Abstract
Most chloroplast outer-membrane proteins are synthesized at their mature size without cleavable targeting signals. Their insertion into the outer membrane is insensitive to thermolysin pretreatment of chloroplasts and does not require ATP. It has therefore been assumed that insertion of outer-membrane proteins proceeds through a different pathway from import into the interior of chloroplasts, which requires a thermolysin-sensitive translocon complex and ATP. Here, we show that a model outer-membrane protein, OEP14, competed with the import of a chloroplast interior protein, indicating that the two import pathways partially overlapped. Cross-linking studies showed that, during insertion, OEP14 was associated with Toc75, a thermolysin-resistant component of the outer-membrane protein–conducting channel that mediates the import of interior-targeted precursor proteins. Whereas almost no OEP14 inserted into protein-free liposomes, OEP14 inserted into proteoliposomes containing reconstituted Toc75 with a high efficiency. Taken together, our data indicate that Toc75 mediates OEP14 insertion, and therefore plays a dual role in the targeting of proteins to the outer envelope membrane and interior of chloroplasts.
INTRODUCTION
Most proteins in chloroplasts are encoded by the nuclear genome and synthesized in the cytosol. Nuclear-encoded chloroplast proteins can be divided into two groups based on the presence or absence of cleavable targeting signals. The first group of proteins is synthesized in the cytosol as precursor proteins with cleavable N-terminal targeting signals called transit peptides. Most of these proteins are imported into the interior of chloroplasts. These proteins share a general import pathway, using a proteinaceous import apparatus located in the chloroplast envelope (for a review, see Schleiff and Soll, 2000). The second group of proteins is synthesized in the cytosol at their mature size without cleavable transit peptides. This group includes most of the outer-membrane proteins identified so far. For most of these outer-membrane proteins, neither thermolysin-sensitive proteins exposed on the chloroplast surface nor ATP are required for their insertion into the outer membrane (Salomon et al., 1990; Li et al., 1991; Ko et al., 1992; Fischer et al., 1994; Bolter et al., 1999). It has generally been assumed that these outer-membrane proteins use an import pathway different from the general import pathway used by interior-targeted precursor proteins.
Many components in the import apparatus mediating the import of interior-targeted precursor proteins have been identified. Components located in the outer membrane are named Toc (translocon at the outer envelope membrane of chloroplasts) proteins and those in the inner membrane are named Tic (translocon at the inner envelope membrane of chloroplasts) proteins (Schnell et al., 1997). The association of Tic and Toc components as a complex can be observed even in the absence of importing precursor proteins (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). Among the Toc components identified, Toc159 and Toc34 have been shown to be the receptors for transit peptides (Perry and Keegstra, 1994; Ma et al., 1996; Kouranov and Schnell, 1997; Sveshnikova et al., 2000). Toc75 is postulated to span the outer membrane with multiple β strands (Schnell et al., 1994; Tranel et al., 1995). A considerable amount of evidence suggests that Toc75 is the major component of the protein-translocating channel in the outer membrane (Hinnah et al., 1997; Reumann et al., 1999). Tic110 is the major Tic component identified. It has an N-terminal membrane anchor and a large stroma-located hydrophilic domain. The stromal domain binds transit peptides; therefore, Tic110 is proposed to be the scaffold for protein translocation across the inner membrane into the stroma (Inaba et al., 2003).
The import process of interior-targeted precursor proteins can be roughly divided into three stages (Chen and Schnell, 1999). The first stage involves an energy-independent and low-affinity interaction between the transit peptide and the Toc complex on the chloroplast surface. Cross-linking studies showed that at this stage precursors are in direct contact predominantly with Toc159 and Toc34 (Perry and Keegstra, 1994; Ma et al., 1996; Kouranov and Schnell, 1997). In the presence of low concentration of ATP and GTP, import proceeds to the second stage with the precursors inserting across the outer membrane through the Toc75 channel and coming into contact with the Tic complex. This stage has been called the early import intermediate or the docking stage (Nielsen et al., 1997; Chen and Schnell, 1999). Cross-linking studies showed that at this stage, precursors are in direct contact predominantly with Toc75 and the Tic components (Ma et al., 1996; Kouranov and Schnell, 1997). In the third stage, the precursor protein is translocated across both membranes into the stroma at the expense of high concentrations of stromal ATP (>1 mM) (Theg et al., 1989).
Much less is known about the mechanism of outer-membrane protein insertion. It has been suggested that proteins insert into the outer membrane through a spontaneous insertion mechanism (Keegstra and Froehlich, 1999) or through interactions with lipids (Schleiff and Klosgen, 2001). A spinach (Spinacia oleracea) outer-membrane protein OEP7 was shown to insert with reverse orientation into liposomes containing the average lipid compositions of chloroplast outer membrane. Correct orientation was achieved by reducing the concentration of charged lipids in the liposomes (Schleiff et al., 2001). In both kinds of liposomes, <5% of the OEP7 molecules presented to the liposomes actually inserted (Schleiff et al., 2001). Pea (Pisum sativum) Toc34 was also shown to insert into protein-free liposomes, and its orientation was independent of charged lipids but was dependent of the size of the hydrophilic domain (Qbadou et al., 2003). Neither of the proposed insertion mechanisms could explain the apparent specificity of chloroplast outer-membrane protein insertion (see below).
Outer-membrane targeting/insertion signals have been identified from the mature regions of several outer-membrane proteins (Wu and Ko, 1993; Li and Chen, 1996, 1997). These targeting/insertion signals are necessary and sufficient to insert these proteins and other passenger proteins specifically into the chloroplast outer membrane. A region within the Arabidopsis thaliana outer-membrane protein AtOEP7 has been shown to be the signal that distinguishes between the chloroplast outer membrane and the endomembrane system (Lee et al., 2001). Furthermore, using OEP14, the pea homolog of AtOEP7, we demonstrated that insertion of OEP14 requires some trypsin-sensitive and N-ethylmaleimide–sensitive protein components. Association and insertion of OEP14 into the chloroplast outer membrane are saturable and competed for the import of another outer-membrane protein (Tu and Li, 2000). These results suggest that specific proteinaceous components exist as insertion sites for OEP14.
In this report, we show that OEP14 was directly cross-linked to Toc75 during insertion, and Toc75 mediated efficient OEP14 insertion into reconstituted proteoliposomes. These data indicate that Toc75 forms the insertion site for OEP14 and therefore plays direct roles in the targeting of proteins to the outer membrane and interior of chloroplasts.
