Abstract
The 65-kD microtubule-associated protein (MAP65) family is a family of plant microtubule-bundling proteins. Functional analysis is complicated by the heterogeneity within this family: there are nine MAP65 genes in Arabidopsis thaliana, AtMAP65-1 to AtMAP65-9. To begin the functional dissection of the Arabidopsis MAP65 proteins, we have concentrated on a single isoform, AtMAP65-1, and examined its effect on the dynamics of mammalian microtubules. We show that recombinant AtMAP65-1 does not promote polymerization and does not stabilize microtubules against cold-induced microtubule depolymerization. However, we show that it does induce microtubule bundling in vitro and that this protein forms 25-nm cross-bridges between microtubules. We further demonstrate that the microtubule binding region resides in the C-terminal half of the protein and that Ala409 and Ala420 are essential for the interaction with microtubules. Ala420 is a conserved amino acid in the AtMAP65 family and is mutated to Val in the cytokinesis-defective mutant pleiade-4 of the AtMAP65-3/PLEIADE gene. We show that AtMAP65-1 can form dimers and that a region in the N terminus is responsible for this activity. Neither the microtubule binding region nor the dimerization region alone could induce microtubule bundling, strongly suggesting that dimerization is necessary to produce the microtubule cross-bridges. In vivo, AtMAP65-1 is ubiquitously expressed both during the cell cycle and in all plant organs and tissues with the exception of anthers and petals. Moreover, using an antiserum raised to AtMAP65-1, we show that AtMAP65-1 binds microtubules at specific stages of the cell cycle.
INTRODUCTION
Most of the plant microtubule-associated proteins (MAPs) discovered so far have structural homologs in other eukaryotes, but not all animal or fungal MAPs are present in plants (Gardiner and Marc 2003; Lloyd et al., 2004). Plant microtubules, although structurally similar to their eukaryotic counterparts, differ in their organization and dynamics. Plant cells have three unique microtubule organizations: the interphase cortical array (consisting of parallel rather than radial microtubules), the preprophase band (which predicts the division plane), and the cytokinetic phragmoplast (consisting of two interdigitating sets of antiparallel microtubules). The radial microtubule array of animal cells is a result of polymerization from a single centrosome juxtaposed to the nucleus, whereas plant microtubules are polymerized from multiple dispersed nucleation sites in the cytoplasm (Chan et al., 2003a). Plant microtubules are also more dynamic (Hush et al., 1994; Yuan et al., 1994; Moore et al., 1997) and persist in the treadmilling state longer than their animal counterparts (Shaw et al., 2003). To understand the molecular basis for these differences and to model the regulation of plant microtubule organization and dynamics, more information on the plant structural and regulatory MAPs is needed.
MAP-65 is the name given to the most abundant group of MAPs, of electrophoretic molecular weight approximating 65 kD, in microtubule preparations from tobacco (Nicotiana tabacum Bright Yellow-2) (Jiang and Sonobe, 1993) and carrot (Daucus carota) (Chan et al., 1996). Corresponding MAP65 cDNAs have been cloned (NtMAP65-1, Smertenko et al., 2000; DcMAP65-1, Chan et al., 2003b), and a gene family of nine members has been identified in Arabidopsis thaliana (Hussey et al., 2002). Biochemically purified MAP65 proteins have been found to bind and bundle microtubules in vitro (Jiang and Sonobe, 1993; Chan et al., 1999). Antibodies raised against biochemically purified tobacco MAP65 decorate all microtubules (Jiang and Sonobe, 1993) but antibodies raised to one isotype recombinant NtMAP65-1 do not (Smertenko et al., 2000). Anti-NtMAP65-1 recognizes only subsets of interphase microtubules and in particular the anaphase spindle midzone and at the midline of the cytokinetic phragmoplast. The overlapping microtubules at the spindle midzone have the same polarity as those in the phragmoplast, and it has been suggested that MAP65 cross-links antiparallel microtubules (Smertenko et al., 2000). In biochemically purified carrot MAP65 preparations there are three electrophoretically separable bands, and only one, the 62-kD band, was found to be present in elongating cells containing only cortical microtubules, indicating that this MAP65 is involved in directional cell expansion (Chan et al., 2003b). Furthermore, it has been suggested that stabilization of the interphase cortical array by MAP65 is necessary for the normal progression of embryogenesis (Smertenko et al., 2003).
A family of nine AtMAP65 genes with predicted open reading frames has been identified in the Arabidopsis Genome Database (Hussey et al., 2002). Cloning the Arabidopsis PLEIADE (PLE) gene revealed that PLE is synonymous with AtMAP65-3 (Hussey et al., 2002; Müller et al., 2004). ple mutants were isolated in genetic screens for defects in root and embryo morphogenesis (Müller et al., 2002; Söllner et al., 2002; Sorensen et al., 2002). To date, six ple alleles have been isolated, and all are recessive and develop short irregular expanded roots with multinucleated cells and incomplete cell walls. AtMAP65-3/PLE localizes in the midzone of overlapping microtubules during cell division and the mutations in ple-1, ple-5, and ple-6 cause C-terminal truncations of the AtMAP65-3/PLE protein (Müller et al., 2004). Thus, the ple phenotypes together with the subcellular localization of MAP65-3/PLE support its essential role for the completion of cytokinesis. Vertebrate PRC1 (Mollinari et al., 2002) and yeast Ase1p (Schuyler et al., 2003) are homologs of MAP65 (Hussey et al., 2002), and either downregulation or deletion of these genes causes the disruption of anaphase/telophase microtubule arrays and the accumulation of multinucleated cells, respectively.
In this article, we characterize the activities and mechanisms of action of AtMAP65-1. We have examined its role in microtubule polymerization, stability, and bundling and also identified the region responsible for microtubule binding by biochemical and genetic analyses. In addition, we show that AtMAP65-1 can dimerize and we identify the dimerization region that strongly suggests that dimerization is necessary for 25-nm cross-bridge formation. Also, we note that AtMAP65-1 is ubiquitously expressed but binds microtubules in a cell cycle–dependant manner, suggesting tight posttranscriptional control on the activity of this protein.
RESULTS
AtMAP65-1 Bundles Microtubules but Does Not Promote Microtubule Polymerization
We have identified an Arabidopsis MAP65, AtMAP65-1 (At5g55230), that encodes a protein that shows 86% similarity to tobacco NtMAP65-1 (Smertenko et al., 2000). The predicted open reading frame encodes a 587–amino acid protein of 65.8 kD molecular mass and a pI of 4.72. We have expressed and purified recombinant AtMAP65-1 in bacteria and used this protein to assess its effects on microtubule polymerization and bundling in vitro.
