Abstract
Fluorescence recovery after photobleaching (FRAP) is a widely used tool for estimating mobility parameters of fluorescently tagged molecules in cells. Despite the widespread use of confocal laser scanning microscopes (CLSMs) to perform photobleaching experiments, quantitative data analysis has been limited by lack of appropriate practical models. Here, we present a new approximate FRAP model for use on any standard CLSM. The main novelty of the method is that it takes into account diffusion of highly mobile molecules during the bleach phase. In fact, we show that by the time the first postbleach image is acquired in a CLSM a significant fluorescence recovery of fast-moving molecules has already taken place. The model was tested by generating simulated FRAP recovery curves for a wide range of diffusion coefficients and immobile fractions. The method was further validated by an experimental determination of the diffusion coefficient of fluorescent dextrans and green fluorescent protein. The new FRAP method was used to compare the mobility rates of fluorescent dextrans of 20, 40, 70, and 500 kDa in aqueous solution and in the nucleus of living HeLa cells. Diffusion coefficients were lower in the nucleoplasm, particularly for higher molecular weight dextrans. This is most likely caused by a sterical hindrance effect imposed by nuclear components. Decreasing the temperature from 37 to 22°C reduces the dextran diffusion rates by ∼30% in aqueous solution but has little effect on mobility in the nucleoplasm. This suggests that spatial constraints to diffusion of dextrans inside the nucleus are insensitive to temperature.
INTRODUCTION
Fluorescence recovery after photobleaching (FRAP) was developed 30 years ago (Peters et al., 1974) as a tool to study the molecular mobility in several media, including aqueous solutions, gels, and living cells. In cell biology, FRAP was originally used to study membrane diffusion of lipids and proteins coupled to fluorophores (Axelrod et al., 1976; Edidin et al., 1976). More recently, noninvasive fluorescent tagging with the green fluorescent protein (GFP) or its variants has stimulated the use of FRAP to measure protein dynamics inside the living cell (Phair and Misteli, 2000; Reits and Neefjes, 2001).
In a typical FRAP experiment a region of the cell is briefly illuminated with a high-intensity laser beam with a wavelength near the excitation peak of a fluorophore. Consequently, most of the fluorophores inside that region loose their fluorescence irreversibly, a phenomenon known as photobleaching. As fluorophores are attached to molecules that move in and out of the bleached region, fluorescence inside that zone increases, and eventually equilibrium is reached. A higher mobility of the molecules results in a shorter time of recovery. From the recovery curve, it is possible to obtain estimates of the diffusion coefficient and immobile fraction (i.e., the percentage of proteins that are unable to move) (for reviews, see White and Stelzer, 1999; Klonis et al., 2002; Carrero et al., 2003).
Early applications of FRAP involved the use of nonscanning microscopes (Axelrod et al., 1976; Soumpasis, 1983; Tsay and Jacobson, 1991; Berk et al., 1993). In most cases, bleaching was performed with a stationary laser beam (either considered to be uniform or Gaussian) focused toward thin samples where diffusion could be considered to be mostly two-dimensional. Fluorescence recovery was then recorded by the fluorescence microscope. To obtain estimates for the diffusion coefficients and immobile fractions, appropriate theoretical models were developed. One of the most widely used models in cell biology was developed by Axelrod et al., 1976 (Calapez et al., 2002; Wei et al., 2003; Shimi et al., 2004). The model assumed an infinite homogenous medium with cylindrical symmetry. Bleaching was considered to be a first-order linear process, taking a negligible amount of time compared with diffusion. It also was assumed that there was no significant delay between bleaching and scanning. The laser was considered to have a Gaussian intensity profile, for both bleaching and observation of recovery. Using these assumptions, the two-dimensional diffusion equation was solved and a formula for fluorescence evolution was found.
In conjunction with GFP tagging of intracellular proteins, the development of confocal laser scanning microscopes (CLSMs) provided cell biologists with an excellent standard tool to perform FRAP experiments. Currently, most commercially available CLSMs are equipped with acoustic-optic tunable filters (AOTF) that allow bleaching arbitrary regions in the sample. During bleaching, the microscope directs a moving excitation laser beam to the sample. The bleach region is thus scanned point by point and line by line, increasing the incident laser intensity inside the pattern defined (for a thorough theoretical explanation of the bleaching process in a CLSM, see Braeckmans et al., 2003).
In the context of CLSM imaging, some assumptions of the models developed for nonscanning microscopes do not hold, because bleaching and monitoring of fluorescence recovery are done in a fundamentally different way.
Despite the widespread use of photobleaching microscopy, quantitative analysis of the data has been limited by lack of appropriate FRAP models for the CLSM. Although two-dimensional (2D) and three-dimensional (3D) models have been developed based on numerical approaches (Wedekind et al., 1996; Kubitscheck et al., 1994, 1998; Cutts et al., 1995; Peters and Kubitscheck, 1999), these have had little practical application in cell biology. A more practical 3D model was devised for use with objective lenses of low numerical aperture (Blonk et al., 1993). However, this method relies on a stationary laser beam for bleaching and a line-scanning beam for recording the fluorescence recovery, a sequence that is not possible in standard commercial CLSMs (Braeckmans et al., 2003). Recently, a new 3D FRAP model that can be easily applied on almost any modern CLSM was reported (Braeckmans et al., 2003). This method was shown to be valid only when a number of assumptions are met by the experimental conditions. Namely, that an objective lens of low numerical aperture is used, and that the bleaching phase is sufficiently short to avoid recovery during bleach.
However, in most cell biological applications of FRAP microscopy, a region of at least 1 μm2 is bleached for hundreds of milliseconds and acquisition of an image takes at least tens of milliseconds (i.e., the bleached region of interest [ROI], which is typically near the center of the image, is not imaged immediately after the end of bleaching). Thus, it is expected that for molecules diffusing at rates as low as 0.2 μm2 s-1, fluorescence recovery will occur before the bleached zone is effectively imaged for the first time. Here, we demonstrate experimentally that this is indeed the case. We therefore devised a novel 2D and 3D approximation that takes into account diffusion during the bleach period. The new FRAP model can be easily programmed in a standard fitting routine and readily applied on most commercial CLSMs, even when high numerical aperture objectives lenses are used.
MATERIALS AND METHODS
Cell Culture
HeLa cells were cultured as monolayers in modified Eagle's medium (MEM) supplemented with 10% fetal calf serum (Invitrogen, Paisley, Scotland). Cells were plated and observed in glass bottom chambers (MatTek, Ashland, MA).
Enhanced green fluorescent protein plasmid (pEGFP) (BD Biosciences Clontech, Palo Alto, CA) for transfection assays was purified using plasmid DNA midi-prep kit (QIAGEN, Hilden, Germany). HeLa subconfluent cells were transiently transfected with 1 μg of purified pEGFP plasmid by using FuGENE6 reagent (Roche Biochemicals, Indianapolis, IN) according to the manufacturer's protocol.
For imaging and microinjection, the medium was changed to Dulbecco's-MEM/F-12 without phenol red supplemented with 15 mM HEPES buffer (Invitrogen).
Dextran Purification
Fluorescein isothiocyanate (FITC)-labeled dextrans with average molecular sizes of 20, 40, and 70 kDa (Sigma-Aldrich, St. Louis, MO) were dissolved to 10 mg/ml in water. One milliliter of each sample was fractionated on a Superdex 200 column (Amersham Biosciences, Piscataway, NJ). For each dextran, the column fractions with higher absorbency were pooled and lyophilized. The 500-kDa FITC-labeled dextran was used without further purification. For imaging, all samples were diluted to 200 μg/ml in water.
Confocal Microscopy and Image Analysis
Live cells and FITC-dextrans in aqueous solution were imaged at either 37 or 22°C maintained by a heating/cooling frame (LaCon, Staig, Germany), in conjunction with an objective heater (PeCon, Erbach, Germany). Images were acquired on an LSM 510 (Carl Zeiss, Jena, Germany) with the Planapochromat 63×/1.4 objective. FITC fluorescence was detected using the 488 laser line of an Ar laser (25% of 25-mW nominal output) in conjunction with a LP 505 filter. For imaging the 3D bleached volume, 500-kDa FITC-dextrans were immobilized in a 15% polyacrylamide gel, and a z-stack series was acquired. In total, 39 images were taken and the distance between each image in the stack was 0.4 μm. Image size was 256 × 256 pixels, and the pixel width was 114 nm. Background was estimated from a 15% polyacrylamide gel devoid of fluorescent molecules.
