Abstract
MicroRNAs (miRNAs) and short interfering RNAs (siRNAs) are small noncoding RNAs that have recently emerged as important regulators of mRNA degradation, translational repression, and chromatin modification. In Arabidopsis thaliana, 43 miRNAs comprising 15 families have been reported thus far. In an attempt to identify novel and abiotic stress regulated miRNAs and siRNAs, we constructed a library of small RNAs from Arabidopsis seedlings exposed to dehydration, salinity, or cold stress or to the plant stress hormone abscisic acid. Sequencing of the library and subsequent analysis revealed 26 new miRNAs from 34 loci, forming 15 new families. Two of the new miRNAs from three loci are members of previously reported miR171 and miR319 families. Some of the miRNAs are preferentially expressed in specific tissues, and several are either upregulated or downregulated by abiotic stresses. Ten of the miRNAs are highly conserved in other plant species. Fifty-one potential targets with diverse function were predicted for the newly identified miRNAs based on sequence complementarity. In addition to miRNAs, we identified 102 other novel endogenous small RNAs in Arabidopsis. These findings suggest that a large number of miRNAs and other small regulatory RNAs are encoded by the Arabidopsis genome and that some of them may play important roles in plant responses to environmental stresses as well as in development and genome maintenance.
INTRODUCTION
Whereas the roles of proteins as regulatory factors in development and adaptation to the environment are well known, the involvement of regulatory small RNA molecules is just emerging (Carrington and Ambros, 2003). MicroRNAs (miRNAs) and short interfering RNAs (siRNAs) are two classes of small noncoding RNAs with important roles in the regulation of gene expression in plants and animals. miRNAs are an abundant class of tiny RNAs that are often found to be conserved evolutionarily across species boundaries and are capable of regulating the expression of protein-coding genes in multicellular eukaryotes (Bartel, 2004). miRNAs are ∼20 to 24 nucleotides in length, single-stranded noncoding RNAs derived predominantly from intergenic regions. miRNA genes are transcribed as long pre-miRNA transcripts ranging from 70 to 300 nucleotides that form fold-back structures in which mature miRNAs reside in either the 5′ or 3′ arm and are processed by DICER (Ambros et al., 2003a; Bartel and Bartel, 2003; Bartel, 2004). miRNAs are known to silence genes posttranscriptionally by targeting cognate mRNAs for degradation (Llave et al., 2002a; Palatnik et al., 2003; Tang et al., 2003) or translation repression (Lee et al., 1993; Reinhart et al., 2000; Aukerman and Sakai, 2003; Chen, 2004).
The miRNA profile of Caenorhabditis elegans is probably the most complete, with the number of validated miRNA genes being ∼95 (Ambros et al., 2003b; Lim et al., 2003). The total number of miRNAs in each organism is unknown but is estimated to represent ∼1% of the number of coding genes (Grad et al., 2003; Lai et al., 2003; Lim et al., 2003; Bartel, 2004). This prediction is based on the fact that miRNAs are derived from evolutionarily conserved hairpin precursor RNAs. The total number of miRNAs in a plant is unknown but could also be ∼1% of the total coding genes (∼250 to 300 miRNAs in Arabidopsis thaliana). Three groups have independently reported miRNAs from Arabidopsis (Llave et al., 2002b; Park et al., 2002; Reinhart et al., 2002) and yet the number of miRNAs identified appears to be far from saturation. So far, only 43 miRNAs falling into 15 families have been reported in Arabidopsis (Bartel and Bartel, 2003; Bartel, 2004). Several of these miRNAs were demonstrated to play critical roles in leaf or flower development by targeting relevant developmental regulators for mRNA cleavage or translational repression (Aukerman and Sakai, 2003; Palatnik et al., 2003; Chen, 2004; Juarez et al., 2004; Kidner and Martienssen, 2004). Consistent with the important roles of miRNAs in the development of plants, the mutants dcl1, hen1-1, and hyl1 that are affected in miRNA accumulation all have severe developmental defects (Lu and Federoff, 2000; Chen et al., 2002; Golden et al., 2002; Schauer et al., 2002; Han et al., 2004; Vazquez et al., 2004). Phenotypic defects in these mutants could be attributed, at least in part, to blocking or lowered levels of miRNA production.
siRNAs are double-stranded 21- to 26-nucleotide small RNAs derived from the processing of long double-stranded RNAs (Waterhouse et al., 2001; Plasterk, 2002). siRNAs were originally discovered in plants undergoing transgene silencing or infection with viruses (Hamilton and Baulcombe, 1999; Zamore et al., 2000). Endogenous small RNAs with characteristic features of siRNAs have been cloned from Arabidopsis (Llave et al., 2002b; Reinhart et al., 2002). Double-stranded RNA may be derived from the transcription of inverted-repeat loci because of converging promoters or may be the product of host- or viral-encoded RNA-dependent RNA polymerases (Dalmay et al., 2000; Mourrain et al., 2000; Aravin et al., 2001; Grishok et al., 2001; Sijen et al., 2001). siRNAs assemble in complexes termed RNA-induced silencing complexes and target homologous RNA sequences for endonucleolytic cleavage, which is referred to as RNA interference in animals, posttranscriptional gene silencing in plants, and quelling in fungi (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Zamore et al., 2000). The biological roles of siRNA-mediated RNA silencing include protection of the genome against mobile DNA elements (Ketting et al., 1999; Tabara et al., 1999; Wu-Scharf et al., 2000) and resistance against viruses (Voinnet, 2001; Waterhouse et al., 2001). In addition to PTGS, siRNAs may be involved in transcriptional gene silencing by directing DNA methylation and chromatin modification (Mette et al., 2000; Aufsatz et al., 2002; Gong et al., 2002; Hamilton et al., 2002; Volpe et al., 2002; Zilberman et al., 2003).
A distinguishing feature of plants is that they are sessile and thus have to cope with, rather than move to avoid, adverse environments. Plants have evolved sophisticated mechanisms to adapt to environmental stresses (Zhu, 2002). Abiotic stresses, such as drought, salinity, and cold, regulate the expression of thousands of genes in plants at both transcriptional and posttranscriptional levels. We are interested in exploring potential roles of regulatory small RNAs in abiotic stress tolerance and gene regulation. This study was performed with two objectives. One is to identify new miRNAs and siRNAs in Arabidopsis, given that only a small number of these small RNAs have been reported, and thus there is likely more to be found. The other objective is to discover miRNAs that might be regulated by abiotic stresses. We constructed a library of small RNAs from seedlings treated with cold, dehydration, salinity, and the plant stress hormone abscisic acid (ABA). Through library sequencing and analysis, we found 15 new families of miRNAs and many novel putative siRNAs. Some of the miRNAs are preferentially expressed in certain tissues, and several are either upregulated or downregulated by abiotic stresses. Our results have important implications for gene regulation under abiotic stresses and also contribute significantly to the long-term goal of having a complete profile of miRNAs and siRNAs in Arabidopsis.
RESULTS
Identification of 15 New Families of miRNAs from Stress-Treated Seedlings
To identify miRNAs that might be regulated by abiotic stress, the expression of known Arabidopsis miRNAs under dehydration, salt, cold, and ABA treatments was tested by RNA gel blot analysis. However, we did not find substantial stress regulation on any of the known miRNAs (see Supplemental Figure C online; data not shown). Therefore, to identify novel miRNAs and miRNAs that might be regulated by abiotic stresses, we constructed a library of small RNA species from pooled seedlings exposed to cold, dehydration, salinity, and ABA. Small RNAs (15 to 26 bp) were recovered by size fractionation on denaturing polyacrylamide gels, eluted from the gel, ligated to 3′ and 5′ adapters, amplified, cloned, and sequenced. A BLAST search against the Arabidopsis database revealed that a major portion (90%) of the ∼2500 cloned sequences appears to be breakdown products from rRNAs, tRNAs, and small nuclear RNAs. The proportion (9%) of putative regulatory small RNAs is slightly lower than published results, presumably because the library was made from seedlings subjected to different abiotic stresses that may cause enhanced breakdown of noncoding RNAs (rRNAs, tRNAs, and nucleolar RNAs). Several clones matched to chloroplast or mitochondrial sequences and may represent degradation or possibly regulatory products of organellar RNAs. A few clones do not have perfect matches to the Arabidopsis genome sequence and appear to be derived from RNAs of fungal/bacterial/viral origin. Thirteen previously published families of miRNAs from Arabidopsis are represented in our sequences. These include miR156, miR157, miR158, miR159, miR161, miR163, miR166, miR168, miR169, miR171, miR172, miR173, and miR319/miRJAW (Llave et al., 2002b; Park et al., 2002; Reinhart et al., 2002; Palatnik et al., 2003). The frequency of any one miRNA appearing in our sequenced library varied from 1 to 30 times (Table 1; see Supplemental Table 1 online).
Table 1.