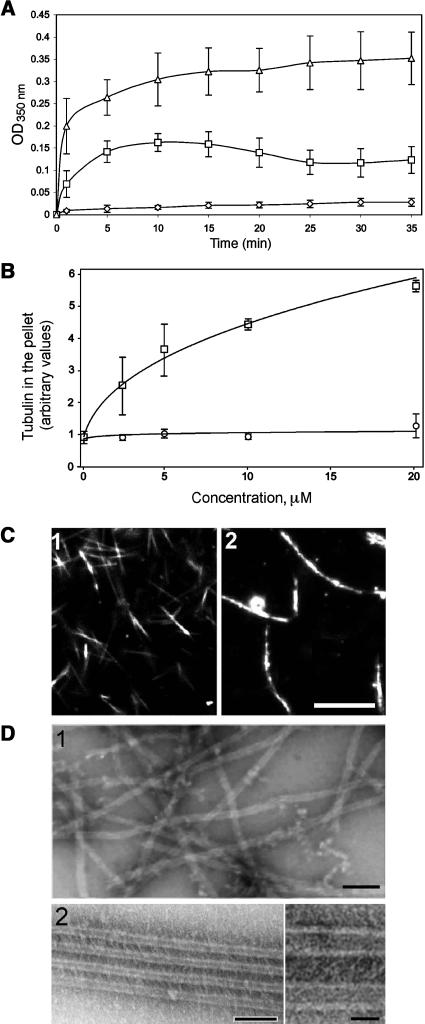
The effect of AtMAP65-1 on microtubule polymerization was assessed using a turbidimetric assay. AtMAP65-1 was added to a MAP-free porcine brain tubulin solution (final concentration 20 μM) at the AtMAP65-1 to tubulin dimer molar ratio of 1:2. The turbidity of the mixture was monitored at 350 nm. AtMAP65-1 induced a dramatic increase in the turbidity of the polymerizing microtubule mixture compared with control (Figure 1A). These data indicate two possibilities: AtMAP65-1 could increase the total amount of microtubule polymer or it could induce bundling of assembled microtubules. Both processes are not necessarily mutually exclusive because bundling can also stabilize microtubules resulting in the increase of the total amount of microtubule polymer by preventing dynamic instability. To distinguish between these two possibilities, we polymerized microtubules in the presence of increasing concentrations of AtMAP65-1 and analyzed the amount of tubulin that cosediments with the AtMAP65-1. If AtMAP65-1 increases the total amount of microtubule polymer, the amount of tubulin in the pellet would increase proportionally to the point of saturation. However, the amount of tubulin in the pellet did not change significantly across the AtMAP65-1 range of 0 to 20 μM (Figure 1B). Taxol, a microtubule stabilizing agent and capable promoter of microtubule polymerization, also increases the turbidity of the tubulin solution (Figure 1A), but in contrast with AtMAP65-1, taxol increased the total amount of tubulin in the pellet in a concentration-dependent fashion (Figure 1B). This result strongly suggests that AtMAP65-1 bundles but does not promote the polymerization of microtubules. This conclusion was further confirmed by an analysis using dark-field microscopy (Figure 1C). Addition of AtMAP65-1 to dynamic tobacco microtubules (shown in panel 1; average length 4.2 ± 1.1 μm, n = 89) caused the formation of long, thick microtubule bundles (panel 2; average length 11.5 ± 4.4 μm, n = 62). Examination of these bundles under the electron microscope showed that they are composed of parallel microtubules separated by 25-nm cross-bridges (Figure 1D). These data suggest that AtMAP65-1 does not promote microtubule polymerization in vitro but bundles polymerized microtubules via the formation of 25-nm cross-bridges.
Figure 1.
AtMAP65-1 Bundles Microtubules.
(A) Turbidity of the 20-μM tubulin solution (diamonds) and 20-μM tubulin solution after the addition of 10 μM AtMAP65-1 (squares) or 10 μM taxol (triangles) monitored at 350 nm and at 32°C.
(B) Amount of tubulin sedimented at 100 000g in the presence of increasing concentration (0 to 20 μM) of AtMAP65-1 (circles) or taxol (squares). Three independent experiments were performed, and the error bars show the standard deviation. The x axis indicates concentration of taxol or AtMAP65-1.
(C) Dark-field microscopy images of microtubules polymerized in a 20-μM tubulin solution in the absence (panel 1) or presence (panel 2) of 10 μM AtMAP65-1. Bar = 10 μm.
(D) Transmission electron microscopy images of samples from the same experiments in (C). Panel 1, microtubules without AtMAP65-1; panel 2, microtubules with AtMAP65-1; the inset shows a higher magnification image of the microtubule bundle formed in the presence of AtMAP65-1. Bar = 100 nm (inset bar = 20 nm).
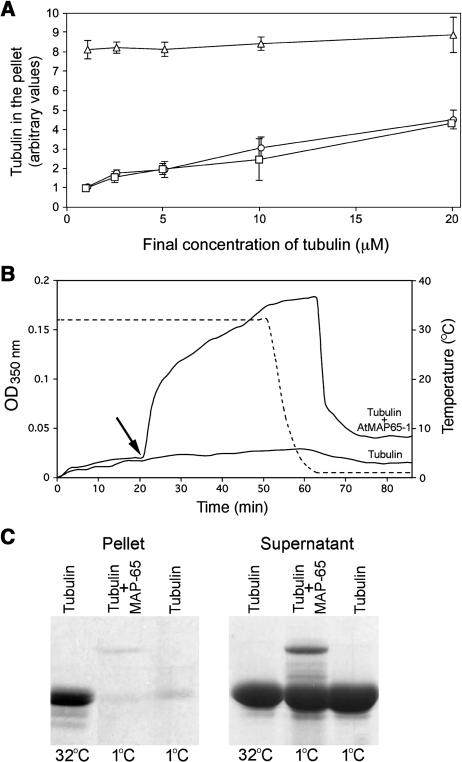
An effect of microtubule bundling can be a reduction in the depolymerization of microtubules, for example, by inhibiting catastrophe. To determine whether AtMAP65-1 affected depolymerization, microtubules were first polymerized at 32°C for 10 min, then AtMAP65-1 protein was added at the tubulin:AtMAP65-1 molar ratio of 2:1. The mixture was diluted with microtubule-polymerizing buffer prewarmed to 32°C, incubated for 10 min, and the microtubules were pelleted at 100,000g and analyzed on SDS-PAGE gels. Taxol at a concentration of 10 μM was used as a positive control to demonstrate the effect of a microtubule stabilizing agent on the amount of tubulin polymer upon isothermal dilution. The results of three independent experiments are presented in Figure 2A. No significant difference in the quantity of tubulin in the supernatant with or without AtMAP65-1 was observed: the total amount of tubulin polymer decreased fivefold with the decrease in final tubulin concentration from 20 to 1 μM. By contrast, the amount of tubulin polymer in the presence of taxol decreased by only 10%.
Figure 2.
AtMAP65-1 Does Not Affect Microtubule Dynamics.
(A) Amount of tubulin sedimented at 100,000g after dilution of the 20-μM tubulin mixture. Squares, tubulin only solution; circles, tubulin with 10 μM AtMAP65-1; triangles, tubulin with 10 μM taxol.
(B) Turbidity of the 20-μM tubulin solution without, and with, the addition of 10 μM AtMAP65-1 (solid lines) at 32°C and after decreasing the temperature. Temperature is indicated by the broken line. The arrow indicates the time at which AtMAP65-1 was added.
(C) Coomassie-stained SDS-PAGE gel of microtubule pellets and supernatants of a 20-μM tubulin solution incubated at 32°C for 10 min, a 20-μM tubulin solution with 10 μM AtMAP65-1 incubated at 32°C for 10 min and then at 1°C for 10 min, and a 20-μM tubulin solution incubated at 32°C for 10 min and then at 1°C for 10 min. The final temperature of the reaction mixtures are indicated below the lanes.
To assess whether AtMAP65-1 changes the stability of microtubules, we analyzed the effect of AtMAP65-1 on the cold-induced depolymerization of microtubules. Here, the turbidity of a tubulin solution (20 μM) was monitored at 350 nm after addition of GTP. Microtubules were polymerized at 32°C for 20 min, then AtMAP65-1 was added at a tubulin:AtMAP65-1 molar ratio of 2:1, and the reaction was allowed to proceed for another 30 min (Figure 2B). The addition of AtMAP65-1 induced an increase in the turbidity of the tubulin solution compared with the control. Subsequently, the temperature of the reaction was decreased to 1°C, temperature readings were collected every 2 min and plotted (shown as the broken line in Figure 2B). The drop in the absorbance indicates cold-induced microtubule depolymerization, and this only occurred when the temperature reached 1°C. However, in the microtubule only and the microtubule:AtMAP65-1 samples, the minimal absorbance reading after microtubule depolymerization was always above the starting point. This would indicate that a small proportion of the microtubules were intrinsically resistant to cold-induced depolymerization. To demonstrate this, the samples were centrifuged at 100000g, and the amount of tubulin in the pellet analyzed. The amount of tubulin in the pellet in both samples was similar (but less than in samples incubated at 32°C), which indicates that the majority of microtubules depolymerized regardless of being bundled by AtMAP65-1 (Figure 2C).