Each FRAP experiment started with three image scans, followed by a bleach pulse of 242 ms on a spot with a diameter of 21 pixel (1.19-μm radius). A series of 97 single section images were then collected at 29.82-ms intervals, and the first image started to be acquired 2 ms after the end of bleaching. Image size was 256 × 30 pixels and the pixel width was 114 nm. For imaging the laser power was attenuated to 2% of the bleach intensity.
For FRAP analysis of intracellular mobility, FITC-dextrans (at 200 μg/ml in water) were microinjected into the nucleus of HeLa cells, as described previously (Almeida et al., 1998). In each time series, the background and nuclear regions were identified using an implementation of the ICM segmentation algorithm (Calapez et al., 2002) in Matlab software (Mathworks, Natick, MA). The average fluorescence in the nucleus T(t) and the average fluorescence in the bleached region I(t) were calculated for each background subtracted image at time t after bleaching. FRAP recovery curves were normalized according to Phair and Misteli (2000))
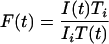 |
1 |
where Ti is the fluorescence in the nucleus before bleaching and Ii is the fluorescence in the bleached region before bleaching. This normalization corrects for the loss of fluorescence caused by imaging. Typically, <10% of the total FITC dextran fluorescence was lost during the bleach pulse. During the postbleaching scanning phase the fluorescence lost was <5% for FITC dextrans.
Image processing routines also outputted the normalized fluorescence profile of the first postbleach image. From the fit with Eq. 5, parameters wM and KM were obtained.
FRAP recovery curves were then fitted with Eq. 10, yielding parameters τD, γ, and K. All fitting procedures were performed with NonLinearRegress function of Mathematica 4.0 (Wolfram Research, Champaign, IL).
RESULTS
Derivation of the Model: Bleaching Profiles Generated by a Scanning Beam Are Approximated by an Exponential of a Gaussian
In a sample of immobile molecules, the theoretical 3D bleach pattern generated by an idealized stationary Gaussian laser beam is an exponential of a Gaussian of the form (Blonk et al., 1993)
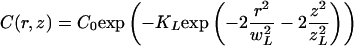 |
2 |
where wL and zL are the laser beam radii in the radial and axial direction, and KL is the bleach efficiency.
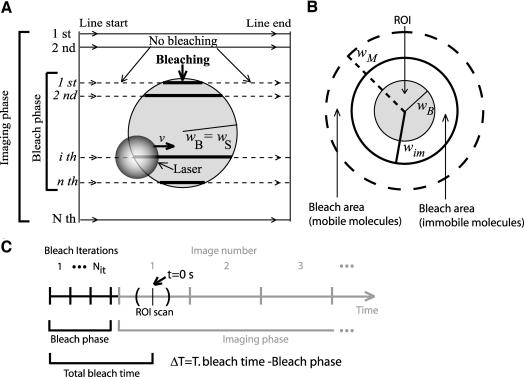
In most commercially available CLSMs, the bleaching laser beam is not stationary (Braeckmans et al., 2003). Rather, these microscopes are equipped with a scanning beam. The user defines the bleach region of interest (ROI), and the laser scans this region line by line. Laser intensity is precisely controlled by an AOTF and is set to a high value only when the laser beam enters the region defined (Figure 1).
Figure 1.
Schematic representation of bleaching in a CLSM. (A) A laser beam scans at a defined speed v a 2D circular region (ROI) with radius wB (comprising a total of n lines, numbered in italic), for Nit iterations. The laser beam has a certain width (depicted as the gray transparent circle). Laser intensity is high inside the user defined region (thick lines) and nearly zero outside (dashed lines). After bleaching, the entire region is imaged. For imaging, the monitoring beam scans the whole image with constant illumination intensity. The resulting image contains the bleached ROI and consists of N lines (numbered in normal lettering), with N>n. Image processing tasks extract fluorescence recovery from a set of pixels of radius wS (usually wS = wB). In this work, all distances are referred to the bleach region geometrical center. (B) Due to the geometry of the laser beam, the effective bleached area is always wider than the user-defined region of interest (ROI). For immobile molecules, the bleached region extends over an area (limited by the thick black line) imposed only by the laser beam width. For mobile molecules, the bleached region will be increasingly wider (dashed line), depending on the molecular mobility rate. (C) Time events in a FRAP experiment. Gray curved lines represent beginning and ending of ROI scan. The tick in the middle of these lines represents the time when the central line of the ROI is scanned. To simplify, this time is used as the image acquisition time of all the pixels in the ROI.
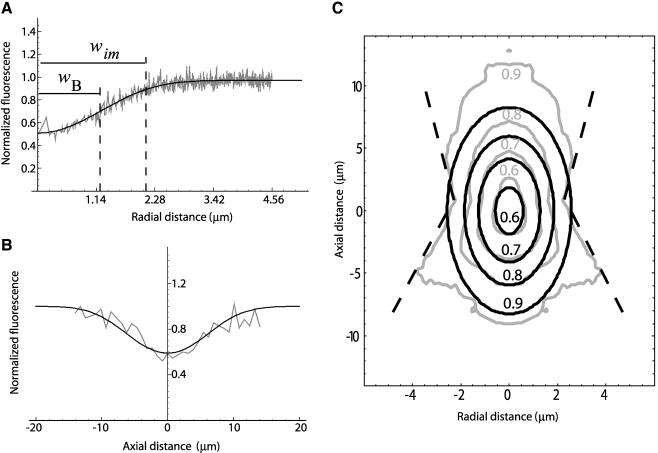
To directly observe the fluorescence profiles generated by bleaching a region of interest with a scanning laser beam, we immobilized FITC-dextrans in a 15% polyacrylamide gel and imaged the bleached volume along the z-direction. For bleaching, we used a small circular region with radius wB (∼1 μm).
As depicted in Figure 2, the observed fluorescence profiles can be approximated by the following equation:
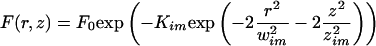 |
3 |
where wim and zim are the extent of the bleached volume in the radial and axial direction, and Kim is the bleach efficiency (the coordinates origin is the center of the bleach region). Only for regions far from the bleach center does this approximation deviate significantly from the experimental data (Figure 2C).
Figure 2.
Characterization of the bleach volume. 500-kDa FITC-dextrans were immobilized in a 15% polyacrylamide gel. Bleaching was performed within a circular ROI with 1.19 μm of radius for 242 ms. Curves shown represent the average of nine independent bleaching experiments. (A) Experimentally obtained fluorescence profiles in the radial direction (gray line) and corresponding best fit with Eq. 3 (black line). (B) Experimentally obtained fluorescence profiles in the axial direction (gray line) and corresponding best fit with Eq. 3 (black line). The fit yielded the following parameters: wim = 2.11 μm, zim′ = 10.83 μm, and Kim = 0.624. Due to the refractive index mismatch between the oil of the objective and the medium (which is considered to have the same refractive index of water) the axial profile width was corrected to zim = zim′/1.14 = 9.5 (Sheppard and Torok, 1997). (C) Contour plot representation of the experimentally obtained bleach volume (gray line) and its corresponding best fit with Eq. 3 (black line). Experimental contours 0.6, 0.7, and 0.8 have approximately ellipsoidal shapes as predicted by Eq. 3. Experimental contour 0.9 strongly deviates from the ellipsoidal shape, and near the focal plane (z = 0) it seems to have a conical shape (dashed line), as expected for a high NA lens.
Noteworthy, Eq. 3 has the same functional form as Eq. 2, indicating that bleaching profiles generated in a standard CLSM approach those produced by an idealized stationary Gaussian laser beam.
