Abstract
Generating specific actin structures via controlled actin polymerization is a prerequisite for eukaryote development and reproduction. We here report on an essential Caenorhabditis elegans protein tetraThymosinβ expressed in developing neurons and crucial during oocyte maturation in adults. TetraThymosinβ has four repeats, each related to the actin monomer-sequestering protein thymosinβ 4 and assists in actin filament elongation. For homologues with similar multirepeat structures, a profilin-like mechanism of ushering actin onto filament barbed ends, based on the formation of a 1:1 complex, is proposed to underlie this activity. We, however, demonstrate that tetraThymosinβ binds multiple actin monomers via different repeats and in addition also interacts with filamentous actin. All repeats need to be functional for attaining full activity in various in vitro assays. The activities on actin are thus a direct consequence of the repeated structure. In containing both G- and F-actin interaction sites, tetraThymosinβ may be reminiscent of nonhomologous multimodular actin regulatory proteins implicated in actin filament dynamics. A mutation that suppresses expression of tetraThymosinβ is homozygous lethal. Mutant organisms develop into adults but display a dumpy phenotype and fail to reproduce as their oocytes lack essential actin structures. This strongly suggests that the activity of tetraThymosinβ is of crucial importance at specific developmental stages requiring actin polymerization.
INTRODUCTION
Actin filament nucleation, elongation, and turnover form the basis of the dynamic remodelling of the actin cytoskeleton that takes place during cellular migration and cytokinesis. Multicellular organisms critically depend on these actin-based motile processes for development, reproduction, host defense, and survival. Many actin-binding proteins affect, in a regulated manner, either actin filament organization or the dynamics of the actin polymerization cycle (Ono et al., 1999; Ono, 2001; Condeelis, 2001; Small et al., 2002; Pollard and Borisy, 2003). The high dynamics of the actin system ultimately results from the interplay between differently functioning actin-binding proteins. Many of these exert their function via a single actin interaction site. However, the activity of a number of actin modulators is generated through combined activity of multiple actin-binding modules within the same molecule. This has evolved via different ways: either by multiplication of a specific module leading to a repeated structure or by domain swapping, i.e., by bringing together different actin-binding modules (Van Troys et al., 1999). Gelsolin is a well-known example where homologous segments have different actin-binding activities and their combined activity is responsible for severing and subsequent capping (Pope et al., 1991; Van Troys et al., 1996a). Multiple differential actin-binding units have also been reported in Ena/VASP proteins (Bachmann et al., 1999; Walders-Harbeck et al., 2002), Calponin Homology-domain containing proteins (Gimona et al., 2002), twinfilin (Ojala et al., 2002), MARCKS (Yarmola et al., 2001), and MIM (Mattila et al., 2003; Yamagishi et al., 2004).
Also the β-thymosin or WH2 module (Van Troys et al., 1996b; Paunola et al., 2002) is found as repeats. In Van Troys et al. (1999), we first reported the sequence of a hypothetical Caenorhabditis elegans protein tetraThymosinβ with the striking feature that it consists of a four times–repeated thymosinβ-module. The actin-binding determinants of this module are clearly delineated: a hexapeptide motif (17LKKTET22 in thymosinβ4) and a hydrophobic cluster in a preceding α-helix (Vancompernolle et al., 1992; Van Troys et al., 1996b; Rossenu et al., 1997; Simenel et al., 2000; Rossenu et al., 2003; Domanski et al., 2004). Given their repeated structures, tetraThymosinβ, Drosophila ciboulot, and the amoebal protein actobindin (harboring four, three, and two repeats, respectively; Lambooy and Korn, 1986; Van Troys et al., 1999; Boquet et al., 2000) form a novel subset of β-thymosins. Both ciboulot and actobindin have been demonstrated to display a similar promotive effect on filament growth as is observed for profilin (Boquet et al., 2000; Hertzog et al., 2002). Indeed, profilin serves as a polymerization catalyst by capturing an actin monomer and ushering the actin onto the growing filament barbed end as a 1:1 profilin:actin complex, whereupon profilin itself is released (Pantaloni and Carlier, 1993; Kang et al., 1999). Ciboulot is suggested to act on elongation via the same mechanism as profilin by forming a 1:1 complex with an actin monomer (Boquet et al., 2000). The complex of the first domain of ciboulot and monomeric actin have very recently been crystallized (Hertzog et al., 2004).
The activity of the larger β-thymosins contrasts that of single repeat β-thymosins that, by forming a 1:1 complex, only function in monomer sequestering. We postulated that the newly acquired functions (e.g., promoting polymerization) are a consequence of the repeated structure. Indeed, in contrast to the reported 1:1 stoichiometry for its homolog ciboulot, we observe formation of tetraThymosinβ complexes with multiple actin monomers. All four tetraThymosinβ repeats are functional actin-binding units and, moreover, have different preferences for monomeric and filamentous actin. The combined action of the repeats is absolutely required for the observed effects on actin polymerization. We provide evidence that tetraThymosinβ is, among other, enriched in developing neuronal structures and maturing oocytes. The observed defects present in tetraThymosinβ-/- organisms demonstrate its essential role in the generation of specific actin structures in vivo.
MATERIALS AND METHODS
TetraThymosinβ Wild-Type and Mutant Proteins—tetraThymosinβ Peptides
The tetraThymosinβ cDNA was amplified via PCR from a tetraThymosinβ EST containing phagemid y326d7, obtained from Y. Kohara (Genome Biology Lab, National Institute of Genetics, Mishima, Japan) and NcoI and BamHI cloned into the bacterial expression vector pET11d (Promega, Madison, WI). From this wild-type (WT) construct, we generated tetraThymosinβ variants carrying one or multiple mutated repeats via one or several consecutive rounds of mutagenesis using the PCR-based QuikChange method (Stratagene, La Jolla, CA). The generated mutations result in substitution of the first four amino acids of the conserved motif of the targeted repeat into four alanines (Table 1). For the N-Cys tetraThymosinβ-construct, needed for surface plasmon resonance (SPR) measurements (see below), a variant 5′-PCR-primer with an additional Cys-codon was used, resulting at the amino acid level in MACAV instead of MAAV as amino terminal sequence. All constructs were checked by sequencing.
Table 1.
Mutations introduced into tetraThymosinβ
| tTβR1: LHST62-65AAAA (in R2),a LKKT100-103AAAA (in R3), LHHV134-137AAAA (in R4) |
| tTβR2: LKKV23-26AAAA (in R1), LKKT100-103AAAA (in R3), LHHV134-137AAAA (in R4) |
| tTβR3: LKKV23-26AAAA (in R1), LHST 62-65 AAAA (in R2), LHHV134-137AAAA (in R4) |
| tTβR4: LKKV23-26AAAA (in R1), LHST 62-65 AAAA (in R2), LKKT100-103AAAA (in R3) |
| tTβR2,3,4: LKKV 23-26 AAAA (in R1) |
| tTβR1,3,4: LHST 62-65 AAAA (in R2) |
| tTβR1,2,4: LKKT100-103AAAA (in R3) |
| tTβR1,2,3: LHHV134-137AAAA (in R4) |
R1, R2, R3, and R4 refer to the repeats 1 to 4; in the mutant nomenclature the repeat(s) indicated in the name are unchanged
Recombinant tetraThymosinβ WT and mutant proteins were obtained after isopropyl-β-thiogalactoside induction of Escherichia coli MC1061 cells harboring the pET11d constructs and plasmid pT7POL26 (Mertens et al., 1995). We purified tetraThymosinβ WT and mutants from bacterial lysates by DEAE anion exchange chromatography in 25 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, and 1 mM dithiothreitol (DTT), followed by hydrophobic interaction chromatography (phenyl sepharose) using 25% saturated (NH4)2SO4, 25 mM Tris-HCl, pH 8.0, as starting conditions. The purified proteins were concentrated via ultrafiltration (Amicon, Beverly, MA), dialyzed against 25 mM Tris-HCl, pH 8.0, 1 mM DTT, 0.02 mM EDTA, and stored on ice.
TetraThymosinβ peptides were chemically synthesized on a model 431A peptide synthesizer (Applied Biosystems, Foster City, CA), purified, and checked for correct mass. Their concentration was determined as in Van Troys et al. (1996b).
Immunofluorescence Microscopy
Wild-type strain N2 was obtained from Caenorhabditis Genetics Center (Minneapolis, MN). Adult worms were stained as described (Finney and Ruvkun, 1990). Worm embryos (obtained by cutting gravid adults; Epstein et al., 1993) and dissected gonads (prepared as in Rose et al., 1997) were attached on poly-lysine–coated slides by freeze-cracking, fixed with methanol at -20°C for 5 min, and stained with antibodies. Rabbit polyclonal anti-tetraThymosinβ antibody raised against full-length recombinant protein at the Centre d'Economie rurale, Laboratoire d'Hormonologie (Marloie, Belgium) was enriched by affinity purification over tetraThymosinβ coupled to CNBr-Sepharose 4B (Pharmacia, Piscataway, NJ). The specificity of the tetraThymosinβ stainings is based on loss of signal in a control experiment using antibody preincubated with resin-coupled recombinant tetraThymosinβ. Mouse monoclonal anti-actin (C4) and mouse monoclonal anti–α-tubulin antibody were from ICN Biomedicals (Costa Mesa, CA) and Amersham Biosciences (Piscataway, NJ), respectively. 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Sigma, St. Louis, MO) was used at 0.1 μg/ml as DNA-stain. Samples were viewed using a Nikon Eclipse TE2000 (Garden City, NY) inverted microscope with a 40× CFI Plan Fluor objective. Images were captured by a SPOT RT Monochrome CCD camera (Diagnostic Instruments, Sterling Heights, MI) and processed by the IPLab imaging software (Scanalytics, Billerica, MA).