Novel miRNAs from Arabidopsis
| miRNA Gene | miRNA Sequence (5′→3′) | Fold-Back | Distance to Nearest Gene | Conserved in Other Plants
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Length | Arm | Chr. | Expression | Rice | Lotus | Medicago | Populus | ||||
| MIR171b | UUGAGCCGUGCCAAUAUCACG (3) | 21 | Yes | 3′ | 1 | 2.80 kb upstream At1g62040 | |||||
| MIR171c | 21 | Yes | 3′ | 1 | 1.27 kb downstream At1g11730 | d | + | + | |||
| MIR319c | UUUGGACUGAAGGGAGCUCC (5) | 20 | Yes | 3′ | 2 | 2.90 kb downstream At2g40800 | d | + | |||
| MIR389a.1 | UUCUAAGUCCAACAUAGCGUA (5) | 21 | Yes | 3′ | 1 | 0.51 kb downstream At1g50050 | d | ||||
| MIR389b.1 | 21 | Yes | 3′ | 2 | 0.66 kb downstream At2g27395 | ||||||
| MIR389b.3 | UUUUAAGUCUAACAUAGCGU (5) | 20 | Yes | 3′ | 2 | 0.70 kb downstream At2g27395 | d | ||||
| MIR390a.2 | CUAUCCAUCCUGAGUUUCAUU (1) | 21 | Yes | 3′ | 2 | 2.50 kb upstream At2g38330 | nd | ||||
| MIR393a | UCCAAAGGGAUCGCAUUGAUCC (30) | 22 | Yes | 5′ | 2 | 2.63 kb downstream At2g39880 | d | + | + | ||
| MIR393b | 22 | Yes | 5′ | 3 | 2.02 kb upstream At3g55735 | ||||||
| MIR397a | UCAUUGAGUGCAGCGUUGAUGU (3) | 22 | Yes | 5′ | 4 | 0.50 kb upstream At4g05110 | nt | + | + | ||
| MIR397b | UCAUUGAGUGCAUCGUUGAUGU (4) | 22 | Yes | 5′ | 4 | 0.81 kb upstream At4g13560 | d | ||||
| MIR398a | UGUGUUCUCAGGUCACCCCUU (3) | 21 | Yes | 3′ | 2 | 0.35 kb upstream At2g03450 | nt | + | + | + | |
| MIR398b | UGUGUUCUCAGGUCACCCCUG (7) | 21 | Yes | 3′ | 5 | 0.10 kb upstream At5g14550 | d | + | + | + | |
| MIR398c | 21 | Yes | 3′ | 5 | 0.40 kb upstream At5g14570 | ||||||
| MIR399b | UGCCAAAGGAGAGUUGCCCUG (2) | 21 | Yes | 3′ | 1 | 1.30 kb downstream At1g63000 | nt | + | + | ||
| MIR399c | 21 | Yes | 3′ | 5 | 1.30 kb downstream At5g62160 | nt | |||||
| MIR399f | UGCCAAAGGAGAUUUGCCCGG (6) | 21 | Yes | 3′ | 2 | 0.85 kb upstream At2g34210 | d | + | + | ||
| MIR400 | UAUGAGAGUAUUAUAAGUCAC (4) | 21 | Yes | 5′ | 1 | 0.31 kb downstream At1g32580 | d | ||||
| MIR401 | CGAAACUGGUGUCGACCGACA (1) | 21 | Yes | 5′ | 4 | 4.10 kb downstream At4g08102 | nd | ||||
| MIR402 | UUCGAGGCCUAUUAAACCUCUG (3) | 22 | Yes | 5′ | 1 | First intron of At1g77230 | d | ||||
| MIR403 | UUAGAUUCACGCACAAACUCG (2) | 21 | Yes | 3′ | 2 | 1.70 kb upstream At2g47280 | d | + | |||
| MIR404 | AUUAACGCUGGCGGUUGCGGCAGC (1) | 24 | Yes | 5′ | 1 | 1.73 kb upstream At1g31360 | nd | ||||
| MIR405a | AUGAGUUGGGUCUAACCCAUAACU (2) | 24 | Yes | 3′ | 2 | 1.44 kb upstream At2g22670 | nd | ||||
| MIR405b | 24 | Yes | 3′ | 3 | 0.50 kb upstream At4g05510 | ||||||
| MIR405c | 24 | Yes | 3′ | 5 | 0.66 kb upstream At5g50720 | ||||||
| MIR405d | 24 | Yes | 3′ | 5 | 0.65 kb upstream At5g50720 | ||||||
| MIR406 | UAGAAUGCUAUUGUAAUCCAG (2) | 21 | Yes | 3′ | 1 | 4.24 kb downstream At1g52180 | nd | ||||
| MIR407a | UUUAAAUCAUAUACUUUUGGU (10) | 21 | Yes | 5′ | 2 | 0.49 kb upstream At2g32700 | d | ||||
| MIR408 | AUGCACUGCCUCUUCCCUGGC (4) | 21 | Yes | 3′ | 2 | Antisense polarity to At2g47020 | d | + | + | + | |
The sequences given represent the longest miRNA sequence identified by cloning. Frequency of cloning is indicated in parentheses after the miRNA sequences. Chromosomal (Chr.) positions and distance to the nearest annotated gene are indicated. miRNA location in the predicted hairpin structure is specified (5′ or 3′ arm). Expression of some of the miRNAs were confirmed by RNA blot analysis; d, nd, and nt denote detected, tested but not detected, and not tested because of a potential cross-hybridization problem, respectively. miRNAs that are conserved in other plants are indicated.
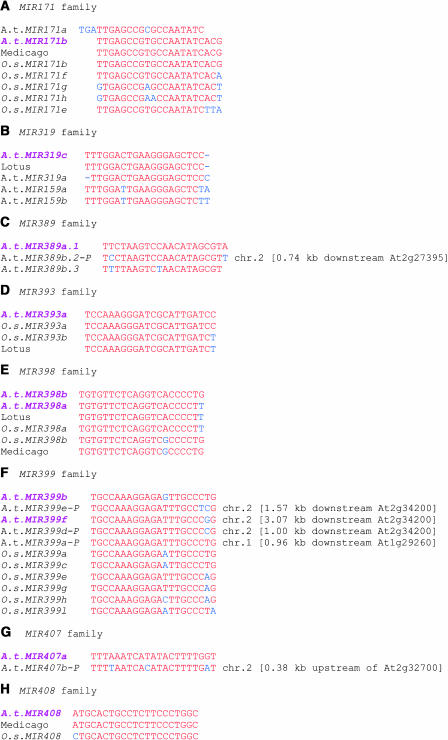
Importantly, we identified many new miRNAs and putative siRNAs. The ability of RNA corresponding to the genomic sequences surrounding the clones, to fold into hairpin miRNA precursors, was applied to distinguish miRNAs from other potential small regulatory RNAs. This criterion, together with phylogenetic conservation of mature miRNAs, is now generally accepted as evidence for the existence of a miRNA (Ambros et al., 2003a). We analyzed miRNA precursors of up to 300 nucleotides for hairpin structures using the mfold program (see Supplemental Figure A online) (Zuker, 2003). The analysis revealed that we have cloned 21 new miRNAs from Arabidopsis that correspond to 29 loci (Table 1). Furthermore, on analyzing the Arabidopsis genome, we were able to predict three homologs of the cloned miRNAs miR399b and miR399f based on sequence similarity, the capacity of their precursor sequences to adopt hairpin structures, and their conservation in rice (Oryza sativa) (Figure 1F). One homolog for each of the cloned miRNAs, miR389a.1 and miR407a, is also predicted in Arabidopsis (Figures 1C and 1G). Taken together, the total number of new miRNAs identified in this study is 26, which come from 34 loci. Twenty-four of the newly identified miRNAs define 15 new families (Table 1, Figure 1). Two of the new miRNAs, miR171b and miR319c, belong to the previously reported miR171 and miR319/miRJAW families, respectively.
Figure 1.
miRNA Families in Arabidopsis and Other Plants (Rice, Lotus, and Medicago).
miRNAs cloned in this study are labeled in pink. The letter P following miRNA loci (miR399, miR389b.2, and miR407b) denotes predicted miRNAs based on similarity with the cloned sequences and the ability of their precursor sequences to form hairpin structures. miR399 family is represented by six loci in the Arabidopsis (A.t.) genome. Only five loci (a, b, c, d, and e) are shown in Figure 1A because the locus d is represented twice with identical sequence (Table 1). Identical and conserved nucleotides are labeled in red. Blue letters represent changes in nucleotide sequences in some family members. The location of the predicted Arabidopsis miRNAs in the chromosomes is shown to the right of the sequence. Sequences were aligned with the ClustalW program (Thompson et al., 1994). All miRNAs from rice (O.s.), Lotus, and Medicago are from computer prediction only.
Genomic Organization of the miRNA Genes
Twenty of the miRNAs identified in this study, including the five predicted miRNAs, correspond to single copy loci in the Arabidopsis genome, whereas the other six miRNAs (miR399b and c, miR393a and b, miR398b and c, miR405a, b, c, and d, miR171b and c, miR389a.1, and miR389b.1) correspond to multiple loci (two to four) that are scattered in the genome (Table 1). Out of the 34 loci, 15, 9, and 5 come from chromosome 2, 1, and 5, respectively, and the remaining five loci are represented in chromosomes 3 and 4. Based on sequence homology and predicted fold-back structures, we were able to identify some miRNA homologs in rice, Lotus, Medicago, and Populus.
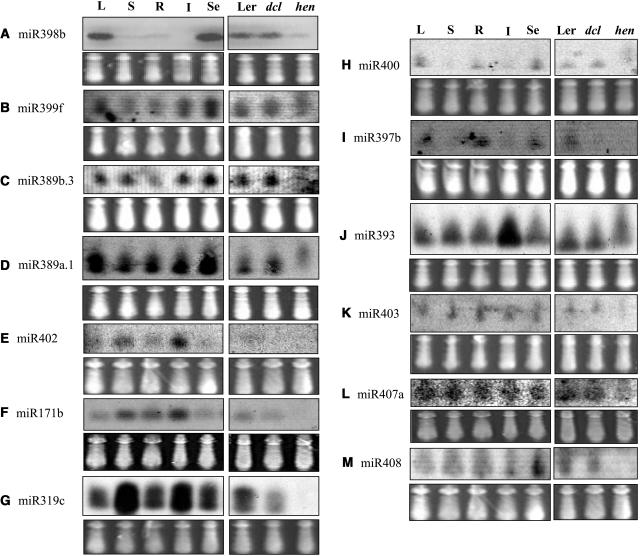
Thirty-two miRNA loci matched with intergenic regions, whereas two loci correspond to either an intron or 3′ untranslated region (UTR) of a gene. miR408 is located in the antisense polarity to the 3′ UTR of At2g47020 (Table 1). miR402 originates from the intron of gene At1g77230, in the same orientation as the pre-mRNA. Small RNAs corresponding to the introns or exons of pre-mRNAs in the same polarity may be miRNAs or may simply be general degradation products. The miRNAs can be ascertained by the capacity of their precursors to form hairpin structures, together with expression studies. The miR402 precursor sequence has the ability to adopt a long and typical fold-back structure (see Supplemental Figure A online). In addition, this miRNA can be detected as a discrete band of 22 bp on RNA gel blots (Figure 5E). These results show that it is an miRNA and not a nonspecific degradation product from the intron.
Figure 5.
