Abstract
Epigenetics is the regulation of gene expression (transcription) in response to changes in the cell environment through genomic modifications that largely involve the non-coding fraction of the human genome and that cannot be attributed to modification of the primary DNA sequence. Epigenetics is dominant in establishing cell fate and positioning during programmed embryonic development. However the same pathways are used by mature postnatal and adult mammalian cells during normal physiology and are implicated in disease mechanisms. Recent research demonstrates that blood flow and pressure are cell environments that can influence transcription via epigenetic pathways. The principal epigenetic pathways are chemical modification of cytosine residues of DNA (DNA methylation) and of the amino tails of histone proteins associated with DNA in nucleosomes. They also encompass the post-transcriptional degradation of mRNA transcripts by non-coding RNAs (ncRNA). In vascular endothelium, epigenetic pathways respond to temporal and spatial variations of flow and pressure, particularly hemodynamic disturbed blood flow, with important consequences for gene expression. The biofluid environment is linked by mechanotransduction and solute transport to cardiovascular cell phenotypes via signaling pathways and epigenetic regulation for which there is an adequate interdisciplinary infrastructure with robust tools and methods available. Epigenetic mechanisms may be less familiar than acute genomic signaling to Investigators at the interface of biofluids, biomechanics and cardiovascular biology. Here we introduce a biofluids / cellular biomechanics readership to the principal epigenetic pathways and provide a contextual overview of endothelial epigenetic plasticity in the regulation of flow-responsive transcription.
Keywords: hemodynamics, epigenetics, endothelial phenotype, gene transcription, disturbed blood flow, mechanotransduction
INTRODUCTION
The mechanical properties of blood flow are ever-present at the luminal cell surface of the vascular endothelium. This biofluid interface plays a central role in the health and pathology of the endothelium through flow-driven cell deformation and mechanotransduction (Davies and Helmke 2010) that has been etensively studied by gene and protein expression, metabolism, and function (Davies 1995, 2009; Chiu et al 2011; Gimbrone and Garcia-Cardena 2016; Rizzo et al 1998) Transcription of the genomic (DNA) code to mRNA (“gene expression”) in preparation for protein synthesis is well described to be mechano-sensitive (Garcia-Cardena et al 2001; Dekker et al 2002; Passerini et al 2004). Cis/trans mechanisms involving distal and proximal control elements and transcription factors, several of which are master regulators of endothelial phenotype, readily respond to hemodynamic patterns in vivo and recapitulated flow characteristics in vitro. However, although it was long recognized that these classical mechanisms are insufficient to completely account for the suppression and activation of genes, elucidation of the prominence in epigenetic transcriptional regulation of non-coding regions of the human genome has only emerged recently. In cardiovascular research, epigenetic mechanisms are now linked to hypercholesterolemia (Schiano et al 2015), hypertension (Friso et al 2015), exercise (Horsburgh et al 2015) and cigarette smoking (Joehanes et al 2016). It is estimated that the epigenomic code accounts for the regulation of 30% of mammalian proteins. In vascular research, mostly microRNA (miRNA) regulation of gene expression has been studied (Zhou et al 2011; Kumar et al 2014). Other epigenetic pathways, however, that regulate chromatin structure are now identified with flow-mediated transcription. Here we introduce epigenetic mechanisms to a readership that may be less familiar with the emerging roles of biofluids and biomechanics in epigenetics, particularly in cardiovascular research, and outline the influence of cell epigenetic status including its plasticity in the regulation of endothelial transcription in flow-sensitive environments (Jiang et al 2014; Davies et al 2014; Dunn et al 2014).
EPIGENETIC MECHANISMS AND BIOFLUID MECHANICS
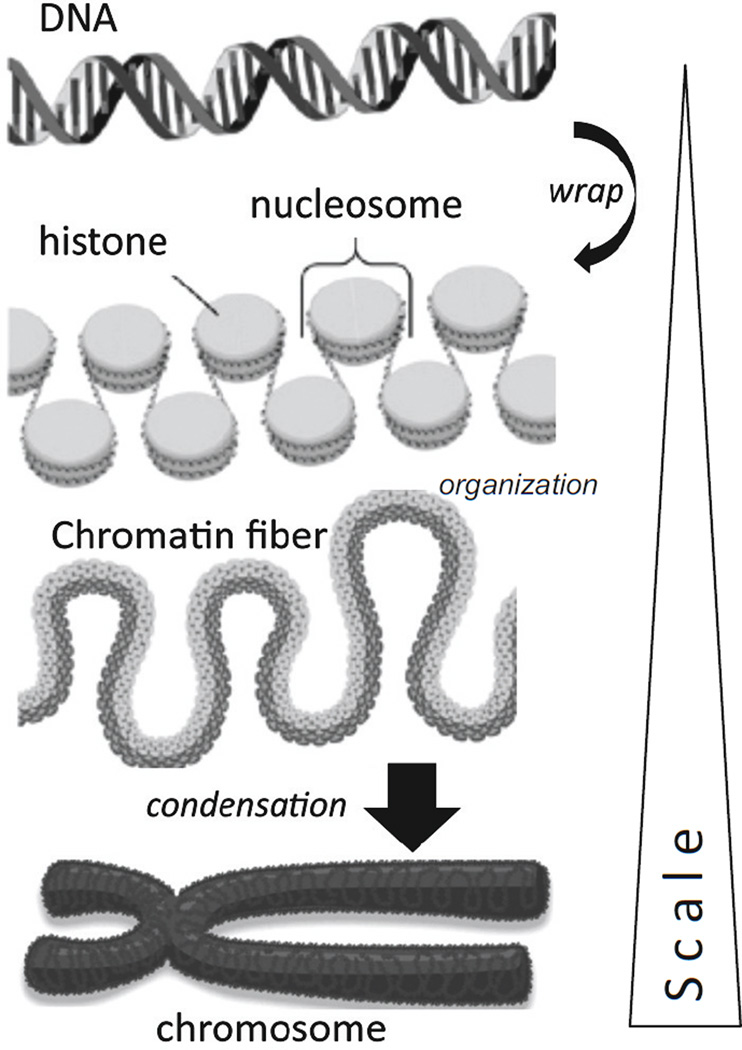
Figure 1 outlines the packaging of DNA into nucleosomes that organize and condense to form the chromosomes in the cell nucleus. In genetic regulation the fixed DNA code contains all of the sequence information and cis/trans elements required to facilitate gene expression. However, a powerful overlay (or epi-layer) of control is provided by flexible epigenetic regulation of gene expression in which chemical modification of DNA and its associated histone proteins occurs without a change to the DNA sequence. The epigenetic tags result in conformational changes of DNA and histones that regulate gene expression. In some cases epigenetic modifications are fixed, stable and inherited, but in other instances they are dynamic and change in response to environmental stimuli. It is increasingly apparent that physical and biomechanical forces, including hemodynamic flow characteristics, are environmental stimuli that induce epigenetic regulation of gene transcription.
