Abstract
We have investigated transforming growth factor beta (TGF-β)–mediated induction of actin stress fibers in normal and metastatic epithelial cells. We found that stress fiber formation requires de novo protein synthesis, p38Mapk and Smad signaling. We show that TGF-β via Smad and p38Mapk up-regulates expression of actin-binding proteins including high-molecular-weight tropomyosins, α-actinin and calponin h2. We demonstrate that, among these proteins, tropomyosins are both necessary and sufficient for TGF-β induction of stress fibers. Silencing of tropomyosins with short interfering RNAs (siRNAs) blocks stress fiber assembly, whereas ectopic expression of tropomyosins results in stress fibers. Ectopic-expression and siRNA experiments show that Smads mediate induction of tropomyosins and stress fibers. Interestingly, TGF-β induction of stress fibers was not accompanied by changes in the levels of cofilin phosphorylation. TGF-β induction of tropomyosins and stress fibers are significantly inhibited by Ras-ERK signaling in metastatic breast cancer cells. Inhibition of the Ras-ERK pathway restores TGF-β induction of tropomyosins and stress fibers and thereby reduces cell motility. These results suggest that induction of tropomyosins and stress fibers play an essential role in TGF-β control of cell motility, and the loss of this TGF-β response is a critical step in the acquisition of metastatic phenotype by tumor cells.
INTRODUCTION
There is solid evidence that the transforming growth factor beta (TGF-β) signaling pathway is a major cellular growth inhibitory and proapoptotic pathway in epithelial, endothelial, hematopoeitic, and other cell types (Roberts and Wakefield, 2003). However, clinical and experimental studies indicate that metastatic cancers of the breast and other tissues express elevated levels of TGF-β that appears to support the metastatic behavior of the tumor cells (Saito et al., 2000; Derynck et al., 2001). This apparent paradox has been associated with a progressive decline in the antitumorigenic function and a gain of protumorigenic activities of TGF-β, including induction of epithelial to mesenchymal transition (EMT) and tumor cell migration and invasion (Derynck et al., 2001; Wakefield and Roberts, 2002). Oncogenic Ras, Src, and ErbB2 as well as alterations in TGF-β signaling mediated by Smads, mitogen-activated protein kinases (Mapks), Rho kinases, and Akt/PKB are thought to contribute to the metastatic phenotype (Derynck and Zhang, 2003; Roberts and Wakefield, 2003).
The actin cytoskeleton plays a central role in the regulation of cellular processes linked to metastasis including cell proliferation, apoptosis, anchorage-independent cell growth, and cell migration and invasion (Pawlak and Helfman, 2001; Jaffe and Hall, 2002). TGF-β induces a rapid reorganization of the actin cytoskeleton, leading to membrane ruffling at the cell edges in both nontumorigenic and tumorigenic epithelial cells, whereas a prolonged incubation with TGF-β results in the formation of stress fibers (Bakin et al., 2002; Edlund et al., 2002). The immediate TGF-β–mediated changes in the actin cytoskeleton have been associated with activation of the Rho family of GTPases, Rac, CDC42, and RhoA (Bakin et al., 2002; Edlund et al., 2002), which control cell motility and invasive phenotypes by regulating organization of actin filaments (Jaffe and Hall, 2002). TGF-β regulates activity of these GTPases in various epithelial cell lines independently of Smad signaling (Bhowmick et al., 2001; Bakin et al., 2002; Edlund et al., 2002). The interplay between Rho-like GTPases regulate both the protrusive and contractile forces required for cell migration, via a combination of actin polymerization, depolymerization, and the interaction of myosin-based motors with actin filaments (Etienne-Manneville and Hall, 2002). Although RhoA contributes to cell migration by inducing actomyosin contractility, RhoA can also inhibit cell movement by stimulating the assembly of stress fibers and focal adhesions associated with the cell substratum (Cox et al., 2001). The TGF-β induction of actin stress fibers has been shown to depend on Smad signaling (Piek et al., 1999b), the RhoA-Rho kinase pathway (Bhowmick et al., 2001), and p38Mapk signaling (Hannigan et al., 1998; Bakin et al., 2002; Edlund et al., 2002). However, the cellular targets regulated by these pathways and their roles in TGF-β regulation of stress fibers and cell motility have not been defined.
Oncogenic transformation mediated by Ras and Src results in the disruption of actin stress fibers and focal adhesions, whereas restoration of actin stress fibers inhibits cell transformation and reduces metastasis (Pawlak and Helfman, 2001). The mechanisms mediating the disruption of stress fibers by the Ras-ERK pathway involve inhibition of the RhoA/ROCK pathway (Sahai et al., 2001; Pawlak and Helfman, 2002a, 2002b; Vial et al., 2003) and repression of actin-binding proteins involved in stabilization of actin filaments including tropomyosins and α-actinin (Pawlak and Helfman, 2001). Thus, the Ras-Erk pathway may modify TGF-β regulation of stress fibers and cell motility through one or both of these mechanisms.
In this study we demonstrate that expression of tropomyosins mediated by Smad and p38Mapk signaling is required for TGF-β regulation of stress fibers and cell motility. We show that the Ras-ERK pathway antagonizes TGF-β induction of stress fibers by suppressing expression of tropomyosins. TGF-β does not modulate cofilin phosphorylation, suggesting that the RhoA-ROCK-LIM kinase-cofilin pathway is not rate limiting. We provide evidence that tropomyosins are both necessary and sufficient for TGF-β induction of stress fibers. We show that expression of tropomyosins in metastatic cells results in stress fibers and reduces cell motility. These results suggest that loss of TGF-β–induced stress fibers is an essential characteristic of a prometastatic conversion of TGF-β function and that regulation of tropomyosin expression is an important component of this response.
MATERIALS AND METHODS
Antibodies, Plasmids, and Other Reagents
TGF-β1 was obtained from R&D Systems (Minneapolis, MN). The following antibodies were obtained from: to Smad2/3 (BD Transduction Laboratories, BD Biosciences, Lexington, KY); rabbit polyclonal to hemaglutinin (HA) epitope (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal antibodies to tropomyosin (TM311), α-actinin, actin, and the Flag epitope (Sigma, St. Louis, MO); to phospho-Smad2, phospho-ERK1/2, phospho-p38Mapk, phospho-ATF2, phospho-HSP27, and HSP27 (Cell Signaling Technology, Beverly, MA). Phalloidin-Alexa Green and phalloidin-Texas Red were from Molecular Probes (Eugene, OR). Fluorescein-labeled anti-HA antibody was from Roche Applied Science (Indianapolis, IN). Plasmids encoding rat HA-tagged TM2 and TM3 isoforms were described previously (Gimona et al., 1995). Inhibitors of p38Mapk (SB202190, SB203580, PD165319), MEK1/2 (PD098059, U0126), JNK (SP600125), Raf kinase, and protein synthesis (cycloheximide) were from Calbiochem (La-Jolla, CA). Phospho-cofilin antibody was provided by Dr. James Bamburg, Colorado State University (Fort Collins, CO).
Cell Culture
Mouse mammary epithelial NMuMG cells, human cervical carcinoma SiHa cells, human breast cancer MDA-MB-231 cells, human epidermoid carcinoma A431 cells, and human kidney HEK293T cells were purchased from American Tissue Culture Collection (ATCC, Manassas, VA). Cells were cultured as recommended by ATCC.
Adenoviral Infection of Cells
Adenoviruses encoding EGFP, Flag-tagged Smads, and the HA-tagged constitutively active TGF-β type I Alk5T204D receptor were produced using HEK-293T cells and stored in aliquots at –80°C. Cells grown on plastic dishes or glass coverslips were incubated for 3 h with supernatant containing adenoviruses at 5–10 MOI. Medium was replenished and cells were grown for additional 24 h before further treatments.
RNA Isolation and cDNA Microarray Analysis
RNA from mouse nontumor mammary epithelial NMuMG cells treated with 2 ng/ml TGF-β1 for 8 and 24 h was extracted as described in Bakin and Curran, (1999). Total RNA from each sample (35 μg) was labeled and hybridized with the NIA 15K Mouse array. Detailed descriptions of labeling and hybridization procedures are available from http://array.mc.vanderbilt.edu/Pages/Protocols/Protocols.htm. The array slides were scanned with an Axon 4000 scanner (Axon Instruments, Foster City, CA) at a resolution of 10 μm. The reference RNA from untreated NMuMG cells was labeled by using cyanine 3-dUTP (Cy3), and the RNA samples from TGFβ1-treated cells were labeled with cyanine 5-dUTP (Cy5). This experiment was performed in triplicate. Data were analyzed using GenePix4.0 software.
Immunoblot Analysis
Cells were incubated in medium containing 5% serum for 24 h before treatment with 2 ng/ml TGF-β1. Cells were lysed in buffer containing 20 mM Tris, pH 7.4, 137 mM NaCl, 1% NP-40, 10% glycerol, 20 mM NaF, 1 mM Na orthovanadate, 1 mM PMSF, 2 μg/ml aprotinin, and 2 μg/ml leupeptin. Immunoblot analyses of protein extracts were performed as described (Bakin et al., 2002).
