Abstract
The Mre11/Rad50/Nbs1 complex is involved in many aspects of chromosome metabolism. Aberrant function of the complex is associated with defects in the DNA checkpoint, double-strand break repair, meiosis, and telomere maintenance. In this article, we report the consequences of Mre11 dysfunction for the stability of mitotic and meiotic chromosomes in Arabidopsis thaliana. Although plants homozygous for a T-DNA insertion in a conserved region of the MRE11 gene are viable, they exhibit growth defects and are infertile. Analysis of mitotic chromosomes prepared from the mutant plants revealed abundant dicentric chromosomes and chromosomal fragments. Fluorescence in situ hybridization showed that anaphase bridges are often formed by homologous chromosome arms. The frequency of chromosome fusions was not reduced in mre11 ku70 double mutants, suggesting that plants possess DNA end-joining activities independent of the Ku70/80 and Mre11 complexes. Cytogenetic examination of pollen mother cells revealed massive chromosome fragmentation and the absence of synapsis in the initial stages of meiosis. The fragmentation was substantially suppressed in mre11 spo11-1 double mutants, indicating that Mre11 is required for repair but not for the induction of Spo11-dependent meiotic DNA breaks in Arabidopsis.
INTRODUCTION
Genomes are continuously exposed to a variety of DNA-damaging or modifying agents from the external environment or from intracellular processes. Because alterations to genetic information incurred by DNA damage can adversely affect cellular metabolism, cells possess elaborate mechanisms that serve to rapidly detect and accurately repair DNA lesions. DNA double-strand breaks (DSBs) are particularly deleterious because they can lead to the loss of extensive parts of chromosomes. The two major DSB-repair pathways present in eukaryotes are nonhomologous end joining (NHEJ) and homologous recombination (HR) (Christmann et al., 2003; Scharer, 2003). NHEJ mediates the ligation of two broken DNA ends, whereas HR uses genetic information from a homologous DNA molecule for error-free repair. The response to DNA damage must be coordinated because the incorrect repair of DSBs can lead to chromosomal instabilities.
Evidence gathered in recent years demonstrates that the Mre11 protein complex, which is composed of Mre11, Rad50, and Nbs1 (Xrs2 in Saccharomyces cerevisiae), plays a central role in the initial stages of DSB repair and the DNA checkpoint (reviewed in Connelly and Leach, 2002; D'Amours and Jackson, 2002). Although the exact molecular mechanisms underlying the function of the Mre11 complex have yet to be determined, it is known that the Nbs1 protein is phosphorylated in response to the presence of DSBs and is thus linked to the signaling of DNA damage (Lim et al., 2000). In addition, biochemical analyses have shown that the Mre11 protein possesses double-stranded and single-stranded nuclease activities, which suggests a role in the resection of DSBs (reviewed in D'Amours and Jackson, 2002). Scanning force microscopy indicates that the human Mre11 and Rad50 proteins can form multimeric complexes at DNA ends and tether linear DNA molecules together (de Jager et al., 2001). This observation implies a structural role of the Mre11 complex in the positioning of DNA molecules during DSB repair.
Mre11 was originally identified in a screen for genes that are essential for meiosis in S. cerevisiae (Ajimura et al., 1993). Meiosis is characterized by two successive rounds of cell division, during which homologous chromosomes are segregated in meiosis I and sister chromatids in meiosis II. The segregation of homologous chromosomes during meiosis I is preceded by HR, which leads to a physical link between homologous chromosomes via chiasmata. Chiasmata formation is essential for the faithful segregation of homologous chromosomes at meiosis I (reviewed in Page and Hawley, 2003; Petronczki et al., 2003). HR is initiated by the formation of programmed DSBs, a process catalyzed by the Spo11 protein (Keeney et al., 1997). The DSBs are then subjected to 5′-3′ resection and repaired through recombination with the homologous chromosome. Mre11 is required for both DSB induction and end processing in S. cerevisiae (Nairz and Klein, 1997; Usui et al., 1998).
The Mre11 complex has also been implicated in other aspects of chromosome maintenance. Studies in S. cerevisiae suggest the direct involvement of the Mre11 complex in at least two DSB repair pathways: NHEJ, which utilizes sequence microhomologies (Ma et al., 2003), and break-induced replication, an HR mechanism that participates in the Rad51-independent repair of DSBs (Signon et al., 2001). Time-course analysis of the distribution of induced DSBs in human nuclei suggests that the Mre11 complex is involved in the clustering of chromosomal domains containing DSBs during G1 phase (Aten et al., 2004). In addition, Mre11 has a crucial function during DNA replication by preventing the accumulation of DSBs, which arise as a consequence of stalled replication forks (Pichierri and Franchitto, 2004). The Mre11 complex also acts as a positive regulator of telomerase on yeast telomeres (Dubrana et al., 2001).
Hypomorphic mutations in the human MRE11 and NBS1 genes lead to an Ataxia telangiectasia-like disorder and Nijmegen breakage syndrome, respectively (Carney et al., 1998; Stewart et al., 1999). These genetic disorders are associated with genome instability and a high incidence of cancer. Deciphering Mre11 function in vertebrates is hampered by the fact that null mutations in any component of the Mre11 complex are lethal (Xiao and Weaver, 1997; Luo et al., 1999; Yamaguchi-Iwai et al., 1999; Zhu et al., 2001).
Several viable mutant lines deficient for the Mre11 and Rad50 proteins have recently been reported in Arabidopsis thaliana. These mutants exhibit an increased sensitivity to DNA damage, aberrant telomere maintenance, and infertility (Gallego et al., 2001; Gallego and White, 2001; Bundock and Hooykaas, 2002), thus confirming evolutionary conserved roles of the Mre11 complex in genome maintenance. These studies did not, however, directly address the impact of the mutations on genome stability. In this article, we present an extensive molecular-cytogenetic analysis of an Arabidopsis mutant line carrying a T-DNA insertion in a conserved region of the MRE11 gene. We show that somatic cells in this mutant exhibit severe chromosomal aberrations, which may be the primary cause of the developmental defects observed in mre11 plants. Furthermore, an analysis of male meiocytes from mre11 plants revealed massive genome fragmentation in the presence of a functional SPO11-1 gene. This result highlights interesting differences in the function of Mre11 in the initial stages of meiotic recombination between yeast and Arabidopsis.