Fitting with Eq. 3 was then tested for different radial values of the bleach region (0.14, 0.26, 0.71, 1.14, and 2.28 μm). Fitting with Eq. 3 was valid for the bleaching spots smaller or equal to 1.14 μm. The values of Kim were contained in a typical range from 0 to 6. For the 2.28-μm bleach region, the fit yielded unexpectedly high values for Kim (∼60). In this region, the postbleach profile is constant for r <2.0 μm, which is consistent with the uniform disk model (Braeckmans et al., 2003). The maximum size of the bleaching region radius, for which fitting Eq. 3 is valid, will depend essentially on the effective size of the bleaching beam spot, which is related with the numerical aperture of the objective used, and should be experimentally determined for each objective. Because the size of the bleaching laser beam is roughly proportional to 1/numerical aperture (NA), lower NA objective lenses are expected to allow the use of larger bleaching regions.
According to Eq. 3, normalized fluorescence inside a region of radius ws then yields (see Appendix A)
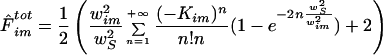 |
4 |
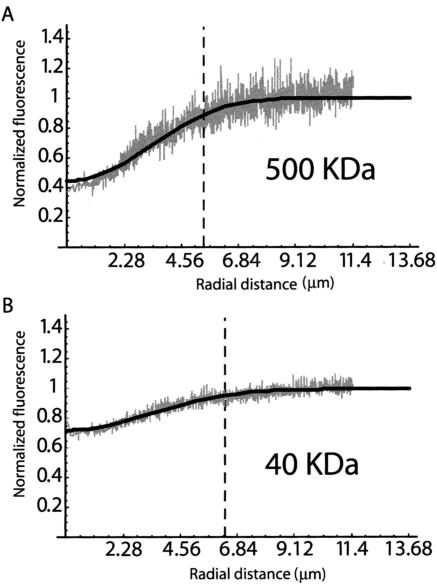
To date, most FRAP models assume that diffusion during bleaching is negligible (Axelrod et al., 1976; Braeckmans et al., 2003).). It also is assumed that all image points are taken at the same time, but in a CLSM pixels are acquired sequentially, so the bleached region is imaged a few milliseconds after the start of image acquisition. For simplicity, the time taken from the start of the bleaching phase until the first effective scan of the ROI will be hereafter referred as total bleach time (Figure 1C). To test whether if diffusion during this period can be neglected in a typical cell biological application of FRAP, we imaged the radial fluorescence profiles of FITC-dextrans immediately after bleaching (Figure 3). Dextrans of 40 and 500 kDa in aqueous solution were used. Assuming that no significant diffusion occurs during total bleach time, fluorescence profiles should be similar for both dextrans. As shown in Figure 3, this is clearly not the case. The 40-kDa dextran molecules diffuse faster than those of 500 kDa and by the time the postbleach profile is acquired by the CLSM, a larger fraction of the 40-kDa bleached molecules have moved away from the original bleach region. Consequently, the fluorescence reduction is less pronounced and the radius of the bleached area is larger.
Figure 3.
Experimentally acquired fluorescence profiles reveal recovery during bleach phase. FITC-dextrans of 500-kDa (A) and 40-kDa (B) were dissolved in water and bleached by a CLSM. The figure depicts the radial fluorescence profiles obtained after bleaching a circular ROI with 1.19 μm of radius during 242 ms (our typical bleaching settings). The gray lines represent the experimentally obtained fluorescence intensity values and the black lines the best fit with Eq. 5. For the 500-kDa-dextran, the fluorescence intensity is reduced from 1 to 0.4, whereas for the 40-kDa-dextran the reduction is from 1 to 0.7. Moreover, the bleached area extends for a radial distance of 5 μm in the case of the 500-kDa-dextran, and 6 μm in the case of the 40-kDa dextran.
For mobile molecules, we found that with the following approximation good fitting results were obtained for the postbleach radial profile:
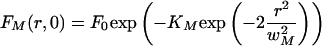 |
5 |
where KM is the bleach efficiency for the mobile molecule and wM is the profile width in the radial direction, and these parameters are dependent on the diffusion coefficient of the species. Thus, diffusion is implicitly taken into account.
Assuming isotropic diffusion, we expect that the best 3D fitting results are obtained with the following approximation:
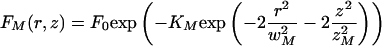 |
6 |
We expect that when D → 0 we have wM → wim, zM → zim and KM → Kim, and thus this approximation approaches Eq. 3.
Derivation of the Model: 3D Recovery after Bleaching
As in most previous models (Tsay and Jacobson, 1991; Blonk et al., 1993; Braeckmans et al., 2003), we consider diffusion as the only relevant transport process and that the medium is homogenous. Thus, after bleaching, the system behaves according to the classical diffusion equation
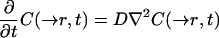 |
7 |
where 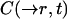 is the concentration of the unbleached fluorophores at position
is the concentration of the unbleached fluorophores at position 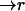 at time t.
at time t.
The partial differential equation above may be written in terms of observed fluorescence and with Eq. 6 as initial value, it has a known analytical solution (Blonk etal., 1993). Normalized fluorescence recovery inside a region of radius wS (which in general can be different from wB) is thus given by (see Appendix B):
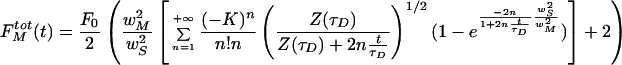 |
8 |
where
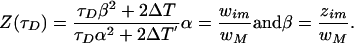 |
The square root term in Eq. 8 takes the finite axial size of the bleached volume into account. If the sample thickness is smaller than that, the system will behave like in a 2D situation. The 2D counterpart of Eq. 8 is found by zim→+∞
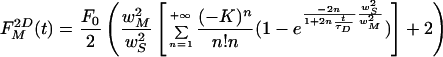 |
9 |
On the other hand, neglecting the 3D correction factor in a thick sample, would result in an overestimation of D.
So far, we assumed that all the molecules are diffusing. But if a fraction γ of molecules are immobile then the normalized recovery curve will be
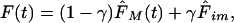 |
10 |
where F̂im is the normalized fluorescence of the immobile molecules as determined in the previous section (Eq. 4), and F̂M is the normalized fluorescence of the mobile fraction (from either Eqs. 8 or 9). The variant using the 3D correction (Eq. 8) will be referred as 3D variant, whereas its 2D counterpart (Eq. 9) will be referred as 2D variant.
This equation computes the fluorescence recovery curve for molecules in which some of them are diffusing while the others are immobile. The first term of Eq. 10 is the contribution of the fluorescence of the mobile fraction to the normalized fluorescence recovery, whereas the second term is a theoretical estimate of the postbleach total fluorescence of the immobile fraction inside the ROI (which is assumed to remain constant during a FRAP experiment).
It should be emphasized that wM refers to the profile width of the mobile population. However, in the presence of an unknown immobile fraction, it is not possible to directly estimate wM because the mobile and immobile populations are indistinguishable in an image. The postbleach concentration profile width of a mixture is always narrower than the purely mobile one (i.e., with γ = 0). Thus, an underestimation of the diffusion coefficient is expected especially in the presence of a high immobile fraction.
To overcome this problem we propose to make the estimation in two steps. In the first step, the recovery curve is fitted with Eq. 10, by using the fluorescence profile width, as shown by the first postbleach image, as a first estimate of wM. Numerical simulations show that immobile fraction estimates are relatively independent of the real value of wM (our unpublished data). Thus the immobile fraction can be estimated with a reasonable degree of accuracy. This immobile fraction value would be used to make an estimation of the real mobile fraction concentration, according to the relation
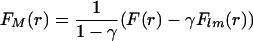 |
11 |
where FM is the mobile fraction fluorescence, F(r) is the observed fluorescence in the first postbleach image and FIm is the postbleach concentration profile of immobilized molecules. Now, the profile width found is a more correct estimate for the mobile fraction. This value is then used in the second run of the fitting procedure with Eq. 10. This two-steps variant of the 3D method will be hereafter referred as 3D variant with immobile fraction (IF). The 2D counterpart will be referred accordingly as the 2D variant with IF.