Northern Blot and Reverse Transcription-Polymerase Chain Reaction Analysis
A cDNA fragment of rat TM3 and a polymerase chain reaction (PCR)-generated fragment of α-actinin1 cDNA spotted on the microarrays (GenBank accession number BG077689) were used as probes for Northern blot analysis. Identity of α-actinin1 and calponin2 cDNAs were verified by DNA sequencing matched through BLAST analysis (www.ncbi.nlm.nih/BLAST/). Total RNA samples (15 μg/lane) obtained from NMuMG cells treated with 2 ng/ml TGF-β1 for 4, 8, and 24 h were subjected to Northern blot analysis as described previously (Bakin and Curran, 1999). Amplification of transcripts was performed using 50 ng/μL of total RNA and one-step reverse transcription (RT)-PCR system from Invitrogen (Carlsbad, CA) according to the manufacturing protocol. Primer sequences: β-actin, accession no. NM_007393, forward: GCTGGTCGTCGACAACGGCTC, reverse: CAAACATGATCTGGGTCATCTTTTC; α-tropomyosin, accession no. NM_024427.2, forward: GCTGGTGTCACTGCAAAAGA, reverse: CCTGAGCCTCCAGTGACTTC; β-tropomyosin, accession no. NM_009416.2, forward: AAGGATGCCCAGGAGAAACT, reverse: CTTCCTTCAGCTGCATCTCC; calponin2, accession no. NM_007725.1, forward: ACCCTGTGGACCTGTTTGAG, reverse: TGGAAGAGTTGTCGCACTTG; PAI-1, accession no. M33960.1, forward: CCACCGACTTCGGAGTAAAA, reverse: GCGTGTCAGCTCGTCTACAG.
Immunofluorescence Microscopy
Cells (105cells/well) were grown in DMEM containing 5% fetal bovine serum (FBS) on glass coverslips (22 × 22 mm) for 24 h before treatment with 2 ng/ml TGF-β1. Cells were fixed with 4% paraformaldehyde and stained as described (Bakin et al., 2002). Actin filaments (F-actin) were stained with phalloidin-Alexa Green or phalloidin-Texas Red, and tropomyosins were visualized using TM311 antibody. Fluorescent images were captured using Zeiss Axiophot upright microscope (Thornwood, NY) and Nikon TE2000-E inverted microscope (Garden City, NY). In some experiments cells were permeabilized with 0.05% Triton X-100 for 10 min followed by fixation and staining.
Wound Closure Assay
The assay was performed as described previously (Bakin et al., 2002). MDA-MB-231 cells (1–2 × 105/well) were seeded in 12-well plates and incubated in serum-free IMEM (Invitrogen) medium for 24 h before wounding with plastic tip across the cell monolayer. Kinase inhibitors were added 1 h before wounding. The cells were left untreated or treated with 2 ng/ml TGF-β1 for 16 h. The wound closure was estimated as the ratio of the remaining wounded area relative to the initial area. Experiments were repeated at least three times.
Transcriptional Assay
NMuMG cells (3 × 104) were seeded in 24-well plates and transfected with 0.16 μg/ml pSBE-Lux containing 12 repeats of Smad binding sequence (provided by J.-M. Gauthier, Laboratoire Glaxo Wellcome, Les Ulis Cedex, France) with 0.002 μg/ml pCMV-Rl (Promega, Madison, WI) using FuGENE6 reagent (Roche Molecular Biochemicals, Indianapolis, IN) according to the manufacturer's protocol. Cells were incubated for 8 h in 0.5% FBS-DMEM before treatment with 1 ng/ml TGF-β1 for 16 h. Firefly luciferase (Luc) and Renilla reniformis luciferase (Rl) activities in cell lysates were determined using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol in a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA). Luc activity was normalized to Rl activity and presented as Relative Luciferase Units. All assays were done in triplicate wells and each experiment was repeated at least twice.
Short Interference RNA Studies
RNA duplexes against human (cat. no. M-003902) and mouse (cat. no. M-004199) Smad4 were obtained from Dharmacon Research, Inc. (Lafayette, CO). RNA duplexes against tropomyosin (target sequence: AAGCAGCTGGAAGATGAGC) were designed using the siDESIGN program at the Dharmacon siDESIGN center. A scramble control RNA duplex labeled with rhodamine was obtained from Qiagen (Chatsworth, CA). Cells were transfected with RNA duplexes using Oligofectamine reagents (Invitrogen) following the manufacturers protocol. The cells were transferred onto glass coverslips or plastic dishes. Forty-eight hours posttransfection, the cells were treated with TGF-β1 for 24 h followed by immunoblot and immunofluorescence analysis.
RESULTS
TGF-β–induced Actin Stress Fiber Formation in Epithelial Cells Requires De Novo Protein Synthesis and p38Mapk
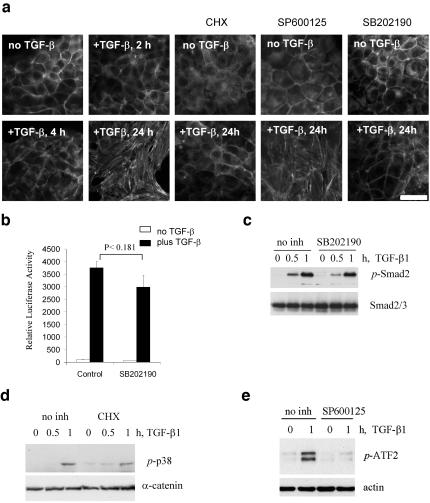
The mechanism of TGF-β–induced stress fiber (SF) formation was characterized in NMuMG mouse mammary epithelial cells. These cells exhibit a cuboidal cell morphology and a cortical organization of actin filaments in adhesion belts. Treatment of the cells with TGF-β1 for 24 h induced formation of actin microfilament bundles (Figure 1). Actin filaments in adhesion belts were not significantly affected in the first 4 h of TGF-β treatment compared with untreated cells (Figure 1a), and SFs were not observed until 8 h after TGF-β addition. SFs were well developed in cells incubated with TGF-β for 24 h. This TGF-β response was blocked by treatment of cells with the p38Mapk inhibitor, SB202190, suggesting involvement of p38Mapk in TGF-β–regulated SFs. This inhibitor did not significantly affect phosphorylation of Smad2 and TGF-β–mediated activation of the Smaddependent luciferase reporter activity (Figure 1, b and c). Similar results were also obtained with two other p38Mapk inhibitors (SB203580 and PD165319; our unpublished results). Treatment of cells with the JNK inhibitor, SP600125, did not block TGF-β–mediated SF formation (Figure 1a), although it effectively blocked phosphorylation of ATF2 (Figure 1e). Expression of kinase-inactive mutant of mitogen-activated protein kinase kinase 6 (MKK6) blocked phosphorylation of p38Mapk and SF formation (Bakin et al., 2000, 2002). In previous studies we have shown that TGF-β does not activate the ERK pathway in NMuMG cells and inhibition of MEK1/2 does not block stress fiber formation. Similar results were obtained for human cervical carcinoma SiHa cells, in which TGF-β induces p38Mapk signaling and SFs (Bakin et al., 2002). To investigate whether this process depends on de novo protein synthesis, cells were treated with cycloheximide, the protein synthesis inhibitor. Cycloheximide blocked SF formation when added as late as 6 h after initiation of TGF-β treatment without inhibition of p38Mapk activation (Figure 1, a and d), but it had no effect after 12 h (our unpublished results). Thus, de novo protein synthesis and p38Mapk activity are required for TGF-β–mediated actin SF formation in epithelial cells.
Figure 1.
TGF-β1–induced actin stress fibers require p38Mapk and a novel protein synthesis. (a) Actin filaments staining with phalloidin-Alexa Green in cells treated with 2 ng/ml TGF-β1 in the absence or presence of inhibitors. Where it is indicated, cells were treated with 10 μg/ml cycloheximide (CHX) starting at 6 h after addition of TGF-β1. Kinase inhibitors (15 μM SP600125, a JNK inhibitor, and 10 μM SB202190, a p38Mapk inhibitor) were added 1 h before addition of TGFβ1. Scale bar, 15 μm. (b) Luciferase activity in NMuMG transfected with Smad-dependent reporter pSBE-Lux and pCMV-Rl vectors and treated with 1 ng/ml TGF-β1 for 16 h in the absence or presence of 15 μM SB202190. Each bar represents the mean ± SD of three wells. P value was determined by t test. The difference in luciferase activity in control and SB202190-treated cells is not statistically significant. (c) Immunoblot analysis of Smad2 phosphorylation in protein extracts (35 μg/well) from NMuMG cells treated with 2 ng/ml TGF-β1 in the absence or presence of 10 μM SB202190. (d) p38Mapk phosphorylation in response to TGF-β1 in protein extracts (35 μg/well) from SiHa cells cotreated with 10 μg/ml cycloheximide (CHX). (e) Inhibition of ATF2 phosphorylation by JNK inhibitors in protein extracts (35 μg/well) from NMuMG cells treated with 2 ng/ml TGF-β1.
