Abstract
In higher plants, photorespiratory Gly oxidation in leaf mitochondria yields ammonium in large amounts. Mitochondrial ammonium must somehow be recovered as glutamate in chloroplasts. As the first step in that recovery, we report glutamine synthetase (GS) activity in highly purified Arabidopsis thaliana mitochondria isolated from light-adapted leaf tissue. Leaf mitochondrial GS activity is further induced in response to either physiological CO2 limitation or transient darkness. Historically, whether mitochondria are fully competent for oxidative phosphorylation in actively photorespiring leaves has remained uncertain. Here, we report that light-adapted, intact, leaf mitochondria supplied with Gly as sole energy source are fully competent for oxidative phosphorylation. Purified intact mitochondria efficiently use Gly oxidation (as sole energy, NH3, and CO2 source) to drive conversion of l-Orn to l-citrulline, an ATP-dependent process. An A. thaliana genome-wide search for nuclear gene(s) encoding mitochondrial GS activity yielded a single candidate, GLN2. Stably transgenic A. thaliana ecotype Columbia plants expressing a p35S∷GLN2∷green fluorescent protein (GFP) chimeric reporter were constructed. When observed by laser scanning confocal microscopy, leaf mesophyll and epidermal tissue of transgenic plants showed punctate GFP fluorescence that colocalized with mitochondria. In immunoblot experiments, a 41-kD chimeric GLN2∷GFP protein was present in both leaf mitochondria and chloroplasts of these stably transgenic plants. Therefore, the GLN2 gene product, heretofore labeled plastidic GS-2, functions in both leaf mitochondria and chloroplasts to faciliate ammonium recovery during photorespiration.
INTRODUCTION
Glutamine synthetase ([GS]; glutamate-ammonia ligase, EC 6.3.1.2) activity is primarily responsible for scavenging ammonia, a highly reactive and cytotoxic metabolite, maintained in dynamic equilibrium with conjugate ammonium cation. In active photosynthetic cells, both chloroplast stromal and cytosolic compartments are weakly alkaline. Then, some 0.5 to 1.0% of total ammonia(ium) exists as free ammonia, the reactive, cytotoxic nucleophile. GS catalyzes the glutamate- and ATP-dependent conversion of toxic ammonia to nontoxic Gln, which in turn serves as amidosubstrate for numerous, anabolic amidotransferase activities, regenerating glutamate. Ammonia scavenging is thus coupled to growth. In higher plants, GS activity is predominantly cytosolic, appropriately so for ammonia scavenging purposes. Conversely, degradative metabolic processes yielding ammonia, such as ureolysis, are also typically cytosolic (Faye et al., 1986). Yet, some ammonia-generating processes are organellar, including, in chloroplasts, ferredoxin-dependent nitrite reduction associated with nitrate uptake and, in mitochondria, Gly decarboxylation associated with photorespiratory C2 cycle activity.
Among higher plants, GS is encoded by a nuclear gene family and not by organellar DNA. In Arabidopsis thaliana, the nuclear gene family encoding GS includes six genes. Of these, five paralogs (GLN1-x) yield cytosolic GS-1 activities, whereas the sixth gene (GLN2; A. thaliana locus 5G35630) yields plastidic GS-2 (Peterman and Goodman, 1991). As demonstrated in pea (Pisum sativum), the orthologous GS-2 nuclear gene is both light inducible and highly expressed in photosynthetic tissues. The GS-2 precursor is synthesized on cytosolic polysomes, taken up by chloroplasts, and is active in stromal fractions (Tingey et al., 1988; Tjaden et al., 1995). In corroborative work, light-inducible, plastidic GS-2 activity was confirmed in various plants, including bean (Phaseolus vulgaris) (Lightfoot et al., 1988) and barley (Hordeum vulgare) (Freeman et al., 1990).
In actively photorespiring, CO2-limited terrestrial plants, leaf mitochondria generate ammonia at extraordinarily high rates, as much as 50-fold those of nitrate/nitrite reduction (Ogren, 1984; Artus et al., 1987). Mitochondrial ammonia is predominantly owed to Gly decarboxylase complex activity, strongly induced in response to physiological CO2 limitation in the light. How is this large pool of photorespiratory ammonia detoxified and reassimilated? As we report here, GS activity is present in light-adapted, highly purified A. thaliana leaf mitochondria. Moreover, these leaf mitochondria are fully competent for oxidative phosphorylation with Gly as substrate.
The A. thaliana nuclear genome carries but a single candidate as source of organellar GS activity, GLN2, which encodes plastidic GS-2 (Peterman and Goodman, 1991). To reassess its subcellular compartmentation in leaves, GLN2 N-terminal trafficking and coding sequences were fused in frame to a modified Aequoria victoria gene encoding green fluorescent protein (mGFP5) as reporter; expression of the chimeric gene was placed under transcriptional control of the (high-level, constitutive) 35S promoter of Cauliflower mosaic virus. Stably transgenic A. thaliana Columbia (Col) derivatives carrying this GLN2∷GFP reporter construct were obtained. In both leaf mesophyll and epidermal tissue, punctate green fluorescence colocalizing with mitochondria was observed by laser scanning confocal microscopy. In immunohybridization experiments, both leaf mitochondria and chloroplasts isolated from transgenic GLN2∷GFP plants showed fusion proteins that immunoreacted with polyclonal anti-GFP serum. Therefore, A. thaliana GLN2 encodes GS-2 precursor(s) able to be translocated to both leaf mitochondria and chloroplasts. Either the GLN2 mature mRNA transcript is alternatively translated to yield multiple, distinct GS-2 precursors or else the N-terminal trafficking peptide of the GS-2 precursor allows its dual trafficking to both chloroplasts and mitochondria.
RESULTS
Highly Purified A. thaliana Leaf Mitochondria Show Gln Synthetase Activity
In previous experiments, functioning A. thaliana ecotype Col-0 leaf chloroplasts were purified using GS activity assay as chloroplast marker (Ludwig, 1993). Elution profiles of Percoll gradients used for chloroplast purification reproducibly yielded GS activity peaks in multiple gradient fractions. To follow up these observations, leaf mitochondria from both light- and dark-adapted A. thaliana Col-0 plants were highly purified by repeated velocity sedimentation in Percoll solutions (see Methods). Under both bright-field and fluorescence microscopy, highly purified leaf mitochondria were essentially homogenous and entirely lacked (<1%) intact chloroplasts. In biochemical tests, these purified leaf mitochondria showed high succinate dehydrogenase specific activities (data not shown).
Light-adapted, purified leaf mitochondria and chloroplasts were lysed by a combination of ultrasonication and osmotic dilution (see Methods). Both soluble (mitochondrial matrix and chloroplast stromal) and envelope fractions were recovered after centrifugation. Total GS activity measured was some 20-fold higher in osmotically lysed mitochondria relative to osmotically protected mitochondria, suggesting substantial GS activity within. Upon further testing, GS activity in matrix (soluble) fractions was present at high levels; GS activity in envelope fractions was absent. Conversely, when root mitochondria were similarly purified (see Methods), little or no GS activity in any mitochondrial subfraction was observed. GS-specific activities in light-adapted leaf mitochondria were reproducibly some fourfold higher when compared with leaf chloroplasts (Table 1).
Table 1.
