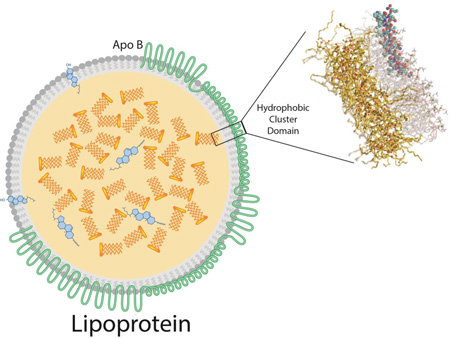
Abstract
Apolipoprotein B (apoB) is a large amphipathic protein that is the structural scaffold for the formation of several classes of lipoproteins involved in lipid transport throughout the body. The goal of the present study was to identify specific domains in the apoB sequence that contribute to its lipid binding properties. A sequence analysis algorithm was developed to identify stretches of hydrophobic amino acids devoid of charged amino acids, which are referred to as hydrophobic cluster domains (HCDs). This analysis identified 78 HCDs in apoB with hydrophobic stretches ranging from 6 to 26 residues. Each HCD was analyzed in silico for secondary structure and lipid binding properties, and a subset was synthesized for experimental evaluation. One HCD peptide, B38, showed high affinity binding to both isolated HDL and LDL, and could exchange between lipoproteins. All-atom molecular dynamics simulations indicate that B38 inserts 3.7 Å below the phosphate plane of the bilayer. B38 forms an unusual α-helix with a broad hydrophobic face and polar serine and threonine residues on the opposite face. Based on this structure, we hypothesized that B38 could efflux cholesterol from cells. B38 showed a 12-fold greater activity than the 5A peptide, a bihelical Class A amphipathic helix (EC50 of 0.2658 vs. 3.188 µM; p<0.0001), in promoting cholesterol efflux from ABCA1 expressing BHK-1 cells. In conclusion, we have identified novel domains within apoB that contribute to its lipid biding properties. Additionally, we have discovered a unique amphipathic helix design for efficient ABCA1-specific cholesterol efflux.
Keywords: Lipoprotein, Peptide, Cholesterol efflux, Amphipathic helix, Molecular Dynamics Simulation, Trilayer
Graphical abstract
INTRODUCTION
Lipoproteins are nanoparticles composed of an outer monolayer of amphipathic lipids surrounding an inner core of non-polar lipids of varying compositions and concentrations, and surface bound stabilizing amphipathic proteins called apolipoproteins. The lipid binding properties of apolipoproteins are critical to the formation and stability of lipoprotein particles. These particles exist in the circulation as several distinct classes, distinguishable from each other based on size, density, and their associated protein and lipid species. Lipoproteins have well-established relationships with the incidence of cardiovascular disease and are routinely measured as biomarkers of cardiovascular risk. Some lipoproteins, such as high density lipoprotein (HDL), are associated with atheroprotection, whereas others, such as very low density lipoprotein (VLDL) and low density lipoprotein (LDL), promote the development of cardiovascular disease (1,2). The current study aims to identify hydrophobic motifs that are important for lipid binding in apolipoprotein B (apoB), the major protein of proatherogenic lipoproteins.
In humans, apoB exists in two distinct forms. ApoB-100 containing lipoproteins, including VLDL and LDL, participate in the transport and delivery of triglyceride and cholesterol respectively, from the liver, where the protein is synthesized, to peripheral tissues. ApoB-100 is a 512 kDa protein composed of 4,563 amino acids and is one of the largest proteins produced in humans (3). ApoB is also produced by the intestine where the post-transcriptional introduction of a stop codon by APOBEC generates a truncated form called apoB-48 (4). This form includes the N-terminal 48% of the apoB-100 sequence and is the major structural protein of chylomicrons. Unlike most other apolipoproteins, apoB is a non-exchangeable apolipoprotein. In other words, it remains bound to the lipoprotein particle, does not exchange between the different lipoprotein classes, and is virtually insoluble in the lipid-free form in aqueous solutions. The formation of apoB containing lipoproteins occurs co-translationally in the endoplasmic reticulum, and, once bound to the nascent lipoprotein particle, apoB does not later dissociate from it (5). Low resolution structures of apoB bound to lipoproteins have recently been obtained by electron microscopy (6–8) and small angle neutron scattering (9). The lipid binding properties of apoB are of significant interest in the study of lipoprotein formation and metabolism, but detailed studies on apoB structure have been extremely difficult due to the inherent problems in working with such a large and highly hydrophobic protein.
Segrest et. al. have proposed a pentapartite modular structure for apoB based on molecular modeling. It consists of an alternating sequence of Class A amphipathic α-helical domains and β-sheet domains (10). Class A amphipathic helices are characterized by a relatively large hydrophobic face with positively charged residues at the polar/non-polar interface and negatively charged residues in the central part of the hydrophilic face (11). These positively charged residues have been proposed to facilitate the interaction of amphipathic peptides with the negatively charged head groups of phospholipids (12). Class A amphiphathic α-helices, which lie superficially on the surface of lipoproteins, are also commonly found on the exchangeable apolipoproteins, such as apoA-I. Although the hydrophobic face of amphipathic α-helices interact with lipid, the affinity for any individual helix is relatively low and thus they are in dynamic equilibrium between the lipid bound and free states (11). When not bound to lipid, exchangeable apolipoproteins maintain their solubility by forming tight helical bundles and shielding the hydrophobic faces of their amphipathic helices (13).
Unlike Class A amphipathic α-helices, β-sheet domains are mostly found on apoB and not on the other types of apolipoproteins (10). Amphipathic β-sheet domains are characterized as containing alternating hydrophobic and hydrophilic amino acids and form relatively large flat planar surfaces by inter-strand hydrogen bonding, with their non-polar face oriented toward the hydrophobic core of a lipoprotein (10). β-sheet domains in apoB have been estimated to have a high average ΔGtransfer energy into lipid as much as -2 kcal/mol per amino acid (14), and have been proposed to account for the non-exchangeability of apoB (15).
Different structural motifs on other proteins besides apolipoproteins have also been shown to bind to lipids. For example, oleosins are a family of proteins found in seeds and contain long hydrophobic loops of more than 70 amino acids that are buried deeply into lipid droplets (16). The exact structure of the hydrophobic loops of the oleosins is not known, but they have been predicted to form α-helical hairpins, with hydrophobic amino acids and a proline-knot turn in the center of the sequence (17). Several other intracellular membrane proteins that are sensitive to the radius of curvature of lipid membranes have also been shown to bind lipids by relatively short hydrophobic loop sequences (18). The vast majority of plasma membrane bound proteins contain at least one or more transmembrane lipid binding domains typically made up of at least 20 hydrophobic amino acids in an α-helical configuration that usually span the membrane in a parallel orientation to the acyl chains of phospholipids (19). Less commonly, transmembrane domains can also be in a β-sheet configuration as part of larger tertiary structure, such as a β-barrel (20). None of these other types of lipid-binding structural motifs, however, have been reported on apoB.
In this study, the apoB sequence was searched for hydrophobic cluster domains containing continuous stretches of at least 6 relatively hydrophobic amino acids that could contribute the lipid binding affinity of apoB. Four such apoB peptides identified by our analysis were synthesized and tested further for their lipid binding affinity. One peptide, B38, consisting of an inner segment of 27 amino acids (B38’) and two 11-mer peptide flanks (or outer segments), was found to readily bind to lipoproteins and was predicted to form a hydrophobic helix parallel to and just below the bilayer/water interface, thus revealing another potential structural motif on apoB that contributes to its lipid binding. The structure and location in the bilayer of B38 was refined using all-atom molecular dynamics (MD) simulations.
EXPERIMENTAL PROCEDURES
Identification of hydrophobic clusters in apolipoprotein B
To allow for computational screening of the apoB amino acid sequence based on hydrophobicity, hydrophobicity scores derived from the octanol:water distribution coefficients for each amino acid were used for analysis (21). All 20 amino acids were also ranked based on relative hydrophobicity with the most hydrophobic amino acid given a value equal to −1 and the most hydrophilic amino acid equal to +1. A table of assigned hydrophobicity scores is provided (Supplemental Table I).