Microtubule Binding Region of AtMAP65-1
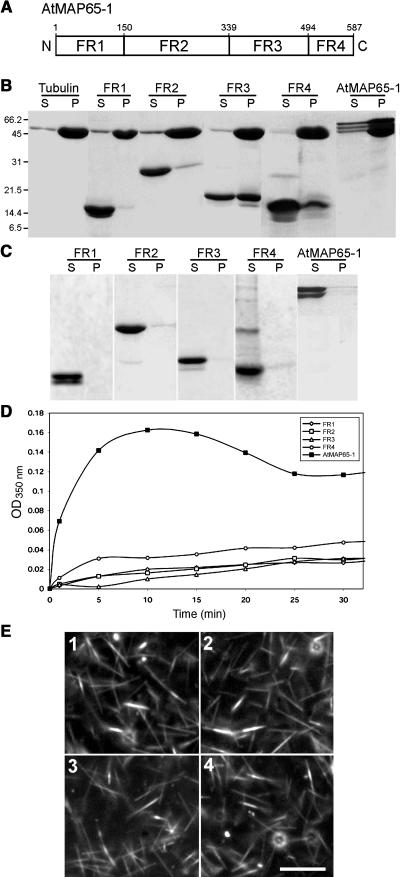
Alignment of all nine AtMAP65 amino acid sequences revealed regions of high sequence conservation. Using this alignment, we divided the AtMAP65-1 protein into four fragments (Figure 3A): fragment 1, amino acids 1 to 150; fragment 2, amino acids 151 to 339; fragment 3, amino acids 340 to 494; fragment 4, amino acids 495 to 587. Fragment 3 includes the most evolutionarily conserved section of MAP65 proteins across different plants, and it is the region of greatest similarity to the functional equivalents from vertebrates, PRC1, and yeast Ase1p (Schuyler et al., 2003). These fragments were subcloned, expressed, and purified from bacteria and then used to assess their microtubule binding capabilities using a cosedimentation assay (Figures 3B and 3C). Fragments 3 and 4, but not fragments 1 and 2, cosedimented with taxol-stabilized microtubules. In controls, none of the fragments pellet significantly after centrifugation in the absence of microtubules. These data indicate that the C-terminal half of AtMAP65-1 harbors the microtubule binding region.
Figure 3.
Identification of the Microtubule Binding Domain of AtMAP65-1.
(A) Diagram showing the positions of the four AtMAP65-1 fragments (FR1, FR2, FR3, and FR4). The numbers indicate the positions of the first and the last amino acids of each fragment in the AtMAP65-1 full-length sequence. N and C indicate the N and C termini, respectively.
(B) Cosedimentation of AtMAP65-1 and fragments 1 to 4 with taxol-stabilized microtubules. Microtubules on their own or as a mixture with AtMAP65-1 or AtMAP65-1 fragments 1 to 4 were centrifuged at 100,000g and then the supernatants (S) and pellets (P) were separated on an SDS-PAGE gel and stained with Coomassie.
(C) Sedimentation of the same AtMAP65-1 and AtMAP65-1 fragments 1 to 4 (as in [B]) in the absence of microtubules. AtMAP65-1 and fragments 1 to 4 were analyzed as in (B).
(D) Effect of AtMAP65-1 and fragments 1 to 4 (each 20 μM) on the turbidity of a 20-μM tubulin solution. The assay was performed at 32°C, and the turbidity was monitored at 350 nm.
(E) Dark-field microsopy images of a 20-μM tubulin solution polymerized in the presence of 10 μM AtMAP65-1 fragments 1 to 4 (panels 1 to 4, respectively). Bar = 1 μm.
We have assessed these four fragments for microtubule bundling activity using a turbidimetric assay. Figure 3D shows that none of the fragments were able to reproducibly increase the turbidity of polymerizing microtubules significantly compared with full-length AtMAP65-1 (Figure 3D) and dark-field microscopy confirms that no bundling occurs (Figure 3E). These results show that the microtubule binding region alone is not sufficient to cause microtubule bundling and form the 25-nm cross-bridges.
Ala420 Is Essential for AtMAP65-1 Interaction with Microtubules
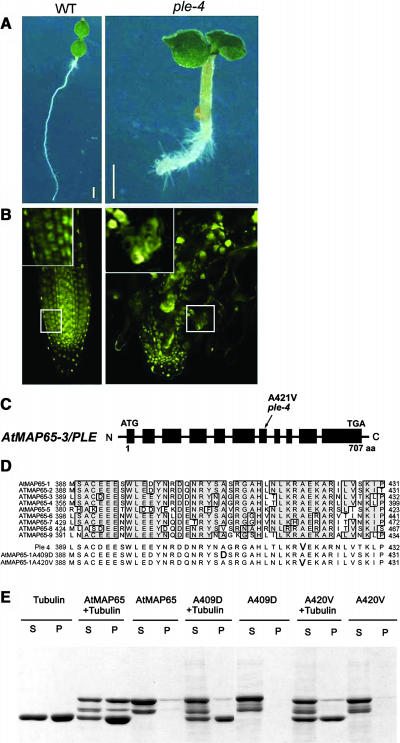
We have recently shown that the Arabidopsis PLE gene is synonymous with AtMAP65-3 (Müller et al., 2004). Recessive ple alleles display irregular expanded roots (Figure 4A) and enlarged multinucleated cells with incomplete cross-walls (Figure 4B) characteristic of a defective cytokinesis (Müller et al., 2002). Whereas ple-1, ple-5, and ple-6 have nonsense mutations, sequencing of the ple-4 alleles revealed a single point mutation that causes the substitution of the Ala421 to a Val (Figure 4C). This amino acid is conserved in all nine AtMAP65 proteins and corresponds to Ala420 in fragment 3 of AtMAP65-1 (Figure 4D).
Figure 4.
Ala409 and Ala420 Are Essential for AtMAP65-1 Binding to Microtubules.
(A) Arabidopsis wild-type and ple-4 mutant allele.
(B) Nuclei in the roots of wild-type and ple-4 7-d-old seedlings visualized with the DNA-specific dye YO-PRO.
(C) The intron (thin line) and exon (thick line) map of AtMAP65-3/PLE, with the position of the A421V mutation in the ple4 allele indicated. aa, amino acids.
(D) Alignment of the most conserved region of all AtMAP65 proteins. The corresponding sequences encoded by the ple4 allele and AtMAP65-1/A409D and AtMAP65-1/A420V mutant proteins are also shown.
(E) Microtubule cosedimentation assay using AtMAP65-1 and AtMAP65-1/A409D and A420V mutants. Supernatants (S) and pellets (P) were separated on an SDS-PAGE gel and stained with Coomassie.
We have mimicked the ple4 mutation in the AtMAP65-1 protein by substituting the corresponding conserved Ala420 for the hydrophobic Val. The mutant protein was unable to bind microtubules (Figure 4E) and did not induce bundling of microtubules in vitro (data not shown). This hydrophobic substitution is likely to have caused some conformational change in the protein. To assess whether Ala420 is solely responsible for microtubule binding, we have chosen another conserved Ala at position 409 for mutation analysis. This Ala is naturally substituted by a Val in AtMAP65-8; thus, we substituted it for a charged amino acid, an Asp. Again the mutated protein did not bind and bundle microtubules. Thus, it is possible that these substitutions induce conformational changes in the protein. To assess whether the region harboring the Ala409 and Ala420 was sufficient to affect microtubule binding, we generated a peptide to a highly conserved region of 25 amino acids in length corresponding to residues 403 to 427 in AtMAP65-1 (DQNRYSASRGAHLNLKRAEKARILV). This peptide was unable to inhibit microtubule binding by AtMAP65-1 even at a molar ratio of 100:1, peptide to AtMAP65-1. These data suggest that the interaction of AtMAP65-1 with microtubules is complex and depends on conserved tertiary structural features.
AtMAP65-1 Forms Dimers
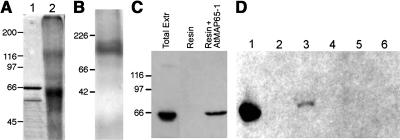
Previously it was suggested that the 25- to 30-nm cross-bridges between microtubules created using a carrot MAP65 enriched protein preparation were unlikely to be generated by monomeric MAP65 molecules (Chan et al., 1999). Therefore, we assessed whether the recombinant AtMAP65-1 could form oligomers. Using two different methods, we show that AtMAP65-1 can form dimers. Firstly, chemical cross-linking of AtMAP65-1 with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) produces a band of ∼130 kD molecular mass on one-dimensional SDS-PAGE (Figure 5A). Secondly, on native acrylamide gel electrophoresis, recombinant AtMAP65-1 runs at a position corresponding to 120 to 140 kD (Figure 5B). In both experiments, the size of the complex indicates an AtMAP65-1 dimer. Moreover, we have immobilized recombinant AtMAP65-1 on a nickel-nitrilotriacetic acid agarose (Ni-NTA) resin column that was then loaded with a total cell extract of a tobacco BY-2 cell line expressing the human influenza hemagglutinin (HA) epitope tagged tobacco equivalent to AtMAP65-1, NtMAP65-1. Immunoblotting of the eluates from control (resin only) and AtMAP65-1 affinity columns with anti-HA antibodies demonstrated that the HA-epitope–tagged NtMAP65-1 interacted with AtMAP65-1 on the column (Figure 5C).