Gln Synthetase Activity in Purified A. thaliana Organelles
| Light/Dark Response
| ||||
|---|---|---|---|---|
| Light Adapted | Dark Adapted
|
|||
| Organelle | 4 h | 8 h | 12 h | |
| Leaf mitochondria | 2.70a | 3.10 | 3.40 | 4.10 |
| Leaf chloroplasts | 0.70 | 0.62 | 0.60 | 0.40 |
| Root mitochondria | 0.10 | <0.10 | <0.10 | <0.10 |
| CO2 Response, Constant Light
| |||||
| Organelle | [CO2] (Pa) | 0 h | 4 h | 8 h | 12 h |
| Leaf mitochondria | 36 ± 2.0 | 4.0 | 4.0 | 4.4 | 4.1 |
| 3 ± 0.3 | 5.7 | 7.1 | 8.1 | ||
| Leaf chloroplasts | 36 ± 2.0 | 0.5 | 0.6 | 0.6 | 0.8 |
| 3 ± 0.3 | 1.8 | 2.1 | 1.7 | ||
μmol min−1 mg protein−1.
To assess whether significant GS activity in leaf or root mitochondrial matrix fractions was masked, GS was partially purified by DEAE-Sepharose chromatography (see Methods). For leaf mitochondria, between 90 and 100% of total GS activity was recovered in column eluates. For light-adapted mitochondrial matrix preparations, GS activity was eluted in two discrete peaks corresponding to different KCl concentrations (data not shown). For dark-adapted mitochondrial matrix fractions, no increase in total GS activity was obtained after DEAE-Sepharose chromatography (data not shown). Therefore, dark-adapted mitochondrial GS activity was neither inhibited nor masked in mitochondrial lysates. Matrix fractions obtained from isolated root mitochondria were also subjected to DEAE-Sepharose chromatography; no significant GS activity in column eluates was observed.
Leaf Mitochondrial GS Activity Is Dark Stable and Induced by CO2 Limitation
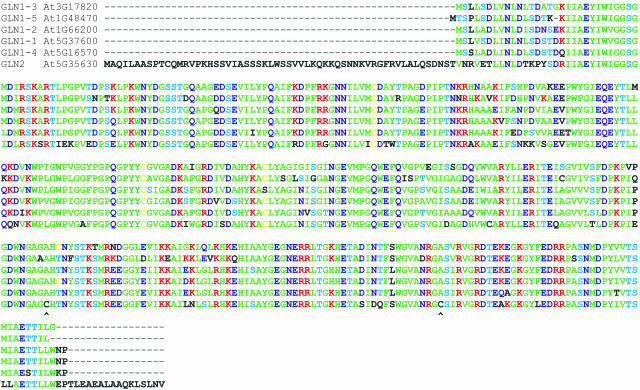
To test diurnal regulation, A. thaliana plants shifted to darkness for varying times were harvested, and leaf mitochondria and chloroplasts were isolated and purified (see Methods). In dark-shifted mitochondria, GS activity was stable and further increased over the 12-h dark period (Table 1). A. thaliana GS-2 possesses two Cys residues, Cys-306 and Cys-371, absent in cytosolic isoforms (Figure 1) but highly conserved in plastidic GS-2 of all tested angiosperms. As evidenced by site-specific mutageneses, both Cys-306 and Cys-371 are critical for light stability; presumably, internal, oxidative, disulfide bond formation between Cys-306 and Cys-371 results in loss of plastidic GS-2 activity (Choi et al., 1999). Conceivably, stable GS activity in dark-shifted A. thaliana mitochondria might be owed to thioredoxin activity, which would both maintain Cys sulfhydryl groups chemically reduced and prevent formation of any internal disulfide bonds. In turn, mitochondrial thioredoxin activity presupposes dark respiration (Motohashi et al., 2001).
Figure 1.
ClustalV Multiple Alignment of A. thaliana GS Gene Products.
To test photorespiratory regulation, A. thaliana plants growing in a 12-h photoperiod at high light intensity were maintained at the end of the normal 12-h light period under continuous light and shifted to either ambient CO2 (36 ± 2.0 Pa) or diminished CO2 (3.0 ± 0.3 Pa) atmosphere (see Methods). At 4-h intervals, plants were collected, and both leaf mitochondria and chloroplasts were purified (see Methods). GS activities from leaf organelles of plants maintained at ambient CO2 were similar over this time course. GS activities from leaf organelles of plants shifted to limiting (3 Pa) CO2 showed marked increases. Chloroplast GS activities jumped almost fourfold in 4 h, whereas leaf mitochondrial activities showed a slower but steady time-dependent increase (Table 1). Therefore, organellar GS activity in both chloroplasts and mitochondria was further induced by photorespiratory stress.
Light-Adapted A. thaliana Leaf Mitochondria Are Fully Competent for Oxidative Phosphorylation
In terrestrial plants, whole-tissue photosynthetic O2 evolution rates measured as a function of light intensity typically show a discontinuity initially thought to result from high light inhibition of mitochondrial respiration (Kok, 1948). Thereafter, corroborative experiments seemed to imply that leaf mitochondria were not fully competent for oxidative phosphorylation in the light (Heber and Heldt, 1981). Subsequently, however, light-adapted mitochondria from photosynthetic tissues were shown capable of rapid Gly-dependent oxidative phosphorylation in pea (Dry et al., 1983) and in barley (Gardeström and Wigge, 1988). As part of the photorespiratory C2 cycle, mitochondrial Gly decarboxylase activity generates ammonium at very high rates (Canvin et al., 1976; Woo and Osmond, 1976; Douce et al., 2001). In principle, mitochondrial GS activity might help reassimilate this ammonium.
To resolve this uncertainty, freshly purified, light-adapted, intact A. thaliana leaf mitochondria were tested for ability to drive the conversion of l-Orn to l-citrulline in a coupled assay system using either Gly or l-malate as an energy source (see Methods). In initial experiments, little activity was observed. However, when coupled assays were supplemented with l-glutamate, ADP, and NAD+, significant activity was observed (Table 2). In principle, these three cosubstrates should be regenerated and thus need be supplied at only catalytic levels. To the contrary, efficient coupling required addition of all three metabolites (l-glutamate, ADP, and NAD+) albeit at submillimolar levels (see Methods). The mitochondrial requirement for l-glutamate, ADP, and NAD+ supplementation may imply leakage of these intermediates during purification and/or assay.
Table 2.
Conversion of l-Orn to l-Citrulline by Purified, Intact Leaf Mitochondria
| Energy, C-Source | l-Citrulline Productiona |
|---|---|
| None | <0.10 |
| Gly | 2.10 |
| l-malate | 0.20 |
| Gly + l-malate | 1.10 |
μmol min−1 mg protein−1.
In coupled assays with intact osmotically protected mitochondria, Gly when added as sole energy, CO2, and N substrate sustained l-Orn –dependent l-citrulline production at high rates (Table 2), whereas added l-malate, or a mixture of both Gly and l-malate, supported this coupled process poorly (Table 2). Also, dark-adapted leaf mitochondria also converted l-Orn to l-citrulline as a Gly-dependent process at similar rates. In all cases, measured l-citrulline was quantitatively exported (Cohen et al., 1987; Cheung et al., 1989). When assay mixtures were pelleted by centrifugation and pellets lysed by osmotic dilution, >95% of total l-citrulline was present in the supernatant fraction, and <5% was present in the mitochondrial pellet. Coupled activity required freshly isolated, osmotically stabilized leaf mitochondria. Even when stored on wet ice, mitochondria lost 50% of coupled activity within several hours; coupled activity was completely lost upon overnight storage, whether mitochondria were stored on wet ice or at −70°C. Gly-dependent conversion of l-Orn to l-citrulline was approximately linear over a 20-min time course at 30°C, implying a steady state coupled process. Indeed, because conversion of l-Orn to l-citrulline slowed in the presence of both Gly and l-malate, efficient coupling was important for high-level activity (Table 2).
