Abstract
The complete sequence of the Arabidopsis thaliana genome revealed thousands of previously unsuspected genes, many of which cannot be ascribed even putative functions. One of the largest and most enigmatic gene families discovered in this way is characterized by tandem arrays of pentatricopeptide repeats (PPRs). We describe a detailed bioinformatic analysis of 441 members of the Arabidopsis PPR family plus genomic and genetic data on the expression (microarray data), localization (green fluorescent protein and red fluorescent protein fusions), and general function (insertion mutants and RNA binding assays) of many family members. The basic picture that arises from these studies is that PPR proteins play constitutive, often essential roles in mitochondria and chloroplasts, probably via binding to organellar transcripts. These results confirm, but massively extend, the very sparse observations previously obtained from detailed characterization of individual mutants in other organisms.
INTRODUCTION
The Arabidopsis thaliana genome sequence uncovered many previously undescribed and often unsuspected genes. The initial analysis of the genome sequence estimated that 31% of Arabidopsis genes were too dissimilar to genes of known function to be attributed putative functions (Arabidopsis Genome Initiative, 2000). Many of the genes in this class fall into families that have greatly expanded in plants or are entirely plant specific. A major challenge for the plant science community is to discover the functions of these genes, given that little or no help can be expected from studies on model microorganisms or metazoa, so useful for more widely conserved gene families. Amongst these families, the largest (∼450 members) and perhaps the most mysterious is defined by the so-called pentatricopeptide repeat (PPR) (Small and Peeters, 2000). PPR proteins make up a significant proportion (∼6%) of the unknown function proteins in Arabidopsis. Only a few very recent articles describe the functional analysis of individual Arabidopsis PPR genes (Hashimoto et al., 2003; Meierhoff et al., 2003; Yamazaki et al., 2004).
Although individual PPR genes were described from mutant studies in yeast and Neurospora many years ago (Manthey and McEwen, 1995; Coffin et al., 1997), the existence of a large family of similar proteins only became apparent with the sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). Two independent partial and sometimes contradictory descriptions of the family and the characteristic motif(s) defining it were offered before the completion of the genome sequence. Small and Peeters coined the name PPR for the major motif based on its similarity to the better-known tetratricopeptide repeat (TPR) motif and briefly described a large protein family containing it (Small and Peeters, 2000). Aubourg et al. described a large Arabidopsis protein family containing multiple modules of different repeated motifs (Aubourg et al., 2000). Subsequently it became obvious that these two families overlapped considerably and that several of the motifs described by Aubourg et al. correspond to PPR motifs or variants thereof. One of the goals of this article is to provide a unified and detailed description of the family from the complete Arabidopsis genome sequence to provide a coherent, stable platform on which future studies can be based.
The few data available point to an involvement of PPR proteins in posttranscriptional processes in organelles. The mutants pet309 (Manthey and McEwen, 1995; Manthey et al., 1998) and cya-5 (Coffin et al., 1997) in yeast and Neurospora crassa are affected in stability and translation of mitochondrial cox1 transcripts. A similar PPR gene in humans has been linked to a genetic disease characterized by COX1 deficiency (Mootha et al., 2003). In a landmark study, Fisk et al. recognized the similarity of the fungal genes to a maize (Zea mays) gene, CRP1, implicated by genetic analysis in processing and translation of plastid pet transcripts (Fisk et al., 1999). Similar effects on plastid transcripts have been subsequently observed in other mutants from Chlamydomonas (Lown et al., 2001), Arabidopsis (Hashimoto et al., 2003; Meierhoff et al., 2003; Yamazaki et al., 2004), and maize (Williams and Barkan, 2003). Further evidence for a role of PPR proteins in organelle gene expression has come from positional cloning of cytoplasmic male sterility (CMS) restorer genes. Rf1 from petunia (Petunia hybrida) (Bentolila et al., 2002), Rfk and Rfo from radish (Raphanus sativus) (Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003), and Rf-1 from rice (Oryza sativa) (Kazama and Toriyama, 2003; Komori et al., 2004) have all been shown to encode PPR proteins. Although the mechanism of action of these proteins is not known, they all share the ability to prevent expression of the protein encoded by the corresponding mitochondrial CMS inducer gene.
The phenotypes of these mutants strongly suggest that PPR proteins act directly or indirectly on RNA, but evidence for RNA binding by these proteins remains very sketchy. Radish p67 and Drosophila bicoid stability factor were both purified as sequence-specific RNA binding proteins (Lahmy et al., 2000; Mancebo et al., 2001), although doubts remain as to whether the bait RNAs used are physiological targets for these proteins. Wheat (Triticum aestivum) p63 (Ikeda and Gray, 1999) and mouse LRP130 (Tsuchiya et al., 2002) have been shown to bind DNA, although LRP130 has also been convincingly shown to bind RNA (Mili and Pinol-Roma, 2003). Hcf152 has been shown to bind the pet transcript affected in the hcf152 mutant and is the best documented case to our knowledge where a PPR protein has been demonstrated to bind its probable natural target RNA (Meierhoff et al., 2003; Nakamura et al., 2003).