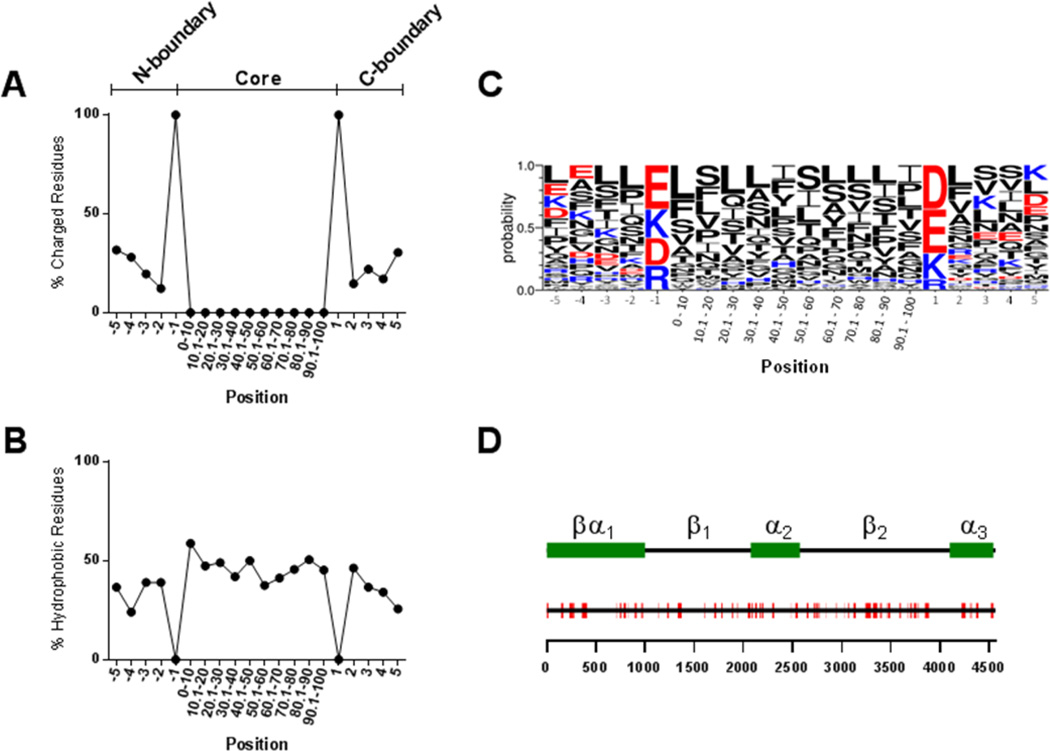
We next performed a hydrophobic “window” analysis of the entire amino acid sequence of apoB. For each position in the apoB sequence, this analysis calculated the sum of hydrophobicity scores for the previous 7 upstream amino acids. The distribution of these scores was examined and the top 5% (most hydrophobic) were selected for further analysis. For each of the top scoring positions, the analysis searched both upstream and downstream until a charged amino acid (D, E, K, or R) was encountered and then included the following four amino acids in each direction. These stretches of hydrophobic residues, bordered by charged residues, were designated as hydrophobic cluster domains (HCD). For definition purposes, each HCD is composed of a “core” and two “boundary” regions (N and C terminal). The core consists of the stretch of only hydrophobic residues and the boundaries consist of the bordering charged residues plus the 4 residues either upstream or downstream of each charged amino acid (Fig. 2A).
Figure 2. Properties of identified HCD sequences.
The distribution of charged (A) and hydrophobic (B) residues along the 78 HCD sequences was analyzed. Positions -1 and 1 represent the upstream and downstream charged boundary residues, respectively. The core region, due to varying lengths among HCD sequences, is represented as equal sized domains each composed of 10% of the sequence length. LOGO analysis was performed to determine the probability of the appearance of specific amino acid residues at each position (C). The positions of the 78 HCD sequences (red) are shown along the full length apoB sequence (black line) (D). The scale below indicates amino acid position within apoB and the legend above indicates the positions of the domains from the pentapartite model of apoB structure (55).
Structural analysis and molecular modeling
Each of the identified HCD sequences was further computationally analyzed for structural and lipid binding properties. LOGO sequence analysis was performed using WebLogo 3.4 (22,23). Helical wheel plots were generated using HeliQuest (24). Structural features of the identified HCD sequences were modeled using PEP-FOLD 2.0 (25). The lipid binding properties of the resulting structures were then analyzed using the position of proteins in membranes (PPM) server (26).
Synthesis of hydrophobic cluster domain containing peptides
Peptides were prepared by solid phase synthesis (Pepmic Co.), purified to >95% by HPLC and confirmed by mass spectrometry. The sequences of the synthesized peptides are presented in Table 1. To preserve the predicted surface orientation and improve the solubility of the HCD peptides, the identified sequences were inserted into a prototypical amphipathic Class A α-helical peptide ELK (27) between two glycine residues, which served as a flexible linker. The term “inner segment” refers to the hydrophobic cluster domain including core and boundary residues (bold text in Table 1). The ELK and GG flexible linker region of the peptides are referred to as the “outer segments”. The inner segment of B38 without the outer segments, denoted by a prime (B38’), was also studied in silico. Note that the native apoB sequence includes B38’ rather than B38. The FLANK peptide, without a hydrophobic loop sequence, was used as a control peptide for all experiments and in silico studies.
Table 1.
Sequences of synthesized peptides.
| Name | Amino acid sequence |
|---|---|
| FLANK | EKLKAKLKEGGEKLKAKLKE |
| B04 | EKLKAKLKEGGLNIKRGIISALLVPPETEEAGGEKLKAKLKE |
| B07 | EKLKAKLKEGGGLSDEAVTSLLPQLIEVSSPGGEKLKAKLKE |
| B38 | EKLKAKLKEGGQELQRYLSLVGQVYSTLVTYISDWWTLGGEKLKAKLKE |
| B61 | EKLKAKLKEGGKATLELSPWQMSALVQVHASQPSSFHDFPDLGGEKLKAKLKE |
Fluorescent labeled FLANK and B38 peptides were synthesized as described above with addition of an N-terminal FITC label.
Circular dichroism spectroscopy
Measurements were recorded using a Jasco J-715 spectropolarimeter across the spectral range from 190 to 240 nm. Peptides were prepared at a final concentration of 0.2 mg/mL in 10 mM phosphate buffer, with or without 20% trifluoroethanol (TFE). During measurements, temperature was maintained at 37 °C. Ellipticity measurements (mdeg) were converted to mean residue ellipticity (θ, deg cm2 dmol−1) (28).
Isothermal titration calorimetry (ITC)
Human LDL and HDL (isolated by ultracentrifugation (29)) and HCD peptides were dialyzed into PBS (150 mM NaCl, 1 mM KH2PO4, 5.6 mM Na2HPO4, pH = 7.4). Peptides were adjusted to 80 µM and lipoproteins adjusted to 8 µM for use in experiments. Calorimetry measurements were performed on a MicroCal iTC200 instrument (Malvern Instruments). The titration involved injection of lipoprotein into the cell containing buffer or the target peptide and consisted of an initial injection of 0.4 uL followed by 35 injections of 1 uL with a delay of 180 seconds between injections. Thermograms were analyzed using NITPIC software (Version 1.1.2) (30).
Cross linking
Experiments were performed using the homo-bifunctional amine reactive crosslinker dimethylpimelimidate hydrochloride (DMP, Thermo Scientific). DMP was incubated with either FLANK or B38 at a 30:1 molar ratio (DMP:peptide) for 2 hours at room temperature in PBS. The crosslinking reaction was quenched with 1 M Tris solution and the crosslinked peptides were analyzed by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and stained with coomassie blue.
Gel filtration chromatography
FITC labeled peptides were incubated with human plasma for 1 hour at 37°C followed by gel filtration chromatography on two Superose 6 columns (GE Healthcare) arranged in series on an AKTA Pure liquid chromatography system (GE Healthcare). Samples were run in standard Tris buffer (10 mM Tris base, 150 mM NaCl, 0.5 mM EDTA, 0.01% sodium azide) at a flow rate of 0.5 mL/min and 0.5 mL fractions were collected. Phospholipid content of the fractions was measured by enzymatic assay (Wako) and labeled peptides were detected using a fluorescent plate reader (ex: 485 nm / em: 535 nm).
Molecular dynamics simulations
FLANK, B38’, and B38 were simulated in fully hydrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) bilayers for a total of 520 ns each. The last 380 ns of all simulations were considered for analyses. Simulations were performed at constant pressure and temperature with a constant number of particles (NPT ensemble). Systems were assembled using the CHARMM-GUI interface (31), and simulated and analyzed using CHARMM 39b2 (32) with the CHARMM 36 all-atom lipid and protein parameters (33–35). 2×64, 2×64, and 2×128 POPCs were used for systems containing FLANK, B38’, and B38, respectively. In each system, two peptides, one per leaflet, were placed on bilayer surfaces with their hydrophobic residues facing the bilayer interior. To minimize interaction across the bilayer, peptides were rotated to be perpendicular to each other. The initial positions of peptides were above the C2 atoms of oleoyl acyl chains. Systems were hydrated using 50 water molecules per lipid. TIP3P water model (36) as modified for CHARMM (37) was used to describe water molecules. 150 mM NaCl was added to systems to mimic the experimental condition. The Lennard-Jones (LJ) parameters of Na+ and Cl− were taken from the CHARMM C36 ion parameters (38) which includes improved Lennard-Jones pairwise distances for Na+ interacting with lipid (39,40).
For computational efficiency, the simulations described above were based on POPC bilayers rather than multilayered lipoproteins. The reliability of results inferred from these simulations was tested by comparing the simulation of the B38’/bilayer complex with a simulation of B38’ bound to a trilayer of POPC leaflets surrounding a layer of triglycerides. Firstly, a peptide-free 2×64 POPC bilayer, hydrated with 5800 water molecules and 150 mM NaCl, was built and simulated. 64 triglycerides (palmitoyl-oleoyl-D-glycerol-linoleoyl) were placed in grid points, separated by 25 Å to make a cube of ~ 75×75×75 Å3, and simulated to reach the target cross-section area of the preceding POPC bilayer (~ 65 Å). The coordinates of the last configuration of POPC simulation were shifted to create a void between the two leaflets where the slab was placed. For the peptide-bound trilayer, the coordinates of the peptide-bound bilayer at 520 ns were shifted to create a void. This way the structure of B38’ was transferred from the bilayer system to the trilayer system. The peptide-free bilayer and trilayer and the peptide-bound trilayer were simulated for 120 ns of which the first 20 ns was considered as the equilibration time and discarded.