A Functional Genomic Analysis of A. thaliana Mitochondrial l-Orn–Dependent l-Citrulline Production
Because mitochondria require additional catalytic activities to convert l-Orn to l-citrulline, a functional genomic analysis was undertaken. In the first step, candidate gene products were identified after systematic searches of the A. thaliana genome. The candidate genes were exclusively of nuclear origin; no putative participating organellar (either mitochondrial or chloroplast) genes were identified. Next, these nuclear gene products were analyzed for organelle targeting N-terminal peptides using the PSORT algorithm (Horton and Nakai, 1996; Nakai, 2000). A collection of A. thaliana nuclear gene products with appropriate mitochondrial presequences (organellar targeting signals) was identified (Table 3). This tabulation included the following mitochondria-targeted activities: Gly decarboxylase, Gly-Ser hydroxymethyltransferase, GS, carbonic anhydrase, Gln-dependent carbamoylphosphate synthetase, and Orn carbamoyltransferase activities (Table 3).
Table 3.
A. thaliana Nuclear Gene Products with Mitochondrial Presequences or Chloroplast Transit Peptides Able to Participate in the Gly-Dependent Interconversion of l-Orn and l-Citrulline
| TAIR Locusa | Gene Product | Trafficking Peptide, N Terminus | pl | M/Sb |
|---|---|---|---|---|
| 5G35630.1 | Gln synthetase | MAQILAASPTCQMRVPKHSSVIASSSKLWS | 6.15 | M/S |
| 1G34320.1 | Carbamoylphosphate synthetase, catalytic | MGGLCSRSSSVNNAPGGTFAHVNGHHLNNN | 6.08 | M |
| 3G27740.1 | Carbamoylphosphate synthetase, glutaminase | MATRTLGFVLPTSLSSQPSFDRRGGGFRVS | 6.17 | M |
| 1G75330.1 | Orn carbamoyltransferase | MAAAMASHVSTARSPALSFSSSSSSFFPGT | 6.31 | M |
| 5G46800.1 | Orn-citrulline transporter | |||
| 1G58180.1 | Carbonic anhydrase I | MAFTLGGRARRLVSATSVHQNGCLHKLQQI | 6.84 | M |
| 5G14740.1 | Carbonic anhydrase II | MVPFWTTVSRNGSSDSETTLQSASKATKQY | 5.91 | M |
| 2G26080.1 | Gly decarboxylase complex P-protein | MERARRLAYRGIVKRLVNETKRHRNGESSL | 7.60 | M |
| 1G32470.1 | Gly decarboxylase complex H-protein | MALRMWASSTANALKLSSSVSKSHLSPFSF | 6.41 | M |
| 1G11860.1 | Gly decarboxylase complex T-protein | MRGGSLWQLGQSITRRLAQSDKKVVSRRYF | 6.49 | M |
| 5G26780.1 | Gly-Ser hydroxymethyltransferase | MALALRRLSSSVKKPISLLSSNGGSLRFMS | 7.06 | M |
| 4G24830.1 | Argininosuccinate synthase | MAEISATSFPSSSSSALVIRSSHNGSLKCQ | 5.61 | S |
| 5G10920.1 | Argininosuccinate lyase | MGAIDLSFSQSLLFSSSRSNLSSSTHRSVS | 5.59 | S |
| 4G08900.1 | Arginase | MSRIIGRKGINYIHRLNSASFTSVSASSIE | 7.21 | S |
M, mitochondrial; S, chloroplast stromal.
A comprehensive search of the A. thaliana genome also yielded a family of five paralogous nuclear genes, GLN1-x, encoding cytosolic GS-1 isoforms. From multiple alignments, these 39-kD proteins all show >83% homology (Figure 1). In addition, a sixth nuclear gene, GLN2, encoding plastidic GS-2 was identified. In contrast with the GS-1 family, GS-2 comprises a 43-kD protein with, uniquely among this family, a 58-residue N-terminal trafficking peptide (Figure 1). Correspondingly, five distinct A. thaliana GS genes were previously identified from cDNA analysis of polyadenylated mRNA preparations. From RNA gel blot analysis of this gene family, GLN2 expression proved both leaf specific and light inducible (Peterman and Goodman, 1991). Indeed, in various terrestrial higher plants, plastidic GS-2 activity is both light inducible and dark unstable (Lightfoot et al., 1988; Tingey et al., 1988; Tjaden et al., 1995).
The PSORT algorithm (Horton and Nakai, 1996; Nakai, 2000) identified the 58-residue N-terminal peptide of the GLN2 precursor as a mitochondrial presequence (Table 3). To further this analysis, the N-terminal trafficking peptides of 13 orthologous plastidic GS-2 precursor isoforms representing various terrestrial plants were likewise analyzed. PSORT certified all 13 as trafficking peptides. Obligated by program instructions to make unambiguous calls on each, PSORT called 8 of the 13 mitochondrial presequences and 5 chloroplast stromal transit peptides. In several cases, PSORT made alternate calls (mitochondrial versus stromal) in response to single residue changes of the N-terminal trafficking peptides (Table 4).
Table 4.
Organellar GS Gene Products (Arranged by Homology)
| Protein ID | Organism | Presequence/Transit Peptide (1 to 30) | pI | M/Sa |
|---|---|---|---|---|
| AAF17703 | Canavalia lineata | MAQILAPSTQWQMRITKTSPNASPVTSNMW | 6.18 | S |
| AAK43833 | Glycine max | MAQILAPSTQWQMRISKSSPNASPITSNMW | 6.18 | S |
| AJFBQD | P. vulgaris | MAQILAPSTQWQMRFTKSSRHASPITSNTW | 6.15 | M |
| AAD28443 | Medicago sativa | MAQILAPSIQCQTRITKTSPLATPISSKMW | 6.15 | S |
| P08281 | P. sativum | MAQILAPSTQWQMRITKTSPCATPITSKMW | 6.18 | S |
| CAA47373 | Nicotiana sylvestris | MAQILAPSGEWQMRMTKSSTDANPLTSKMW | 6.02 | M |
| AF019561 | Daucus carota | MAQILAPSVQWQMRFTKNSTEVSSMTSKMW | 6.18 | M |
| CAA51280 | Brassica napus | MAQILAASPTCQMRLTKPSSIASSKLWNSV | 6.15 | M |
| AAB20558 | A. thaliana | MAQILAASPTCQMRVPKHSSVIASSSKLWS | 6.15 | M |
| AAD31898 | Mesembryanthemum crystallinum | MAQILAPSLQVKMSITKSSTPASSMLPNTW | 6.15 | M |
| AAK07678 | Beta vulgaris | MAQILAPNMQCQMKLSKSLTNSMIPNSWTS | 6.04 | M |
| CAA46724 | Zea mays | MAQAVVPAMQCRVGVKAAAGRVWSAGRTRT | 6.18 | M |
| P14655 | Oryza sativa | MAQAVVPAMQCQVGAVRARPAAAAAAAGGR | 5.84 | S |
PSORT (Horton and Nakai, 1996; Nakai, 2000). M, mitochondrial; S, chloroplast stromal.