Hence, whereas the functional data available on PPR proteins are rather coherent, they remain very sparse and, as we shall demonstrate, almost completely confined to one subgroup of the whole family. The second goal of the work described here was to obtain family-wide data, using high-throughput functional genomics technology where possible, to expand our fragmentary knowledge of the roles of these proteins.
RESULTS
Identification of the PPR-Encoding Genes in the Arabidopsis Genome
With the completion of the sequencing of the Arabidopsis genome, we were able to search for all the PPR genes in Arabidopsis. We first used the alignment of 2357 previously identified PPR motifs (Small and Peeters, 2000) to obtain a consensus model using the HMMER package. This PPR signature matrix was subsequently used to search for PPR motifs in the six reading frames of the whole Arabidopsis genome. Because PPR motifs are highly degenerate, a fairly large number of false positive hits were expected. However, because PPR motifs are almost always observed in proteins as tandem repeats, we could remove most random uninteresting matches by omitting all orphan PPR motifs that were found farther than 200 nucleotides from any other PPR motif. The 6371 PPR motifs retained formed 526 clusters, each of which comprises a putative PPR gene. Each PPR motif cluster was then investigated in detail by manually analyzing the positions and reading frames of the PPR motifs compared with (1) Arabidopsis Genome Initiative (AGI) open reading frame (ORF) models in the same region, (2) sequenced cDNAs or ESTs matching to the same region, and (3) predicted N-terminal mitochondrial and plastid targeting sequences within potential coding sequence in the region. From this analysis, 441 putative PPR ORF models were constructed (i.e., ∼80 motif clusters were discarded or fused with other clusters). As of Release 5.0 of The Institute for Genomic Research (TIGR) annotations of Arabidopsis proteins, 447 of the AGI ORFs are annotated as PPR-containing proteins based on matches with the PFAM profile PF01535 or Interpro profile IPR002885. Three other AGI models are annotated as homologs of known PPR proteins. Of the two sets of ORF models, 326 are identical (i.e., our manual analysis agreed with the AGI model) (see Supplemental Table S1 online). Twenty-four AGI models do not have equivalents in our set; nine because we do not consider them to be PPR-containing proteins by our criteria (they lack tandem motifs matching our HMMER matrices) and 15 because we consider them to be probable pseudogenes (they would require multiple unlikely introns and/or frameshifts to produce a model resembling other members of the family). Six of our models have no AGI equivalent and correspond to genes apparently overlooked during the AGI annotation or considered to be pseudogenes. In three more cases (At1g62910, At5g08310, and At5g40410), we split AGI models into two to form six new models because we felt that the TIGR annotations had fused tandemly duplicated PPR genes. We also found six other cases of probable fusions of ORF models, this time between PPR genes and members of completely unrelated families. In all, 115 of our models differ in at least some respects from the corresponding AGI model. It should be noted that in very few of these cases are molecular data available that can be used to decide between the two discordant models. Our choice has been generally made by comparison with other genes in the family and a general familiarity with these proteins. This manually annotated version of the PPR family from the Arabidopsis genome is available in the supplemental data online.
The PPR-Related Motifs
The PPR motifs are usually present in proteins as tandem arrays of about a dozen repeats. In about one half of the family, these PPR motifs are adjacent to each other without gaps between the motifs. In the other half of the family, we observed regular gaps between PPR motifs of ∼65 to 70 amino acids. We extracted and aligned the fragments found separating PPR motifs, and these alignments were used to define models using the HMMER package. This analysis allowed us to define two new motifs of 31 and 35 to 36 amino acids. These consensus sequences were compared with PPR consensus (Figure 1) showing that the two motifs are clearly related to the PPR motif. In particular, the secondary structure predictions for the two motifs strongly suggested a pair of α-helices as already proposed for the PPR motif (Small and Peeters, 2000). These two motifs were named the PPR-like S (for short) and PPR-like L (for long) motifs. The L motifs in particular are variable in size and sequence, and several variants could be defined. The two major variants, described in Figure 1, have been used in this study. The HMMER matrices for these motifs were used to search for PPR-like motifs in the six frames of the Arabidopsis genome (as described earlier for the PPR motif). Because all these PPR-like motifs are related, many overlapping or coincident matches were obtained. In these cases, the arrangement of nonoverlapping motifs giving the highest cumulative hmmsearch scores was retained (see Supplemental Table S2 online). After removing redundant motifs in this way, 806 L-type and 1304 S-type motifs were found in >300 proteins to add to the 3340 P-type classical PPR motifs. It is noticeable that these motifs were only found in previously identified PPR proteins, reinforcing the idea that these motifs are highly related to PPR motifs. By contrast, these motifs were not found in all the PPR proteins identified in Arabidopsis. Slightly less than half of the PPR proteins present an organized pattern of triple motifs (P-L-S) repeated along the protein sequence. This subclass of the PPR family corresponds to the Arabidopsis plant combinatorial and modular protein (AtPCMP) family, previously discovered by bioinformatic analysis of the Arabidopsis genome (Aubourg et al., 2000). They described 127 proteins characterized by a combinatorial organization of A, B, and C motifs. These motifs, identified on the basis of sequence similarity rather than structural considerations, match to parts of the PPR and PPR-related motifs that we describe here (Figure 1). The A, B, and C motifs correspond to the N-terminal part of the L motif, an internal peptide of the S motif, and a peptide overlapping with S and PPR motifs, respectively. Aubourg et al. considered that the sequence similarity between PCMP proteins and PPR proteins was not significant, but the structural and organizational similarities highlighted by this genome-wide study strongly suggest that they are related and belong to the same superfamily.