Figure 1.
In the nucleus, DNA wraps around histone proteins to form linked nucleosomes that organize into chromatin fibers that condense to form a chromosome.
Main pathways
Gene expression can be turned on or off by three types of epigenetic pathways that operate to promote or inhibit mRNA synthesis or selectively degrade mRNA once it is made (Jaenisch and Bird 2003; Matouk and Marsden 2014; Turgeon et al 2014). Each epigenetic pathway is distinct but interacts with the others ensuring integration into a complex hierarchy of gene regulation:
DNA methylation: methylation of cytosine residues in DNA, often at or in proximity to a gene promoter,
Histone code: posttranslational modifications to histone amino-terminal tails, particularly acetylation that alters the compaction of DNA coiled around histones,
Non-coding RNAs: post-transcriptional targeting of mRNA by short and long ncRNAs (microRNAs and lncRNAs respectively) that facilitate transcript degradation. Since ncRNAs make up >97% of the transcriptome, there is vast potential for dynamic epigenomic*regulation of gene expression, including through biomechanical stimulation of the epigenomic code.
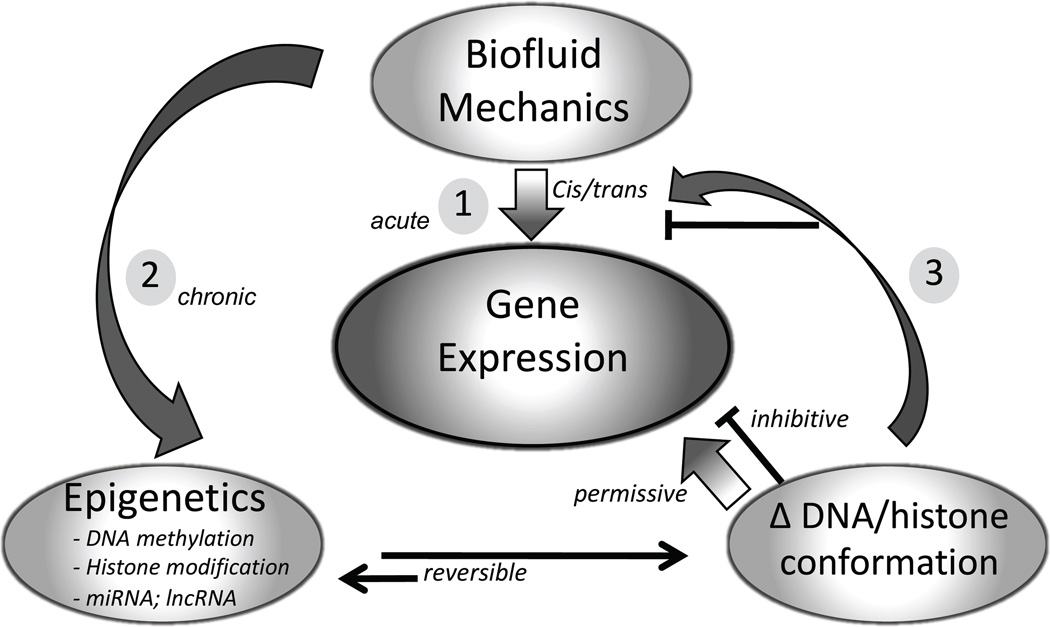
Figure 2 summarizes the overlayering of biomechanics-induced epigenetic changes onto genetic regulation of gene expression. Acute transcriptional responses to biomechanics (e.g. early response genes) are regulated by the activation of existing cis and trans factors. However a sustained stimulus alters the epigenetic equilibrium, changes the conformation of DNA and histones, and regulates access of transcription factors and other cis/trans regulators to their cognate DNA binding sites. The resulting regulation may be permissive (gene expression) or inhibitive (gene silencing).
Figure 2.
Summary of mechano-regulation of gene expression by epigenetic modification of DNA/histone biophysical conformation. Regulation of transcription occurs by cis/trans mechanisms at any existing state of epigenetic status (1). However, mechanotransduction can also induce changes in epigenetic status (2) for selective genes – usually over longer periods (24h+ for flow). The resulting DNA/histone state directly silences/permits transcription or indirectly regulates upstream molecules (3) involved in cis/trans gene expression. The net effect is epigenetic oversight of transcription spread throughout the genome.
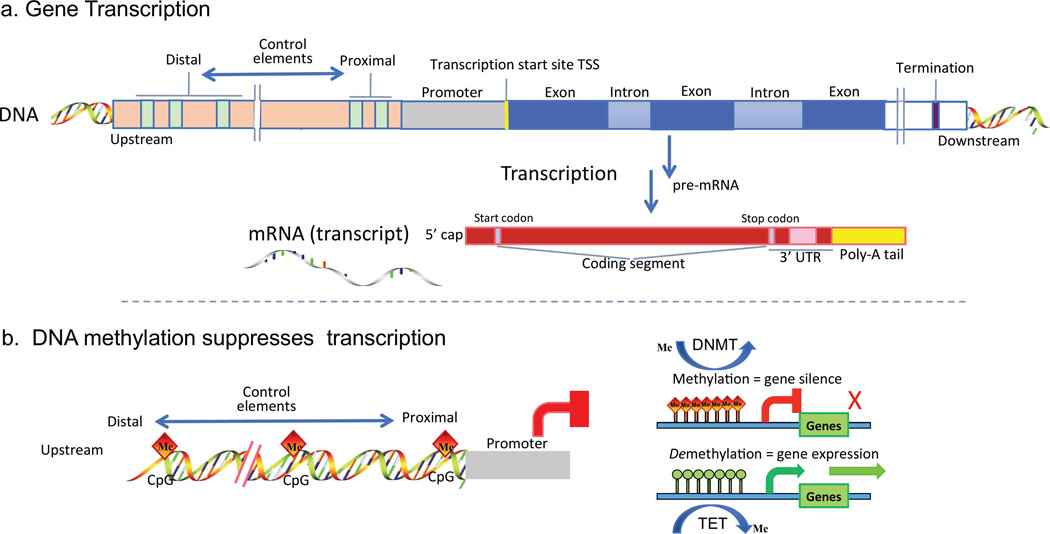
Figure 3a outlines the transcription of DNA to mRNA after which the transcript is normally further processed by the cell; regulation of this step is where epigenetic pathways converge.