RESULTS
Molecular and Morphological Characterization of the mre11-3 Mutant
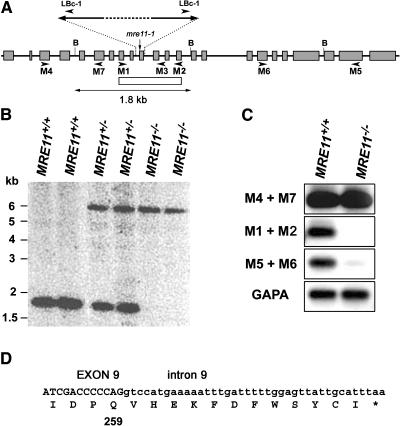
To examine the role of Mre11 in genome maintenance, we obtained an Arabidopsis line with a disruption of the MRE11 gene from the SIGnAL collection of Arabidopsis T-DNA insertion lines (Alonso et al., 2003). The insertion was annotated within the 10th intron with the left border oriented toward the 3′ end of the MRE11 gene (line SALK_054418; Figure 1A). Plants homozygous and heterozygous for the T-DNA insertion were identified by multiplex PCR with primers M1, M3, and LBc-1. The T-DNA disruption of the MRE11 gene was further verified by DNA gel blot hybridization with an MRE11 probe spanning the insertion site (Figure 1B). DNA gel blot analysis with a probe derived from the T-DNA indicated that the disruption is composed of several adjacent T-DNA molecules (data not shown). To precisely map the border between the T-DNA insertion and the 5′ end of the gene, we performed PCR reactions with primer M1 in combination with primers specific for the left border or right border of the T-DNA. A specific PCR product was obtained with primers M1 and LBc-1 (data not shown). Sequence analysis of the PCR product revealed that the other border of the insertion site is located in the 9th intron, which results in a deletion of the entire 10th exon of the MRE11 gene (Figure 1A). We refer to this mutation, which disrupts the gene in the region coding for the conserved phosphoesterase motif IV, as the mre11-3 allele.
Figure 1.
Molecular Characterization of the mre11-3 Allele.
(A) Schematic representation of the mre11-3 allele with the T-DNA disruption. T-DNA insertion sites and orientation of left borders are indicated. The gray boxes represent exons, and the white box defines the region used as a probe for DNA gel blot hybridization. The positions of the primers (arrowheads) and BglII restriction sites (B) are marked. The T-DNA insertion in the mre11-1 allele is indicated by an arrow.
(B) DNA gel blot hybridization of BglII-digested genomic DNA with the MRE11 probe results in the production of ∼1.8-kb and ∼6-kb bands from the wild-type and mre11-3 alleles, respectively.
(C) Analysis of MRE11 expression by semiquantitative RT-PCR. Total RNAs prepared from flowers were subjected to cDNA synthesis by reverse transcription. MRE11 transcripts were amplified by 25 cycles of PCR with indicated sets of primers. Control RT-PCR with primers specific for glyceraldehyde-3-phosphate dehydrogenase A (GAPA) were performed for 20 cycles. The PCR products were detected by DNA gel blot hybridization.
(D) The DNA sequence of the junction between the MRE11 cDNA and T-DNA insertion site obtained by RT-PCR. A translation of the predicted protein is included.
Expression analysis of the MRE11 gene in plants homozygous for the mre11-3 allele by semiquantitative RT-PCR demonstrated the absence of an mRNA species, which could have arisen by the splicing out of the entire T-DNA insertion site (Figure 1C, primers M1 + M2). Very weak expression of an MRE11 mRNA downstream of the insertion is apparently initiated from a promoter within the T-DNA (Figure 1C, primers M5 + M6). The level of mRNA expression from the 5′ end of the gene in mre11-3 plants was, however, comparable to the expression of the full-length mRNA in wild-type cells (Figure 1C, primers M4 + M7). Sequence analysis of the cDNA species, amplified by RT-PCR with primers M1 and LBc-1, predicts that the mre11-3 allele produces a truncated protein comprised of the first 259 out of 720 amino acids of the full-length Mre11 protein as a TAA stop codon is encountered in the 9th intron (Figure 1D). We expect that the function of this truncated Mre11 protein is severely compromised, if not completely aborted.
MRE11+/− plants were indistinguishable from wild-type plants, and their self-fertilization produced homozygous mutants in the expected 1:3 ratio (χ2 = 0.29; n = 42). Although mre11-3 mutants can be germinated and grown in soil, they exhibit a variety of developmental defects and are infertile. The severity of defects varied between individual plants. Some mutants were somewhat smaller than their wild-type siblings and had slightly wrinkled leaves (Figure 2A), whereas others displayed a dwarf phenotype, which is characterized by miniature rosettes with small malformed leaves (Figures 2A and 2B). The leaves were asymmetric and lobed, usually having rough sectors on their surface that were rarely accompanied with yellowish necrotic fields (Figures 2D and 2E). Scanning electron microscopy showed that the shoot apical meristem was opened and up to 10 times larger in mre11-3 mutants than in wild-type plants, in which the apical dome is almost completely enclosed by emerging leaf primordia (Figures 2F and 2G). Despite the changes in the structure of the shoot apex, there was no significant alteration in leaf phyllotaxy or inflorescence fasciation.
Figure 2.
Developmental Defects in mre11-3 Mutants.
(A) Wild-type and mre11-3 plants 5 weeks after germination.
(B) A dwarf mre11-3 plant.
(C) A tert plant from the 8th generation of mutants.
(D) and (E) A rosette (D) and cauline leaves (E) from mre11-3 and wild-type plants.
(F) and (G) Scanning electron micrographs of shoot apical meristems from 2-week-old seedlings of wild-type (F) and mre11-3 plants (G). Bars = 80 μM.
Genome Instability Is Associated with Mre11 Dysfunction in Somatic Cells
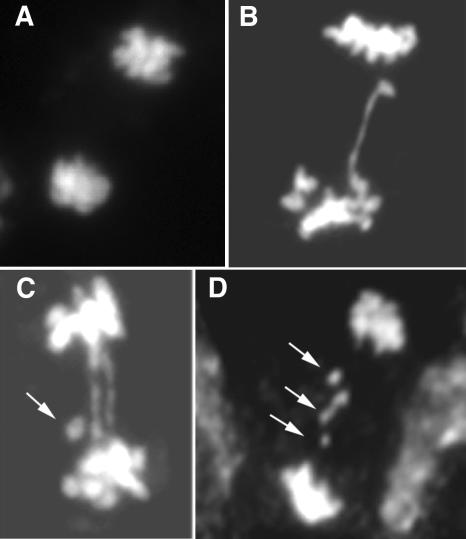
The phenotype of mre11-3 plants resembles the developmental aberrations observed in late generations of telomerase reverse transcriptase (tert) mutants, which have lost functional telomeres (cf. Figures 2B and 2C) (Riha et al., 2001). Because the growth defects in tert mutants are associated with chromosomal rearrangements (Riha et al., 2001; Siroky et al., 2003), we performed cytogenetic analysis in mre11-3. The small size of Arabidopsis metaphase chromosomes precludes their detailed morphological characterization. We therefore assessed the frequency of chromosomal aberrations during anaphase. At this stage, dicentric chromosomes form anaphase bridges, and acentric fragments can be recognized as chromatin bodies lagging between the separating daughter nuclei. In wild-type nuclei, only one anaphase bridge was observed out of 288 anaphases, whereas in mre11-3 ∼16% of cells possessed one or two anaphase bridges (Figure 3, Table 1). Acentric fragments were detected in ∼2% of Mre11-deficient anaphases (Table 1). These data suggest that mre11-3 mutants suffer ongoing genome destabilization, which may contribute to the observed aberrant vegetative growth.