Expression Patterns of miRNAs Cloned from Arabidopsis.
RNA gel blots of total RNA isolated from different tissues and 2-week-old seedlings were probed with labeled oligonucleotides. The blots also included RNA from dcl1-9 and hen1-1 mutants and their wild type, Landsberg erecta. The samples are leaf (L), stem (S), root (R), inflorescence (I), 2-week-old-seedlings (Se), Landsberg erecta (Ler), dcl1-9 (dcl), and hen1-1 (hen). The tRNA and 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and serve as loading controls.
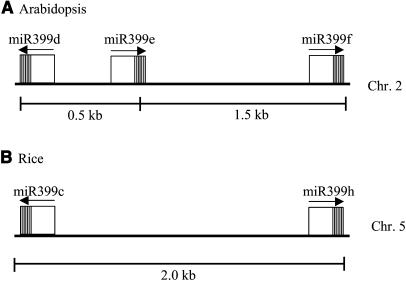
miR399 and its homologs correspond to six genomic loci (a, b, c, d, e, and f; b and c are identical in sequence but come from two different loci [Table 1]) in Arabidopsis with identical sequences or only a slight change in sequence. A sequence identical to miR399f is also found in rice, but the rice precursor sequence does not appear to form a fold-back structure; therefore, it cannot be ascertained as miRNA. However, rice has nine other loci with sequences that differ from the Arabidopsis miR399f in only one or two nucleotides (Figure 1F). These six loci from Arabidopsis and nine loci from rice all have the ability to form fold-back structures. Arabidopsis miR399 loci, miR399f, miR399d, and miR399e are located within a 2.0-kb region (Figure 2), and miR399f and miR399d are oriented in opposite directions. This organization of the two miRNAs within 2.0 kb and in opposite orientations is also conserved in rice (Figure 2). The only difference is that an additional locus miR399e exists in Arabidopsis in the same orientation as miR399f, but this is absent in rice. The conserved arrangement of these miRNAs suggests that some important aspects of their regulation may also be evolutionarily conserved.
Figure 2.
Conserved Genomic Organization of the miR399 miRNA Cluster in Arabidopsis and Rice.
miR399 loci d, e, and f are located within a 2.0-kb region on chromosome 2 in Arabidopsis, and part of this cluster is conserved in rice (chromosome 5). The hairpin precursor is indicated as a box, and the position of the miRNA in the precursor is shown as thick vertical lines. Arrow on top of the box indicates the orientation of miRNAs.
Although miR389 has three loci in Arabidopsis, only two loci (on chromosomes 1 and 2) are able to adopt fold-back structures (Figures 3A and 3B). The predicted fold-back structure of miR389b.1 from chromosome 2 may give rise to two miRNAs that belong to the same family, one each from the 5′ and 3′ arms (Figure 3B). However, only one of the two (from the 3′ arm, miR389b.1) is represented in the library, and the other (on the 5′ arm, miR389b.2) is predicted based on sequence identity. Because miR389b.1 and its predicted homolog on the 5′ arm are not located directly opposite to each other (Figure 3B), this represents a novel type of miRNA gene organization.
Figure 3.
Predicted Fold-Back Structures of miR389a.1 Precursors with Flanking Sequences on Chromosome 1 and Chromosome 2.
(A) Chromosome 1.
(B) Chromosome 2.
The fold-back structure shown in (B) is expected to yield two miRNAs, one each on 3′ and 5′ arms. The cloned miRNA sequence is labeled in red, and the predicted one is labeled in blue.
Conserved miRNA Gene Families
Based on sequence similarity, the miRNAs were grouped into families. Five of the miRNA families (miR399, miR393, miR398, miR171b, and miR408) are highly conserved in rice (Table 1, Figure 1). Some are also conserved in Lotus and/or Medicago or Populus (Table 1, Figure 1). The miR399 family is represented by six members in Arabidopsis and nine members in rice (Figure 1F) and is one of the largest miRNA families in plants. miR171b is represented by one and seven members in Arabidopsis and rice, respectively, with slight changes in nucleotide sequence (Figure 1A). The miR398 family has two members each in Arabidopsis and rice (Figure 1E). miR393 is represented by one member in Arabidopsis and two members in rice (Table 1, Figure1D). Two members of miR389a.1 and miR407 with slight change in sequence exist in Arabidopsis (Figures 1C and 1G). In some cases, the number of family members is greater in rice than in Arabidopsis. For example, miR156 (Reinhart et al., 2002) and miR171b and miR399 (this study) are represented by more family members in rice.
One miRNA (miR319c) was represented by five clones in our library, and the sequence is highly similar to the previously cloned miR319/miRJAW and miR159 (Reinhart et al., 2002; Palatnik et al., 2003). miRJAW has two genomic loci, one each on chromosomes 3 and 5 (Palatnik et al., 2003). However, miR319c is derived from a distinct locus on chromosome 2 with one nucleotide change. miR319c and miRJAW have the same size (20 nucleotides), and the only difference is that miR319c has an additional U at its 5′ end. The presence of an additional U in miR319c makes it more similar to miR159 because both have three uridines at the 5′ end. This may explain why we detected two equally strong signals at 20- and 21-nucleotide size positions using miR319c as a probe (Figure 5G). The miR319c probe likely cross-hybridizes with miR159, thus the 20- and 21-bp signals appear to correspond to miR319c and miR159, respectively (Figure 5G). This is in contrast with the miR319a/miRJAW probe, which detects a stronger signal at its expected size of 20 bp and a weaker signal at 21 bp corresponding to miR159 (Palatnik et al., 2003). The miR319c sequence is conserved in Lotus (Table 1, Figure 1B).
Predicted Targets of the miRNAs
To predict potential targets of the miRNAs identified in this study, we searched various databases for Arabidopsis mRNAs with sequence complementarity to the miRNAs, allowing up to three mismatches without any gap (Rhoades et al., 2002) and treating G:U pair as a match. In some cases, there is no predictable target with three or fewer mismatches, and the best match is regarded as a putative target (Table 2). Complementarity to the 5′ end of the miRNA has been given due importance while predicting a target (Bartel, 2004). This complementarity requirement includes a 7-nucleotide perfect or near perfect match near the 5′ terminus of the miRNA (Lai et al., 2003; Lewis et al., 2003; Stark et al., 2003; Doench and Sharp, 2004). The 3′ region of the miRNA is less critical (Bartel, 2004) but can modulate its activity under certain circumstances (Doench and Sharp, 2004).
Table 2.
Predicted Targets of Newly Identified miRNAs
| miRNA | Target Genes with Number of Mismatches in Parentheses | Target Protein(s) |
|---|---|---|
| MIR171b and c | At2g45160(2), At4g00150(2), and At3g60630(2) | Scarecrow-like transcription factors |
| MIR389a.1 | At5g18040(1), At5g18065(1), At4g29760(2), At1g51670(2), and At4g29770(3) | Unknown proteins |
| MIR389b.3 | At4g28670(2) | Protein kinase |
| MIR390a.2 | At3g49250(3), At5g14050(3), and At2g39480(3) | Unknown protein; WD-40 repeat protein; ABC transporter |
| MIR393 | At3g26810(2), At1g12820(2), At3g62980(2), At3g26830(2), and At4g03190(3) | TIR1 (an F-box protein) family |
| MIR397a | At2g29130(2) and At2g38080(2) | Laccases |
| MIR397b | At3g60250(2), At2g29130(3), and At2g38080(3) | Casein kinase II; laccases |
| MIR398a | At3g15640(3) and At1g08050(5) | Cytochrome c oxidase; zinc finger (C3HC4-type ring finger) protein |
| MIR398b | At3g15640(4) and At1g08050(4) | Cytochrome c oxidase; Zinc finger (C3HC4-type ring finger) protein |
| MIR399b and c | At2g33770 and At4g00170(3) | Ubiquitin conjugating enzyme (UCE); vesicle-associated membrane protein |
| MIR399f | At2g33770 (zero to two mismatches, up to four target sites) | Putative UCE2 |
| MIR400 | At1g06580(0), At1g62720(1), At1g62670(1), At1g62590(2), At1g22960(2), At1g20300(2), At2g31400(2), At5g39710(2), At2g31400(2), At3g16010(3), and At4g19440(3) | PPR proteins |
| MIR401 | At2g06095(1), At3g42350(2), and At2g13270(3) | Unknown proteins |
| MIR402 | At4g34060(1) | ROS1-like, a putative DNA glycosylase |
| MIR403 | At3g07780(5) | Unknown protein |
| MIR404 | At1g07650(1) | LRR-TM protein kinase |
| MIR405 | At5g48280(3) | Unknown protein |
| MIR406 | At2g42510(3) and At1g54380(3) | Spliceosomal proteins |
| MIR407a | At1g17770(3), At3g13100(3), At5g49870(3), and At1g12010(3) | SET domain protein; ABC transporter; lectin; ACC oxidase |
| MIR407b | At1g54870(3) | Short-chain dehydrogenase/reductase |
| MIR408 | At2g47020(0) and At2g02850(2) | Peptide chain release factor; plantacyanin |
For each potential target gene, the number of mismatches between the miRNA and the mRNA is indicated in parentheses.
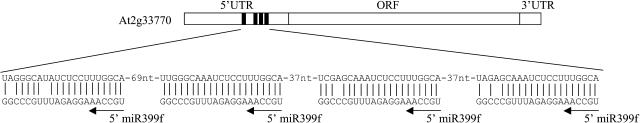
Our analysis predicted 51 potential targets for 21 of the newly identified miRNAs (Table 2). The plant miRNA target sites are predominantly found within the protein-coding segment of the mRNAs (Rhoades et al., 2002), whereas animal miRNAs appear to primarily target the 3′ UTRs (Enright et al., 2003; Lewis et al., 2003; Stark et al., 2003). Consistent with the earlier findings in Arabidopsis (Rhoades et al., 2002), 47 of our predicted target sites fall in the open reading frames (ORFs). Five of the predicted target sites lie in the 3′ UTR regions (At1g07650 [miR404], At1g12010 and At5g49870 [miR407a], At1g54870 [miR407b], and At2g47020 [miR408]). Interestingly, two miRNAs (miR399 and miR398) exhibit complementarity to the 5′ UTR regions of their respective targets. miR399 appears to target four sites in the 5′ UTR of At2g33770 (a putative ubiquitin conjugating enzyme E2) (Figure 4), whereas the miR398 family is likely to target the 5′ UTR of At3g15640 (a member of cytochrome c oxidase).