Figure 3.
a. Gene transcription. Outline of mRNA synthesis (transcription) from a typical DNA region illustrating the coding region (blue), upstream promoter region (green) and proximal and distal cis/trans control elements. Single strand mRNA is copied from the coding region, the introns are removed from pre-mRNA and the exon information with start and stop codons are capped and tailed to form a discrete transcript (mature mRNA). b. DNA methylation. Methylation of cytosines of cytosine-guanosine dinucleotides (CpG) at or proximal to a promoter in DNA block transcription. The methylated DNA interacts differently with the transcript machinery of the cell. DNMT enzymes coordinate the methylation step in concert with binding protein complexes (not shown). For some genes, silencing by DNA methylation may be reversible (demethylation) by enhanced TET enzyme activity.
1. DNA methylation
DNA methylation adds a methyl group to the 5-carbon of cytosine residues (5-mC) in DNA, usually at cytosine-phosphate–guanine dinucleotides (CpG) sites. Figure 3b outlines methylation of a limited number of upstream CpG sites in the proximal promoter and/or distal DNA regions resulting in suppression of gene transcription. The decoration of DNA by methyl groups changes its conformation and mechanical properties (Severin et al 2011) altering interaction with the transcriptional machinery of the cell. CpG sites tend to be more prevalent near gene promoters upstream of exon coding regions. Unmethylated CpG permits gene transcription; conversely, methylation of cytosines results in gene silencing.
Central regulators of DNA methylation
CpG sites are methylated by DNA methyltransferase (DNMT) enzymes. Three active mammalian methyltransferases DNMT1, 3A and 3B promote methylation which is reversed by DNA demethylation through the TET (tet methylcytosine dioxygenase) gene pathway. Flow-induced changes in DNMT1 and 3A have recently been reported (Davies et al 2014; Dunn et al 2014; Jiang et al 2014, 2015b; Zhou et al 2011). DNMT1 is primarily a maintenance enzyme to ensure fidelity of methylation patterns during somatic cell division whereas DNMT3A and 3B regulate de novo DNA methylation. While the methylation or non-methylated state of each cytosine/CpG site is a binary event, the overall status of a cytosine DNA region is a composite methylation score of multiple cells. Evaluation of the contribution that methylation of each cytosine (or cluster of CpGs) exerts on gene transcription can be assessed experimentally, e.g. for KLF4 (Jiang et al 2014), or surveyed on a chromosome-wide basis (Weber et al 2005, Jiang et al 2015a). Within disturbed flow regions in vivo, an inverse relationship between hypermethylation and mRNA expression generally exists when the CpG sites are upstream of the promoter but not when intragenic cytosines are methylated (Jiang et al 2015a).
Flow-mediated DNA methylation
Disturbed vs undisturbed flow regions in vivo predict atherosusceptibility or atheroprotection, respectively (Davies et al 2009; Chiu and Chien 2011). In the context of epigenetics, differential DNA methylation regions (DMR) map to vascular sites of disturbed and undisturbed flow (Jiang et al 2015a). These differences can be recapitulated in vitro(Jiang et al 2014) and, importantly, can be induced by surgical manipulation of flow in vivo (Dunn et al 2014). Complete methylated DNA immunoprecipitation sequencing (MeDIP-seq) of in vivo arterial endothelium associated with disturbed (vs undisturbed) flow in pigs (Jiang et al 2015a) identified differentially methylated DNA enrichment in exons and 5’UTR sequences of annotated genes, approx 60 of which are linked to cardiovascular disease. Thus dynamic DNA methylation appears to be a genome-wide epigenetic mechanism of transcriptional control related to the local site-specific biomechanical stresses of flow. Furthermore, disturbed flow promoted DNA hypermethylation that resulted in transcriptional suppression in human endothelial cells in culture (Davies et al 2014; Dunn et al 2014; Jiang et al 2014, 2015a). Collectively these studies in pigs and mice and cultured cells identified endothelial genes and their promoters as DMR-sensitive epigenomic targets associated with site-specific disturbed flow.
2. Histone remodeling
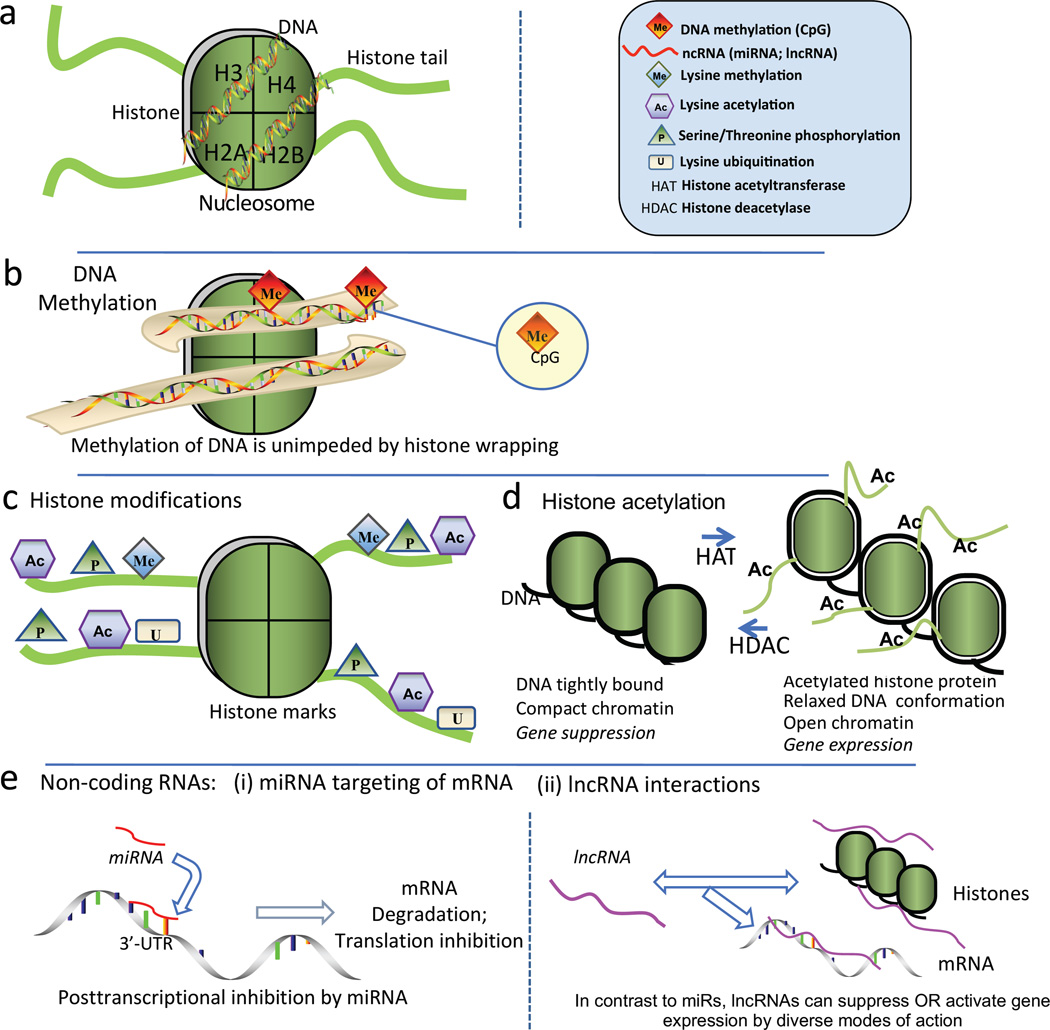
In the cell nucleus chromatin DNA is wrapped around proteins called histones to form a composite unit, the nucleosome (Figure 4a), in which ~146 bp of DNA is wrapped around each histone octameric core (Kornberg and Lorch 1999). Whereas DNA methylation proceeds equally effectively in DNA that is wound around histones (Figure 4b) or in the linear arrangements of DNA between nucleosomes (cf. Figure 3b). The histone amino terminal domains extend from the core as ‘tails’ that can be modified by lysine acetylation and methylation, serine phosphorylation and lysine ubiquitination and sumoylation (Figure 4c). Modification of amino acid residues of histone tails profoundly changes the biophysical association between DNA and histone protein influencing its compaction state. The degree of compaction or relaxation of DNA wound around histones is instrumental in allowing or preventing access to the DNA by transcription factors, repressors, activators and other cofactors of cis/trans transcriptional regulation.