In Stably Transgenic A. thaliana Plants Expressing a Chimeric GLN2∷GFP, Mitochondria of Leaf Mesophyll and Epidermal Tissues Show GFP Fluorescence
Might the GLN2 gene encode the measured leaf mitochondrial GS activity? To test this hypothesis, stably transgenic A. thaliana plants expressing a GLN2∷GFP5 reporter (Haseloff et al., 1997) were constructed as follows. In vitro, an ∼800-np (545-np coding + 240-np intron) fragment of A. thaliana Col-0 genomic DNA encoding the first 181 residues of the GS-2 precursor was amplified by PCR (see Methods). This PCR fragment was then spliced into pCAMBIA1302 yielding an in-frame GLN2∷mGFP5 translational fusion whose expression was driven by the 35S promoter of Cauliflower mosaic virus. This ligation mixture was used to transform Escherichia coli DH5α′ competent cells to kanamycin resistance, and the desired recombinant plasmid, pUVS5, was identified by PCR analysis (see Methods). Both plasmid pUVS5 and its parent, pCAMBIA1302, were used to electroporate Agrobacterium tumefaciens AGL1 (Lazo et al., 1991), again selecting kanamycin resistance; the A. tumefaciens AGL1/pUVS5 derivative was verified by PCR analysis. In vivo, inflorescence meristems of A. thaliana Col-0 plants were inoculated with induced A. tumefaciens AGL1/pUVS5 or AGL1/pCAMBIA1302 cultures by vacuum infiltration (Bechtold et al., 1993). T1 generation seed was collected from self-fertilized inoculated plants and germinated aseptically on defined medium containing hygromycin-B (see Methods). Resistant seedlings were identified, transplanted to soil, and propagated normally. Ten independent stably transgenic A. thaliana lines (USBL 1-10) carrying the GLN2∷GFP5 construct derived from pUVS5 were identified, analyzed, and propagated. Ten independent stable transgenic A. thaliana lines (USBL 11C-20C) carrying GFP derived from the pCAMBIA1302 vector were similarly identified and propagated.
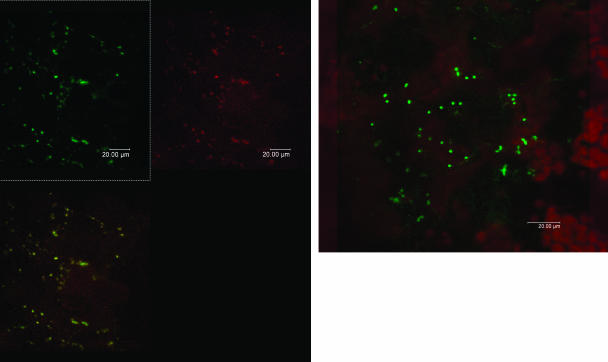
Stably transgenic lines USBL1 and USBL2 carrying the GLN2∷mGFP5 construct and line USBL12C carrying the unadulterated GFP reporter (lacking N-terminal GLN2 sequences) were propagated and harvested in the light, and leaf tissue was examined by laser scanning confocal microscopy (see Methods). In ventral epidermal cells (Figure 2A) and mesophyll cells (Figure 2B) of line USBL1, strong, punctate, green fluorescence was evident. Green fluorescence originated from subcellular ellipsoids some 1 μ in length. Leaf mesophyll cells also showed punctate green fluorescence, whose size and shape and subcellular distribution was consistent with mitochondria. By contrast, mesophyll chloroplasts showed red autofluorescence (Figure 2B) but no green fluorescence, possibly owed to quenching by accessory photopigments. In USBL12C plants carrying the unadulterated GFP reporter, green fluorescence was diffuse not punctate. When microscopic sections of line USBL1 leaves carrying the GLN2∷mGFP5 construct were then infused 15 min with a 50-nM aqueous solution of MitoTracker Red dye (Molecular Probes, Eugene, OR), colocalization of green and red punctate fluorescence (Haseloff, 1999) was observed. Therefore, mitochondria were inferred as source of punctate green fluorescence in leaves of line USBL1 plants. Consequently, A. thaliana GS-2 precursor is targeted to mitochondria.
Figure 2.
Confocal Laser Scanning Microscopy of Transgenic A. thaliana Leaf Tissue Expressing Chimeric GLN2∷GFP.
(A) Leaf epidermal tissue showing punctate green fluorescence.
(B) Leaf mesophyll tissue showing green fluorescence in mitochondria and red autofluorescence in chloroplasts.
In Immunoblots of Transgenic A. thaliana Plants, Chimeric GLN2∷GFP Is Present in Both Leaf Mitochondria and Chloroplasts
Both mitochondria and chloroplasts were highly purified from leaf tissue of light-adapted A. thaliana T1 generation plants corresponding to transgenic lines USBL1, USBL2, and USBL12C (see Methods). Soluble protein-enriched fractions for both organelles were obtained after DEAE-Sepharose chromatography and step elution with high-ionic-strength buffer (see Methods). Protein-enriched matrix and stromal samples were analyzed for αGFP cross-reacting material by immunoblotting using conventional protein gel blot analysis (see Methods).
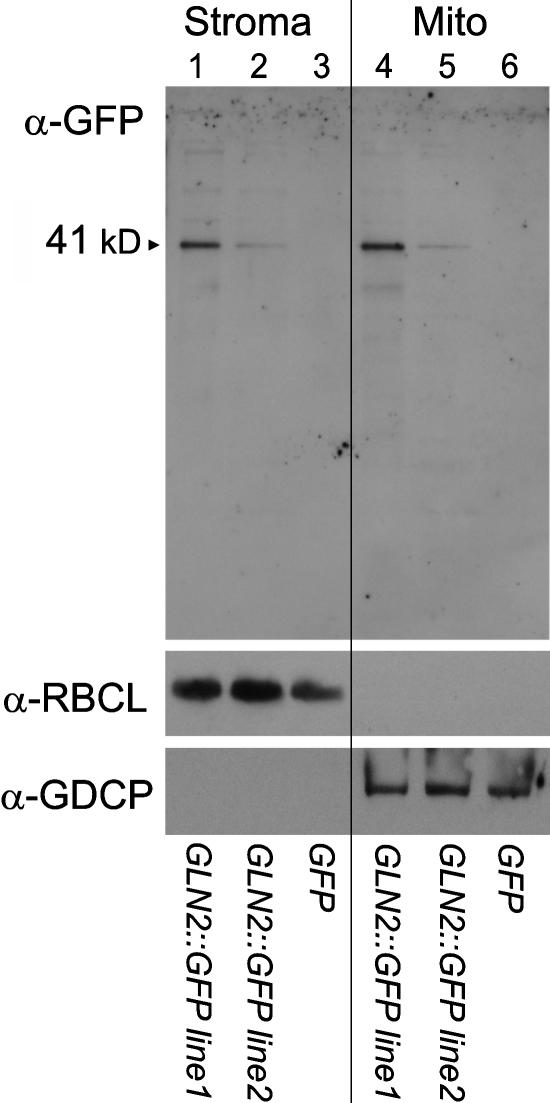
In stably transgenic lines USBL1 and USBL2 expressing the chimeric GLN2∷GFP5 reporter, both stromal and matrix fractions showed an αGFP cross-reacting protein. The measured molecular mass, 41 kD, of the αGFP cross-reacting protein was that expected for chimeric GLN2∷mGFP5 protein (Figure 3). In the two independent lines, absolute levels of chimeric GLN2∷GFP varied markedly. By contrast, in transgenic line USBL12C carrying the GFP reporter absent any N-terminal fusion, αGFP cross-reacting material was absent in both protein-enriched matrix and stromal fractions. To assess for cross-contamination of purified chloroplasts and mitochondria, chloroplast stromal fractions and mitochondrial matrix fractions were also challenged with either stromal-specific or matrix-specific antisera. Chloroplast stromal fractions of all strains cross-reacted with a polyclonal antiserum prepared against purified, H. vulgare ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Figure 3). Strong signals corresponding to Rubisco large subunit (54 kD) and weak signals from small subunit (20 kD) were both observed. Mitochondrial matrix preparations did not detectably cross-react with α-Rubisco serum. Conversely, mitochondrial matrix fractions of all strains specifically cross-reacted with matrix-specific polyclonal antiserum prepared against A. thaliana Gly decarboxylase complex P-protein (104 kD); chloroplast stromal fractions did not (Figure 3). Therefore, chimeric GLN2∷GFP was present in both leaf mitochondria and chloroplasts of stably transgenic plants.