RNA Interference Experiments and Gene Knockout
For RNAi we either microinjected dsRNA (0.5 μl 1.0 mg/ml) of the entire coding region of tetraThymosinβ generated by aMEGAscript T7-kit (Ambion, Austin, TX) in adult C. elegans or used soaking or feeding. The latter was performed as described in Ono and Ono (2002) using E. coli HT115(DE3) harboring plasmid L4440 (provided by Dr. A. Fire, Carnegie Institution of Washington, Baltimore, MD). The tth-1 null allele gk43 was generated by the C. elegans Reverse Genetics Core Facility (University of British Columbia, Canada). This allele was maintained with the szT1 balancer in the strain VC115 (+/szT1 [lon-2 (e678)] I; tth-1 (gk43)/szT1 X). tth-1 (gk43) carries a deletion of 715 base pairs in F08F1.8 (cosmid F08F1 coordinates 18199–18914 inclusive). This strain was outcrossed three times with no attempt of recombination within X chromosome. Genomic DNA fragments of the tth-1 gene were amplified from a single worm as described (Barstead and Waterston, 1991) by two rounds of PCR using nested sets of primers (with 5′-GCCCCATTATTGTTTTTTCACC and 5′-CCTCGGCGACATCTGAAATG as first round, IR (5′-CCACTTATCGGTGACTAACA) and IL (5′-GACTCCGAACATCGAAAACA) as second round primers). To determine the tetraThymosinβ protein levels, the lysate of 50 worms, obtained by incubating at 97°C for 2 min in 2% SDS, 80 mM Tris-HCl, 5% β-mercaptoethanol, 15% glycerol, 0.05% bromophenol blue, pH 6.8, was analyzed by SDS-PAGE (15%) and subjected to Western blot as described previously (Ono and Ono, 2002) with anti-tetraThymosinβ antibody. Anti-actin antibody was used to monitor equal loading levels of the lysates.
Reverse Transcription-PCR
Total RNA (1 μg) from WT worms was used as template for amplifying RNA for act-1 (a muscle actin gene), F08F1.3 and tth-1 using the SuperScript III One-Step RT-PCR System with Platinum TaqDNA polymerase (Invitrogen, Carlsbad, CA). As sense and antisense primers we used for act-1: 5′-GATCGAATTCTCTTCTATGTGTGACGACGAGGTTGC and 5′-GATCGCTAGCTTAGAAGCACTTGCGGTGAACGATG, for F08F1.3: 5′-TGCGTTTTGTATACATCTTCCTGG and 5′-ACTTTTCAAATCTAATGATGGCAGGTT, and for tth-1: 5′-TGGCTGCCGTCACCGAACTT and 5′-ATTGAGCTTCAGTGACGCGGACAT.
Northern Blot
Embryo, four larval stages (L1 to L4) and adult N2 C. elegans were collected according to Sulston and Hodgkin (1988). Ten micrograms total RNA of each of these stages, prepared using RNeasy (Qiagen, Valencia, CA), was separated on a 1.5% agarose gel with 5% formaldehyde, transferred to nylon membrane (ZetaProbe, Bio-Rad, Richmond, CA) and UV-cross-linked. Northern blotting was performed according to Eyckerman et al. (1999).
G- and F-actin–binding Assays
Ca-ATP-G-actin was prepared from rabbit skeletal muscle (Spudich and Watt, 1971) and further purified via Sephacryl S-300 (Pharmacia) in G-buffer (5 mM Tris-HCl, pH 7.7, 0.1 mM CaCl2, 0.2 mM ATP, 0.2 mM DTT, 0.01% NaN3). We assayed for monomer binding of tetraThymosinβ using band shift in nondenaturing PAGE (Safer, 1989). Actin monomer binding was also assayed using G-actin labeled with 7-chloro-4-nitrobenzeno-2-oxa-1,3-diazole (NBD) after N-ethylmaleimide treatment according to Detmers et al. (1981). Emission spectra of 1.5 μM NBD-actin in the presence of increasing concentrations of tetraThymosinβ WT in G-buffer were recorded in a Hitachi F4500 spectrophotometer (Woodbury, NY; using 470 nm as excitation wavelength). The fluorescence change at the emission maximum (535 nm) induced by binding of tetraThymosinβ to actin is Δ(delta)-fluorescence: the measured fluorescence—the fluorescence of 1.5 μM NBD-actin in the absence of ligand. Alternatively, Δ-fluorescence at 535 nm was measured for a set of samples in which the NBD-actin concentration was changed so that the ratio of NBD-actin to tetraThymosinβ WT was increased up to 4:1. In this experiment, tetraThymosinβ was constant at 4 μM.
F-actin binding of tetraThymosinβ protein was monitored using cosedimentation: samples containing varying concentrations, as indicated, of prepolymerized actin were incubated for 1 h at RT with WT (40 μM) or mutant (60 μM) tetraThymosinβ and subsequently spun at 100,000 × g (Beckman airfuge, Berkeley, CA) for 20 min at RT. A control sample with only tetraThymosinβ was used to correct for the small amount of self-aggregated and hence self-sedimented tetraThymosinβ.
For binding of peptides (that mimic the tetraThymosinβ repeats) to monomeric or filamentous actin, zerolength cross-linking reagents (1-ethyl-3(3-dimethylaminopropyl)carbodiimide [EDC, Sigma] and sulfo-N-hydroxysuccinimide [sulfo-NHS, Pierce, Rockford, IL]) were added to incubated mixtures of the peptides (increasing concentrations as indicated in figure) and a constant amount of G-actin or prepolymerized F-actin (10 μM), respectively (as in Van Troys et al., 1996a, 1996b). For the F-actin-peptide interactions the crosslinking step was followed by sedimentation (100,000 × g, 10 min, at RT). The cross-linking step is included given the potentially high off rates of the peptides (based on data for WT tetraThymosinβ; see SPR measurements). In addition, using this strategy for F-actin interaction, we prevented having to correct the amount of F-actin bound peptide for the free peptide that is aspecifically captured in the actin pellet upon sedimentation. The concentration of actin and tetraThymosinβ proteins or peptides used in these assays is shown in figures or figure legends. Binding was evaluated by analyzing the samples of sedimentation and/or cross-linking on SDS-mini slab gels (Bio-Rad) followed by Coomassie staining and densitometry.
Surface Plasmon Resonance
The N-Cys-tetraThymosinβ variant, carrying a unique cysteine residue, was labeled using LC-biotin-iodoacetamide (Pierce) following instructions of the manufacturer. After removal of free label by desalting, the biotinylated tetraThymosinβ was coupled in HBS buffer to a streptavidine-coated chip (SA) mounted in a BiacoreX (Pharmacia). The system was washed with G-buffer supplemented with 150 mM NaCl. G-actin in G-buffer, to which we added 150 mM NaCl immediately before injection, was passed in different concentrations over the chip using a flow of 5 μl/min. The salt was added to prevent nonspecific binding (probed on a nontetraThymosinβ coated chip [blank] analyzed in parallel) and did not induce actin polymerization within the time span of the measurement. The blank corrected increase in response units (RU) is proportional to the amount of complex formed and the stoichiometry (n) of the tetraThymosinβ-G-actin interaction under these conditions can be calculated using the equation:
 |
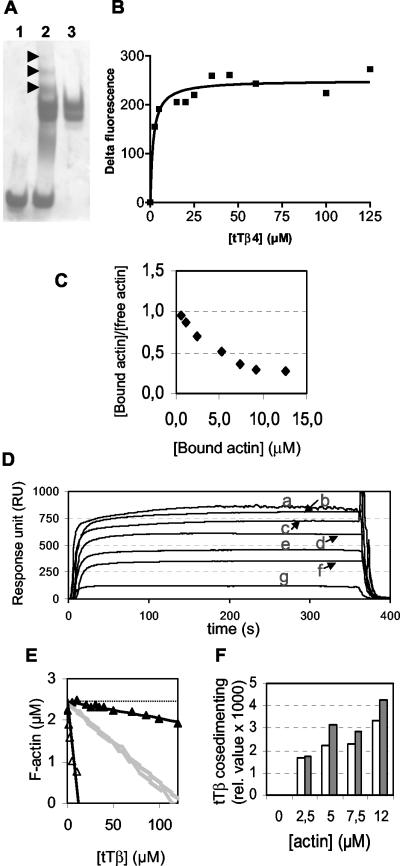
with MW the molecular weights of the respective proteins and Rc the RU-signal corresponding to the amount of coupled tetraThymosinβ, in casu 118 RU. Using this value, Rmax will equal either 310, 620, 929, or 1240 RU, depending on a stoichiometry n of 1, 2, 3, or 4, respectively. The observation that the RU values measured for the highest G-actin concentrations converge to a maximal read-out (see Figure 7B) indicates that the experimental RU increase of 864 RU for 60 μM (curve a, Figure 7B, corresponding to n = 2.8) approaches Rmax and that n equals three, i.e., three actin monomers vs. one tetraThymosinβ. The overall affinity for the formation of this complex (∼15 μM) is derived from the G-actin concentration yielding Rmax/2. This corresponds to the G-actin concentration for half fractional occupancy (C50%).
Figure 7.