Figure 4.
a. Nucleosome structural elements. DNA is wrapped around each histone octomer protein that has basic amino acid-rich tail domains. Each core contains two copies of the histones H2A, H2B, H3 and H4, the amino terminal domains of which extend from the core as basic amino acid-rich ‘tails’ that can be modified, principally by acetylation and methylation (see Figure 4c) b. Proximal promoter DNA methylation at CpG islands results in gene silencing by transcriptional repression (cf. Fig 4b). c. Posttranslational ‘marks’ on N-terminal histone tails constitute a histone code; they include lysine acetylation and methylation, serine phosphorylation and lysine ubiquitination and sumoylation that remodel chromatin by changing histone protein-DNA association to influence transcriptional regulation. d. Chromatin remodeling by HATs and HDACs. Histone acetylation relaxes tightly wound compact DNA to enable access of transcriptional regulators. HDACs deacetylate to restore compact chromatin and prevent transcription. e. Posttranscriptional RNA-based mechanisms include short (microRNAs) and long noncoding RNAs (lncRNAs). e[i].miRNAs target the 3’-untranslated region of mRNA promoting transcript degradation and also translation destabilization. e[ii]. lncRNA facilitate changes of histone methylation through complex mechanisms that can result in enhanced or repressed gene expression depending upon the histone mark. Modified from Jiang et al (2015b).
The most dynamic (and reversible) regulation of histone-wound DNA relaxation is lysine acetylation that allows access of transcriptional regulators to cis-DNA binding domains (Figure 4d) presumably by neutralizing the strong positive charge of lysine. In contrast deacetylated histone tightly condenses the chromatin creating a barrier to small molecule regulators and preventing DNA transcription (Shahbazian and Grunstein 2007).
Central regulators of histone acetylation
Histone acetylases (HAT) and deacetylases (HDAC) catalyze the net acetylation state usually in conjunction with multi-protein complexes (Figure 4d).
The other dominant histone tail modification is the methylation of lysine and arginine, which is regulated by histone-N-methyltransferases. Combinations of particular methylated histone tail residues and their degree of methylation (histone “marks”) can promote or prevent transcription by histone-DNA compaction. Histone methylation may be independent of DNA methylation at the promoter region (Roh et al 2008). The net effect of these dynamic histone modifications, all of which are posttranslational, is to control accessibility to the nucleosomal DNA by other regulatory proteins, cofactors and transcriptional inhibitors.
Flow-mediated histone remodeling
The molecular basis for epigenetic histone modification by shear stress in endothelial cells was established in cultured human umbilical vein endothelial cells (Illi et al 2003); activation of histone H3 phosphorylation and H3/H4 acetylation by flow (vs no-flow) was increased by an HDAC inhibitor. However, to recapitulate in vivo flow characteristics (where flow is rarely stationary) comparisons of pulsatile disturbed vs undisturbed flow are necessary. Disturbed flow increased HDAC3 expression leading to protection against apoptosis by forming a complex with the serine/threonine protein kinase Akt, thereby promoting cell survival (Zampetaki et a 2010). A more comprehensive survey of HDACs in disturbed flow demonstrated upregulated expression and nuclear accumulation of HDAC1/2/3 and HDAC5/7 whereas undisturbed flow promoted nuclear export of HDACs 5 and 7 (Lee et al 2012). The protective antioxidant transcription factor NF-E2–related factor 2 (Nrf2) was deacetylated by association with HDAC1/2/3 in disturbed flow (Fledderus et al 2008), and a NAD-dependent HDAC, Sirtuin 1 (SIRT1), that protects against oxidative stress prevalent in disturbed flow regions in vivo was shown to associate with endothelial NOS (NOS3), promoting its availability (Chen et al 2010).
3. MicroRNA and lncRNA -associated gene regulation
RNA derived from non-coding sequences in the transcriptome (ncRNAs) can regulate gene expression by targeting newly synthesized mRNA for degradation. Short 20–26 nucleotide microRNAs (miRNA) and long non-coding RNAs (lncRNA; >200 nucleotides) are prominent examples.