RESULTS
OEP14-His6 Competed with the Import of an Interior-Targeted Precursor Protein
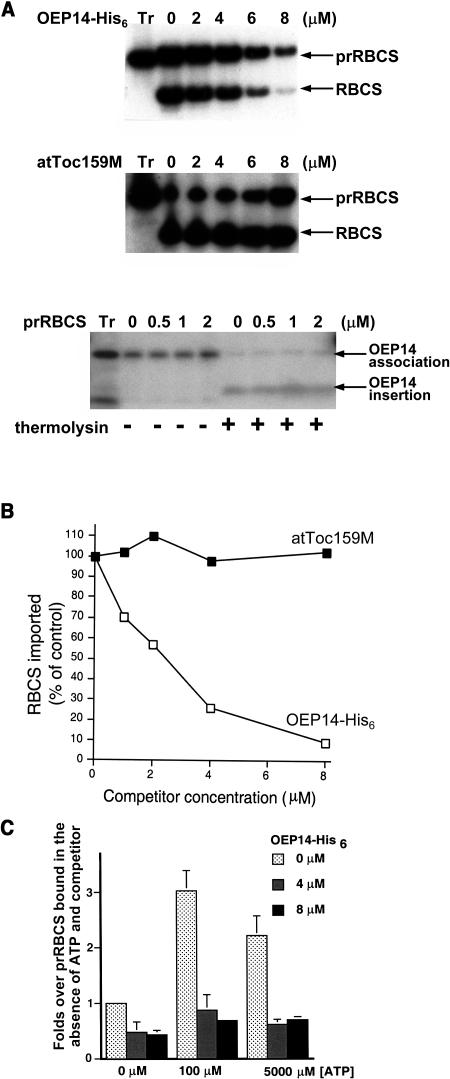
To identify the insertion site of OEP14, we overexpressed OEP14 in Escherichia coli with a C-terminal His6 tag. The recombinant protein is called OEP14-His6. We have previously shown that OEP14-His6 inserts into chloroplasts with the same efficiency as in vitro–translated OEP14 and competes with the import of another outer-membrane protein (Tu and Li, 2000). As a first step in OEP14 insertion site identification, we tested whether OEP14-His6 competed with the import of an interior-targeted precursor protein to determine whether these two pathways share components. As shown in Figure 1, excess OEP14-His6 could compete with the import of the precursor to the small subunit of ribulose bisphosphate carboxylase (prRBCS). As the concentration of unlabeled OEP14-His6 increased in the import reactions, the import of prRBCS decreased significantly (Figure 1A, top panel). To rule out the possibility that the competition resulted from nonspecific aggregation of OEP14 on the chloroplast surface, we performed the same competition experiment with the C-terminal membrane-associated domain of Arabidopsis Toc159 (atToc159M). This truncated protein can attach to the surface of chloroplasts with the same efficiency as other outer-membrane proteins but cannot insert into the outer membrane (Muckel and Soll, 1996; Smith et al., 2002). It had no effect on the import of prRBCS (Figure 1A, middle panel). We also tested if prRBCS could compete with the insertion of OEP14 into the outer membrane. The concentrations tested were lower because prRBCS was purified from E. coli inclusion bodies and solubilized with urea, which lyses chloroplasts at a high concentration. We used up to 2 μM, for which prRBCS competition with interior-targeted precursor proteins has been observed to be effective (Tranel et al., 1995). Interestingly, at these concentrations prRBCS had no effect on either the association of OEP14 (measured by the amount of full-length OEP14 associated with repurified chloroplasts after the import reaction, including both the OEP14 bound on the chloroplast surface and the OEP14 inserted into the outer membrane; Tu and Li, 2000) or the insertion of OEP14 into the outer membrane (measured by the amount of the 4-kD membrane-protected fragment produced by the thermolysin posttreatment) (Figure 1A, bottom panel). It is possible that OEP14 can use multiple components or a component in multiple forms for insertion into the outer membrane, and only one of the components or one of the forms is shared with prRBCS.
Figure 1.
Import of prRBCS Was Competed by OEP14-His6 at the Docking Stage.
(A) Import of prRBCS was competed by OEP14-His6 but not by atToc159M, and overexpressed prRBCS failed to compete with OEP14. [35S]prRBCS was incubated with chloroplasts and increasing concentrations of unlabeled OEP14-His6 (top panel) or atToc159M (middle panel). [35S]OEP14 was incubated with chloroplasts and increasing concentrations of unlabeled prRBCS (bottom panel). Half of the chloroplasts were further treated with thermolysin to reveal the amount of OEP14 inserted into the outer membrane. The concentration of cold OEP14-His6, atToc159M, or prRBCS is indicated above the lane. Tr, in vitro–translated [35S]prRBCS or [35S]OEP14 used for the import reactions.
(B) Quantification of the gels shown in (A). The amount of mature RBCS imported in the absence of unlabeled OEP14-His6 or atToc159M was set as 100%.
(C) Competition occurred at the docking stage. Energy-depleted [35S]prRBCS and chloroplasts were incubated with increasing concentrations of unlabeled OEP14-His6, and the reaction was supplemented with different concentrations of ATP. The amount of prRBCS associated with chloroplasts under each condition was quantified. The amount of prRBCS bound in the absence of ATP and unlabeled OEP14-His6 was set as 1. Data points represent mean (±se) of three independent experiments.
We further investigated which stage of prRBCS import was competed by OEP14-His6. Import reactions were performed at three different ATP concentrations to support initial surface association (0 μM ATP), early import intermediate formation (or docking, 100 μM ATP), and stroma translocation (5 mM ATP). OEP14 had little effect on initial surface association of prRBCS but almost totally competed out the early import intermediates produced after adding 100 μM ATP (Figure 1C). The amount of mature RBCS imported at 5 mM ATP was reduced to the same extent as prRBCS (data not shown; Figure 1A), indicating the reduction resulted from the inhibition of prRBCS docking. The competition results indicate that insertion of OEP14 shares some components with the import of prRBCS at the stage of early import intermediate formation. Previous results have shown that at this stage prRBCS is associated with the entire Tic/Toc complex (Akita et al., 1997) and is in direct contact predominantly with Toc75 and the Tic complex (Kouranov and Schnell, 1997).
OEP14-His6 Was Associated with Tic/Toc Translocon Components during Insertion
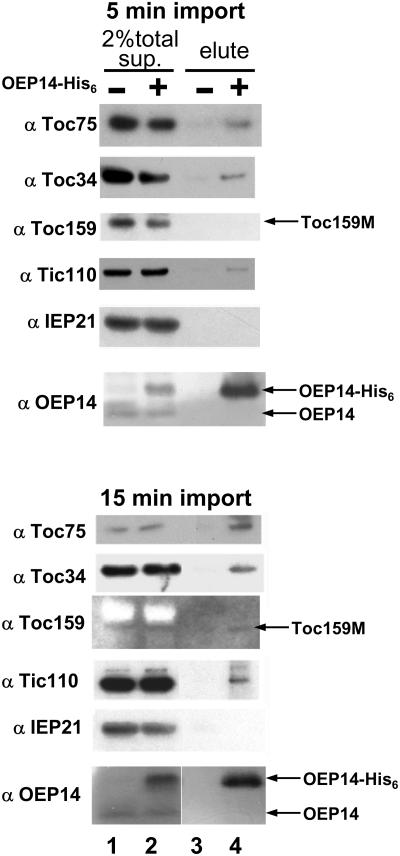
To further identify components mediating OEP14-His6 insertion, we performed chemical cross-linking experiments with the homobifunctional and cleavable cross-linker dithiobis-9-succinimidylpropionate (DSP). Chloroplasts were incubated with OEP14-His6 for 5 or 15 min, reisolated, and treated with DSP. For a control, chloroplasts were incubated with a truncated Toc34, Toc34(1-258)His6 (Sun et al., 2002), in which the outer-membrane insertion signal has been deleted, and the truncated protein cannot insert into the outer membrane (Li and Chen, 1997). Total chloroplast membranes were isolated and solubilized. Protein complexes containing OEP14-His6 were purified from the solubilized supernatant fraction by metal-affinity resin and analyzed by SDS-PAGE and immunoblots. In agreement with the competition results, Toc75, Toc34, and Tic110 copurified with OEP14-His6 (Figure 2). We observed the same cross-linking pattern when import was performed for 5 (Figure 2, top half) or 15 min (bottom half), except the amount of Toc75 copurified was higher at 15 min. Only the 54-kD M domain of Toc159 (Toc159M) was present in the total solubilized membrane fraction, and specific copurification of this fragment with OEP14-His6 was difficult to detect. The inner-membrane protein IEP21 and endogenous OEP14, two proteins that are not in the Tic/Toc translocon, did not copurify with OEP14-His6. This result showed that OEP14 specifically associated with the Tic/Toc translocon components during its insertion into the outer membrane.