Figure 5.
AtMAP65-1 Can Form Dimers.
(A) AtMAP65-1 control (lane 1) and EDC cross-linked AtMAP65-1 (lane 2) separated on an SDS-PAGE gel and stained with Coomassie.
(B) AtMAP65-1 run on a nondenaturing acrylamide gel and stained as in (A).
(C) Immunoblot probed with anti-HA epitope. Lane 1, a BY-2 cell total protein extract expressing NtMAP65-1:HA epitope tag; lane 2, 0.5 M NaCl eluate from control Ni-NTA column; lane 3, 0.5 M eluate from AtMAP65-1 affinity column.
(D) Immunoblot probed with anti-HA epitope. Lane 1, a BY-2 cell total protein extract expressing NtMAP65-1:HA epitope tag; lanes 2 to 5, 0.5 M NaCl eluates from AtMAP65-1 fragments 1 to 4 affinity columns; lane 6, 0.5 M NaCl eluate from control Ni-NTA column.
The numbers on the left of each gel are molecular masses of markers in kD.
AtMAP65-1 Dimerization Region
We have used this column AtMAP65-1 binding method to determine which fragment in the AtMAP65-1 was capable of interacting to form the dimer. We prepared affinity columns with fragments 1, 2, 3, and 4 (see above). The columns were again loaded with the BY-2 cell extracts expressing the HA-tagged NtMAP65-1, eluted with 0.5 M NaCl, and the washes immunoblotted with anti-HA. The results show that fragment 2 (i.e., amino acids 151 to 339) had the highest affinity for NtMAP65-1 (Figure 5D). These data show that fragment 2 in the N-terminal half of the protein, which does not bind microtubules, is involved in dimerization of MAP65.
AtMAP65-1 Is Expressed throughout the Cell Cycle
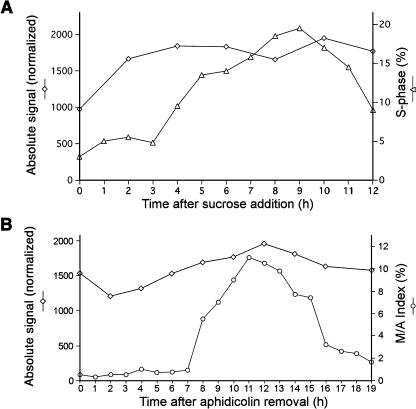
The expression pattern of AtMAP65-1 through the cell cycle was analyzed using publicly available Affymetrix microarray data of two synchronization experiments using Arabidopsis tissue culture cells (Menges et al., 2003). In the first experiment, the cell cycle was arrested in G1 by sucrose deprivation and restarted by addition of sucrose. In the second, the cells were blocked in S-phase using the DNA synthesis inhibitor aphidicolin, and washing out this inhibitor resumed the cell cycle. The level of AtMAP65-1 transcript changed 1.43- and 1.62-fold in each experiment, respectively, which is far below known cell cycle regulated transcripts (e.g., 14.9- and 97-fold for Cyclin A1 and Cyclin B1 transcripts, respectively). The expression profile for the other members of the AtMAP65 gene family was different from AtMAP65-1, which suggests that there was no cross-reactivity between the different isotypes on the same array (data not shown). The results of both experiments demonstrate that the amount of AtMAP65-1 transcript does not change significantly throughout the cell cycle (Figures 6A and 6B).
Figure 6.
Level of AtMAP65-1 Transcript through the Cell Cycle.
The level of AtMAP65-1 transcript was quantified with Affymetrix microarrays as described by Menges et al. (2003).
(A) The Arabidopsis suspension culture cells were synchronized by sucrose starvation, and mRNA samples were collected and measured after sucrose addition to the medium. Diamonds, level of AtMAP65-1 transcript; triangles, percentage of cells in S-phase.
(B) The cells were synchronized with the DNA synthesis inhibitor aphidicolin, and samples for mRNA quantification were collected after the herbicide was washed off. Diamonds, level of AtMAP65-1 transcript; open circles, percentage of cells in metaphase/anaphase (M/A).
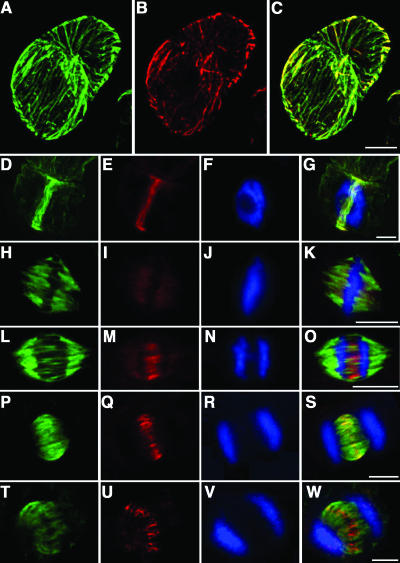
We prepared a polyclonal antiserum against AtMAP65-1 and examined the localization of this protein through the cell cycle. During interphase, AtMAP65-1 binds cortical microtubules (Figures 7A to 7C). In M-phase, AtMAP65-1 locates to the preprophase band (Figures 7D to 7G); in the anaphase spindle, AtMAP65-1 localized to the microtubule overlap zone of the two half spindles (Figures 7L to 7O). In the phragmoplast, AtMAP65-1 was concentrated at the midzone where vesicles guided by microtubules coalesce to form the cell plate (Figures 7P to 7W). No strong staining of AtMAP65-1 was observed on the microtubules of the metaphase spindle, though staining within the spindle zone is apparent, indicating that the protein is present (Figures 7H to 7K). These data indicate that microtubule binding by AtMAP65-1 is regulated through the cell cycle.
Figure 7.
AtMAP65-1 Localization through the Cell Cycle.
Arabidopsis suspension culture cells in interphase ([A] to [C]), preprophase ([D] to [G]), metaphase ([H] to [K]), anaphase ([L] to [O]), telophase ([P] to [S]), and cytokinesis ([T] to [W]) were stained fortubulin (green channel), AtMAP65-1 (red channel), and DNA (blue channel). (C), (G), (K), (O), (S), and (W) are merged images where the yellow coloration corresponds to tubulin and AtMAP65-1 colocalization. Bars = 10 μm.
AtMAP65-1 Expression Program
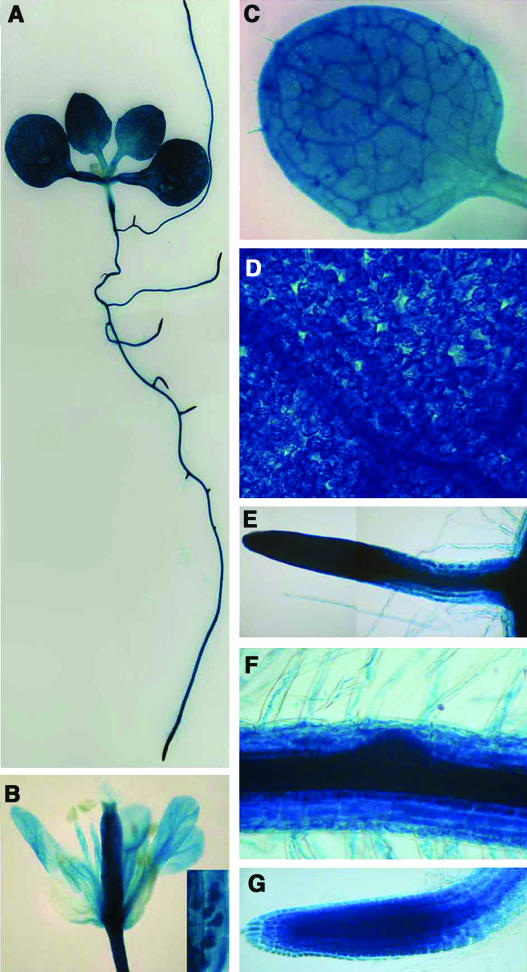
The expression program of AtMAP65-1 in various organs and tissues was determined by transforming Arabidopsis with an AtMAP65-1 promoter:β-glucuronidase (GUS) reporter gene transcriptional fusion construct. Seven transformed lines were analyzed, and segregation analysis revealed that each line contained one insertion of the AtMAP65-1:GUS construct. GUS activity was detected in all organs and tissues with the exception of sepals and anthers (Figures 8A to 8G), and this pattern of expression was similar in all seven lines.