Micro-RNAs (miRNAs)
miRNAs, are highly-conserved small RNAs that regulate post-transcriptional gene expression by binding to target mRNA, promoting mRNA degradation and/or inhibiting translation of the protein-coding genes (Ambros 2004) as outlined in Figure 4e[i]. Pairing between cognate mRNA 3’ UTR and miRNA 5’ regions is centered on seed nucleotides (2–7nt) that primarily determine miRNA target selection (Bartel 2009). Individual miRNAs fine-tune the synthesis of many genes. Given the widespread scope but modest repression of transcriptomes by individual miRNAs, net cell phenotypes likely result from the coordinated actions on multiple targets by single miRNAs or integrated regulation of key pathways by multiple miRNAs (Liang 2009). Consequently there are many interactions between miRNAs and epigenetic DNA methylation and chromatin remodeling (Chuang and Jones 2007). For example, miRNAs can be involved in establishing DNA methylation by targeting methyltransferases DNMT1, 3a, and 3b (Rajewsky 2006). In turn, DNMT inhibitors such as 5-azacytosine that inhibit DNA methylation can affect miRNA expression, e.g. upregulation of miR127 (Saito et al 2006). Similar reciprocities were reported with respect to chromatin structure through miRNA regulation of key histone modifiers such as HDAC4; at the same time 4-phenylbutyric acid, an HDAC inhibitor, upregulated some miRNAs. Collectively, these miRNA epigenetic interactions represent complex dynamic regulation at multiple levels. It is likely that many of the flow-sensitive miRNAs identified to date contribute to epigenetic chromatin structure-function via mechanotransduction-specific DNA methylation and histone modifications.
Flow-mediated miRNAs
MicroRNAs have been extensively studied in the endothelium especially in inflammation, atherogenesis and angiogenesis and have been shown to play important roles in flow-induced endothelial cell responses both in vivo and in vitro (Fang et al 2010, 2012; Zhou et al 2011; Kumar et al 2014). The co-localization of disturbed flow and endothelial activation by the pro-inflammatory transcription factor NFkB is highly relevant to the identification of miRNAs −10a, −92a, −146, −155 and −181b as negative regulators of the NFkB signaling pathway (Fang et al 2010; Cheng et al 2014). Although miRNAs exist as a diverse interactive network, the loss/gain of function of only a single miRNA can lead to quite focused physiological effects in vitro and in vivo (Daniel et al 2014) suggesting a narrower therapeutic potential than might be expected.
Long noncoding RNAs (lncRNAs)
There is currently much interest in epigenetic regulation by a diverse population of lncRNAs of which there are several thousand in the mammalian genome. It is estimated that each annotated gene is overlapped by multiple lncRNA isoforms. There is no common mechanism of action and they can both promote and suppress transcription by interactions with DNA, mRNA and histones (Figure 4e[ii]). lncRNA ‘archetypes’ include cis and trans targeting, enhancement, decoy, scaffold, conformational and coactivation/corepressor mechanisms (Lee 2012). The association of lncRNA with cardiovascular diseases (Uchida and Dimmeler 2015) attests to widespread involvement in heart and blood vessel pathophysiology at multiple levels. In an endothelial study, in vitro silencing of the lncRNA MALAT1 promoted change from a proliferative to a migratory endothelial phenotype while MALAT1 deletion or inhibition in vivo reduced vascular growth (Michalik et al 2014). The role of lncRNAs in endothelial inflammatory pathways is currently unknown despite a strong impetus to further identify the lncRNAs involved in controlling the vascular inflammatory response (Kumar et al 2014).
lncRNAs in Flow
In contrast to miRNAs, lncRNAs are only recently under investigation in flow mechanotransduction and without publications to date. An abstract (Hartung et al 2014) reported that simple laminar flow vs no-flow in vitro) regulates metalloprotease AMZ2 expression via ‘lncRNA2’ binding to the repressive chromatin mark H3K27me3 (methylation of lysine 27 within histone H3).
Despite current challenges in functional interpretation confounded by isoforms and multiple interactions, intense interest in lncRNA will likely yield important principles about their role in endothelial cell regulation.
DISCUSSION
Variation in genomic DNA sequence alone incompletely explains risk factor associations with cardiovascular diseases, suggesting a significant role for epigenetics (Webster et al 2013); tying into this concept is hemodynamics as a risk factor for atherosusceptibility (Davies et al 2013; Gimbrone and Garcia-Cardena 2016). Epigenetic sensitivity of cells to the biomechanical environment is intrinsic to an understanding of mechanotransduction mechanisms.
Flow studies suggest a mechano-sensitive physiological regulatory role for DNA methylation in addition to its well-studied functions in development and differentiation, many of which are permanent. However, it is currently unknown if the DNA methylation status of most endothelial genes is fixed in vivo during development or whether the flow responses are readily reversible and/or inheritable. For example, while a methylome ‘snapshot’ of pig endothelial HOX genes in disturbed flow regions revealed most to be differentially methylated compared to undisturbed flow (Jiang et al 2015a), only HoxA5 changed its methylation score in vivo in mice following the surgical induction of disturbed flow in a region of previously undisturbed flow (Dunn et al, 2014). Therefore it is conceivable that only a subset of genes demonstrate epigenetic plasticity to hemodynamics, an important consideration that is addressable by systematic combined MeDIP-sequencing/RNA-sequencing of flow-exposed cells in vitro and further manipulation of flow in vivo. To date, only a small number of gene subsets responsive to hemodynamic characteristics have been identified from in vivo manipulations and MeDIP studies of the methylome. In contrast to DNA methylation, most histone modifications are rapid and reversible, particularly histone acetylation and methylation which elicit dynamic epigenetic regulation; determining the time constants under flow conditions of chromatin state change (through histone marks) is an attractive quest complementary to DNA methylation. Endothelial flow studies of posttranscriptional regulation of mRNA by ncRNAs, the third arm of epigenetic regulation, are most active for small miRNAs with rapidly increasing interest in lncRNA as more tools become available. Each of these mechanisms is likely involved in the short and long term maintenance and adaptability of endothelial phenotypes in the cardiovascular system with important epigenetic implications for the initiation and development of disease through strong biophysical and biomechanical regulatory input.