TGF-β Up-regulates Expression of Genes Encoding Actinbinding Proteins
To identify TGF-β target genes that mediate actin remodeling, we compared gene expression profiles in NMuMG cells before and after treatment with TGF-β1 for 24 h using mouse cDNA microarrays. The results indicate that expression of 62 genes changed more than twofold after treatment with TGF-β1. Among these genes TGF-β stimulated expression of several genes encoding actin-binding proteins including tropomyosins (TM), α-actinin1, and calponin2 (Table 1). These proteins are known to be involved in the assembly of stable actin microfilament bundles (Ayscough, 1998; Danninger and Gimona, 2000; Tseng et al., 2002; Hossain et al., 2003). The α-tropomyosin and β-tropomyosin genes encoding high-molecular-weight tropomyosins were up-regulated 2–2.6-fold and were represented by two and three cDNA clones, respectively. Interestingly, Tpm3 and Tpm4 genes encoding low-molecular-weight tropomyosins were not regulated by TGF-β (Table 1).
Table 1.
Regulation of genes encoding actin-binding proteins by TGF-β1 in NMuMG cells
| Fold Change ±SE
|
||||
|---|---|---|---|---|
| Gene | Accession no. of cDNA clones | TGF-β1 | TGF-β1 + SB202190 | |
| 1 | Actn1 | BG077689 | 3.03 ± 0.66 | 2.20 ± 0.18 |
| 2 | Actn1 | BG065930 | 2.94 ± 0.48 | 2.37 ± 0.48 |
| 3 | α-Tropomyosin | BG086016 | 1.81 ± 0.39 | 1.48 ± 0.09 |
| 4 | α-Ttropomyosin | BG079039 | 2.15 ± 0.57 | 1.30 ± 0.04 |
| 5 | β-Tropomyosin | BG076419 | 2.48 ± 0.09 | 1.52 ± 0.20 |
| 6 | β-Tropomyosin | BG073088 | 2.37 ± 0.36 | 1.37 ± 0.36 |
| 7 | β-Tropomyosin | BG087093 | 2.83 ± 0.43 | 1.83 ± 0.48 |
| 8 | Tpm3 | BG077822 | 1.28 ± 0.12 | 1.38 ± 0.19 |
| 9 | Tpm3 | BG078017 | 1.22 ± 0.08 | 1.40 ± 0.08 |
| 10 | Tpm3 | BG064681 | 1.28 ± 0.07 | 1.40 ± 0.08 |
| 11 | Tpm3 | BC029186 | 1.16 ± 0.15 | 0.91 ± 0.11 |
| 12 | Tpm4 | BG077934 | 0.96 ± 0.11 | 0.90 ± 0.18 |
| 13 | Tpm4 | AW537534 | 1.34 ± 0.12 | 1.46 ± 0.22 |
| 14 | Cnn2 (H2) | BG079442 | 3.00 ± 0.25 | 3.11 ± 0.29 |
cDNA microarray analysis was performed using total RNA from NMuMG cells, which were untreated or treated with 2 ng/ml TGF-β1 for 24 h in the absence or presence of 10 μM SB202190. The data represent an average of three independent experiments.
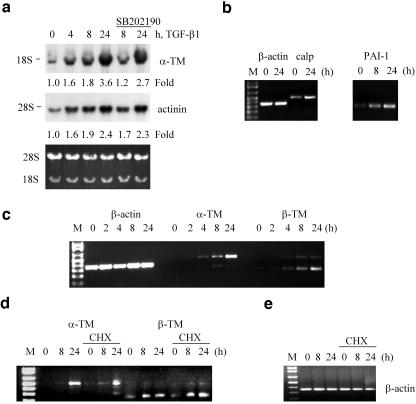
Treatment with a p38Mapk inhibitor suppressed induction of tropomyosins by 30–45% without a significant effect on calponin2 (Table 1). Northern blot analysis with rat TM3 cDNA, a product of the α-TM gene, revealed a 1.6-fold increase in the TM mRNA levels at 4 h reaching a 3.6-fold induction at 24 h of TGF-β1 treatment (Figure 2a). Similar regulation was observed for α-actinin (Figure 2a). Cotreatment with a p38Mapk inhibitor reduced by 35% the induction of α-TM mRNA, without a significant effect on α-actinin1 (Figure 2a), suggesting that p38Mapk is involved in tropomyosin gene expression. Using RT-PCR we confirmed TGF-β–mediated regulation of calponin2 and that PAI-1, a known TGF-β-target gene, is regulated with kinetics similar to the newly identified TGF-β target genes (Figure 2b). The regulation of highly conserved α-TM and β-TM genes was further confirmed using RT-PCR with isoform specific primers (Figure 2c), because tropomyosin sequences are conserved. The specificity was also confirmed using cDNA clones for TM1, TM2, and TM3 (our unpublished results). Examination of α- and β-TM mRNA levels in the cells cotreated with cycloheximide showed that TGF-β induction of these genes was not affected by cycloheximide, indicating that these genes are directly regulated by the TGF-β signaling pathway (Figure 2, d and e). Thus, we have identified tropomyosins α-actinin1 and calponin2 as novel TGF-β target genes that may account for TGF-β regulation of actin filament dynamics.
Figure 2.
TGF-β regulates expression of genes encoding actin-binding proteins in NMuMG cells. (a) Northern blot analysis of TM2/3 and α-actinin1 mRNA levels in total RNA samples (15 μg/lane) from NMuMG cells treated with 2 ng/ml TGF-β1 in the absence or presence of 10 μM SB202190. Blots were quantified using PhosphorImager. Bottom panel shows ethidium bromide staining of total RNA. (b–e) Analysis of β-actin, PAI-1, calponin2, and α- and β-tropomyosin transcripts by PCR with reverse transcription in total RNA samples from NMuMG cells treated with 2 ng/ml TGF-β1. Where it is indicated, cells were treated with 10 μg/ml cycloheximide (CHX) 1 h before addition of TGF-β1.
TGF-β Up-regulates Expression of Tropomyosins and Induces Phosphorylation of HSP27 in Epithelial Cells
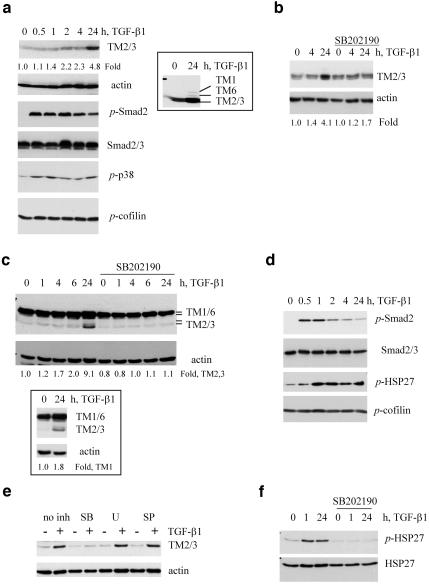
In vertebrates, more than 10 different isoforms of high-molecular-weight tropomyosins are expressed from α- and β-TM genes and by alternative RNA splicing (Pittenger et al., 1994). Tropomyosins form α-helical coil-coil dimers that bind along the length of the actin filaments interacting with 6–7 actin monomers are thought to be essential for the assembly and stabilization of actin filaments. (Ayscough, 1998). In this study we used the TM311 mAb recognizing tropomyosin 1 (TM1), a product of the β-TM gene, and tropomyosin isoforms 2, 3, and 6 (TM2,3,6), products of the α-TM gene (Temm-Grove et al., 1998). Immunoblot analysis with TM311 antibody followed by reblotting with anti-β-actin mAb showed that TGF-β induced a 4.8-fold increase in TM2 and TM3 in NMuMG cells (Figure 3a). Induction of TM1 and TM6 was also detected but required longer film exposures (Figure 3a, insert), suggesting that TM2 and TM3 are the main tropomyosin isoforms regulated by TGF-β at the protein level in NMuMG cells. The difference in the regulation of protein and mRNA levels of TM1 in response to TGF-β1 is not obvious and may be related to a tight regulation of α/β tropomyosin isoforms (Robbins, 1998). Analysis of Smad2 and p38Mapk phosphorylation in the same cells showed activation of Smad and p38 signaling at 30 min and a sustained level for at least 24 h (Figure 3a). Inhibition of p38Mapk significantly reduced the induction of tropomyosins without affecting basal level expression (Figure 3b), suggesting a more profound effect of p38Mapk on tropomyosin protein than on mRNA (see Figure 2a). Inhibition of p38Mapk did not block phosphorylation of Smad2 and Smad-dependent transcription (Figure 1, b and c) and did not affect expression of calponin2 (Table 1).
Figure 3.
TGF-β regulates tropomyosins and HSP27 phosphorylation in epithelial cells. (a and b) Immunoblot analysis of actin and tropomyosins as well as phosphorylation of Smad2, HSP27, and cofilin using phosphospecific antibodies in NMuMG cells treated with 2 ng/ml TGF-β1. Where it is indicated 10 μM SB202190 was added 1 h before TGF-β treatment. Inset shows induction of TM1 and TM6 on a longer exposed film of the same immunoblot. (c–f) Immunoblot analysis of tropomyosin expression and phosphorylation of Smad2, p38Mapk, and cofilin in SiHa cells treated with 2 ng/ml TGF-β1. Where it is indicated, the cells were cotreated 1 h before addition of TGF-β with 10 μM SB202190 (SB), 5 μM U0126 (U), or 15 μM SP600125 (SP). Insert shows immunoblot with TM311 antibody using the same protein extracts as in c. Fold difference of tropomyosin levels relative to actin was estimated using NIH ImageJ software (http://rsb.info.nih.gov/nih-image/).