TetraThymosinβ binds multiple actin monomers and has both sequestering and F-actin-binding capacity. (A) Native gel electrophoresis of 12 μM G-actin without (lane 1) or with 24 μM tetraThymosinβ (lane 2), only 24 μM tetraThymosinβ (lane 3). Arrowheads point at additional bands observed in the mixture as a result of complex formation. (B) Fluorescence change induced by binding of tetraThymosinβ to NBD-labeled actin monomers measured at the emission maximum (535 nm). The Δ-fluorescence is calculated as the fluorescence of the NBD-actin (1,5 μM)-tetraThymosinβ mixture minus the fluorescence of NBD-actin (1.5 μM). Nonlinear fitting is performed using GraphPad Prism software. (C) Scatchard analysis of fluorescence measurements of NBD-actintetraThymosinβ complexes (excitation 470 nm, emission 535 nm) obtained by incubating 4 μM tetraThymosinβ with increasing concentrations of NBD-actin (up to 16 μM the actin-tetraThymosinβ ratio is maximally 4). This fluorescence was for each sample corrected for the fluorescence of NBD-actin in the absence of tetraThymosinβ. Delta fluorescence is translated into μM bound actin using the plateau value obtained in A. A nonlinear correlation is observed in this Scatchard Plot. (D) Response unit increases obtained in SPR measurements using tetraThymosinβ coupled to the sensor chip over which different monomeric actin concentrations were passed: 60 μM (a), 40 μM (b), 30 μM (c), 20 μM (d), 15 μM (e), 10 μM (f), 5 μM (g); see Materials and Methods and Results for details on derivation of stoichiometry and affinity. (E) The F-actin decrease at steady state (2.5 μM total actin concentration) is plotted as a function of tetraThymosinβ concentration using actin filaments with gelsolin-capped (▵) or elongation competent free barbed ends (▴). The calculated curves (in gray) show the hypothetical decreases for the latter condition if only sequestering would occur for complexes of tetraThymosinβ-actin with stoichiometries 1:1 (top) to 1:3 (bottom); see text. The dashed black horizontal line is the reference value for F-actin concentration in the absence of tetraThymosinβ. (F) Cosedimentation of tetraThymosinβ WT and mutant (TβR3) with F-actin in function of increasing total actin concentration. The amounts were derived from densitometry of pellet fractions analyzed on Coomassie-stained SDS mini-slab gels. The results shown are corrected for the small amount of spontaneously sedimenting tetraThymosinβ WT or mutant in a control sample without actin (0.7 and 0.8% of added WT and mutant, respectively). TetraThymosinβ WT (white bars) and mutant tTβR3 (having only repeat 3 intact; gray bars) were preincubated at the indicated concentration with prepolymerized actin filaments before spinning.
Actin Polymerization Assays
Actin was labeled on Cys 374 using N-pyrenyl-iodoacetamide (Kouyama and Michashi, 1981). Pyrene fluorescence was measured on a Hitachi F4500 spectrophotometer using 365 and 388 nm as excitation and emission wavelength, respectively. We induced polymerization of G-ATP-actin (10 μM, 10% labeled) in absence or presence of tetraThymosinβ WT (2 μM) or variants (2–25 μM as indicated) by the addition of 100 mM KCl, 1 mM MgCl2 and followed fluorescence increase as a function of time.
We assessed the sequestering capacity of tetraThymosinβ WT, mutants, or peptides based on steady state measurements of F-actin (2.5 μM, 10% labeled) as described in Van Troys et al. (1996b) but using gelsolin capped actin filaments (molar ratio gelsolin:actin, 1/200). The decrease in fluorescence, correlated with the amount of actin complexed by tetraThymosinβ, is used to calculate a Kdseq as in Van Troys et al. (1996b) but taking into account the different possible stoichiometries of the sequestered complex tetraThymosinβ-(actin)n-complex with n between 1 and 3 (i.e., using the equation that the slope of the decrease in F-actin vs. the total tetraThymosinβ concentration is given by n CMC/(Kd + CMC) with CMC the critical monomer concentration). We performed similar measurements using uncapped filaments at final actin concentrations of 2.5 μM. The hypothetical data corresponding to pure sequestering in the presence of actin filaments with free barbed ends were derived as in Hertzog et al. (2002).
The putative promotive activity of WT or mutant tetraThymosinβ on filament elongation is measured from induced CMC-shifts at equilibrium in the presence and absence of thymosinβ4 as in Pantaloni and Carlier (1993).
RESULTS
A Thymosinβ Homologue with Repeated Structure in C. elegans
In search of a thymosinβ homologue in C. elegans, we discovered a predicted protein of 151 amino acids (accession number NM_077029, gi:17551491, www.wormbase.org; Van Troys et al., 1999) that consists of four stretches of 30–47 amino acids each with similarity to thymosinβ4. We named this protein tetraThymosinβ and the gene tth-1. Figure 1 shows the four repeats aligned to human thymosinβ4 and to the repeats of the analogous proteins actobindin from A. castellanii and ciboulot from Drosophila melanogaster. This alignment shows that the two regions important for actin interaction in thymosinβ4, a hexapeptide motif and a preceding hydrophobic triplet (Van Troys et al., 1996b), are well conserved in the tetraThymosinβ repeats. All four tetraThymosinβ motifs, even the most diverse motif LHSTPV in the second repeat, could in principle allow actin-binding based upon an extensive mutagenesis study of phage displayed thymosin β4 (Rossenu et al., 2003) yielding insight in the plasticity of the hexapeptide motif.
Figure 1.
Primary structure alignment of C. elegans tetraThymosinβ and family members. The four C. elegans tetraThymosinβ (NM_077029) repeats (R1 to R4) are aligned to human thymosinβ4 (Hs Tβ4, NP_004193) and to the repeats of Acanthamoeba castellanii actobindin (Ac acto, P18281) and Drosophila melanogaster ciboulot (Dm cib, NP_525065). The actin-binding determinants, as defined for thymosinβ4 (Van Troys et al., 1996b), are in bold (motif [underlined], hydrophobic triplet [*]). The hydrophobic triplet indicated in ciboulot domain 1 (R1) is a subset of the hydrophobic residues involved in the contact with actin (Hertzog et al., 2004). The residues boxed in this sequence are the proposed determinants in the barbed end elongation promoting activity (Hertzog et al., 2004). The sequences highlighted in gray in tetraThymosinβ correspond to the synthetic peptides (r1, r2, r3, and r4), used as individual repeat mimics (see Figure 8A). Together the four peptides comprise 100 of the 151 tetraThymosinβ residues.
TetraThymosinβ Is Present in Specific Actin-rich Structures at Various Stages during Development
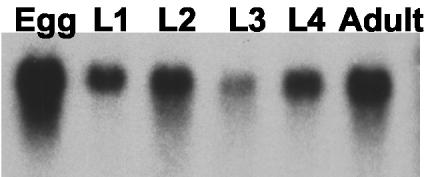
Expression of the predicted tetraThymosinβ protein was confirmed by a Western Blot of total lysate of adult C. elegans using antibodies raised against purified recombinant protein (see Materials and Methods); this revealed a single band of similar size as the recombinant protein (our unpublished data). A Northern Blot of different developmental stages (Figure 2) shows that tetraThymosinβ mRNA is present throughout the worm's lifespan. To gain insight in the physiological processes tetraThymosinβ may participate in, we studied tissue distribution and intracellular localization using immunofluorescence microscopy. The localization of tetraThymosinβ is shown in oocytes (in dissected gonads of adult worms, Figure 3), in embryos (Figure 4A) and in whole adult organisms (Figure 4B). We tested several different fixation conditions and found that the optimal conditions to preserve the signals for tetraThymosinβ were cold methanol (-20 C for 5 min) for embryos and dissected gonads and a mixture of 4% formaldehyde and 50% methanol (on ice for 60 min) for whole-mount staining of adult worms. Preincubation of the antibody with tetraThymosinβ coupled to Sepharose beads eliminated immunofluorescent signals (unpublished results), indicating that the signals were specific for tetraThymosinβ. TetraThymosinβ was maternally expressed and in the adult gonad (Figure 3) diffusely present in the cytoplasm of the oocytes, with strong staining at the cell cortex (Figure 3A) where also actin is present (Figure 3, B and C). In the distal arm of the gonad, tetraThymosinβ localized to the inside edges of the membrane cubicles surrounding germ cell nuclei (Figure 3A). Here, tetraThymosinβ did not localize to the lateral side of the membrane cubicles where actin is arranged in a “honeycomb” structure (Strome, 1986). Staining of actin at the inside edges of the cubicle was weak (Figure 3B), but this is likely due to fixation with methanol that does not preserve microfilaments very well. In two-cell stage embryos (Figure 4Aa), tetraThymosinβ was enriched in the cell-cell contact where actin was also concentrated. The tetraThymosinβ signal was clearly apparent until the four-cell stage (Figure 4Ab) and became weaker later on (unpublished data). At the comma stage (∼290 min after the first cell division) (Figure 4A, c–e), tetraThymosinβ staining was observed at the developing nerve ring that is the largest axonal bundle in the nematode body and that positions around the isthmus of the pharynx. TetraThymosinβ was evident in this region even before a clear actin signal was apparent (Figure 4Ac). In larvae and adults (Figure 4B), immunofluorescence yielded a diffuse but specific staining of the entire worm body with enrichment at the intestinal tract and spermatheca (Figure 4Ba). At these stages, in contrast to what is observed in developing embryo (see above), no prominent signal was observed in the head region where the nerve ring is located (Figure 4B, b, d, and f). These immunostaining patterns were reproducible in three or more experiments and in 20 or more worms or embryos.
Figure 2.
TetraThymosinβ mRNA is present throughout development. Signals obtained by Northern blot using a tetraThymosinβ specific probe for the different developmental stages of C. elegans: embryo, Egg; four larval stages, L1–L4; and adult. A representative experiment is shown.
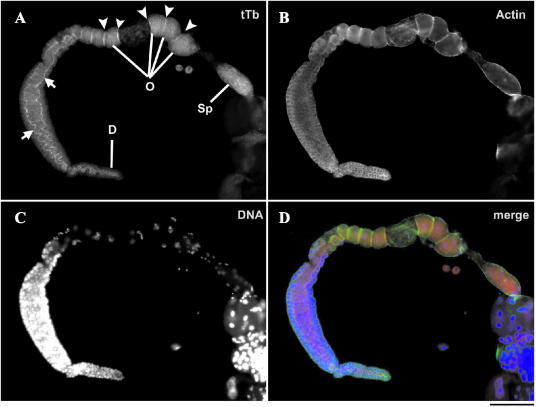
Figure 3.