DNA methylation, chromatin remodeling by histone modification and ncRNA transcriptional regulation are driven by enzymatic activation (DNMTs, HDACs, HATs) that in turn are themselves subject to complex regulation that appear to be directly influenced by flow/biomechanics. A major challenge is to define the mechanisms of flow specificity of such fundamental widespread regulators in the cell. Measurement of the effects of specific molecular knockdown are useful but not definitive and potential therapeutic inhibitors of, for example, DNMTs do not discriminate between DNMT1 and 3A both of which have been demonstrated to be inducers of flow-responsive DNA methylation (Zhou et al 2011; Jiang et al 2014; Dunn et al 2015). As in flow-mediated endothelial mechanotransduction, there are epigenetic flow responses that converge with, as well as parallel, classic pathways of cell metabolism or occupy critical positions as pathway nodes. Since they are integrated into overall cell regulation, their relative importance will emerge from hierarchical analyses under normal and stressed conditions. In addition to Hox genes for example, inflammatory pathway genes, Kruppel-like Factors (KLFs), anti-oxidant transcription factor (Nrf2) and ER-stress responders are of topical interest.
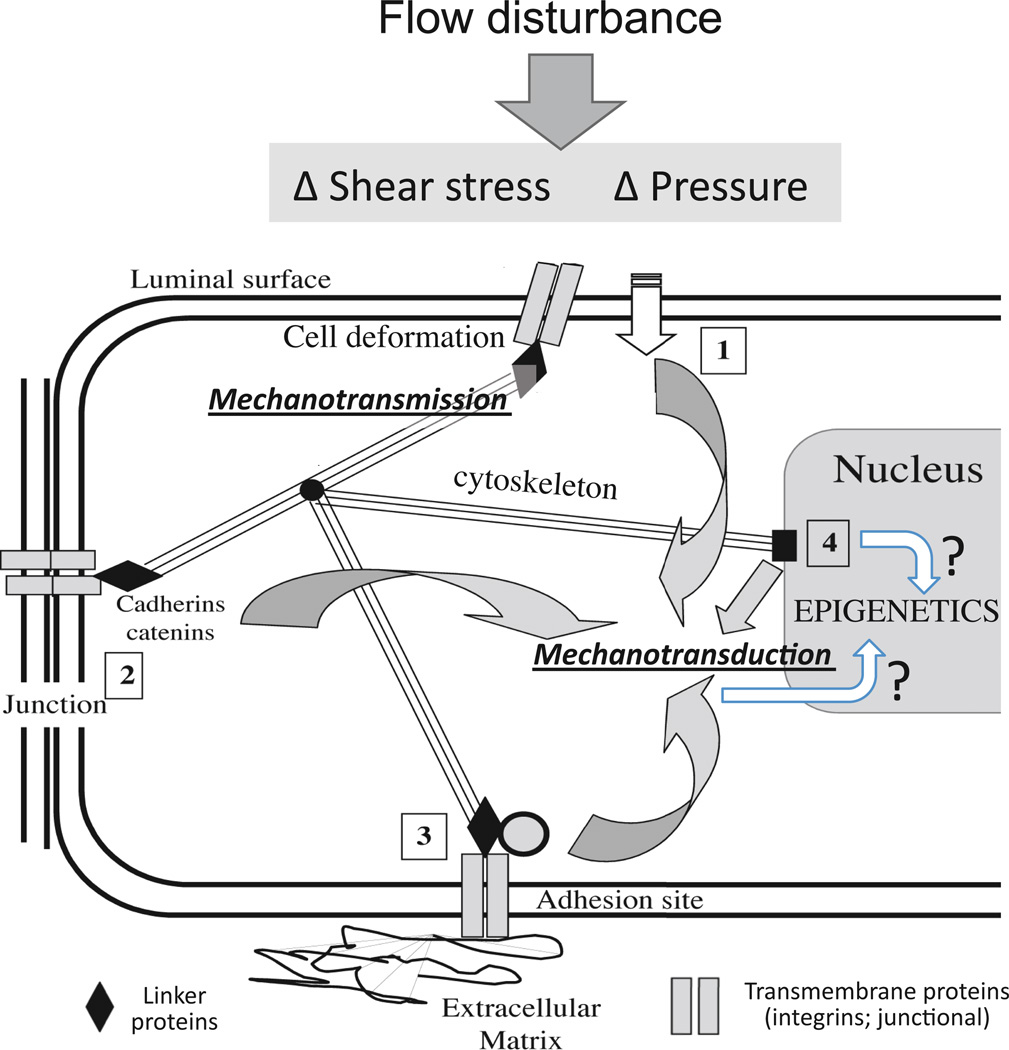
The ability of the endothelium to respond to disturbed flow by sensing different hemodynamic forces may reside in subcellular spatio-temporal mechanotransduction criteria transmitted throughout the cell by the cytoskeleton and converted to chemical/metabolic responses (mechanotransduction) at multiple decentralized subcellular locations (Davies 2009; Davies and Helmke 2010; Figure 5) including transmission of stress to the nucleus (Maniotis et al 1997). Changes in the epigenome could be directly attributable to strain-induced nuclear structural changes and/or be secondary to mechanotransduction at other subcellular locations (Dolmetsch et al 2001) including the viscoelastic cytoskeleton itself (Janmey 1998). While flow–induced stresses are directly transmitted to the cytoskeleton and distributed widely throughout the cell (Davies and Helmke 2010), the measurements are microscale; the conversion from stress/strain to cell chemistry remains unknown and may occur at the nanoscale as conformational changes in single proteins; for example nucleic acid polymers can be considered as prestressed structures (Ingber 1998).
Figure 5.
In a decentralized model of flow-induced endothelial mechanotransduction (Davies 1995), the cytoskeleton plays a central role in mechanotransmission of tensional changes throughout the cell that distributes forces to multiple subcellular locations. Mechanical forces at the luminal surface (1), cell junctions (2), adhesion sites (3) and nuclear structures (4) can elicit mechanotransduction responses. The mechanotransduction sites are not mutually exclusive and are biochemically interconnected.
Epigenetic status (which is entirely nuclear) is influenced by two broad mechanisms (marked by ? in the figure): - (i) by the local deformation of nuclear structure (Maniotis et al 1997) or (ii) by downstream second messengers in extranuclear mechanotransduction pathways that enter the nuclear space (Dolmetsch et al 2001). It is notable that cell fate decisions in differentiating stem cells can be manipulated by extracellular substrates and/or stiffness changes that alter nuclear mechanics (Buxboim et al 2010). The powerful dependence of cell fate decisions upon DNA methylation suggests a strong link regulation of transcription by nuclear mechanics through epigenetic pathways. Alternatively cytoplasmic second messengers from plasma membrane receptors, junctional, cytosketal and adhesion site mechanotransduction are free to move into the nucleus to regulate transcription by activating DNMTs, HATs, HDACs and other epigenetic enzymes. Modified from Davies (2009).