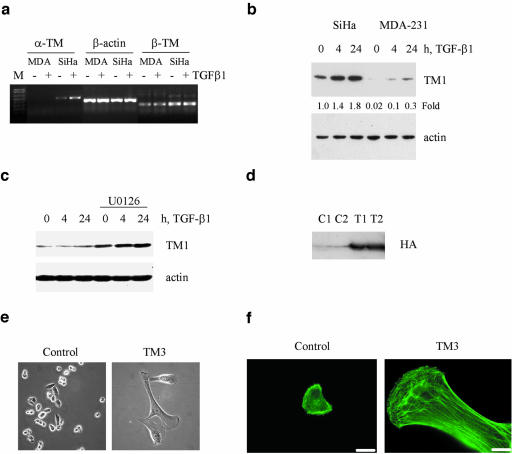
TGF-β also mediated up-regulation of TMs and p38Mapk signaling in human cervical carcinoma SiHa cells (Figure 3, c–f), which respond to TGF-β with SFs (Bakin et al., 2002). TGF-β stimulated a nearly twofold increase in TM1 and a ninefold increase in TM2/3 levels (Figure 3c). A comparable TGF-β–mediated induction of TMs and stress fiber formation were also observed in A549 lung epithelial cells (our unpublished results). SiHa cells express relatively low basal levels of TM2/3/6, but a high basal level of TM1 (Figure 3c). p38Mapk inhibitors blocked this up-regulation of TM2/3 (Figure 3, c and e), whereas inhibitors of MEK1/2 (U0126) and JNK, SP600125, did not (Figure 3e). Induction of the α-TM gene in SiHa cells at the mRNA level was confirmed by RT-PCR (see Figure 8a). We found that the activation of the Smad pathway and Smad3 levels were noticeably lower in SiHa cells (Figure 3d) than in NMuMG cells (Figure 3a). This may explain the moderate regulation of tropomyosins in SiHa cells compared with NMuMG cells and support the notion that Smads are involved in TGF-β–mediated regulation of tropomyosins.
Figure 8.
TGF-β and ERK signaling differentially regulates expression of tropomyosins. (a) Analysis of α-TM and β-TM transcripts by RT-PCR in total RNA samples from MDA-MB-231 (MDA) and SiHa cells treated with 2 ng/ml TGF-β1 for 24h. (b) Immunoblot analysis of tropomyosin expression in protein extracts (35 μg/lane) from SiHa and MDA-MB-231 cells treated with 2 ng/ml TGF-β1. (c) Tropomyosin protein expression in MDA-MB-231 cells cotreated with TGF-β1 and 5 μM U0126 for 24h. (d) Detection of HA-tagged rat TM3 with anti-HA antiserum in protein extracts from two independent transfections of MDA-MB-231 cells with expression vector encoding HA-tagged rat TM3 (T1 and T2) or a control empty vector (C1 and C2). (e) Phase-contrast images show flattening and size increase in TM3-transfected MDA-MB-231 cells compared with control cells. (f) Immunofluorescence images show a marked increase in actin stress fibers in TM3-transfected MDA-MB-231 cells. Scale bar, 20 μM. Fold differences in tropomyosin levels relative to actin were estimated using NIH ImageJ software.
The small heat shock protein HSP27 is a downstream target of p38Mapk signaling (Huot et al., 1998). The HSP27 phosphorylation by p38Mapk-MAPKAP2/3 signaling at three serine residues increases a pool of HSP27 tetramers that facilitate actin polymerization (Huot et al., 1998; Hedges et al., 1999; Rogalla et al., 1999). Using phospho-HSP27 antibodies we found that TGF-β stimulates a sustained phosphorylation of HSP27 in SiHa cells (Figure 3d). This is blocked by the p38Mapk inhibitor (Figure 3f). The actin filament dynamics is also controlled by the RhoA/ROCK/LIM-kinase pathway that regulates actin depolymerizing activity of ADF/cofilin by phosphorylation of a conserved serine3 in ADF/cofilin (Bamburg, 1999). Examination of cofilin phosphorylation in NMuMG (Figure 3a) and SiHa cells (Figure 3d) with phospho-Ser3–specific antibodies showed that levels of cofilin phosphorylation did not change in response to TGF-β during stress fiber formation. These results indicate that TGF-β induction of stress fiber formation in epithelial cells is accompanied with an increase in expression of actin-binding proteins and p38Mapk-HSP27 signaling without a significant regulation of the ROCK/LIM-kinase/cofilin pathway.
The Smad Signaling Pathway Mediates Regulation of Tropomyosin Expression by TGF-β
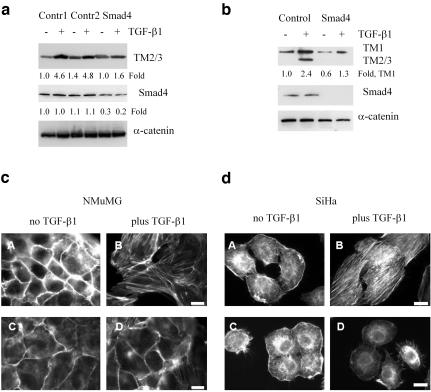
To examine the involvement of Smads in TGF-β–induced expression of tropomyosins and SFs, we transfected NMuMG and SiHa cells with short interfering RNA duplexes (siRNA) against Smad4. Transfection of siRNAs significantly reduced Smad4 protein levels and TGF-β–induced expression of TM2/3 in NMuMG (Figure 4a). A more effective action of siRNAs was observed in SiHa cells where TM2/3 expression was prevented and TM1 level was reduced by 40–55% (Figure 4b). Staining of actin filaments with phalloidin-Alexa Green demonstrated a significant reduction in TGF-β–induced SFs in both cells lines transfected with siRNAs to Smad4 (Figure 4, c and d, panels C and D), compared with control siRNA (Figure 4, c and d, panels A and B). These results support a model that Smad signaling mediate induction of tropomyosin expression in response to TGFβ leading to formation of SFs in epithelial cells.
Figure 4.
Smad signaling is required for TGF-β–induced expression of tropomyosins and stress fiber formation in epithelial cells. (a and b) Immunoblot analysis of tropomyosins and Smad4 in NMuMG cells (a) and SiHa cells (b) transfected with siRNAs against Smad4. (c and d) Actin filament staining with phalloidin-Alexa Green in NMuMG and SiHa cells transfected with control scramble siRNA (A and B) or siR-NAs to Smad4 (C and D). The cells were treated with 2 ng/ml TGF-β1 for 24 h. Scale bar, 10 μm. Fold differences in tropomyosin and Smad4 levels relative to α-catenin were estimated using NIH ImageJ software.
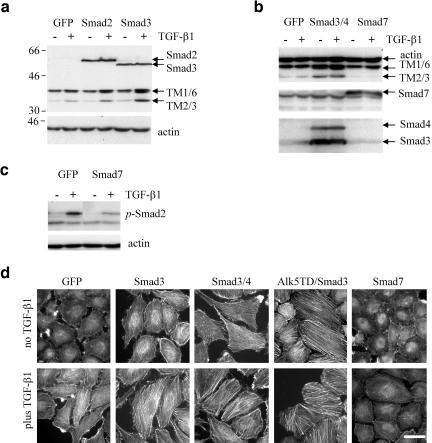
To test the contribution of specific Smads in the regulation of TMs and SFs, we used adenovirus-mediated expression of cDNAs encoding individual Smads in SiHa cells. These cells express low levels of Smad3 and Smad4 compared with NMuMG cells (Figure 3, a and d; Lee et al., 2001). Flagtagged Smad2 and Smad3 were expressed at comparable levels in SiHa cells infected with Smad-encoding adenoviruses (Figure 5, a and b). Smad3 significantly increased TGF-β–induced expression of tropomyosins compared with control EGFP-encoding adenovirus, whereas Smad2 exhibited only a moderate effect (Figure 5a). Coinfection with adenoviruses encoding Smad3 and Smad4 resulted in a marked increase of TMs even in the absence of added cytokine (Figure 5b), suggesting that Smad3 and Smad4 mediate TGF-β–regulated expression of tropomyosins. We next examined tropomyosin regulation by Smad7, an inhibitor of TGF-β signaling mediated by Smad2 and Smad3 (Massague, 1998). Expression of Smad7 inhibited TGF-β–induced expression of TM2/3 (Figure 5b) and phosphorylation of Smad2 (Figure 5c), whereas Smad6, an antagonist of bone morphogenic protein (BMP) signaling, had no effect (our unpublished results). In parallel, we examined actin filaments in SiHa cells infected with the adenoviral constructs (Figure 5d). As predicted, Smads that mediated enhancement of TM expression also increased SFs. Coexpression of Smad3 and Smad4 markedly increased SFs in the absence of exogenous TGF-β, which were further enhanced by the cytokine suggesting that other signaling events may also be involved in the assembly of stress fibers. Coexpression of constitutively active TGF-β type I receptor, Alk5T204D, and Smad3 resulted in SFs independent of exogenous TGF-β. Finally, expression of Smad7 significantly inhibited TGF-β–induced SF assembly (Figure 5e). These results demonstrate that Smad3 and Smad4 mediate TGF-β–induced expression of tropomyosins and SF formation.