Figure 4.
Schematic Representation of the At2g33770 mRNA Showing the 5′ UTR, ORF, and 3′ UTR Regions.
Predicted miRNA targeting sites in the 5′ UTR region are shown (black rectangles). The enlarged portion shows the pairing between miR399f and the target sites.
We found that if more than one mRNA is predicted as the target of a miRNA, these targets tend to belong to the same gene family. For instance, miR400 is predicted to target more than 10 genes of the pentatricopeptide repeat (PPR) protein family; miR393 appears to target five genes belonging to the TIR1 (an F-box protein important for signaling by the phytohormone auxin) family; miR406 has the potential to target two members of spliceosome-related genes; miR397 appears to target two members of the laccase family; and miR389a.1 is capable of targeting four unknown proteins in the same family (Table 2). miR319c, a homolog of miR319/miRJAW, is likely to target the Teosinite branched1, cycloidea, and proliferating cell nuclear antigen factors family of transcription factors (Palatnik et al., 2003). miR399 family members are predicted to target the same gene, At2g33770 (ubiquitin conjugating enzyme E2). miRNAs that are represented by more than one member may recognize a consensus target sequence and, hence, might act on a common target or they might have tissue/stage-specific roles. In addition, different members of an miRNA family with slight changes in nucleotide sequence might also target different genes. For example, miR399b miRNA displays complementarity with a gene encoding a vesicle-associated membrane protein (At4g00170) in addition to the common target (At2g33770) of this family of miRNAs. miR397a is likely to target members of the laccase family, whereas another family member, miR397b, displays better complementarity with another gene (At3g60250, a casein kinase). Therefore, a slight change in miRNA sequence may alter its target preferences, distinguishing its role from that of other members of the same family.
The previously reported Arabidopsis miR161 is predicted to target PPR genes At1g06580(3), At1g62720(3), and At1g62670(3) (numbers in parenthesis are mismatches) (Rhoades et al., 2002). We have discovered an miRNA, miR400, that is different from miR161 and yet it displays perfect or near perfect complementarity with some of the predicted targets of miR161 [At1g06580(0), At1g62720(1), and At1g62670(1)]. This suggests that two completely different miRNAs may target the same gene. The attack on one gene by more than one miRNA could be a matter of cooperative regulation. Alternatively, it may indicate a redundant control of target gene expression by multiple miRNA genes or the deployment of different miRNAs for the regulation of a target in a developmental- or tissue-specific manner. Indeed, miR161 and miR400 have different spatial expression patterns. miR161 is strongly expressed in the stem and inflorescence (Reinhart et al., 2002), whereas miR400 is almost below the detection limit in stem and inflorescence but is moderately expressed in leaves and roots (Figure 5H). The PPR motif has been proposed to mediate macromolecular interactions. More than 400 genes encoding PPR proteins have been reported in Arabidopsis, and the majority are predicted to reside in either the chloroplast or mitochondrion (Small and Peeters, 2000). Because the PPR gene family is very large and diverse, it is conceivable that diverse miRNAs might be required for their regulation. Interestingly, PPR proteins have not been reported from animals.
Some of the miRNA complementary sites are highly conserved in different members of the same gene family in Arabidopsis or in mRNA targets from different plant species (Table 3). For instance, miR393 complementary sites are highly conserved in F-box proteins in Arabidopsis, rice, and maize (Zea mays) (Table 3). miR397 is predicted to target laccases, and the target sites are conserved in Arabidopsis, rice, tobacco (Nicotiana tabacum), and Populus (Table 3). miR408 may target mRNAs encoding plantacyanins, and the complementary sites are conserved in rice, maize, chickpea (Cicer arietinum), and spinach (Spinacia oleracea) (Table 3). The complementary sites of miR389a.1 are conserved in mRNAs that encode proteins belonging to the same family (Table 3). The complementary sites are also conserved in spliceosomal proteins, which are predicted to be targeted by miR406. The conservation in the target sequence is observed not only at the amino acid level but also at the nucleotide level, including sequences at codon position 3 (Table 3). Furthermore, the sequence conservation in the target genes is markedly higher in the section corresponding to the 5′ end of the miRNAs (Table 3).
Table 3.
miRNA Complementary Sites in Potential mRNA Targets Conserved in Arabidopsis and Other Plant Species
| Target Gene | miRNA/mRNA | Peptide Sequence |
|---|---|---|
| CCU AGU UAC GCU AGG GAA ACC U 5′ miR393 | ||
| At1g12820(2) | GaA aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| At3g26830(2) | GaA aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| At3g26810(2) | GaA aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| At3g62980(2) | GaG aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| At4g03190(3) | GaG aCc AUG CGA UCC CUU UGG A | ETMRSLWM |
| ZmAY105491(2) | GaG aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| ZmAY109095(2) | GaG aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| OsAK121600(2) | GaG aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| OsAK072338(2) | GaG aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| OsAK100862(2) | GaG aCA AUG CGA UCC CUU UGG A | ETMRSLWM |
| UG UAG UUG CGA CGU GAG UUA CU 5′ miR397a | ||
| At2g29130(2) | ua AUC AAU GCU GCA CUC AAU GA | LINAALND |
| At2g38080(2) | ua GUC AAC GCU GCA CUU AAU GA | LVNAALNE |
| OsNM-190623(3) | uC AUC AAC GCU GCA gUC AAU Gu | IINAAVNV |
| OsNM-190556(1) | uC AUC AAC GCU GCG CUC AAU GA | LINAALND |
| OsNM-192960(3) | uC AUC AAC GCU GCA gUC AAc GA | IINAAVND |
| PtY13773(3) | ua AUC AAC GCU GCA CUC AAc GA | LINAALND |
| NtU43542(1) | uC AUC AAC GCU GCG CUC AAU GA | VINAALNE |
| CG GUC CCU UCU CCG UCA CGU A 5′ miR408 | ||
| At2g02850(2) | cC aAG GGA AGA GGC AGU GCA U | AKGRGSAS |
| OsAK107381(2) | cC CAG GGA AGA GGC AGU GCA g | AQGRGSAA |
| ZmAY108638(2) | cU CAG GGA AGA GGC AGU GCG a | AQGRGSAT |
| SoU76296(1) | GU CAG GGA AGA GGC AGU GCA a | GQGRGSAR |
| CaAJ012693(2) | cU UuG GGA AGA GGC AGU GCA U | ALGRGSAL |
| G CAC UAU AAC CGU GCC GAG UU 5′ miR171b | ||
| At2g45160(2) | a GgG AUA UUG GCG CGG CUC AA | QGILARLN |
| At3g60630(2) | a GgG AUA UUG GCG CGG CUC AA | QGILARLN |
| At4g00150(2) | g GgG AUA UUG GCG CGG CUC AA | QGILARLN |
| OsAK100757(2) | a GaG AUA UUG GCG CGG CUC AA | REILARLN |
| OsAK120317(2) | a GaG AUA UUG GCG CGG CUC AA | REILARLN |
| OsAK101035(1) | C GaG AUA UUG GCG CGG CUC AA | REILARLN |
| OsAK106868(2) | g GaG AUA UUG GCG CGG CUC AA | REILARLN |
| OsNM-197667(1) | C GaG AUA UUG GCG CGG CUC AA | REILARLN |
| AT GCG AUA CAA CCU GAA UCU U 5′ miR389a.1 | ||
| At5g18065(1) | cA CGC UAU GUU GGA CUU AGG G | SRYVGLRD |
| At5g18040(1) | cA CGC UAU GUU GGA CUU AGG G | SRYVGLRD |
| At4g29760(2) | cA CaC UAU GUU GGA CUU AGA G | THYVGLRD |
| At4g29770(2) | cA CGa UAU GUU GGA CUU AGA G | TRYVGLRD |
| GAC CUA AUG UUA UCG UAA GAU 5′ miR406 | ||
| At1g54380(3) | gau GAU UAC AAU AGC AUU UUG | DDYNSIL |
| At2g42510(3) | gac GAU UAC AAU AGC AUU UUG | DDYNSIL |
For each gene, the nucleotide sequence of the miRNA complementary site is broken into codons corresponding to the reading frame of the mRNA. miRNA 5′ end is indicated. Mismatches are shown in lower case, and the number of mismatches is indicated in parentheses. The peptide sequence of the miRNA complementary site is shown. At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays; Nt, Nicotiana tabacum; Ca, Cicer arietinum; So, Spinach oleracea; Pt, Populus trichocarpus.
Tissue and Developmental Expression Patterns of miRNAs
Preferential expression of an miRNA in specific tissues or developmental stages may suggest a role in development for the miRNA. We investigated the tissue distribution of the newly identified miRNAs by RNA gel blot analysis on total RNA samples from various tissues of mature plants as well as young seedlings. We were able to detect a signal of the expected size for 13 miRNAs identified through cloning (Figure 5). The validation of other members of miR399, miR398, miR397, and predicted members of miR389 and miR407 were not attempted because of potential cross-hybridization. Some of the small RNAs were not detected on RNA gel blots but are still considered here as miRNAs because they were cloned and their precursors can adopt a hairpin structure. The miRNAs that could not be detected on RNA gel blots appeared only once in our library. A correlation was reported previously between the number of times a miRNA appeared in the library and its expression level. miRNAs identified in just one or a few clones have been difficult to detect on RNA gel blots, whereas those isolated many times are more easily detected (Grad et al., 2003; Lim et al., 2003; Kim et al., 2004). Some miRNAs may be expressed at low levels that are below the experimental detection threshold by RNA gel blot analysis, perhaps because of restricted tissue/cell-type expression (Grad et al., 2003; Lim et al., 2003; Kim et al., 2004).