Lennard-Jones potentials were terminated at 12 Å, with a smoothing function operating between 8 and 12 Å. Electrostatics were evaluated using particle-mesh Ewald (41) with ca. 1 grid point per Å, a sixth-order spline interpolation for the complementary error function, a real-space cutoff of 12 Å, and a κ value of 0.32 Å−1. Temperature and pressure were maintained 310 K and 1 bar using the Nose-Hoover thermostat (42,43) and Langevin barostat, respectively. The masses of temperature and pressure pistons were 20% and 2% of the systems’ masses, respectively. The pressure was applied semi-isotropically, i.e., the extension of the simulation box in the direction of bilayer normal (z) and bilayer plane (XY) could vary independently. Trajectories were generated with a leapfrog Verlet algorithm with a time step of 1 fs. All bonds to hydrogen atoms were constrained using the SHAKE algorithm (44).
Simulation Analyses
The peptide insertion depth is defined as the vertical distance between centers of mass of backbones of peptides and the plane defined by the phosphorous atoms. The peaks of electron density profiles were used to calculate the depths. Electron density profiles along the bilayer normal (z-axis) were obtained by dividing the simulation box into slabs of 0.2 Å thickness. To determine if a hydrogen bond exists, the following criterion was used: r < 3 Å and α < 30°, where r is the acceptor-donor distance and α is the acceptor-donor-hydrogen angle.
In vitro cholesterol efflux assay
Cholesterol efflux studies were performed as described previously (45). Briefly, BHK-mock (control) and BHK cells stably transfected with human ABCA1 or ABCG1 were incubated for 18 hours with 1 mCi/ml of [3H] cholesterol in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum. After the incubation, cells were washed and the media was replaced with serum-free DMEM containing 0.1 mg/ml fatty acid–free bovine serum albumin. Cholesterol efflux was measured after the addition of media containing peptides or PBS. After 18 hours, media was collected and filtered (Whatman, 24 well 25-mmpore size; GE Healthcare) and cells were lysed in 0.5 ml of 0.1% SDS and 0.1 N NaOH. Radioactive counts in media and cell fractions were measured by liquid scintillation counting on a Perkin Elmer MicroBeta 1450, and results are expressed as the percentage of total counts effluxed.
Solubilization of DMPC vesicles
1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) multilamellar vesicles (1 mg/ml) were prepared by re-suspension of dried DMPC in PBS and vortexing for 5 minutes. Changes in light absorbance at 432 nm upon addition of peptides (final concentration of 100 ng/ml) were recorded every 5 seconds for 60 mins at 24°C with shaking to prevent settling (fast setting). The effect of the peptides on solution turbidity was compared with a negative control solution containing only PBS.
Red blood cell lysis assay
Human red blood cells (RBCs) were washed 5 times in PBS containing 5% glucose (pH = 7.4). Cellular toxicity of peptides was evaluated by incubating RBCs with peptides at various concentrations (range: 0 – 100 µM) for 2 hours at room temperature, followed by centrifugation at 1000×g for 5 minutes to pellet cells. The supernatant was collected and absorbance was measured at 450 nm. Triton X-100 (1%) was used to completely lyse RBCs (100% lysis control).
RESULTS
Hydrophobic Cluster Domain Sequence Analysis
The amino acid sequence of human apoB was searched for hydrophobic cluster domains (HCDs) characterized by continuous stretches of hydrophobic residues flanked on either end by charged residues. This analysis reported hydrophobicity window scores for 4557 positions in the apoB sequence. After analyzing the distribution of these scores, the top 5% (most hydrophobic) were chosen for further analysis, resulting in 78 highly hydrophobic clusters of interest. For each position, the analysis searched both upstream and downstream until the first charged residues were encountered and then included an additional 4 boundary residues in each direction. The sequence information for all identified hydrophobic cluster domains and their positions within the apoB sequence are provided in Supplemental Table II.
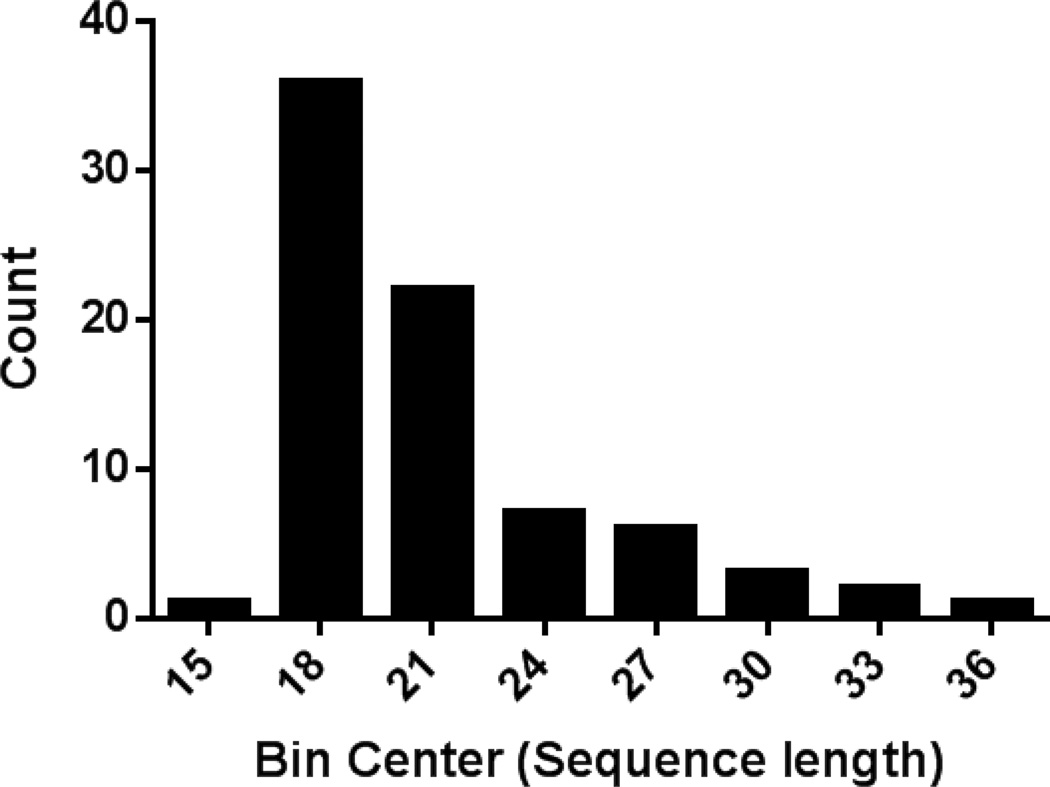
The mean length of the 78 HCDs was 21 residues. The histogram in Fig. 1 displays the distribution of sequence lengths of the HCDs. The majority of HCDs, about 75%, were between 17 and 22 residues long, and the longest was 36 residues (Fig. 1). None of them, however, were long enough to span the diameter of an LDL particle, which would require 200 amino acids if in a helical conformation. One of the longest hydrophobic cluster domains (30 residues) was the amino terminal signal peptide of apoB (residue numbers 1–30) and was excluded from further analysis.
Figure 1. Histogram showing the distribution of HCD sequence lengths.
The lengths of identified HCD sequences (core + boundary residues) were determined and analyzed according to size distribution. Bin centers are presented on the x-axis and bin width was set to 3. The number of sequences whose length fell within a specified bin is indicated on the y-axis.
Analysis of the overall distribution of the charged and hydrophobic residues in the 78 putative hydrophobic clusters is presented in Fig. 2. It is clear that the identified hydrophobic clusters share a common sequence arrangement, by having a central hydrophobic domain of variable length (Core) flanked on both ends by a charged residue (Fig. 2A–B). Excluding the initial charged boundary residues, the mean hydrophobicity of the other 4 boundary residues was less than the central region. The most common residues in the charged boundary positions were negatively charged, Glu (34%) and Asp (28%), followed by the positive residues Lys (24%) and Arg (14%) (Fig. 2C). The charged boundary residues were analyzed to evaluate the frequency of +/+, −/−, or +/− combinations of boundary resides in the 78 sequences. A Chi-square test indicated a significant discrepancy in the expected distribution of boundary residue combinations with significant enrichment of −/− over +/+ combinations (35% vs. 10%, p<0.01) (Supplemental Figure 1).
The most predominant amino acid found in the hydrophobic cluster domains was Leu (15.3%) followed by Ser (12.2%) and Ile (9.4%) (Fig. 2C). When compared to their overall frequency in apoB (Leu 11.7%, Ser 8.6%, Ile 6.3%), these amino acids appear to be enriched in the identified hydrophobic clusters. Although charged amino acids (Lys, Arg, Asp, and Glu) were by definition excluded from the HCD, 31% of the residues within the hydrophobic clusters were uncharged polar amino acids (e.g. Ser, Thr, Asn, Gln). Examination of the relative distribution of each amino acid along the hydrophobic clusters did not reveal obvious enrichment at any location along the length of the identified sequences. This was also done for those residues that can promote turns, such as Pro, Gly, Asn, and Ser, but there was not a predominance of these residues in the center of the hydrophobic clusters, suggesting that the majority of these HCD sequences do not have a central turn. In terms of the location of the hydrophobic cluster domains within apoB, they were found throughout the apoB sequence (Fig. 2D) and did not show distinct clustering among regions as defined by the pentapartite model of apoB (10). Altogether, HCD sequences comprise 36% of the total apoB sequence.