Figure 8.
AtMAP65-1 Gene and Protein Expression in Arabidopsis Tissues and Organs.
GUS activity in the tissues and organs of transgenic Arabidopsis harboring an AtMAP65-1 promoter:GUS gene fusion construct.
(A) Whole seedling.
(B) Flower (inset shows higher magnification of the ovules).
(C) Leaf.
(D) Leaf mesophyl cells.
(E) Lateral root.
(F) Initiation point of the lateral root.
(G) Primary root tip.
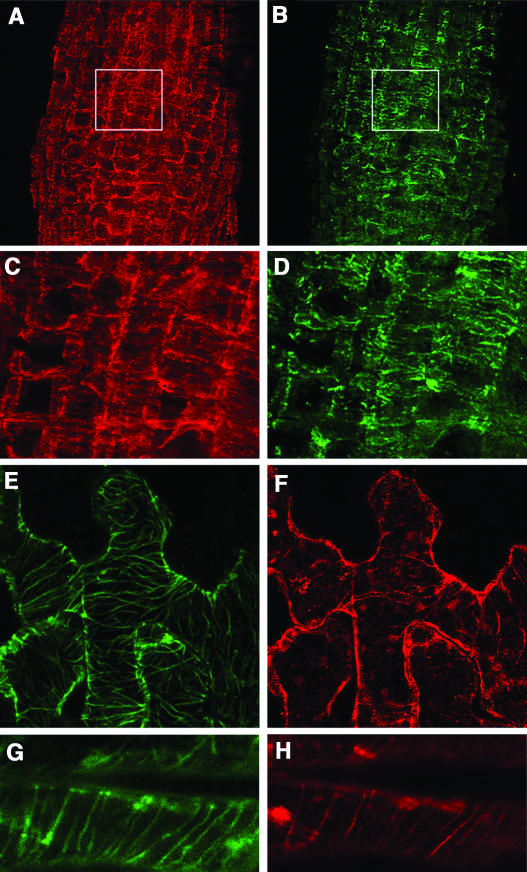
Knowing that AtMAP65-1 is expressed in all plant tissues, we analyzed the subcellular localization of AtMAP65-1 by immunofluorescence. AtMAP65-1 protein was bound to subsets of microtubules in the cells of root epidermis (Figures 9A and 9B and higher magnification in 9C and 9D), cotyledon (Figures 9E and 9F), and hypocotyl (Figures 9G and 9H).
Figure 9.
Immunolocalization of AtMAP65-1 in Plant Tissues.
Root epidermal cells stained with anti tubulin (A) and anti AtMAP65-1 (B) antibodies. A higher magnification of the boxed areas in (A) and (B) is shown in (C) and (D). Immunostaining of microtubules ([I] and [K]) and AtMAP65-1 ([J] and [L]) in cotyledons ([I] and [J]) and hypocotyl ([K] and [L]) epidermal cells.
DISCUSSION
Here, we have shown that a recombinant Arabidopsis MAP65, AtMAP65-1, can bind and bundle microtubules. However, it does not promote microtubule polymerization or stabilize microtubules against cold-induced depolymerization. AtMAP65-1 bundles the microtubules into a regular lattice structure and forms cross-bridges of 25 nm. Fragmentation of AtMAP65-1 and analysis of mutants revealed that the microtubule binding region was in the C-terminal half and that Ala residues at positions 409 and 420 play key roles. Moreover, an AtMAP65-1 dimerization region was found in the N-terminal half. AtMAP65-1 is expressed throughout the cell cycle and in the majority of plant organs. AtMAP65-1 binds microtubules in a cell cycle–specific manner.
AtMAP65-1 Cross-Bridges Microtubules
Recombinant AtMAP65-1 bundles microtubules forming cross-bridges of 25 nm in length but does not increase the total amount of tubulin polymer, suggesting that bundling of microtubules does not directly affect the frequency of catastrophes or rescues. One can imagine a condition when either the frequency of catastrophe and rescue might increase or decrease similarly or that tubulin turnover might change for the microtubules bundled by AtMAP65-1. In either case, microtubule dynamics might be different but the total amount of tubulin polymer under steady state conditions of microtubule polymerization will remain the same. Nonetheless, this equilibrium will change when the steady state conditions are affected. An increase in the dynamics of microtubules resulting from either high rescue and catastrophe frequencies or a rapid tubulin turnover will cause a faster rate of microtubule depolymerization and a decrease in the total amount of tubulin polymer when the concentration of tubulin is below the critical assembly point. If the dynamics of microtubules is decreased either because of low frequencies of rescue and catastrophe or slow tubulin turnover, the total amount of tubulin polymer will be less vulnerable to the decrease of tubulin concentration. During the isothermal dilution of microtubules (Figure 2A), the total amount of tubulin polymer was not affected by bundling with AtMAP65-1 producing identical curves for samples containing tubulin with or without AtMAP65-1. By contrast, when taxol, which promotes microtubule polymerization and decreases tubulin turnover, was added we observed a significant increase in the amount of tubulin polymer and a decrease in the microtubule depolymerization when the tubulin concentration was below the critical assembly point. These data suggest that AtMAP65-1 is unlikely to have a significant effect on microtubule dynamics in vitro.
The fact that bundling does not decrease microtubule dynamics is consistent with the rapid recovery by treadmilling of bundled green fluorescent protein (GFP)-tubulin–tagged microtubules after photobleaching (Shaw et al., 2003) and the movement of AtEB1a:GFP-tagged microtubules within bundles (Chan et al., 2003a). It would therefore seem that the principal role of AtMAP65-1 would be to form a lattice network than to stabilize the microtubules per se. Of the nine AtMAP65 proteins, AtMAP65-1 has the greatest sequence identity to NtMAP65-1 and falls in the same phylogenetic clade (Hussey et al., 2002). The biochemical activity of AtMAP65-1 corresponds to the in vivo experimental data on NtMAP65-1 where the effect of cold on BY-2 cell microtubules was examined. In these experiments, microtubules in BY-2 cells were depolymerized by cold, and their reformation was followed as the temperature was increased to 25°C (Smertenko et al., 2000). The recovery of the microtubules occurred independently of MAP65 binding, and MAP65 binding was only observed after the microtubules were polymerized, suggesting that NtMAP65-1 was not involved in the promotion of microtubule polymerization but in the crossbridging of microtubules once formed. These in vivo data from tobacco correspond with the in vitro data described here for AtMAP65-1.
The crossbridging of microtubules by AtMAP65-1 is similar to that observed using carrot MAP65 enriched preparations (Chan et al., 1999). The carrot MAP65 preparation contained three electrophoretically separable bands, minor 60- and 68-kD bands, and a predominant (85%) 62-kD band (Chan et al., 1999). These protein bands were subsequently analyzed by mass spectral analysis: the 68 and 60 kD were shown to disappear when the carrot suspension culture stopped dividing, leaving the 62-kD species as the sole detectable MAP65 in elongating cells containing only cortical microtubules (Chan et al., 2003b). Peptide sequencing and sequencing the cDNA (DcMAP65-1) established that carrot MAP62 was most similar to AtMAP65-1 and NtMAP65-1 than to any other known MAP65 (Chan et al., 2003b). Because MAP-62 was biochemically purified, it is not known whether it is posttranslationally modified. From the data presented in this study we can conclude that mixtures of MAP65 isoforms are not required to promote bundling and that the single unmodified gene product of AtMAP65-1 is capable of forming the 25-nm cross-bridges.