Figure 3.
Immunoblot Analysis of Partially Purified Chloroplast Stromal and Mitochondrial Matrix Proteins.
Proteins were isolated from transgenic A. thaliana lines, separated by SDS-PAGE, and probed with polyclonal α-GFP serum (stromal fractions, lanes 1 to 3; matrix fractions, lanes 4 to 6). As controls, polyclonal H. vulgare α-Rubisco serum (α-RBCL) and polyclonal A. thaliana α-Gly decarboxylase P-protein (α-GDCP) serum were also tested. A. thaliana lines USBL1 and USBL2 carry p35S∷GLN2∷GFP; A. thaliana line USBL12c carries p35S∷GFP only. Molecular mass values (kD) are as indicated.
DISCUSSION
Previously, GS-2 isoforms in terrestrial plants were known as plastidic. As demonstrated in this work with A. thaliana, GS-2 precursor is also imported by leaf mitochondria. In A. thaliana, the GLN2 coding sequence was determined from cDNA synthesized from polyadenylated mRNA. In multiple cDNA clones, no heterogeneity was detected (Peterman and Goodman, 1991). Conceivably, a discrete polyadenylated mRNA species might be alternatively translated or posttranslationally modified to yield multiple distinct precursors. Alternatively, the N-terminal trafficking peptide of a single GS-2 precursor constitutes both a mitochondrial presequence and a chloroplast transit peptide. Indeed, such ambiguity would help rationalize why trafficking peptides, in general, do not show absolute sequence conservation (Heins et al., 1998). In A. thaliana, there exist several precedents for dual targeting of proteins (Peeters and Small, 2001), including nuclear genes encoding mitochondrial and plastid histidyl-tRNA synthetase (Akashi et al., 1998) and mitochondrial and plastidic RNA polymerase (Hedtke et al., 2000).
Using reference databases of many hundreds of bona fide peptide sequences representing various higher, terrestrial plants, trafficking algorithms such as PSORT (Horton and Nakai, 1996; Nakai, 2000) and Target-P (Emanuelsson et al., 2000) have ascertained rules, generalizations, and tendencies—none absolute—that collectively influence protein trafficking. Given an inferred trafficking peptide, these algorithms are programmed to specify precise targeting destinations (PSORT) or to assign probabilities of potential targeting to multiple destinations (Target-P). By these criteria, the inferred N-terminal trafficking peptide of the presumed A. thaliana GS-2 precursor is ambiguous, showing properties of both plant mitochondrial presequences and chloroplast transit peptides. Indeed, among a larger set of 13 organellar GS-2 isoforms representing diverse angiosperms, all inferred trafficking peptides are similarly ambiguous—archetypal for neither mitochondria nor chloroplast import. Mitochondrial presequences are typically characterized by amphipathic α-helices and isoelectric points exceeding pH 6.20, whereas chloroplast transit peptides are more acidic, with isoelectric points typically below pH 6.0. As a family, the inferred plant organellar GS-2 family trafficking peptides are also indeterminate in this regard (Table 4).
In A. thaliana, leaf organellar GS-2 activity is further induced in response to physiological CO2 limitation. Increased GS-2 levels likely facilitate assimilation of ammonium produced by photorespiratory phosphoglycolate (C2) cycle activity (Beckmann et al., 1997). Ammonium released by mitochondrial Gly decarboxylase complex activity may then be directly reassimilated as mitochondrial Gln. In principle, Gln, as a neutral molecule, might shuttle to chloroplasts by passive transport for conversion to glutamate via ferredoxin-dependent glutamate synthase (l-Gln-oxoglutarate amidotransferase, EC 1.4.7.1) activity (Somerville and Ogren, 1980). In turn this glutamate might be exported to leaf peroxisomes as substrate for aminotransferase activity, regenerating photorespiratory Gly. Alternatively, mitochondrial Gln might be further metabolized in a carbamoylphosphate-dependent process (Ludwig, 1993). Necessarily, mitochondrial Gln also serves as anabolic substrate—directly, in the case of mitochondrial protein synthesis. As demonstrated here, mitochondrial Gln also directly drives conversion of mitochondrial l-Orn to l-citrulline, for export and subsequent conversion to l-Arg in chloroplasts. As such, l-citrulline synthesis is ATP dependent, and coupled activity implies that light-adapted leaf mitochondria are fully competent for oxidative phosphorylation.
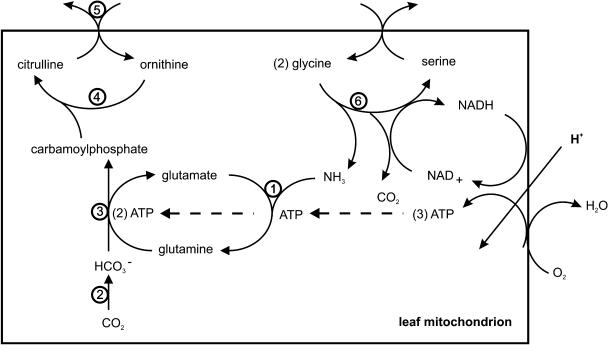
In mitochondria of actively photorespiring leaf tissue, Gly combustion generates reduced pyridine nucleotide (NADH). In principle, this NADH might be shuttled to leaf peroxisomes as substrate for reduction of hydroxypyruvate to glycerate by glycerate dehydrogenase activity (Raghavendra et al., 1998). However, such shuttle activity is antithetical to the organellar division of labor in photosynthetically active tissues. In the light, chloroplasts assume primary responsibility for export of reducing power to the cytosol via a triose phosphate/phosphoglycerate shuttle (Stocking and Larson, 1969). Indeed, leaf mitochondrial oxidative phosphorylation rates are stimulated by export of excess reducing equivalents generated by chloroplast photoelectron transport at high light intensities (Krömer et al., 1993). Correspondingly, as demonstrated here, mitochondrial Gly oxidation is able to drive conversion of mitochondrial l-Orn to l-citrulline as a tightly coupled process. In actively photorespiring tissues, O2 is relatively abundant and CO2 is limited. Gly-driven oxidative phosphorylation should yield some three ATP per Gly consumed. In the conversion of l-Orn to l-citrulline, one ATP is consumed in the synthesis of Gln, and two ATP are consumed in the synthesis of carbamoylphosphate, allowing tight coupling (Figure 4).