Figure 2.
OEP14-His6 Was Cross-Linked to Translocon Components during Insertion.
OEP14-His6 was incubated with chloroplasts for 5 (top half) or 15 min (bottom half). Intact chloroplasts were reisolated and treated with DSP. Chloroplasts were lysed, and total membranes were isolated and solubilized. The clarified supernatant (sup., lanes 1 and 2) was purified with TALON resin (lanes 3 and 4) and analyzed by SDS-PAGE and immunoblots with antibodies labeled on the left. Lanes 1 and 3, mock import with the truncated outer-membrane protein Toc34(1-258), which does not associate with chloroplasts. Toc159M, the 54-kD degradation fragment of Toc159.
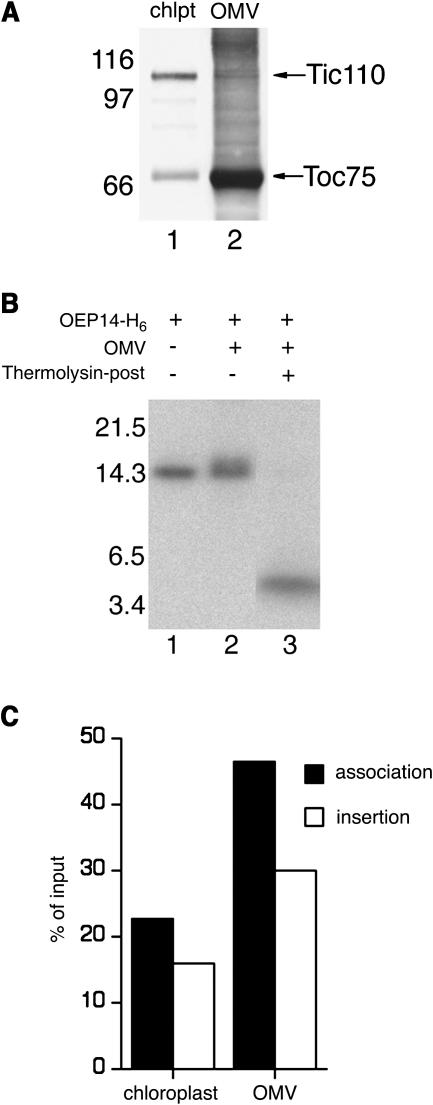
To investigate whether components of the outer envelope membrane were sufficient for insertion of OEP14, we performed OEP14 import with highly purified outer-membrane vesicles (OMVs). Immunoblotting of the vesicles demonstrated that they were enriched in the outer-membrane marker Toc75 and devoid of the inner-membrane marker Tic110 (Figure 3A, compare lanes 1 and 2). When the OMVs were used for import assays, OEP14-His6 could still insert into these vesicles (Figure 3B). Both the association of OEP14 with OMV (Figure 3B, lane 2) and the insertion of OEP14 (Figure 3B, lane 3) were highly efficient. In fact, the insertion efficiency was higher than that observed with intact chloroplasts (Figure 3C). It is possible that components required for OEP14 insertion were present in a higher concentration in the OMV preparation than in intact chloroplasts. A regular outer-membrane-vesicle import reaction contained outer-membrane proteins from ∼40 mL of chloroplasts (at a concentration of 1 mg/mL chlorophyll) compared with 50 μL of chloroplasts of the same concentration in a regular chloroplast import reaction. In any event, these results suggested that components of the outer membrane were sufficient for OEP14 import and Tic110, or other inner-membrane proteins were not required.
Figure 3.
OEP14-His6 Could Insert into Purified OMVs.
(A) Purity of the isolated OMVs. Total proteins of chloroplasts (chlpt, 9 μg) and isolated OMVs (2.5 μg) were resolved by SDS-PAGE. Immunobloting was performed using antibodies against pea Toc75 and Tic110.
(B) Import of OEP14-His6 into isolated OMVs. Lane 1, [35S]OEP14-His6 used in the import reactions. Lane 2, OEP14-His6 associated with the OMVs. Lane 3, same as lane 2 except the OMVs were further treated with thermolysin (thermolysin-post) to reveal the amount of OEP14-His6 inserted into the vesicles. Molecular masses of marker proteins are labeled at left.
(C) Efficiencies of OEP14 import into chloroplasts and isolated OMVs. In vitro–translated OEP14 was imported into isolated chloroplasts or OMVs. The amounts of OEP14 associated or inserted were quantified and plotted as percentage of total [35S]OEP14-His6 added to the import reactions.
Toc75 Was in Close Proximity to the Insertion Signal during OEP14 Insertion
We next investigated which components of the Toc complex directly interacted with OEP14. We used the heterobifunctional and cleavable crosslinker N-[4-(p-azidosalicylamido) butyl]-3'-(2'-pyridyldithio) propionamide (APDP), which can be labeled with 125I and attached specifically to Cys residues of the substrate protein. After photoactivation of APDP by UV irradiation, cross-linked adducts are formed between the substrate protein and proteins in its vicinity. APDP can then be cleaved by reducing agents, and 125I would then be transferred to the proteins crossed-linked to the substrate protein.
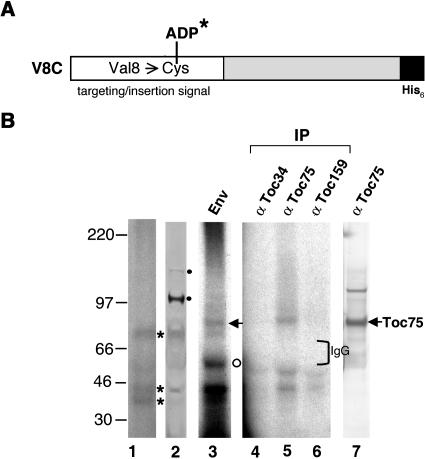
Because there is no Cys residue in OEP14-His6, Val of residue 8 within the outer-membrane targeting/insertion signal of OEP14 (Li and Chen, 1996) was mutagenized to a Cys to create the V8C mutant (Figure 4A). The Cys replacement and the APDP attachment had no effect on the insertion of OEP14-His6 (data not shown). The V8C mutant was produced and purified from E. coli. The protein was conjugated with [125I]-APDP (V8C-[125I]-ADP) and used for chloroplast import and cross-linking experiments. OEP14 has a tendency to form multimers upon association with chloroplasts (data not shown). Without enrichment by isolating the envelope fraction, the only 125I-labeled products clearly visible when total chloroplast proteins were analyzed were the V8C-[125I]-ADP multimers (Figure 4B, lane 1, asterisks). They were specifically recognized by anti-OEP14 antibodies (Figure 4B, lane 2). They could also result from cross-linking of V8C-[125I]-ADP to endogenous OEP14 multimers that were not resolved by SDS-PAGE. When only envelope membranes of the reisolated chloroplasts were analyzed, two additional cross-linked products of 55 and 75 kD were observed (Figure 4B, lane 3). The identity of the 55-kD protein (Figure 4B, lane 3, open circle) was unknown. A 55-kD protein has previously been observed to cross-link to precursor of ferredoxin using the same cross-linker (Rensink et al., 2000). The 75-kD protein (Figure 4B, lane 3, arrow) was specifically immunoprecipitated by anti-Toc75 antibodies, suggesting that this protein was Toc75 (lane 5). Immunoblots confirmed that this protein comigrated with endogenous Toc75 (lane 7). Anti-Toc159 and anti-Toc34 antibodies did not immunoprecipitate any 125I-labeled Toc159 or Toc34, indicating that these two proteins were not in the vicinity of OEP14 during insertion. The V8C-[125I]-ADP label transfer cross-linking data suggested that Toc75 was in the vicinity of OEP14 during OEP14 insertion.