Predicted Secondary Structure of Hydrophobic Clusters
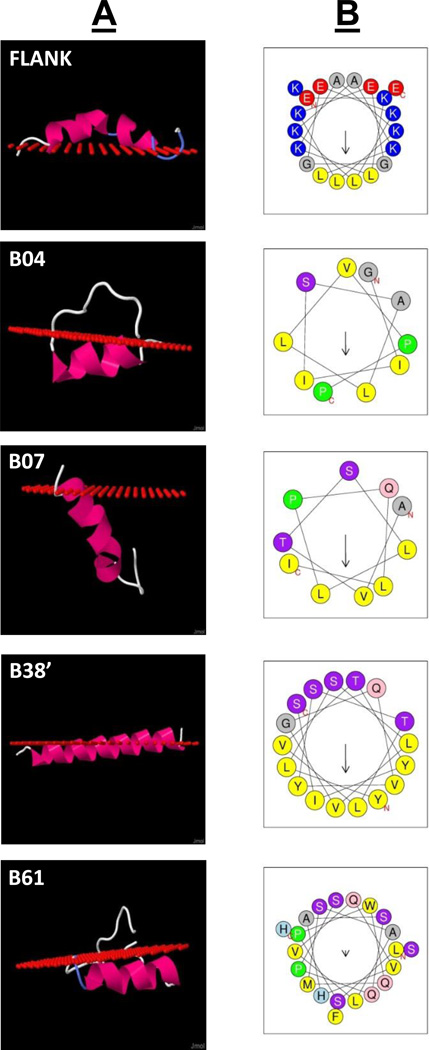
To examine the structure of the HCD peptides and how they interact with lipids, the secondary structure of each HCD (core + boundary residues) was predicted using PepFold (25). The interaction of these peptides with lipids was also modeled, using the Position of Proteins in Membranes (PPM) server (26). From the 78 identified hydrophobic clusters, four peptides with high predicted ΔG for lipid association (i.e. B04, B07, B38, and B61) were selected for synthesis and further analysis. All of the peptides were predicted to form α-helices (Fig. 3A). Furthermore, in each case, except B07, all were predicted to deeply embed in the membrane below the first carbon of the acyl chains of phospholipids and in an orientation parallel to the membrane surface. None of the peptides were predicted to have a turn. Helical plots of the core region of each peptide helped explain its orientation in the membrane (Fig. 3B). Although charged residues were excluded from the sequence analysis in the identification of the hydrophobic clusters, other polar residues were not. As can be best seen in B38, there is a cluster of polar residues on one face of the helix, which is oriented toward the aqueous interface. B07 is predicted to have more like a transmembrane orientation in the lipid surface, and there is not a tight clustering of polar residues at any section of the helix. The predicted physical and lipid binding properties of the 4 peptides are shown in Table 2. Refinements of structures of FLANK, B38’, and B38 are presented in the MD simulation results below.
Figure 3. Predicted lipid interaction of selected HCD peptides.
The positional interaction of synthesized HCD peptides and the FLANK peptide with a lipid surface were predicted using the Position of Proteins in Membranes server (Column A; red spheres indicate level of phospholipid acyl chain carbonyl carbon atoms). Column B displays helical wheel plots for the hydrophobic core domains of each HCD peptide (arrows indicate directionality and magnitude of hydrophobic moment).
Table 2.
Calculated lipid binding properties of native apoB hydrophobic cluster domain and FLANK peptides.
| Peptide | Sequence | ΔG transfer (kcal/mol) |
Length | ΔG per residue (kcal/mol) |
Hydrophobicity | H Moment | Charge |
|---|---|---|---|---|---|---|---|
| FLANK | EKLKAKLKEGGEKLKAKLKE | −2.6 | 20 | −0.13 | −0.153 | 0.444 | 4 |
| B04 | LNIKRGIISALLVPPETEEA | −8.4 | 20 | −0.42 | 0.474 | 0.187 | −1 |
| B07 | GLSDEAVTSLLPQLIEVSSP | −10.7 | 20 | −0.54 | 0.531 | 0.227 | −3 |
| B38' | QELQRYLSLVGQVYSTLVTYISDWWTL | −16.6 | 27 | −0.61 | 0.701 | 0.478 | −1 |
| B61 | KATLELSPWQMSALVQVHASQPSSFHDFPDL | −10.4 | 31 | −0.34 | 0.512 | 0.224 | −2 |
In addition to the 4 selected peptides, Fig. 3A–B also displays the predicted structure of a symmetric helical peptide (FLANK), which was designed to form a Class A amphipathic helix composed mostly of E, L, and K residues. This peptide was designed to be a generic host in which sequences containing hydrophobic clusters can be inserted between the central flexible glycine residues to increase the solubility of the HCD sequences and to help mimic the likely orientation of the hydrophobic cluster sequences within the apoB sequence. Compared to the other peptides, the FLANK peptide without a hydrophobic insert is predicted to be more peripherally associated with the lipid surface and to have a lower binding energy (ΔG) (Table 2). These estimates are similar to what has been previously described for other Class A amphipathic helices in terms of their lipid binding properties (11).
CD Spectroscopy of HCD Peptides
The four HCD peptides were synthesized by inserting their hydrophobic sequences in the flexible glycine hinge region in the FLANK peptide (see Methods). Helicity was measured by circular dichroism in the presence of buffer only, POPS vesicles, or trifluoroethanol (TFE, 20%) to simulate a lipid environment. In aqueous solution, only B38 displayed α-helical secondary structure (40.1% helicity), whereas B04, B07, B61, and FLANK all produced spectra consistent with random coil structure (Supplemental Fig. 2). When added to POPC vesicles, B38 showed a slight increase in helicity (42.4% vs 40.1% helicity). It is possible that the small magnitude of this effect could be due to the proportion of B38 actually bound to vesicles under the conditions measured. All other peptides displayed no change in secondary structure profile and remained as random coil. When TFE was added, all peptides produced classic α-helical CD spectra (FLANK − 38%, B04 − 43%, B07 − 40%, B38 − 59%, and B61 − 60% helicity) consistent with the structural models in Fig. 3.
Lipid Interaction of HCD Peptides
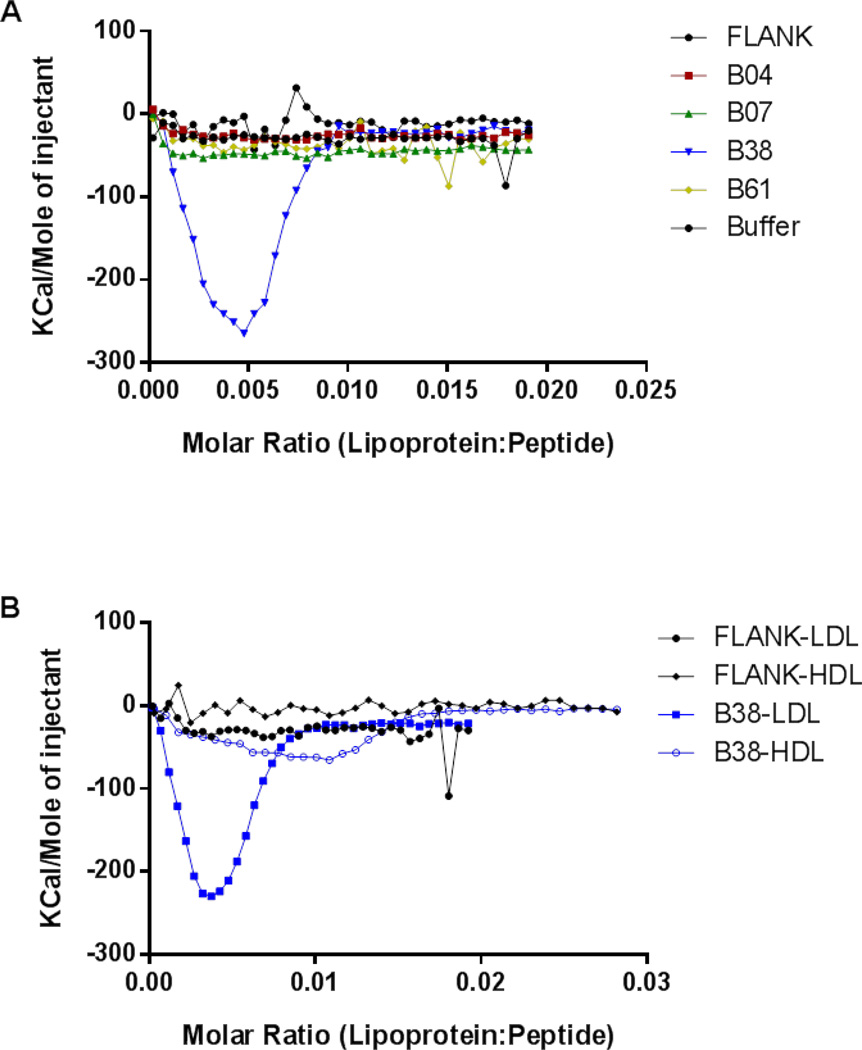
Isothermal titration calorimetry (ITC) was used to examine the thermodynamics of the binding interaction between the HCD peptides and isolated human lipoproteins. This data indicated that B38 was the only peptide with significant binding affinity for LDL (Fig. 4A) and hence the remaining analysis was done just for B38. B38 was also found to bind to high density lipoprotein (HDL) but with lower affinity than LDL. FLANK did not appear to bind strongly to either lipoprotein (Fig. 4B).
Figure 4. Isothermal titration calorimetry measurements of HCD peptides binding to lipoproteins.
To measure the heat energy associated with the binding of the HCD peptides to lipoprotein particles, ultracentrifugally isolated human lipoproteins (80 uM) were injected into the calorimeter cell containing peptide (8 uM) and binding energy was constantly monitored. Isotherms of HCD peptides binding to isolated human LDL (A). Isotherms of FLANK and B38 peptides binding to isolated human LDL or HDL (B).