Most of the previously known miRNAs in Arabidopsis are strongly expressed in floral tissues (Llave et al., 2002b; Park et al., 2002; Reinhart et al., 2002). This pattern is mirrored by miR319c, which is expressed at moderate levels in leaves, roots, and young seedlings but is very strongly expressed in stems and inflorescence (Figure 5G). Similarly, miR393 is ubiquitously detected in all samples but is strongest in the inflorescence, indicating that this miRNA might be important in silencing genes in reproductive tissues (Figure 5J). Also, miR402 and miR171b appear to have relatively lower levels of expression in leaves and young seedlings than in other tissues (Figures 5E and 5F).
However, other miRNAs in this study have quite divergent expression patterns. The most striking is that of miR398b, which could not be detected in the inflorescence (Figure 5A) and is preferentially expressed in leaves and young seedlings that consist mostly of young leaves. Similarly, miR399f, miR389b.3, and miR389a.1 are expressed more strongly in leaves and young seedlings, although they are also expressed in stems, roots, and inflorescence (Figures 5B to 5D). miR400 and miR397b show low but detectable expression in leaves, roots, and young seedlings but are undetectable in stems and inflorescence tissues (Figures 5H and 5I). miR403 and miR407 appear to be uniformly expressed in all tissues (Figures 5K and 5L). miR408 is strongly expressed in seedlings but weakly in all the tissues from adult plants (Figure 5M).
We also examined the expression of the miRNAs in dcl1-9 and hen1-1 mutant seedlings and compared the mutants to their wild-type background (Landsberg erecta ecotype). Consistent with the important roles of DCL1 (Reinhart et al., 2002) and HEN1 (Park et al., 2002) in miRNA accumulation, most of the miRNAs show substantially reduced expression in hen1-1, and several display reduced expression in dcl1-9 (Figure 5). The miRNAs that could be detected in the hen1 mutant are slightly longer than in the wild-type plants (Figure 5). We were unable to detect precursors for the miRNAs, even in the dcl1 and hen1 mutants. The inability to detect precursors of Arabidopsis miRNAs has been noted before (Park et al., 2002; Reinhart et al., 2002). That the dcl1 mutation does not impair the expression of many of the miRNAs may be because the Arabidopsis genome has several Dicer-like genes that might be functionally redundant for some of the miRNAs.
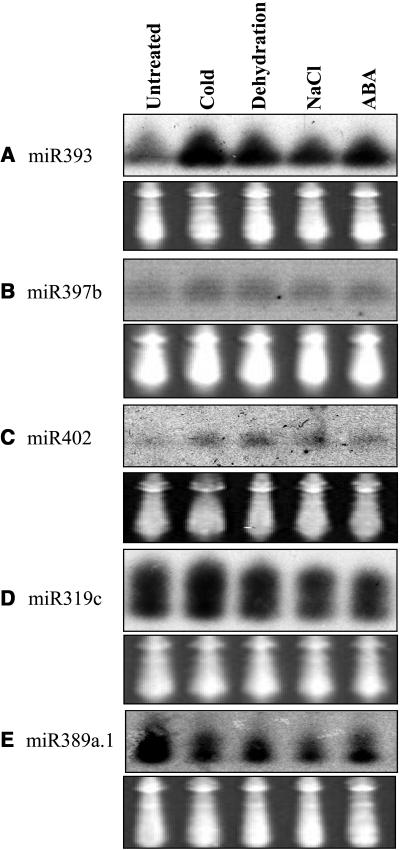
Abiotic Stress Regulation of miRNAs
To test whether the expression of any of the miRNAs is regulated by abiotic stresses, RNA gel blot analysis was performed on 2-week-old seedlings grown on MS-agar plates without stress treatment or treated with cold (0°C for 24 h), 100-μM ABA for 3 h, or dehydration (seedlings in agar plates were exposed to dry air), or seedlings were transferred to a blotting paper saturated with 300-mM NaCl for 5 h. We found that the expression of several of the miRNAs is either upregulated or downregulated by one or more of the stress treatments (Figure 6). miR393 is strongly upregulated by cold, dehydration, NaCl, and ABA treatments (Figure 6A). miR397b and miR402 are slightly upregulated by all the stress treatments, whereas miR319c appears to be upregulated by cold but not dehydration, NaCl, or ABA (Figures 6B to 6D). Interestingly, miR389a.1 appears to be downregulated by all of the stress treatments (Figure 6E).
Figure 6.
Regulation of miRNA Expression by Abiotic Stresses.
Shown are RNA gel blots of total RNA isolated from 2-week-old seedlings untreated or treated with different abiotic stresses. The tRNA and 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and serve as loading controls.
Other Novel Endogenous Small RNAs
Large censuses of small RNA cDNA libraries have revealed a significant class of other 21- to 22-nucleotide RNAs (Reinhart and Bartel, 2002; Ambros et al., 2003b; Aravin et al., 2003). These small RNAs differ from miRNAs in that they do not arise from hairpin precursors. Nevertheless, they display a strong size bias that suggests regulated processing by a Dicer-like activity. Furthermore, unlike some miRNAs, these small RNAs are generally not conserved phylogenetically. The recent discovery of a large class of tiny noncoding RNAs from C. elegans that share some features with miRNAs is an example for exceptional cases like this (Ambros et al., 2003b). These tiny noncoding RNAs are not even conserved within Caenorhabditis species (Ambros et al., 2003b). Another class of small RNAs, repeat-associated silencing RNAs, has been reported from Drosophila (Aravin et al., 2003). Repeat-associated silencing RNAs were noticed to be derived potentially from genomic regions rich in repeats (Aravin et al., 2003).
In this study, we have identified 102 endogenous small RNAs (putative siRNAs) that cannot be designated as miRNAs (Table 4). The putative siRNAs represent a heterogeneous class with respect to size and origin. The size ranged between 15 and 25 nucleotides, although the majority are in the range of 20 to 24 nucleotides. The small RNAs do not exhibit nucleotide preferences at their 5′ end. The majority have a single locus in the Arabidopsis genome, although some (28) correspond to multiple loci (2 to 64). Twelve of the small RNAs corresponding to multiple loci appear to be derived from repeat-rich regions or centromeric regions (Table 4). Two small RNAs (P22-H04 and P43-A12) are derived from putative retrotransposons. The vast majority is found in intergenic regions, but some match to coding regions and introns in the antisense polarity. We consider those in the sense polarity as possible degradation products. Some of the putative siRNAs have perfect complementarity with transposases, LOB, AIG1, 23s rRNA, F-box protein, or pre-mRNA splicing factor. Some siRNAs display complementarity with their putative target genes with one to three mismatches. For instance, P78-C02 is complementary to At2g27152 (AAO3, a drought stress-inducible ABA biosynthesis enzyme) with two mismatches, whereas P52-B05 is complementary with a putative lipoxygenase with three mismatches. Some of these small RNAs may actually be miRNAs but are not considered as miRNAs at this time because the precursor sequences cannot adopt a fold-back structure using the mfold program. We were unable to predict targets for the majority of the putative siRNAs identified. These small RNAs might target promoter regions of protein coding genes for transcriptional gene silencing (Park et al., 2002; Ambros et al., 2003b; Finnegan and Matzke, 2003; Bartel, 2004). The existence of diverse and vast numbers of endogenous small RNAs suggest that they may play important roles in the silencing of endogenous genes, in addition to their proposed roles in defense against transposons and viral invasion (Voinnet, 2002; Ambros et al., 2003b).
Table 4.