Figure 4.
Functional Coupling in Leaf Mitochondria.
Indicated activities are GS (1), carbonic anhydrase (2), carbamoylphosphate synthetase (3), Orn carbamoyltransferase (4), Orn/citrulline antiporter (5), and Gly decarboxylase complex (6). Also represented is oxidative phosphorylation driven by Gly decarboxylase complex activity.
In A. thaliana, l-citrulline exported by mitochondria is presumably taken up by chloroplasts and converted to l-Arg via stromal argininosuccinate-synthetase and -lyase activities. Nuclear genes encoding these stromal activities, as well as that of chloroplast (catabolic) arginase activity, which regenerates l-Orn, have been identified (Table 3). Notably, the A. thaliana nuclear genome also encodes a mitochondrial arginase isoform (Table 3), which activity would also yield l-Orn as substrate for conversion to l-citrulline.
Nuclear genomes of analyzed terrestrial plants carry single GLN2 genes encoding GS-2. (However, several Glycine species may carry multiple GLN2 genes; J.J. Doyle, unpublished results.) The same is true for other nuclear genes whose products are seemingly dual targeted. Gene duplication events, which would allow independent evolution of distinct mitochondrial and plastid isoforms, would seem unambiguous (Vision et al., 2000). Alternatively, differential transcription, splicing, and/or translation of a single gene to yield multiple precursors might confer multiple targeting but limit optimization of mature protein(s) for different working environments. Partition of function resulting from gene duplication events that might be maladaptive, however, were dual targeting driven by coevolution of mitochondria and plastids in terrestrial plants (Kadowaki et al., 1996; Elo et al., 2003).
METHODS
Growth and Purification of Arabidopsis thaliana Leaf Mitochondria and Chloroplasts
A. thaliana ecotype Col-0 plants were grown at high light intensity (250 μE s−1 m2) in a 12-h photoperiod, 23°C days and 16°C nights. For physiological shift experiments to enhance photorespiration, plants were shifted to a separate, high-light growth chamber and continuously sparged with a mixture of air scrubbed of CO2 by passing through 2 M NaOH solution and then blended with measured trace amounts of compressed CO2. Prevailing growth chamber CO2 concentrations were then measured using a LiCor 6262 CO2 analyzer (LiCor Biosciences, Lincoln, NE). As needed for leaf organelle purification purposes, 4-week-old plants were shifted to constant light or dark conditions for 48 h before harvest.
Intact chloroplasts were purified from leaf tissue as previously described (Ludwig, 1993). Leaf mitochondria were purified from rosettes of either light- or dark-adapted plants. Leaf tissue (500 g) was suspended in buffered saline on ice and washed (30 s) by bath ultrasonication. Tissue was rinsed in ice water, spun dry, chopped into small pieces with a small food processor, and suspended in 4 volumes of HB (50 mM Tris-Cl, pH 7.6, 0.1 mM DTT, 0.2% BSA [fraction 5], 1.0% polyvinyl pyrrolidone 40, and 0.35 M sorbitol) for 15 min on ice. Leaf tissue was homogenized (Polytron) 5 s in 10-mL portions, and the homogenate was filtered through double layers of cheesecloth and Miracloth. Filtrate was centrifuged (3000g) 10 min at 4°C. The supernatant was recovered and again centrifuged (16,000g) 30 min at 4°C. The crude mitochondrial pellet was resuspended (1:1 volume HB) using a Teflon pestle and ground glass mortar and slowly diluted (10 min) with 100 mL of WB (50 mM Tris-Cl, pH 7.6, 0.1 mM DTT, 0.1% BSA, and 0.35 M sorbitol); both centrifugation steps were then repeated. Mitochondrial pellets were resuspended in 20 mL of WB and centrifuged a third time (16,000g) for 30 min at 4°C. Pellets were again resuspended in 20 mL of WB on ice, and 5 mL portions were layered onto 18:23:40% (w/v) Percoll solutions in WB in the ratio of 8:10:8 volume. Percoll gradients were centrifuged (12,000g) for 60 min at 4°C (Beckman JA-13 rotor). The visible beige mitochondria band (below the 23:40% Percoll interface) was collected and slowly (15 min) diluted on ice with 5 volumes of WB. Highly purified mitochondria were pelleted by centrifugation (20,000g) for 30 min at 4°C. To remove residual Percoll solution, this final washing step was repeated. For whole mitochondria assays, preparations were stored on ice and used immediately. For enzyme partial purifications, mitochondrial pellets were stored at −80°C.
Partial Purification of GS from Purified A. thaliana Leaf Mitochondria
Frozen purified mitochondria were resuspended in an equal volume of buffer A (10 mM Tris-Cl, pH 7.5, 5 mM MgCl2, 0.1 M KCl, 0.1 mM EDTA, and 0.1 mM DTT) and lysed by tip ultrasonication using three 15-s bursts on ice. Lysates were centrifuged (16,000g) for 30 min at 4°C. Supernatants (matrix fractions) were loaded onto an 80-mL DEAE-Sepharose 6B (Pharmacia; fast-flow) column equilibrated with buffer A and chromatographed at 4°C. Loaded columns were washed with 1 volume of buffer A, and bound material was eluted with a 0.1- to 0.8-M KCl gradient (400 mL) in buffer A. Fractions (2 mL) were assayed for both GS activity and total protein (Bradford, 1976).
GS Activity
GS activity was assayed as the ADP-dependent conversion of l-Gln to γ-glutamylhydroxamate (Shapiro and Stadtman, 1970). Assay mixtures comprised 40 mM imidazole-Cl, pH 7.0, 30 mM l-Gln, 3 mM MnCl2, 0.4 mM ADP, 20 mM sodium arsenate, and 60 mM NH2OH in a final volume of 1.5 mL. Reactions were started by adding enzyme fractions and were incubated 15 min at 30°C. Reactions were stopped by adding 1.0 mL of a mixture of 2.6% FeCl3·6H2O, 4% trichloroacetic acid, and 1 N HCl. Ferric γ-glutamylhydroxamic acid was measured by spectrophotometric absorbance at 540 nm.
Conversion of l-Orn to l-Citrulline in Coupled, Intact Mitochondria
Intact, highly purified A. thaliana leaf mitochondria were assayed for l-Orn –dependent production of l-citrulline (Ness and Weiss, 1985; Davis, 1986). Freshly isolated mitochondria were resuspended in an equal volume of WB, and 0.05-mL samples were mixed on ice with 0.45 mL MRB (50 mM Tris-Cl, pH 7.4, 7 mM MgCl2, 15 mM NaCl, 2 mM EDTA, 5 mM KxPO4, and 0.3 mM sorbitol) supplemented with 1.0 mM l-glutamate, 0.2 mM ADP, and 0.2 mM NAD+. Reaction mixtures (13 × 100-mm tubes) were supplemented with C and/or N source as described (see text) and initiated with 5 mM l-Orn. Reaction tubes were transferred to a roller drum and incubated at 29°C with rolling aeration. At indicated times, 0.1-mL samples were removed and added to 0.1 volume of 50-mM sodium azide on ice. Samples were centrifuged at 16,000g for 30 min at 4°C. Supernatants were recovered, diluted with 0.1 mL of 17.5% trichloroacetic acid, and mixed on ice. Samples were centrifuged at 13,000g for 10 min at 4°C. Supernatants were recovered and diluted with 0.2 mL of 75% concentration H3PO4, 25% concentration H2SO4, and 0.025 mL of 3% diacetyl-monooxime. Samples were mixed, boiled 15 min in water, cooled to ambient temperature, and centrifuged at 13,000g for 1 min. l-citrulline oxime was measured by spectrophotometric absorbance at 480 nm.