Localization of tetraThymosinβ in dissected adult gonad of C. elegans. Immunolocalization of tetraThymosinβ (A) and actin (B) in the dissected adult gonad (D, distal tip; Sp, spermatheca; O, oocytes). A merge of both images with DAPI staining (C) is shown in D (red, tetraThymosinβ; green, actin; and blue, DNA; scale bar, 50 μm). TetraThymosinβ localizes to the cytoplasm and the cortex (arrowheads in A) of the oocytes and to the inside edges of the membrane cubicles (arrows in A) surrounding germ cell nuclei.
Figure 4.
Localization of tetraThymosinβ in C. elegans embryo, larvae, and adults. (A) (a–e) Immunolocalization of tetraThymosinβ (left) and actin (middle) at different phases of embryonic development (scale bar, 10 μm). In the right panel, colocalization is visualized (red, tetraThymosinβ; green, actin; blue, DAPI staining). Arrows and stars indicate the tetraThymosinβ staining at cell-cell contacts and at the site of the nerve ring, respectively. (B) Localization of tetraThymosinβ in WT young adults; anti-tetraThymosinβ antibody (a and b) and DAPI for DNA (c and d); scale bar, 50 μm. (e and f) Merged images of tetraThymosinβ (red) and DNA (blue). The entire worm is shown in a, c, and e; the head region is enlarged in b, d, and f. TetraThymosinβ is diffusely detected in the entire body (a) and enriched in the spermatheca (a, Sp), the lumen of the intestine (a, arrow), and the posterior bulb of the pharynx (b, arrowhead). Only a part of the intestine was stained because of incomplete penetration of the antibody. Adult nerve ring was identified as a region where DAPI staining was weak (d, arrow), but it was not stained by anti-tetraThymosinβ antibody (f, arrow).
TetraThymosinβ-/- Organisms Show Defects in Oocyte Maturation
To further assess the importance of tetraThymosinβ in vivo, we first used RNA interference, by feeding, soaking, or microinjection, to inhibit protein expression in live animals. However, this technique did not result in reduced tetraThymosinβ levels as judged by Western blot, and consequently we did not observe an altered phenotype in the treated worms or in their progeny. This is consistent with the result of a large-scale RNA-interference project (Kamath et al., 2003).
We however obtained a tetraThymosinβ mutant, tth-1 (gk43), that has a deletion of 715 base pairs in the 5′-upstream region (Figure 5, A and B). By Western Blot, the tetraThymosinβ protein was undetectable in the tth-1 (gk43) homozygotes (Figure 5C), indicating that the deletion removes an essential part of the tetraThymosinβ promoter sequence. Analogously, staining of mutant worm with anti tetraThymosinβ antibody was negative. It should be noted that this deletion is located within 4-kb upstream of the adjacent predicted gene F08F1.3 with unknown function. Therefore, we could not a priori exclude the possibility that the deletion also affected expression of this neighboring gene. However, the mRNA of F08F1.3 (expected size: 650 base pairs) is not present in WT adult worms as demonstrated in Figure 5D showing a reverse transcription-PCR analysis on total RNA of WT worms. In accordance, reports by others also suggest that F08F1.3 may not be a functional gene: i) no Expressed Sequence Tag clones have been isolated for this gene (www.wormbase.org), ii) the ORFeome project was not able to amplify cDNA for this gene (Reboul et al., 2003) and (iii) RNAi of this gene causes no detectable phenotype (Kamath et al., 2003). Together, these observations strongly suggest that the deletion in tth-1 (gk43) specifically affects expression of tth-1. The tth-1 (gk43) homozygotes died as late larvae (L3 or L4) or young adults displaying a dumpy phenotype (Figure 5E). Staining of mutant worms with rhodamine-phalloidin revealed no remarkable abnormalities in actin-rich tissues including pharynx, body wall muscle, vulva, and spermatheca (our unpublished results). Therefore, the specific cause of the dumpy phenotype is not clear. In addition, the tth-1 mutants showed a maternal-effect lethal phenotype. The F1 tth-1 homozygotes from the tth-1/+ heterozygous parents survived to late larvae or adults but produced only dead eggs or arrested embryos. Mating the F1 tth-1 homozygous hermaphrodites with WT males did not rescue lethality (unpublished data). A more detailed phenotypic analysis of dissected gonads (Figure 6) revealed that mutant eggs that were ovulated from the ovary were significantly deformed in the spermatheca and uterus (Figure 6, D and F, arrow). In WT worms, actin (Figure 6A, arrows) and tetraThymosinβ (Figure 3A, arrows) are concentrated at the cortex of oocytes in the ovary. However, in the mutant oocytes, actin is uniformly distributed in the cytoplasm and not concentrated at the cortex (Figure 6B, arrows). Because methanol fixation, used for the stainings in Figure 6 is not optimal to preserve microfilaments, we also fixed dissected gonads with 4% formaldehyde, stained with rhodamine-phalloidin and confirmed that 0% of WT (n = 23) and 74% of tth-1 mutants (n = 23) showed reduced actin filaments at the cortices of the oocytes. Localization of tetraThymosinβ to the oocyte cortex (Figure 3A) in WT organisms and the tth-1 mutant phenotype suggest that tetraThymosinβ is required in assembly of the cortical actin network necessary for oocyte rigidity. As a result, mutant oocytes do not endure the ovulation process that involves vigorous contraction of the ovarian myoepithelial cells (Hubbard and Greenstein, 2000).
Figure 5.
Characterization of the tth-1 mutant. (A) Genomic structure of the tth-1 gene. Exons 1–3 are indicated by open boxes. The tth-1 (gk43) deletion is located in the upstream region of the gene. IR and IL indicate the location of PCR primers used in B. (B) Genomic DNA fragments of the tth-1 gene from a WT or tth-1 (gk43) homozygote were amplified by PCR using primers IR and IL. DNA size markers in kb are shown on the left. (C) Protein levels of tetraThymosinβ and actin in WT or tth-1 (gk43) homozygotes were examined by Western blot using anti-tetraThymosinβ and anti-actin antibody. (D) Reverse transcription-PCR of mRNAs for act-1 (1.1 kb), F08F1.3 (predicted size, 650 base pairs), and tth-1 (450 base pairs) in WT worms. DNA size markers in kb are shown on the left. (E) Morphology of WT or tth-1 (gk43) homozygous adult worms (scalebar, 0.2 mm). The tth-1 (gk43) homozygotes have a dumpy phenotype.
Figure 6.
Morphology and cytoskeletal organization of gonads from WT and tth-1-mutant adult. Dissected adult gonads (D, distal tip; Sp, spermatheca; O, oocytes) from a WT (A, C, and E) or tth-1 (gk43) homozygote (B, D, and F) were stained for actin (A and B) and tubulin (C and D) (scalebar, 50 μm). Merged images of actin (red) and tubulin (green) are shown in E and F. Actin is accumulated at the cortex of the oocytes (A, arrows) in WT but absent in the tth-1 mutant (B, arrows). The fine filamentous actin network observed in B is derived from the myoepithelial cells of the ovary, not from the oocytes. In the tth-1 mutant, deformed oocytes are found in the spermatheca (D and F, arrow).
TetraThymosinβ Binds Multiple Actin Monomers and Has Both Sequestering and Filament Binding Capacity
In view of its important role in the nematode, it is essential to gain insight in the effect of tetraThymosinβ on actin and actin dynamics using in vitro analysis. In line with its homology with thymosinβ4, it is expected that tetraThymosinβ interacts with actin monomers but because tetraThymosinβ consists of a fourfold repeat of the β-thymosin module, complexes containing multiple actins could potentially be formed. The purified protein partly shifts monomeric α-skeletal muscle actin in native PAGE and intriguingly, several extra bands are already apparent in the mixture (arrowheads, Figure 7A) indicative of complexes with different stoichiometries. TetraThymosinβ can be chemically crosslinked to actin monomers using a zero-length cross-linker and it retains C. elegans G-actin from total worm lysates using affinity chromatography (our unpublished results). TetraThymosinβ binding to NBD-labeled monomeric actin (1.5 μM) induces a dose-dependent increase in fluorescence at the emission maximum of the fluorophore (Figure 7B). To compare this tetraThymosinβ activity with that reported for its homologues (ciboulot, actobindin), we used nonlinear fitting of the data assuming a 1:1 stoichiometry. The thus obtained value 1.72 μM (SD 0.51 μM) is comparable; however, below we present several experiments indicating that the stoichiometry is not one to one. Indeed, doing an inverse experiment, i.e., increasing the concentration of NBD-actin over tetraThymosinβ, yields data that the actin-tetraThymosinβ interaction is not a simple binding event. In a Scatchard plot the data obtained under these conditions cannot be fitted by linear regression (Figure 7C). This nonlinearity is caused by the presence of multiple binding sites (either of different affinity or showing cooperativity) and hence indicative of a stoichiometry larger than 1:1. For this reason, we additionally performed SPR measurements (Biacore) in which monomeric actin is allowed to interact with tetraThymosinβ that is coupled to a sensor chip (Figure 7D) as a means to determine the stoichiometry of complex formation. The observed increase in response units (ΔRU) is a measure for the tetraThymosinβ-actin complex formed and this signal increases at higher actin concentrations (Figure 7D). As outlined in detail in Materials and Methods, this SPR analysis demonstrates that at least three actin monomers can simultaneously interact with one tetraThymosinβ molecule. This is clearly in contrast with conclusions made for the homologous protein ciboulot where a 1:1 stoichiometry is proposed (Boquet et al., 2000; Hertzog et al., 2002, 2004). The steepness of the slopes of the binding curves suggests that on and off rates are fast. In this assay the overall affinity of tetraThymosinβ for actin monomers over the different contributing binding sites is ∼15 μM (C50%-value based on the theoretical maximal response (Rmax) most closely corresponding to the highest measured ΔRU and in casu corresponding with an 1:3 complex). This is consequently an overall Kd for three G-actin molecules binding to tetraThymosinβ and this, together with buffer differences (note that the NaCl concentration is higher than in the NBD-actin interaction assay, see Materials and Methods), may account for the higher Kd-value compared with the one described above. Given the multimodular structure of tetraThymosinβ, the results on stoichiometry (presented in Figure 7, C and D) are consistent with the concept that more than one repeat interacts with actin.