Figure 3.
Genome Instability in Mitotic Cells from mre11-3 Plants.
Anaphase spreads were prepared from pistils stained with 4′,6-diamidino-2-phenylindole (DAPI) and visualized by epifluorescence microscopy.
(A) Wild-type anaphase.
(B) and (C) Examples of mre11-3 anaphases with one (B) and two (C) bridges.
(D) Several fragments lagging between separating daughter nuclei. Acentric fragments are indicated by arrows.
Table 1.
Genome Instabilities in mre11-3 and mre11-3 ku70 Mutants
| Anaphases Scored | Percentage with Bridges | Percentage with Fragments | |
|---|---|---|---|
| Wild type | 288 | 0.3 | 0.0 |
| mre11-3 | 483 | 16.2 | 1.7 |
| ku70 | 285 | 0.0 | 0.0 |
| mre11-3 ku70 | 446 | 18.8 | 6.3 |
Because the NHEJ DNA repair pathway is believed to be responsible for the formation of dicentric chromosomes (Bertuch, 2002), we decided to determine whether the chromosome fusions observed in Mre11-deficient plants depend on the Ku70/80 heterodimer, which is essential for the initial stages of NHEJ. We have previously demonstrated that the Ku70 deficiency does not lead to chromosomal instability in Arabidopsis (Riha et al., 2002; Table 1). We generated mre11-3 ku70 double mutants by genetic crossing and analyzed them for the presence of anaphase bridges. The mre11-3 ku70 plants were viable and morphologically resembled mre11-3 single mutants. Furthermore, the frequency of bridges in mre11-3 ku70 double mutants was similar to the frequency observed in mre11-3 plants (Table 1), thus demonstrating that the Ku70 protein is not essential for the formation of chromosome fusions. Nevertheless, a slight increase in the incidence of acentric fragments (Table 1) indicates that the Ku complex contributes to the repair of DSBs, which arise in Mre11-deficient cells.
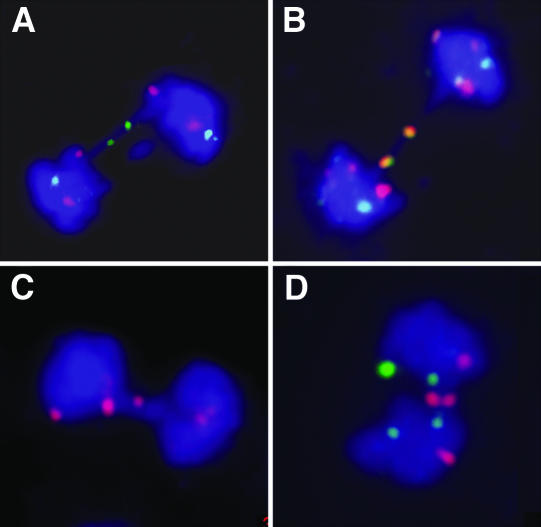
To learn more about the processes leading to chromosome fusions, we performed fluorescence in situ hybridization (FISH) analysis of anaphase bridges with unique probes derived from BACs. Because multicolor FISH technology, with probes that distinguish every individual chromosome arm, has yet to be developed for Arabidopsis, we sequentially hybridized anaphase spreads with a set of BACs that were labeled with different dyes (Figure 4). Hybridization with a combination of various BACs from intercalary and terminal parts of chromosomes permitted us to identify the chromosome arms in 75 out of the 170 analyzed bridges from mre11-3 plants. Interestingly, 33 of the analyzed bridges were decorated with two signals of the same probe, hence suggesting a fusion of homologous chromosomes or sister chromatids (Figures 4A and 4B). We also detected 12 bridges with a signal from BAC probes derived from the most distal parts of the chromosome arms, which suggests the involvement of chromosome ends in fusions (Figures 4C and 4D).
Figure 4.
FISH Analysis of Anaphase Bridges.
Anaphase bridges were hybridized with one, two, or three differently labeled BAC probes and counterstained with DAPI.
(A) and (B) Examples of symmetric chromosome fusions.
(A) Bicolor hybridization with BACs F8J2 (intercalary position at chromosome 3, green) and T4P13 (chromosome 3, terminal, red).
(B) Tricolor hybridization with BACs F2I11 (chromosome 5, intercalary, red), F1N20 (chromosome 4, intercalary, green), and F8J2 (chromosome 3, intercalary, yellow).
(C) Hybridization of the terminally localized BAC probe F7J8 (chromosome 5, red) to the center of the bridge suggests chromosome end-to-end fusion.
(D) Two anaphase bridges containing signals from the BAC F11L15 (red), which is localized at the terminus of the long arm of chromosome 2. Green signals represent BAC T5J17 (terminal position at the long arm of chromosome 4).
The Sterility of mre11-3 Plants Is a Result of Genome Fragmentation during Meiosis
Although the severity of vegetative growth defects varies amongst individual mre11-3 plants, all mutants are sterile and siliques are devoid of seeds (Table 2). The necessity of the Mre11 protein for fertility was further illustrated by the absence of normal pollen in mre11-3 mutants (Table 3). Three nuclei, one diffuse vegetative nucleus and two bright sperm nuclei, can usually be detected by DAPI staining in wild-type pollen grains. In contrast with the wild type, mre11-3 pollen grains were much smaller and were either empty or only contained traces of diffuse chromatin (Table 3).
Table 2.
Fertility of mre11-3 and mre11-3 spo11-1-1 Mutants
| Siliques Scored | Total Number of Seeds | Seeds per Silique | |
|---|---|---|---|
| Wild type | 10 | 564 | 56.40 |
| mre11-3 | 140 | 0 | 0.00 |
| spo11-1-1 | 30 | 62 | 2.01 |
| mre11-3 spo11-1-1 | 160 | 5 | 0.03 |
Table 3.
Frequency of Trinucleate Pollen in mre11-3 and mre11-3 spo11-1-1 Mutants
| Pollen Scored | Trinucleate | Empty | Aberranta | |
|---|---|---|---|---|
| Wild type | 550 | 97.8% | 1.1% | 1.1% |
| mre11-3 | 730 | 0.0% | 62.3% | 37.7% |
| spo11-1-1 | 697 | 13.8% | 75.2% | 11.0% |
| mre11-3 spo11-1-1 | 512 | 2.5% | 46.7% | 50.8% |
Pollen grains containing diffuse traces of chromatin.