Figure 4.
Toc75 Was Cross-Linked to OEP14-[125I]-ADP.
(A) Schematic representation of OEP14-His6 mutant V8C. The targeting/insertion signal (open box) and the hexa-histidine tag (filled box) are shown.
(B) Label-transfer cross-linking experiment with V8C-[125I]-ADP. Chloroplasts were incubated with V8C-[125I]-ADP, and cross-linking was activated by UV irradiation. Chloroplasts were pelleted, and a small portion was analyzed by SDS-PAGE and viewed by autoradiography (lane 1) or by immunobloting decorated with anti-OEP14 antibodies (lane 2). Asterisks indicate the V8C-[125I]-ADP multimers, which were also recognized by the anti-OEP14 antibodies. Closed circles indicated nonspecific cross-hybridization by the antibodies. Envelope membranes (Env) were isolated from the rest of the chloroplasts. A small portion of the membranes was analyzed by SDS-PAGE and autoradiography (lane 3). The open circle indicates the 55-kD protein of unknown identity. The rest of the membranes were solubilized and immunoprecipitated (IP) with antibodies against Toc34, Toc75, and Toc159 (lanes 4 to 6). Lane 7 shows an immunoblot of total chloroplast proteins probed with anti-Toc75 antibodies. The position of the IgG heavy chain is marked by a bracket. Molecular masses of marker proteins are labeled at left.
Toc34 and Toc159 Were Not Required for OEP14-His6 Import
Both Toc34 and Toc159 have a thermolysin-sensitive domain exposed on the chloroplast surface (Kessler et al., 1994; Seedorf et al., 1995). It is well documented that OEP14 import is not affected by thermolysin pretreatment of chloroplasts (Li et al., 1991; Tu and Li, 2000). Therefore, the cytosolic domains of Toc159 and Toc34 are most likely not essential for OEP14 import. To further investigate whether Toc34 and Toc159 play any major role in OEP14 import, we used Arabidopsis mutants that are disrupted in genes encoding atToc159, atToc34, or atToc33 (see below) to check if the import of OEP14 is affected in these mutants. Arabidopsis has three homologs of pea Toc159: atToc159, atToc132, and atToc120 (Bauer et al., 2000; Jackson-Constan and Keegstra, 2001). Among them, atToc159 is most similar to pea Toc159. Two Toc34 homologs are present in Arabidopsis: atToc34 and atToc33. Both of them share ∼60% identity to pea Toc34. Knockout mutants of atToc33, atToc159, and atToc34 have been identified (Jarvis et al., 1998; Bauer et al., 2000; Constan et al., 2004).
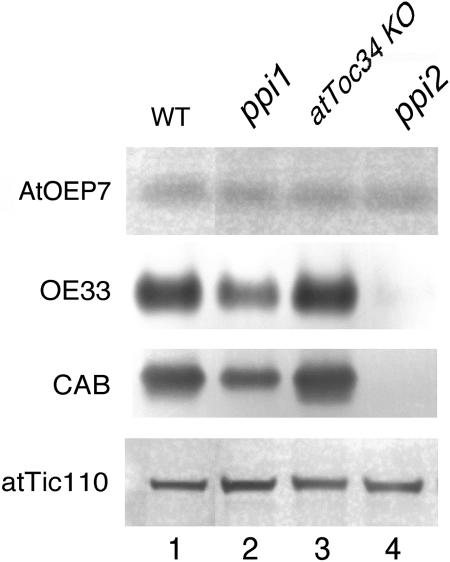
We did not isolate chloroplasts from these mutants and compare their import efficiencies directly because the atToc159 knockout mutant is albino. We could only compare the steady state levels of various chloroplast proteins in these mutants. Antibodies against the Arabidopsis homolog of pea OEP14, AtOEP7, were prepared and used in immunoblots to detect AtOEP7 in cell extracts of the wild-type and mutant plants. As shown in Figure 5, although the amounts of chloroplast interior proteins (e.g., chlorophyll a/b binding protein [CAB] and the 33-kD oxygen-evolving complex protein [OE33]) were decreased when atToc159 or atToc33 was mutated, the amount of AtOEP7 was normal in the mutant. The atToc34-knockout mutant was wild type in appearance and had a normal amount of all the chloroplast proteins analyzed. These results indicated that, whereas the absence of atToc159 and atToc33 resulted in reduced amounts of chloroplast interior proteins, it did not affect the steady state level of AtOEP7. We cannot use the same approach to test the role of Toc75 because a Toc75 knockout mutant is lethal (D.J. Schnell, unpublished results).
Figure 5.
The Amount of AtOEP7 Was Normal in Arabidopsis Knockout Mutants of atToc159, atToc33, and atToc34.
Immunobloting analysis of AtOEP7, CAB, OE33, and atTic110 in various mutants. Total protein extracts were prepared from 14-d-old seedlings. For analysis of AtOEP7 and atTic110, 40 μg of proteins were used. For analysis of OE33 and CAB, 2 and 1 μg of proteins were used, respectively. KO, knockout mutant lines.
