.A. (1990). A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development 108, 289-298. [DOI] [PubMed] [Google Scholar]
- Yoon, Y.J., and Mowry, K.L. (2004). Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131, 3035-3045. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Houston, D.W., King, M.L., Payne, C., Wylie, C., and Heasman, J. (1998). The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell 94, 515-524. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., et al. (1999). Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech. Dev. 88, 101-106. [DOI] [PubMed] [Google Scholar]
- Zhao, W.M., Jiang, C., Kroll, T.T., and Huber, P.W. (2001). A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 20, 2315-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., and King, M.L. (1996a). Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development 122, 2947-2953. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., and King, M.L. (1996b). RNA transport to the vegetal cortex of Xenopus oocytes. Dev. Biol. 179, 173-183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.