We analyzed whether the observed interaction of tetraThymosinβ with actin monomers results in a monomer sequestering effect. When actin filaments are capped at their barbed ends, the addition of increasing amounts of tetraThymosinβ leads to a strong decrease in filamentous actin (Figure 7E, ▵). This indicates that, under these conditions, tetraThymosinβ binds actin monomers and shifts the equilibrium to the monomeric form, similarly as observed for thymosinβ4. From the slope of the decrease in F-actin plotted vs. tetraThymosinβ concentration (Figure 7E), we calculated an overall affinity that varies from 10 to 28 μM, depending on the possible stoichiometry of the sequestered (actin)n-tetraThymosinβ complex (n = 1, 2 or 3, see above). At present it is unknown whether these values are the resultant of independent or cooperative binding of the repeats. This value thus forms an indication of the overall sequestering capacity of this protein.
The single repeat protein thymosinβ4 is a pure sequestering agent, independent whether the actin filaments with which the actin-thymosinβ4 pool is in equilibrium, have capped or free barbed ends (Pantaloni and Carlier, 1993). In contrast, profilin only effectively sequesters actin monomers in the presence of barbed end capped filaments (Pantaloni and Carlier, 1993; Kang et al., 1999), a property used to demonstrate its ability to deliver actin monomers to free barbed ends and hence promote barbed end elongation. A same effect has been described for tetraThymosinβ homologues ciboulot and actobindin (Boquet et al., 2000; Herzog et al., 2002). Similar as in Hertzog et al. (2002), we performed the same assay as above but now using filaments with free barbed ends in order to determine whether the sequestering activity of tetraThymosinβ is dependent on the status of the filament ends. Figure 7E (▴) shows that using filaments with free ends, the equilibrium amount of F-actin decreases much less as a function of tetraThymosinβ concentration. If, under this condition, tetraThymosinβ were a pure sequestering agent (with a Kd of 10 μM (n = 1) to 28 μM (n = 3), see above), a decrease in F-actin should be observed that only differs from the one when barbed ends are capped as a result of the different critical monomer concentration when barbed ends are free. We theoretically calculated this decrease that should accompany pure sequestering activity in the presence of free barbed ends (Figure 7E, gray lines, n varies from 1 [top gray curve] to 3 [bottom gray curve]). It is evident that this is significantly different from the experimental decrease observed (▴; note this conclusion is independent of the assumed stoichiometry for the sequestered complex). In line with Herzog et al. (2002), this may be indicative of a barbed end elongation promoting activity for tetraThymosinβ (see Discussion).
In cosedimentation experiments, tetraThymosinβ also interacts with actin filaments. In function of increasing F-actin concentration more tetraThymosinβ is cosedimenting (Figure 7F, white bars). We calculated that 1.2, 2.3, 4.0, and 13.0% of the total tetraThymosinβ is cosedimenting in the presence of 2.5, 5.0, 7.5, and 12 μM F-actin, respectively, whereas only 0.7% sediments in the absence of actin. An estimate, based on relative intensities of the actin and tetraThymosinβ bands, as derived from densitometry of the Coomassiestained gel on which the samples were analyzed (taking size differences into account), suggests that tetraThymosinβ binds to the filament in a molar ratio varying from 1:5–1:10 to actin protomers for the tetraThymosinβ-actin ratio's used here. This low ratio may point at a relatively weak interaction or alternatively suggests that tetraThymosinβ targets only ATP-protomers at the barbed end of the actin filaments in line with its effect on polymerization at this end (see also Discussion). The significance of the F-actin interaction is also shown using individual tetraThymosinβ repeats and mutant tetraThymosinβ proteins (see below).
All Four TetraThymosinβ Repeats Can Interact with Actin
We set out to determine the contribution of each repeat of the full-length tetraThymosinβ protein in the multiple effects on actin described above and hence to understand why this protein has a repeated structure. Initially, we determined which repeats function in G- or F-actin binding. The derived stoichiometry of the tetraThymosinβ-G-actin complex already indicates that multiple tetraThymosinβ repeats contribute to the formation of a large tetraThymosinβ-actin complex. It does however not allow evaluating the contribution of each of the different tetraThymosinβ repeats. To gain more insight in this, we initially studied four peptides (r1–r4, Figure 1) expected to mimic each repeat in actin binding based on the known binding determinants of thymosinβ4 (Van Troys et al., 1996b). All four peptides can be EDC-cross-linked to G-actin in a concentration-dependent manner, albeit to different extents, with r3 showing lowest efficiency (unpublished data). This is informative about relative affinities of the repeats although differences in crosslinking efficiency cannot be ruled out. As the intact protein has affinity for actin filaments, we also analyzed F-actin interaction under physiological salt conditions using cross-linking of the individual peptides to prepolymerized actin followed by sedimentation. Two of the four tetraThymosinβ peptides, namely r2 and r3, interact with protomers in the actin filament (Figure 8A), already at the lowest peptide concentrations tested. In contrast for the repeat 1 and 4 peptides, we do not observe significant cross-linking to F-actin, proving the specificity of the method. Note that, under these polymer conditions, peptide r4-G-actin cross-linking was again observed, combined with an increase in G-actin (Figure 8A, supernatant), which suggests a strong sequestering activity for peptide r4. The latter additionally suggests that the actin interaction for repeat r3 is most likely not the effect of dynamic incorporation of G-actin-peptide cross-linked complexes into the filament but rather bona fide F-actin binding by this repeat. Under these conditions, repeat 2 cross-links both to F-actin and in a lower extent to G-actin (see supernatant fraction, Figure 8A).
Figure 8.
The four repeats of tetraThymosinβ display differences in G- and F-actin interaction and cooperate in sequestering and inhibition of salt-induced polymerization.(A) Synthetic peptides r1 to r4 (see Figure 1) were incubated with 10 μM prepolymerized actin before EDC-cross-linking and subsequently spun. SDS-PAGE analysis of equal amounts of supernatantia (S, containing G-actin, G-actin-peptide cross-link, and free peptide) and of pellets (P, containing F-actin and peptide that cross-links to F-actin subunits) are shown for each peptide concentration; FA is a control sample with only F-actin; marker proteins (M) are 43 and 67 kDa. The actin-peptide cross-link migrates at slightly higher molecular weight than actin (45 vs. 42 kDa). (B) Change in pyrene fluorescence corresponding to the amount of gelsolin capped F-actin at steady state (actin concentration 2.5 μM) is plotted as a function of tetraThymosinβ concentration (WT or mutants tTβR1, tTβR2, tTβR3, and tTβR4 having only the indicated repeat intact as indicated above the graph; see also Table 1). If these mutants are inactive, the starting fluorescence will not change (dotted gray line). (C) Salt-induced actin polymerization is followed by recording pyrene fluorescence in time for actin (10 μM, control), actin (10 μM) incubated with 2 μM tetraThymosinβ WT, or incubated with 25 μM mutant tTβR1,tTβR2,tTβR3,ortTβR4. (D) Concentration-dependent effect of the strongest sequestering mutant tTβR4 on salt-induced polymerization (as in Figure 8C); 10 μM actin control (gray), actin in the presence of 2, 10, 20, and 25 μM tTβR4 (black curves). (E) Salt-induced polymerization of 10 μM actin in absence (control) or presence of tetraThymosinβ WT or mutants. The experiment is performed as in Figure 8C but the mutants TβR2,3,4 (a), TβR1,3,4 (b), TβR1,2,4 (c), and TβR1,2,3 (d) having only one repeat mutated (as indicated, see also Table 1) are used at 2 μM as is WT tetraThymosinβ.
The Repeats Cooperate during Actin Sequestering and Inhibition of Polymerization
Given that two of four repeats display F-actin–binding capacity, we investigated the contribution of each repeat in the sequestering activity of tetraThymosinβ. Therefore we performed the sequestering assay with capped filament ends (Figure 8B) using four tetraThymosinβ full-length mutants in which each time three of four repeats were inactivated by mutation of their motif sequences (Table 1: tTβR1–tTβR4, the repeat indicated in the name is unchanged). These mutant proteins display similar cross-linking characteristics as their peptide counterparts r1–r4 (unpublished data). Figure 8B shows that these mutants display significant differences in sequestering capacities. tTβR4 is the strongest sequester of the four mutants, although it is less efficient than WT. Next to tTβR4 only tTβR1 has sequestering activity; hence, the WT sequestering affinity is likely due to the combined activities of these two repeats. In contrast, mutants tTβR3 and tTβR2 have completely lost their sequestering capacity and intriguingly increase fluorescence as a function of concentration. For tTβR3, this is consistent with the r3-F-actin interaction (see above) and possibly with F-actin stabilization by this repeat at steady state. Figure 7F shows that this mutant tTβR3 also cosediments with actin filaments (gray bars, corresponding to 1.4, 3.1, 4.9, and 16.4% of total tTβR3 in the presence of 2.5, 5.0, 7.5, and 12 μM actin, respectively; 0.8% of total tTβR3 sediments in the absence of actin). We note that, performing the same sequestration experiment as in Figure 8B with the peptide mimetics r1–r4 yielded identical results except for the second repeat (unpublished results), because peptide r2 still has weak sequestering activity. Hence, repeat 2 apparently has a mixed G- and F-actin–binding phenotype (see also Figure 8A). The result obtained for tTβR2 (Figure 8B), may thus be the effect of (weak) F-actin–binding capacity of repeat 2 enhanced by residual F-actin–binding present in repeat 3 via residues outside the mutated motif.