Toc75 Mediated OEP14 Insertion into Proteoliposomes
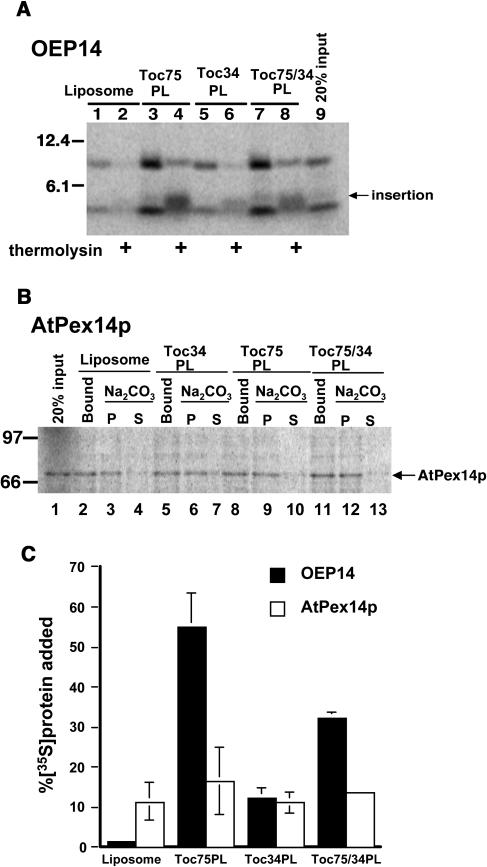
To positively demonstrate that Toc75 mediates OEP14 insertion, we reconstituted Toc75, Toc34, or both proteins into proteoliposomes (Wallas et al., 2003) and assayed their ability to mediate OEP14 insertion. As shown in Figure 6A, a low amount of OEP14 bound to protein-free liposomes and was almost completely removed by thermolysin posttreatment (Figure 6A, lanes 1 and 2), indicating that little insertion had occurred. This low efficiency is similar to that observed for insertion of spinach OEP7 into protein-free liposomes containing the average lipid composition of chloroplast outer membrane (Schleiff et al., 2001). By contrast, with proteoliposomes containing Toc75, >40% of the OEP14 added to the reaction associated and inserted into the proteoliposomes (Figures 6A, lanes 3 and 4, and 6C). Because the N-terminal Met is the only Met present in OEP14, the appearance of the 4-kD fragment indicated that the N-terminal portion of OEP14 was protected from thermolysin, and OEP14 had inserted into the Toc75 proteoliposomes in the correct Nin-Cout orientation (Li and Chen, 1996). Toc34 alone mediated an insertion efficiency slightly higher than the background (Figure 6A, lane 6). Adding back Toc75 together with Toc34 restored the insertion efficiency (Figure 6A, lane 8). The proteoliposomes containing both Toc75 and Toc34 had a lower insertion efficiency than proteoliposomes containing Toc75 alone, which may indicate that OEP14 preferentially inserts into the outer membrane via free Toc75 that is not associated with other Toc components. Interestingly, the efficiency of OEP14 insertion into proteoliposomes containing both Toc75 and Toc34 was almost the same as that of insertion into isolated OMVs (Figure 3C). As a control, the same experiment was performed with the peroxisomal membrane protein AtPex14p (Figure 6B). Similar to OEP14, AtPex14p is also an integral membrane protein (Hayashi et al., 2000) and inserted into the peroxisome membrane posttranslationally. An equally low level of nonspecific insertion was observed with protein-free liposomes and proteoliposomes containing Toc proteins, supporting that the high insertion efficiency mediated by Toc75 was specific for OEP14. The proteoliposome reconstitution experiments demonstrated that Toc75 was sufficient to mediate membrane insertion of OEP14.
Figure 6.
Toc75 Mediates Insertion of OEP14.
(A) Insertion of OEP14 into various proteoliposomes. [35S]OEP14 was incubated with liposomes without any protein or proteoliposomes (PL) containing Toc75, Toc34, or both proteins (Toc75/34 PL). Half of the reaction was further treated with thermolysin to reveal the amount of OEP14 inserted. Lane 9, 20% [35S]OEP14 added to each reaction.
(B) Insertion of AtPex14p into various proteoliposomes. [35S]AtPex14p was incubated with liposomes or various proteoliposomes as in (A). Half of the reaction was further extracted with 0.1 M Na2CO3 to reveal the amount of AtPex14p inserted. Lane 1, 20% [35S]AtPex14p added to each reaction.
(C) Quantitative analysis of insertion (thermolysin-resistant fragment of OEP14 and alkaline-extracted pellet fraction of AtPex14p) from two experiments, including those presented in (A) and (B) with error bars.
DISCUSSION
Spontaneous insertion has been generally assumed to be the import mechanism for most chloroplast outer-membrane proteins (Bruce, 1998; Keegstra and Cline, 1999; Schleiff and Klosgen, 2001). However, the identification of targeting signals and the requirements for specific proteinaceous insertion sites suggested otherwise (Li and Chen, 1996; Tu and Li, 2000; Lee et al., 2001). In this report, we further showed that OEP14 was associated with translocon components during import. However, Toc159, Toc34, and Tic110 were not necessary for OEP14 import. Copurification of these components with OEP14-His6 is most likely because of preassembly of the translocon complex (Akita et al., 1997; Kouranov et al., 1998). On the other hand, Toc75 was in close proximity to the insertion signal of OEP14. We have previously shown that OEP14 import requires a trypsin-sensitive and thermolysin-resistant component (Tu and Li, 2000). Toc75 is trypsin sensitive and thermolysin resistant (Schnell et al., 1994; Tu and Li, 2000). Finally, proteoliposomes containing purified Toc75 mediated membrane insertion of OEP14 with a high efficiency. Therefore, we conclude that Toc75 is the major component mediating OEP14 insertion. Interestingly, a bacterial homolog of Toc75, Neisseria meningitidis Omp85, recently has been shown to be important for bacterial outer-membrane protein assembly (Voulhoux et al., 2003). Therefore, the role of Toc75 in mediating the insertion of proteins into the chloroplast outer membrane could be an evolutionarily conserved function.
In mitochondria, outer-membrane proteins are also synthesized at their mature size without cleavable targeting signals. Their import is also generally insensitive to protease pretreatment of mitochondria (Neupert, 1997). However, it has been shown that import of the outer-membrane protein TOM20 is inhibited by antibodies against TOM40, which is the insertion pore of the TOM complex for importing mitochondrial matrix proteins (Schneider et al., 1991). The results suggest that TOM20 bypasses the surface receptors, which are required for matrix protein import, and inserts directly into TOM40. Furthermore, import of other mitochondrial outer-membrane proteins, including porin, TOM6, TOM7, and TOM40 itself, has also been shown to require the TOM core complex (Rapaport and Neupert, 1999; Dembowski et al., 2001; Krimmer et al., 2001). Taken together, these results suggest that the import pathways of mitochondrial outer-membrane proteins and interior proteins converge at the general insertion pore, especially at TOM40. Similar phenomena have also been observed in bacteria. The E. coli SRP- and Sec-dependent export pathways converge at the SecYEG translocon (Valent et al., 1998). Here, we provide evidence to show that a similar situation occurs in chloroplasts. Toc75 not only functions as the translocation channel for chloroplast interior proteins but also mediates the insertion of outer-membrane proteins.
Even though both groups of proteins use Toc75 for import, this does not necessarily mean that they bind to identical sites on Toc75. Similar to the situation in mitochondria, the membrane insertion signals of chloroplast outer-membrane proteins are clearly distinct from the stroma-targeting transit peptides. Therefore, it is possible that two different sites on Toc75 are involved in binding the two different groups of signals. This may explain why a higher concentration of OEP14 is needed to achieve the same competition effect as that of an interior-targeted precursor protein (Tranel et al., 1995). Binding of outer-membrane proteins to Toc75 may result only in a steric hindrance effect, rather than a direct competition on the binding of interior-targeted precursor proteins to Toc75. Recently, a single-point mutation in the mitochondrial TOM40 channel has been shown to affect only import of inner-membrane carrier proteins but not matrix or outer-membrane proteins (Gabriel et al., 2003). Therefore, TOM40 appears to have different regions for interacting with proteins of different submitochondrial locations.
It has been shown that ∼50% of Toc75 molecules in the outer membrane are not associated with Toc34 and Toc159 (Kouranov et al., 1998). Because OEP14 insertion only requires Toc75 but not other Toc components identified so far, it is possible that OEP14 not only interacts with Toc75 in the Toc complex but also with free Toc75 not assembled into a Toc complex (Figure 7). This may explain why OEP14 insertion was not competed by prRBCS (Figure 1A). Even when all the Toc complex-assembled Toc75 is occupied by prRBCS, the remaining free Toc75 is still sufficient to mediate the insertion of OEP14 added to an in vitro import experiment. However, we could not exclude the possibility that prRBCS failed to compete with OEP14 because prRBCS was solubilized in urea and therefore was not in its native conformation, or the concentration of prRBCS used was not high enough.