Figure 5.
Smads mediate TGF-β–induced tropomyosin expression and stress fiber formation. (a and b) Tropomyosin expression in cells infected with adenoviruses encoding EGFP (GFP) and Flag-tagged Smad2, Smad3, Smad4, and Smad7. Cells were treated with 2 ng/ml TGF-β1 for 24 h. (c) Inhibition of TGF-β1–induced phosphorylation of Smad2 by adenoviral expression of Smad7 in SiHa cells. (d) Actin filaments staining in SiHa cells infected with adenoviruses: EGFP, Smad3, Smad4, Smad7, constitutively active Alk5T204D, or their combinations. The cells were treated with 2 ng/ml TGF-β1 for 24 h. Scale bars, 15 μm.
Tropomyosins Are Required for SF Formation in Response to TGF-β
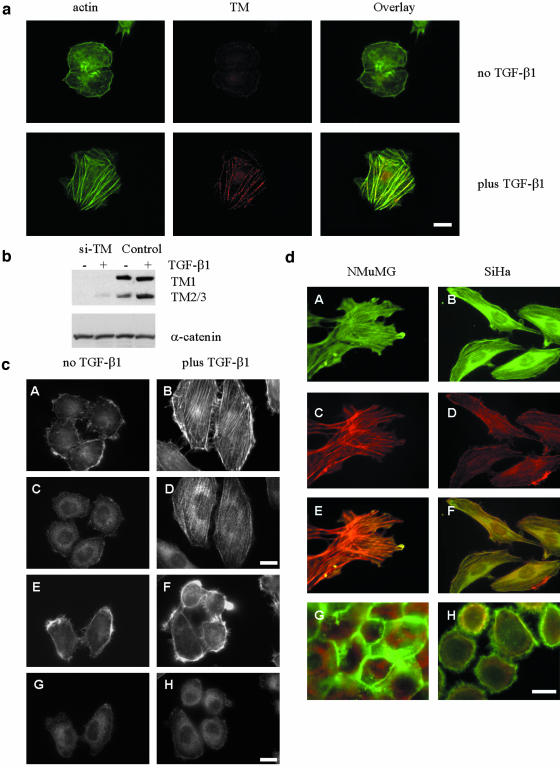
We confirmed that tropomyosins are localized with stable actin filaments resistant to Triton treatment. SiHa cells treated with TGF-β for 24 h were first incubated with Triton X-100 and then fixed with 4% paraformaldehyde. Actin filaments were detected with phalloidin and tropomyosins with the TM311 antibody. Immunofluorescence microscopy showed that tropomyosins were localized along the actin microfilaments in a periodical pattern (Figure 6a).
Figure 6.
Tropomyosins are required for TGF-β–induced stress fiber formation. (a) Localization of tropomyosins (TM) to stable actin filaments (actin) resistant to 0.05% Triton X-100 treatment in SiHa cells untreated or treated with TGF-β1 for 24 h. Scale bar, 10 μM. (b) Suppression of tropomyosin expression in SiHa cells transfected with siRNA against TMs (si-TM) compared with a scrambled control. (c) Actin filaments (A, B, E, and F) and tropomyosin (C, D, G, and H) in SiHa cells, transfected with siRNA against tropomyosins (E–H) and a scrambled control siRNA (A–D). The cells were treated with 2 ng/ml TGF-β1 for 24 h. Scale bar, 10 μM. (d) Actin filaments in NMuMG and SiHa cells expressing HA-tagged TM3. Cells were stained with phalloidin-Texas Red and fluorescein-labeled anti-HA antibody (A–F). Overlay images are shown in panels E and F. Panels G and H show actin filaments and tropomyosins (TM311 antibody) in cells transfected with empty vector control. Scale bar, 15 μm.
To examine whether tropomyosins are required for TGF-β–mediated SF formation, SiHa cells were transfected with siRNA duplexes against tropomyosins (si-TMs) or a scrambled siRNA control. si-TMs effectively suppressed basal and TGF-β–induced expression of tropomyosins (Figure 6b). TGF-β induced SFs in cells transfected with a scrambled siRNA control (Figure 6c, panels A and B), whereas SFs were significantly reduced by si-TMs (Figure 6c, panels E and F). In control cells, TGF-β induced elongation of cells and localization of tropomyosins to actin filaments, whereas in the si-TM cells this response was significantly reduced (Figure 6c, panels C and D and G and H). A complementary experiment tested the gain-of-function by transfection of NMuMG and SiHa cells with expression vector for rat HA-tagged TM3. Expression of TM3 alone, without TGF-β treatment, was sufficient to induce SFs in both cell lines similar to cells treated with TGF-β1 (Figure 6d, red, panels C and D). Staining with fluorescein-labeled anti-HA antibody showed colocalization of HA-tagged TM3 with actin filaments (Figure 6d, panels A and B, and overlay). These results indicate that tropomyosins are both necessary and sufficient for TGF-β–induced stress fiber formation.
TGF-β Does Not Induce Stress Fibers but Stimulates Cell Migration in Metastatic Cells
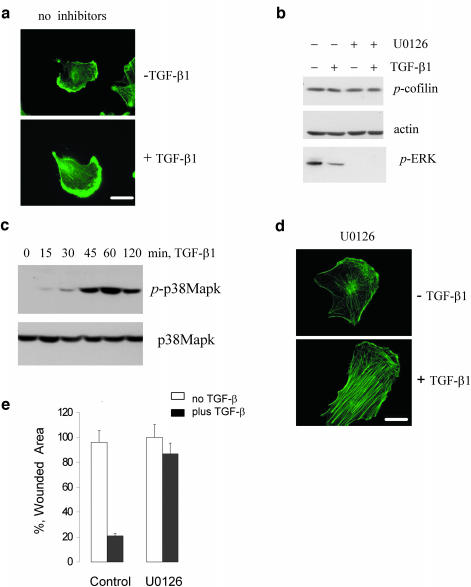
Actin filaments are dynamic structures and stabilization of actin filaments limits cell movement. TGF-β induces stress fibers in NMuMG and SiHa cells and cells from both lines fail to migrate in response to TGF-β in a wound closure assay (our unpublished results). TGF-β has been shown to stimulate migration of metastatic breast cancer MDA-MB-231 cells in wound closure assay (Bakin et al., 2002). We hypothesized that TGF-β–mediated stress fiber response is altered in MDA-MB-231 cells. Accordingly, treatment of MDA-MB-231 cells with TGF-β did not result in the formation of stress fibers (Figure 7a). MDA-MB-231 cells express TGF-β receptors, Smad factors, and respond to TGF-β1 with activation of Smad, p38Mapk signaling, and regulation of gene expression (Bakin et al., 2002; Dumont et al., 2003 and Figure 7c). It has been reported that MDA-MB-231 cells have constitutively active Ras-ERK signaling (Kozma et al., 1987; Ogata et al., 2001), which may through the repression of the ROCK/LIM-kinase/cofilin pathway affect SF formation (Sahai et al., 2001; Pawlak and Helfman, 2002b). Thus, we examined phosphorylation of cofilin, a target of LIM kinase, in MDA-MB-231 cells. The immunoblot showed a relatively high basal level of the cofilin phosphorylation that was not modulated by TGF-β. Treatment of these cells with the MEK inhibitor did not affect the basal cofilin phosphorylation but blocked phosphorylation of ERK1/2 (Figure 7b). However, MEK inhibition significantly enhanced TGF-β–induced stress fiber formation (Figure 7d) and blocked TGF-β–mediated cell migration (Figure 7e). The TGF-β regulation of stress fibers was also restored by MEK inhibitor PD098059 and by inhibition of Raf kinase (our unpublished results). These results suggest that the ERK pathway suppresses TGF-β–mediated stress fiber formation in epithelial cells through a mechanism mediated by a pathway other than the ROCK/LIM-kinase/cofilin pathway.
Figure 7.
TGF-β and ERK signaling differentially regulate stress fiber formation. (a) Actin filaments staining in MDA-MB-231 cells treated with TGF-β1 for 24 h. (B) Phosphorylation of cofilin and ERK1/2 in MDA-MB-231 cells cotreated with TGF-β1 for 24 h and 5 μM U0126. (c) Immunoblot analysis of p38Mapk phosphorylation in MDA-MB-231 cells treated with 2 ng/ml TGF-β1. (d) Actin filaments staining in MDA-MB-231 cells cotreated with TGF-β1 and 5 μM U0126 for 24 h. (e) Wound closure in MDA-MB-231 cells treated with TGF-β1 in the absence or presence of 5 μM U0126. The experiment was done in triplicates and repeated at least two times. Scale bar, 20 μM.
