Abstract
The utility of cytostatic antiangiogenic agents (AA) in cancer chemotherapy lies in their combination with cytotoxic chemotherapeutic agents. Clinical combinations of AA with microtubule targeting agents (MTAs) have been particularly successful. The discovery, synthesis and biological evaluations of a series of 7-benzyl-N-substituted-pyrrolo[3,2-d]pyrimidin-4-amines are reported. Novel compounds which inhibit proangiogenic receptor tyrosine kinases (RTKs) including vascular endothelial growth factor receptor-2 (VEGFR-2), platelet-derived growth factor receptor-β (PDGFR-β) and epidermal growth factor receptor (EGFR), along with microtubule targeting in single molecules are described. These compounds also inhibited blood vessel formation in the chicken chorioallantoic membrane (CAM) assay, and some potently inhibited tubulin assembly (with activity comparable to that of combretastatin A-4 (CA)). In addition, some of the analogs circumvent the most clinically relevant tumor resistance mechanisms (P-glycoprotein and β-III tubulin expression) to microtubule targeting agents (MTA). These MTAs bind at the colchicine site on tubulin. Two analogs displayed two to three digit nanomolar GI50 values across the entire NCI 60 tumor cell panel and one of these, compound 7, freely water soluble as its HCl salt, afforded excellent in vivo antitumor activity against an orthotopic triple negative 4T1 breast cancer model and was superior to doxorubicin.
Keywords: antiangiogenic, combination chemotherapy, receptor tyrosine kinase inhibitors, multitargeted agents, microtubule targeting agents
Graphical abstract
1. Introduction
Angiogenesis is the process of formation of new blood vessels from pre-existing vasculature. When a tumor grows beyond 2 mm3 it requires nutrients and oxygen for maintenance and growth and initiates the process of acquiring additional blood supply.1 The activation of receptor tyrosine kinases (RTKs) regulates signal transduction from the extracellular domain of endothelial cells to the nucleus2 and represents the most important factor for triggering angiogenesis. Under hypoxic conditions, the tumor secretes several proangiogenic factors.1 The principal mediator of angiogenesis is the proangiogenic vascular endothelial growth factor (VEGF) and its RTK, VEGFR-2.3 Rapid regrowth of tumor vasculature after the removal of VEGFR-2 inhibitors from the treatment regimen attests to their cytostatic mechanism of action and also promotes metastases.4, 5 Thus, VEGFR-2 inhibitors are known to cause the regression of impaired tumor blood vessels but transiently normalize the surviving vessels, which are less leaky and are more like normal blood vessels. VEGFR-2 inhibitors transiently increase blood flow in the surviving vasculature.6 Administration of a cytotoxic agent during this transient phase of tumor vasculature “normalization” provides improved delivery of the drug to the tumor via the surviving vasculature. 6, 7 In combination chemotherapy, dosing separate antiangiogenic and cytotoxic agents may miss the timing window of the transient normalization.8 Single molecular entities with both antiangiogenic and cytotoxic attributes allow the cytotoxicity to be manifested as soon as the antiangiogenic effect and vasculature normalization occur. Such single agents could circumvent pharmacokinetic problems of multiple agents, avoid drug-drug interactions, and could be used at lower doses to minimize toxicities and tumor cell resistance than two or more agents dosed separately in combination chemotherapy.
Microtubule targeting agents (MTAs) are among the most successful anticancer agents in clinical use. These compounds produce tumor cytotoxic effects by interfering with microtubule dynamics or by stabilizing or destabilizing tubulin polymerization.9 Several clinical trials are ongoing for a variety of cancers including ovarian, breast, and lung, among others, and involve FDA approved receptor tyrosine kinase inhibitors (RTKIs), as well as those in development, and cytotoxic MTAs. These combinations include paclitaxel, docetaxel, or vinorelbine with RTKIs bevacizumab, vandetanib, gefitinib, sorafenib, dasatinib or famitinib among others.10
It has been our interest to design single entities with RTKI attributes and cytotoxic mechanisms of action.11-17 Our choice of MTAs as the mechanism for the cytotoxic component was predicated, in part, on the finding that half of all human tumors have mutations in the p53 gene and p53 status effects vulnerability of tumor cells for cell cycle arrest. The most effective drugs in cell lines with p53 gene mutations are MTAs.18, 19 Additionally, there is clinical evidence for the success of RTKIs in combination with MTAs.20-22
Development of resistance to chemotherapeutic agents is one of the major hallmarks of cancer.23 Clinically significant mechanisms of resistance to MTAs, especially to paclitaxel, are overexpression of the multidrug resistance protein P-glycoprotein (Pgp) and the β-tubulin isoform βIII.24 With RTKIs, resistance to VEGFR-2 inhibition is associated with increased platelet-derived growth factor receptor-β (PDGFR-β) expression in tumor endothelial cells, increased recruitment of pericytes to tumor vasculature, and increases in other proangiogenic factors.25 Similarly, epidermal growth factor receptor (EGFR) inhibition can lead to tumor VEGFR-2 up-regulation, and this subsequently promotes tumor growth signaling independent of EGFR and thus contributes to tumor resistance of EGFR inhibitors.26, 27 The effect of EGFR inhibition can also be partially overcome by activation of PDGFR-β.26, 28 Due to the complexity of angiogenic pathways, disrupting a single pathway of angiogenesis may not result in significant clinical success. Multiple RTKs are co-activated in tumors and redundant inputs drive and maintain downstream signaling, thereby limiting the efficacy of therapies targeting single RTKs.26- 28 Moreover, high intratumoral heterogeneity has been reported with different subpopulations producing distinct growth factors. 29-31 Thus, targeting a single RTK could be less effective due to subpopulations of cells that are either not affected by the drug mechanism and possess or acquire a greater drug resistance.32 Hence, targeting multiple angiokinases such as VEGFR-2, PDGFR-β and/or EGFR maximizes the proportion of angiogenic signaling that is effectively targeted. If such a multiple angiokinase inhibitor, which targets the principal angiogenic growth factor receptors, could be structurally designed to include cytotoxic activity by incorporating the effects of a MTA in single molecular entities, multi-agent combination chemotherapy potential in single molecules could be realized. Such agents would be expected to thwart RTK resistance mechanisms involving redundant angiogenic pathways and also provide a cytotoxic MTA to effectively kill tumor cells.
We recently reported the preclinical evaluation of 7-benzyl-N-substituted-pyrrolo[3,2-d]pyrimidin-4-amines as MTAs with VEGFR-2 inhibition.15, 33 In this study, we extend our original reports to the design, discovery and preclinical evaluation of 7-benzyl-N-substituted-pyrrolo[3,2-d]pyrimidin-4-amines with MTA and triple angiokinase (VEGFR-2, PDGFR-β, EGFR) inhibitory activities in single molecules.
2. Rationale
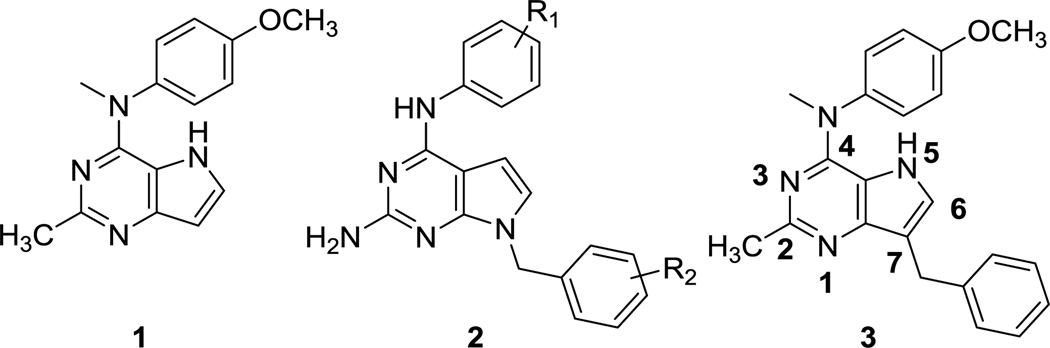
We previously reported that compound 1 (Fig. 1) containing the pyrrolo[3,2-d]pyrimidine scaffold afforded potent MTAs.34 The on-going clinical trials of antiangiogenic agents in combination with tumor-cytotoxic MTAs prompted the design and development of single agents with both VEGFR-2 and tubulin inhibitory activities. As an initial study we appended the 7-benzyl group, which dictates RTK inhibitory activity in substituted 7-benzyl pyrrolo[2,3-d]pyrimidines of general structure 235 (Fig. 1), to the pyrrolo[3,2-d]pyrimidine scaffold of 1. Our hybrid design to structurally engineer VEGFR-2 inhibitory activity into our existing MTA 1 afforded 3, a single agent with potent activities against both VEGFR-2 and tubulin assembly.15 Compound 3 reduced tumor size and vascularity in two flank xenograft models [the BLBC MDA-MB-435 and U251 glioma models] and in a 4T1 triple negative breast orthotopic allograft model, without overt toxicity to animals.15 A structure-activity study suggested that the methyl group attached to the nitrogen bridge in the 4-position and the 4’-methoxy group in 3 were crucial for inhibition of tubulin and removal of either of these moieties resulted in a complete loss of the ability of the compound to inhibit tubulin assembly and also decreased VEGFR-2 inhibition in cells.15
Figure 1.
Lead compounds
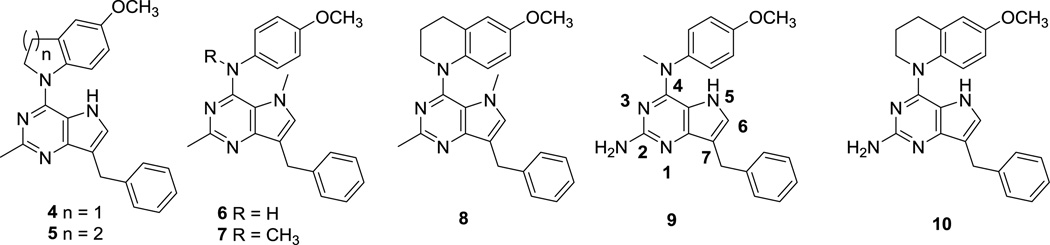
In this study we explore the effect of conformational changes in compound 3 on the RTK and tubulin inhibitory properties to extend our previous studies of agents which inhibit multiple RTKs including VEGFR-2, PDGFR-β, and EGFR.11, 36, 37 Thus, it was of interest to explore the effect of structural variations in 1 on activity against the RTKs VEGFR-2, PDGFR-β and EGFR, in addition to cytotoxic MT effects with the goal of identifying single molecules as MTAs with multiple angiokinase inhibitory potential. The clinical successes of the triple angiokinase inhibitor nintedanib in combination with docetaxel38–41 makes a compelling argument for triple angiokinase inhibitors with MTAs in single molecular entities. Compounds 4 – 8 (Fig. 2) were designed as conformationally restricted analogs of 3. Compounds 4 and 5 incorporate the bicyclic 6-methoxy-tetrahydroquinoline or the bicyclic 5-methoxy-dihydroindole respectively onto the 4-position of the 2-methyl pyrrolo[3,2-d]pyrimidine. Compared to the N-methylaniline, these bicyclics incorporated on the 4-position restrict the rotation of the phenyl ring, thus affording more rigid structures than 3 while maintaining the phenyl and alkyl substitutions on the N4 as in 3. Compounds 6 −8 were designed to determine whether the addition of a 5-methyl group is conducive to microtubule targeting and RTKI activities. The 5-methyl group could allow additional hydrophobic interactions with the targets and further restrict rotation around the N-phenyl bond, thus providing further conformational rigidity which could increase potency.
Figure 2.