Validation of the Model by using Numerical Simulations
Considering diffusion as the only relevant transport process and assuming homogeneity of the medium, the evolution of the concentration of unbleached fluorophore is described by the following partial differential equation accounting for diffusion during bleach:
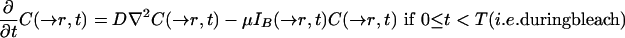 |
12 |
where 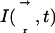 is the 3D laser intensity distribution at time t of the bleaching beam, and μ the fluorophore bleach rate constant. To check the influence of the intensity distribution on parameter estimation several configurations were used: stationary Gaussian beam, scanning Gaussian beam, modified Gaussian beam (Kubitscheck et al., 1998) and a “stationary” intensity distribution derived from experimental data obtained with immobile molecules. The modified Gaussian beam approximates the true illumination profile by
is the 3D laser intensity distribution at time t of the bleaching beam, and μ the fluorophore bleach rate constant. To check the influence of the intensity distribution on parameter estimation several configurations were used: stationary Gaussian beam, scanning Gaussian beam, modified Gaussian beam (Kubitscheck et al., 1998) and a “stationary” intensity distribution derived from experimental data obtained with immobile molecules. The modified Gaussian beam approximates the true illumination profile by
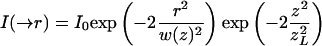 |
13 |
where
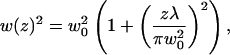 |
13a |
with λ being the wavelength of the laser and w0 its radial width at z = 0.
The characteristic recovery time is
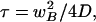 |
13b |
For diffusion to be neglected, the total bleaching time should be at least 15 times smaller than the characteristic recovery time (Meyvis et al., 1999). In simulations employing the “stationary” intensity distribution, the total bleaching time chosen was 240 ms. For slowly diffusing molecules (<0.1 μm2 s-1), this time is insignificant in comparison with the characteristic recovery time. In this situation, the stationary intensity distribution derived from the experimental data provides a more realistic bleaching profile.
After bleaching, the system behaves according to the classical diffusion equation
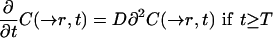 |
14 |
The following boundary conditions are assumed to be valid:
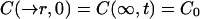 |
15 |
The radius of the bleach region (∼1 μm) is considered to be much smaller than the size of the compartment under study (i.e., ∼6 μm of radius in a HeLa cell nucleus).
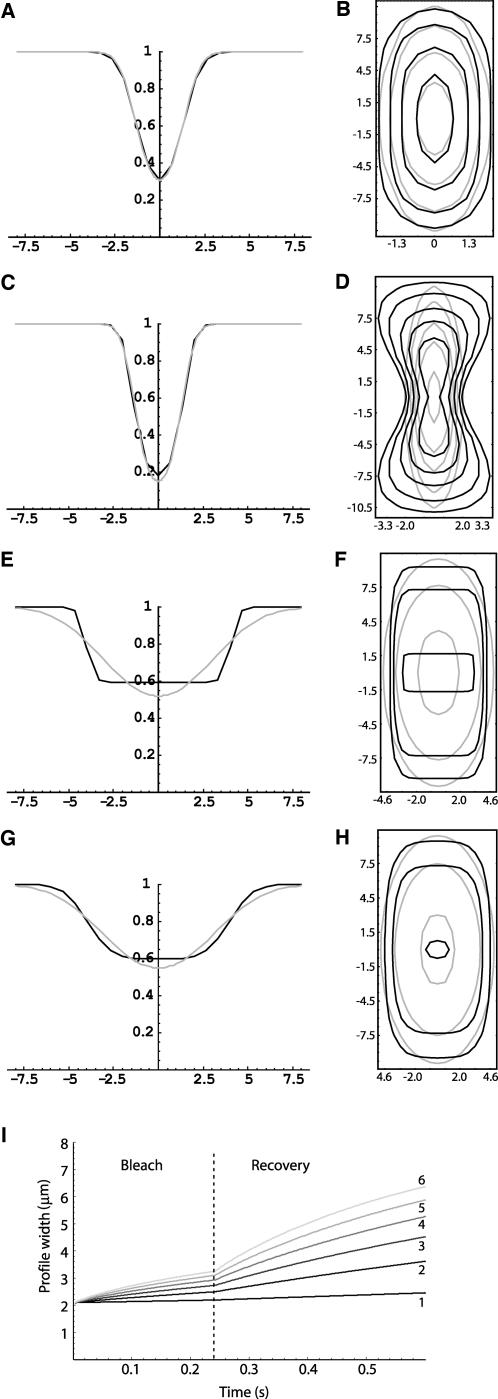
First, we used simulations to assess how the scanning laser beam shape influenced the postbleach profiles (for simulation details, see Appendix C). Results indicate that the bleach profile generated by a scanning Gaussian laser is still properly approximated by Eq. 6 if the size of the bleach ROI is smaller or comparable to the size of the Gaussian bleaching laser (Figure 4, A and B). A scanning beam deviating strongly from a Gaussian will generate accordingly a non-Gaussian bleach pattern (Figure 4, C and D). Together with the experimental measurements of the bleached volume, simulations further support the idea that, for our purposes, the bleaching laser beam can be considered to be approximately Gaussian, even when using a high NA lens.
Figure 4.
Gaussian approximation for simulated postbleach profiles in scanning configurations. Black line is the simulation result and the gray line the corresponding best fit with either Eq. 5 (for the radial profile) or Eq. 6 (for the contour plot). (A and B) The bleach ROI has approximately the same size as the bleaching laser beam. The resultant bleached volume is properly fitted by the exponential of a Gaussian. (C and D) If the scanning bleaching laser spot deviates strongly from a Gaussian, the corresponding bleached volume will also deviate from Eq. 6 (D), although the radial profile is still properly fitted by Eq. 5 (C). (E and F) The bleach ROI is 5 times larger than the bleaching Gaussian laser beam. Diffusion coefficient was set to 0 μm2 s-1. The postbleach profiles approach the uniform disk model (15) deviating strongly from the Gaussian approximation. (G and H) The same situation as before, but now D = 10 μm2 s-1. Due to diffusion, the postbleach profiles approach the Gaussian approximation. (I) Fluorescence profile width evolution during a FRAP experiment for different values of the diffusion coefficients: 1) 0.5, 2) 2.5, 3) 4.5, 4) 6.5, 5) 8.5, and 6) 10.5 μm2 s-1.
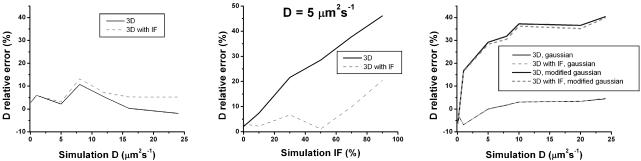
Figure 5 summarizes simulation results, comparing the performance of the two 3D variants of the new method for several configurations. For the stationary Gaussian configuration, in A, for the diffusion coefficients range tested (between 0.1 and 24 μm2 s-1), the maximum error in the estimates is ∼12% (for D = 10 μm2 s-1), decreasing for higher diffusion coefficients. For high diffusion coefficients (D > 5 μm2 s-1), the 3D variant has a better performance than the 3D variant with IF. In B, the influence of the immobile fraction is tested. It is shown that the estimates error grows continuously for the 3D variant, whereas for the 3D variant with IF the error is <10% until γ = 70%. In C, the influence of the 3D shape of the illuminating beam in scanning configurations is tested. If the beam deviates strongly from the Gaussian approximation, the error on the estimated D is much larger than in the Gaussian case. The error for the Gaussian beam is relatively small, as the bleach pattern generated by this beam could be effectively approximated by an exponential of a Gaussian function. This situation is similar to what was obtained experimentally, so we expect that when the new method is applied to a real situation its estimation performance is also similar.
Figure 5.
Comparison between the 3D and the 3D with IF variants for estimating diffusion coefficients and immobile fractions. Relative error is defined as the difference between simulated and estimated parameters divided by the simulated parameter. Underestimation of the parameter yields a positive value of the error whereas a negative value reflects an overestimation. (A and B) The 3D method (black line) and the 3D with IF method (dashed gray line) in stationary Gaussian laser configuration. (A) Plot of relative error on the estimated diffusion coefficient versus the simulated diffusion coefficient (immobile fraction was set to zero). (B) Plot of relative error on the estimated diffusion coefficient versus the simulated immobile fraction, considering a diffusion coefficient of 5 μm2 s-1. (C) Performance comparison in a Gaussian laser configuration and modified Gaussian laser configuration (immobile fraction was set to zero).