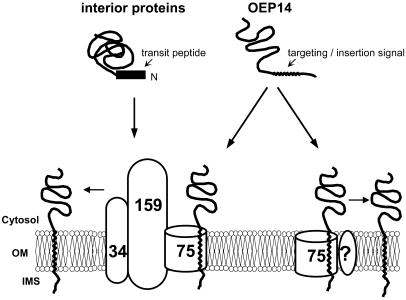
Figure 7.
A Model for OEP14 Insertion.
Whereas interior-targeted precursor proteins use the Toc complex for import, OEP14 may use Toc75 by itself (or associated with some yet-unidentified components designated by a question mark), or Toc75 in the Toc complex, for its insertion into the chloroplast outer membrane. OM, outer membrane; IMS, intermembrane space.
Biogenesis pathways for membrane proteins can usually be divided into three stages: (1) targeting to the membrane, (2) insertion into the membrane, and (3) assembly into the final, lipid-embedded functional structure (Rapaport, 2002). For example, for the biogenesis of most mitochondrial outer-membrane proteins, the second stage is the insertion of these proteins into the TOM40 channel. The third stage will be the release of the proteins from the channel to the surrounding lipid bilayer (Rapaport, 2002). Because nothing is known about the function of OEP14, we can only divide OEP14 import into two stages: initial contact with the membrane surface and insertion. We have observed the same cross-linking pattern with the Toc components when OEP14 import was performed for 5 or 15 min, except that the amount of Toc proteins copurified with OEP14 was higher when import was performed for 15 min (Figure 2). Because endogenous OEP14 is not part of the Toc complex, retention of OEP14 in the Toc75 channel 15 min after import suggests that lateral release of OEP14 into the surrounding lipid bilayer is a rate-limiting step in OEP14 biogenesis.
OEP14 can still bind to the surface of chloroplasts even when Toc75 is almost completely digested by trypsin (Tu and Li, 2000). It is possible that OEP14 nonspecifically sticks to the chloroplast surface when Toc75 is impaired by trypsin digestion. It is also possible that OEP14 can specifically bind to lipid components of the outer membrane. Chloroplast membranes contain several unique lipids, such as digalactosyl diacylglycerol, monogalactosyl diacylglycerol, and sulfolipids (Douce and Joyard, 1990). Although it has been shown that neither digalactosyl diacylglycerol nor monogalactosyl diacylglycerol is likely to be involved in OEP14 import (Chen and Li, 1998; Tu and Li, 2000), it is still possible that sulfolipids or other lipids may play a role in the binding of OEP14 to the outer membrane. We propose that proteinaceous insertion sites, like Toc75, provide the insertion with the high efficiency and specificity required in vivo. Further work is needed to determine if there are other proteins or lipids involved in OEP14 insertion.
METHODS
Plasmid Construction and Sources of Antibodies
OEP14 mutant V8C was generated by PCR-based site-directed mutagenesis on plasmid pET22b-OEP14 (Tu and Li, 2000) using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). To construct pET22b-Toc75, a fragment containing the entire coding region of pea (Pisum sativum) Toc75 precursor was amplified from p214 (Tranel et al., 1995) by PCR using primers that added an NdeI site to the N terminus and a SacI site to the C terminus of the PCR fragment. The PCR fragment was subcloned into the NdeI/SacI site of pET22b (Novagen, Madison, WI). To construct pET22b-AtOEP7, the AtOEP7 coding region was directly amplified from Arabidopsis thaliana genomic DNA using primers that added an NdeI site to the N terminus and an EcoRI site to the C terminus of the PCR fragment. The PCR fragment was subcloned into the NdeI/EcoRI site of pET22b. For overexpression of atToc159M, the corresponding coding region was excised with PstI and EcoRI from Arabidopsis EST clone H9H12. The PstI/EcoRI fragment, which encodes the C-terminal 54-kD region of atToc159 from residues 1224 to 1503, was cloned into the PstI/EcoRI site of pRSET-B (Invitrogen, Carlsbad, CA). Antibodies against Toc75, Toc34, Toc159, Tic110, IEP21, and OEP14 were generated against Escherichia coli overexpressed and purified full-length pea Toc75 precursor (described above), pea Toc34(1-258)His6 (Sun et al., 2002), atToc159(1224-1503) (M domain described above), Arabidopsis Tic110 residue 431 to 1016, pea inner membrane protein IEP21 (Kouranov et al., 1998), and pea OEP14 with an N-terminal His6 tag, respectively. OEP14-related proteins were all soluble when overexpressed in E. coli and were purified as described (Tu and Li, 2000). Arabidopsis Tic110(431-1016), atToc159M, and pea Toc75 precursor were purified as inclusion bodies as described (Perry and Keegstra, 1994) except proteins were solubilized in 8 M urea instead of 6 M guanidine-HCl.
Import Competition and Import into OMVs
Chloroplasts were isolated from 9- to 11-d-old pea (P. sativum cv Little Marvel) seedlings as described (Perry et al., 1991). Competition of prRBCS by atToc159M and OEP14-His6 under standard import conditions were performed as described (Tu and Li, 2000). For competition of OEP14 by prRBCS, prRBCS was overexpressed and purified from E. coli as described (Perry and Keegstra, 1994) except the inclusion bodies were solubilized by 8 M urea in 50 mM Hepes-KOH, pH 7.6, 10 mM MgCl2, and 2 mM DTT. All reactions contained the same concentration of urea (for atToc159M and prRBCS) or imidazole (for OEP14-His6) as in the reaction with the highest amount of competitors. For OEP14-His6 competition with prRBCS under different concentration of ATP, in vitro–translated [35S]prRBCS was filtered through a Sepharose G-25 column (Olsen et al., 1989), and isolated chloroplasts were incubated on ice in the dark for 2 h to deplete ATP. Import was initiated by adding 50 μL of chloroplasts to a 100-μL reaction mixture containing 5 μL of [35S]prRBCS and various amounts of cold OEP14-His6 and ATP in import buffer (50 mM Hepes-KOH, pH 8.0, and 300 mM sorbitol). Import was performed for 15 min at room temperature under a dim green safelight.
OMVs were isolated by differential centrifugation on a linear sucrose gradient as described (Keegstra and Yousif, 1988). For OEP14-His6 import into OMVs, a 100-μL reaction was performed by incubating OMVs (40 μg of proteins) with 5 μL of purified [35S]OEP14-His6 (Tu and Li, 2000) or in vitro–translated [35S]OEP14 in import buffer at room temperature for 15 min. The reaction was terminated by adding cold import buffer. The OMVs were pelleted by centrifugation at 100,000g for 45 min and resuspended in import buffer. Thermolysin posttreatment was performed by resuspending the vesicles in 200 μg/mL of thermolysin and 1 mM CaCl2 and incubating at 4°C for 30 min. The vesicles were reisolated through a 0.23 M sucrose cushion.