Structures of compounds 4–10
Compounds 9 and 10 were designed to study the effect of a 2-amino group on multiple RTK inhibition. We42 previously reported that a 2-amino group on fused pyrimidine ring systems improves hinge region binding in the ATP-binding site of RTKs and provides multiple-RTK inhibition. Compounds 9 and 10 replace the 2-methyl group in the dual acting, VEGFR-2 inhibiting, MTAs 3 and 5, respectively, with a 2-amino group that could potentially increase binding to the hinge region of EGFR and/or PDGFR-β via hydrogen bonds, without loss of MT and VEGFR-2 inhibitory activities.
To further support our structural modifications, we carried out molecular modeling studies using MOE 2015.10, in which the proposed compounds 3–10 were docked in the x-ray crystal structures of tubulin, 43 VEGFR-244 and EGFR45 and a homology model of PDGFR-β.11
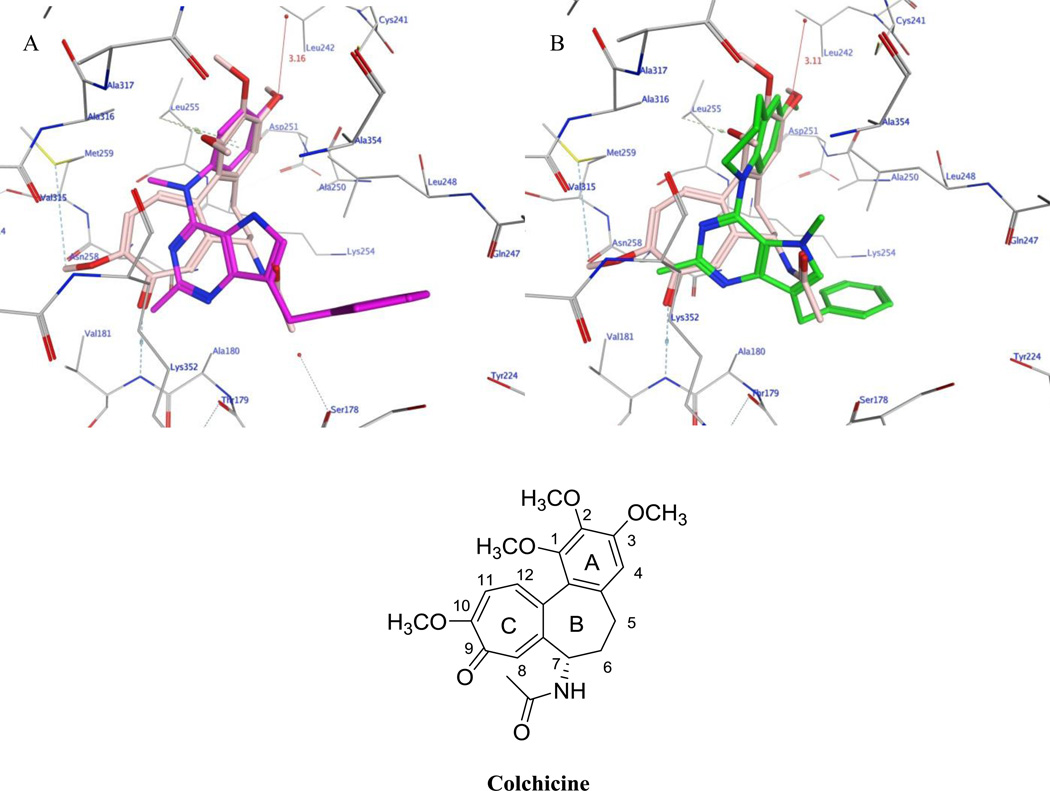
Compounds 3–10 were docked in the x-ray crystal structure of the colchicine site in tubulin (PDB: 4O2B, 2.3 Å),43 using the parameters specified in the experimental section. Multiple low energy conformations were obtained on docking. As representative examples, figure 3 A shows the docked conformation of 3 (magenta), superimposed on the co-crystallized ligand, colchicine (pink). The pyrrolo[3,2-d]pyrimidine scaffold of 3 forms hydrophobic interactions with Alaα180, Valα181, Leuβ248, Asnβ258 and Lysβ352 and partially occupies the region where the C-ring of colchicine binds. 43 The benzylic group lies in the pocket lined by residues Serα178, Glnβ247, Leuβ248 and Lysβ352. The N4-methyl interacts with Leuβ255, Asnβ258, Metβ259 and Alaβ316. The phenyl group of 3 superimposes on the A ring of colchicine and interacts with Cysβ241, Leuβ248, Alaβ250, and Leuβ255 and the 4’-methoxy interacts with Cysβ241, Leuβ242 and Leuβ255. In addition, the oxygen atom of the 4’-methoxy group lies within H-bonding distance with a water molecule (3.16 Å for 3 and 3.11 Å for 8, figure 3B) present in the crystal structure in the vicinity of Cysβ241. The best docked pose of 3 had a score of −8.82 kcal/mol. Figure 3B shows the docked pose of 8 (green), superimposed on the crystallized ligand colchicine (pink). It’s mode of binding in tubulin was similar to that of 3 and had a better docked score of −9.50 kcal/mol compared to 3. Compounds 4–10, when similarly docked in the colchicine site of tubulin, had scores that ranged between −8.82 and −9.50 kcal/mol and exhibited similar modes of binding. These docking scores suggest that the designed analogs 4–10 should have attributes of MTAs comparable to or better than those of 3.
Figure 3.
A) Superimposition of the docked pose of 3 (magenta) and colchicine (pink) in tubulin (PDB ID: 4O2B).43B) Superimposition of docked pose of 8 (green) and colchicine (pink) in tubulin (PDB ID: 4O2B).43
Since the design objective was to provide multiple RTKIs as well as MTAs in single molecules, it was also of interest to dock compounds 4–10 in the RTKs VEGFR-2, EGFR and the homology model of PDGFR-β with 3 as the lead compound. Figure 4A shows the docked conformation of 3 (magenta) superimposed on the co-crystallized ligand axitinib (pink) in the crystal structure of VEGFR-2. 44 The pyrrolo[3,2-d]pyrimidine scaffold of 3 form hydrophobic interactions with Val848, Lys868, Val916, Cys1045 and Phe1047. The N5-H of pyrrole ring undergoes H-bonding interaction with backbone carbonyl of Asp1046. The 7-benzyl group interacts with Leu840, Val848, Ala866, Leu1035 and Phe1047 and lies in the region occupied by the indazole group of axitinib. The N4-methyl of 3 is oriented towards Leu889 and Val914. The N-phenyl group of 3 interacts with residues Leu889, Val899, Cys1045 and Asp1046. The best docked pose of 3 had a score of −6.98 kcal/mol. Figure 4B shows the docked pose of 9 (green), as a representative example, superimposed on the co-crystallized ligand axitinib (pink). The mode of binding of 9, in VEGFR-2 was similar to that of 3, in addition the 2-amino of 9 formed H-bond interactions with the backbone carbonyl of Ala866 and had a docked score of −7.77 kcal/mol, somewhat better than the value obtained with 3. Compounds 4–10, when similarly docked into the crystal structure of VEGFR-2, had scores ranging between −5.33 to −7.77 kcal/mol and exhibited similar modes of binding as 3 and 9. These results indicate that, like 3, analogs 4–10 should inhibit VEGFR-2.
Figure 4.
A) Docked pose of 3 (magenta) in the binding site of axitinib (pink) in VEGFR-2 (PDB ID: 4AG8).44B) Docked pose of 9 (green) in the binding site of axitinib (pink) in VEGFR-2 (PDB ID: 4AG8).44 H-bond highlighted in red circle.
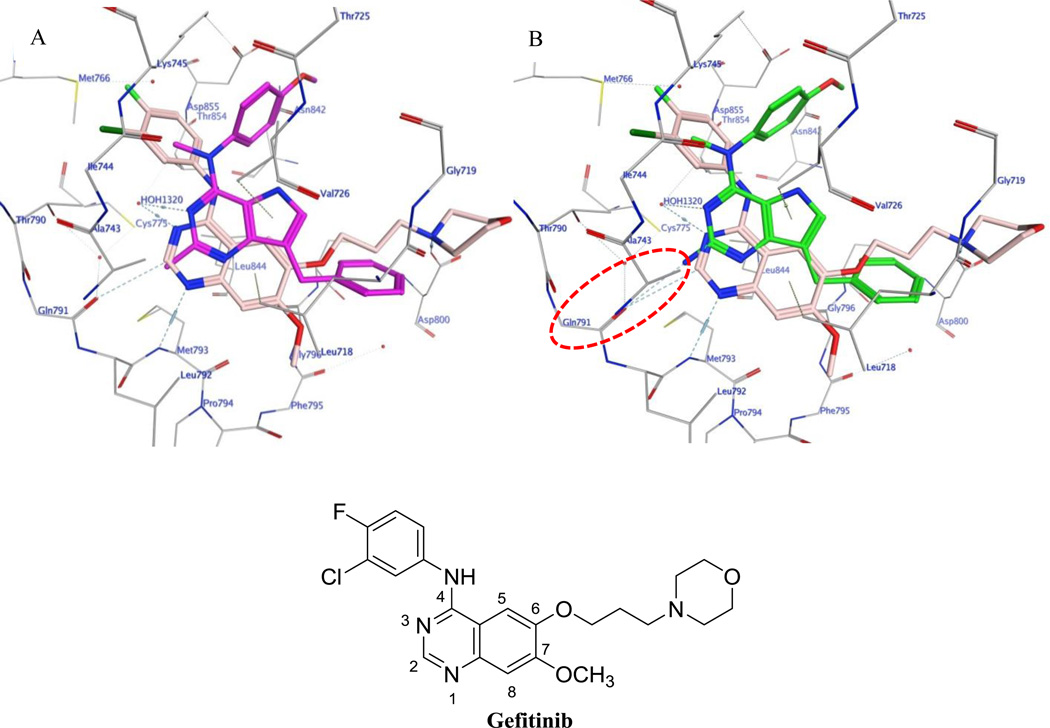
Figure 5A and 5B shows the docked conformations of 3 (magenta) and 9 (green), respectively, superimposed on the co-crystallized ligand gefitinib (pink) in the crystal structure of EGFR. 45 The pyrrolo[3,2-d]pyrimidine scaffold of 3 and 9 form hydrophobic interactions with Leu718, Val726, Ala743, Thr790, Met793 and Leu844. The N3 atom in both 3 and 9 undergo a H-bonding interaction with HOH1320. As predicted in our rationale, the 2-amino group in 9 makes an additional H-bonding interaction in the hinge region with the carbonyl backbone of Gln791. This additional bonding is anticipated to increase the potency of 9 as compared to 3. The benzylic group of 3 and 9 lies in the region occupied by the morpholinopropoxy side chain of gefitinib and interacts with Leu718, Gly796 and Leu844. The N4-methyl group of 3 and 9 interacts with Lys745 and Thr790, whereas the 4’-methoxyanilino group undergoes hydrophobic interactions with the side chain carbons of Lys745, Thr854 and Asp855. The best docked pose of 3 had a score of −6.97 kcal/mol and, for 9, the score was −7.08 kcal/mol. Compounds 4–10 when docked into EGFR had scores ranging between −6.68 to −7.30 kcal/mol and exhibited modes of binding similar to those of 3 and 9 indicating that these analogs are expected to possess EGFR inhibitory properties.
Figure 5.
A) Docked pose of 3 (magenta) in the binding site of gefitinib (pink) in EGFR (PDB ID: 4WKQ).45B) Docked pose of 9 (green) in the binding site of gefitinib (pink) in EGFR (PDB ID: 4WKQ).45 H-bond highlighted in red circle.