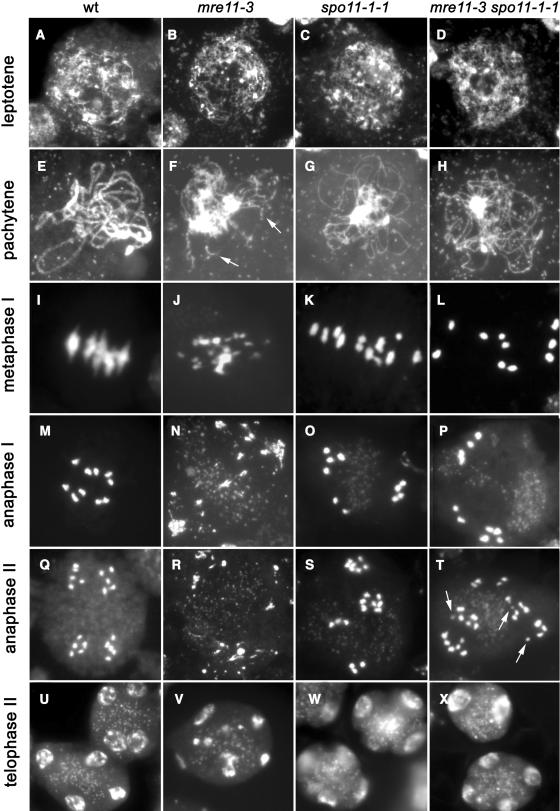
To examine the meiotic role of Mre11 in Arabidopsis, we followed the progression of meiosis in pollen mother cells (PMCs) derived from mre11-3 mutants. In wild-type male meiocytes, chromosomes can first be detected at leptotene, when they begin to condense and form granular and thread-like structures (Figure 5A). Homologous chromosomes begin to synapse during zygotene and chromosomes are fully synapsed in pachytene, forming looped ribbon-like structures (Figure 5E) (Ross et al., 1996; Armstrong and Jones, 2003). Chromosomes continue to condense during diplotene and diakinesis, and homologs are held together via chiasmata and sister chromatid cohesion. Paired chromosomes can be detected as five bivalents at metaphase I (Figure 5I).
Figure 5.
Meiosis in Wild-Type, mre11-3, spo11-1-1, and mre11-3 spo11-1-1 Mutants.
Meiotic spreads were prepared from PMCs and stained by DAPI. Genotypes are marked on the top, and corresponding stages of meiosis are indicated on the left (see text for further details). (F), (G), and (H) represent mid-prophase stages in mutants that correspond to the wild-type zygotene-pachytene. Chromosome fragments in mre11-3 mutants are indicated by arrows in (F). Arrows in (T) indicate fragments detected in mre11-3 spo11-1-1 mutants at anaphase II.
Analysis of PMCs from mre11-3 plants showed that mutants lack a regular prophase I and that all the subsequent stages of meiosis are severely impaired. Although leptotene appeared normal in our cytogenetic analysis (Figure 5B), wild type–looking pachytene was never observed. Instead, nuclei with fragmented chromosome threads occurred in mre11-3 cells at the mid-prophase stage that corresponds to the wild-type zygotene-pachytene (Figure 5F). The extent of chromosome fragmentation became more pronounced as chromatin continued to condense in the subsequent stages of meiosis. Typically, a large number of small fragments but no regular bivalents or univalents were observed at the stage that corresponds to metaphase I (Figure 5J). At this stage, the chromosomal fragments frequently clustered into a disorganized chromatin mass and chromatin bodies of different sizes. Despite the massive genome fragmentation, meiosis progressed into anaphase I and then into meiosis II (Figures 5N and 5R). This process eventually led to the formation of polyads containing microspores with unequal amounts of DNA (Figure 5V).
The chromosome threads detected at the zygotene-pachytene stage in mre11-3 PMCs lacked the double structure organization that is characteristic of synapsed chromosomes in wild-type pachytene (cf. Figures 5E and 5F), thus suggesting a failure to synapse homologous chromosomes in the absence of Mre11 function. To confirm this observation, we performed bicolor FISH analysis with BAC probes specific to unique loci on chromosomes 1 and 4. All of the wild-type pachytene nuclei examined (n = 6) contained a single FISH signal for each locus, illustrating a perfect alignment of homologous chromosomes (Figure 6A). Out of the 41 zygotene-pachytene nuclei analyzed from mre11-3 plants, colocalization of the chromosome 1 and chromosome 4 signals occurred in only six and three meiocytes, respectively. The detection of two separate signals in 89% of the zygotene-pachytene nuclei (Figure 6B) indicates that the Mre11 complex plays an important role in the synapsis of homologous chromosomes in Arabidopsis.
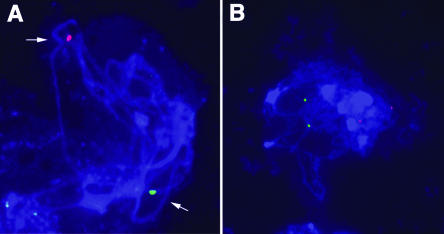
Figure 6.
Aberrant Chromosome Pairing in mre11-3 Mutants Analyzed by FISH.
Meiotic spreads from PMCs of wild-type (A) and mre11-3 (B) plants corresponding to the pachytene stage were hybridized with BACs F6F3 (chromosome 1, terminal, red) and F1N10 (chromosome 4, intercalary, green). The paired signals indicated by arrows in the wild-type pachytene indicate synapsis of homologous chromosomes.
Mre11 Is Required for the Repair but Not for the Induction of Spo11-Dependent DSBs
To ascertain whether Spo11 function is involved in the chromosome fragmentation observed in prophase I in Mre11-deficient plants, we generated mre11-3 spo11-1-1 double mutants. Arabidopsis has three SPO11 homologs, and it is the Spo11-1 paralog that appears to be essential for meiotic recombination (Grelon et al., 2001). Despite severe perturbation in chromosome synapsis and HR, spo11-1-1 mutants still produce an average of two seeds per silique (Table 2) (Grelon et al., 2001). Inactivation of the Spo11-1 protein partially rescued the complete sterility of mre11-3 mutants, but the yield of seeds in mre11-3 spo11-1-1 double mutants was almost two orders of magnitude lower than in spo11-1-1 plants (Table 2). In addition, mre11-3 spo11-1-1 double mutants regained the capacity to produce a small fraction of trinucleate pollen grains with one vegetative and two sperm nuclei. The number of these trinucleate grains was, however, five times lower than in the spo11-1-1 single mutant (Table 3).
We next examined the impact of the spo11-1-1 mutation on the progression of meiosis in Mre11-deficient plants. Meiosis in spo11-1-1 mutants is characterized by the presence of univalents at metaphase I and by their random segregation during anaphase I (Figures 5K and 5O) (Grelon et al., 2001). In contrast with the mre11-3 plants, the integrity of the chromosomes in mre11-3 spo11-1-1 mutants was preserved, and 10 univalents could be regularly detected at metaphase I (Figure 5L). The suppression of genome fragmentation in the spo11-1-1 background was already apparent in pachytene as the chromosomes formed loops as opposed to the short chromosome threads typical for mre11-3 PMCs (cf. Figures 5F and 5H). The presence of univalents in metaphase I and anaphase I in both spo11-1-1 and mre11-3 spo11-1-1 mutants indicates that the observed genome disintegration in the absence of Mre11 is caused by the failure to repair Spo11-induced DSBs. Furthermore, this result implies that mre11-3 plants are proficient in the initiation of meiotic DSBs.