We also tested the method with the experimentally obtained bleach profile, for diffusion coefficients ranging from 0.01 μm2 s-1 to 0.1 μm2 s-1. Both 3D methods yielded similar estimates and diffusion coefficients were slightly overestimated by 5–9%.
Simulations were performed with a bleach ROI radius much larger (5×) than the radial extent of the bleaching laser. For the example depicted in Figure 4, G and H, the error on the diffusion coefficients estimate was 6%. One reason for this low error is that the fluorescence profiles, due to diffusion, rapidly approach a curve similar to Eq. 6, approaching the 3D method assumptions. For a bleach time small compared with characteristic diffusion time (i.e., for D <0.50 μm2 s-1, in our case), the bleaching profile obtained (Figure 4, E and F) is consistent with the predictions from the uniform disk model (Braeckmans et al., 2003). This should be the method of choice in these conditions.
Our previous methodology based on a model proposed by Axelrod et al., 1976, underestimated significantly diffusion coefficients, especially when simulating highly mobile molecules. This method yielded the best results for simulations performed under its assumptions (our unpublished data). This also shows that both numerical simulations and fitting routines were working properly.
Experimental Validation of the method
Assuming that dextrans in aqueous solution have a dynamical behavior similar to random walking particles, we estimate their diffusion coefficient (D) using the Stokes-Einstein equation:
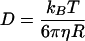 |
16 |
where kB is the Boltzmann constant, T the absolute temperature, η the viscosity of the medium, and R the hydrodynamic radius of the molecule. At 37°C (310 K), water has a viscosity of 0.6915 mPa.s and at 22°C (295 K) its viscosity is 0.955 mPa.s. Because dextran molecules are not perfect spheres, the theoretical values must be considered as an approximation of the expected diffusion rates.
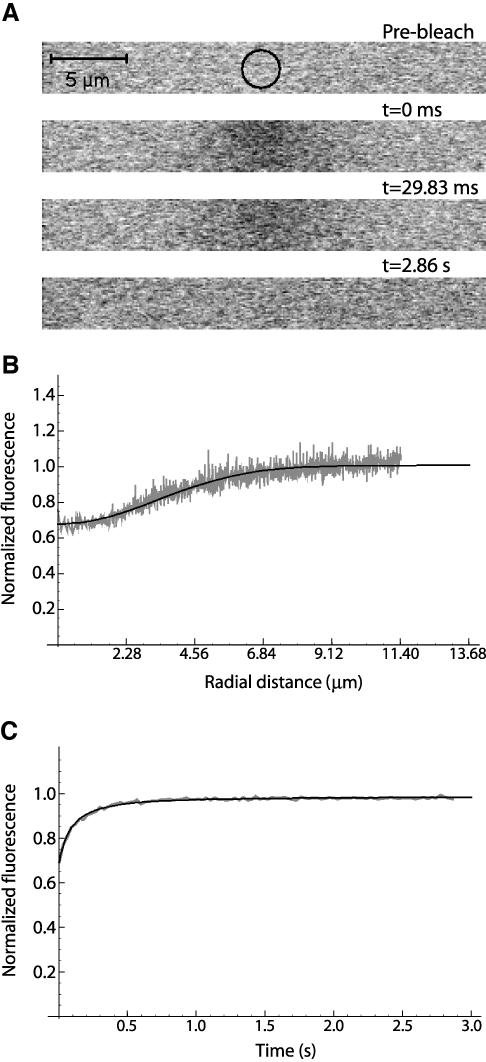
First, we used a method based on Axelrod formula (Axelrod et al., 1976) and that has been widely used to perform quantitative FRAP in cell biology (Phair and Misteli, 2000, Calapez et al., 2002; Wei et al., 2003, Shimi et al., 2004), to determine the diffusion coefficient of a FITC-labeled 20-kDa dextran in aqueous solution. The estimated value was 21 μm2 s-1, contrasting with an expected value of 97 μm2 s-1 (as calculated from the Stokes-Einstein equation). Clearly, this method is underestimating the diffusion coefficient of fast-moving molecules. Next, we estimated the diffusion coefficient values for each dextran by using Eq. 10 (3D variant), as sample thickness is of the order of ∼100 μm, larger than the axial extent of the bleached volume (∼9 μm). Nearly complete fluorescence recovery for all the FITC-dextrans was observed (>95% of the initial fluorescence), which is consistent with previous reports (Seksek et al., 1997). Table 1 shows, for dextrans of different sizes dissolved in water, the theoretically expected values of diffusion coefficients and the experimentally obtained values. As indicated by the ratio between expected and estimated values, the diffusion coefficients measured by the new FRAP model are always very close to the theoretical predictions (Figure 6).
Table 1.
Experimental data obtained for the mobility of dextrans in water
| 37°C
|
22°C
|
||||||
|---|---|---|---|---|---|---|---|
| Mw (kDa) | R (Å)a | Theoretical D (μm2s–1) | Experimental 3D D (μm2s–1) | Ratio | Teoretical D (μm2s–1) | Experimental 3D D (μm2s–1) | Ratio |
| 20 | 33 | 97.5 | 79.9 ± 5.2 | 0.82 | 70.6 | 63.43 ± 3.9 | 0.90 |
| 40 | 45 | 73 | 59.0 ± 4.6 | 0.80 | 52.9 | 51.5 ± 2.3 | 0.96 |
| 70 | 60 | 54.2 | 48.5 ± 2.6 | 0.89 | 39.3 | 43.7 ± 1.3 | 1.11 |
| 500 | 133b | 24.6 | 32.8 ± 1.9 | 1.33 | 17.8 | 23.2 ± 1.1 | 1.30 |
Ratio is the quotient between the experimentally estimated and the theoretically expected diffusion coefficients at each temperature. Diffusion coefficients were estimated using the 3D method.
Values reported in Sigma-Aldrich; supplier data online
This value was extrapolated from the radius values reported in supplier's data using the relation R = C + A M1/3, where C = –1.81 Å and A = 0.19
Figure 6.
FITC-labeled dextrans of 70 kDa were dissolved in water (200 μg/ml) and analyzed by FRAP at 22°C. (A) Prebleach image, first scan after bleach (t = 0.0 ms), second scan after bleach (t = 29.8 ms), and the last scan (t = 2.86 s). (B) Fluorescence profile from the first postbleach scan (gray) and the corresponding best fit (black). (C) FRAP recovery curve (gray) with the corresponding fit with 3D variant (black).
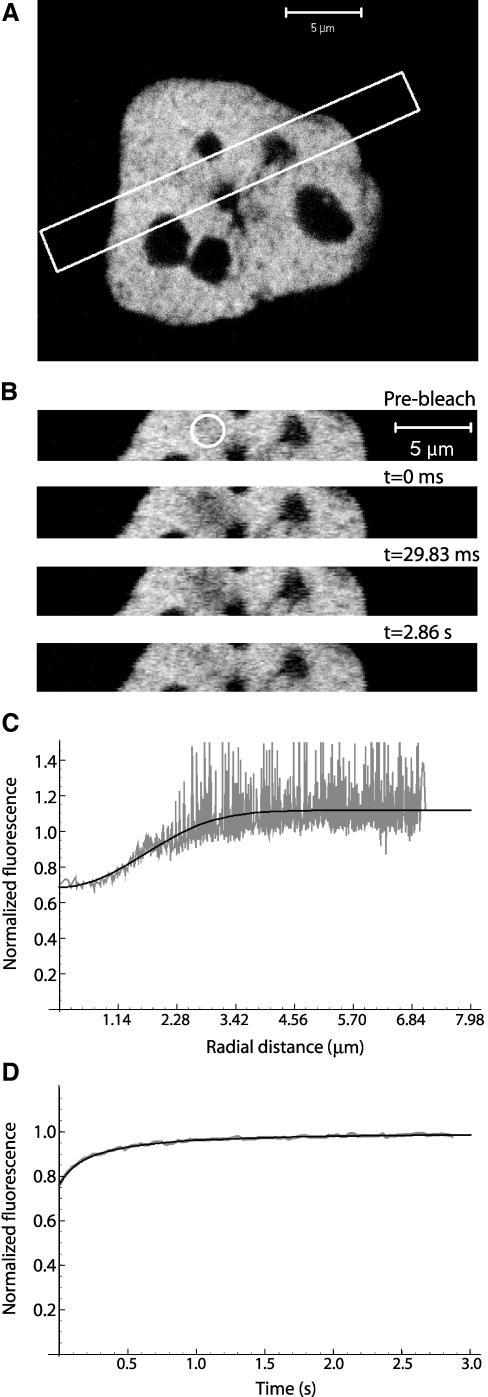
Having experimentally validated the model, the different dextrans were then microinjected into the nucleus of HeLa cells. As the average thickness of a HeLa cell in the z-direction (∼5 μm) is almost 2 times smaller than the axial extent of the bleached volume, the 2D variant of the model was selected (Eq. 9). The diffusion coefficient values and IF values estimated in cells maintained at either 37 or 22°C are shown in Table 2 (also see Figure 7). As can be seen from the ratio between values estimated in water and in cell nucleus at 22 or 37°C, the mobility impairment inside the nucleus is higher for the higher molecular weight dextrans.