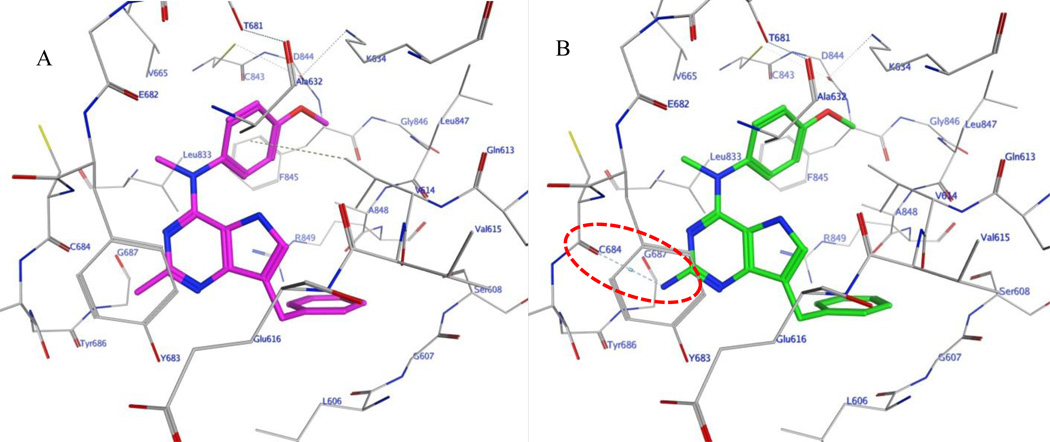
In the absence of an x-ray crystal structure, docking of compounds 3 and 4–10 was carried out in our validated homology model of PDGFR-β.11 The docked conformations of 3 (magenta) and 9 (green) in the homology model of PDGFR-β are shown in figure 6A and 6B, respectively, as representative examples. The pyrrolo[3,2-d]pyrimidine scaffold of 3 and 9 form hydrophobic interactions with Val614, Tyr683 and Gly687. The benzylic group interacts with residues Leu606, Gly607, Val614 and Ala848. The N4-methyl group lies in a pocket lined by residues Val665, Cys684 and Leu833 and the 4’-methoxyaniline moiety undergoes hydrophobic interactions with Val614, Lys634, Ala632, Leu833, Phe845 and Ala848. The 2-amino group in 9 makes an additional H-bonding interaction in the hinge region with the carbonyl backbone of Cys684 as compared with 3, as predicted.
Figure 6.
A) Docked pose of 3 (magenta) in the homology model of PDGFR-β.11B) Docked pose of 9 (green) in the homology model of PDGFR-β. H-bond highlighted in red circle.
3. Chemistry
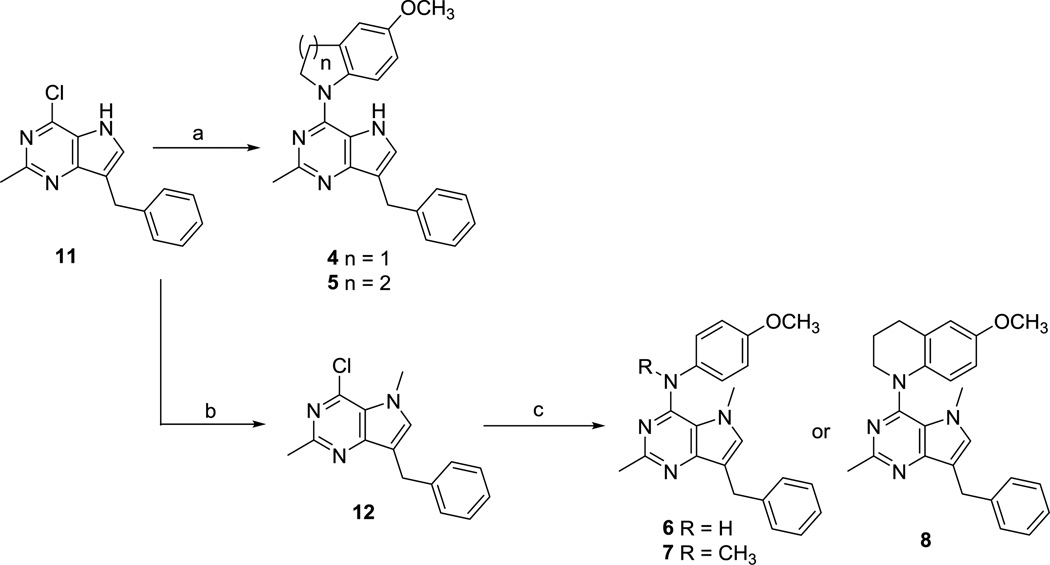
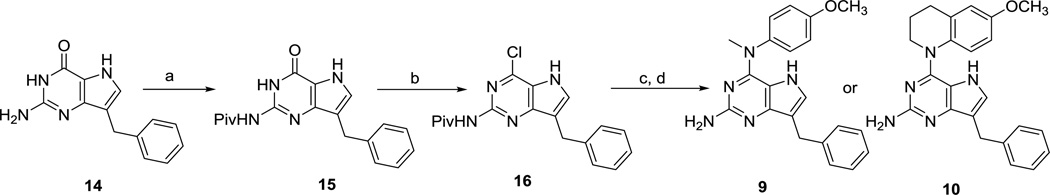
The key intermediate 7-benzyl-4-chloro-2-methyl-5H–pyrrolo[3,2-d]pyrimidine 1115 (Scheme 1) was synthesized from benzaldehyde as previously described.15 Nucleophilic aromatic substitution reactions of 11 with appropriately substituted amines in isopropanol at reflux afforded compounds 4 and 5 in 76 and 79% yields, respectively. Compound 11 was deprotonated with sodium hydride and methylated with methyl bromide to afford the N5-methylated pyrrolo[3,2-d]pyrimidine 12, which, on treatment with appropriately substituted anilines in isopropanol at reflux afforded compounds 6 and 7 in 74 and 79% yields, respectively. Treatment of 12 with 6-methoxy-1,2,3,4-tetrahydroquinoline gave 8 in 76% yield. Displacement reactions with the bulky nucleophile 6-methoxy-1,2,3,4-tetrahydroquinoline required longer reaction times as compared to other amines for complete disappearance of starting material, as estimated from TLC.
Scheme 1.
Reagents and conditions: (a) 5-methoxyindoline or 6-methoxy-1,2,3,4-tetrahydroquinoline, isopropanol, reflux, 8–12 h (76–79%); (b) NaH, CH3Br, 3 h (84%); (c) appropriately substituted aromatic amine, isopropanol, reflux, 4–24 h (74–79%)
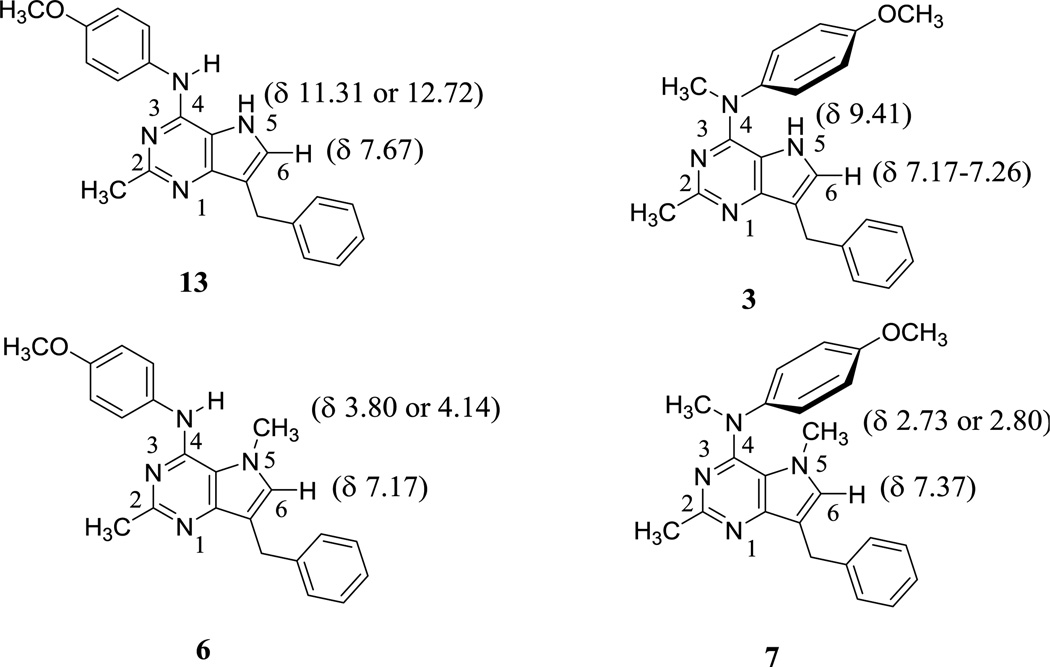
Comparison of 1H NMRs (in DMSO-d6) of the 4-N-CH3 compounds 3 and 7 with their corresponding 4-NH analogs 1315 and 6, respectively, suggests a conformational preference of the 4-anilino moiety. As shown in figure 7, the chemical shift of the N5-H proton in 3, compared with that in 13, indicates that this proton in 3 is in the shielding zone of the 4-methoxyphenyl ring and adopts the conformation shown in figure 7 in DMSO-d6, with the phenyl ring positioned over the N5-H proton. Similarly, comparing the chemical shift of the N5-CH3 protons in 7 with that in 6 suggests a deshielding and a similar conformational preference for 7 in DMSO-d6, as shown in figure 7 with the phenyl ring positioned above the N5-CH3. These conformational preferences from solution 1H-NMR in DMSO-d6 corroborate the molecular modeling conformations in the docked poses of 3 and 7 in tubulin (Fig. 3) and 3 and 9 in VEGFR-2, EGFR and PDGFR-β (Fig. 4, Fig. 5 and Fig. 6, respectively), suggesting that not only are these conformations the most stable in solution but are also the preferred bound conformations in tubulin as well as RTKs.
Figure 7.
Comparison of 1H NMRs of 3, 6, 7 and 13
Chlorination of the 2-amino-4-oxo-pyrrolo[3,2-d]pyrimidine 1446 (Scheme 2) gave poor yields (<20%). The subsequent aniline displacement reactions also did not proceed to completion, and a separation of the product from the starting materials was tedious and required extensive column chromatography. Hence, compound 14 (Scheme 2) was first protected with trimethylacetic anhydride to provide amide 15 in 92% yield. The pivaloyl protected 4-oxo pyrrolo[3,2-d] pyrimidine 15, with significantly improved solubility in organic solvents compared to 14, was chlorinated with phosphorus oxychloride to generate the 4-chloro-pyrrolo[3,2-d]pyrimidine 16, which was subjected to nucleophilic aromatic substitution reactions using the appropriately substituted amines, followed by base catalyzed amide deprotection to afford 9 and 10 in 66 and 68% yields, respectively.
Scheme 2.
Reagents and conditions: (a) trimethylacetic anhydride, 100 °C, 2 h (92%); (b) POCl3, 110 °C, 3 h; (c) 4-methoxy-N-methylaniline or 6-methoxy-1,2,3,4-tetrahydroquinoline, isopropanol, reflux, 4–16 h; (d)1 N NaOH, CH3OH, reflux, 10 h (66–68%).