Detailed analysis of the PMCs revealed that chromosome fragmentation was not completely suppressed in mre11 spo11-1-1 mutants because we observed chromosomal fragments at the end of the first meiotic division. Altogether, 4.6% of PMCs (n = 87) at metaphase I/anaphase I contained such fragments. The incidence of fragmentation dramatically increased in metaphase II/anaphase II, where 92% of PMCs (n = 87) presented with acentric fragments. However, the extent of fragmentation was substantially lower than in the mre11-3 single mutants because the overall morphology of chromosomes was preserved, and only a few fragments per meiocyte were detected (Figure 5T). The residual chromosomal fragmentation, which is especially obvious in the second meiotic division, possibly contributes to the lower fertility of mre11-3 spo11-1-1 double mutants compared with the spo11-1-1 plants (Tables 2 and 3).
DISCUSSION
The Mre11 complex is involved in several fundamental aspects of chromosome maintenance, and its inactivation is lethal for vertebrate cells (Xiao and Weaver, 1997; Luo et al., 1999; Yamaguchi-Iwai et al., 1999). In this study, we describe the consequences of Mre11 dysfunction on chromosome behavior in both somatic and meiotic cells from Arabidopsis. Homozygous mre11-3 mutants are viable, but they exhibit pleiotropic defects in growth and development, which are largely consistent with the morphological phenotypes previously described for the mre11-1 allele (Bundock and Hooykaas, 2002). Whereas both alleles possess T-DNA insertions at almost identical positions in the conserved phosphodiesterase motif IV (see Figure 1A), the mre11-1 mutation appears to be more severe than the mre11-3 disruption. Only the fittest mre11-1 plants are able to develop to maturity when grown in soil (Bundock and Hooykaas, 2002), whereas most of the mre11-3 mutants produced inflorescent bolts with flowers, and the fittest plants were almost indistinguishable from the wild type before flowering. In addition, the mre11-3 allele is recessive, and mutants do not exhibit extensive necrosis. The discrepancy between the observed phenotypes is probably because of the overexpression of the truncated protein in mre11-1 mutants. The substantially increased level of 5′ transcript from the MRE11 gene and the semidominant nature of the mre11-1 allele (Bundock and Hooykaas, 2002) support this explanation.
The morphology of Mre11-deficient plants, including the enlargement of shoot apical meristem, is reminiscent of phenotypes observed in late generation telomerase mutants (Riha et al., 2001). Because both mutations are associated with chromosomal aberrations (Riha et al., 2001; Siroky et al., 2003; this article), we propose that ongoing chromosome destabilization and chronic genotoxic stress are the underlying causes of the observed developmental defects in mre11-3 and tert mutants.
The Mre11 Complex and Genome Stability in Somatic Cells
A link between aberrant Mre11 complex function and genome instability has been established in every organism in which it has been studied. Chicken cells, conditionally null for Mre11, display a pronounced increase in spontaneous chromosomal breaks and apoptosis within 5 d of inactivation of the MRE11 gene (Yamaguchi-Iwai et al., 1999). Chromosome instability manifested by chromosome fusions and chromosomal breaks was also observed in murine cells carrying the hypomorphic mutations Rad50S/S or Mre11ATLD1/ATLD1 (Bender et al., 2002; Theunissen et al., 2003). The incidence of gross chromosomal rearrangements, including terminal deletions and translocations, was ∼600-fold increased in yeast strains deficient for Mre11, Rad50, or Xrs2 (Chen and Kolodner, 1999). The occurrence of dicentric chromosomes and acentric fragments in the Arabidopsis mre11-3 mutant further demonstrates the conserved role of the Mre11 complex in the maintenance of genome integrity.
Which mechanisms lead to the spontaneous chromosomal aberrations in cells that lack a fully functional Mre11 complex? Dicentric chromosomes could arise from a NHEJ reaction that fuses internal DSBs located on different chromatids or from end-to-end chromosome fusions caused by aberrant telomere function. The presence of FISH signals from subterminal BACs on the anaphase bridges in mre11-3 plants suggests that a fraction of fusions might be derived from partial telomere dysfunction in the absence of Mre11. A telomeric role of the Mre11 complex in Arabidopsis has been implicated by a recent finding on telomere extension in Mre11-deficient plants (Bundock and Hooykaas, 2002) as well as by the rapid loss of telomeres in a rad50 mutant cell suspension culture (Gallego and White, 2001). In addition, chromosome end-to-end associations that retain telomeric DNA at the fusion point were observed in the Rad50S/S mouse (Bender et al., 2002).
The presence of acentric fragments in mre11-3 mutants argues that chromosome end-to-end fusions may only account for some of the aberrations. The observation that an elevated level of chromosomal rearrangements occurs in yeast replication mutants suggests that aberrant DNA replication is the major source of genome instabilities in eukaryotic cells (Chen and Kolodner, 1999). The replication fork can collapse when it encounters a DNA lesion or an unusual secondary structure, or when it collides with protein-DNA adducts, such as another replication fork or RNA polymerase. Replication is reestablished through recombination intermediates in a process that may include DSB formation (reviewed in Kuzminov, 2001; McGlynn and Lloyd, 2002). It has been proposed that the Mre11 complex is involved in the repair of collapsed replication forks. Mre11 colocalizes with stalled replication forks in human cell lines (Franchitto and Pichierri, 2002), and frog extracts depleted of Mre11 accumulate DSBs during DNA replication (Costanzo et al., 2001). The dicentric chromosomes and acentric fragments in mre11-3 plants may thus result from aberrant repair of DSBs that occur during DNA replication. It is tempting to speculate that the high incidence of homologous fusions in Mre11-deficient Arabidopsis reflects the ligation of sister chromatids after the collapse of a replication fork.
The occurrence of dicentric chromosomes in the mre11-3 ku70 double mutant demonstrates that Ku-dependent NHEJ is not essential for the chromosome fusions in Mre11-deficient plants. Two major NHEJ pathways appear to exist in yeast: one includes the Ku heterodimer and ligase IV, whereas the other depends on the Mre11 and Rad1/Rad10 complexes (Ma et al., 2003). The high incidence of chromosome fusions in mre11-3 ku70 plants is consistent with our recent finding that the absence of Ku70 and Mre11 does not abort fusions of critically short telomeres in telomerase-deficient Arabidopsis (Riha and Shippen, 2003; Heacock et al., 2004). These observations imply that redundant NHEJ repair pathways exist in Arabidopsis.