Suppression of TGF-β–regulated Tropomyosin Expression by Ras-ERK Signaling in Metastatic MDA-MB-231 Cells
The Ras-ERK pathway has been implicated in suppression of tropomyosins and disruption of the actin cytoskeleton (Ljungdahl et al., 1998; Shields et al., 2002). We next examined whether the inability of TGF-β to induce stress fibers in MDA-MB-231 cells is associated with alteration of tropomyosin expression or function by Ras-ERK signaling. We compared expression of tropomyosins in MDA-MB-231 cells and SiHa cells, which show a low basal level of ERK phosphorylation. RT-PCR and immunoblot analysis showed that MDA-MB-231 cells express significantly less of TM1 mRNA and protein and undetectable levels of TM2/3 in comparison to SiHa cells (Figure 8, a and b). Treatment of MDA-MB-231 cells with the MEK inhibitor U0126 reduced ERK phosphorylation (Figure 7b), increased TGF-β–induced expression of TM1 (Figure 8c), and restored stress fibers (Figure 7d). Similar results were obtained with a Raf kinase inhibitor (our unpublished results). These data suggest that Raf-ERK signaling down-regulates a basal and TGF-β–regulated expression of tropomyosin. Our findings also indicate that the α-tropomyosin gene is silenced in MDA-MB-231 cells.
We next examined whether ectopic expression of TM3, a product of the α-tropomyosin gene, will affect SFs and cell migration. MDA-MB-231 cells were transfected with expression vector encoding rat HA-tagged TM3 (Figure 8d) and analyzed for changes in cell morphology and actin filament assembly. Phase contrast images showed that TM3 expressing cells have a significant increase in cell size and a flatter more well-spread morphology compared with the refractile appearance of the parental cells or the control cells transfected with an empty vector (Figure 8e). Expression of TM3 markedly increased SFs in MDA-MB231 (Figure 8f) and inhibited cell motility assessed in the wound closure assay (our unpublished results). Interestingly, ectopic expression of either TM3 or TM2 inhibited proliferation of MDA-MB-231 cells increasing a number of multinucleated cells. We are currently developing inducible model to study effect of TMs on motility and growth of MDA-MB-231 cells. It has been also reported that overexpression of TM1 in MDA-MB-231 cells inhibits growth and motility of MDA-MB-231 cells (Raval et al., 2003). Thus, overexpression of tropomyosins in metastatic MDA-MB-231 cells results in stress fibers and reduces cell motility. Collectively, the data presented above demonstrate that the Ras-ERK pathway inhibits TGF-β induction of stress fibers by suppressing expression of tropomyosins.
DISCUSSION
The molecular mechanism(s) underlying the prometastatic conversion of the TGF-β function is a major focus of current investigation by many research groups (reviewed in Derynck and Zhang, 2003; Roberts and Wakefield, 2003). In this study we found that the ability of TGF-β to induce stress fibers and, therefore, to control cell migration is significantly compromised in metastatic breast carcinoma cells. We provide evidence that tropomyosins are critical cellular components of Smad/p38Mapk-dependent actin stress fiber assembly in response to TGF-β in epithelial cells. We further show that the Ras-ERK pathway antagonizes TGF-β induction of tropomyosins and stress fibers. The restoration of tropomyosin expression results in stress fibers and reduces cell motility. These studies provide a direct causal link between TGF-β regulation of stress fibers and control of cell motility. These results suggest that the loss of the TGF-β stress fiber response in tumor cells is a critical step in prometastatic conversion of the TGF-β function.
We have investigated the mechanism of TGF-β regulation of actin filament dynamics and cell motility in normal and tumor epithelial cells. In untransformed epithelial cells TGF-β can rapidly induce membrane ruffling and actin polymerization at the cell edges, whereas a prolong incubation with TGF-β results in the formation of stable actin filament bundles (stress fibers; Bakin et al., 2002; Edlund et al., 2002). We found that inhibition of either de novo protein synthesis or p38Mapk blocked TGF-β induction of stress fibers, suggesting that novel transcription/translation and p38Mapk signaling are required for TGF-β–mediated stress fiber formation. Consistent with these findings expression of kinase-inactive p38Mapk inhibited TGF-β induction of actin stress fibers (Bakin et al., 2002). Here we show that concomitantly with stress fibers TGF-β induced a sustained activation of p38Mapk signaling and phosphorylation of HSP27 (Figure 3), a downstream target of the p38Mapk-MAPKAP kinase 2 pathway (Stokoe et al., 1992). Phosphorylated HSP27 and the triple aspartate mutant mimicking HSP27 phosphorylation at Ser15, Ser78, and Ser82 form small oligomers and tetramers that facilitate de novo actin polymerization (Huot et al., 1998; Hedges et al., 1999; Rogalla et al., 1999). We found that although inhibition of de novo protein synthesis blocked stress fibers, it did not affect p38Mapk signaling (Figure 1d), suggesting that p38Mapk-HSP27 signaling is required but not sufficient for TGF-β induction of stress fibers. This notion is also supported by previous studies that have suggested involvement of Smad signaling in stress fiber formation (Piek et al., 1999a).
We identified several TGF-β target genes including α- and β-tropomyosins, α-actinin1, and calponin2 encoding actinbinding proteins implicated in the assembly of stress fibers. Among these genes, tropomyosins (TMs) have been shown to play a critical role in the stress fiber assembly by stabilizing actin filaments and preventing access of actin-severing factors gelsolin and ADF/cofilin to filamentous actin (Pawlak and Helfman, 2001). We found that TGF-β specifically up-regulates expression of α- and β-TM genes encoding high-molecular-weight TMs and does not regulate Tpm3 and Tpm4 genes encoding low-molecular-weight TMs (Table 1). The expression of TMs correlated with the ability of TGF-β to induce stress fibers in several epithelial cell lines including NMuMG, SiHa, and A549 cells. Moreover, suppression of TM expression with siRNAs completely blocked TGF-β induction of stress fibers in mouse and human cell lines, whereas ectopic expression of TM2 and TM3, products of the α-tropomyosin gene, was sufficient to induce stress fibers even in the absence of the cytokine (Figures 6 and 7). This is the first demonstration that TMs play an essential role in TGF-β–induced stress fiber formation in mouse and human epithelial cell lines.
The TGF-β induction of TM expression depends on p38Mapk and Smad signaling. Inhibition of p38Mapk blocked expression of TM proteins without a significant effect on TM mRNA levels. These results suggest that p38Mapk is involved in posttranscriptional control of TM expression, although a recent study has implicated p38Mapk in regulation of TM mRNA in intestinal epithelial cells (Shields et al., 2002). Silencing of Smad4 with siRNAs suppressed tropomyosin expression and blocked stress fiber formation (Figure 4), whereas adenoviral expression of Smad factors showed that Smad3 and Smad4 are required for the induction of tropomyosins and formation of stress fibers in epithelial cells (Figure 5). Importantly, inhibitory Smad7, but not Smad6, blocks TGF-β induction of TM expression and stress fiber formation. These results demonstrate that Smad3/Smad4 and p38Mapk are required for TGF-β–induced TM expression and stress fiber formation in epithelial cells.
Tropomyosins have been implicated in regulation of actin filament dynamics and control of cell motility (Pawlak and Helfman, 2001). Early studies have found that cell transformation by oncogenic Ras and Src leads to down-regulation of tropomyosins and disruption of actin stress fiber filaments (Leonardi et al., 1982; Hendricks and Weintraub, 1984). Subsequently, it has been shown that ectopic expression of tropomyosins in Ras-transformed fibroblasts restores stress fibers and significantly reduces cell motility and cell growth (Takenaga and Masuda, 1994; Braverman et al., 1996; Gimona et al., 1996; Janssen and Mier, 1997). The importance of tropomyosins in the control of tumor invasion and metastasis is highlighted by several studies indicating that high-grade tumors of breast, prostate, bladder, and brain express significantly lower levels of tropomyosins that that of normal tissues (Franzen et al., 1996; Wang et al., 1996; Hughes et al., 2003; Raval et al., 2003). Thus, tropomyosins and thereby stress fibers may play a critical role in the TGF-β control of tumor invasion and metastasis. In support of this idea, we found that TGF-β induction of tropomyosins and stress fibers is markedly reduced in metastatic breast cancer MDA-MB-231 cell line (Figure 7). MDA-MB-231 cells express constitutively active Ras-ERK signaling (Kozma et al., 1987; Ogata et al., 2001) that has been implicated in down-regulation of tropomyosins and disruption of actin stress fibers (Ljungdahl et al., 1998; Shields et al., 2002). We found that pharmacological inhibition of the Raf-ERK pathway significantly increased basal and TGF-β–induced levels of TM1, restored TGF-β induction of stress fibers, and inhibited cell motility without any effect on phosphorylation of cofilin (Figures 7 and 8). These results can be attributed at least in part to changes in Smad signaling. In fact, recent studies have shown that the Ras-ERK pathway attenuates Smad signaling by affecting the subcellular localization of Smad2 and Smad3 (Kretzschmar et al., 1999) and by inducing a proteasome-mediated degradation of Smad4 (Saha et al., 2001). We also found that the α-TM gene is not expressed in MDA-MB-231 cells. Our unpublished data indicate that the CpG island in the proximal promoter of the human α-TM gene is methylated. The significance of this finding is currently under investigation. Importantly, ectopic expression of TM3 in MDA-MB-231 cells resulted in stress fibers (Figure 8) and severely affected cell motility. This finding is consistent with studies in rat NRK 1569 cells and mouse NIH-3T3 cells (Gimona et al., 1996; Janssen and Mier, 1997). Our previous results suggest that Ras-ERK signaling does not affect TGF-β induction of membrane ruffling at the leading edge (Bakin et al., 2002). Thus, Ras-ERK signaling suppresses TGF-β induction of tropomyosin expression and stress fibers leading to more motile and invasive phenotype.