4. Biological evaluations and discussion
4.1 Tubulin Assembly and RTK- inhibitory effects
Inhibition of tubulin assembly
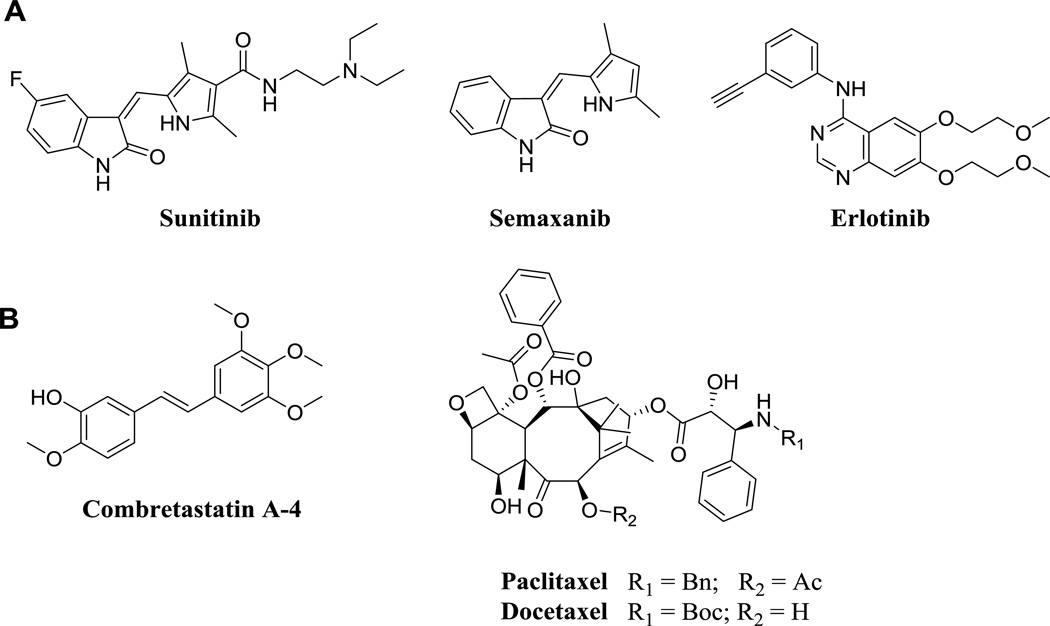
Quantitative studies were conducted to evaluate the effects of compounds 4 – 10 on the polymerization of purified bovine brain tubulin (Table 1). As inhibitors of tubulin assembly, these compounds were compared with combretastatin A-4 (CA, figure 8), the active metabolite of the water soluble prodrug combretastatin A-4 phosphate (Zybrestat, Fosbretabulin), which has orphan drug status for ovarian cancer.47 Compounds 5, 7 and 8 were effective and potent inhibitors of bovine tubulin assembly, with activity comparable to that of CA. Compound 5, the conformationally restricted tetrahydroquinoline analog of 3, was almost 7-fold more potent than 3. In contrast with 5, conformational restriction using a 5-membered ring (compound 4) was not favorable to inhibition of tubulin assembly. Compound 7, the N5-methyl analog of 3, was the most potent inhibitor of tubulin assembly in this series. It was almost 44-fold more inhibitory than 3 and almost 7-fold more inhibitory than 5, indicating that N5-methylation is highly conducive for a strong interaction with tubulin. Clearly N5-methylation of 3 to 7 affords further conformational restriction for the N-phenyl ring in addition to providing hydrophobic interactions (Leuβ248 and Alaβ354) with the N5-methyl moiety instead of a hydrogen donor at the pyrrole N5 in 3. In contrast, compound 8, the N5-methyl analog of 5, wherein the N-phenyl bond is completely restricted via a tertrahydroquinoline ring, was only 3.6-fold more potent against tubulin assembly than 5. However, compound 8 with added conformational restriction via a N5-methyl over 5 was highly potent and only 2-fold less potent than the most potent analog 7. As with previously described pyrimidine fused bicyclic and tricyclic compounds, 15, 34, 48–50 the methyl group attached to the 4-nitrogen bridge is crucial for inhibition of tubulin assembly. Removal of this N-methyl group (6 compared with 7) resulted in significant loss of tubulin inhibitory activity. The 2-aminopyrrolo[3,2-d]pyrimidine analogs 9 and 10 (as free bases) were less potent in the tubulin assembly inhibitory assay as compared to the corresponding 2-methylpyrrolo[3,2-d]pyrimidine analogs 3 and 5, respectively, indicating that the 2-amino was less important than the 2-methyl moiety for an interaction with tubulin. This, along with the results for 9, indicates the importance of conformational restriction via the N5-methylation in both 7 and 8 as being important for a strong interaction with tubulin. However, further increase in conformational restriction, as in 4 with the 5-methoxydihydroindole, abolished the interaction with tubulin. Thus for these analogs, limited conformational restriction is an important facet for potent interactions with tubulin.
Table 1.
Inhibition of tubulin assembly, [3H]colchicine binding and cellular VEGFR-2, EGFR and PDGFR-β RTKs
| COMPOUND | tubulin assembly IC50 (µM) ± SD |
[3H]colchicine binding % Inhibition ± SD |
VEGFR-2 IC50 (nM) ± SD |
EGFR IC50 (nM) ± SD |
PDGFR-P IC50 (nM) ± SD |
A431 Cytotoxicity IC50 (nM) ± SD |
|
|---|---|---|---|---|---|---|---|
| 5 µM inhibitor | 1 µM inhibitor | ||||||
| 3-HCl15 | 21 ± 1 | ND | ND | 21.3 ± 3.2 | 23.6 ± 5.8 | >200 | 9.2 ± 0.8 |
| 4-HCl | > 20 | ND | ND | >200 | 167.1 ± 27.6 | >200 | ND |
| 5-HCl | 3.3 ± 0.3 | 43 ± 3 | ND | 38.7 ± 7.1 | 33.6 ± 5.6 | 90.3 ± 18.3 | 40.1 ± 5.6 |
| 6-HCl | > 20 | ND | ND | 26.7 ± 4.6 | 2.6 ± 0.42 | 7.2 ± 0.9 | 2.3 ± 0.3 |
| 7-HCl | 0.48 ± 0.008 | 92 ± 0.2 | 73 ±3 | 33.0 ± 5.0 | 2.3 ± 0.30 | 10.3 ± 1.7 | 1.2 ± 0.07 |
| 8-HCl | 0.91 ± 0.03 | 72 ± 2 | ND | 32.9 ± 4.9 | 4.9 ± 0.6 | 30.2 ± 7.2 | 8.1 ± 0.8 |
| 9 | > 20 | ND | ND | 27.3 ± 5.1 | 1.1 ± 0.2 | 8.1 ± 0.91 | 0.7 ± 0.09 |
| 10 | 13 ± 1 | ND | ND | 84.3 ± 3.9 | 47.1 ±5.0 | >200 | ND |
| CA | 1.2 ± 0.01 | 99 ± 0.3 | 90 ± 3 | ND | ND | ND | ND |
| semaxinib | ND | ND | ND | 12.9 | ND | ND | ND |
| sunitinib | ND | ND | ND | 18.9 ± 2.7 | ND | 83.1 ± 10.1 | ND |
| erlotinib | ND | ND | ND | ND | 1.2 ± 0.2 | ND | ND |
| doxorubucin | ND | ND | ND | ND | ND | ND | 1.35 ± 0.03 |
ND: not determined
Figure 8.
Standards used in biological assays: A) RTKIs B) MTAs
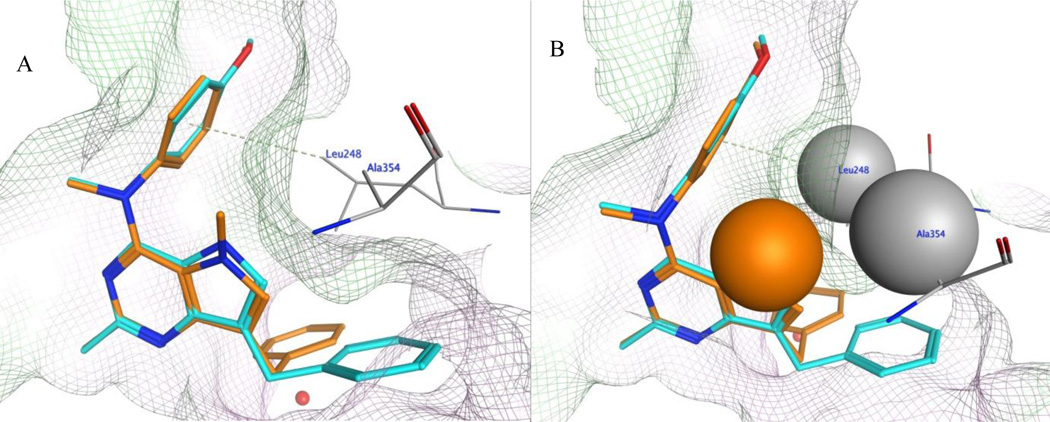
In order to rationalize the 40–fold improvement in the inhibition of tubulin assembly when comparing the activities of 3 and 7, their docked poses in the colchicine site of tubulin were compared. Figure 9A and 9B shows the superimposed docked conformations of 3 and 7, respectively, in the colchicine site. In figure 9, compound 7 retains all the other interactions at the binding site as discussed above, and, in addition, the N5-methyl group of 7 undergoes additional hydrophobic interactions with Leuβ248 and Alaβ354. The best docked pose of 7 had a score of −9.21 kcal/mol. An additional reason for the overall improved binding of 7 over 3 can be attributed to the further conformational restriction imposed on the rotation of the C4-N bond due to the presence of the N5-methyl moiety in 7.
Figure 9.
A) Superimposition of docked pose of 3 (cyan) and 7 (orange) in the colchicine site of tubulin (PDB ID: 4O2B).43B) Space fill view of selected residues (N5-methyl of compound 7, Leuβ248 and Alaβ354) in the colchicine site of tubulin (PDB ID: 4O2B).43 The green surface indicates hydrophobic regions and the pink surface indicates hydrophilic region in the pocket.
A conformational search carried out by a 1°rotation about C4-N bond using Sybyl-X 2.1.151 to compare the number of low energy conformations generated within 5 kcal/mol for 3 afforded 111 conformers, whereas for 7 only 53 conformers were possible. Similar improvement in binding affinity by introducing methyl groups has been recently reviewed in the literature and has been attributed to a combination of conformational, hydrophobic and desolvation effects.52 It was also observed that 6 generated 112 conformers, whereas 7 generated 53 conformers. Thus, the increased rotational flexibility about the C4-N bond might be one of the contributing factors responsible for the loss of inhibitory activity against tubulin assembly for compound 6.
Compounds 5, 7 and 8, which showed potent tubulin inhibitory activities, were evaluated for their ability to inhibit the binding of radiolabelled colchicine to tubulin (Table 1). These compounds inhibited [3H]colchicine binding by percentages proportional to their tubulin assembly IC50s, indicating that they are probably colchicine site MTAs, as is CA.53
Inhibition of RTKs
Compounds 4 – 10 were evaluated for their activity against RTKs that are overexpressed by tumor and endothelial cells (Table 1). Compounds 5 – 9 have potencies comparable with standards sunitinib (clinically used) and semaxinib against VEGFR-2, although these compounds are less potent than 3 (Table 1). The EGFR cellular potency of 5, the conformationally restricted tetrahydroquinoline analog of 3, is comparable to 3. The N5-methyl analogs 6 – 8, have significantly improved EGFR potencies compared to 3 and are also comparable with clinically used erlotinib against EGFR. Compound 9, the 2-amino analog of 3, is the most potent analog against EGFR and indicates the important contribution of the 2-amino moiety to EGFR inhibitory potency. Although compound 5 was not significantly potent against PDGFR-β, the N5-methyl analogs 6 – 8, along with 9, have potencies better than the clinically used standard sunitinib against PDGFR-β. Replacing the 2-methyl group of 3 with a 2 amino group (compound 9) resulted in better activity against both EGFR and PDGFR-β. These activities were comparable to those of standard erlotinib and better than sunitinib, respectively, perhaps due to improved binding to the hinge region of the ATP-pocket of these RTKs, as predicted by our molecular modeling studies. Compounds 6 – 9 showed potent inhibition of A431 tumor cells comparable to that obtained with doxorubicin.
4.2 Effect on Pgp overexpression and βIII-tubulin mediated resistance to paclitaxel
The ability of MTAs 5, 7 and 8 to overcome βIII-tubulin drug resistance was evaluated by using an isogenic HeLa cell line pair (Table 2). While paclitaxel was 3-fold less potent in the βIII-tubulin overexpressing cell line than in the wild type HeLa cells, compounds 5, 7 and 8 inhibited both cell lines with equal potency without regard to their expression of βIII-tubulin.
Table 2.