The Role of Mre11 in Meiotic Recombination
In this study, we show that the sterility of mre11-3 plants is a result of genome disintegration during the initial stages of meiotic prophase I. Acentric fragments have also been detected in plants deficient for the MEI1/MCD1 and CDC45 genes. The chromosome fragmentation in such mutants is likely of premeiotic origin because it does not depend on Spo11 (Grelon et al., 2003; Stevens et al., 2004). In contrast with mei1 and cdc45 mutants, most of the genome fragmentation was suppressed by the spo11-1 mutation in mre11-3 plants. This observation suggests that Spo11 may not require Mre11 for DSB induction in Arabidopsis. The ability of mre11-3 plants to form DSBs is surprising because this process is dependent on the Mre11 complex in budding yeast, Caenorhabditis elegans, and Coprinus cinereus (Cao et al., 1990; Nairz and Klein, 1997; Gerecke and Zolan, 2000; Chin and Villeneuve, 2001). Although mre11-3 plants can putatively express the N-terminal third of the Mre11 protein, it is unlikely that this peptide would provide enough residual function for DSB induction. Mutational analysis of the MRE11 gene in S. cerevisiae showed that its function in DSB initiation is distinct from its end processing function and is located in the C-terminal part of the protein (Nairz and Klein, 1997; Usui et al., 1998). Truncation of the last 136 C-terminal amino acids results in severe defects in DSB formation and in meiotic recombination in yeast (Usui et al., 1998). By contrast, Arabidopsis mre11-2 mutants in which the last 191 amino acids of the Mre11 protein are deleted are fertile (Bundock and Hooykaas, 2002). This implies that no meiotic function is associated with the C terminus in plants. The same may also be the case in mammals because the Mre11ALTD1/ALTD1 mutation that results in a 75–amino acid truncation from the C terminus is not associated with gross meiotic defects in mice (Theunissen et al., 2003).
Elimination of Spo11-1 function does not completely abolish DSB formation in mre11-3 spo11-1-1 mutants, and residual chromosome fragmentation is particularly obvious during the second meiotic division. The high incidence of acentric fragments at metaphase II/anaphase II could be caused by chromosome breakage after the first meiotic division. Another more likely explanation is that the fragments represent chromatids that were broken either premeiotically or during an early stage of meiosis. A portion of the breaks can be attributed to premeiotic DNA replication, but the much lower occurrence of fragments in somatic cells of mre11-3 plants argues that the majority of the breaks are specifically associated with meiosis. This raises the possibility that one of the other two Arabidopsis Spo11 paralogs (Hartung and Puchta, 2000, 2001) participates in the formation of meiotic DSBs. This hypothesis is supported by the observation that meiotic recombination is not fully aborted, and rare chiasmata can be detected in spo11-1 mutants (Grelon et al., 2001). Sister chromatids are held together by cohesin until anaphase I, when its cleavage along chromosome arms allows resolution of chiasmata and separation of homologous chromosomes (Klein et al., 1999; Buonomo et al., 2000). Cohesin degradation between broken chromatids would lead to the liberation of fragments during anaphase I. However, the fragments in mre11-1 spo11-1-1 mutants appeared rather after the first meiotic division. This might reflect the physiological preservation of cohesin in broader chromosomal regions in Arabidopsis or a delay of cohesin destruction in asynaptic mutants. The latter idea is supported by an obsrvation that in the absence of chiasmata, anaphase I occurs independently of cohesin degradation in S. cerevisiae (Buonomo et al., 2000).
The Spo11-1–dependent genome fragmentation in mre11-3 mutants is consistent with a role of the Mre11 complex in the repair of meiotic DSBs (reviewed in Roeder, 1997; Zickler and Kleckner, 1999). Several genes assumed to act in DSB repair downstream of Spo11-dependent break induction have recently been studied in Arabidopsis. At least three members of the RecA family of recombination proteins appear to be involved in Arabidopsis meiosis. Plants lacking a functional XRCC3 protein display normal meiosis up to the pachytene stage, but later stages exhibit severe chromosome fragmentation (Bleuyard and White, 2004). Mutation of the DMC1 gene results in the almost complete absence of recombination and synapsis (Couteau et al., 1999). Although dmc1 single mutants do not exhibit any fragmentation, downregulation of the RAD51 gene by RNA interference in dmc1 plants results in entangled chromosomes, anaphase bridges, and acentric fragments (Siaud et al., 2004). A similar phenotype was also observed in plants with suppressed expression of the Arabidopsis BRCA2 ortholog (Siaud et al., 2004). Fragmentation of meiotic chromosomes has also been described in plants deficient for the ATM and AHP2 genes that are believed to play a role in meiotic recombination (Garcia et al., 2003; Schommer et al., 2003).
Although meiosis was profoundly disturbed in these recombination mutants, none of them exhibited as severe meiotic phenotypes as mre11-3 plants. This observation is supported by the fact that mre11-3 mutants do not produce any seeds, although residual fertility is associated with spo11-1, xrcc3, dmc1, ahp2, and atm mutations (Couteau et al., 1999; Grelon et al., 2001; Garcia et al., 2003; Schommer et al., 2003; Bleuyard and White, 2004). mre11-3 mutants are unable to efficiently synapse chromosomes that undergo severe fragmentation in early stages of meiosis. This finding clearly demonstrates that Mre11 operates very early in DSB repair, probably immediately after the induction of DSBs by Spo11. Because yeast mre11S and rad50S mutants accumulate unprocessed DSBs (Cao et al., 1990; Nairz and Klein, 1997), the Mre11 complex is assumed to be involved in the formation of single-stranded 3′ tails that are required for strand invasion (Haber, 1998).
The synapsis of homologous chromosomes depends on DSB formation in yeast and mouse. The absence of chromosome pairing in spo11-1-1 mutants revealed that initiation of DSBs is also important for synapsis in Arabidopsis (Grelon et al., 2001). The lack of efficient chromosome synapsis in mre11-3 mutants indicates that not only induction but also initial stages of HR are important for stable chromosome pairing in Arabidopsis. Similar observations have been made in yeast (Nairz and Klein, 1997) and C. cinereus (Gerecke and Zolan, 2000). Interestingly, synapsis does not appear to be affected in plants deficient for the XRCC3 protein, which is assumed to act in the later stages of HR (Bleuyard and White, 2004). Although the data obtained in spo11-1-1 and mre11-3 mutants clearly established that early steps of HR are a requirement for stable associations of homologous chromosomes in Arabidopsis, the mechanisms governing homology search are still poorly understood. The recent identification of the phs1 gene that coordinates homology pairing and synapsis in maize (Pawlowski et al., 2004) promises that the molecular characterization of plant meiosis will shed novel insights into these complex processes.
METHODS
Generation of mre11-3 ku70 and mre11-3 spo11-1-1 Double Mutants
Seeds of the mre11-3 Arabidopsis thaliana SALK_054418 line (N554418) were obtained from Nottingham Arabidopsis Stock Centre (Nottingham, UK). Because mre11-3 mutants are sterile, the mre11-3 allele was maintained via the self-fertilization of heterozygous plants. mre11-3 ku70 double mutants were generated by crossing plants heterozygous for the tert and ku70 mutations (Riha and Shippen, 2003) with MRE11+/− plants. mre11-3 ku70 double mutants were identified by PCR among the F2 progeny derived from the F1 plants heterozygous for all three genes. mre11-3 spo11-1-1 double mutants were obtained from crosses between SPO11-1-1+/− (Grelon et al., 2001) and MRE11+/− plants. Double mutants were identified by PCR of the F2 population obtained by selfing F1 plants heterozygous for both genes. All plants were grown at 22°C under a 16/8-h light/dark photoperiod.