Identification of Microtubule Binding and Dimerization Regions of AtMAP65-1
The AtMAP65-1 microtubule binding region is in the C-terminal half of the protein and two amino acids, Ala420 and Ala409, are essential structural elements in the microtubule:AtMAP65-1 interaction. The C-terminal half of the protein was divided into fragments 3 and 4. Fragment 3 harbors sequence that was most conserved in all nine AtMAP65 genes. Fragment 4 contains the most divergent sequences across the MAP65 family and is highly charged (pI of 10.47). Both fragments 3 and 4 bound microtubules, although it cannot be discounted that the high charge of fragment 4 might be responsible for nonspecific binding. Fragment 3 is not only conserved between plant MAP65 proteins, but sequence within this fragment also exhibits strong similarity with mammalian PRC1 and yeast Ase1p especially within a 25–amino acid region (37.5%; Schuyler et al., 2003). Mutation of a conserved amino acid within this short sequence, Ala420 Val, diminished MAP65 microtubule binding. This mutation was mimicked in AtMAP65-1 based on the sequence of the cytokinesis defective ple-4 allele. Here, we show that changing the conserved Ala to a more hydrophobic amino acid disturbs the structure to such an extent that microtubule binding is greatly reduced. Mutating a second Ala, at residue 409, in the conserved 25–amino acid sequence also diminished microtubule binding. By synthesizing a peptide covering the whole 25–amino acid conserved motif, we were able to perform competition studies where we allowed AtMAP65-1 and the synthetic peptide to compete for the microtubule binding site. These studies demonstrated that we could not inhibit the AtMAP65-1 microtubule interaction in this way, which strongly suggests that sequences other than this 25–amino acid conserved motif, within fragment 3, are structurally important for microtubule binding. It is possible that the microtubule binding site depends on several points of contact and requires a specific tertiary structure as is known for MAP2/tau.
The amino terminal half of AtMAP65-1 harbors a dimerization region, and dimerization may be important for 25-nm cross-bridge formation. The N-terminal half of AtMAP65-1 was divided into two sections, and the fragment encompassing residues 151 to 339 was found to bind another MAP65. Consideration of the structure of the cross-bridges between microtubules created by carrot MAP65s and the size of MAP65 led to the suggestion that MAP65 is unlikely to cross-bridge as a single molecule (Chan et al., 1999). The fact that neither the microtubule binding region nor the dimerization region alone were capable of microtubule bundling strongly suggests that these cross-bridges are formed by MAP65 dimers, with the C-terminal halves binding adjacent microtubules and the N-terminal halves being responsible for MAP65:MAP65 interaction.
AtMAP65-1 Binding to Microtubules Is Cell Cycle Specific
AtMAP65-1 is expressed constitutively through the cell cycle, but the protein binds only subsets of microtubules in a cell cycle–dependent manner. The main difference in the localization of AtMAP65-1 in cells compared with AtMAP65-3/PLE (Müller et al., 2004) and its homologs in animals and fungi is that AtMAP65-1 (like NtMAP65-1; Smertenko et al., 2000) also binds interphase cortical microtubules (Pellman et al., 1995; Jiang et al., 1998). However, mammalian PRC1 is capable of binding and bundling microtubules in interphase as overexpression causes the disruption of normal microtubule organization and the appearance of circular filaments around the nuclei (Mollinari et al., 2002). Bundling of microtubules in interphase in plant cells has been suggested to be important for the formation of the interphase cortical array (Shaw et al., 2003). As the cells enter M-phase, AtMAP65-1 does decorate the preprophase band but not the metaphase spindle. It appears to bind microtubules once more at the midzone of the anaphase spindle where the phragmoplast forms. In the phragmoplast it is present where the microtubules overlap at the midline. Because the AtMAP65-1 transcript is present throughout the cell cycle, the microtubule binding activity of AtMAP65-1 must be under posttranscriptional control.
Motif searches have revealed a cyclin destruction box and several phosphorylation sites, including a cyclin-dependent kinase (CDK) phosphorylation site (Hussey et al., 2002). AtMAP65-1 can be phosphorylated by a protein extract prepared from 3-d-old Arabidopsis tissue culture cells, and the phosphorylation can be inhibited by roscovitine, suggesting that CDK might be responsible for this activity. However, phosphorylated AtMAP65-1 did cosediment with microtubules and did increase the turbidity of a tubulin solution (data not shown). Interestingly, a phosphorylation mimic of PRC1 (the mammalian AtMAP65 homolog) where the CDK sites were mutated to Glu was still capable of binding and bundling microtubules in the same manner as the nonphosphorylatable protein (Mollinari et al., 2002). Thus, phosphorylation on its own is not the sole factor that controls the microtubule:AtMAP65-1 interaction, and unraveling the signaling pathways that regulate this process will be the next crucial step in understanding the control of this MAP.
METHODS
Cloning of AtMAP65-1 and Expression of Recombinant Proteins
The cDNA for AtMAP65-1 was identified by comparison of NtMAP65-1a sequence with The Arabidopsis Information Resource database. The cDNA clone 109M12T7 was obtained from the ABRC (Columbus, OH). Sequencing the clone revealed that the first four amino acids were missing, and this was confirmed by rapid amplification of cDNA ends PCR. A full-length cDNA was constructed using a primer that incorporated the first four amino acids and using a codon usage optimized for bacterial expression (i.e., 5′-ATGGCGGTGACA-3′). The sequence of the full-length cDNA was verified by sequencing. Single point mutations in the AtMAP65-1 sequence were introduced using the QuickChange site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands) according to the manufacturer's recommendations.
Full-length AtMAP65-1, A420V, and A409D mutants and fragments 1 to 4 were cloned into NdeI/XhoI-digested pET28a vectors (Novagen, Madison, WI). The restriction sites were added to the inserts by PCR. The sequence of each clone was verified by sequencing. The cDNAs were expressed in Escherichia coli [BL21(DE3), BL21(DE3) Rosetta, or BL21(DE3)pLysS] and each recombinant protein incorporated a 6xHis tag on the N terminus. Bacterial cultures were grown in LB-broth medium supplemented with 100 μg/mL of kanamycin and 35 μg/mL of chloramphenicol at 30°C until the culture reached an optical density of 0.4 to 0.6 measured at 600 nm. Then protein expression was induced by the addition of 1 mM isopropylthio-β-galactoside. At the same time, 50 μg/mL of kanamycin and 15 μg/mL of chloramphenicol were added. The induction was allowed to progress for 2 to 6 h. The bacteria were pelleted at 4000g for 5 min, resuspended in protein extraction buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, and 5 mM β-mercaptoethanol) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/mL of leupeptin, and 10 μg/mL of pepstatin A), and sonicated (Soniprep 150; Sanyo, Herts, UK; 10 pulses at amplitude of 26 μm, the length of each pulse was 1 s/mL of extraction buffer). The bacterial lysates were centrifuged at 30,000g for 30 min, filtered through 20-μm nitrocellulose membranes, and applied to columns containing 1 to 2 mL of Ni-NTA agarose resin (Qiagen, Crawley, UK). The columns were washed three times with protein extraction buffer containing (1) 20 mM imidazole, (2) 40 mM imidazole, and (3) 60 mM imidazole. The specifically bound proteins were eluted with 200 mM imidazole, and proteins were dialyzed overnight against MTSB buffer (0.1 M Pipes, pH 6.8, 2 mM EGTA, 2 mM MgSO4, 2 mM DTT, 50 mM NaCl, and 10% glycerol).
Tissue Culture Conditions
The Arabidopsis thaliana suspension culture was maintained at 27°C in MS medium containing 30 g/L of sucrose and supplemented with 0.5 mg/L of naphthalene acetic acid and 0.05 mg/L kinetin. The tobacco (Nicotiana tabacum Bright Yellow-2) BY-2 cell line expressing NtMAP65-1 with a C-terminal HA-epitope tag was grown in MS medium containing 30 g/L of sucrose and supplemented with 0.2 mg/L 2,4-D and 100 mg/L of kanamycin.
Microtubule Cosedimentation Assay
Tubulin was isolated from porcine brain by two assembly/disassembly cycles followed by phosphocellulose chromatography as described by Shelanski et al. (1973). Tubulin, AtMAP65-1 wild-type and mutant proteins, fragments 1 to 4, and synthetic peptide were centrifuged at 150,000g for 15 min at 2°C to remove any aggregates. For the assays with nonstabilized microtubules, tubulin at a concentration of 20 μM was mixed with the specified concentration (see Results) of recombinant AtMAP65-1, incubated at 32°C or 1°C for 15 min, and centrifuged at 100,000g for 15 min (Beckman TLX ultracentrifuge, rotor TLA120.1; Fullerton, CA). The supernatant was collected and the pellet was washed and resuspended in SDS-PAGE sample buffer. For the experiments with stable microtubules, microtubules were polymerized with 10 μM taxol, mixed with recombinant proteins, incubated for 10 min at 32°C, and centrifuged at 100,000g for 15 min at 32°C. In all the negative controls (i.e., when the microtubules were not added to the proteins), the reaction mixture was supplemented with MTSB buffer containing 10 μM taxol.