Inhibition of growth of β-III and Pgp overexpressing cells.
| COMPOUND | WT HeLa IC50 (µM) ± SD |
β-III overexpressing HeLa IC50 (µM) ± SD |
Parental OVCAR-8 IC50 (µM) ± SD |
Pgp overexpressing NCI/ADR-RES IC50 (µM) ± SD |
|---|---|---|---|---|
| 5·HCl | 250 ± 0 | 250 ± 0 | 120 ± 4 | 70 ± 10 |
| 7·HCl | 14 ± 2 | 14 ± 1 | ||
| 8·HCl | 60 ± 0 | 58 ± 2 | ||
| CA | 1.8 ± 0.4 | 2.5 ± 0.7 | 1.3 ± 0.6 | 1.3 ± 0.6 |
| Paclitaxel | 5.3 ± 2 | 16 ± 1 | 5.0 ± 1 | 2,500 ± 0 |
| Docetaxel | 4.0 ± 2 | 13 ± 4 |
The ability of 5 to circumvent Pgp-mediated drug resistance was evaluated by using an ovarian cancer cell line pair (Table 3). In this cell line pair, paclitaxel, which is a well-known Pgp substrate, was 500-fold less potent in the Pgp overexpressing cell line than in the wild type OVCAR-8 cells. Surprisingly, compound 5 is about 1.4-fold more potent in the Pgp overexpressing cell line. Hence, compound 5 could have advantages over some clinically useful microtubule targeting drugs, such as paclitaxel, docetaxel, vinblastine and vinorelbine, since 5 is a poor substrate for transport by Pgp.
Table 3.
Tumor cell inhibitory activity (NCI) GI50 (10−8 M) for 5 and 7
| Panel/ Cell line | GI50 (10−8 M) Compound |
Panel/ Cell line |
GI50 (10−8 M) Compound |
Panel/ Cell line | GI50 (10−8 M) Compound |
Panel/ Cell line | GI50 (10−8 M) Compound |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Leukemia | 5·HCl | 7·HCl | Colon Cancer | 5·HCl | 7·HCl | Melanoma | 5·HCl | 7·HCl | Renal Cancer | 5·HCl | 7·HCl |
| CCRF-CEM | 22.6 | 3.35 | COLO 205 | 23.2 | 2.63 | LOX IMVI | 52.1 | 5.88 | 786 – 0 | 93.4 | 9.32 |
| HL-60(TB) | 22.2 | 2.68 | HCC-2998 | 31.8 | 7.11 | MALME-3M | 33.0 | 1.77 | A498 | 19.7 | 2.02 |
| K-562 | 11.1 | 3.37 | HCT-116 | 36.8 | 3.50 | M14 | 25.2 | 2.95 | ACHN | 83.0 | 17.2 |
| MOLT-4 | 34.5 | 5.08 | HCT-15 | 36.6 | 3.81 | MDA-MB-435 | 34.1 | 2.14 | CAKI-1 | 34.1 | 5.66 |
| RPMI-8226 | 26.6 | 3.69 | HT29 | 3.51 | SK-MEL-2 | 2.86 | RXF 393 | 21.2 | 2.54 | ||
| SR | 92.2 | 3.38 | KM12 | 36.6 | 4.45 | SK-MEL-28 | 36.8 | 4.87 | SN12C | 61.8 | 8.87 |
| NSCLC | SW-620 | 37.0 | 3.84 | SK-MEL-5 | 32.5 | 5.42 | TK10 | 12.3 | |||
| A549/ATCC | 40.6 | 4.40 | CNS Cancer | UACC-257 | 10.8 | 7.82 | UO-31 | 39.0 | 20.9 | ||
| EKVX | 46.3 | SF-268 | 72.3 | 35.0 | UACC-62 | 65.9 | 47.5 | Prostate Cancer | |||
| HOP-62 | 37.9 | 4.78 | SF-295 | 19.2 | 3.04 |
Ovarian cancer |
PC-3 | 4.96 | |||
| HOP-92 | 18.9 | 4.71 | SF-539 | 26.0 | 2.54 | IGROVI | 44.0 | 9.58 | DU-145 | 38.3 | 4.44 |
| NCI-H226 | 13.6 | 30.4 | SNB-19 | 54.9 | 5.47 | OVCAR-3 | 28.5 | 3.52 | Breast Cancer | ||
| NCI-H23 | 32.0 | 4.58 | SNB-75 | 18.2 | 2.78 | OVCAR-4 | 46.8 | MCF7 | 24.8 | 3.33 | |
| NCI-H322M | 36.2 | 7.36 | U251 | 31.6 | 4.43 | OVCAR-5 | 54.3 | 26.9 | MDA-MB- 231/ATCC |
64.1 | 6.37 |
| NCI-H460 | 33.7 | 3.87 | OVCAR-8 | 48.7 | 4.91 | HS 578T | 58.2 | 6.23 | |||
| NCI-H522 | 31.6 | 2.39 | NCI/ADR-RES | 26.5 | 3.28 | BT-549 | 75.5 | 4.82 | |||
| SK-OV-3 | 42.3 | 4.54 | T-47D | 12.8 | |||||||
| MDA-MB-468 | 15.3 | 3.01 | |||||||||
4.3 Antiproliferative effects in NCI 60 tumor panel
Compounds 5 (a tubulin and VEGFR-2 inhibitor) and 7 (a tubulin, VEGFR-2, EGFR and PDGFR-β inhibitor) were selected for tumor cytotoxicity evaluation in the NCI 60 tumor cell line panel (Table 3). Both compounds show 2- to 3-digit nanomolar GI50 values across all tumor types, with 7 more active than 5. Antiangiogenic activities of these compounds are not pertinent in these cell culture assays since there is no angiogenesis involved in these proliferation studies.
4.4 Antiangiogenic effects in CAM Assay
Compounds 6 – 9 were evaluated for their effects on blood vessel formation in the chicken chorioallantoic membrane (CAM) antiangiogenic activity assay against standards sunitinib and erlotinib (Table 4). Compounds 6, 7 and 9, which inhibit VEGFR-2, EGFR and PDGFR-β, were found to have strong antiangiogenic activities, comparable with those obtained with the standards.
Table 4.
Chorioallantoic membrane assay
| IC50 [µM] ± SD | |
|---|---|
| 6·HCl | 8.7 ± 0.9 |
| 7·HCl | 2.2 ± 0.4 |
| 8·HCl | 26.1 ± 4.3 |
| 9 | 2.3 ± 0.3 |
| sunitinib | 1.3 ± 0.07 |
| erlotinib | 29.1 ± 1.9 |
4.5 Effect of 7·HCl on primary tumor growth and lung metastasis in the 4T1 orthotopic breast tumor model
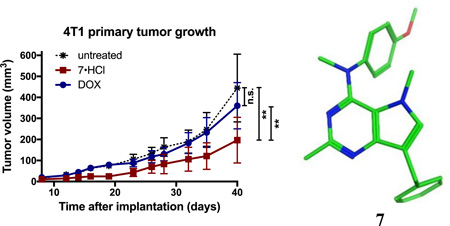
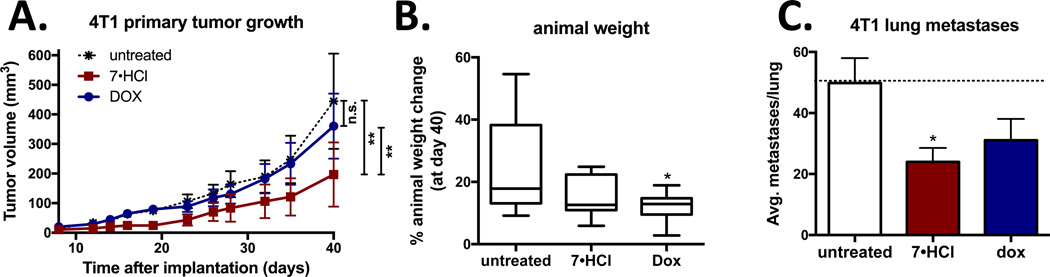
Compound 7·HCl was selected for in vivo studies, since it is a unique compound with cytotoxic MTA effects and triple angiokinase (PDGFR-β, VEGFR-2 and EGFR) inhibitory mechanisms of action in a single entity. The ability of 7·HCl in reducing tumor growth and metastasis was evaluated in a relevant triple negative breast cancer model in which metastasis can also be assessed. The 4T1 triple negative orthotopic allograft model was used because it mimics human metastatic progression and does not require the use of immunodeficient mice or Matrigel. Treatment with 7·HCl significantly reduced primary tumor growth as compared to untreated control. In addition, 7·HCl significantly reduced tumor growth compared to doxorubicin treatment, which did not have a significant effect on tumor volume (Fig 7A). Compound 7·HCl-treated mice had similar weight gain as the untreated control group, indicating that 7·HCl lacks systemic toxicity. In contrast, doxorubicin-treated mice showed significantly less weight gain compared to untreated control animals (Fig 7B). Next, the effect of 7·HCl on lung metastasis was evaluated by counting the number of GFP positive cell clusters. Compound 7·HCl reduced the average number of lung metastases better than did doxorubicin (Fig 7C), which is an important finding since most cancer patients succumb to metastatic disease rather than the primary tumor.54 Compound 7 serves as a lead analog for further preclinical development, analog design and synthesis for potential use in triple negative breast cancer.
5. Summary
A series of 7-benzyl-N-substituted-pyrrolo[3,2-d]pyrimidin-4-amines were synthesized and evaluated as designed multiangiokinase inhibitors and as MTAs in single molecular entities. We discovered a compound as a MTA with VEGFR-2 inhibitory activity (compound 5), compounds with VEGFR-2, PDGFR-β and EGFR triple angiokinase inhibitory activities (compounds 6 and 9) and compounds as MTAs and VEGFR-2, PDGFR-β and EGFR triple angiokinase inhibitors (compounds 7 and 8). These are compounds with novel combination chemotherapeutic potential in single molecules. In addition the solution conformation of active compounds 3 and 7 among others, gleaned from the 1HNMR studies suggests that these solution conformations are similar to the bound conformations determined from molecular modeling in all four targets, tubulin and RTKs. Compounds 5 and 7 displayed antiproliferative activities of two or three digit nanomolar GI50 values across the entire NCI 60 tumor cell panel. Compounds 6, 7 and 9 also inhibited blood vessel formation in the CAM assay. The cytotoxic effects of some of these compounds are independent of the overexpression of Pgp (5, 7 and 8) and βIII-tubulin (5). In vivo, in an orthotopic mouse 4T1 allograft triple negative breast tumor model, compound 7 reduced tumor growth and lung metastases and was superior to doxorubucin in this regard. Compound 7 was devoid of systemic toxicity to the animals and is identified for further preclinical and analog development for potential use in triple negative breast cancer.
6. Experimental section
6.1 Synthesis
7-benzyl-4-(5-methoxyindolin-1-yl)-2-methyl-5H-pyrrolo[3,2-d]pyrimidine hydrochloride (4·HCl)
7-benzyl-4-chloro-2-methyl-5H–pyrrolo[3,2-d]pyrimidine 1115 (0.1 g, 0.39 mmol) and 5-methoxyindoline (58 mg, 0.48 mmol) were dissolved in isopropanol (15 mL) and heated at reflux for 12 h, and the solvent was evaporated in vacuo. The crude material obtained was purified by column chromatography and eluted with 1% (v/v) MeOH/CHCl3. Fractions containing the product (TLC) were pooled, and the solvent was evaporated to afford 5 (109 mg, 76%). TLC Rf 0.4 (MeOH: CHCl3; 1:20). The product obtained was dissolved in a minimum amount of ethyl acetate, and diethyl ether (10 mL) was added to the solution. HCl gas was bubbled through the solution for 2–3 mins. The precipitate obtained was collected by filtration and washed with diethyl ether to afford 4·HCl. mp 293–294 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm 2.67 (s, 3 H, CH3) 2.70 – 2.80 (m, 2 H, CH2) 3.79 (s, 3 H, CH3) 4.11 (s, 4 H, CH2) 6.77 – 6.82 (m, 1 H, Ar) 6.92 – 6.96 (m, 1 H, Ar) 7.12 – 7.24 (m, 2 H, Ar) 7.28 – 7.34 (m, 4 H, Ar) 7.44 – 7.48 (m, 1 H, Ar) 10.88 (s, 1 H, exch, NH) 14.36 (s, 1 H, exch, HCl) Anal. Calcd. for C23H22N4O·HCl: C, 67.89; H, 5.70; N, 13.77; Cl, 8.71. Found C, 67.99; H, 5.66; N, 13.75; Cl, 8.72.