Analyzing the effect of this mutant series on salt-induced actin polymerization (Figure 8C) confirms the functional differences between the repeats. WT tetraThymosinβ at 2 μM completely inhibits polymerization of 10 μM actin during the time span of the experiment, although polymerization reaches its plateau value after overnight incubation. This inhibitory activity is weaker or lost for every mutant tetraThymosinβ protein tested that contains only one intact repeat because only higher concentrations of the mutants generate an effect (shown in a concentration-dependent manner for tTβR4, Figure 8D). Consistent with its stronger sequestering activity, tTβR4 is the most efficient of this mutant series. A same concentration of the mutants tTβR1 or tTβR2 also inhibit salt-induced actin polymerization but always to a lesser extent than WT tetraThymosinβ. tTβR3 at 25 μM appears to have no inhibitory effect on polymerization indicating this mutant has lost the sequestering capacity, consistent with its observed F-actin interaction.
Using the same assay, we tested a second series of mutants in which only one of four repeats was inactivated (tTβR2,3,4, tTβR1,3,4, tTβR1,2,4 to tTβR1,2,3, Table 1, the indicated repeats are unchanged). Figure 8E shows that none of these mutants, tested like WT at 2 μM, displays WT activity. This again indicates that all repeats participate in actin binding and that none of the repeats is dispensable to obtain the WT inhibitory effect on actin polymerization.
We cannot exclude that the decreased effects of the mutants on actin are in part induced by altered folding of intact repeats when adjacent repeats are mutated. However the observations that each of the repeats also functions under its individual form of peptide mimetic is a strong argument against a determining role for folding differences in the mutant activities.
Taken together, the data obtained with peptide mimetics, with mutants with only one repeat intact or with those that have one repeat inactivated, demonstrate that in none of the cases a single repeat displays WT activity.
Full Effect of tetraThymosineβ on the G-/F-actin Balance at Steady State Using Uncapped Filaments Requires the Combined Activity of All Repeats: Repeat 3 Is Essential but Not Sufficient for This Activity
TetraThymosinβ, like ciboulot and actobindin, behaves differently with respect to actin filaments with free or capped barbed ends (see above). This difference has been proposed to result from a positive effect on barbed end elongation and to act via a same mechanism as profilin, i.e., by forming a 1:1 complex with actin (Hertzog et al., 2002). Our results above however show the functionality of multiple actin-binding repeats in the tetraThymosinβ-actin interaction. To gain further insight in the underlying mechanism, we performed for WT tetraThymosinβ and selected mutants the in vitro assay presented by Pantaloni and Carlier (1993) in which a possible barbed end elongation promoting effect of the protein of interest is derived from the nonadditive shift in critical monomer concentration (CMC) of actin polymerization observed in the additional presence of the sequestering agent thymosinβ4. For WT tetraThymosinβ (Figure 9A) this nonadditive CMC-shift is indeed clearly observed: The CMC-shift induced by tetraThymosinβ in the presence of thymosinβ4 is much smaller than the shift for thymosinβ4 (Figure 9A, detailed view in Figure 9B). In Figure 9C, the effect of tetraThymosinβ mutant tTβR1,2,4 on the CMC shift is shown using the same assay. This shows that the effect of thymosinβ4 and of this mutant, that lacks the filament binding repeat 3, on the CMC appears additive: the WT effect is lost. We also tested tetraThymosinβ variants, tTβR2,3,4, tTβR1,3,4, and tTβR1,2,3, in which only repeat 1, 2, and 4, respectively, are mutated (Table 2: these data for tetraThymosinβ WT, and all four mutants were determined independently of the data presented in Figure 9). Note that for these three mutants, the nonadditive effect on CMC shift in the presence of thymosinβ4 is still observed but less pronounced. Thus, our data indicate that repeat 3 is essential and repeats 1, 2, and 4 all three contribute (to more or less similar extent) in generating the WT effect in this assay.
Figure 9.
All four tetraThymosinβ repeats combine to produce the WT effect on G-/F-actin balance at steady state in the presence of actin filaments with free barbed ends. (A and B) Determination of shifts in critical monomer concentration (ΔCMC) vs. pure actin: actin (gray, closed gray triangle), actin with 2 μM WT tetraThymosinβ (light blue), actin with 5 μM thymosinβ4 (green), and in presence of both tetraThymosinβ (2 μM) and thymosinβ4 (5 μM) (dark blue). In B the part of graph A indicated by the gray box is enlarged to better show the differences in induced CMC-shift. For each curve, the CMC, corresponding to the actin concentration at which the fluorescence starts increasing, is determined from the intercept of linear fitting through all data points with higher than basal G-actin fluorescence (correlation coefficients shown). The shift in critical monomer concentration induced by both tetraThymosinβ WT and thymosinβ4 is already smaller than the one induced by thymosinβ4 alone demonstrating a nonadditive effect on the CMC for tetraThymosinβ and thymosinβ4 (see text). (C) Same experiment as in A and B, but using tetraThymosinβ mutant tTβR1,2,4 that is mutated only in repeat 3 (actin (gray, closed gray triangle), actin with 4 μM tTβR1,2,4 (light blue), actin with 5 μM thymosinβ4 (green), or with both tTβR1,2,4 (4 μM) and thymosinβ4 (5 μM) (dark blue). Here ΔCMCtTβR1,2,4+Tb4 is larger than ΔCMCTβ4 indicating an additive effect. Only part of the data points is shown, and the lines shown are the results of linear regression analysis (least square method) of all data points (same number as in A) yielding R2 values larger than 0.92 as indicated.
Table 2.
Effect of WT and mutant tetraThymosinβ on the induced CMC shift in the presence or absence of thymosinβ4
| activity (%)a | |
|---|---|
| WT tetraThymosinβ | 100 |
| tTβR2,3,4 | 50 |
| tTβR1,3,4 | 60 |
| tTβR1,2,4 | 0 |
| tTβR1,2,3 | 40 |
The ratio of CMC shifts Δ2/Δ1 for WT is set to 100%. Δ1: CMC-shift induced by tetraThymosinβ alone, Δ2: CMC-shift induced by tetraThymosinβ in the presence of thymosinβ4 corrected for the shift induced by thymosinβ4 alone (ΔCMCtTβ+Tβ4 – ΔCMCTβ4)
DISCUSSION
The in vivo data on tetraThymosinβ presented in this work suggest an active role for this C. elegans protein in actin-based motile processes in cells. Our data show that tetraThymosinβ is strikingly present in embryonic neuronal tissue, i.e., in the developing nerve ring. Expression of tetraThymosinβ is however absent in the adult nerve ring, indeed suggesting that its presence in these neuronal cells is mainly required during formation of the neuronal bundle, a process driven by actin polymerization. Similarly, ciboulot is important in fly brain metamorphosis (Boquet et al., 2000). tth-1(gk43)-animals, having no detectable levels of tetraThymosinβ, cannot survive. Some of the phenotypes of the knock out animals are consistent with a crucial role of tetraThymosinβ in specific actin-related processes during the worm life cycle. Most notably, formation of the cortical actin in oocytes during their maturation appears dependent on tetraThymosinβ. Deletion of tetraThymosinβ causes defects in this actin structure and prevents the normal ovulation process and consequently reproduction. It is intriguing that both tetraThymosinβ mRNA and protein can be detected at all developmental stages, whereas the observable phenotypes detected in knockouts are only limited: a dumpy phenotype and fragile oocytes. The maternal-effect lethality of the tth-1 mutation suggests that the maternal supply of the tetraThymosinβ protein from the heterozygous parents is sufficient for many morphogenetic processes in the homozygous mutants. It is also possible that the tetraThymosinβ activity can in a number of processes be compensated for by the activity of other, simultaneously expressed actin-binding proteins. Prime candidates herein may be the three profilin isoforms present in C. elegans (Polet et al., unpublished results). In D. melanogaster, overexpression of chicadee, the fly profilin, indeed rescues the brain defects caused by lack of ciboulot (Boquet et al., 2000). One of the C. elegans profilins PFN-1 is expressed in early embryos and required for cytokinesis (Severson et al., 2002), and the other two profilins are expressed in many other cell types (Polet et al., unpublished results).
TetraThymosinβ expression patterns in WT worms and the defects observed in tth-/- homozygotes reveal its crucial role in actin polymerization based processes. This is underscored by biochemical data presented here that show tetraThymosinβ to be a true member of a subgroup within a family of actin-binding proteins, the multirepeat β-thymosins, that we previously defined (Van Troys et al., 1999). Like the other members ciboulot and actobindin (Boquet et al., 2000; Hertzog et al., 2002), tetraThymosinβ is characterized by the ability to sequester actin monomers only when actin filament barbed ends are capped (Figure 7E) and to induce a nonadditive CMC-shift in the presence of a pure sequestering agent (thymosinβ4) when filaments are uncapped (Figure 9A). Taken together, this suggests that, next to a sequestration function in cells, tetraThymosinβ-actin complexes may under given conditions participate in filament formation.