The Rho-like GTPases, RhoA, Rac1, and CDC42, have been implicated in TGF-β mediated stress fiber formation (Moustakas and Stournaras, 1999; Bhowmick et al., 2001; Bakin et al., 2002; Edlund et al., 2002). These GTPases through RhoA-ROCK/Rho-kinase and Rac/CDC42-Pak signaling can activate LIM kinases that negatively regulate ADF/cofilins by phosphorylating a conserved Serine3 in ADF/cofilins (Gungabissoon and Bamburg, 2003). ADF/cofilins regulate the turnover rates of actin filaments by promoting the dissociation of actin filaments into monomers (Bamburg, 1999). Thus, Rho-like GTPases through LIM kinases may contribute to stress fiber formation by inhibiting actin depolymerization. In this study we found that phosphorylation of cofilin was not modulated by TGF-β in three different cell lines, suggesting that the ROCK/LIM-kinase/cofilin pathway is not a target in TGF-β induction of stress fibers. Tropomyosins bound to filamentous actin prevent access of ADF/cofilins to actin filaments, thereby stabilizing actin filaments and reducing actin dynamics (Ono and Ono, 2002). In addition to tropomyosins, calponin2 and α-actinin1, two other TGF-β targets identified in this study, have been also implicated in stabilization of actin filaments (Panasenko and Gusev, 2001; Gimona et al., 2003). It remains to be determined whether, in addition to tropomyosins, calponins and α-actinin play a role in formation of stress fibers in response to TGF-β by blocking ADF/cofilins and gelsolin from binding to actin filaments.
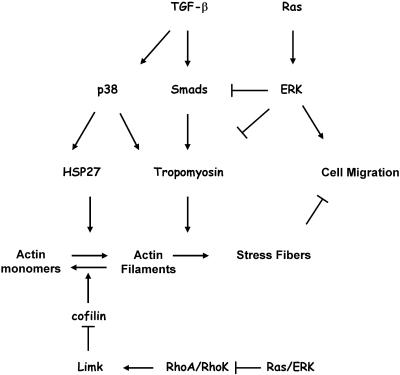
Our studies demonstrate an important role of tropomyosins in TGF-β regulation of stress fibers and cell migration (Figure 9). ERK signaling may inhibit TGF-β induction of stress fibers by suppressing Smad-dependent expression of tropomyosins. In addition, ERK signaling may affect stress fibers by disabling the RhoA/ROCK pathway (Pawlak and Helfman, 2002a, 2002b; Sahai et al., 2001; Vial et al., 2003). The suppression of tropomyosin expression by oncogenic Src (Hendricks and Weintraub, 1984) may also contribute to the cooperation of TGF-β and Src in tumorigenesis (Sieweke et al., 1990). Thus, our study support an idea that the acquisition of metastatic phenotype by tumor cells results from the action of oncogenes and tumor suppressor genes regulating cell proliferation and survival (Bernards and Weinberg, 2002). Our results suggest that loss of TGF-β induction of stress fibers is an essential characteristic of a prometastatic conversion of TGF-β function and restoration of this response represents a potential target for the development of effective antimetastatic therapies.
Figure 9.
Opposing roles of the TGF-β and Ras-ERK signaling pathways in the regulation of actin filament dynamics and cell motility.
Acknowledgments
We thank Kohei Miyazono for adenoviral constructs, James Bamburg for phospho-cofilin antibody, Shawn Levi and Braden Boone for help with microarray studies, Lei Quan for RT-PCR analysis of tropomyosin isoforms in human cell lines, and Irwin Gelman, Mark DeCaesteker, and Heinz Baumann for critical reading the manuscript. This work was supported by Public Health Service (PHS) grant R01 CA-95263 and U.S. Army Medical Research and Materiel Command grant DAMD17–02-01–0602 (to A.V.B.), and PHS R01 grant CA-83182 (to D.M.H.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–04–0353. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–04–0353.
Abbreviations used: TGF-β, transforming growth factor beta; Mapk, mitogene-activated protein kinase; TM, tropomyosin; siRNA, short-interfering RNA. NMuMG cells that were untreated or treated with 2 ng/ml TGF-β1 for 24 h in the absence or presence of 10 μM SB202190. The data represent an average of three independent experiments. Actn1, α-actinin1, Tpm3, tropomyosin3; Tpm4, tropomysin4; Cnn2 (H2), calponin2 (H2).
References
- Ayscough, K.R. (1998). In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol. 10, 102-111. [DOI] [PubMed] [Google Scholar]
- Bakin, A.V., and Curran, T. (1999). Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 283, 387-390. [DOI] [PubMed] [Google Scholar]
- Bakin, A.V., Rinehart, C., Tomlinson, A.K., and Arteaga, C.L. (2002). p38 mitogen-activated protein kinase is required for TGF{beta}-mediated fibroblastic transdifferentiation and cell migration. J. Cell Sci. 115, 3193-3206. [DOI] [PubMed] [Google Scholar]
- Bakin, A.V., Tomlinson, A.K., Bhowmick, N.A., Moses, H.L., and Arteaga, C.L. (2000). Phosphatidylinositol 3-kinase function is required for TGFbeta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275, 36803-36810. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R. (1999). Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185-230. [DOI] [PubMed] [Google Scholar]
- Bernards, R., and Weinberg, R.A. (2002). A progression puzzle. Nature 418, 823. [DOI] [PubMed] [Google Scholar]
- Bhowmick, N.A., Ghiassi, M., Bakin, A.V., Aakre, M., Lundquist, C.A., Engel, M., Arteaga, C.L., and Moses, H.L. (2001). TGFb mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 12, 27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman, R.H., Cooper, H.L., Lee, H.S., and Prasad, G.L. (1996). Antioncogenic effects of tropomyosin: isoform specificity and importance of protein coding sequences. Oncogene 13, 537-545. [PubMed] [Google Scholar]
- Cox, E.A., Sastry, S.K., and Huttenlocher, A. (2001). Integrin-mediated adhesion regulates cell polarity and membrane protrusion through the Rho family of GTPases. Mol. Biol. Cell 12, 265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danninger, C., and Gimona, M. (2000). Live dynamics of GFP-calponin: isoform-specific modulation of the actin cytoskeleton and autoregulation by C-terminal sequences. J. Cell Sci. 113, 3725-3736. [DOI] [PubMed] [Google Scholar]
- Derynck, R., Akhurst, R.J., and Balmain, A. (2001). TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 29, 117-129. [DOI] [PubMed] [Google Scholar]
- Derynck, R., and Zhang, Y.E. (2003). Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577-584. [DOI] [PubMed] [Google Scholar]
- Dumont, N., Bakin, A.V., and Arteaga, C.L. (2003). Autocrine Transforming Growth Factor-beta Signaling Mediates Smad-independent Motility in Human Cancer Cells. J. Biol. Chem. 278, 3275-3285. [DOI] [PubMed] [Google Scholar]
- Edlund, S., Landstrom, M., Heldin, C.H., and Aspenstrom, P. (2002). Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell 13, 902-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- Franzen, B., Linder, S., Uryu, K., Alaiya, A.A., Hirano, T., Kato, H., and Auer, G. (1996). Expression of tropomyosin isoforms in benign and malignant human breast lesions. Br. J. Cancer 73, 909-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona, M., Kaverina, I., Resch, G.P., Vignal, E., and Burgstaller, G. (2003). Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell 14, 2482-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona, M., Kazzaz, J.A., and Helfman, D.M. (1996). Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc. Natl. Acad. Sci. USA 93, 9618-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona, M., Watakabe, A., and Helfman, D. (1995). Specificity of dimer in tropomyosins: influence of alternatively spliced exons on homodimer and heterodimer assembly. Proc. Natl. Acad. Sci. USA 92, 9776-9780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungabissoon, R.A., and Bamburg, J.R. (2003). Regulation of growth cone actin dynamics by ADF/cofilin. J. Histochem. Cytochem. 51, 411-420. [DOI] [PubMed] [Google Scholar]
- Hannigan, M., Zhan, L., Ai, Y., and Huang, C.K. (1998). The role of p38 MAP kinase in TGF-beta1-induced signal transduction in human neutrophils. Biochem. Biophys. Res. Commun. 246, 55-58. [DOI] [PubMed] [Google Scholar]
- Hedges, J.C., Dechert, M.A., Yamboliev, I.A., Martin, J.L., Hickey, E., Weber, L.A., and Gerthoffer, W.T. (1999). A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration. J. Biol. Chem. 274, 24211-24219. [DOI] [PubMed] [Google Scholar]