1-(7-benzyl-2-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline hydrochloride (5·HCl)
Compound 6·HCl (synthesized from 11 and 6-methoxy-1,2,3,4-tetrahydroquinoline as described for 4·HCl): yield = 79%; TLC Rf 0.4 (MeOH: CHCl3; 1:20). mp 262–264 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm 1.97 – 2.05 (m, 2 H, CH2) 2.67 (s, 3 H, CH3) 2.76 – 2.82 (m, 2 H, CH2) 3.79 (s, 3 H, CH3) 4.11 (s, 4 H, CH2) 6.77 – 6.82 (m, 1 H, Ar) 6.92 – 6.96 (m, 1 H, Ar) 7.12 – 7.24 (m, 2 H, Ar) 7.28 – 7.34 (m, 4 H, Ar) 7.44 – 7.48 (m, 1 H, Ar) 10.88 (s, 1 H, exch, NH) 14.36 (s, 1 H, exch, HCl) Anal. Calcd. for C24H24N4O·HCl·0.25H2O: C, 67.75; H, 6.04; N, 13.17; Cl, 8.33. Found C, 67.84; H, 6.21; N, 12.95; Cl, 8.06.
7-benzyl-4-chloro-2,5-dimethyl-5H-pyrrolo[3,2-d]pyrimidine (12)
7-benzyl-4-chloro-2-methyl-5H–pyrrolo[3,2-d]pyrimidine 11 (1 g, 3.88 mmol) was dissolved in dimethylformamide (20 mL), and sodium hydride (140 mg, 5.82 mmol) was added under nitrogen atmosphere. The mixture was stirred for 15 min, after which time no further production of hydrogen gas was observed. Methyl bromide (0.4 mL, 7.37 mmol) was added, and the reaction mixture was stirred for 3 h. The reaction was quenched by addition of water and ethyl acetate was added. The organic layer was collected, washed with brine and dried over sodium sulfate. A silica gel plug was made, and the crude material was purified by column chromatography (CHCl3: MeOH; 100:1 v/v) to give an off-white solid (890 mg, 84%) TLC Rf 0.5 (CH3OH: CHCl3; 1:25). mp, 145–147 °C 1H NMR (400 MHz, DMSO-d6) δ ppm 2.62 (s, 3 H, CH3) 4.01 (s, 3 H, CH3) 4.03 (s, 2 H, CH2) 7.17 (td, J = 5.65, 2.76 Hz, 1 H, 6-CH) 7.26 – 7.30 (m, 4 H, C6H5) 7.64 (s, 1 H, C6H5)
7-benzyl-N-(4-methoxyphenyl)-2,5-dimethyl-5H-pyrrolo[3,2-d]pyrimidin-4-amine hydrochloride (6·HCl)
Compound 6·HCl (synthesized from 12 and p-anisidine as described for 4·HCl): yield = 79%; TLC Rf 0.54 (CH3OH: CHCl3; 1:20). mp, 291–292 °C; 1H NMR (DMSO-d6): δ 2.57 (s, 3 H, 2-CH3) 3.80 (s, 3 H, NCH3) 4.14 (s, 3 H, OCH3) 4.17 (s, 2 H, CH2) 6.97 – 7.06 (m, 2 H, Ar) 7.19 – 7.23 (m, 1 H, Ar) 7.26 – 7.36 (m, 4 H, Ar) 7.44–7.53 (m, 2 H, Ar) 7.61 (s, 1 H, 6-CH) 9.52 (br, 1 H, exch, NH) 14.70 (s, 1 H, exch, HCl). Anal. Calcd. for C22H22N4O·HCl: C, 66.91; H, 5.87; N, 14.19; Cl, 8.98. Found C, 66.88; H, 5.86; N, 14.07; Cl, 8.84.
7-benzyl-N-(4-methoxyphenyl)-N,2,5-trimethyl-5H-pyrrolo[3,2-d]pyrimidin-4-amine hydrochloride (7·HCl)
Compound 7·HCl (synthesized from 12 and 4-methoxy-N-methylaniline as described for 4·HCl): yield = 74%; TLC Rf = 0.6 (CH3OH: CHCl3; 1:20). mp, 186–187 °C; 1H NMR (400 MHz, DMSO-d6) 5 2.73 (s, 3 H, CH3) 2.80 (s, 3 H, CH3) 3.63 (s, 3 H, CH3) 3.78 (s, 3 H, CH3) 4.08 (s, 2 H, CH2) 7.00 (d, J = 9.03 Hz, 2 H, Ar) 7.22 (d, J = 5.52 Hz, 1 H, Ar) 7.25 – 7.33 (m, 4 H, Ar) 7.37 (s, 1 H, Ar) 8.33 (s, 1 H, Ar) 14.63 (s, 1 H, exch, HCl) Anal. Calcd. for C23H24N4O·HCl: C, 67.55; H, 6.16; N, 13.70; Cl, 8.67. Found C, 67.41; H, 6.20; N, 13.59; Cl, 8.61.
1-(7-benzyl-2,5-dimethyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline hydrochloride (8·HCl)
Compound 8·HCl (synthesized from 12 and 6-methoxy-1,2,3,4-tetrahydroquinoline as described for 4·HCl): yield = 76%; TLC Rf 0.68 (CH3OH: CHCl3; 1:20) mp, 130–132 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm 2.07 (t, J = 6.53 Hz, 2 H, CH2) 2.72 (s, 3 H, CH3) 2.79 – 2.88 (m, 2 H, CH2) 2.97 (s, 3 H, CH3) 3.72 – 3.78 (m, 3 H, CH3) 3.96 – 4.06 (m, 2 H, CH2) 4.13 (s, 2 H, CH2) 6.68 – 6.72 (m, 1 H, Ar) 6.79 (d, J = 9.03 Hz, 1 H, Ar) 6.91 (d, J = 2.76 Hz, 1 H, Ar) 7.22 (td, J = 5.84, 2.64 Hz, 1 H, Ar) 7.30 – 7.35 (m, 4 H, Ar) 7.50 (s, 1 H, Ar) 14.71 (s, 1 H, exch, HCl). Anal. Calcd. for C25H26N4O·HCl: C, 69.03; H, 6.26; N, 12.88; Cl, 8.15. Found C, 69.10; H, 6.25; N, 12.84; Cl, 8.17.
N-(7-benzyl-4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-2-yl)pivalamide (15)
A suspension of 2-amino-7-benzyl-3H–pyrrolo[3,2-d]pyrimidin-4(5H)-one 1446 (1.25 g, 5.20 mmol in 20 mL of trimethylacetic anhydride was heated at 100 °C for 2 h. The resulting mixture was cooled, diluted with hexanes and filtered. The solid obtained was resuspended in water, and an ammonia solution was added to adjust the pH to 8. Ethyl acetate was added, and the organic layer was separated and dried over sodium sulfate, and the solvent was evaporated to afford 1.55 g (92%) of 14 as a white solid. TLC Rf= 0.47 (CH3OH /CHCl3, 1:20); mp 185–187°C 1H NMR (400 MHz, DMSO-d6) δ ppm 1.25 (s, 9 H, (CH3)3) 3.95 (s, 2 H, CH2) 7.11 (s, 1 H, Ar) 7.13 −7.19 (m, 1 H, Ar) 7.20 – 7.30 (m, 4 H, Ar) 10.77 (br, 1 H, exch, NH) 11.88 (br, 1 H, exch, NH) 12.09 (br, 1 H, exch, NH) Anal. Calcd. for C18H20N4O2·H2O: C, 63.14; H, 6.48; N, 16.36. Found C, 63.17; H, 6.24; N, 16.30.
7-benzyl-N4-(4-methoxyphenyl)-N4-methyl-5H-pyrrolo[3,2-d]pyrimidine-2,4-diamine (9)
Phosphorus oxychloride (30 mL) was added to N-(7-benzyl-4-oxo-4,5-dihydro-3H–pyrrolo[3,2-d]pyrimidin-2-yl)pivalamide 15 (1.5 g, 4.62 mmol). The reaction mixture was heated at reflux with stirring in an anhydrous atmosphere for 3 h. The dark-orange solution was cooled to room temperature and the solvent removed under vacuum to afford a brown gum. Aqueous ammonium hydroxide was added to the residue at 0 °C with vigorous stirring. The pH was adjusted to 8, and the resulting precipitate was collected by filtration, washed with water, and dried in vacuo to afford 1.15 g (73%) of crude 16. Compound 16 (0.15 g, 0.44 mmol) and 4-methoxy-N-methylaniline (75 mg, 0.55 mmol) were dissolved in isopropanol (15 mL) and heated at reflux for 4 h, after which the solvent was evaporated in vacuo. The residue was dissolved in methanol (15 mL), 1 N NaOH (2mL) was added, and the reaction mixture was heated to reflux for 10 h, after which the solvent was evaporated in vacuo. The crude material obtained was purified by column chromatography and eluted with 1% (v/v) MeOH/CHCl3. Fractions containing the product (TLC) were pooled, and the solvent was evaporated to afford 9 (128 mg, 66%). TLC Rf 0.48 (MeOH: CHCl3; 1:10). mp 167–169 °C; 1H NMR (400 MHz, DMSO-d6) 5 ppm 3.38 (s, 3 H, CH3) 3.80 (s, 3 H, CH3) 3.81 (s, 2 H, CH2) 5.46 (s, 2 H, exch, NH2) 6.71 (d, J = 3.26 Hz, 1 H, Ar) 6.97 – 7.02 (m, 2 H, Ar) 7.13 (m, J = 4.30 Hz, 1 H, Ar) 7.20 – 7.25 (m, 6 H, Ar) 7.86 (d, J = 2.76 Hz, 1 H, exch, NH) Anal. Calcd. for C21H21N5O: C, 70.17; H, 5.89; N, 19.48. Found C, 70.11; H, 5.96; N, 19.42.
7-benzyl-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-5H-pyrrolo[3,2-d]pyrimidin-2-amine (10)
Compound 10 (synthesized from crude 16 and 6-methoxy-1,2,3,4-tetrahydroquinoline as described for 9): yield = 68%; TLC Rf 0.6 (CH3OH: CHCl3; 1:10). mp 239–240 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm 1.85 – 1.94 (m, 2 H, CH2) 2.71 – 2.81 (m, 2 H, CH2) 3.73 (s, 3 H, OCH3) 3.86 (s, 4 H, CH2) 5.55 (br, 2 H, exch, NH2) 6.64 – 6.70 (m, 1 H, Ar) 6.75 – 6.81 (m, 2 H, Ar) 6.88 – 6.92 (m, 1 H, Ar) 7.12 – 7.18 (m, 1 H, Ar) 7.26 (s, 4 H, Ar) 9.36 (br, 1 H, exch, NH) Anal. Calcd. for C23H23N5O: C, 71.67; H, 6.01; N, 18.17. Found C, 71.90; H, 5.99; N, 18.11.