While the binding energy measurements in these experiments support the interaction between B38 and lipoprotein particles, the thermal energy curves obtained display a U-shaped curve rather than the typical sigmoidal curve associated with protein-protein binding interactions measured by ITC. This type of unusual ITC thermogram has been reported previously in other lipid binding peptides and is generally attributed to altered lipid phase or rapid initial dissolution of aggregated peptides (46). Because hydrophobic peptides are well known to self-associate, we hypothesize that there may be an opposing energy from the dissolution of the B38 oligomers in the presence of LDL. This hypothesis is supported by crosslinking data, which demonstrate the presence of B38 oligomers in aqueous solution (Supplemental Fig. 3). The lack of significant binding energy in ITC experiments when B38 is added to the buffer control (no LDL) indicates that dilution effect by itself does not generate significant energy and the presence of lipoprotein facilitates the energetically favorable transition from oligomer to the lipoprotein bound state.
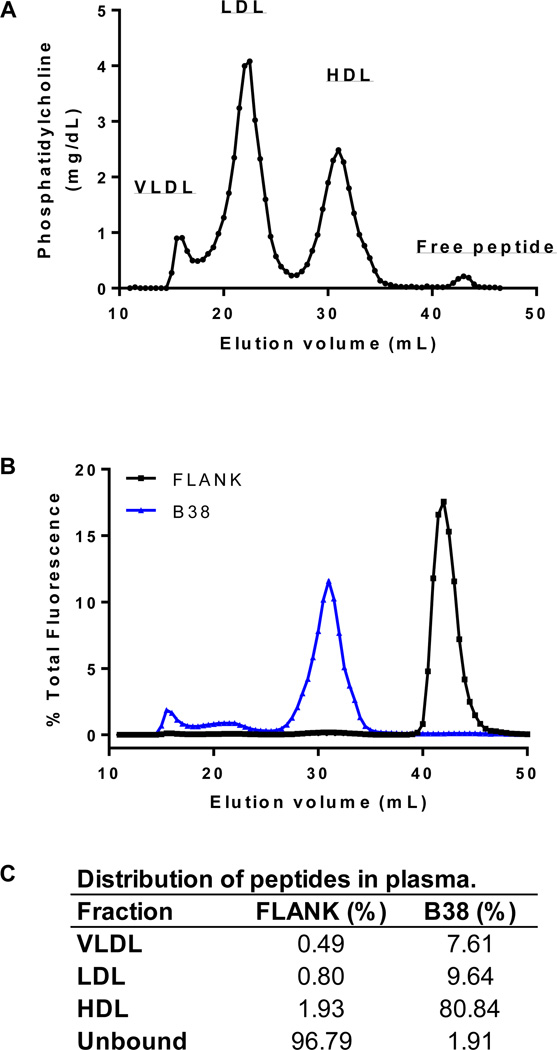
To further examine the binding of these peptides to lipoproteins in the context of whole plasma, fluorescent labeled FLANK or B38 (50 µM) were incubated with human plasma (200 µL) and then lipoproteins and free peptide were separated by gel filtration chromatography. Collected fractions were analyzed for phospholipid content to determine the elution patterns for lipoproteins (Fig. 5A) and fluorescence was used to track the distribution of FLANK and B38 (Fig. 5B). The percentage of total peptide associated with each of the lipoprotein classes is presented in Figure 5C. Overall, B38 displayed significantly greater affinity for lipoproteins than FLANK, 98.1% vs. 3.2% of total peptide bound to lipoproteins, respectively. Furthermore, B38 was mostly associated with HDL (80.8%) with only a relatively small percentage bound to the apoB-containing lipoproteins (7.6 % on VLDL and 9.6% on LDL).
Figure 5. Binding of fluorescent labeled B38 or FLANK to lipoproteins in plasma.
Fluorescent labeled FLANK or B38 peptide was incubated with human plasma prior to separation by size-exclusion chromatography. The resulting distributions of phosphatidylcholine (A) and fluorescent peptide (B) are displayed. Elution positions of the major plasma lipoprotein classes (i.e. VLDL, LDL, and HDL) are indicated. The quantitative distribution of fluorescent FLANK and B38 among lipoprotein classes is presented as percent of total fluorescence associated with each peak (C).
All-atom MD simulations of peptide-lipid interactions
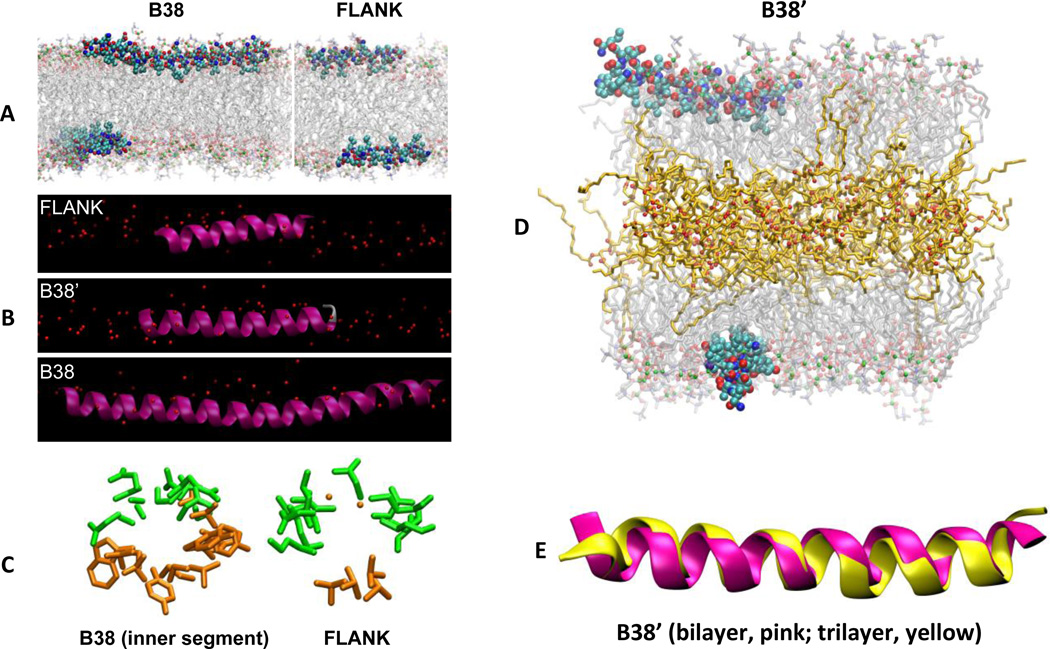
The results from MD simulations provide atomic resolution detail of the interaction of B38 and the membrane surface. Figure 6A shows snapshots of simulations of B38 and FLANK. The electron density profiles (Supplemental Fig. 5) indicate that B38’, B38, and FLANK are 3.5 Å, 3.7 Å, and 1.7 Å below the phosphorus atoms of POPC lipids, respectively. The inner segment of B38 is closer to the hydrophobic core of bilayer than its outer segments (Supplemental Fig. 5, red and blue lines). That is, B38 is slightly depressed toward the bilayer interior (Fig. 6B), with the bend occurring at Gly22. The outer segments of B38 (Supplemental Fig. 5, blue line) insert deeper than FLANK (Supplemental Fig. 5, black line). This shows the ability of the inner segment of B38 not only to bury itself under the phospholipid head groups, but also to pull its coupled outer segments toward the center of the bilayer. Figure 6C shows the view along the axes of helices of FLANK and the inner segment of B38. These snapshots are consistent with the helical wheel diagrams in Fig. 3.
Figure 6. All-atom MD simulations of FLANK and B38 in a POPC bilayer.
Simulation snapshots of the peptides in their average orientations in three different representations. (A) B38 (left) and FLANK (right) in CPK representation with carbons light blue, oxygens red, and nitrogens blue. Acyl chains and head groups of lipids are represented by transparent lines and balls, respectively, with phosphorus atoms green. Water molecules and hydrogens are omitted for clarity. (B) FLANK, B38’, and B38 in the lipid surface (red spheres denote phosphorus). (C) “Helical-wheel” representations of peptides looking along the helical axes of B38 and FLANK. Only side chains (excluding hydrogen atoms) are shown for clarity. Hydrophilic and hydrophobic residues are shown in green and orange, respectively. These helical wheels, which are obtained using MD simulations, confirm those of Fig. 3, which are obtained using the Position of Proteins in Membranes server. (D) Simulation snapshot of B38’ interacting with a trilayer at 120 ns. Triglyceride acyl chains and oxygens are represented by orange lines and red balls, respectively. (E) Superimposed structures of B38’ in bilayer (pink) and trilayer (yellow). The structures correspond to last configurations of bilayer (520 ns) and trilayer (120 ns) systems.
The inner segment of B38 made fewer hydrogen bonds (H-bonds) with lipids than its outer boundary sequences. As Supplemental Table III shows, the average number of H-bonds per residue of the inner segment of B38 is less than half of that of its outer boundary sequences (0.12 vs 0.28). Both the outer segments of B38 and FLANK make H-bonds mostly with phosphate groups, which is consistent with the observation that they are closer to phosphorus atoms than to C2 atoms of oleoyl acyl chains (Supplemental Fig. 5). Compared to FLANK and the outer segment of B38, the inner segment of B38 has more H-bonds with carbonyl groups and fewer bonds with phosphate, consistent with deeper membrane insertion. The most significant contributions to H-bonds between the inner segment of B38 and POPC lipids came from ARG16, TYR25, TRP36, TYR31, SER19, GLN15, TYR17, TRP35, SER33, SER26, and GLN12, in order from largest to smallest. Being more exposed to water, both FLANK and outer segments of B38 make more H-bonds with water than the inner segment of B38 does. The ability of the outer segments of B38 to make H-bonds with POPC lipids or water is less than the ability of FLANK. This is due to the re-orientation of the outer segments of B38 caused by the inner segment of B38.