Cross-Linking and Affinity Purification
For cross-linking with DSP (Pierce, Rockford, IL), 800 μL of chloroplasts were incubated with 40 μg of OEP14-His6 or 144 μg of Toc34(1-258)His6, and import buffer was added to a final volume of 2.4 mL. No ATP was added to the reaction. After import at room temperature for 5 or 15 min, intact chloroplasts were reisolated, DSP was added to a final concentration of 0.25 mM, and cross-linking was performed at 4°C for 15 min. The reaction was terminated by adding Gly to a final concentration of 50 mM and further incubated at 4°C for 15 min to quench the free DSP. Chloroplasts were reisolated, washed with import buffer, and lysed in 2 mL of hypotonic buffer (25 mM Hepes-KOH, pH 8.0, and 4 mM MgCl2). The total membrane fraction was collected by centrifugation at 100,000g for 45 min. Solubilization was performed by resuspending the membranes with 1 mL of solubilization buffer (25 mM Hepes-KOH, pH 8.0, 200 mM KCl, 2 mM imidazole, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM pepstatin, and 1 mM leupeptin) and incubated at 4°C for 10 min, followed by centrifugation at 100,000g for 5 min. The supernatant was diluted with an equal volume of solubilization buffer without Triton X-100, mixed with 50 μL of TALON resin (Clontech, Palo Alto, CA), and incubated overnight at 4°C with constant shaking. The resin was washed four times with 1 mL of solubilization buffer and one time with solubilization buffer containing 5 mM imidazole. The bound proteins were eluted with 60 μL of elution buffer (solubilization buffer plus 200 mM imidazole). Samples were analyzed by SDS-PAGE, blotted onto polyvinylidene difluoride membranes, and detected by horseradish peroxidase–conjugated secondary antibody using the SuperSignal West Femto chemiluminescence kit (Pierce). For each antibody, all samples were run on the same gel and developed for the same amount of time.
[125I]-APDP was custom labeled by NEN (Boston, MA). V8C proteins (25 nmol) were overexpressed and purified from E. coli and were reduced by incubating the proteins with 2% β-mercaptoethanol at 37°C for 1.5 h and filtered through a 1-mL Sepharose G-25 column to remove the β-mercaptoethanol. The filtered protein was incubated with 230 μCi of [125I]-APDP at 4°C in the dark for 2.5 h, and the reaction mixture was filtered through the Sepharose G-25 column to remove free [125I]-APDP. Chloroplasts (6 mg of chlorophyll) were incubated with 40 μg of V8C-[125I]-ADP, and import buffer was added to a final volume of 18 mL. After import at room temperature in the dark for 15 min, the chloroplasts were transferred to a 100-mm Petri dish plate and irradiated from 1.5 cm above with a hand-held UV transilluminator (Spectroline, Westbury, NY) at 254 nm for 20 min with gentle shaking and then collected by centrifugation at 3,000g for 3 min. Chloroplasts were lysed hypertonically, and total envelope membranes were collected as described (Keegstra and Yousif, 1988). Envelope membranes were resuspended in 1 mL of TE buffer (10 mM Tricine-HCl, pH 7.5, and 2 mM EDTA) containing 2% β-mercaptoethanol, incubated at room temperature for 15 min to cleave the cross-linker, recovered by ultracentrifugation, and washed with 1 mL of TE buffer. For immunoprecipition, the envelopes were solubilized with 0.5 mL of IP buffer (25 mM Hepes-KOH, pH 8.0, 50 mM KCl, 2% Triton X-100, 10% glycerol, 1 mM PMSF, 1 mM pepstatin, and 1 mM leupeptin) at 4°C for 10 minutes, followed by centrifugation at 100,000g for 5 minutes. The supernatant was diluted with 0.5 mL of HKG buffer (25 mM Hepes-KOH, pH 8.0, 50 mM KCl, and 10% glycerol), mixed with 20 μL of antiserum at 4°C for 2 h, followed by incubating with 30 μL of protein A resin (Pierce) at 4°C overnight with constant shaking. The resin was washed with 0.5 mL of IP buffer (1% Triton X-100) four times and directly eluted with SDS-PAGE sample buffer.
Insertion of OEP14 into Proteoliposomes
Reconstitution of pea Toc75 and pea Toc34 into proteoliposomes was performed as described (Wallas et al., 2003). Ten microliters of liposomes or proteoliposome were mixed with 12 μL of in vitro–translated [35S]OEP14 or AtPex14p in import buffer in a final volume of 150 μL. Import was performed at 26°C for 15 min. The reaction was stopped by adding 850 μL of ice-cold import buffer and divided into two 500-μL aliquots and pelleted. For experiment involving OEP14, the proteoliposomes were resuspended in import buffer either without or with 50 μg/mL of thermolysin and incubated for 30 min on ice in the dark. The thermolysin treatment was terminated by addition of EDTA, and the proteoliposomes were collected by centrifugation. For experiments involving AtPex14p, the proteoliposomes were resuspended in import buffer (the bound portion) or 0.1 M Na2CO3, pH 11.5, incubated on ice for 15 min, and then collected by centrifugation. The supernatant of the Na2CO3-treated liposomes was saved. The pellets from both treatments were resuspended in import buffer. Proteins in all samples were precipitated with 80% acetone and analyzed by SDS-PAGE.
Miscellaneous Procedures
Total plant extracts from various Arabidopsis mutants were prepared from 14-d-old seedlings grown on MS medium by homogenization in extraction buffer (300 mM Tris-HCl, pH 8.5, 8% SDS, 1 mM EDTA, 1 mM PMSF, 1 mM pepstatin, and 1 mM leupeptin). Samples were analyzed by SDS-PAGE, blotted onto polyvinylidene difluoride membranes, and detected by horseradish peroxidase–conjugated secondary antibody. All electrophoresis was performed as described (Tu and Li, 2000). For analysis of cross-linked complexes, the Mops running buffer was used instead of the Mes running buffer. Dried gels were used for quantification using the PhophorImager SP (Molecular Dynamics, Sunnyvale, CA).
Acknowledgments
We thank Ken Keegstra and Diane Jackson-Constan for the atToc34 knockout mutant line, Laura Olsen for the AtPex14p cDNA clone, and ABRC for the EST clone H9H12. This work was supported by grants to H.-m.L. from the National Science Council (NSC 90-2321-B001-006) and the Academia Sinica of Taiwan and to D.J.S. from the U.S. National Institutes of Health (GM61893).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hsou-min Li (mbhmli@ccvax.sinica.edu.tw).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023952.