Immunoblot Analysis
Purified chloroplasts and/or mitochondria were lysed by tip ultrasonication on ice, and membrane fractions were pelleted by centrifugation at 16,000g for 30 min at 4°C. Supernatants were loaded onto individual, single-use DEAE-Sepharose 6B minicolumns equilibrated in buffer A, and loaded columns were washed with two volumes of buffer A plus 0.1 M KCl. Bound protein was step eluted with buffer A plus 0.8 M KCl and concentrated/desalted by centrifugation/ultrafiltration (Centricon YM-10; Millipore, Bedford, MA) at 5000g for 60 min at 20°C. From the retentates, which comprised protein-enriched stromal or matrix fractions, 20 μg of total protein samples were boiled 10 min in loading buffer (0.04 M Tris-Cl, pH 6.8, 0.1 M DTT, 2% SDS, 10% glycerol [v/v], and 0.025% bromophenol blue) and centrifuged 5 min at 13,000g at 20°C. Supernatants, along with protein molecular weight tracking standards, were loaded onto SDS-polyacrylamide slab gels comprising a 3.9% stack, pH 6.8, and a 10% running gel, pH 8.8, and were subjected to electrophoresis in Tris-Gly buffer, pH 8.4, containing 0.1% SDS (Laemmli, 1970). The resulting running gel was blotted onto nitrocellulose membranes (ProTran, 0.2-μ pore, Schleicher and Schüll) by electrophoretic transfer in ETB (0.025 mM Tris-Cl, 0.19 M Gly, and 20% methanol, pH 8.4) at 30 mA and 4°C overnight. Membranes were washed and blocked in TTBS solution (0.025 M Tris-Cl, pH 7.4, 0.15 M NaCl, 0.0015 M KCl, and 0.1% Tween-20) 1 h at 20°C. A commercial (BD Laboratories) polyclonal antiserum prepared against purified GFP was used. As experimental controls for cross-contamination, a stromal-specific antiserum prepared against purified barley (Hordeum vulgare) Rubisco and a mitochondrial matrix-specific antiserum prepared against purified A. thaliana Gly decarboxylase complex P-protein were used. Test polyclonal antisera were added at 1:1,000 dilution and incubated 1 h at 20°C. Membranes were washed 1 h at 20°C with six changes TTBS buffer and subsequently treated 1 h at 20°C with a 1:5,000 dilution of a secondary horseradish peroxidase conjugate serum. Membranes were again washed 1 h at 20°C with six changes TTBS buffer. Luminescence detection was performed with a commercial (SuperSignal West Pico, Perbio) luminol-enhancer substrate mixture and captured by autoradiography.
Isolation of Transgenic A. thaliana Lines Carrying Chimeric GLN2 Translationally Fused in Frame to mGFP5 as Fluorescent Reporter
A chimeric gene comprising proximal GLN2 trafficking and coding sequences fused in frame to GFP was constructed in plasmid pCAMBIA1302. Two synthetic oligodeoxynucleotides, AtGS1 (5′-CCATGGCTCAGATCTTAGCAGC-3′) and AtGS2 (5′-CCATGGGAACCTCGCCAGAGACCTTC-3′), were used to amplify an ∼800-np fragment from A. thaliana ecotype Col-0 genomic DNA as template using thermostable DNA polymerase in conventional (0.05-mL reaction, 30 cycles, and 2-min elongation at 72°C) PCR. Amplified DNA fragment was mixed 3:1 mole:mole with purified pCAMBIA1302, digested to completion with added NcoI endonuclease, and purified by commercial minicolumn silicagel chromatography (Qiagen, Valencia, CA) to remove both primer and digested oligodeoxynucleotides. The column eluate (50 μL) was warmed to 70°C and slowly cooled to ambient temperature. An equal volume of 2× RL buffer (132 mM Tris-HCl, pH 7.6, 20 mM MgCl2, 2 mM DTT, and 15% polyethylene glycol 8000), 1 mM ATP, and 100 Weiss units T4 DNA polymerase were added and the ligation reaction allowed to run 10 min at ambient temperature. Ligation reaction samples (1 μL) were used to transform Escherichia coli DH5α′ competent cells by standard procedures. E. coli transformants were selected by growth on LB medium containing kanamycin (25 μg mL−1) at 37°C. From candidate transformants, plasmid DNA was purified and used as template for standard PCR reactions primed with both oligodeoxynucleotides AtGS1 and AtGS2. Putative recombinant plasmids were confirmed by digesting candidate plasmid DNAs with NcoI endonuclease followed by analytical agarose gel electrophoresis; appearance of an ∼800-np DNA fragment (545-np coding + 240-np intron) verified insert DNA. Using AtGS1 and GFP2 as PCR primers, in-frame orientation was verified by successfully amplifying an ∼800-np fragment. The resulting plasmid (pRV5) carried a chimeric reporter gene comprising the proximal half of the presumed GLN2 coding sequence fused in frame at codon 182 to the entirety of the GFP coding sequence as carried by pCAMBIA1302.
Purified plasmid DNA from strain E. coli DH5α/pRV5 was used in electroporation experiments with Agrobacterium tumefaciens AGL1 (Lazo et al., 1991). The resulting strain A. tumefaciens AGL1/pRV5 was confirmed, as described for the parental E. coli DH5α/pRV5 strain, cultured, induced, and used to inoculate A. thaliana ecotype Columbia flowering plants by vacuum infiltration (Bechtold et al., 1993). Seeds from self-fertilized, inoculated plants were germinated aseptically on phytological agar containing WPM salts (Haughn and Somerville, 1986) supplemented with hygromycin-B (50 mg L−1). Fluorescent seedlings were identified, transplanted to soil mix, and propagated by standard procedures. For analytical verification, total DNA of leaves from candidate lines was isolated, purified, and tested as template for ability to amplify an ∼1-kb chimeric GLN2∷GFP DNA fragment in standard PCR reactions using AtGS1 and AtGFP2 as primers. All fluorescent seedlings tested positive in this assay. Identified transgenic lines were self-fertilized and propagated.
Laser Scanning Confocal Microscopy
Confocal microscopy was performed on a Leica confocal microscope fitted with 63× 1.4–numerical aperture plan apochromat objectives and a 25-nm krypton-argon ion laser. Excitation was set at 488 nm, and multichannel emissions were obtained for fluorescein, rhodamine, and Cy5 filter sets. GFP fluorescence was measured as emission at 522 nm; chlorophyll autofluoresence was measured as emission at 645 nm. Interchannel crossover was negligible. Images were analyzed using MetaMorph software (Universal Imaging, Downington, PA).
Acknowledgments
We thank Blake Riggs and Uyen Tram for help with confocal microscopy, David Oliver and Ray Zielinski for gifts of antisera, and Craig Atkins for useful discussions. This work was supported by grants to R.L. from the U.S. National Science Foundation and U.S. National Institutes of Health.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Robert A. Ludwig (ludwig@biology.ucsc.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022046.