Table 2.
Experimental data obtained for the mobility of dextrans in HeLa cell nucleus
| 37°C
|
22°C
|
|||||
|---|---|---|---|---|---|---|
| Mw (kDa) | Experimental 2D (μm2s–1) | Ratio Dwater/Dnuc | IF | Experimental 2D (μm2s–1) | Ratio Dwater/Dnuc | IF |
| 20 | 11.0 ± 1.8 | 7.3 | 0.9 ± 0.2 | 10.8 ± 2.5 | 5.9 | 0.0 ± 0.4 |
| 40 | 10.5 ± 1.7 | 5.6 | 0.6 ± 0.2 | 6.5 ± 1.5 | 7.8 | 0.0 ± 0.4 |
| 70 | 5.9 ± 0.7 | 8.2 | 0.0 ± 0.3 | 4.6 ± 1.1 | 9.5 | 4.6 ± 0.5 |
| 500 | 1.7 ± 0.3 | 19.3 | 0.0 ± 1.0 | 1.5 ± 0.3 | 15.5 | 5.5 ± 1.3 |
Diffusion coefficients were estimated using the 2D method. Ratio is the quotient between the diffusion coefficients measured in water and in the nucleus at each temperature. IF is the immobile fraction expressed in percentage.
Figure 7.
FITC-labeled dextrans of 70 kDa were dissolved in water (200 μg/ml), microinjected into HeLa cell nuclei and analyzed by FRAP at 22°C. (A) The white rectangle shows the zone selected for scanning during the experiment. A smaller observation zone allows quicker imaging, enhancing the temporal resolution of a FRAP experiment. (B) Prebleach image, first scan after bleach (t = 0.0 ms), second scan after bleach (t = 29.8 ms), and the last scan (t = 2.86 s). (C) Fluorescence profile from the first postbleach scan (gray) and the corresponding best fit (black). (D) FRAP recovery curve (gray) with the corresponding fit with 2D variant (black).
Finally, using the 2D variant of the new method we estimated a diffusion coefficient of 33.3 ± 3.6 μm2 s-1 for GFP in the HeLa cell nucleus, with an IF of 1.0 ± 0.1%. This is consistent with recently reported values of 22 ± 2 μm2 s-1 at 22°C (with an of IF = 4.0 ± 0.3%) (Potma et al., 2001) and 32 ± 4 μm2 s-1 at 37°C (Beaudouin, 2003).
DISCUSSION
A new approximate FRAP model for use on any standard CLSM is presented here. The novelty of this method is that it takes into account diffusion during bleach and is valid for use with objective lenses with high NA. The method can be readily applied by anyone familiar with a CLSM, as the mathematical expressions are straightforwardly programmed with standard fitting programs.
The GFP is a 28-kDa protein with a Stokes' radius of 2.35 nm (Ribbeck and Görlich, 2001). This corresponds to an expected diffusion coefficient in water at 37°C of 139.2 μm2 s-1. Confocal spot photobleaching recovery measurements performed with a specifically modified microscopic system yielded GFP diffusion coefficients of 87 ± 3 μm2 s-1 in aqueous solution and 24 ± 2 μm2 s-1 in the cytoplasm of Dictyostelium discoideum cells kept at room temperature (Potma et al., 2001). In our previous work, we used a standard CLSM to perform quantitative FRAP, and we estimated an apparent diffusion coefficient of 5 μm2 s-1 for GFP in the nucleus of HeLa cells (Calapez et al., 2002). We believe that this value was significantly underestimated because the mathematical model applied considered that fluorescence recovery during the bleach phase is negligible, an assumption that is not met by the experimental conditions in the case of highly mobile molecules (Figure 3). Using the same CLSM instrument and the new FRAP model, we estimate a diffusion coefficient of 33.3 ± 3.6 μm2 s-1 for GFP in the nucleus of HeLa cells maintained at 37°C.
To develop a more accurate practical approach that can be readily applied by cell biologists interested in performing quantitative FRAP analysis with a standard CLSM, we first analyzed experimentally the fluorescence profile generated by bleaching a region of interest with a scanning laser beam. FITC-labeled dextrans were immobilized in a polyacrylamide gel, and the bleached volume was imaged along the z-direction. The results show that, for bleaching spots up to ∼1 μm in radius, the observed fluorescence profiles can be effectively approximated by the theoretical models that consider bleaching generated by a stationary Gaussian laser. Our data thus validate the application of the Gaussian approximation to postbleach radial fluorescence profiles generated with the CLSM, as assumed previously (Calapez et al., 2002). However, our results further indicate that in order to apply the Gaussian approximation the bleach region should not exceed ∼1 μm in radius (the maximum radius for which fitting is valid depends on the numerical aperture of the objective lens). For much larger bleach regions, a better approximation is the uniform disk profile, as predicted from the model proposed by Braeckmans et al., 2003.
To date, most FRAP models assume that both bleaching and image acquisition are sufficiently fast to avoid diffusion during those periods (Braeckmans et al., 2003). As a rule, for diffusion to be neglected, the total bleaching time should be at least 15 times smaller than the recovery time (Meyvis et al., 1999). If we consider a molecule that diffuses at 0.5 μm2 s-1 and a bleaching region with a radius of 1 μm, then the characteristic recovery time will be ∼0.5 s. The bleaching time should therefore be <33 ms. In practice, most cell biological FRAP applications using a similar bleach ROI size use bleach periods of ∼100 ms or longer (Phair and Misteli, 2000; Shimi et al., 2004). One may therefore predict that in most FRAP experiments the assumption that negligible fluorescence recovery occurs during bleaching is not correct.
A direct demonstration that before the first postbleach image is acquired significant diffusion takes place (namely, for a 40-kDa dextran) is depicted in Figure 3. Diffusion during bleach is obviously less important for slower moving molecules (i.e., a 500-kDa dextran).
Using the approximations mentioned above. it was possible to derive analytical formulas for the normalized fluorescence recovery curve. The three-dimensional correction is important for measurements in bulk solutions. In cellular measurements, it will contribute significantly only if the cell thickness is larger than the axial extent of the bleach volume. In a thin sample, the 2D variant should be used instead.
For simultaneously high values of the immobile fraction and of the diffusion coefficient the capability of the 3D (or 2D) method to correctly estimate these parameters is reduced. This happens because the mobile fraction concentration profile is not directly identified, being wM estimated from the fluorescence profile of the mixture of the mobile and immobile fractions. Thus, a higher level of immobilized molecules leads to an underestimation of the mobile fraction profile radius, and consequently, to an underestimation of the diffusion coefficient. A way to circumvent this problem was devised, by making a fitting procedure (with Eq. 10) in two steps. The first run would be used to estimate the immobile fraction only, computing then the mobile fraction fluorescence profile (Eq. 11), and in the second run corrected profile width values would be used. It is important to highlight that the approach describe here requires a new calibration every time there is a change in bleaching disk radius, laser beam power, or fluorophore.
To be sure that these expressions were adequate for analysis of real FRAP experiments, we generated recovery curves from the simulations and fitted those curves with the formula. The estimated diffusion coefficients and immobile fractions were always close to the parameters used in the simulations (Figure 5).