Other Endogenous Small RNAs from Arabidopsis
| Clone ID | Sequence (5′→3′) | Length | Chromosomal Number | Number of Origins | Genomic Location | Putative Target(s) |
|---|---|---|---|---|---|---|
| P9-A09 | UUAAUCCACCGUUUCAAUACGG | 22 | All | 49 | Intergenic | –a |
| B06-B06 | AUGAUGUAGAUCUCUCACUGAGA | 23 | 1 | 1 | Intergenic | – |
| P11-F11 | GUUUUUGCGUUAGUUUGCGG | 20 | 2, 3, and 4 | 3 | Intergenic | – |
| P15-A04 | UAAAUUAAUCAUAGUCUACAC | 21 | 3 | 2 | Intergenic | – |
| P16-C07 | UUGGCGCAUCUGCCGAUCC | 19 | 3 | 1 | Intergenic | – |
| P17-H03 | AUGUAAUCCAAUAUAUAUGGAGAG | 24 | 3 | 1 | Intergenic | – |
| P18-B03 | GGAGUCUGACAUGUGUGCAA | 20 | 5 | 1 | Intergenic | – |
| P18-H11 | UUGGAAGAUCGAGUAGACGCC | 21 | 3 | 1 | Intergenic | – |
| P22-H04 | CUUAGAAGUCGCCGAUUCGAA | 21 | 2 | 1 | Intergenic/retrotransposon, SINE-like | – |
| P24-E07 | UUGAGUUGAGUUACCCAUUU | 20 | 1 | 1 | Intergenic | – |
| P24-H08 | CGAUUAUUGCGUUCACACUAC | 21 | 2, 3, and 4 | 9 | Six intergenic, three intron At2g23720, At4g09380, and At4g28970 | – |
| P26-B01 | GUGUCGUCUAAAUAGAGAG | 19 | 5 | 1 | Antisense to At5g40050 | At5g40050 (F-box protein) |
| P26-G10 | CUCUGGUGGAAGCACGAUGAGG | 22 | 5 | 1 | Antisense to At5g37370 | At5g37370 (pre-mRNA splicing factor) |
| P27-G10 | AUAGAAACCUGACAUGUGUC | 20 | 1 | 1 | Intergenic | – |
| P29-B02 | AUGUGGCCCAUAUAAGUGGCAU | 22 | 4 | 1 | Intergenic | – |
| P30-B07 | GGUCGCAUUUGCCAAAAGCAAGU | 22 | 1 | 1 | Intergenic | – |
| P30-D08 | CAACUGCUGUGGAGUUUGGUUGAU | 24 | 1 | 1 | Intergenic | – |
| P34-E11 | UUUGGCAAUAUGCAAUGGUGGACU | 24 | 2, 3, 4, and 5 | 5 | Intergenic | – |
| P37-A03 | UCUUCCCGUUGGUGUGCGCCC | 21 | 2, 3, 4, and 5 | 8 | Intergenic | – |
| P37-E11 | UGAUUCUUGGUCUGACCAUG | 20 | 2 | 1 | Antisense to At2g14660 | At2g14660 (unknown protein) |
| P40-H11 | AUCACACAAGAAAAUGACAU | 20 | All | 25 | Intergenic | – |
| P42-A10 | CAUAUCGUUAAUAAAUGUUAG | 21 | 2 | 1 | Intergenic | – |
| P43-A12 | AGGAGGUUUUGGACCGAAGCC | 21 | 1 | 1 | Intergenic/retrotransposon, SINE-like | – |
| P43-C01 | UCAACCUUUUUCCGACGCCAGUGUA | 25 | 4 | 1 | Intergenic | – |
| P43-F05 | CAUGGUGUAGCCAAAGUCCAU | 20 | All | 64 | Centromeric region | At2g07981 (unknown protein) |
| P43-E06 | AGACGUCGGCGGGGGCCUCGGGAA | 24 | 2 and 3 | 2 | Intergenic | – |
| P44-C04 | UAUGUUUACUACAACUC | 17 | 4 | 1 | Intergenic | – |
| P45-A01 | CGAGGAAAGGCUUAGUGUGAA | 21 | 2 | 1 | Intergenic | – |
| P46-G11 | UUUGGUUGCGAUGAUGAU | 18 | 1 | 1 | Intergenic | – |
| P47-E12 | AUAAAUUUACUCCGGCGACC | 20 | 1 | 1 | Intergenic | – |
| P49-F06 | UUUAAGGGCCGUCCUGAACAUAAU | 24 | 2 | 1 | Intergenic | – |
| P48-C06 | AAAGUGUGAUCGUGGAAU | 18 | All | 51 | Intergenic | – |
| P4-F11 | GUGGUAACUGGUAUUCAUUUUUU | 23 | 3 | 1 | Intergenic | – |
| P50-C08 | CUUGGCAUGUGAUACCUUUUUGGA | 24 | 3 and 5 | 54 | Centromeric region | – |
| P52-B05 | CAGCUAAUAAAAGUCUGUCGUA | 22 | 1, 4, and 5 | 3 | Intergenic | At1g72520 (putative lipoxygenase) |
| P52-H12 | ACCGAGGACGAGGUCGUUGAC | 21 | 2 | 1 | Intergenic | – |
| P53-C02 | AACUCGUGUGAUUGUGACACGCGU | 24 | 5 | 1 | Intergenic | – |
| P54-C07 | UUUGUCGACGGCAUUUUAAAA | 21 | 1 | 1 | Intergenic | – |
| P55-F01 | CCUGCCAAAACGGUCUC | 17 | 2 | 1 | Intergenic | – |
| P56-B02 | AAUAGGAGAAUGUACCAUAUGACC | 24 | All | 18 | Intergenic | – |
| P56-H09 | UGUCGCGCUUUUCCCAUUACC | 21 | 4 | 1 | Antisense to At4g09940 | At4g09940 (AIG1, avirulence-induced protein) |
| P58-C11 | GAUAAACCAGGAGGAAUAAGA | 21 | 2 | 1 | Antisense to At2g44090 (intron) | – |
| P59-C11 | CCACCAACCUACCAAAUUCAGACU | 24 | 5 | 1 | Intergenic | – |
| P60-C08 | UUUCAGGAGGUCGAGUGACUUC | 21 | 1 | 1 | Intergenic | – |
| P61-D02 | UUUAUUGGACCGUUUUUCACCCA | 23 | 5 | 1 | Intergenic | – |
| P61-H07 | GUCAGAAUUCUACGCAGAUAGAAC | 24 | 4 | 1 | Intergenic | – |
| P62-G09 | CUAGAUCAUCUGUUAGCACAC | 21 | 3 | 1 | Intergenic | – |
| P63-F06 | GCCUUCGUAGCGGCGAAUUAAAGA | 24 | 4 | 1 | Intergenic | – |
| P64-B12 | UUGAAGAUAGAGAGCGGUAGAG | 22 | 2 | 1 | Intergenic | – |
| P66-F10 | AGAUAGGGUCCGGCGG | 16 | 1 | 1 | Antisense to At1g70985 | At1g70985 (extensin-like) |
| P67-C05 | AAUUUACGGUGUGCGUACAUGUGU | 24 | 3 | 1 | Intergenic | – |
| P67-G03 | AACGUCCGUGUCCCGAGUUAGGGA | 24 | 3 | 1 | Intergenic | – |
| P69-A11 | AUUAAAUAAAUAUGGUGCACUCUU | 24 | All | 48 | Intergenic | – |
| P71-B07 | UACUAAUACGAAUCACUAAA | 20 | 1 | 1 | Intergenic | – |
| P73-A04 | UGAACCUCAAAAACAUAGUAUGC | 23 | 5 | 1 | Intergenic | – |
| P73-G06 | CGAUGAAAUGCCUAAGAGAAUUU | 23 | 2 | 1 | Intergenic | – |
| P74-C01 | UUGGAGAUGGUCUAAAACUCU | 21 | 4 | 1 | Intergenic | – |
| P74-D02 | UUGGAGGGUCCUGCUUUCGAGUG | 23 | 1 | 1 | Intergenic | – |
| P74-G10 | GCAGAUCUUGGUGGUAG | 17 | 2 and 3 | 1 | Intergenic | – |
| P74-E12 | UUGGACGCACCAUUU | 15 | 4 | 1 | Intergenic | – |
| P75-B05 | AUAGUCAUACAGAAAAGUAAUA | 22 | 2 | 1 | Intergenic | – |
| P77-A08 | CAUAUAUCCCAAACCUAGAGUGA | 23 | 5 | 1 | Intergenic | – |
| P78-A02 | UAGAAUAUGAGCUCU | 15 | 5 | 1 | Intergenic | – |
| P78-C02 | UGGAACAUCACCUACUGGUGG | 21 | 3 and 5 | 2 | Antisense to At3g43090 (intron) | At2g27150 (aldehyde oxidase 3 [AAO3]) |
| P79-H10 | GCCCGGGAACCGCCUUGG | 18 | 3 | 1 | Intergenic | – |
| P81-H06 | UGAAGACGUGAUCGAGUGUCC | 21 | All | 29 | Centromeric regions | At1g36670 and At1g37050 (transposases); At4g05636, At4g05640, At5g32161, and At1g41650 (hypothetical proteins) |
| P82-B07 | CUUCGAGCGAAGGCUCGGGUC | 21 | 2 | 1 | Intergenic | – |
| P85-F04 | GAGUUGUAUAUGUUUUGUUGA | 21 | All | 51 | Intergenic | – |
| P86-E10 | CCAUGUGAGAGACUA | 17 | 3 and 1 | 1 | Intergenic | – |
| P87-C02 | AUAGACUUAGUGAGGUGUAUCGGG | 24 | 1 | 2 | Intergenic | – |
| P87-E07 | UGUGCCCAAGAUGGGAGGGAGA | 22 | 4 | 1 | Intergenic | – |
| P89-H02 | GUUUUGACCGGAGCCUUCUCAAA | 22 | 1 | 1 | Intergenic | – |
| P89-H03 | CUAUCCAUCCUGAGUUUC | 18 | 2 | 1 | Intergenic | – |
| P90-C03 | UAGCUCAGGUAAGUCGAUCGU | 21 | 5 | 1 | Junction of intron and exon At5g13740 | – |
| P90-H01 | UUUAAUAGGAGAAUGUACCAUAU | 23 | All | 18 | Intergenic | – |
| P91-B11 | UUUGAUUUAGUGGGUAAAGCC | 22 | 1, 2, 3, and 5 | 6 | Intergenic | – |
| P91-G03 | UUUCACAAAACCUUGACCUCAAGG | 24 | 1 and 3 | 2 | Intergenic | – |
| P91-H05 | GUCGAAUAUGACUUGAUCUCAUGU | 24 | 1, 2, and 5 | 6 | Intergenic | – |
| P92-A05 | AUGGAUUGUCAAAGUCAAAUGUGG | 24 | Chr | 14 | Intergenic | – |
| P92-B09 | CUCCCUUCGGGGUAAAGCCCUA | 22 | 1 and 5 | 3 | Intergenic | – |
| P92-F02 | UUUCUUGUGUCGAUA | 15 | 1 | 1 | Intergenic | – |
| P93-H01 | UAGAGGUGUCAGAAA | 15 | 1, 2, and 3 | 3 | Intergenic | – |
| P94-G03 | AAUGAAAGCACCAAUUGAUAA | 21 | 3 | 3 | Intergenic | – |
| P94-H07 | AUAACAGGCCUAGGCCCAAUUA | 22 | 1, 2, 3, and 5 | 18 | Intergenic | – |
| P96-G12 | AGAAUCUUGAUGAAA | 15 | 3 | 2 | Intergenic | At5g64900 (unknown protein) |
| P96-H07 | UAUAACUCCUUAUCACCA | 18 | 3 | 1 | Intergenic | – |
| P97-B12 | UGAGGGUUCCGGGAGGACCC | 20 | 2 | 1 | Antisense to At2g40470 | At2g40470 (lateral organ boundaries domain protein 15 [LOBD15]) |
| P97-F09 | AUCCGUGAUGGAUAUUCCGGGAUG | 24 | 5 and 3 | 2 | Intergenic | – |
| P98-H09 | GUGCAUGUUAGUGAUGUGGAGGAG | 24 | 1 | 1 | Intergenic | – |
| P98-G06 | UUGAUAGUGACUACACUCGG | 20 | 1 | 1 | Intergenic | – |
| P99-A12 | UUCGAAUCCUACUUGGGGAG | 20 | 4 | 1 | Intergenic | – |
| P99-B05 | CCAGAGACGAGGCAA | 15 | 2 | 1 | Intergenic | – |
| P99-B10 | GUUGAAAUAAGCGUAG | 16 | 5 | 1 | Intergenic | – |
| P63-G11 | AUUAAAUAAAUAUGGUGCACUCUU | 24 | 1, 2, and 5 | 6 | Intergenic | – |
| P48-C06 | CAAUCCUCCUACCACUUGGUC | 21 | 1 | 1 | Antisense to At1g79280 | At1g79280 (unknown protein) |
| P96-F02 | UAUCCCGGCCCCUCUCGGGUC | 21 | 5 | 1 | At5g62520 | At5g62530 (P5C dehydrogenase) |
| P12-H02 | ACGCAAUUGUUCGAUAG | 17 | 1 | 1 | Intergenic | – |
| P17-G10 | AGGAGGGUUUCGUGAGCCUGUU | 22 | 5 | 1 | Intergenic | – |
| P67-H01 | CCGCAGUUACAAUAUACUC | 19 | 3 | 1 | Antisense to At3g03560 | At3g03560 (unknown protein) |
| P61-F02 | AUGUUGACUAAUCUGGCUAAC | 21 | Chloroplast | 1 | Chloroplast | At2g07739 (unknown protein) |
| P78-G11 | UGGCUGUCUCUGCACCCCUA | 20 | 2, chloroplast, and mitochondria | 3 | Intergenic | At2g07707 (23s rRNA) |
The small RNA sequence, length, chromosomal origin(s), and potential targets are indicated.