DNA Analyses
DNA from flowers of individual plants was extracted according to Cocciolone and Cone (1993). DNA gel blot analysis of the MRE11 locus was performed with 1 μg of genomic DNA digested with BglII. A 32P-labeled genomic fragment amplified by PCR with primers M1 (5′-CCAATGGATGAGGCCTGAAGTT-3′) and M2 (5′-CCAATGGGAGTTTGATCTCTGA-3′) was used as a probe for hybridization. Plants were genotyped by PCR with two gene-specific primers flanking insertion sites and with a primer specific for the left border of the T-DNA. Primers M1 and M3 (5′-GTCTGCCACCACCATAAC AT-3′) and LBc-1 (5′-TGGACCGCTTGCTGCAACTCT-3′) were used for the MRE11 locus. A 702-bp product was generated from the wild-type allele and a 477-bp product from the mre11-3 allele. Primers SPO-1 (5′-CGTGTCCGAGCATAAAGCTTATGA-3′), SPO-2 (5′-CCCTTTGGTTTATCAGAGCTGCAT-3′), and LB-BAR2 (5′-TGTGCCAGGTGCCCACGGAATAGT-3′) were used for the SPO11-1 locus, resulting in 943-bp and ∼500-bp products for the wild-type and spo11-1-1 alleles, respectively. The KU70 locus was genotyped as described previously (Riha et al., 2002).
RNA Extraction and RT-PCR
Total RNA was extracted from young inflorescences using the TriReagent solution (Sigma, St. Louis, MO). MRE11 and GAPA cDNAs were synthesized as follows: 5 pmol of primers M7 (5′-ACACCAGAACCACCAAGAACCAT-3′), M2, M5 (5′-CGTCATCGTCTTTGCTAC TGAGTA-3′), LBc-1, and GAPA-2 (5′-CAACTCTCTGTGAGTAACCCCAT-3′) were incubated with 1 μg of total RNA at 65°C for 5 min. Reverse transcription was performed with 100 units of M-MLV reverse transcriptase (Promega, Madison, WI) at 45°C for 50 min. One-tenth of the cDNA was used for PCR amplification. PCR products were analyzed by DNA gel blot hybridization with gene-specific probes.
Analysis of Meiotic and Mitotic Chromosomes
Mitotic anaphases were obtained from pistils of unopened floral buds. Meitotic figures were prepared from anthers of young floral buds. Whole inflorescences were fixed in ethanol:acetic acid (3:1) and transferred to 0.01 M citrate buffer, pH 4.6, and dissected pistils or anthers were transferred to an enzyme mixture containing 0.5% (w/v) cellulase Onozuka R-10 (Serva, Heidelberg, Germany) and 0.5% (w/v) pectolyase (Sigma) in citrate buffer. The enzyme mixture was replaced by citrate buffer after ∼2 h of incubation at 37°C, and slides were prepared by a standard squashing technique in a drop of 60% acetic acid. Slides were stained with 2 μg/mL of DAPI and mounted in Vectashield antifade solution (Vector Laboratories, Burlingame, CA). Mitotic and meiotic nuclei were examined by a Zeiss Axioplan epifluorescence microscope (Jena, Germany) equipped with a CCD camera (Photometrics, Tucson, AZ) using IPLab Spectrum software (Scanalytics, Fairfax, VA).
FISH Analysis
Whole BAC clones were used as probes for in situ hybridization. The clones were labeled either with Cy3-dUTP (Amersham Biosciences, Buckinghamshire, UK) or Spectrum Green-dUTP (Vysis, Downers Grove, IL) or with a combination of both dyes by nick translation. DAPI-stained chromosome spreads were first examined by epifluorescence microscopy, the coordinates of anaphases were recorded, and slides were subjected to FISH. In situ hybridization was performed as described previously (Siroky et al., 2003). One, two, or three differently labeled BAC probes were usually used for hybridization. To prevent damage to the delicate pachytene threads, the denaturation of meiotic slides was performed in 60% formamide in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 72°C for 90 s. After hybridization and stringent washings, the preparations were analyzed under the Olympus AX 70 fluorescent microscope (Tokyo, Japan) with separate filter sets for DAPI, Cy3, and Spectrum Green dyes, photographed with a monochrome AxioCam MR CCD camera (Zeiss), and visualized by ISIS software (MetaSystems, Altussheim, Germany) as RGB overlays. Mitotic slides could be used for two or three rounds of FISH with different sets of BAC probes.
The following BAC clones were obtained from ABRC (Columbus, Ohio) and used as probes for FISH: F6F3 (AC023628), F26G16 (AC009917), F9M8 (AC083859), F17J6 (AC079279), F5I6 (AC018848), T2G17 (AC006081), F5H14 (AC006234), T9I22 (AC006340), T9J23 (AC006072), F11L15 (AC007927), T4P13 (AC008261), T22N4 (AC010676), F8J2 (AL132969), F16M2 (AL138648), F21I2 (AF147261), F1N20 (AL022140), T22F8 (AL050351), T5J17 (AL035708), F7J8 (AL137189), and F2I11 (AL360314).
Acknowledgments
We thank Franz Klein, Josef Loidl, Maria Siomos, Peter Schloegelhofer, and Dorothy Shippen for critical reading of the manuscript and stimulating discussion. We also thank Mathilde Grelon for providing spo11-1-1 mutants and Paula Bombosi for assistance with scanning electron microscopy. This work was supported by grants from the Austrian Science Foundation (P16405) to K.R. and from the Grant Agency of the Czech Republic (522/03/0354) as well as by the Grant Agency of the Czech Academy of Sciences (A6004304) to J.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Karel Riha (karel.riha@gmi.oeaw.ac.at).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022749.