References
- Akita, M., Nielsen, E., and Keegstra, K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, J., Chen, K., Hiltbunner, A., Wehrli, E., Eugster, M., Schnell, D., and Kessler, F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207. [DOI] [PubMed] [Google Scholar]
- Bolter, B., Soll, J., Hill, K., Hemmler, R., and Wagner, R. (1999). A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J. 18, 5505–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, B.D. (1998). The role of lipids in plastid protein transport. Plant Mol. Biol. 38, 223–246. [PubMed] [Google Scholar]
- Chen, L.J., and Li, H.M. (1998). A mutant deficient in the plastid lipid DGD is defective in protein import into chloroplasts. Plant J. 16, 33–39. [DOI] [PubMed] [Google Scholar]
- Chen, X., and Schnell, D.J. (1999). Protein import into chloroplasts. Trends Cell Biol. 9, 222–227. [DOI] [PubMed] [Google Scholar]
- Constan, D., Patel, R., Keegstra, K., and Jarvis, P. (2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38, 93–106. [DOI] [PubMed] [Google Scholar]
- Dembowski, M., Kunkele, K.-P., Nargang, F.E., Neupert, W., and Rapaport, D. (2001). Assembly of Tom6 and Tom7 into the TOM core complex of Neurospora crassa. J. Biol. Chem. 276, 17679–17685. [DOI] [PubMed] [Google Scholar]
- Douce, R., and Joyard, J. (1990). Biochemistry and function of the plastid envelope. Annu. Rev. Cell Biol. 6, 173–216. [DOI] [PubMed] [Google Scholar]
- Fischer, K., Weber, A., Arbinger, B., Brink, S., Eckerskorn, C., and Flugge, U.-I. (1994). The 24 kDa outer envelope membrane protein from spinach chloroplasts: Molecular cloning, in vivo expression and import pathway of a protein with unusual properties. Plant Mol. Biol. 25, 167–177. [DOI] [PubMed] [Google Scholar]
- Gabriel, K., Egan, B., and Lithgow, T. (2003). Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22, 2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T., and Nishimura, M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19, 5701–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah, S.C., Hill, K., Wagner, R., Schlicher, T., and Soll, J. (1997). Reconstitution of a chloroplast protein import channel. EMBO J. 16, 7351–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, T., Li, M., Alvarez-Huerta, M., Kessler, F., and Schnell, D.J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278, 38617–38627. [DOI] [PubMed] [Google Scholar]
- Jackson-Constan, D., and Keegstra, K. (2001). Arabidopsis genes encoding components of the chloroplast protein import apparatus. Plant Physiol. 125, 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, P., Chen, L.J., Li, H.M., Peto, C.A., Fankhauser, C., and Chory, J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Froehlich, J.E. (1999). Protein import into chloroplasts. Curr. Opin. Plant Biol. 2, 471–476. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Yousif, A.E. (1988). Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 118, 316–325. [Google Scholar]
- Kessler, F., Blobel, G., Patel, H.A., and Schnell, D.J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Ko, K., Bornemisza, O., Kourtz, L., Ko, Z.W., Plaxton, W.C., and Cashmore, A.R. (1992). Isolation and characterization of a cDNA clone encoding a cognate 70-kDa heat shock protein of the chloroplast envelope. J. Biol. Chem. 267, 2986–2993. [PubMed] [Google Scholar]
- Kouranov, A., Chen, X., Fuks, B., and Schnell, D.J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov, A., and Schnell, D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139, 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmer, T., Rapaport, D., Ryan, M.T., Meisinger, C., Kassenbrock, K.K., Blachly-Dyson, E., Forte, M., Douglas, M.G., Neupert, W., Nargang, F.E., and Pfanner, N. (2001). Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 152, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., Kim, D.H., Kim, Y.-W., and Hwang, I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.M., and Chen, L.J. (1996). Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell 8, 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.M., and Chen, L.J. (1997). A novel chloroplastic outer membrane-targeting signal that functions at both termini of passenger polypeptides. J. Biol. Chem. 272, 10968–10974. [PubMed] [Google Scholar]
- Li, H.M., Moore, T., and Keegstra, K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y., Kouranov, A., LaSala, S.E., and Schnell, D.J. (1996). Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckel, E., and Soll, J. (1996). A protein import receptor of chloroplasts is inserted into the outer envelope membrane by a novel pathway. J. Biol. Chem. 271, 23846–23852. [DOI] [PubMed] [Google Scholar]
- Neupert, W. (1997). Protein import into mitochondria. Annu. Rev. Biochem. 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Nielsen, E., Akita, M., Davila-Aponte, J., and Keegstra, K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, L.J., Theg, S.M., Selman, B.R., and Keegstra, K. (1989). ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 246, 6724–6729. [PubMed] [Google Scholar]
- Perry, S.E., and Keegstra, K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, S.E., Li, H.M., and Keegstra, K. (1991). In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol. 34, 327–344. [DOI] [PubMed] [Google Scholar]
- Qbadou, S., Tien, R., Soll, J., and Schleiff, E. (2003). Membrane insertion of the chloroplast outer envelope protein, Toc34: Constrains for insertion and topology. J. Cell Sci. 116, 837–846. [DOI] [PubMed] [Google Scholar]
- Rapaport, D. (2002). Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 27, 191–197. [DOI] [PubMed] [Google Scholar]
- Rapaport, D., and Neupert, W. (1999). Biogenesis of Tom40, core component of the Tom complex of mitochondria. J. Cell Biol. 146, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink, W.A., Schnell, D.J., and Weisbeek, P.J. (2000). The transit sequence of ferredoxin contains different domains for translocation across the outer and inner membrane of the chloroplast envelope. J. Biol. Chem. 275, 10265–10271. [DOI] [PubMed] [Google Scholar]
- Reumann, S., Davila-Aponte, J., and Keegstra, K. (1999). The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: Identification of a cyanobacterial homolog. Proc. Natl. Acad. Sci. USA 96, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, M., Fischer, K., Flugge, U.-I., and Soll, J. (1990). Sequence analysis and protein import studies of an outer chloroplast envelope polypeptide. Proc. Natl. Acad. Sci. USA 87, 5778–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff, E., and Klosgen, R.B. (2001). Without a little help from “my” friend: Direct insertion of proteins into chloroplast membranes? Biochim. Biophys. Acta 1541, 22–33. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., and Soll, J. (2000). Travelling of proteins through membranes: Translocation into chloroplasts. Planta 211, 449–456. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., Tien, R., Salomon, M., and Soll, J. (2001). Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell 12, 4090–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, H., Sollner, T., Dietmeier, K., Eckerskorn, C., Lottspeich, F., Trulzsch, B., Neupert, W., and Pfanner, N. (1991). Targeting of the master receptor MOM19 to mitochondria. Science 254, 1659–1662. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J., Blobel, G., Keegstra, K., Kessler, F., Ko, K., and Soll, J. (1997). A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol. 7, 303–304. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J., Kessler, F., and Blobel, G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., Waegemann, K., and Soll, J. (1995). A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 7, 401–411. [DOI] [PubMed] [Google Scholar]
- Smith, M.D., Hiltbrunner, A., Kessler, F., and Schnell, D.J. (2002). The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 159, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.J., Forouhar, F., Li, H.M., Tu, S.L., Yeh, Y.H., Kao, S., Shr, H.L., Chou, C.C., Chen, C., and Hsiao, C.D. (2002). Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nat. Struct. Biol. 9, 95–100. [DOI] [PubMed] [Google Scholar]
- Sveshnikova, N., Soll, J., and Schleiff, E. (2000). TOC 34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl. Acad. Sci. USA 97, 4973–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg, S.M., Bauerle, C., Olsen, L.J., Selman, B.R., and Keegstra, K. (1989). Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264, 6730–6736. [PubMed] [Google Scholar]
- Tranel, P.J., Froehlich, J., Goyal, A., and Keegstra, K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14, 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, S.L., and Li, H.M. (2000). Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell 12, 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, Q.A., Scotti, P.A., High, S., de Gier, J.-W.L., von Heijne, G., Lentzen, G., Wintermeyer, W., Oudega, B., and Luirink, J. (1998). The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17, 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux, R., Bos, M., Geurtsen, J., Mols, M., and Tommassen, J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. [DOI] [PubMed] [Google Scholar]
- Wallas, T., Smith, M., Sanchez-Nieto, S., and Schnell, D. (2003). The roles of Toc34 and Toc75 in targeting the Toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 278, 44289–44297. [DOI] [PubMed] [Google Scholar]
- Wu, C., and Ko, K. (1993). Identification of an uncleavable targeting signal in the 70-kilodalton spinach chloroplast outer envelope membrane protein. J. Biol. Chem. 268, 19384–19391. [PubMed] [Google Scholar]