The effect of noise was tested by generating several curves with the same level of Gaussian noise added to a simulated recovery curve. This procedure led to the conclusion that noise has a relevant role in the quality of the estimates, introducing some variability in the estimation. This was especially noticeable when simulating molecules with high diffusion coefficients. The best way to improve the estimation was to average the highest number possible of experiments.
As a first biological application, the new FRAP method was used to compare the diffusion rates of different size macromolecules in aqueous solution and in the nucleus of living HeLa cells. FITC-dextrans were either directly imaged in solution or microinjected into the nucleus. A rather homogeneous fluorophore distribution was observed, suggesting that the dextrans spread freely throughout both the aqueous sample and the nucleoplasm. In aqueous solution, the ratios between the diffusion coefficient values theoretically expected from the Stokes-Einstein equation and those estimated by FRAP both at 37 and 22°C were close to 1. As expected, in the nucleoplasm the diffusion coefficient values decreased relative to water. Noteworthy, higher molecular weight dextrans were proportionally more retarded in the nucleoplasm, at 37 and 22°C, than smaller molecular weight molecules (Table 2). This means that nucleoplasm deviates from an ideal liquid behavior, with the effective viscosity increasing with the size of the molecules. A similar observation was reported for FITC-dextrans injected into the eye vitreous, but not for dextrans diffusing in cystic fibrosis sputum (Braeckmans et al., 2003), where the decrease in diffusion coefficient values of the FITC-dextrans seemed rather independent of their size. In contrast with the aqueous sputum, the vitreous is composed of a meshwork formed by polymers of hyaluronic acid. Most probably, this meshwork causes a sterical hindrance that is stronger for larger molecules (Braeckmans et al., 2003). A similar situation is likely to occur in the nucleoplasm, where sterical hindrance is caused by the meshwork composed of chromatin and nonchromatin nuclear components. Most interestingly, the sterical hindrance effect inside the nucleus is similar at 37 and at 22°C (1.7± 0.3 and 1.5 ± 0.3 μm2 s-1 for 500-kDa dextrans). Yet, decreasing the temperature from 37 to 22°C reduces the expected mobility rate of the dextrans in aqueous solution by ∼30% (from 32.8 ± 1.9 to 23.2 ± 1.1 μm2 s-1 for 500-kDa dextrans). We have previously observed that when cells are depleted of ATP or incubated at 22°C, messenger ribonucleoprotein (mRNP) particles show significantly reduced mobility rates in the nucleus, whereas large-molecular-weight dextrans are not much affected (Calapez et al., 2002). The results reported here reinforce the view that spatial constraints to diffusion of dextrans inside the nucleus are insensitive to temperature and therefore energy independent. In contrast, energy-dependent processes are possibly involved in facilitating the diffusion of mRNP complexes in transit to the cytoplasm (Calapez et al., 2002; Carmo-Fonseca et al., 2002).
Inside the cell, binding events are expected to slow down the diffusion dynamics of macromolecules. In the case that the binding reaction is faster than the typical times involved in the diffusive process, an effective diffusion coefficient can be defined (Crank, 1975). Such effective diffusion coefficient integrates the absolute diffusion coefficient and the rates of association and dissociation of the macromolecule. However, further work is needed to address the complex interplay between macromolecular diffusion and binding events that takes place in the living cell (Phair et al., 2004; Sprague et al., 2004).
Acknowledgments
We are particularly indebted to Henrique Pereira who provided the original idea for the present work and encouraged further research. We also are thankful to José Rino (Institute of Molecular Medicine, University of Lisbon, Lisbon, Portugal) for stimulating discussions and to Gonçalo Cabrita (Instituto Superior Técnico, Universidade Técnica de Lisboa) and Rita Lemos (Instituto de Technologia Química e Biológica, Universdade Nova de Lisboa) for help in gel filtration of dextrans. We further acknowledge Inês Condado (IMM, Universidade de Lisboa) for help with tissue culture and Alexandre Calapez (IST, Universdade Técnica de Lisboa) for the image processing programs. Dr. Ernst H.K. Stelzer (European Molecular Biology Laboratory, Heidelberg, Germany) is gratefully acknowledged for fruitful discussions on this work. J.B. is supported by a fellowship from Fundação para a Ciência e Tecnologia, Portugal (BD/21605/99). This work was supported by grants from Fundação para a Ciência e Tecnologia (POCTI/36511/BME/2000) and the European Commission (QLG2-CT-2001-01554 and LSHG-CT-2003-503259).
Appendix
A. Immobile Fraction: Fluorescence Inside a Disk of Radius wS
As pointed out in previous works, integrating the fluorescence signal over a set of pixels improves the signal to noise ratio (Klonis et al., 2002; Braeckmans et al., 2003). Accordingly, we averaged our fluorescence signal over a circular region of radius wS from the images taken at the focal plane (z = 0). wS in general can be different from wB. The normalized fluorescence is thus
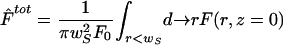 |
A.17 |
where F(r, z) is the fluorescence at the point (r, z) and F0 is the prebleach fluorescence.
For immobile molecules, if bleaching is performed on a small bleaching region (i.e., when fitting with Eq. 3 is valid) we have
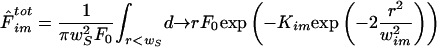 |
A.17a |
By expanding the exponential as a series yields And finally, by performing the integrations for each term of the series, we get Eq. 4 as a result.
B. Mobile Fraction: Fluorescence Recovery Inside a Disk of Radius wS
In any fluorescence microscope, an image created by an object is the convolution of the fluorophore concentration and the point spread function (PSF) of the system (Berk et al., 1993). If the system behaves according to the classical diffusion equation, we have from the convolution properties
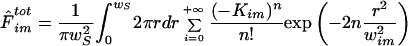 |
A.17b |
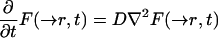 |
A.18 |
Using as initial condition the approximation suggested by Eq. 7
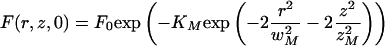 |
A.19 |
where t = 0 is the time of the first postbleach scan of the bleached zone. Considering the medium as much larger than the bleached spot, the boundary condition is
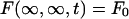 |
A.20 |
The solution to a similar partial differential equation is found elsewhere (Blonk et al., 1993), yielding
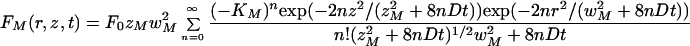 |
A.21 |
Applying Eq. A.17, the normalized fluorescence recovery inside a region of radius wS is thus given by
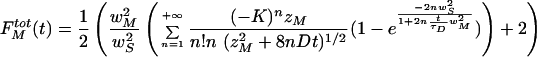 |
A.22 |
where
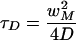 |
A.22a |
Because during a FRAP experiment we are acquiring 2D images, it is not possible to measure zM directly. We estimated this parameter by using an empirical approach. From the simulations, we observed that the broadening of the fluorescence profile during bleaching is much slower than after its end (Figure 4I). Thus, a rough estimate for the radial fluorescence profile width immediately after bleaching is given by wim (and zim for the axial profile).
After bleaching, by the time the bleached region is imaged the expected theoretical profile width should be approximately (similarly to Jain et al., 1990):
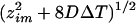 |
A.23 |
where ΔT is the difference between total bleach time and the bleach phase (Fig. 1). As expected, this formula states that the speed by which profiles spread is higher for faster diffusive rates. To correct for slight mismatches between the real width and the estimated width in the axial direction, we multiply Eq.A.7 by the ratio between real (wM) and estimated values in the radial direction. So zM becomes:
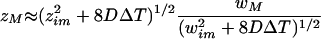 |
A.24 |
Note that in this expression we use experimentally measurable parameters: wim, zim (both determined from bleaching experiments with immobilized dextrans) and zM (determined from the first image after bleaching).