An important additional consideration is that convective transport in disturbed flow is significantly changed. Site-specific flow separation in arteries creates transients vortices of average lower flow velocity/shear stress within which solute and particle retention times are significantly longer than in undisturbed flow. The artery wall is constantly producing reactive oxidant species (ROS) such as free radicals and other potent intermediary metabolites. Active peptides and hormones generated from precursors in the blood are rapidly metabolized at the vessel surface. This may be why disturbed flow ROS is higher in flow separated regions than in nearby undisturbed flow regions (Civelek et al 2011) creating an unbalanced local environment that may influence epigenetic pathways. Oxidative damage is known to target assembly complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands (O’Hagan et al 2011). Since the NFkB pathway is also more active in disturbed flow (Passerini et al 2004) and Rel/p65 is able to recruit DNMTs to specific genome loci (Liu et al 2012), these pathways may influence DNMT enrichment and DNA methylation.
FUTURE PERSPECTIVE
The epigenomic code therefore has the potential to figure prominently in disease susceptibility and pathogenesis arising from environmental influences independent of, or in concert with, mutations and single nucleotide polymorphisms (SNP) linked to multifactorial diseases, including hypertension and cardiovascular disease. The role of disturbed flow as a hemodynamic regulator of epigenomic DNA methylation-mediated and histone-mediated gene expression may play a prominent role in endothelial vascular function and dysfunction. Nevertheless, the cellular mechanisms that link hemodynamics and biomechanical stress to the epigenomic code remain relatively unexplored.
A mature infrastructure now exists to support such studies. Lowering of barriers between traditional disciplines has led to collaborative and integrated research in well established interdisciplinary departments, institutes and centers that also provide formal training at the graduate student and postdoctoral levels. In addition to bioengineering departments, most mechanical and chemical engineering and materials science departments are heavily involved in biology and biomedical projects through collaborative or integrated (Center) mechanisms. There is complementary integration of engineering and physical sciences into academic medicine where the practice and teaching of rigorous quantitation alongside descriptive biomedicine has been significantly enhanced and where engineering approaches are embedded in research. Challenges that arise in epigenetic research are addressable because of resources and tools that were previously unavailable. These include affordable next generation sequencing facilities, sophisticated high resolution MRI applied to 4D flow as well as to tissue structure, the computing power for bioinformatics analyses of sequencing datasets and for complex computational modeling of dynamic systems, and great precision in gene editing of cells both in vitro and in vivo. The formation of national and international epigenomic consortia such as the ENCODE (ENCyclopedia Of DNA Elements) Project (Abbott 2010; National Human Genome Research Institute et al 2004) facilitate academic interactions.
Given the associative relationships between risk factors and cardiovascular disease, the interplay of mechanotransduction and epigenomic regulation of vascular phenotypes - with the potential for novel therapeutic interventions - warrants further investigation.
Acknowledgments
Supported by NIH grant P01 HL62250 from the National Heart Lung & Blood Institute (PFD, EM, CJS), NIH grant K25 HL107607 (JMJ), AHA Postdoctoral Fellowship 13POST14070010 (Y-ZJ), and the Robinette Foundation Endowed Professorship in Cardiovascular Medicine (PFD). We thank Dr. Maggie McCormick for helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Epigenomics: The global analysis of epigenetic changes across the entire genome
CONFLICT OF INTEREST
The authors have no conflicts of Interest to declare concerning the contents of this manuscript.
REFERENCES
- Abbott A. Project set to map marks on genome. Nature. 2010;463:596–597. [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Ivanovska IL, Discher DE. Matrix elasticity, cytoskeletal forces and physics of the nucleus: how deeply do cells ‘feel’ outside and in? Journal of Cell Science. 2010;123:297–308. doi: 10.1242/jcs.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Peng I-C, Cui X, Li Y-S, Chien S, Shyy JY-J. Shear stress, SIRT1 and vascular homeostasis. Proceedings National Academy of Sciences USA. 2010;107:10268–10273. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HS, Njock M-S, Khyzha N, Dang LT, Fish JE. Noncoding RNAs regulate NF-κB signaling to modulate blood vessel inflammation. Frontiers in Genetics. 2014;5:422. doi: 10.3389/fgene.2014.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiological Reviews. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatric Research. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Davies PF. Coronary artery endothelial transcriptome in vivo: identification of endoplasmic reticulum stress and enhanced reactive oxygen species by gene connectivity network analysis. Circulation Cardiovascular Genetics. 2011;4:243–252. doi: 10.1161/CIRCGENETICS.110.958926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Penzkofer D, Teske R, Dutzmann J, Koch A, Bielenberg W, Bonauer A, Boon RA, Fischer A, Bauersachs J, van Rooij E, Dimmeler S, Sedding DG. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovascular Research. 2014;103:564–572. doi: 10.1093/cvr/cvu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PF. Flow-mediated endothelial mechanotransduction. Physiological Reviews. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PF. Hemodynamic Shear Stress and the Endothelium in Cardiovascular Pathophysiology. Nature Clinical Practice Cardiovascular Medicine. 2009;6:16–26. doi: 10.1038/ncpcardio1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PF, Helmke BP. Endothelial Mechanotransduction. In: Mofrad MRK, Kamm RD, editors. Cellular Mechanotransduction: Diverse Perspectives from Molecules to Tissues. Cambridge University Press; 2010. pp. 20–60. [Google Scholar]
- Davies PF, Civelek M, Fang Y, Fleming I. The atherosusceptible endothelium: Endothelial phenotypes in complex hemodynamic shear stress regions in vivo. Cardiovascular Research. 2013;99:315–327. doi: 10.1093/cvr/cvt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PF, Manduchi E, Stoeckert CJ, Jimenez JM, Jiang Y-Z. Emerging topic: flow-related epigenetic regulation of the atherosusceptible endothelial phenotype through DNA methylation. Vascular Pharmacology. 2014;62:88–93. doi: 10.1016/j.vph.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, Fan Y, Jordan IK, Jo H. Flow-dependent epigenetic DNA methylation regulates gene expression and atherosclerosis. Journal of Clinical Investigation. 2014;124:3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proceedings National Academy of Sciences USA. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Davies PF. Site-Specific MicroRNA-92a Regulation of Kruppel-Like Factors 4 and 2 in Atherosusceptible Endothelium. Arteriosclerosis Thrombosis & Vascular Biology. 2012;32:979–987. doi: 10.1161/ATVBAHA.111.244053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fledderus JO, Boon RA, Volger OL, Hurttila H, Ylä-Herttuala S, Pannekoek H, Levonen AL, Horrevoets AJ. KLF2 primes the antioxidant transcription factor Nrf2 for activation in endothelial cells. Arteriosclerosis Thrombosis & Vascular Biology. 2008;28:1339–1346. doi: 10.1161/ATVBAHA.108.165811. [DOI] [PubMed] [Google Scholar]