In addition to the peptide-bound bilayers described above, three systems were simulated to ascertain the differences in peptide conformation and orientation that might be induced by the multilayered lipoprotein environment: pure POPC bilayer, a trilayer consisting of POPC leaflets surrounding a layer of triglyceride, and a B38’/trilayer complex. Figure 6D shows the snapshot of the peptide/trilayer complex at 120 ns. The triglycerides mostly, though not entirely, occupy the interleaflet space; electron density profiles of the peptide-free system indicate that 9.5 mol % of triglycerides penetrate into the POPC hydrophobic region (Supplemental Fig. 6). This relatively small amount of interdigitation of triglycerides and POPC acyl chains has two consequences. Firstly, the area-per-POPC increases from 66.5 ± 0.2 Å2 in the bilayer to 68.0 ± 0.2 Å2 in the trilayer (approximately 2%) (Supplemental Fig. 7). Secondly, as the analysis of deuterium order parameter (Scd) shows (Supplemental Fig. 8), POPC acyl chains become more ordered in the trilayer. Similar simulations and analyses of trilayers made of POPC and cholesteryl esters showed an increase of 1% in the surface area and a solubility of 8 mol % of the esters, but no change in the ordering of lipids (47).
Due to the ordering effect of triglycerides on POPC lipids, B38’ insertion into the trilayer is approximately 1 Å shallower than in the bilayer. (See the time series in Supplemental Fig. 9, where B38’ is 2.2 Å below the phosphorus atoms in the trilayer and 3.5 Å in the bilayer.)
Despite different insertion depths, B38’ adopts the same secondary structure in the bilayer and trilayer (Fig. 6E). Time series of the percentage of α-helical residues is shown in the Supplemental Fig. 10. The average helical content of B38’ is 85 ± 1 % and 87 ± 1 % in the bilayer and trilayer, respectively. The structural stability of B38’ was further confirmed by the Root Mean Square (RMS) analysis of the backbone of B38’ with respect to a reference structure. As evident from the Supplemental Fig. 11, the RMS of B38’ in the trilayer remained consistent throughout the 120 ns-long simulation.
Exchangeability of B38 among lipoprotein classes
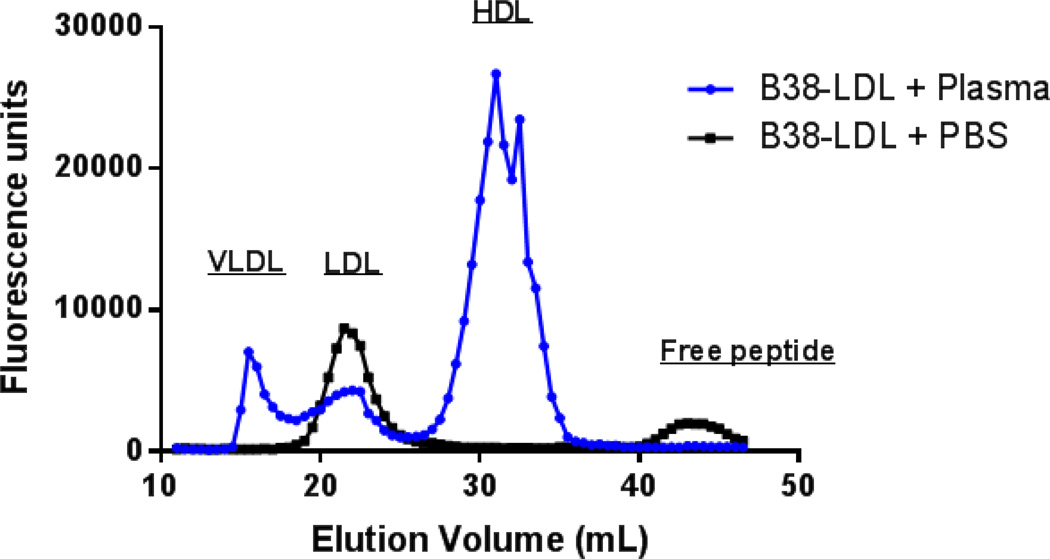
To assess the exchangeability of the B38 peptide between lipoproteins, fluorescent labeled B38 peptide was incubated with isolated human LDL and unbound peptide was removed by gel filtration chromatography. The B38-LDL was then incubated with human plasma for 1 hour at 37 °C and the transfer of fluorescent peptide to other lipoproteins was measured. This experiment showed the appearance of fluorescence signal in all lipoprotein fractions after incubation with the B38-LDL, indicating that the B38 peptide, unlike apoB, is exchangeable between lipoproteins (Fig. 7).
Figure 7. Exchangeability of B38 peptide bound to LDL.
To examine the exchangeability of the B38 peptide among lipoproteins, FITC labeled B38 peptide was co-incubated with isolated human LDL for 1 hour at 37 °C and LDL was repurified by FPLC to remove unbound peptide. Fractions containing LDL were then pooled and concentrated before incubation with either PBS or human plasma for 1 hour at 37 °C. These samples were again separated by FPLC and fluorescence distribution across collected fractions was measured. Elution positions of major lipoprotein classes are indicated.
B38 is a potent stimulator of ABCA1 mediated cholesterol efflux
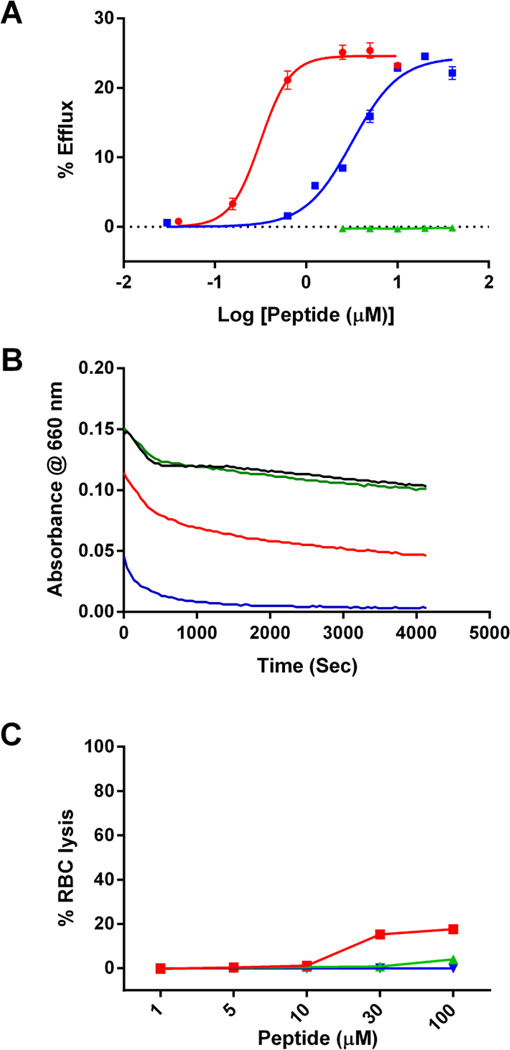
The helical wheel plot of B38 (Fig. 3) suggests that this peptide exists as an amphipathic α-helix, a common motif that can promote cellular cholesterol efflux. In contrast to the usual helices found on exchangeable apolipoproteins, the B38 amphipathic helix has a very broad hydrophobic face (56% of B38 circumference) and a relatively strong hydrophobic moment (µH = 0.478) (Table 2). Additionally, the hydrophilic face is composed mostly of uncharged polar residues (i.e. serine and threonine). Although apoB itself does not promote cholesterol efflux from cells, we hypothesized based on its structure and high lipid affinity that the B38 peptide would be capable of cholesterol efflux (48). As a comparator, the efflux capacity of the well-studied apolipoprotein mimetic peptide “5A” was also measured (48,49). The results indicate that B38 has a 12-fold lower EC50 (0.2658 vs. 3.188 µM; p < 0.0001) for cholesterol efflux from BHK cells transfected with ABCA1 (Fig. 8A). In contrast, the FLANK was ineffective in promoting cholesterol efflux. None of the tested peptides had significant capacity for efflux from Mock or ABCG1 transfected BHK cells (Supplemental Fig. 4).
Figure 8. ABCA1 mediated cholesterol efflux to FLANK, B38, and 5A peptides.
(A) The ability of the FLANK (green) and B38 (red) peptides to accept radiolabeled cholesterol from ABCA1 expressing BHK cells was measured over a range of peptide concentrations and compared against a well-studied apolipoprotein mimetic peptide (5A, blue) with potent cholesterol efflux capacity. Percent efflux was calculated as the amount of radiolabel detected in the media, after 18 hour incubation with peptide, divided by the total amount of radiolabel in the system (media + cells) * 100. (B) DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) clearance assay was performed to evaluate the ability of PBS buffer (black), FLANK (green), B38 (red), and 5A (blue) to solubilize lipid vesicles. Peptides (100 ng/mL were incubated with DMPC vesicles at 24 °C and light absorbance at 432 nm was recorded every 5 seconds for a total of 60 minutes (C) Red blood cell lysis assay was performed to evaluate cytotoxicity of FLANK (green), B38 (red), and 5A (blue). 1% triton solution was used as a 100% lysis control.
Some peptides with high capacity for cholesterol efflux have detergent-like properties that can cause them be cytotoxic due to disruption of the cell membrane. The detergent properties of B38, were evaluated using a 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) vesicle clearance assay (Fig. 8B). FLANK (green line) did not solubilize DMPC vesicles; however, both B38 (red line) and 5A (blue line) rapidly cleared the solution. Finally, to evaluate potential toxicity of B38, a red blood cell lysis assay was performed (Fig. 8C). FLANK (green line) and 5A (blue line) did not cause red cell lysis, B38 (red line) caused a small amount of lysis at concentrations above 30 uM, although no visual evidence of cytotoxicity was observed during the cholesterol efflux experiments.