6.2 Molecular modeling
Docking of compounds 3–10 was carried out in the published x-ray crystal structure of colchicine in tubulin (PDB: 4O2B, 2.3 Å), axitinib in VEGFR-2 (PDB: 4AG8, 1.95 Å), gefitinib in EGFR (PDB: 4WKQ, 1.85 Å) and in a homology model of PDGFR-β using Molecular Operating Environment (MOE 2015.10).55 For tubulin, VEGFR-2 and EGFR, the crystal structures were obtained from the protein database and imported into MOE 2015.10. The proteins were then prepared using the QuickPrep function and the Amber10:EHT forcefield for energy minimization under default settings. Structure preparation using QuickPrep function utilizes interactive ligand modification and energy minimization in the active site of the selected flexible receptor. It deletes distant solvents, adds hydrogens, installs tethers, calculates charges and performs refinement of the system. Ligands were sketched using the builder function in MOE and minimized using Amber10:EHT forcefield. The ligands were docked in the binding site of VEGFR-2 using the default settings in the docking protocol. The placement was carried using Triangle Matcher and scored using London dG. The refinement was carried out using Rigid Receptor and scored using GBVI/WSA dG. The ligands were docked in the binding site of tubulin, EGFR and PDGFR-β using a different docking protocol to include the key interactions with water molecules in the binding site (for tubulin and EGFR). The placement was carried using Triangle Matcher and scored using London dG. The refinement was carried out using Induced Fit, the side chain was set free, cutoff was increased to 8 Å and radius offset was set to 0.6. For preparation of tubulin, prior to setting up the protein for QuickPrep, chains C, D, E and F were deleted to reduce the time for protein preparation. After the preparation of the protein, Ca2+, Mg2+, GDP, GTP and all other bound ligands except for colchicine were deleted. To validate our docking studies, the native ligands colchicine, axitinib and gefitinib were re-docked into the binding sites of tubulin, VEGFR-2 and EGFR, respectively, using the same set of parameters as described above. The rmsd of the best docked pose of colchicine in tubulin, axitinib in VEGFR-2 and gefitinib in EGFR were 0.358 Å, 0.506 Å and 0.853 Å, respectively, thus validating the docking using MOE.
6.3 Biological evaluation methods
6.3.1 Tubulin assembly assay
Bovine brain tubulin was purified as described previously.56 The control compound CA, a potent inhibitor of colchicine binding to tubulin,53 was generously provided by Dr. George R. Pettit, Arizona State University.
The tubulin assembly assay, described in detail previously,57 was performed with 1.0 mg/mL (10 µM) tubulin, 0.8 M monosodium glutamate (pH 6.6 in 2 M stock solution with HCl), 0.4 mM GTP, 4% (v/v) DMSO, and varying compound concentrations to determine IC50 values. All components except GTP were preincubated for 15 min at 30 °C. The reaction mixtures were chilled on ice, and the GTP was added. Samples were transferred to cuvettes held at 0 °C in Beckman DU-7400 and 7500 recording spectrophotometers equipped with electronic temperature controllers. After establishing sample baselines at 0 °C, the temperature was jumped to 30 °C over about 30 sec, and turbidity development was monitored at 350 nm for 20 min. The IC50 was defined as the compound concentration inhibiting the extent of assembly at 20 min by 50%, with values determined by interpolation between experimental concentrations.
6.3.2 Assay for inhibition of colchicine binding to tubulin
Each reaction mixture contained 0.1 mg/mL (1.0 µM) tubulin, 5 µM [3H]colchicine (from Perkin-Elmer), 5% DMSO, compound concentrations as indicated, and additional components that strongly stabilize the colchicine binding activity of tubulin.58, 59 Samples were incubated for 10 min at 37 °C, at which point the reaction in the control reaction mixture is 40–60% complete. Samples were diluted with ice-cold water, and each sample was filtered through a stack of two DEAE-cellulose filters (from Whatman), which were extensively washed with water. The bound colchicine was quantitated by liquid scintillation counting.
6.3.2 Phosphotyrosine ELISA
Cells used were tumor cell lines naturally expressing high levels of VEGFR-2 (U251), PDGFR-β (SF-539) and EGFR (A431). Expression levels at the RNA level were derived from the NCI Developmental Therapeutics Program (NCI-DTP) web site describing molecular target information. Briefly, cells at 60–75% confluence were placed in serum-free medium for 18 h to reduce background phosphorylation. Cells were always >98% viable by trypan blue exclusion. Cells were then pretreated for 60 min to obtain dose-response data, using concentrations of 1.4–100 µM compound, followed in ⅓ log increments by 100 ng/mL VEGF, PDGF-BB or EGF for 10 min. The reaction was stopped, and cells permeabilized by quickly removing the media from the cells and adding ice-cold Tris-buffered saline (TBS) containing 0.05% Triton X-100, protease inhibitor cocktail and tyrosine phosphatase inhibitor cocktail. The TBS solution was then removed and cells fixed to the plate for 30 min at 60 °C with a further incubation in 70% ethanol for 30 min. Cells were exposed to a blocking solution (TBS with 1% BSA) for 1 h, washed, and then a horseradish peroxidase (HRP)-conjugated phosphotyrosine (PY) antibody was added overnight. The antibody was removed, and the cells were washed again in TBS, exposed to an enhanced luminol ELISA substrate (Pierce Chemical EMD, Rockford, IL), and light emission was measured using a UV Products (Upland, CA) BioChemi digital darkroom. Data were graphed as a percent of cells receiving growth factor alone and IC50 values were determined from two to three separate experiments (n = 8–24) using non-linear regression dose-response analysis with Prism 5.0 software (GraphPad, San Diego, CA). In each case, the activity of a positive control inhibitor did not deviate more than 10% from the IC50 values listed in the text.
6.3.2 Cell proliferation analysis of tumor cells
The OVCAR-8 and Pgp overexpressing NCI/ADR-RES cell lines were generously provided by the drug screening group of the Developmental Therapeutics Program, NCI. The wild-type and β-III overexpressing HeLa cells were generous gifts, respectively, of Dr. Richard F. Ludueña and Dr. Susan L. Mooberry. The OVCAR-8 and NCI/ADR-RES cells were grown in RPMI 1640 medium with 5% fetal bovine serum at 37 °C in a 5% CO2 atmosphere for 96 h in the presence of varying compound concentrations. The HeLa cells were grown in MEM supplemented with Earle’s salts, nonessential amino acids, 2 mM L-glutamine, and 10% fetal bovine serum at 37 °C in a 5% CO2 atmosphere for 96 h in the presence of varying compound concentrations. In all cultures, the DMSO concentration was 0.5%. Protein was the parameter measured by the sulforhodamine B technique, and the IC50 was defined as the compound concentration causing a 50% reduction in the increase in cell protein as compared with cultures without compound addition.60
6.3.4 CAM assay of angiogenesis
The CAM assay is a standard assay for testing antiangiogenic agents. The CAM assay used in these studies was modified from a procedure by Sheu61 and Brooks,62 as described previously.63 Briefly, fertile leghorn chicken eggs (CBT Farms, Chestertown, MD) were incubated for 10 days. The proangiogenic factors human VEGF-165 and bFGF (100 ng each) were then added at saturation to a 6 mm microbial testing disk (BBL, Cockeysville, MD) and placed onto the CAM by breaking a small hole in the superior surface of the egg. Antiangiogenic compounds were added 8 h after the VEGF/bFGF at saturation to the same microbial testing disk and embryos allowed to incubate for an additional 40 h. After 48 h, the CAMs were perfused with 2% paraformaldehyde/3% glutaraldehyde containing 0.025% Triton X-100 for 20 sec, excised around the area of treatment, fixed again in 2% paraformaldehyde/3% glutaraldehyde for 30 min, placed on Petri dishes, and a digitized image taken using a dissecting microscope (Wild M400; Bannockburn, IL) at 7.5X and a SPOT enhanced digital imaging system (Diagnostic Instruments, Sterling Heights, MI). A grid was then added to the digital CAM images and the average number of vessels within 5–7 grids counted as a measure of vascularity. Sunitinib and semaxanib were used as a positive control for antiangiogenic activity. Data were graphed as a percent of CAMs receiving bFGF/VEGF only and IC50 values calculated from two to three separate experiments (n = 5–11) using non-linear regression dose-response relation analysis.
6.3.5 4T1 triple negative mammary orthotopic allograft model
The maximum tolerated dose (MTD) was determined in BALB/cJ mice as described64 and found to be 50 mg/kg for 7·HCl (three times weekly) and 1 mg/kg for doxorubicin (once weekly). Both compounds were dissolved in 5% Pharmasolve and 5% Solutol HS in normal saline. The efficacy experiment was performed as previously described.65 Briefly, 4T1-Luc2-GFP, dual luciferase/GFP tagged cells, (PerkinElmer, Waltham, MA) were implanted in the mammary fat pad #4 of 8-week-old female BALB/cJ mice at 7,500 cells in 100 µL phosphate-buffered saline with 1 mM EDTA. One week after implantation, the presence of GFP positive tumor cells was confirmed, and mice were treated intraperitoneally with 7·HCl (three times weekly) or doxorubicin (once weekly). Tumor size was measured with Vernier calipers three times weekly and tumor volume calculated using the ellipsoidal formula [0.52*(length*width*depth)]. To assess the presence of lung metastases at the end of the experiment, animals were euthanized through cervical dislocation while under isoflurane sedation, according to AAALAS guidelines. Lungs were excised and imaged with a Leica Model Z16 APO fluorescence dissecting microscope (GFP filter), which was also used to count metastases per lung.
Figure 10. Treatment with 7·HCl decreased primary tumor growth in the 4T1 orthotopic breast model.
(A) 4T1-luc2-GFP tumor bearing BALB/cJ mice were treated with 7·HCl (IP 50 mg/kg, three times weekly), or doxorubicin (dox) (IP 1 mg/kg, weekly) and tumor volumes were determined. Statistical analysis was performed with two-way ANOVA repeated measures post test. (B) Animal weights were graphed as percent weight change at day 40 over the starting weight. Statistical analysis was performed with one-way ANOVA. (C) At the end of the experiment, animals were euthanized and lungs removed and number of metastases per lung counted and graphically represented. n =9 – 10, *P<0.05, **P<0.01 versus untreated control group.
Acknowledgments
We gratefully acknowledge the NCI for performing the in vitro antitumor evaluation in their 60 tumor cell line panel. This work was supported, in part, by the National Institutes of Health and NCI grants CA136944 (A.G.) and CA142868 (A.G.), NSF equipment grant for NMR instrumentation (NMR: CHE 0614785) and Duquesne University Adrian Van Kaam Chair in Scholarly Excellence (A.G.).