A putative target does not exist.
DISCUSSION
Small noncoding RNAs have recently emerged as important regulators of both transcriptional and posttranscriptional gene silencing. The identification of the entire set of small RNAs from an organism is of fundamental importance and would lay the foundation for understanding gene regulation involving small RNAs. The endeavor is as important as mining genes that code for proteins. The establishment of a comprehensive list of miRNAs from any organism will be instrumental for not only gene regulation studies but also for genome organization (e.g., transposon activity and chromatin assembly), phylogenetic comparison, comparative studies of development, and other evolutionary analysis. By sequencing a library of small RNAs from seedlings treated with different abiotic stresses, we are able to add 15 new families of miRNAs to the previously published 15 families in Arabidopsis. In addition, we identified new members of the previously reported miR171 and miR319 families (Table 1, Figure 1).
Some miRNAs are encoded by single copy genes, others by multiple genes. Most miRNA genes come from intergenic regions, but a sizable minority of miRNAs (about one-quarter of the human miRNA genes) occurs in the introns of protein coding genes, preferentially in the same orientation as the mRNAs (Bartel, 2004). All miRNAs identified in this study are intergenic in origin with two exceptions: miR402 is derived from the first intron of At1g77230, and miR408 is in the antisense polarity to the 3′ UTR of At2g47020. The majority of the miRNAs described in this study are located in the 3′ arm of the fold-back structure (Table 1). Most miRNAs have a 5′ U preference that is similar to earlier reports (Bartel and Bartel, 2003; Bartel, 2004), although a few miRNAs begin with a 5′ A or C. Analysis of the thermodynamic profiles of miRNAs from plants revealed that the strand having low internal stability at the 5′ end is retained within the RNA-induced silencing complex (Khvorova et al., 2003). Some of the miRNAs reported in this study conform to the features described by Khvorova et al. (2003) and Schwarz et al. (2003), although some show considerable variability (see Supplemental Table 2 online).
Ten of the miRNAs that we have identified have clear orthologs or homologs in rice, Lotus, Medicago, or Populus, suggesting that they conduct important and perhaps conserved functions. On the other hand, many of the miRNAs are not conserved. These nonconserved miRNAs may play more species-specific roles.
Computational prediction of putative targets for miRNAs in plants has been facilitated by their perfect or near perfect complementarity to target genes (Rhoades et al., 2002). By contrast, bioinformatic approaches to identifying mRNA targets for animal miRNAs are complicated by the partial complementarity between an miRNA and its target (Lewis et al., 2003). Sixty-one targets, predominantly transcription factors, were predicted previously in Arabidopsis (Rhoades et al., 2002; Bartel and Bartel, 2003). The predicted targets that are not transcription factors include PPR mRNAs by miR161, DCL-1 by miR162, and Argonaute by miR168 (Rhoades et al., 2002). In this study, we were able to predict 51 genes that are likely targeted by the miRNAs described here (Table 2). These targets include metabolic enzymes (laccases and cytochrome c oxidase), enzymes in the ubiquitination pathway (ubiquitin conjugating enzyme E2 and TIR1/ubiquitin ligase), transcription factors (Scarecrow-like transcription factors), signal transduction components (protein kinase, casein kinase, and leucine-rich repeat–transmembrane [LRR-TM] kinase), and genes involved in RNA processing (spliceosomal proteins and PPR proteins), protein synthesis (peptide chain release factor), and transcriptional gene silencing [DNA glycosylase and Su(var)3-9, Enhancer-of-zeste, Trithorax (SET) domain proteins]. Our predicted targets appear to be involved in more diverse biological processes and functions and not predominantly in transcriptional regulation as predicted in earlier studies (Rhoades et al., 2002).
It was hypothesized that a complementary site in the coding region of an mRNA may lead to the cleavage of the target, whereas target sites located in 3′ UTRs might attenuate translation (Bartel, 2004). Consistent with this view, Arabidopsis miRNAs that target ORFs have largely been shown to be involved in the cleavage of target mRNAs (Llave et al., 2002a; Kasschau et al., 2003; Palatnik et al., 2003; Xie et al., 2003; Vazquez et al., 2004). Perhaps as an exception, Arabidopsis miR172 represses translation despite having a complementary site within the ORF of its target mRNA, AP2 (Aukerman and Sakai, 2003; Chen, 2004). The complementary sites for the animal miRNAs have been predicted to reside in the 3′ UTRs of target genes (Lewis et al., 2003; Bartel, 2004). This bias might reflect a mechanistic preference for translational repression over target cleavage. Only in a few cases are 3′ UTRs predicted as the target sites for plant miRNAs. For example, the complementary sites for Arabidopsis miRNAs, miR156/157, miR156, and miR169, reside in the 3′ UTRs of At1g53160, At2g33810, At1g17590, and At1g54160, respectively (Rhoades et al., 2002). The target sites for five of the predicted targets described here are within the 3′ UTR regions.
Until now, no miRNA from animals or plants has been predicted to target the 5′ UTR of a gene. In this work, we find that two miRNA families (miR399 and miR398) are likely to target 5′ UTRs. miR398 family members are complementary to the 5′ UTR of At3g15640 (cytochrome c oxidase). The miR399 has multiple complementary sites (four in Arabidopsis; Figure 4) within the 5′ UTR of At2g33770 (putative ubiquitin conjugating enzyme E2). Future functional studies on the miR398 and miR399 families of miRNAs may reveal a novel mode of action of these miRNAs on their targets. The presence of multiple miRNA complementary sites is known in animal targets (Lee et al., 1993; Wightman et al., 1993; Lai, 2003; Bartel, 2004). The presence of multiple target sites within the same target implies that in some cases more than one miRNA complementary site is required in the target gene for its effective downregulation. Indeed, it has been shown that increasing the number of miRNA/siRNA binding sites increases the degree of repression of a target gene (Doench et al., 2003).
The targeting of the 3′ UTR of At2g47020 by miR408 is particularly interesting. The translation of At2g47020 mRNA may be repressed by miR408. At2g47020 is predicted to encode a peptide chain release factor important in mRNA translation. This suggests that the translational machinery may be targeted by an miRNA for translational regulation. The targeting of a ROS1-like DNA glycosylase and a SET domain protein by miR402 and miR407 miRNAs, respectively, is also very interesting. ROS1 is a DNA glycosylase/lyase required for the repression of siRNA-triggered transcriptional gene silencing by DNA demethylation (Gong et al., 2002). SET domain proteins are also important in transcriptional gene silencing by functioning in histone Lys methylation (Malagnac et al., 2002). These, together with previous evidence for the targeting of DCL1 by an miRNA (Xie et al., 2003), point to an important role of miRNAs in the regulation of machineries involved in gene silencing.
The concentration and activities of many cellular proteins are regulated by the ubiquitin pathway. Ubiquitination requires a cascade of three enzymatic activities for activating (E1), conjugating (E2), and ligating (E3) ubiquitin to a substrate. In this study, we have identified two miRNAs (miR399 and miR393) that may target components (ubiquitin conjugating enzyme E2 and ubiquitin ligase/TIR1) of the ubiquitination pathway. The result suggests a role for miRNAs in regulating the process of ubiquitination. Fungal laccases are well known for their roles in morphogenesis, pathogenicity, and lignin degradation (Mayer and Christopher, 2002). The physiological roles of higher plant laccases are relatively unknown despite their existence as multigene families (Mayer and Christopher, 2002). The identification of miRNAs (miR397 family) that can target laccases provides a unique tool to investigate the function of laccases in higher plants. Similarly, the identification of miR389a.1 that is predicted to target four unknown proteins in the same family (Tables 2 and 3) will be valuable for investigating the function of these proteins. Overexpression of miR389a.1 or miR389b.1 may be an effective way to silence these genes (either by cleavage or translational repression) and thus reveal their function.
Several of the miRNAs discovered in this study are either upregulated or downregulated by abiotic stresses, suggesting that they may be involved in stress-responsive gene expression and stress adaptation. Consistent with this notion, the Arabidopsis hen1-1 and dcl1-9 mutants that are impaired in the production of some miRNAs are hypersensitive to abiotic stresses (our unpublished data). Stress-induced or upregulated miRNAs are expected to target negative regulators of stress responses or positive regulators of processes that are inhibited by stresses (e.g., cell division and expansion). On the other hand, stress downregulated miRNAs may repress the expression of positive regulators and/or stress upregulated genes. The significance of miR389a.1 miRNA downregulation by ABA and several stresses is unclear because this miRNA appears to target several genes encoding unknown proteins. The stress upregulation of miR393, which targets TIR1, suggests that stress may cause increased TIR1 mRNA degradation or translational repression. Because TIR1 is a positive regulator of auxin signaling by promoting the degradation of Aux/IAA proteins through ubiquitination (Dharmasiri and Estelle, 2002), the miRNA inhibition of TIR1 would downregulate auxin signaling and seedling growth. Thus, the upregulation of miR393 may contribute to the inhibition of plant growth under stress conditions. Future analysis of TIR1 mRNA and protein levels in plants with altered miR393 expression, or expressing miR393-resistant forms of TIR1 mRNA, will help to test this hypothesis.