DISCUSSION
Prior to this study, two types of secondary structure were reported to account for the lipid binding properties of apoB, namely β-sheets and Class A amphipathic helices (10). The main finding from this study is that short hydrophobic cluster domains also contribute to the lipid binding of apoB. The identified HCDs were variable in length and were largely composed of hydrophobic amino acids, such as Leu and Ile, plus some polar amino acids, such as Ser and Thr. The majority of these sequences were predicted to have α-helical secondary structure. However, these did not display the typical distribution of charged residues associated with traditional Class A alpha α-helices. The HCDs were located throughout the apoB sequence and comprised over one third of the total apoB sequence.
Based on the HCD sequences with high predicted ΔGtransfer for lipid binding, four HCD peptides were synthesized and experimentally tested. Only one synthesized peptide, B38, showed significant lipoprotein binding capacity (Fig. 4). This sequence is located between residues 2,530 – 2,556 in apoB and based on CD spectroscopy the B38 peptide formed an α-helix in aqueous buffer (Supplemental Fig. 2). This peptide had a relatively large hydrophobic face covering more than half of the circumference of the helix, which most likely accounts for its ability for spontaneous binding to preformed lipoproteins. B38 also contains several polar Ser and Thr residues, which were clustered on the hydrophilic face of the helix, facing toward the aqueous solvent when bound to a lipid bilayer (Fig. 3). Thus, this peptide is similar to a Class A amphipathic helix except for the larger hydrophobic face and the presence of uncharged polar residues rather than charged residues on the hydrophilic face (11). Most likely because of these two differences, B38 was predicted to insert deeper into a phospholipid bilayer than Class A amphipathic helices, such as the FLANK peptide (Fig. 3).
MD simulations show a deeper position of B38 than FLANK within bilayers, which agrees with the predictions from Pep-Fold, and a lower ability to make H-bonds with lipids. This indicates that the binding of B38 to lipids is due to its hydrophobicity rather than specific intermolecular interactions between the peptide and the membrane surface. Otherwise, FLANK would have had a higher binding affinity because it makes more H-bonds. That is, entropic effects play more important roles than enthalpic effects in the binding of B38 to lipids. Overall, MD simulations indicate that B38, which does not contain residues that would be predicted to form a tight turn, can nevertheless adopt a conformation that allows a shallow curve, resulting in a deeper penetration of its HCD region into the lipid bilayer.
MD simulation confirms the predicted α-helical structure of B38’ by PepFold, although the position in MD is closer to the phosphate groups. This is not surprising because the membrane representation in PepFold is simpler than in an all-atom model. There is a larger discrepancy between structural results for FLANK (compare Fig. 3 and Fig. 6B). Although the presence of two glycine residues in FLANK can ease the rotation of dihedral angles, no obvious kinking of the peptide was observed throughout the 500 ns MD simulation of FLANK.
There are limitations, however, to the extent that current simulations can elucidate peptide-lipoprotein interactions. Firstly, the periodic boundary conditions enforce bilayers to be flat, whereas lipoproteins are curved. Curvature can lead to surface defects, which in principle can alter the binding mode of peptides. It is computationally intensive to simulate a whole lipoprotein to capture the curvature and not feasible at this time. Secondly, while the simulations of trilayers and bilayers yielded nearly identical conformations for B38’ and qualitatively similar depths of burial and orientations, a more exhaustive study of trilayer systems with components including cholesterol and cholesterol esters will be required to understand these systems.
The other synthesized HCD peptides (and presumably many of the identified hydrophobic regions with a relatively small predicted ΔG for lipid binding) did not bind to lipoproteins. Nevertheless, these regions of apoB could still collectively contribute to the overall lipid binding property of apoB. One possible explanation is that although these hydrophobic sequences do not readily insert into an existing lipoprotein surface, they may readily bind to lipid during lipoprotein formation. This is supported by our current knowledge of the formation of apoB containing lipoproteins, which indicates that an accessory protein, microsomal triglyceride transfer protein (MTP) is required for proper insertion of apoB into a lipid droplet during the lipidation process of apoB (50). Although individual HCD peptides (i.e. B38) are exchangeable among lipoproteins, it is likely that the presence of a large number of these domains within a protein would have a cumulative effect and collectively contribute to the non-exchangeable behavior of apoB. Additionally, the dispersion of these hydrophobic domains could generate flexibility of the apoB protein as the particle on which it resides undergoes lipolysis and experiences dramatic reductions in size. Such flexibility is likely restricted by the rest of apoB sequence, such as the rigid amphipathic β-sheet domains that compose a significant portion of the apoB sequence.
The oligomerization of B38 in aqueous solution is not a surprising finding. It is common for amphipathic peptides and proteins to aggregate in an orientation that provides shielding of their hydrophobic residues from the aqueous surroundings. Many exchangeable apolipoproteins accomplish this through the formation of helical bundles in the absence of lipid (13,51). B38 was found in forms ranging from a monomer to as many as 14 peptides per oligomeric complex. In contrast, the Flank peptide was only found as monomer and dimer in aqueous solution. The reason for this difference may be due to the larger hydrophobic face of the B38 helix, thus requiring more peptides for effective shielding of hydrophobic residues. These findings also likely account for the α-helical nature of B38 in aqueous solution in the absence of lipid (or TFE). Like the other HCD peptides tested, many short amphipathic α-helical peptides are in the random coil state in aqueous solution and require association with lipid to adopt an α-helical conformation.
In ITC experiments, B38 displays a total binding energy for LDL that is 2.7 times greater than for HDL suggesting that the peptide prefers to bind to LDL over HDL. However, when B38 was added to human plasma and lipoprotein binding was analyzed after FPLC separation, the majority of the peptide was found associated with HDL (81%) and much less was bound to apoB containing lipoproteins (17%). One likely factor contributing to this apparent discrepancy is that there is a dramatic difference in the plasma concentrations of these two lipoprotein classes with HDL being about 35 times greater (by particle number) than LDL. The effect of this, and the smaller diameter of HDL particles, is that the total surface area available on HDL could actually be significantly greater than that on LDL given equal plasma phospholipid contents of the two lipoproteins. Another contributing factor could be the possibility that other plasma components have some influence on B38 binding to lipoproteins in the FPLC experiments compared to the ITC experiments, where isolated lipopoproteins were used.
In addition to its lipid binding properties, the B38 peptide is capable of ABCA1-dependent cholesterol efflux from cells. Although it is not likely that this peptide is performing this activity in vivo, particularly within the context of full length apoB, it is surprising that B38 effluxes cholesterol with 12-fold higher efficiency than the apolipoprotein A-I mimetic peptide, 5A (52,53). The structural basis for this feature of B38 is not clear, although the relatively large size of the hydrophobic face, and the nature of the hydrophilic face, consisting of mostly serine and threonine, may hold some clues. A similar type of amphipathic helix containing polar but not charged residues has been described with membrane curvature sensing peptides (54), which gives these peptides flexibility in binding to different size vesicular structures. Our lab has also previously shown that peptides with larger hydrophobic faces are in general superior in cholesterol efflux (27). It is interesting to note that although B38 has more potent cholesterol efflux capacity than 5A, B38 does not clear DMPC vesicles as rapidly suggesting a lower overall capacity for detergent-like solubilization of lipids and a higher specificity of B38 as an ABCA1 specific lipid acceptor, consistent with the data in supplemental figure 4. It is sometimes a concern that highly hydrophobic peptides with strong detergent like properties may cause toxicity due to cell lysis making them unavailable for use as research or therapeutic agents. B38 did show some minor tendency toward hemolysis at the highest concentrations tested.
In conclusion, the hydrophobic cluster domains identified in this study represent a novel lipid binding motif on apoB and are distinct from traditional amphipathic α-helix and β-sheet lipid-binding domains. Our findings indicate that these HCDs, in the form of individual peptides, can be transferred from LDL to other lipoproteins in vitro, however, it is likely that the cumulative effect of these domains comprises a significant contribution to apoB’s non-exchangeable nature, while also allowing structural flexibility. More detailed studies of these domains may yield new insight into the mechanisms of lipid accumulation by apoB and the formation of apoB containing lipoproteins. Additionally, the unexpected cholesterol efflux properties of B38 provide a new foundation for designs of future peptides for stimulating reverse cholesterol transport.
Supplementary Material
Highlights.
Identification of a novel lipid-binding motif within apolipoprotein B.
Molecular dynamics simulations of peptide-lipid surface interactions.
The B38 peptide promotes ABCA1 specific cholesterol efflux.