Abbreviations
- AA
antiangiogenic agents
- MTA
microtubule targeting agents
- VEGFR-2
vascular endothelial growth factor receptor-2
- PDGFR-β
platelet-derived growth factor receptor-β
- CAM
chorioallantoic membrane
- CA
combretastatin A-4
- Pgp
P-glycoprotein
- EGFR
epidermal growth factor receptor
- TBS
Tris-buffered saline
- HRP
horseradish peroxidase
- PY
phosphotyrosine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer
The content of this paper is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
References
- 1.Carmeliet P, Jain RK. Nature. 2000;407:249. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Jain RK. Nature. 2011;473:298. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibuya M. J. Biochem. 2013;153:13. doi: 10.1093/jb/mvs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancuso MR, Davis R, Norberg SM, O’Brien S, Sennino B, Nakahara T, Yao VJ, Inai T, Brooks P, Freimark B, Shalinsky DR, Hu-Lowe DD, McDonald DM. J. Clin. Invest. 2006;116:2610. doi: 10.1172/JCI24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Zhang Y, Iwamoto H, Hosaka K, Seki T, Andersson P, Lim S, Fischer C, Nakamura M, Abe M, Cao R, Skov PV, Chen F, Chen X, Lu Y, Nie G, Cao Y. Nat. Commun. 2016;7:12680. doi: 10.1038/ncomms12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain RK. J. Clin. Oncol. 2013;31:2205. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizawa Y, Ogawara K-i, Fushimi A, Abe S, Ishikawa K, Araki T, Molema G, Kimura T, Higaki K. Mol. Pharm. 2012;9:3486. doi: 10.1021/mp300318q. [DOI] [PubMed] [Google Scholar]
- 8.Winkler F, Kozin SV, Tong RT, Chae SS, Booth MF, Garkavtsev I, Xu L, Hicklin DJ, Fukumura D, di Tomaso E, Munn LL, Jain RK. Cancer cell. 2004;6:553. doi: 10.1016/j.ccr.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Jordan MA, Wilson L. Nat. Rev. Cancer. 2004;4:253. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 10.Clinical trials on clinicaltrials.gov. (accessed in Sept 2016) in which RTKIs were being used in combination with MTAs: NCT01898117; NCT02319577; NCT02898077; NCT00820170; NCT02364362; NCT01606878; NCT02326285; NCT01974440; NCT01683994; NCT02378389; NCT01939054; NCT02191059. [Google Scholar]
- 11.Gangjee A, Zaware N, Raghavan S, Ihnat M, Shenoy S, Kisliuk RL. J. Med. Chem. 2010;53:1563. doi: 10.1021/jm9011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangjee A, Zhao Y, Ihnat MA, Thorpe JE, Bailey-Downs LC, Kisliuk RL. Bioorg. Med. Chem. 2012;20:4217. doi: 10.1016/j.bmc.2012.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangjee A, Li W, Lin L, Zeng Y, Ihnat M, Warnke LA, Green DW, Cody V, Pace J, Queener SF. Bioorg. Med. Chem. 2009;17:7324. doi: 10.1016/j.bmc.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangjee A, Zeng Y, Ihnat M, Warnke LA, Green DW, Kisliuk RL, Lin F-T. Bioorg. Med. Chem. 2005;13:5475. doi: 10.1016/j.bmc.2005.04.087. [DOI] [PubMed] [Google Scholar]
- 15.Gangjee A, Pavana RK, Ihnat MA, Thorpe JE, Disch BC, Bastian A, Bailey-Downs LC, Hamel E, Bai R. ACS Med. Chem. Lett. 2014;5:480. doi: 10.1021/ml4004793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Raghavan S, Ihnat M, Hamel E, Zammiello C, Bastian A, Mooberry SL, Gangjee A. Bioorg. Med. Chem. 2015;23:2408. doi: 10.1016/j.bmc.2015.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Raghavan S, Ihnat M, Thorpe JE, Disch BC, Bastian A, Bailey-Downs LC, Dybdal-Hargreaves NF, Rohena CC, Hamel E, Mooberry SL, Gangjee A. Bioorg. Med. Chem. 2014;22:3753. doi: 10.1016/j.bmc.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, Weinstein JN, Friend S, Fornace AJ, Kohn KW. Cancer Res. 1997;57:4285. [PubMed] [Google Scholar]
- 19.Wang Z, Sun Y. Transl. Oncol. 2010;3:1. doi: 10.1593/tlo.09250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bello E, Taraboletti G, Colella G, Zucchetti M, Forestieri D, Licandro SA, Berndt A, Richter P, D’Incalci M, Cavalletti E, Giavazzi R, Camboni G, Damia G. Mol. Cancer Ther. 2013;12:131. doi: 10.1158/1535-7163.MCT-12-0275-T. [DOI] [PubMed] [Google Scholar]
- 21.Bar J, Onn A. Exp. Opin. Pharmacother. 2008;9:701. doi: 10.1517/14656566.9.5.701. [DOI] [PubMed] [Google Scholar]
- 22.Ulahannan SV, Brahmer JR. Cancer Invest. 2011;29:325. doi: 10.3109/07357907.2011.554476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Nat. Rev. Cancer. 2013;13:714. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 24.Ganguly A, Cabral F. BBA - Rev. Cancer. 2011;1816:164. doi: 10.1016/j.bbcan.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burger RA. Gynecol. Oncol. 2011;121:230–238. doi: 10.1016/j.ygyno.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 26.Ellis PM, Al-Saleh K. Crit. Rev. Oncol. Hemat. 2012;84:47. doi: 10.1016/j.critrevonc.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Socinski MA. Cancer Treat. Rev. 2011;37:611. doi: 10.1016/j.ctrv.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Patel RY, Leung H. Curr. Pharm. Des. 2012;18(19):2672–2679. doi: 10.2174/138161212800626148. [DOI] [PubMed] [Google Scholar]
- 29.Denison TA, Bae YH. J. Control. Release. 2012;164:187. doi: 10.1016/j.jconrel.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher R, Pusztai L, Swanton C. Br. J. Cancer, BJC. 2013;108:479. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marusyk A, Almendro V, Polyak K. Nat. Rev. Cancer. 2012;12:323. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 32.Turner NC, Reis-Filho JS. Lancet Oncol. 2012;13:e178. doi: 10.1016/S1470-2045(11)70335-7. [DOI] [PubMed] [Google Scholar]
- 33.Towner RA, Ihnat M, Saunders D, Bastian A, Smith N, Pavana RK, Gangjee A. BMC Cancer. 2015;15:522. doi: 10.1186/s12885-015-1538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangjee A, Pavana R, Li W, Hamel E, Westbrook C, Mooberry S. Pharm. Res. 2012;29:3033. doi: 10.1007/s11095-012-0816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangjee A, Zaware N, Raghavan S, Yang J, Thorpe JE, Ihnat MA. Bioorg. Med. Chem. 2012;20:2444. doi: 10.1016/j.bmc.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gangjee A, Kurup S, Ihnat MA, Thorpe JE, Disch B. Bioorg. Med. Chem. 2012;20:910. doi: 10.1016/j.bmc.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gangjee A, Zaware N, Raghavan S, Disch BC, Thorpe JE, Bastian A, Ihnat MA. Bioorg. Med. Chem. 2013;21:1857. doi: 10.1016/j.bmc.2013.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popat S, Mellemgaard A, Fahrbach K, Martin A, Rizzo M, Kaiser R, Griebsch I, Reck M. Future Oncol. 2014;11:409. doi: 10.2217/fon.14.290. [DOI] [PubMed] [Google Scholar]
- 39.Reck M, Kaiser R, Mellemgaard A, Douillard J-Y, Orlov S, Krzakowski M, von Pawel J, Gottfried M, Bondarenko I, Liao M, Gann C-N, Barrueco J, Gaschler-Markefski B, Novello S. Lancet Oncol. 2014;15:143. doi: 10.1016/S1470-2045(13)70586-2. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto I, Miyazaki M, Takeda M, Terashima M, Azuma K, Hayashi H, Kaneda H, Kurata T, Tsurutani J, Seto T, Hirai F, Konishi K, Sarashina A, Yagi N, Kaiser R, Nakagawa K. J. Thorac. Oncol. 2015;10:346. doi: 10.1097/JTO.0000000000000395. [DOI] [PubMed] [Google Scholar]
- 41.Quintela-Fandino M, Urruticoechea A, Guerra J, Gil M, Gonzalez-Martin A, Marquez R, Hernandez-Agudo E, Rodriguez-Martin C, Gil-Martin M, Bratos R, Escudero MJ, Vlassak S, Hilberg F, Colomer R. Br. J. Cancer. 2014;111:1060. doi: 10.1038/bjc.2014.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gangjee A, Namjoshi OA, Ihnat MA, Buchanan A. Bioorg. Med. Chem. Lett. 2010;20:3177. doi: 10.1016/j.bmcl.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prota AE, Danel F, Bachmann F, Bargsten K, Buey RM, Pohlmann J, Reinelt S, Lane H, Steinmetz MO. J. Mol. Biol. 2014;426:1848. doi: 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 44.McTigue M, Murray BW, Chen JH, Deng Y-L, Solowiej J, Kania RS. Proc. Natl. Acad. Sci. USA. 2012;109:18281. doi: 10.1073/pnas.1207759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yosaatmadja Y, Squire CJ, McKeage M, Flanagan JU. [Google Scholar]
- 46.Elliott AJ, Morris PE, Petty SL, Williams CH. J. Org. Chem. 1997;62:8071. doi: 10.1021/jo971062j. [DOI] [PubMed] [Google Scholar]
- 47.Ji Y-T, Liu Y-N, Liu Z-P. Curr. Med. Chem. 2015;22:1348. doi: 10.2174/0929867322666150114163732. [DOI] [PubMed] [Google Scholar]
- 48.Gangjee A, Zhao Y, Lin L, Raghavan S, Roberts EG, Risinger AL, Hamel E, Mooberry SL. J. Med. Chem. 2010;53:8116. doi: 10.1021/jm101010n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Raghavan S, Ihnat M, Thorpe JE, Disch BC, Bastian A, Bailey-Downs LC, Dybdal-Hargreaves NF, Rohena CC, Hamel E, Mooberry SL, Gangjee A. Bioorg. Med. Chem. 2014;22:3753. doi: 10.1016/j.bmc.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gangjee A, Zaware N, Devambatla RKV, Raghavan S, Westbrook CD, Dybdal-Hargreaves NF, Hamel E, Mooberry SL. Bioorg. Med. Chem. 2013;21:891. doi: 10.1016/j.bmc.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.SYBYL-X 2.1.1, Tripos International, 1699 South Hanley Rd. St. Louis, Missouri, 63144, USA [Google Scholar]
- 52.Schönherr H, Cernak T. Angew. Chem. Int. Ed. 2013;52:12256. doi: 10.1002/anie.201303207. [DOI] [PubMed] [Google Scholar]
- 53.Lin CM, Ho HH, Pettit GR, Hamel E. Biochemistry. 1989;28:6984. doi: 10.1021/bi00443a031. [DOI] [PubMed] [Google Scholar]
- 54.Weigelt B, Peterse JL, van ‘t Veer LJ. Nat. Rev. Cancer. 2005;5:591. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 55.Molecular Operating Environment (MOE), 2015.10. 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: Chemical Computing Group Inc; 2016. [Google Scholar]
- 56.Hamel E, Lin CM. Biochemistry. 1984;23:4173. doi: 10.1021/bi00313a026. [DOI] [PubMed] [Google Scholar]
- 57.Hamel E. Cell Biochem. Biophys. 2003;38:1. doi: 10.1385/CBB:38:1:1. [DOI] [PubMed] [Google Scholar]
- 58.Verdier-Pinard P, Lai J-Y, Yoo H-D, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Mol. Pharmacol. 1998;53:62. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 59.Hamel E, Lin CM. BBA - Gen. Sub. 1981;675:226. doi: 10.1016/0304-4165(81)90231-2. [DOI] [PubMed] [Google Scholar]
- 60.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. J. Natl. Cancer Inst. 1990;82:1107. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 61.Sheu JR, Fu CC, Tsai ML, Chung WJ. Anticancer Res. 1998;18:4435. [PubMed] [Google Scholar]
- 62.Brooks PC, Montgomery AM, Cheresh DA. Methods Mol. Biol. 1999;129:257. doi: 10.1385/1-59259-249-X:257. [DOI] [PubMed] [Google Scholar]
- 63.Marks MG, Shi J, Fry MO, Xiao Z, Trzyna M, Pokala V, Ihnat MA, Li PK. Biol. Pharm. Bull. 2002;25:597. doi: 10.1248/bpb.25.597. [DOI] [PubMed] [Google Scholar]
- 64.Bastian A, Thorpe JE, Disch BC, Bailey-Downs LC, Gangjee A, Devambatla RK, Henthorn J, Humphries KM, Vadvalkar SS, Ihnat MA. J. Pharm. Exp. Ther. 2015;353:392. doi: 10.1124/jpet.114.220335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey-Downs LC, Thorpe JE, Disch BC, Bastian A, Hauser PJ, Farasyn T, Berry WL, Hurst RE, Ihnat MA. PloS one. 2014;9:e98624. doi: 10.1371/journal.pone.0098624. [DOI] [PMC free article] [PubMed] [Google Scholar]