Microtubule Polymerization Assay
Pig brain tubulin dimer was used at a final concentration of 20 μM in all assays. Tubulin solution was stored at −80°C. Before each assay, tubulin was rapidly thawed at 37°C, incubated on ice for 5 min, and centrifuged at 200,000g for 10 min to pellet polymerized microtubules and tubulin aggregates. The supernatant was diluted to a 30 μM tubulin stock with MTSB. The blank was set up with the cuvette containing 100 μL of 30 μM tubulin in MTSB. Then GTP was added up to a final concentration of 1 mM followed by recombinant AtMAP65-1, mutants, or fragments 1 to 4. The final reaction volume was adjusted to 150 μL with MTSB, and the turbidity of the reaction was monitored at 350 nm and at 32°C using a Helios β spectrophotometer equipped with Unicam Peltier temperature control unit (Thermospectronic, Basingstoke, UK). Each experiment was repeated three times and the average (in the case of Figure 1A) or an example from three series of experiments (in the case of Figure 3D) is presented.
Isothermal Dilution of Microtubules
To polymerize microtubules, 20 μM tubulin in MTSB was incubated at 32°C for 10 min. Thereafter, a 50-μL aliquot was kept separately, while the rest of the solution was diluted with MTSB buffer prewarmed at 32°C and incubated at this temperature for 10 min. Alternatively, 10 μM AtMAP65-1 or taxol was added to the microtubules before incubation at 32°C. The samples were centrifuged at 100,000g for 10 min, and the amount of tubulin in the pellet was analyzed by SDS-PAGE electrophoresis.
Microtubule Bundling Assay
The dark-field microscopy analysis was performed with 20 μM tubulin in MTSB. AtMAP65-1 was added to the tubulin solution at a final concentration of 10 μM and MTSB up to a final volume of 20 μL. The reaction mixture was incubated for 5 min at room temperature, and 5 μL of the mixture was examined with an Olympus BX50 microscope (Tokyo, Japan) equipped with UplanAop 100×/1.35 objective, 100-W mercury bulb, dark-field condenser, and Hamamatsu C4742 black and white CCD camera (Herts, UK).
For the visualization of cross-bridges, microtubules were polymerized with recombinant protein and applied to formvar-coated and ionized copper grids. The grids were negatively stained with 1% aqueous solution of uranyl acetate and examined using a JEOL transmission electron microscope (Welwyn Garden City, UK).
Chemical Cross-Linking
Cross-linking was performed using EDC (Doi et al., 1987). EDC was added up to the final concentration of 5 mM in a 20-μM solution of AtMAP65-1 in MTSB and incubated at room temperature for 1 h. The reaction was stopped by addition of SDS-PAGE sample buffer, and then the mixture was boiled for 3 min and the sample separated on a 7.5% acrylamide gel.
AtMAP65-1 Promoter GUS Fusions
A fragment of 1612 bp upstream of the translation start codon was amplified by PCR using the forward primer 5′-CAGCAATTCTCCGGAGAACT-3′ and the reverse primer 5′-GCGGAATCAGAAGGTTTCCT-3′. The PCR fragment was cloned into the pGEMT-E vector (Promega, Southampton, UK) and its sequence verified and then subcloned into the PΔgusBin19 vector (Topping et al., 1991). The final construct was transformed into Agrobacterium tumefaciens strain C58C3 cells by electroporation. Arabidopsis plants were transformed using the dipping method (Clough and Bent, 1998). The positives were selected on half-strength MS medium containing 0.6% agar, 50 μg/mL of kanamycin, and 200 μg/mL of augmentin (SmithKline Beecham Pharmaceuticals, Slough, UK). Positives were transferred to soil and seeds collected. The T2 generation was used for the histochemical localization of GUS activity.
Plant samples were vacuum infiltrated for 30 min with substrate solution (100 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl glucuronide) and incubated at 37°C for 12 h. Plant tissues were cleared using the chloral-lacto-phenol method (Barthels et al., 1997) and examined using Leica MZ125 (Wetzlar, Germany) and Zeiss Axioscope (Jena, Germany) microscopes equipped with Photometrix CoolSnap CF color CCD camera (Nippon Ropper, Chiba-shi, Japan).
Antibodies and Immunostaining
The AtMAP65-1 antiserum was raised in rabbits using the full-length recombinant AtMAP65-1 protein as immunogen. Recombinant protein (250 μg) was used for each boost, and four boosts over a period of 3 months were performed. Antiserum was collected 10 d after the final boost and tested by immunoblotting. The AtMAP65-1 antiserum identified the AtMAP65-1 recombinant protein, and a single band on a 1-D gel total protein extract of Arabidopsis tissue culture cells (data not shown).
For immunostaining, Arabidopsis suspension culture cells were separated from the tissue culture medium using 100 mesh nylon cloth and fixed for 30 min at room temperature with 4% paraformaldehyde in 0.1 M Pipes, pH 6.8, 5 mM EGTA, 2 mM MgCl2, and 0.4% Triton X-100. The fixative was washed away with PBS buffer, and cells were treated for 5 min at room temperature with the solution of 0.8% macerozyme R-10 and 0.2% pectolyase Y-23 in 0.4 M mannitol, 5 mM EGTA, 15 mM Mes, pH 5.0, 1 mM PMSF, 10 μg/mL of leupeptin, and 10 μg/mL of pepstatin A. Then the cells were washed in PBS buffer and attached to poly-l-Lys–coated cover slips. The cover slips were then incubated for 30 min in 1% (w/v) BSA in PBS and incubated for 1 h with primary antibody. The primary antibodies used were rabbit anti-AtMAP65-1 diluted 1:500 and mouse antitubulin DM1A diluted 1:200 (Sigma, Dorset, UK). The specimens were then washed three times for 10 min in PBS and incubated for 1 h with secondary antibodies: goat anti-mouse tetramethylrhodamine isothiocyanate conjugates and anti-rabbit fluorescein isothiocyanate conjugates; both antibodies were diluted 1:200. After washing in the PBS buffer, the specimens were mounted in Vectashied (Vector Laboratories, Burlingame, CA) mounting medium and examined with a Zeiss 510 confocal microscope.
For Arabidopsis root immunostaining, the roots were fixed for 1 h in the same fixative solution as for tissue culture cells, then attached to poly-l-Lys–coated slides and treated for 10 min with 2% (w/v) Dricelase (Sigma) in 0.4 M mannitol, 5 mM EGTA, 15 mM Mes, pH 5.0, 1 mM PMSF, 10 μg/mL of leupeptin, 10 μg/mL of pepstatin A, and 10 μg/mL of Nα-p-tosyl-l-lysine chloromethyl ketone. Then, roots were incubated with antibodies, mounted, and examined as described above with the modification that the primary antibodies were applied overnight and the secondary antibodies for 3 h.
For immunostaining the hypocotyl and the cotyledons, Arabidopsis seedlings were fixed for 1 h in the same mixture as for tissue culture cells but supplemented with 0.2% (v/v) glutaraldehyde. Then cotyledons and hypocotyl were rapidly immersed into liquid nitrogen, shattered to rupture the cell walls, and incubated with antibodies as described above for roots.
Protein Electrophoresis and Immunoblotting
For the quantification of recombinant proteins in Figures 1B and 2A, the protein samples were separated on a 7.5% one-dimensional SDS-PAGE. The gels were stained with Coomassie Brilliant Blue and scanned using a flat bed scanner. The amount of protein on the gel was quantified using NIH image software version 1.62 (National Institutes of Health, Bethesda, MD). Each experiment was repeated three times. For the quantification of tubulin polymer in the pellet, local values for the background were estimated and subtracted from the protein band values. Then the data from the three replicates were normalized by mean: the intensity for each band in a particular replicate was divided by the mean intensity for all bands in the replicate. The data were then averaged across the replicates. The final number represents an arbitrary value.