Acknowledgments
This project was funded by the National Heart, Lung, and Blood Institute Division of Intramural Research, and utilized the computational resources of the NIH HPC Biowulf and NHLBI LoBoS clusters. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The authors would also like to acknowledge the NHLBI Biophysics Core Facility for training and access to instruments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Kannel WB, Castelli WP, Gordon T. Cholesterol in the prediction of atherosclerotic disease. New perspectives based on the Framingham study. Ann Intern Med. 1979;90:85–91. doi: 10.7326/0003-4819-90-1-85. [DOI] [PubMed] [Google Scholar]
- 2.Austin MA, Hokanson JE, Edwards KL. Hypertriglyceridemia as a cardiovascular risk factor. Am J Cardiol. 1998;81:7B–12B. doi: 10.1016/s0002-9149(98)00031-9. [DOI] [PubMed] [Google Scholar]
- 3.Cladaras C, Hadzopoulou-Cladaras M, Nolte RT, Atkinson D, Zannis VI. The complete sequence and structural analysis of human apolipoprotein B-100: relationship between apoB-100 and apoB-48 forms. EMBO J. 1986;5:3495–3507. doi: 10.1002/j.1460-2075.1986.tb04675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anant S, MacGinnitie AJ, Davidson NO. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270:14762–14767. [PubMed] [Google Scholar]
- 5.Rutledge AC, Su Q, Adeli K. Apolipoprotein B100 biogenesis: a complex array of intracellular mechanisms regulating folding, stability, and lipoprotein assembly. Biochem Cell Biol. 2010;88:251–267. doi: 10.1139/o09-168. [DOI] [PubMed] [Google Scholar]
- 6.Ren G, Rudenko G, Ludtke SJ, Deisenhofer J, Chiu W, Pownall HJ. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proc Natl Acad Sci U S A. 2010;107:1059–1064. doi: 10.1073/pnas.0908004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar V, Butcher SJ, Oorni K, Engelhardt P, Heikkonen J, Kaski K, Ala-Korpela M, Kovanen PT. Three-dimensional cryoEM reconstruction of native LDL particles to 16A resolution at physiological body temperature. PLoS One. 2011;6:e18841. doi: 10.1371/journal.pone.0018841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Atkinson D. Immuno-electron cryo-microscopy imaging reveals a looped topology of apoB at the surface of human LDL. J Lipid Res. 2011;52:1111–1116. doi: 10.1194/jlr.M013946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johs A, Hammel M, Waldner I, May RP, Laggner P, Prassl R. Modular structure of solubilized human apolipoprotein B-100. Low resolution model revealed by small angle neutron scattering. J Biol Chem. 2006;281:19732–19739. doi: 10.1074/jbc.M601688200. [DOI] [PubMed] [Google Scholar]
- 10.Segrest JP, Jones MK, De Loof H, Dashti N. Structure of apolipoprotein B-100 in low density lipoproteins. J Lipid Res. 2001;42:1346–1367. [PubMed] [Google Scholar]
- 11.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. Journal of lipid research. 1992;33:141–166. [PubMed] [Google Scholar]
- 12.Mishra VK, Palgunachari MN, Segrest JP, Anantharamaiah GM. Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. The Journal of biological chemistry. 1994;269:7185–7191. [PubMed] [Google Scholar]
- 13.Tubb MR, Silva RA, Fang J, Tso P, Davidson WS. A three-dimensional homology model of lipid-free apolipoprotein A-IV using cross-linking and mass spectrometry. The Journal of biological chemistry. 2008;283:17314–17323. doi: 10.1074/jbc.M800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Small DM. Interfacial properties of amphipathic beta strand consensus peptides of apolipoprotein B at oil/water interfaces. J Lipid Res. 2004;45:1704–1715. doi: 10.1194/jlr.M400106-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Martin DD, Genter E, Wang J, McLeod RS, Small DM. Surface study of apoB1694-1880, a sequence that can anchor apoB to lipoproteins and make it nonexchangeable. J Lipid Res. 2009;50:1340–1352. doi: 10.1194/jlr.M900040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang AH. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996;110:1055–1061. doi: 10.1104/pp.110.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander LG, Sessions RB, Clarke AR, Tatham AS, Shewry PR, Napier JA. Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta. 2002;214:546–551. doi: 10.1007/s004250100655. [DOI] [PubMed] [Google Scholar]
- 18.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 19.Rath A, Deber CM. Protein structure in membrane domains. Annual review of biophysics. 2012;41:135–155. doi: 10.1146/annurev-biophys-050511-102310. [DOI] [PubMed] [Google Scholar]
- 20.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochimica et biophysica acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Wolfenden R. Experimental measures of amino acid hydrophobicity and the topology of transmembrane and globular proteins. J Gen Physiol. 2007;129:357–362. doi: 10.1085/jgp.200709743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crooks GE, Hon G, Chandonia J-MM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic acids research. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 25.Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37:W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza W, Stonik JA, Murphy A, Demosky SJ, Sethi AA, Moore XL, Chin-Dusting J, Remaley AT, Sviridov D. Structure/function relationships of apolipoprotein a-I mimetic peptides: implications for antiatherogenic activities of high-density lipoprotein. Circ Res. 2010;107:217–227. doi: 10.1161/CIRCRESAHA.110.216507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers JK, Pace CN, Scholtz JM. Helix propensities are identical in proteins and peptides. Biochemistry. 1997;36:10923–10929. doi: 10.1021/bi9707180. [DOI] [PubMed] [Google Scholar]
- 29.Chapman MJ, Goldstein S, Lagrange D, Laplaud PM. A density gradient ultracentrifugal procedure for the isolation of the major lipoprotein classes from human serum. J Lipid Res. 1981;22:339–358. [PubMed] [Google Scholar]
- 30.Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of computational chemistry. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 32.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. Journal of computational chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. Journal of chemical theory and computation. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacKerell AD, Feig M, Brooks CL. Improved treatment of the protein backbone in empirical force fields. Journal of the American Chemical Society. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 35.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. The journal of physical chemistry. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- 37.Durell SR, Brooks BR, Ben-Naim A. Solvent-Induced Forces between Two Hydrophilic Groups. The Journal of Physical Chemistry. 1994;98:2198–2202. [Google Scholar]
- 38.Noskov SY, Roux B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. Journal of molecular biology. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venable RM, Luo Y, Gawrisch K, Roux B, Pastor RW. Simulations of anionic lipid membranes: development of interaction-specific ion parameters and validation using NMR data. The journal of physical chemistry. B. 2013;117:10183–10192. doi: 10.1021/jp401512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Roux Bt. Simulation of Osmotic Pressure in Concentrated Aqueous Salt Solutions. The Journal of Physical Chemistry Letters. 2010;1:183–189. [Google Scholar]
- 41.Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98:10089. [Google Scholar]
- 42.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Physical review. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 43.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. The Journal of Chemical Physics. 1984;81:511. [Google Scholar]
- 44.Ryckaert J-P, Ciccotti G, Berendsen H. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 45.Amar MJ, Sakurai T, Sakurai-Ikuta A, Sviridov D, Freeman L, Ahsan L, Remaley AT. A novel apolipoprotein C-II mimetic peptide that activates lipoprotein lipase and decreases serum triglycerides in apolipoprotein E-knockout mice. J Pharmacol Exp Ther. 2015;352:227–235. doi: 10.1124/jpet.114.220418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrushchenko VV, Aarabi MH, Nguyen LT, Prenner EJ, Vogel HJ. Thermodynamics of the interactions of tryptophan-rich cathelicidin antimicrobial peptides with model and natural membranes. Biochimica et biophysica acta. 2008;1778:1004–1014. doi: 10.1016/j.bbamem.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 47.Koivuniemi A, Heikelä M, Kovanen PT, Vattulainen I, Hyvönen MT. Atomistic simulations of phosphatidylcholines and cholesteryl esters in high-density lipoprotein-sized lipid droplet and trilayer: clues to cholesteryl ester transport and storage. Biophysical journal. 2009;96:4099–4108. doi: 10.1016/j.bpj.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amar MJ, D'Souza W, Turner S, Demosky S, Sviridov D, Stonik J, Luchoomun J, Voogt J, Hellerstein M, Remaley AT. 5A apolipoprotein mimetic peptide promotes cholesterol efflux and reduces atherosclerosis in mice. J Pharmacol Exp Ther. 2010;334:634–641. doi: 10.1124/jpet.110.167890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabet F, Remaley AT, Segaliny AI, Millet J, Yan L, Nakhla S, Barter PJ, Rye KA, Lambert G. The 5A apolipoprotein A-I mimetic peptide displays antiinflammatory and antioxidant properties in vivo and in vitro. Arterioscler Thromb Vasc Biol. 2010;30:246–252. doi: 10.1161/ATVBAHA.109.200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 51.Melchior JT, Walker RG, Morris J, Jones MK, Segrest JP, Lima DB, Carvalho PC, Gozzo FC, Castleberry M, Thompson TB, Davidson WS. An Evaluation of the Crystal Structure of C-terminal Truncated Apolipoprotein A-I in Solution Reveals Structural Dynamics Related to Lipid Binding. The Journal of biological chemistry. 2016 doi: 10.1074/jbc.M115.706093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw JA, Bobik A, Murphy A, Kanellakis P, Blombery P, Mukhamedova N, Woollard K, Lyon S, Sviridov D, Dart AM. Infusion of reconstituted high-density lipoprotein leads to acute changes in human atherosclerotic plaque. Circ Res. 2008;103:1084–1091. doi: 10.1161/CIRCRESAHA.108.182063. [DOI] [PubMed] [Google Scholar]
- 53.Javaheri A, Kolansky DM, Cuchel M. Reconstituted high-density lipoprotein therapies: a cause for optimism. Arterioscler Thromb Vasc Biol. 2014;34:1800–1802. doi: 10.1161/ATVBAHA.114.304156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drin G, Casella J-F, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic α-helical motif for sensing membrane curvature. Nature Structural & Molecular Biology. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 55.Segrest JP, Jones MK, Mishra VK, Anantharamaiah GM, Garber DW. apoB-100 has a pentapartite structure composed of three amphipathic alpha-helical domains alternating with two amphipathic beta-strand domains. Detection by the computer program LOCATE. Arterioscler Thromb. 1994;14:1674–1685. doi: 10.1161/01.atv.14.10.1674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.