C. Numerical Simulations
To obtain simulated recovery curves, Eqs. 12 and 14 were solved numerically in the three spatial dimensions (x,y,z) plus time, by a finite-differencing scheme implemented in Mathematica 4.0 (Wolfram Research). The user controls fluorophore and bleaching beam parameters (the beam width in the axial and radial direction, the laser intensity, the size of the bleach ROI, the number of lines, the time taken per line, the start and the end points of the scanning lines, and the bleach rate constant), the time of bleach phase, the dynamical properties of the molecule (D, γ), acquisition parameters (time between images, the number of bleach iterations, the start of imaging phase and the total duration of the simulated FRAP experiment) and numerical parameters (the grid points number in each spatiotemporal direction, and the location of the boundaries). The bleach region was located at the center of the volume, and the boundaries were set to a distance at least two times larger than the bleaching laser width in that direction. The grid points number was selected to obtain stable numerical solutions.
Simulations parameters chosen are close to those of a real FRAP experiment (see Materials and Methods). For details, see below.
The time between images was set to 30 ms (an image takes 28 ms and the next image starts to be scanned after 2 ms), and there is a time delay between the end of bleach and the start of imaging of 2 ms. The bleached zone is, on average, in the center of the imaging area, so the first postbleach image of the bleached area starts 28/2 + 2 ms = 16 ms after the end of bleaching.
In the scanning laser configurations, the time taken for each line is 1 ms. The bleach region is a circle of 1.19-μm radius (containing 20 scanning lines). This region is scanned for two iterations, for a total bleaching phase time of 40 ms. Although in practice we use longer bleach times, this is still longer than the characteristic recovery time of fast diffusing molecules. The choice of smaller times in these simulations is due to computer memory constraints.
Laser parameters were chosen to get a bleach immobile profile similar (in terms of spatial extension) to the experimentally obtained for dextrans immobilized in a polyacrylamide gel.
Usually, the set of pixels from which fluorescence recovery is computed is the same as the bleach ROI defined. According to the new method its size is independent of bleach ROI defined, changing its size had no significant influence on the results (our unpublished data). If the radius of the monitoring zone is increased the recovery curve becomes sigmoidal, which was also predicted from the analytical expression.
Gaussian noise was added to the simulated recovery curves and fluorescence profiles. The fluorescence profiles were fitted with either Eq. 3 (for immobile molecules) or Eq. 6 (for mobile molecules). The recovery curves were fitted with Eq. 10 by using NonlinearRegress, yielding estimates for KM, τD, and γ.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–06–0496. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–06–0496.
References
- Almeida, F., Saffrich, R., Ansorge, W., and Carmo-Fonseca, M. (1998). Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142, 899-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod, D., Koppel, D.E., Schlessinger, J., Elson, E., and Webb, W.W. (1976). Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 16, 1055-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin, J. (2003). Structural and molecular dynamics of nuclear proteins revealed by fluorescence microscopy. Ph.D. dissertation. Available at http://library.embl-heidelberg.de/pdf/beaudouin.pdf.
- Berk, D.A., Yuan, F., Leunig, M., and Jain, R.K. (1993). Fluorescence photobleaching with spatial Fourier analysis: measurement of diffusion of lightscattering media. Biophys J. 65, 2428-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonk, J.C.G., Don, A., Van Aalst, H., and Birmingham, J.J. (1993). Fluorescence photobleaching recovery in the confocal scanning light microscope. J. Microsc. 169, 363-374. [Google Scholar]
- Braeckmans, K., Peeters, L., Sanders, N.N., De Smedt, S.C., and Demeester, J. (2003). Three-dimensional fluorescence recovery after photobleaching with the confocal scanning laser microscope. Biophys J. 85, 2240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calapez, A., Pereira, H.M., Calado, A., Braga, J., Rino, J., Carvalho, C., Tavanez, J.P., Wahle, E., Rosa, A.C., and Carmo-Fonseca, M. (2002). The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J. Cell Biol. 159, 795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Platani, M., and Swedlow, J.R. (2002). Macromolecular mobility inside the cell nucleus. Trends Cell Biol. 12, 491-495. [DOI] [PubMed] [Google Scholar]
- Carrero, G., McDonald, D., Crawford, E., de Vries, G., and Hendze, M.J. (2003). Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods 29, 14-28. [DOI] [PubMed] [Google Scholar]
- Crank, J. (1975). The Mathematics of Diffusion, 2nd ed. Clarendon Press, Oxford, United Kingdom.
- Cutts, L.S., Roberts, P.A., Adler, J., Davies, M.C., and Melia, C.D. (1995). Determination of localized diffusion coefficients in gels using confocal scanning laser microscopy. J Microsc. 180, 131-139. [Google Scholar]
- Edidin, M., Zagyansky, Y., and Lardner, T.J. (1976). Measurement of membrane protein lateral diffusion in single cells. Science 191, 466-468. [DOI] [PubMed] [Google Scholar]
- Jain, R.K., Stock, R.J., Chary, S.R., and Rueter, M. (1990). Convection and diffusion measurements using fluorescence recovery after photobleaching and video image analysis: in vitro calibration and assessment. Microvasc. Res. 39, 77-93. [DOI] [PubMed] [Google Scholar]
- Klonis, N., Rug, M., Harper, I., Wickham, M., Cowman, A., and Tilley, L. (2002). Fluorescence photobleaching analysis for the study of cellular dynamics. Eur. Biophys. J. 31, 36-51. [DOI] [PubMed] [Google Scholar]
- Kubitscheck, U., Wedekind, P., and Peters, R. (1994). Lateral diffusion measurement at high spatial resolution by scanning microphotolysis in a confocal microscope. Biophys J. 67, 948-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitscheck, U., Wedekind, P., and Peters, R. (1998). Three-dimensional diffusion measurements by scanning microphotolysis. J. Microsc. 192, 126-138. [DOI] [PubMed] [Google Scholar]
- Meyvis, T.K.L., De Smedt, S.C., Van Oostveldt, P., and Demeester, J. (1999). Fluorescent recovery after photobleaching: a versatile tool for mobility and interaction measurements in pharmaceutical research. Pharm. Res. 16, 1153-1162. [DOI] [PubMed] [Google Scholar]
- Peters, R., and Kubitscheck, U. (1999). Scanning microphotolysis: three-dimensional diffusion measurement and optical single-transporter recording. Methods 18, 508-517. [DOI] [PubMed] [Google Scholar]
- Peters, R., Peters, J., Tews, K.H., and Bahr, W. (1974). A microfluorimetric study of translational diffusion in erythrocyte membranes. Biochem. Biophys. Acta 367, 282-294. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., Scaffidi, P., Elbi, C., Vecerová, J., Dey, A., Ozato, K., Brown, D.T., Hager, G., Bustin, M., and Misteli, T. (2004). Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24, 6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potma, E.O., de Boeij, W.P., Bosgraaf, L., Roelofs, J., van Haastert, P.J.M., and Wiersma, D.A. (2001). Reduced protein diffusion rate by cytoskeleton in vegetative and polarized Dyctyostelium cells. Biophys. J. 81, 2010-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits, E.A., and Neefjes, J.J. (2001). From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3, E145-147. [DOI] [PubMed] [Google Scholar]
- Ribbeck, K., and Görlich, D. (2001). Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksek, O., Biwersi, J., and Verkman, A.S. (1997). Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 138, 131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi, T., Koujin, T., Segura-Totten, M., Wilson, K.L., Haraguchi, T., and Hiraoka, Y. (2004). Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J. Struct. Biol. 147, 31-41. [DOI] [PubMed] [Google Scholar]
- Sheppard, C.J.R., and Torok, P. (1997). Effects of specimen refractive index on confocal imaging. J. Microsc. 185, 366-374. [Google Scholar]
- Soumpasis, D.M. (1983). Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, B.L., Pego, R.L., Stavreva, D.A., and McNally, J.G. (2004). Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86, 3473-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay, T.T., and Jacobson, K.A. (1991). Spatial Fourier analysis of video photobleaching measurements–Principles and optimization. Biophys. J. 60, 360-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind, P., Kubitscheck, U., Heinrich, O., and Peters, R. (1996). Line-scanning microphotolysis for diffraction-limited measurements of lateral diffusion. Biophys. J. 71, 1621-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X., Henke, V.G., Strübing, C., Brown, E.B., and Clapham, D.E. (2003). Real-time imaging of nuclear permeation by EGFP in single intact cells. Biophys. J. 84, 1317-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J., and Stelzer, E. (1999). Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 9, 61-65. 3 [DOI] [PubMed] [Google Scholar]