For immunoblotting, proteins were transferred from the gel onto nitrocellulose membranes and probed with mouse monoclonal anti HA-epitope diluted 1:100 (Sigma). The secondary anti-rabbit or anti-mouse horseradish peroxidase conjugates were used at a dilution of 1:2000 and detected using the enhanced chemiluminescence kit (Amersham Biosciences, Buckinghamshire, UK).
The native acrylamide gel with AtMAP65-1 protein in Figure 5B was done according to Zabala and Cowan (1992).
Affinity Columns
Recombinant AtMAP65-1 protein or fragments 1 to 4 were dialyzed against NET buffer (100 mM Tris, pH 7.5, 100 mM NaCl, and 1 mM EDTA) and bound to the column loaded with Ni-NTA resin (Qiagen). Approximately 50 mg of total protein were used in each case. About 300 to 500 mg of miniprotoplast (prepared according to the method of Jiang and Sonobe, 1993) total protein isolated from the BY-2 cell line expressing NtMAP65-1a with a C-terminal HA-epitope tag was run through the column. The column was washed with 20 bed volumes of NET buffer containing 200 mM NaCl, 40 mM imidazole, and 5 mM β-mercaptoethanol. The interacting proteins were eluted with 0.5 M NaCl in NET buffer.
Phenotypic, Molecular Genetic, and Microscopic Analysis
For the analysis of the root morphogenesis phenotype, ple mutants were cultivated on vertical nutrient agar plates containing 1× MS salt mixture and 4.5% sucrose, pH 5.7, in 1% agar. Nuclei were stained with YO-PRO (Molecular Probes, Eugene, OR) on fixed roots and analyzed with a confocal scanning laser microscope (Leica TCS SP2) as described (Müller et al., 2002). Allelism of the ple-4 mutant was confirmed by genetic crosses and sequencing.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number NM_124905.
Acknowledgments
We would like to thank Marylin Vantard (Centre National de la Recherche Scientifique, Grenoble) for useful discussions, Margit Menges and Jim Murray (University of Cambridge) for providing prepublished Affymetrix data, S. Müller, I. Kreuzer, and G. Resch for technical assistance, and Toni Slabas for providing the Arabidopsis suspension culture cells. We are specially obliged to Farhah Assaad and Wolfgang Lukowitz for providing the ple-4 allele. We would like to thank the ABRC for the AtMAP65-1 cDNA clone and the Arabidopsis genome sequencing project. A.P.S., S.F., C.W.L., and P.J.H. are funded by the Biotechnology and Biological Research Council UK. D.K. is supported by the European Union human potential program (TIPNET HPRN-CT-2002-00265). H.-Y.C. is funded by an Overseas Research Studentship. This work was also funded by grants of the FWF Austrian Science Fund to M.-T.H. (project numbers P14477-B04 and P16410-B12). V.W. is funded by the DOC fellowship of the Austrian Academy of Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Patrick J. Hussey (p.j.hussey@durham.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023937.
References
- Barthels, N., et al. (1997). Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell 9, 2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Calder, G.M., Doonan, J.H., and Lloyd, C.W. (2003. a). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5, 967–971. [DOI] [PubMed] [Google Scholar]
- Chan, J., Jensen, C.G., Jensen, L.C.W., Bush, M., and Lloyd, C.W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA 96, 14931–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Mao, G., Smertenko, A., Hussey, P.J., Naldrett, M., Bottrill, A., and Lloyd, C.W. (2003. b). Identification of a MAP65 isoform involved in directional expansion of plant cells. FEBS Lett. 534, 161–163. [DOI] [PubMed] [Google Scholar]
- Chan, J., Rutten, T., and Lloyd, C. (1996). Isolation of microtubule-associated proteins from carrot cytoskeletons: A 120 kDa map decorates all four microtubule arrays and the nucleus. Plant J. 10, 251–259. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Doi, Y., Higashida, M., and Kido, S. (1987). Plasma gelsolin binding sites on the actin sequence. Eur. J. Biochem. 164, 89–94. [DOI] [PubMed] [Google Scholar]
- Gardiner, J., and Marc, J. (2003). Putative microtubule-associated proteins from the Arabidopsis genome. Protoplasma 222, 61–74. [DOI] [PubMed] [Google Scholar]
- Hush, J.M., Wadsworth, P., Callaham, D.A., and Hepler, P.K. (1994). Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J. Cell Sci. 107, 775–784. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., Hawkins, T.J., Igarashi, H., Kaloriti, D., and Smertenko, A. (2002). The plant cytoskeleton: Recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50, 915–924. [DOI] [PubMed] [Google Scholar]
- Jiang, C.J., and Sonobe, S. (1993). Identification and preliminary characterization of a 65kDa higher-plant microtubule-associated protein. J. Cell Sci. 105, 891–901. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Jimenez, G., Wells, N.J., Hope, T.J., Wahl, G.M., Hunter, T., and Fukunaga, F. (1998). PRC1: A human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell 2, 877–885. [DOI] [PubMed] [Google Scholar]
- Lloyd, C.W., Hussey, P.J., and Chan, J. (2004). Microtubules and microtubule-associated proteins. In The Plant Cytoskeleton in Cell Differentiation and Development, P.J. Hussey, ed (Oxford: Blackwell Publishing), pp. 3–31.
- Menges, M., Hennig, L., Gruissem, W., and Murrey, J.A.H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423–442. [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman, J.P., Jiang, W., Schoehn, G., Hunter, T., and Margolis, R.L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R.C., Zhang, M., Cassimeris, L., and Cyr, R.J. (1997). In vitro assembled plant microtubules exhibit a high state of dynamic instability. Cell Motil. Cytoskeleton 38, 278–286. [DOI] [PubMed] [Google Scholar]
- Müller, S., Fuchs, E., Ovecka, M., Wysocka-Diller, J., Benfey, P.N., and Hauser, M.-T. (2002). Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 130, 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., Smertenko, A., Wagner, V., Heinrich, M., Hussey, P.J., and Hauser, M.-T. (2004). The plant microtubule associated protein, AtMAP65-3/PLE, is essential for cytokinetic phragmoplast function. Curr. Biol. 14, 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman, D., Bagget, M., Tu, H., and Fink, G.R. (1995). Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 13, 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., Liu, J.Y., and Pellman, D.J. (2003). The molecular function of Ase1p: Evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.L., Kamyar, R., and Ehrhardt, D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. [DOI] [PubMed] [Google Scholar]
- Shelanski, M.L., Gaskin, F., and Cantor, C.R. (1973). Microtubule assembly in absence of added nucleotides. Proc. Natl. Acad. Sci USA 70, 765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko, A., Saleh, N., Igarashi, H., Mori, H., Hauser-Hahn, I., Jiang, C.J., Sonobe, S., Lloyd, C.W., and Hussey, P.J. (2000). A new class of microtubule-associated proteins in plants. Nat. Cell Biol. 2, 750–753. [DOI] [PubMed] [Google Scholar]
- Smertenko, A.P., Bozhkov, P.V., Filonova, L.H., von Arnold, S., and Hussey, P.J. (2003). Re-organisation of the cytoskeleton during developmental programmed cell death in Picea abies embryos. Plant J. 33, 813–824. [DOI] [PubMed] [Google Scholar]
- Söllner, R., Glasser, G., Wanner, G., Somerville, C.R., Jurgens, G., and Assaad, F.F. (2002). Cytokinesis-defective mutants of Arabidopsis. Plant Physiol. 129, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, M.B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jurgens, G., Somerville, C., Lepiniec, L., and Berger, F. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129, 5567–5576. [DOI] [PubMed] [Google Scholar]
- Topping, J.F., Wei, W.B., and Lindsey, K. (1991). Functional tagging of regulatory elements in the plant genome. Development 112, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Yuan, M., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Dynamic reorientation of cortical microtubules, from transverse to longitudinal in living plant cells. Proc. Natl. Acad. Sci. USA 91, 6050–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala, J.C., and Cowan, N.J. (1992). Tubulin dimer formation via the release of α- and β-tubulin monomers from multimolecular complexes. Cell Motil. Cytoskeleton 23, 222–230. [DOI] [PubMed] [Google Scholar]