References
- Ajimura, M., Leem, S.H., and Ogawa, H. (1993). Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Armstrong, S.J., and Jones, G.H. (2003). Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J. Exp. Bot. 54, 1–10. [DOI] [PubMed] [Google Scholar]
- Aten, J.A., Stap, J., Krawczyk, P.M., van Oven, C.H., Hoebe, R.A., Essers, J., and Kanaar, R. (2004). Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303, 92–95. [DOI] [PubMed] [Google Scholar]
- Bender, C.F., Sikes, M.L., Sullivan, R., Huye, L.E., Le Beau, M.M., Roth, D.B., Mirzoeva, O.K., Oltz, E.M., and Petrini, J.H. (2002). Cancer predisposition and hematopoietic failure in Rad50S/S mice. Genes Dev. 16, 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A.A. (2002). Telomeres: The molecular events driving end-to-end fusions. Curr. Biol. 12, R738–R740. [DOI] [PubMed] [Google Scholar]
- Bleuyard, J.Y., and White, C.I. (2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., and Hooykaas, P. (2002). Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14, 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S.B., Clyne, R.K., Fuchs, J., Loidl, J., Uhlmann, F., and Nasmyth, K. (2000). Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387–398. [DOI] [PubMed] [Google Scholar]
- Cao, L., Alani, E., and Kleckner, N. (1990). A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carney, J.P., Maser, R.S., Olivares, H., Davis, E.M., Le Beau, M., Yates iii, J.R., Hays, L., Morgan, W.F., and Petrini, J.H. (1998). The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477–486. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Kolodner, R.D. (1999). Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23, 81–85. [DOI] [PubMed] [Google Scholar]
- Chin, G.M., and Villeneuve, A.M. (2001). C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 15, 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann, M., Tomicic, M.T., Roos, W.P., and Kaina, B. (2003). Mechanisms of human DNA repair: An update. Toxicology 193, 3–34. [DOI] [PubMed] [Google Scholar]
- Cocciolone, S.M., and Cone, K.C. (1993). Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, J.C., and Leach, D.R. (2002). Tethering on the brink: The evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27, 410–418. [DOI] [PubMed] [Google Scholar]
- Costanzo, V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D., and Gautier, J. (2001). Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8, 137–147. [DOI] [PubMed] [Google Scholar]
- Couteau, F., Belzile, F., Horlow, C., Grandjean, O., Vezon, D., and Doutriaux, M.P. (1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11, 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and Jackson, S.P. (2002). The Mre11 complex: At the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell Biol. 3, 317–327. [DOI] [PubMed] [Google Scholar]
- de Jager, M., van Noort, J., van Gent, D.C., Dekker, C., Kanaar, R., and Wyman, C. (2001). Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Dubrana, K., Perrod, S., and Gasser, S.M. (2001). Turning telomeres off and on. Curr. Opin. Cell Biol. 13, 281–289. [DOI] [PubMed] [Google Scholar]
- Franchitto, A., and Pichierri, P. (2002). Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 157, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., Jeanneau, M., Granier, F., Bouchez, D., Bechtold, N., and White, C.I. (2001). Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 25, 31–41. [DOI] [PubMed] [Google Scholar]
- Gallego, M.E., and White, C.I. (2001). RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V., Bruchet, H., Camescasse, D., Granier, F., Bouchez, D., and Tissier, A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke, E.E., and Zolan, M.E. (2000). An mre11 mutant of Coprinus cinereus has defects in meiotic chromosome pairing, condensation and synapsis. Genetics 154, 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon, M., Ghislaine, G., Daniel, V., and Georges, P. (2003). The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 35, 465–475. [DOI] [PubMed] [Google Scholar]
- Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J.E. (1998). The many interfaces of Mre11. Cell 95, 583–586. [DOI] [PubMed] [Google Scholar]
- Hartung, F., and Puchta, H. (2000). Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, F., and Puchta, H. (2001). Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene 271, 81–86. [DOI] [PubMed] [Google Scholar]
- Heacock, M., Spangler, B., Riha, K., Puizina, J., and Shippen, D.E. (2004). Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 23, 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., Giroux, C.N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Klein, F., Mahr, P., Galova, M., Buonomo, S.B., Michaelis, C., Nairz, K., and Nasmyth, K. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103. [DOI] [PubMed] [Google Scholar]
- Kuzminov, A. (2001). DNA replication meets genetic exchange: Chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98, 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, D.S., Kim, S.T., Xu, B., Maser, R.S., Lin, J., Petrini, J.H., and Kastan, M.B. (2000). ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404, 613–617. [DOI] [PubMed] [Google Scholar]
- Luo, G., Yao, M.S., Bender, C.F., Mills, M., Bladl, A.R., Bradley, A., and Petrini, J.H. (1999). Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.L., Kim, E.M., Haber, J.E., and Lee, S.E. (2003). Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23, 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn, P., and Lloyd, R.G. (2002). Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3, 859–870. [DOI] [PubMed] [Google Scholar]
- Nairz, K., and Klein, F. (1997). mre11S a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11, 2272–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S.L., and Hawley, R.S. (2003). Chromosome choreography: The meiotic ballet. Science 301, 785–789. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W.P., Golubovskaya, I.N., Timofejeva, L., Meeley, R.B., Sheridan, W.F., and Cande, W.Z. (2004). Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89–92. [DOI] [PubMed] [Google Scholar]
- Petronczki, M., Siomos, M.F., and Nasmyth, K. (2003). Un menage a quatre: The molecular biology of chromosome segregation in meiosis. Cell 112, 423–440. [DOI] [PubMed] [Google Scholar]
- Pichierri, P., and Franchitto, A. (2004). Werner syndrome protein, the MRE11 complex and ATR: Menage-a-trois in guarding genome stability during DNA replication? Bioessays 26, 306–313. [DOI] [PubMed]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha, K., and Shippen, D.E. (2003). Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., Watson, J.M., Parkey, J., and Shippen, D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G.S. (1997). Meiotic chromosomes: It takes two to tango. Genes Dev. 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Ross, K.J., Fransz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Scharer, O.D. (2003). Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. Engl. 42, 2946–2974. [DOI] [PubMed] [Google Scholar]
- Schommer, C., Beven, A., Lawrenson, T., Shaw, P., and Sablowski, R. (2003). AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 36, 1–11. [DOI] [PubMed] [Google Scholar]
- Siaud, N., Dray, E., Gy, I., Gerard, E., Takvorian, N., and Doutriaux, M.P. (2004). Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23, 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signon, L., Malkova, A., Naylor, M.L., Klein, H., and Haber, J.E. (2001). Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21, 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siroky, J., Zluvova, J., Riha, K., Shippen, D.E., and Vyskot, B. (2003). Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma 112, 116–123. [DOI] [PubMed] [Google Scholar]
- Stevens, R., Grelon, M., Vezon, D., Oh, J., Meyer, P., Perennes, C., Domenichini, S., and Bergounioux, C. (2004). A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, G.S., Maser, R.S., Stankovic, T., Bressan, D.A., Kaplan, M.I., Jaspers, N.G., Raams, A., Byrd, P.J., Petrini, J.H., and Taylor, A.M. (1999). The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577–587. [DOI] [PubMed] [Google Scholar]
- Theunissen, J.W., Kaplan, M.I., Hunt, P.A., Williams, B.R., Ferguson, D.O., Alt, F.W., and Petrini, J.H. (2003). Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11ATLD1/ATLD1 mice. Mol. Cell 12, 1511–1523. [DOI] [PubMed] [Google Scholar]
- Usui, T., Ohta, T., Oshiumi, H., Tomizawa, J., Ogawa, H., and Ogawa, T. (1998). Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95, 705–716. [DOI] [PubMed] [Google Scholar]
- Xiao, Y., and Weaver, D.T. (1997). Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25, 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., Sonoda, E., Sasaki, M.S., Morrison, C., Haraguchi, T., Hiraoka, Y., Yamashita, Y.M., Yagi, T., Takata, M., Price, C., Kakazu, N., and Takeda, S. (1999). Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Petersen, S., Tessarollo, L., and Nussenzweig, A. (2001). Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11, 105–109. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and Kleckner, N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754. [DOI] [PubMed] [Google Scholar]