- Hendricks, M., and Weintraub, H. (1984). Multiple tropomyosin polypeptides in chicken embryo fibroblasts: differential repression of transcription by Rous sarcoma virus transformation. Mol. Cell. Biol. 4, 1823-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M.M., Hwang, D.Y., Huang, Q.Q., Sasaki, Y., and Jin, J.P. (2003). Developmentally regulated expression of calponin isoforms and the effect of h2-calponin on cell proliferation. Am. J. Physiol. Cell Physiol. 284, C156-C167. [DOI] [PubMed] [Google Scholar]
- Hughes, J.A., Cooke-Yarborough, C.M., Chadwick, N.C., Schevzov, G., Arbuckle, S.M., Gunning, P., and Weinberger, R.P. (2003). High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pediatric and adult CNS tumors. Glia 42, 25-35. [DOI] [PubMed] [Google Scholar]
- Huot, J., Houle, F., Rousseau, S., Deschesnes, R.G., Shah, G.M., and Landry, J. (1998). SAPK2/p38-dependent F-actin reorganization regulates early membrane blebbing during stress-induced apoptosis. J. Cell Biol. 143, 1361-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A.B., and Hall, A. (2002). Rho GTPases in transformation and metastasis. Adv. Cancer Res. 84, 57-80. [DOI] [PubMed] [Google Scholar]
- Janssen, R.A., and Mier, J.W. (1997). Tropomyosin-2 cDNA lacking the 3′ untranslated region riboregulator induces growth inhibition of v-Ki-ras-transformed fibroblasts. Mol. Biol. Cell 8, 897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma, S.C., Bogaard, M.E., Buser, K., Saurer, S.M., Bos, J.L., Groner, B., and Hynes, N.E. (1987). The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res. 15, 5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar, M., Doody, J., Timokhina, I., and Massague, J. (1999). A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 13, 804-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Cho, Y.S., Shim, C., Kim, J., Choi, J., Oh, S., Zhang, W., and Lee, J. (2001). Aberrant expression of Smad4 results in resistance against the growth-inhibitory effect of transforming growth factor-beta in the SiHa human cervical carcinoma cell line. Int. J. Cancer 94, 500-507. [DOI] [PubMed] [Google Scholar]
- Leonardi, C.L., Warren, R.H., and Rubin, R.W. (1982). Lack of tropomyosin correlates with the absence of stress fibers in transformed rat kidney cells. Biochim. Biophys. Acta 720, 154-162. [DOI] [PubMed] [Google Scholar]
- Ljungdahl, S., Linder, S., Franzen, B., Binetruy, B., Auer, G., and Shoshan, M.C. (1998). Down-regulation of tropomyosin-2 expression in c-Jun-transformed rat fibroblasts involves induction of a MEK1-dependent autocrine loop. Cell Growth Differ. 9, 565-573. [PubMed] [Google Scholar]
- Massague, J. (1998). TGF-beta signal transduction. Annu. Rev. Biochem. 67, 753-791. [DOI] [PubMed] [Google Scholar]
- Moustakas, A., and Stournaras, C. (1999). Regulation of actin organisation by TGF-beta in H-ras-transformed fibroblasts. J. Cell Sci. 112, 1169-1179. [DOI] [PubMed] [Google Scholar]
- Ogata, H., Sato, H., Takatsuka, J., and De Luca, L.M. (2001). Human breast cancer MDA-MB-231 cells fail to express the neurofibromin protein, lack its type I mRNA isoform and show accumulation of P-MAPK and activated Ras. Cancer Lett. 172, 159-164. [DOI] [PubMed] [Google Scholar]
- Ono, S., and Ono, K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko, O.O., and Gusev, N.B. (2001). Mutual effects of alpha-actinin, calponin and filamin on actin binding. Biochim. Biophys. Acta 1544, 393-405. [DOI] [PubMed] [Google Scholar]
- Pawlak, G., and Helfman, D.M. (2001). Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 11, 41-47. [DOI] [PubMed] [Google Scholar]
- Pawlak, G., and Helfman, D.M. (2002a). MEK mediates v-Src-induced disruption of the actin cytoskeleton via inactivation of the Rho-ROCK-LIM kinase pathway. J. Biol. Chem. 277, 26927-26933. Epub 2002 May 14. [DOI] [PubMed] [Google Scholar]
- Pawlak, G., and Helfman, D.M. (2002b). Post-transcriptional down-regulation of ROCKI/Rho-kinase through an MEK-dependent pathway leads to cytoskeleton disruption in Ras-transformed fibroblasts. Mol. Biol. Cell 13, 336-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek, E., Heldin, C.H., and Ten Dijke, P. (1999a). Specificity, diversity, and regulation in TGF-beta superfamily signaling. FASEB J. 13, 2105-2124. [PubMed] [Google Scholar]
- Piek, E., Moustakas, A., Kurisaki, A., Heldin, C.H., and ten Dijke, P. (1999b). TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 112, 4557-4568. [DOI] [PubMed] [Google Scholar]
- Pittenger, M.F., Kazzaz, J.A., and Helfman, D.M. (1994). Functional properties of non-muscle tropomyosin isoforms. Curr. Opin. Cell Biol. 6, 96-104. [DOI] [PubMed] [Google Scholar]
- Raval, G.N., Bharadwaj, S., Levine, E.A., Willingham, M.C., Geary, R.L., Kute, T., and Prasad, G.L. (2003). Loss of expression of tropomyosin-1, a novel class II tumor suppressor that induces anoikis, in primary breast tumors. Oncogene 22, 6194-6203. [DOI] [PubMed] [Google Scholar]
- Robbins, J. (1998). Alpha-tropomyosin knockouts: a blow against transcriptional chauvinism. Circ. Res. 82, 134-136. [DOI] [PubMed] [Google Scholar]
- Roberts, A.B., and Wakefield, L.M. (2003). The two faces of transforming growth factor {beta} in carcinogenesis. Proc. Natl. Acad. Sci. USA 100, 8621-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalla, T. et al. (1999). Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J. Biol. Chem. 274, 18947-18956. [DOI] [PubMed] [Google Scholar]
- Saha, D., Datta, P.K., and Beauchamp, R.D. (2001). Oncogenic Ras represses transforming growth factor-beta /Smad signaling by degrading tumor suppressor Smad4. J. Biol. Chem. 276, 29531-29537. [DOI] [PubMed] [Google Scholar]
- Sahai, E., Olson, M.F., and Marshall, C.J. (2001). Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20, 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, H., Tsujitani, S., Oka, S., Kondo, A., Ikeguchi, M., Maeta, M., and Kaibara, N. (2000). An elevated serum level of transforming growth factorbeta 1 (TGF-beta 1) significantly correlated with lymph node metastasis and poor prognosis in patients with gastric carcinoma. Anticancer Res. 20, 4489-4493. [PubMed] [Google Scholar]
- Shields, J.M., Mehta, H., Pruitt, K., and Der, C.J. (2002). Opposing roles of the extracellular signal-regulated kinase and p38 mitogen-activated protein kinase cascades in Ras-mediated downregulation of tropomyosin. Mol. Cell. Biol. 22, 2304-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieweke, M.H., Thompson, N.L., Sporn, M.B., and Bissell, M.J. (1990). Mediation of wound-related Rous sarcoma virus tumorigenesis by TGF-beta. Science 248, 1656-1660. [DOI] [PubMed] [Google Scholar]
- Stokoe, D., Engel, K., Campbell, D.G., Cohen, P., and Gaestel, M. (1992). Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 313, 307-313. [DOI] [PubMed] [Google Scholar]
- Takenaga, K., and Masuda, A. (1994). Restoration of microfilament bundle organization in v-raf-transformed NRK cells after transduction with tropomyosin 2 cDNA. Cancer Lett. 87, 47-53. [DOI] [PubMed] [Google Scholar]
- Temm-Grove, C.J., Jockusch, B.M., Weinberger, R.P., Schevzov, G., and Helfman, D.M. (1998). Distinct localizations of tropomyosin isoforms in LLC-PK1 epithelial cells suggests specialized function at cell-cell adhesions. Cell Motil. Cytoskeleton 40, 393-407. [DOI] [PubMed] [Google Scholar]
- Tseng, Y., Schafer, B.W., Almo, S.C., and Wirtz, D. (2002). Functional synergy of actin filament cross-linking proteins. J. Biol. Chem. 277, 25609-25616. [DOI] [PubMed] [Google Scholar]
- Vial, E., Sahai, E., and Marshall, C.J. (2003). ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell 4, 67-79. [DOI] [PubMed] [Google Scholar]
- Wakefield, L.M., and Roberts, A.B. (2002). TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 12, 22-29. [DOI] [PubMed] [Google Scholar]
- Wang, F.L., Wang, Y., Wong, W.K., Liu, Y., Addivinola, F.J., Liang, P., Chen, L.B., Kantoff, P.W., and Pardee, A.B. (1996). Two differentially expressed genes in normal human prostate tissue and in carcinoma. Cancer Res. 56, 3634-3637. [PubMed] [Google Scholar]