We show that all repeats can interact with actin; two out of four are also able to interact with actin filaments. Importantly, mutants with only one intact repeat score positive in at least one actin-binding assay: repeat 3 in cosedimentation, repeats 1 and 4 in sequestration and inhibition of polymerization, repeat 2 also inhibits salt-induced polymerization. In addition, the activities of mutants with three of four repeats intact, never mimic full WT activity. This allows concluding that each tetraThymosinβ repeat is functional. Moreover, we here show that the WT protein can bind at least three monomers simultaneously, most probably via repeats 1, 2, and 4. Similar to thymosinβ4 (Safer et al., 1997), tetraThymosinβ likely adopts an extended conformation (it is largely unstructured as a free protein, our unpublished results) enabling the formation of complexes with varying stoichiometry. Analogously to our results, actobindin is shown to contain two functional actin-binding modules (Vancompernolle et al., 1991) and to complex an actin dimer (Bubb et al., 1994). For the homolog ciboulot, however, a 1:1 complex with G-actin was proposed (Boquet et al., 2000) and during revision of our manuscript Hertzog et al. (2004) reported that only domain 1 of this three-repeat–containing homolog interacts with actin. However our SPR data and the NBD-actin interaction assay independently indicate that tetraThymosinβ binds multiple actin molecules. This is further corroborated by cross-linking of individual repeats and by kinetic data on both sets of tetraThymosinβ mutants. The discrepancy with the data on ciboulot (Hertzog et al., 2004) may arise from evolutionary differences including the number of domains and variations in amino acid sequence (see also below). Indeed the fourth domain of tetrathymosinβ, that has a clear sequestering activity (Figure 8B), is absent in ciboulot. In addition, the observed lack of actin binding by individual ciboulot repeats 2 and 3 (Hertzog et al., 2004) may arise from differently chosen domain definitions, mainly with regard to the inclusion of the complete hydrophobic triplet as was done in the tested tetraThymosinβ peptides and mutants used here. We note that N-terminal shortening of the peptide repeats of these proteins may result both in destabilizing the N-terminal actin interaction helix as well as in reducing the contact surface.
Based on the here-derived stoichiometry and the fact that at least three repeats have sequestering abilities, it can be understood that substoichiometric amounts of tetraThymosinβ allow the efficient inhibition of actin polymerization (as observed in Figure 8C). Mainly the mutants affected in sequestering (i.e., lacking functional repeats 1, 2, or 4) display strongly reduced inhibitory potential. This suggests that the observed inhibition results from actin monomer sequestration by forming a large actin-tetraThymosinβ complex. It can however not be excluded that the inhibition is in part mediated by the fact that tetraThymosinβ repeats may associate with one or both actin protomers at the pointed end of filaments and influence growth at this end. Such capping ability— described for ciboulot and actobindin to only affect pointed end growth and not depolymerization (Hertzog et al., 2002)— could contribute to the observed deceleration of salt-induced actin polymerization in the presence of tetraThymosinβ.
To yield its effects on actin, the multirepeat nature of the tetraThymosinβ protein is fully exploited: Multiple repeats function in monomer binding and/or sequestering and, in addition, we demonstrate that repeat 3 and, to a smaller extent, repeat 2 are F-actin–interacting modules. Intriguingly, for repeat 3, this gain in function is accompanied by a complete loss in sequestering ability. The action of repeat 2 is less outspoken: depending on the assay used its activity appears more similar to the action of either repeat 1 or 3. The structural basis of these functional differences between the highly similar repeats is not obvious from their primary structure. Thymosinβ4 can also be cross-linked to F-actin, albeit only at very high concentrations (Carlier et al., 1996). Hence, F-actin–binding by this module is not unprecedented (see also Ballweber et al., 2002). Note that, in the nonsequestering, F-actin–binding and thus functionally most divergent, repeat 3, the hexapeptide motif is identical to the one of thymosin β4, whereas in repeat 4, the pure sequestering module, the motif is more diverse. Differences in activity and preferences for G- or F-actin as binding partner for the repeats may thus originate from amino acids both inside and outside the motif. Vancompernolle et al. (1992) showed that elevated concentrations of a thymosin β4 mutant with a single amino acid exchange in the central motif could induce actin polymerization. By contrast the barbed end elongation activity of ciboulot has been contributed to two specific pairs of amino acids present in domain 1 (boxed in Figure 1; Hertzog et al., 2004). Introduction of similar mutations in thymosin β4 indeed conferred an analogous barbed end elongation activity (Hertzog et al., 2004). With regard to tetraThymosinβ and in light of presented differences in its activities, we note that these two suggested determinants are never present in combination in any of the four repeats. Taken together these observations of differential activities for WH2-modules may have significant implications in interpreting the function of other WH2-repeat–containing proteins for which actin monomer binding is usually unequivocally assumed.
Mutant tTβR3, which contains only the F-actin–binding repeat, does not affect salt-induced polymerization at a concentration more than 10 fold higher than the one at which WT is completely inhibitory. This suggests that, if pointed end capping is contributing to the observed effect of WT, repeat 3 is not the capping repeat at this filament end. Alternatively, the F-actin interaction of this repeat, and hence of the intact protein, may be important in the possible effect on barbed end growth. Indeed, only a mutant defective in this repeat has completely lost the desequestering ability in the presence of thymosin β4 that is observed for WT tetraThymosinβ (Figure 9, Table 2). This strongly suggests that F-actin binding by this repeat in the intact tetraThymosinβ is required for the full efficiency of this effect. This does however not rule out that other individual repeats contribute to barbed end elongation in a similar way as reported for ciboulot D1 (Hertzog et al., 2004). Combining the activities of the repeats of tetraThymosinβ, a putative delivery system may be imaginable in which the repeat 3–F-actin interaction may bring the actin monomers, bound to the other repeats, in proximity of the growing filament end and makes them available for incorporation. Consequently, loss of functionality of the sequestering repeats 1, 2, and 4 should reduce the WT-effect, as is shown from measuring desequestration from the thymosin-actin pool (Table 2). As yet we have no evidence that this protein would preferably target barbed ends of actin filaments. However, the fact that all homologues of tetraThymosinβ bind ATP-G-actin more strongly (Hertzog et al., 2002) and the notion that tetraThymosinβ displays F-actin binding may agree with a model in which repeat 3 could putatively serve as a targeting domain to these ends. The combined presence of an F-actin interaction site and actin monomer recruitment sites is not unprecedented: it is also reported, e.g., for VASP and WASP (Bachmann et al., 1999; Walders-Harbeck et al., 2002). For these proteins, the in vivo relevance hereof is underscored by a novel model for actin-based motility (Dickinson and Purich, 2002; Dickinson et al., 2002). The clamped filament elongation or “load-lock and fire” model basically works on two main requirements: a direct interaction (clamp or lock) between the growing filament and the motile machinery and the possibility of direct transfer of monomers to the growing end from multiple actin monomer recruitment sites. In this light and based on its F-actin– and multiple G-actin–binding sites, tetraThymosinβ putatively contains “load and fire” machinery. Insight in local subcellular tetraThymosinβ concentrations or in its putative recruitment to the polymerization zone in cells will be needed to support this.
In conclusion, in contrast to earlier reports on tetraThymosinβ homologues, our experimental findings demonstrate that the repeats present in tetraThymosinβ together form a full functional unit that binds multiple actin molecules. Although the details of the combined action of the repeats are still elusive, we demonstrate that they all contribute to the effects of the intact protein on actin polymerization. This forms a basis for understanding the essential presence of these multirepeat proteins in diverse evolutionary species as well as the conservation within the repeats. Exploiting its repeated structure renders this protein of crucial importance in generating specific cellular actin structures in vivo.
Acknowledgments
We thank M. Goethals and ir. J. Van Damme for peptide synthesis and mass spectrometry and A. Anyanful for preparing total RNA from WT worms. M.V.T. is postdoctoral fellow of F.W.O.-Vlaanderen. N.D. is a predoctoral fellow from the IWT. This work was supported by BOF-GOA 2051401 to J.V. and C.A., by grants from the National Science Foundation (MCB-0110464) and the National Institutes of Health (AR48615) to S.O., and F.W.O. grant G.0007.03 and a grant of the “Geneeskundige Stichting Koningin Elisabeth” to C.A.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0225. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0225.
Abbreviations used: CMC, critical monomer concentration; EDC, 1-ethyl-3(3-dimethylaminopropyl)carbodiimide; RU, response unit; SPR, surface plasmon resonance; Tβ4, thymosinβ4; tTβ, tetraThymosinβ; WH, WASP homology; WT, wild type.