References
- Akashi, K., Grandjean, O., and Small, I. (1998). Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 431, 39–44. [DOI] [PubMed] [Google Scholar]
- Artus, N., Somerville, S.C., and Somerville, C.R. (1987). The biochemistry and cell biology of photorespiration. CRC Crit. Rev. Plant. Sci. 4, 121–147. [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Beckmann, K., Dzuibany, C., Biehler, K., Fock, H., Hell, R., Migge, A., and Becker, T.W. (1997). Photosynthesis and fluorescence quenching, and the mRNA levels of plastidic glutamine synthetase or of mitochondrial serine hydroxymethyltransferase (SHMT) in the leaves of the wild-type and of the SHMT-deficient stm mutant of Arabidopsis thaliana in relation to the rate of photorespiration. Planta 202, 379–386. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Canvin, D.T., Lloyd, N.D., Fock, H., and Przybylla, K. (1976). Glycine and serine metabolism and photorespiration. In CO2 Metabolism and Plant Productivity, R.H. Burris and C.C. Black, eds (Baltimore, MD: University Park Press), pp. 161–176.
- Cheung, C.W., Cohen, N.S., and Raijman, L. (1989). Channeling of urea cycle intermediates in situ in permeabilized hepatocytes. J. Biol. Chem. 264, 4038–4044. [PubMed] [Google Scholar]
- Choi, Y.A., Kim, S.G., and Kwon, Y.M. (1999). The plastidic glutamine synthetase activity is directly modulated by means of redox change at two unique cysteine residues. Plant Sci. 149, 175–182. [Google Scholar]
- Cohen, N.S., Cheung, C.W., and Raijman, L. (1987). Channeling of extramitochondrial ornithine to matrix ornithine transcarbamylase. J. Biol. Chem. 262, 203–208. [PubMed] [Google Scholar]
- Davis, R.H. (1986). Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol. Rev. 50, 280–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce, R., Bourguignon, J., Neuburger, M., and Rebeille, F. (2001). The glycine decarboxylase system: A fascinating complex. Trends Plant Sci. 6, 167–176. [DOI] [PubMed] [Google Scholar]
- Dry, J.B., Day, D.A., and Wiskich, J.T. (1983). Preferential oxidation of glycine by the respiratory chain in pea leaf mitochondria. FEBS Lett. 158, 154–158. [Google Scholar]
- Elo, A., Lyznik, A., Gonzalez, D.O., Kachman, S.D., and Mackenzie, S.A. (2003). Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Faye, L., Greenwood, J.S., and Chrispeels, M. (1986). Urease in jack bean seeds is a cytosolic protein. Planta 168, 579–585. [DOI] [PubMed] [Google Scholar]
- Freeman, J., Marquez, A., Wallsgrove, R.M., Saarelainen, R., and Forde, B.G. (1990). Molecular analysis of barley mutants deficient in chloroplast glutamine synthetase. Plant Mol. Biol. 14, 297–311. [DOI] [PubMed] [Google Scholar]
- Gardeström, P., and Wigge, B. (1988). Influence of photorespiration on ATP/ADP ratios in the chloroplasts, mitochondria, and cytosol, studied by rapid fractionation of barley (Hordeum vulgare) protoplasts. Plant Physiol. 88, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58, 139–151. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn, G., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430–438. [Google Scholar]
- Heber, U., and Heldt, H.W. (1981). The chloroplast envelope: Structure, function and role in leaf metabolism. Annu. Rev. Plant Physiol. 32, 139–168. [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins, L., Collinson, I., and Soll, J. (1998). The protein translocation apparatus of chloroplast envelopes. Trends Plant Sci. 3, 56–61. [Google Scholar]
- Horton, P., and Nakai, K. (1996). A probabilistic classification system for predicting the cellular localization sites of proteins. Int. Syst. Mol. Biol. 4, 109–115. [PubMed] [Google Scholar]
- Kadowaki, K., Kubo, N., Ozawa, K., and Hirai, A. (1996). Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 15, 6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Kok, B. (1948). A critical consideration of the quantum yield of Chlorella-photosynthesis. Enzymologia 13, 1–36. [Google Scholar]
- Krömer, S., Malmberg, G., and Gardeström, P. (1993). Mitochondrial contribution to photosynthetic metabolism. Plant Physiol. 102, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lazo, G.R., Stein, P.A., and Ludwig, R.A. (1991). A transformation competent genome library in Arabidopsis. Biotechnology 9, 963–971. [DOI] [PubMed] [Google Scholar]
- Lightfoot, D.A., Green, N.K., and Cullimore, J.V. (1988). The chloroplast-located glutamine synthetase of Phaseolus vulgaris L.: Nucleotide sequence, expression in different organs and uptake into isolated chloroplasts. Plant Mol. Biol. 11, 191–202. [DOI] [PubMed] [Google Scholar]
- Ludwig, R.A. (1993). Arabidopsis chloroplasts dissimilate L-arginine and L-citrulline for use as N source. Plant Physiol. 101, 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, K., Kondoh, A., Stumpp, M.T., and Hisabori, T. (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. USA 98, 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K. (2000). Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54, 277–344. [DOI] [PubMed] [Google Scholar]
- Ness, S.A., and Weiss, R.L. (1985). Carbamoyl-phosphate synthetases from Neurospora crassa. Immunological relatedness of the enzymes from Neurospora, bacteria, yeast, and mammals. J. Biol. Chem. 260, 14355–14362. [PubMed] [Google Scholar]
- Ogren, W.L. (1984). Photorespiration pathways: Regulation and modification. Annu. Rev. Plant Physiol. 35, 415–442. [Google Scholar]
- Peeters, N., and Small, I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541, 54–63. [DOI] [PubMed] [Google Scholar]
- Peterman, T.K., and Goodman, H.M. (1991). The glutamine synthetase gene family of Arabidopsis thaliana: Light-regulation and differential expression in leaves, roots and seeds. Mol. Gen. Genet. 230, 145–154. [DOI] [PubMed] [Google Scholar]
- Raghavendra, A.S., Reumann, S., and Heldt, H.W. (1998). Participation of mitochondrial metabolism in photorespiration. Reconstituted system of peroxisomes and mitochondria from spinach leaves. Plant Physiol. 116, 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, B.M., and Stadtman, E.R. (1970). Glutamine synthetase (E. coli). Meth. Enzymol. 17A, 910–922. [Google Scholar]
- Somerville, C.R., and Ogren, W.L. (1980). Inhibition of photosynthesis in Arabidopsis thaliana mutants lacking leaf glutamate synthase. Nature 286, 257–259. [Google Scholar]
- Stocking, C.R., and Larson, S. (1969). A chloroplast cytoplasmic shuttle and the reduction of extraplastid NAD. Biochem. Biophys. Res. Commun. 37, 278–282. [DOI] [PubMed] [Google Scholar]
- Tingey, S., Tsai, F., Edwards, J., Walker, E., and Coruzzi, G. (1988). Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J. Biol. Chem. 263, 9651–9657. [PubMed] [Google Scholar]
- Tjaden, G., Edwards, J.W., and Coruzzi, G.M. (1995). Cis elements and trans-acting factors affecting regulation of a nonphotosynthetic light-regulated gene for chloroplast glutamine synthetase. Plant Physiol. 108, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Woo, K.C., and Osmond, C.B. (1976). Glycine decarboxylation in mitochondria isolated from spinach leaves. Aust. J. Plant Physiol. 3, 771–785. [Google Scholar]