The identification of endogenous siRNAs provides insights into the complexity of RNA-guided regulation. In this study, we have also uncovered a large number (102) of putative endogenous siRNAs that were not described previously, and these are distributed across all five chromosomes. We tested several of these small RNAs but were unsuccessful in detecting them on RNA gel blots, likely reflecting their low abundance. Unlike miRNAs, only a few of these small RNAs were represented by more than one clone in the sequenced library. These putative siRNAs are not conserved in other plants, which suggests species-specific endogenous RNA silencing mechanisms. The identification of unique intergenic regions as the sites of origin for most of the siRNAs indicated that they might be expressed independently. However, where multiple loci were identified for an siRNA, the origin was ambiguous. In plants, siRNAs have been suggested to mediate repression of retroelements via histone methylation as well as asymmetric DNA methylation (Zilberman et al., 2003) and to mediate systemic silencing (Hamilton et al., 2002). Our finding that several of the siRNAs might target Athila-related genes suggests that the taming of transposable elements is of critical importance for the plant genome. The production of siRNAs involved in controlling the mobility of transposable elements may also operate in Drosophila (Aravin et al., 2003). In addition, our identification of small RNAs that might target putative LRR-TM kinase, 23s rRNA, AAO3, P5CDH, lipoxygenase, or LOB genes raises the possibility that a wide range of protein coding and noncoding genes may be subjected to silencing by endogenous siRNAs.
Several of the miRNAs reported in this study (miR402, miR403, miR397a, miR171b, and a predicted miRNA, miR389b.2) have also been identified by an NSF 2010 project, which can be accessed through the Arabidopsis Small RNA Project Web site of the Carrington lab (http://cgrb.orst.edu/smallRNA/). Despite the sequencing by the NSF 2010 project of several libraries from different genetic backgrounds and tissues, the overlap with ours is minimal. This suggests that the cloning of miRNAs in Arabidopsis is not saturated, and more miRNAs have yet to be identified. Therefore, continued efforts in the future are needed to identify the complete set of miRNAs and other small RNAs from Arabidopsis.
METHODS
Cloning of miRNAs from Arabidopsis
Total RNA was isolated from different samples (Arabidopsis thaliana ecotype Columbia; untreated, treated with cold stress 0°C for 24 h, 300 mM NaCl for 5 h, dehydration for 10 h, or 100-μM ABA for 3 h). Cloning of miRNAs was performed as described (Elbashir et al., 2001; Llave et al., 2002a). In brief, ∼600 μg of RNA was resolved through two lanes on a denaturing 15% polyacrylamide gel. Labeled DNA oligonucleotides were used as size standards. A gel fragment spanning the size range of 15 to 26 nucleotides was excised, and RNA was eluted overnight with 0.4 M NaCl at 4°C. The RNA was recovered by ethanol precipitation, dephosphorylated, and again ethanol precipitated. Then, the small RNAs were ligated sequentially to 5′ and 3′ RNA/DNA chimeric oligonucleotide adapters (Dharmacon Research, Boulder, CO). The 3′ adapter oligonucleotide (5′-pUUUaaccgcatccttctcx-3′; uppercase, RNA; lowercase, DNA; p, phosphate; x, inverted deoxythymidine) (Elbashir et al., 2001) possessing a 5′ monophosphate and a 3′ inverted deoxythymidine to prevent self ligation, was then ligated to the dephosphorylated small RNA. The ligation product was recovered from the gel, 5′ phosphorylated, and the RNA was recovered by ethanol precipitation. Next, the 5′ adapter (5′tactaatacgactcactAAA; uppercase, RNA; lowercase, DNA) was ligated to the phosphorylated ligation product as described above. The ligation products from the second ligation reaction were excised and eluted from the gel. Reverse transcription reaction was performed using the RT primer (5′-TTTTCTGCAGAAGGATGCGGTTAAA-3′; bold, PstI site). This was followed by PCR using the reverse (RT primer) and forward (5′-AAACCATGGTACTAATACGACTCACTAAA-3′; bold, NcoI site) primers. The PCR product was purified by phenol/chloroform extraction, ethanol precipitated, and digested with PstI and NcoI. After additional phenol/chloroform extraction and ethanol precipitation, the digested products were ligated to a PstI-NcoI–linearized pGEM-T Easy vector using T4 DNA ligase. The inserts from individual colonies were amplified by T7 and SP6 primers and sequenced. The sequences were subsequently processed and used for BLAST analysis against the Arabidopsis sequences in The Arabidopsis Information Resource database (http://www.arabidopsis.org/) and other plant sequences in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/BLAST/).
Prediction of Fold-Back Structures
Fold-back secondary structures were predicted with the computer program RNA fold, using the Zuker algorithm (Zuker, 2003). The cloned RNA was folded with flanking sequence in three contexts, either 300 bp of upstream sequence plus 20 to 30 bp of downstream sequence or vice versa or 150 bp upstream and 150 bp downstream of the cloned sequence.
RNA Gel Blot Analysis
Total RNA was isolated from 2-week-old seedlings (Columbia ecotype) grown on MS nutrient-agar plates (untreated, treated with cold stress 0°C for 24 h, 300 mM NaCl for 5 h, dehydration for 10 h, or 100-μM ABA for 3 h). Total RNA was also isolated from different tissues (stem, leaf, inflorescence, and root) using the TRIZOL reagent (Invitrogen, Life Technologies, Carlsbad, CA). Forty micrograms of total RNA was loaded per lane and resolved on a denaturing 15% polyacrylamide gel and transferred electrophoretically to Hybond N+ (Amersham, Piscataway, NJ) membranes. Membranes were UV cross-linked and baked for 1 h at 80°C. DNA oligonucleotides complementary to small RNA sequences were end labeled with γ-32P-ATP using T4 polynucleotide kinase (Invitrogen). Blots were prehybridized for at least 1 h and hybridized overnight using PerfectHYB Plus buffer (Sigma, St. Louis, MO) at 38°C. Blots were washed three times (two times with 2× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 0.1% SDS for 20 min and once with 1× SSC and 0.1% SDS for 20 min) at 50°C. The membranes were briefly air dried and then exposed to a phosphorimager or autoradiographed.
Supplementary Material
Acknowledgments
We thank Xuemei Chen for providing hen1-1 and dcl1-9 seeds and Becky Stevenson for technical assistance during the course of this work. We also thank Shou-Wei Ding and Andre Jaggendorf for critical reading of the manuscript. We thank Anastasia Khavorova and Annaleen Vermeulen for their help in analyzing thermodynamic profiles of miRNAs. This work was supported by National Institutes of Health Grant R01GM0707501 and National Science Foundation Grant IBN-9808398 to J.K.Z.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022830.
References
- Ambros, V., et al. (2003. a). A uniform system for microRNA annotation. RNA 9, 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. (2003. b). MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13, 807–818. [DOI] [PubMed] [Google Scholar]
- Aravin, A., Lagos-Quintana, M., Yalcin, A., Zavalon, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Aufsatz, W., Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J.M. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21, 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-Like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Liu, J., Cheng, Y., and Jia, D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, S., and Estelle, M. (2002). The role of regulated protein degradation in auxin response. Plant Mol. Biol. 49, 401–409. [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. (2003). siRNAs can function as miRNAs. Genes Dev. 17, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., and Sharp, P.A. (2004). Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5, R1.1–R1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., and Matzke, M.A. (2003). The small RNA world. J. Cell Sci. 116, 4689–4693. [DOI] [PubMed] [Google Scholar]
- Golden, T.A., Schauer, S.E., Lang, J.D., Pien, S., Mushegian, A.R., Grossniklaus, U., Meinke, D.W., and Ray, A. (2002). SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130, 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z., Morales-Ruiz, T., Ariza, R.R., Roldan-Arjona, T., David, L., and Zhu, J.-K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814. [DOI] [PubMed] [Google Scholar]
- Grad, Y., Aach, J., Hayes, G.D., Reinhart, B.J., Church, G.M., Ruvkun, G., and Kim, J. (2003). Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., and Baulcombe, D. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Han, M.-H., Goud, S., Song, L., and Federoff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C.P. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. [DOI] [PubMed] [Google Scholar]
- Kasschau, K., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., and Plasterk, R.H. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., and Jayasena, S.D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through Argonaute1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Kim, J., Krichevsky, A., Grad, Y., Hayes, G.D., Kosik, K.S., Church, G.M., and Ruvkun, G. (2004). Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. USA 101, 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.C. (2003). MicroRNAs: Runts of the genome assert themselves. Curr. Biol. 13, R925–R936. [DOI] [PubMed] [Google Scholar]
- Lai, E., Tomancak, P., Williams, R.W., and Rubin, G.M. (2003). Computational identification of Drosophila microRNA genes. Genome Biol. 4, R42.1–R42.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. (2003). The microRNAs of Caenorhabditis elegans. Genes Dev. 17, 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M., and Carrington, J.C. (2002. b). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002. a). Cleavage of Scarecrew-like mRNA targets directed by a class of Arabidopsis microRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lu, C., and Federoff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac, F., Bartee, L., and Bender, J. (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21, 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, A.M., and Christopher, R.C. (2002). Laccase: New functions for an old enzyme. Phytochem. 60, 551–556. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., Aufstaz, W., van der Winden, J., Matzke, M., and Matzke, A. (2000). Transcriptional gene silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R.H. (2002). RNA silencing: The genome's immune system. Science 296, 1263–1265. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., and Bartel, D.P. (2002). Small RNAs correspond to centromere heterochromatic repeats. Science 297, 1831. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M., Reinhart, B., Lim, L., Burge, B., Bartel, B., and Bartel, D. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H., and Fire, A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Small, I., and Peeters, N. (2000). The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russel, R.B., and Cohen, S.M. (2003). Identification of Drosophila microRNA targets. PLoS Biol. 1, 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2001). RNA silencing as a plant immune system against viruses. Trends Genet. 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2002). RNA silencing: Small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5, 444–451. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf, D., Jeong, B., Zhang, C., and Cerutti, H. (2000). Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-Box RNA helicase. Science 290, 1159–1163. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.