References
- Bachmann, C., Fischer, L., Walterand, U., and Reinhard, M. (1999). The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 274, 23549-23557. [DOI] [PubMed] [Google Scholar]
- Ballweber, E., Hannappel, E., Huff, T., Stephan, H., Haener, M., Taschner, N., Stoffler, D., Aebi, U., and Mannherz, H.G. (2002). Polymerisation of chemically cross-linked actin:thymosin beta(4) complex to filamentous actin: alteration in helical parameters and visualisation of thymosin beta(4) binding on F-actin. J. Mol. Biol. 315, 613-625. [DOI] [PubMed] [Google Scholar]
- Barstead, R.J., and Waterston, R.H. (1991). Vinculin is essential for muscle function in the nematode. J. Biol. Chem. 114, 715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquet, I., Boujemaa, R., Carlier, M.-F., and Preat, T. (2000). Ciboulot regulates actin assembly during Drosophila brain metamorphosis. Cell 102, 797-808. [DOI] [PubMed] [Google Scholar]
- Bubb, M.R., Lewis, M.S., and Korn, E.D. (1994). Actobindin binds with high affinity to a covalently cross-linked actin dimer. J. Biol. Chem. 269, 25587-25591. [PubMed] [Google Scholar]
- Carlier, M.-F. et al. (1996). Tbeta 4 is not a simple G-actin sequestering protein and interacts with F-actin at high concentration. J. Biol. Chem. 271, 9231-9239. [DOI] [PubMed] [Google Scholar]
- Condeelis, J. (2001). How is actin polymerization nucleated in vivo? Trends Cell Biol. 11, 288-293. [DOI] [PubMed] [Google Scholar]
- Detmers, P., Weber, A., Elzinga, M., and Stephens, R.E. (1981). 7-Chloro-4-nitrobenzeno-2-oxa-1,3-diazole actin as a probe for actin polymerization. J. Biol. Chem. 256, 99-105. [PubMed] [Google Scholar]
- Dickinson, R.B., and Purich, D.L. (2002). Clamped-filament elongation model for actin-based motors. Biophys. J. 82, 605-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, R.B., Southwick, F.S., and Purich, D.L. (2002). A direct-transfer polymerization model explains how the multiple profilin-binding sites in the actoclampin motor promote rapid actin-based motility. Arch. Biochem. Biophys. 406, 296-301. [DOI] [PubMed] [Google Scholar]
- Domanski, M., Hertzog, M., Coutant, J., Gutsche-Perelroizen, I., Bontems, F., Carlier, M.-F., Guittet, E., and van Heijenoort, C. (2004). Coupling of folding and binding of thymosin beta4 upon interaction with monomeric actin monitored by nuclear magnetic resonance. J. Biol. Chem. 279, 23637-23645. [DOI] [PubMed] [Google Scholar]
- Epstein, H.F., Casey, D.L., and Ortiz, I. (1993). Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J. Cell Biol. 122, 845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyckerman, S., Waelput, W., Verhee, A., Broekaert, D., Vandekerckhove, J., and Tavernier, J. (1999). Analysis of Tyr to Phe and fa/fa leptin receptor mutations in the PC12 cell line. Eur. Cytokine Netw. 10, 549-556. [PubMed] [Google Scholar]
- Finney, M., and Ruvkun, G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895-905. [DOI] [PubMed] [Google Scholar]
- Gimona, M., Djinovic-Carugo, K., Kranewitter, W.J., and Winder, S.J. (2002). Functional plasticity of CH domains. FEBS Lett. 513, 98-106. [DOI] [PubMed] [Google Scholar]
- Hertzog, M., Yarmola, E.G., Didry, D., Bubb, M.R., and Carlier, M.-F. (2002). Control of actin dynamics by proteins made of beta-thymosin repeats: the actobindin family. J. Biol. Chem. 277, 14786-14792. [DOI] [PubMed] [Google Scholar]
- Hertzog, M. et al. (2004). The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117, 611-623. [DOI] [PubMed] [Google Scholar]
- Hubbard, E.J., and Greenstein, D. (2000). The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev. Dyn. 218, 2-22. [DOI] [PubMed] [Google Scholar]
- Huff, T., Muller, C.S., Otto, A.M., Netzker, R., and Hannappel, E. (2001). beta-Thymosins, small acidic peptides with multiple functions. Int. J. Biochem. Cell Biol. 33, 205-220. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S. et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- Kang, F., Purich, D.L., and Southwick, F.S. (1999). Profilin promotes barbedend actin filament assembly without lowering the critical concentration. J. Biol. Chem. 274, 36963-36972. [DOI] [PubMed] [Google Scholar]
- Kouyama, T., and Mihashi, K. (1981). Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Eur. J. Biochem. 114, 33-38. [PubMed] [Google Scholar]
- Lambooy, P.K., and Korn, E.D. (1986). Purification and characterization of actobindin, a new actin monomer-binding protein from Acanthamoeba castellanii. J. Biol. Chem. 261, 17150-17155. [PubMed] [Google Scholar]
- Mattila, P.K., Salminen, M., Yamashiro, T., and Lappalainen, P. (2003). Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 278, 8452-8459. [DOI] [PubMed] [Google Scholar]
- Mertens, N., Remaut, E., and Fiers, W. (1995). Tight transcriptional control mechanism ensures stable high-level expression from T7 promoter-based expression plasmids. Biotechnology 13, 175-179. [DOI] [PubMed] [Google Scholar]
- Ojala, P.J., Paavilainen, V.O., Vartiainen, M.K., Tuma, R., Weeds, A.G., and Lappalainen, P. (2002). The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol. Biol. Cell 13, 3811-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., Baillie, D.L., and Benian, G.M. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S. (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments J. Cell Biol. 152, 1313-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., and Ono, K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni, D. and Carlier, M.-F. (1993). How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell 75, 1007-1014. [DOI] [PubMed] [Google Scholar]
- Paunola, E., Mattila, P.K., and Lappalainen, P. (2002). WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 513, 92-97. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and Borisy, G.G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453-465. [DOI] [PubMed] [Google Scholar]
- Pope, B., Way, M., and Weeds, A.G. (1991). Two of the three actin-binding domains of gelsolin bind to the same subdomain of actin. Implications of capping and severing mechanisms. FEBS Lett. 280, 70-74. [DOI] [PubMed] [Google Scholar]
- Reboul, J. et al. (2003). C. elegans ORFeome version 1. 1, experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34, 35-41. [DOI] [PubMed] [Google Scholar]
- Rose, K.L., Winfrey, V.P., Hoffman, L.H., Hall, D.H., Furuta, T., and Greenstein, D. (1997). The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 192, 59-77. [DOI] [PubMed] [Google Scholar]
- Rossenu, S., Dewitte, D., Vandekerckhove, J., and Ampe, C. (1997). A phage display technique for a fast, sensitive, and systematic investigation of protein-protein interactions. J. Protein Chem. 16, 499-503. [DOI] [PubMed] [Google Scholar]
- Rossenu, S., Leyman, S., Dewitte, D., Peelaers, D., Jonckheere, V., Van Troys, M., Vandekerckhove, J., and Ampe, C. (2003). A phage display based method for determination of relative affinities of mutants; application to the actin binding motifs in thymosin beta 4 and Villin headpiece. J. Biol. Chem. 278, 16642-16650. [DOI] [PubMed] [Google Scholar]
- Safer, D. (1989). An electrophoretic procedure for detecting proteins that bind actin monomers. Anal. Biochem. 178, 32-37. [DOI] [PubMed] [Google Scholar]
- Safer, D., Sosnick, T.R., and Elzinga, M. (1997). Thymosin β4 binds actin in an extended conformation and contacts both the barbed and pointed end. Biochemistry 36, 5806-5816. [DOI] [PubMed] [Google Scholar]
- Severson, A.F., Baillie, D.L., and Bowerman, B. (2002). A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12, 2066-2075. [DOI] [PubMed] [Google Scholar]
- Simenel, C., Van Troys, M., Vandekerckhove, J., Ampe, C., and Delepierre, M. (2000). Structural requirements for thymosin beta4 in its contact with actin. Eur. J. Biochem. 267, 3530-3538. [DOI] [PubMed] [Google Scholar]
- Small, J.V., Stradal, T., Vignal, E., and Rottner, K. (2002). The lamellipodium: where motility begins. Trends Cell Biol. 12, 112-120. [DOI] [PubMed] [Google Scholar]
- Spudich, J.A., and Watt, S. (1971). The regulation of rabbit skeletal muscle contraction. J. Biol. Chem. 246, 4866-4871. [PubMed] [Google Scholar]
- Strome, S. (1986). Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J. Cell Biol. 103, 2241-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J., and Hodgkin, J. (1988). Methods. In: The Nematode Caenorhabditis elegans, ed. W.B. Woods, Cold Spring Harbor, NY: Cold Spring Harbor Press, 81-121
- Vancompernolle, K., Vandekerckhove, J., Bubb, M.R., and Korn, E.D. (1991). The interfaces of actin and Acanthamoeba actobindin. Identification of a new actin-binding motif. J. Biol. Chem. 266, 15427-15431. [PubMed] [Google Scholar]
- Vancompernolle, K., Goethals, M., Huet, C., Louvard, D., and Vandekerckhove, J. (1992). G- to F-actin modulation by a single amino acid substitution in the actin binding site of actobindin and thymosinβ4. EMBO J. 11, 4739-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys, M., Dewitte, D., Goethals, M., Vandekerckhove, J., and Ampe, C. (1996a). Evidence for an actin binding helix in gelsolin segment 2; have homologous sequences in segments 1 and 2 of gelsolin evolved to divergent actin binding functions? FEBS Lett. 397, 191-196. [DOI] [PubMed] [Google Scholar]
- Van Troys, M., Dewitte, D., Goethals, M., Carlier, M.-F., Vandekerckhove, J., and Ampe, C. (1996b). The actin binding site of thymosin beta 4 mapped by mutational analysis. EMBO J. 15, 201-210. [PMC free article] [PubMed] [Google Scholar]
- Van Troys, M., Vandekerckhove, J., and Ampe, C. (1999). Structural modules in actin-binding proteins: towards a new classification. Biochim. Biophys. Acta 1448, 323-348. [DOI] [PubMed] [Google Scholar]
- Walders-Harbeck, B., Khaitlina, S.Y., Hinssen, H., Jockusch, B.M., and Illenberger, S. (2002). The vasodilator-stimulated phosphoprotein promotes actin polymerisation through direct binding to monomeric actin. FEBS Lett. 529, 275-280. [DOI] [PubMed] [Google Scholar]
- Yamagishi, A., Masuda, M., Ohki, T., Onishi, H., and Mochizuki, N. (2004). A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279, 14929-14936. [DOI] [PubMed] [Google Scholar]
- Yarmola, E.G., Edison, A.S., Lenox, R.H., and Bubb, M.R. (2001). Actin filament cross-linking by MARCKS: characterization of two actin-binding sites within the phosphorylation site domain. J. Biol. Chem. 276, 22351-22358. [DOI] [PubMed] [Google Scholar]