- Friso S, Carvaial CA, Farde E, Olivieri O. Epigenetics and arterial hypertension: the challenge of emerging evidence. Translational Res. 2015;165:154–165. doi: 10.1016/j.trsl.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Garcia-Cardeña G, Comander J, Anderson KR, Blackman BR, Gimbrone MA. Biomechanical activation of vascular endothelium as a determinant of its phenotype. Proceedings National Academy of Sciences USA. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimbrone MA, Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung CT, Michalik KM, You X, Chen W, Zeiher AM, Boon RA, Dimmeler S. Regulation and function of the laminar flow-induced non-coding RNAs in endothelial cells. Circulation. 2014;130:A18783. (Abstract) [Google Scholar]
- Horsburgh S, Robson-Ansley P, Adams R, Smith C. Exercise and inflammation-related epigenetic modifications: focus on DNA methylation. Exerc Immunol Rev. 2015;21:26–41. [PubMed] [Google Scholar]
- Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circulation Research. 2003:93155–93161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- Ingber DE. The architecture of life. Scientific American. 1998;278:48–57. doi: 10.1038/scientificamerican0198-48. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Janmey PA. The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiological Reviews. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. [DOI] [PubMed] [Google Scholar]
- Jiang Y-Z, Jiménez JM, Ou K, McCormick ME, Davies PF. Differential DNA methylation of endothelial Kruppel-like Factor 4 (KLF4) promoter in response to hemodynamic disturbed flow in vitro and in vivo. Circulation Research. 2014;115:32–43. doi: 10.1161/CIRCRESAHA.115.303883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-Z, Manduchi E, Stoeckert CJ, Davies PF. Arterial endothelial methylome: Differential DNA methylation in atherosusceptible disturbed flow regions in vivo. BMC Genomics. 2015a;16:506. doi: 10.1186/s12864-015-1656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-Z, Manduchi E, Jimenez JM, Davies PF, et al. Endothelial epigenetics in biomechanical stress: Disturbed flow-mediated epigenomic plasticity in vivo and in vitro. Arteriosclerosis Thrombosis & Vascular Biology. 2015b;35:1317–1326. doi: 10.1161/ATVBAHA.115.303427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Brody JA, Dhingra R, Yousefi P, et al. Epigenetic signatures of cigarette smoking. Circulation Cardiovasc Genet. Sep 20. 2016 pii: CIRCGENETICS.116.001506. 2016 doi: 10.1161/CIRCGENETICS.116.001506. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kreuz S, Fischle W. Oxidative stress signaling to chromatin in health and disease. Epigenomics. 2016;8:843–862. doi: 10.2217/epi-2016-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kim CW, Simmons RD, Jo H. Role of flow-selective microRNAs in endothelial dysfunction and atherosclerosis. Arteriosclerosis Thrombosis & Vascular Biology. 2014;34:2206–2216. doi: 10.1161/ATVBAHA.114.303425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-Y, Lee CI, Lin TE, Lim SH, Zhou J, Tseng Y-C, Chien S, Chiu J-J. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proceedings National Academy of Sciences USA. 2012;109:1967–1972. doi: 10.1073/pnas.1121214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Liang M. MicroRNA: a new entrance to the broad paradigm of systems molecular medicine. Physiolological Genomics. 2009;38:113–115. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y1, Mayo MW, Nagji AS, Smith PW, Ramsey CS, Li D, Jones DR. Phosphorylation of RelA/p65 promotes DNMT-1 recruitment to chromatin and represses transcription of the tumor metastasis suppressor gene BRMS1. Oncogene. 2012;31:1143–1154. doi: 10.1038/onc.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proceedings National Academy of Sciences USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circulation Research. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circulation Research. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- National Human Genome Research Institute et al. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini AG, Polacek DC, Shi C, Francesco NM, Manduchi E, Grant GR, Pritchard WF, Powell S, Chang GY, Stoeckert CJ, Jr, Davies PF. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proceedings National Academy of Sciences USA. 2004;101:2482–2487. doi: 10.1073/pnas.0305938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. MicroRNA target predictions in animals. Nature Genetics. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rizzo V, McIntosh DP, Oh P, Schnitzer JE. In situ flow activates endothelial nitric oxide synthase in luminal caveolae of endothelium with rapid caveolin dissociation and calmodulin association. Journal of Biological Chemistry. 1998;273:34724–34729. doi: 10.1074/jbc.273.52.34724. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proceedings National Academy of Sciences USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Schiano C, Vietri MT, Grimaldi V, Picascia A, De Pascale MR, Napoli C. Epigenetic-related therapeutic challenges in cardiovascular disease. Trends Pharmacol Sci. 2015;36:226–35. doi: 10.1016/j.tips.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Severin PMD, Zou X, Gaub HE, Schulten K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Research. 2011;39:8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annual Reviews in Biochemistry. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Turgeon PJ, Sukumar AN, Marsden PA. Epigenetics of cardiovascular disease: a new ‘beat’ in coronary artery disease. Medical Epigenetics. 2014;2:37–52. doi: 10.1159/000360766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Dimmeler S. Long non-coding RNAs in cardiovascular diseases. Circulation Research. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Webster ALH, Yan MS-C, Marsden PA. Epigenetics and cardiovascular disease. Canadian Journal of Cardiology. 2013;29:46–57. doi: 10.1016/j.cjca.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Zampetaki A, Zeng L, Margariti A, Xiao Q, Li H, Zhang Z, Pepe AE, Wang G, Habi O, deFalco E, Cockerill G, Mason JC, Hu Y, Xu Q. Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation. 2010;121:132–142. doi: 10.1161/CIRCULATIONAHA.109.890491. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lim SH, Chiu J-J. Epigenetic regulation of vascular endothelial biology/pathobiology and response to fluid shear stress. Cell and Molecular Bioengineering. 2011;4:560–578. [Google Scholar]