Figure 1.
Alignment of the Four Most Abundant PPR-Related Motifs in Arabidopsis in Comparison with the PFAM PF01535 PPR Motif.
Consensus sequences were obtained using the HMMER package based on alignments of thousands of Arabidopsis motifs. The PPR consensus used by PFAM (and obtained using motifs from a variety of organisms) is almost identical to our consensus except shifted by two amino acids, such that it overlaps the first helix of the following motif. Residues in capital letters are more highly conserved within each motif. Residues in bold are conserved between PPR-related motifs. The underlined sequences indicate the correspondences to the motifs A (underlined three times), B (underlined once), and C (underlined twice) described by Aubourg et al. (2000). Motif C overlaps adjacent S and P motifs. The shaded boxes indicate the maximum extent of the predicted α-helical regions.
PPR proteins contain from 2 to at least 26 copies of the PPR motif or its variants, with an average of 12 motifs per protein, usually in a single unbroken tandem array that forms at least two-thirds of the protein sequence. There are clear indications that polymorphism within the family can be because of deletion or duplication of tandem repeats, and this, coupled with the difficulty in obtaining unambiguous alignments given the number of repetitions of similar sequence, makes sequence homology–based phylograms of the family difficult or impossible to interpret.
Identification of C-Terminal Domains
Many PPR proteins contain sequences unrelated to PPR motifs either before or after the tandem arrays of PPR motifs. The sequences before are mostly comprised of organelle targeting sequences and show little sequence similarity. However, the sequences following the PPR motifs show considerable sequence conservation in many cases. To characterize C-terminal motifs in the PPR family, we aligned the polypeptide sequences C-terminal of the last PPR-related motif from all of the Arabidopsis PPR proteins. This alignment allowed us to define three motifs (E, E+, and DYW) that were defined in more detail using the HMMER package (Figure 2). The E, E+, and DYW motifs were found in 191, 145, and 87 Arabidopsis proteins, respectively. They roughly correspond to the E, F-G, and H motifs defined by Aubourg et al. (2000) from 120 AtPCMP proteins. Despite the fact that these new motifs are not related in sequence to the PPR motifs, the three motifs were only found in members of the PPR family and not in any other Arabidopsis protein with the exception of At1g47580, which contains a lone DYW motif. Interestingly, they were only found in the subfamily defined by P-L-S repeats. The relative organization of the three C-terminal motifs follows several quasi-absolute rules: (1) the motifs are never observed in multiple copies in the same protein; (2) when the motifs are observed in the same protein they are ordered colinearly E – E+ – DYW, DYW being the C-terminal tripeptide; (3) proteins carrying the DYW motif almost always have preceding E and E+ motifs; similarly, proteins carrying E+ motif always have a preceding E motif. The identification of these motifs allowed us to define four subclasses in the P-L-S subfamily: (1) proteins that do not have any of the three C-terminal motifs, (2) proteins with the E motif alone, (3) proteins with E and E+ motifs, and (4) proteins with the E, E+, and DYW motifs (Figure 3), named the P-L-S, E, E+, and DYW subclasses, respectively (see Supplemental Table S2 online). These subclasses of the Arabidopsis PPR family contain 6, 47, 60, and 87 proteins, respectively (Figure 3).
Figure 2.
HMMER-Derived Consensus Sequences of C-Terminal Motifs Present in PPR Proteins.
The alignments employed for the E, E+, and DYW motifs contained 184, 148, and 85 sequences, respectively. The best conserved residues are in capital letters; bold, underlined amino acids are completely invariant. For the DYW motif, the DYW triplet (or a closely related sequence) forms the C terminus of the protein.
Figure 3.
Motif Structure of Arabidopsis PPR Proteins.
Typical structures of proteins from each of the principal subfamilies and subgroups are shown. The structures are purely indicative, and the number and even order of repeats can vary in individual proteins. The number of proteins falling into each subgroup is shown.
In contrast with PPR motifs that appear to be TPR-related motifs (Small and Peeters, 2000), the three C-terminal motifs do not resemble any previously described polypeptide motif. The E and E+ motifs are highly degenerate motifs, but the high conservation in the amino acid sequences and the presence of invariant Cys and His in the DYW motif may imply a catalytic function for this domain (Aubourg et al., 2000).
Genomic Organization
PPR genes are fairly evenly distributed throughout the 10 chromosome arms, with little in the way of obvious clusters. The densest grouping of PPR genes lies on the long arm of chromosome 1 (around 23 megabases) where 19 genes and several probable pseudogenes lie within little more than one megabase (data not shown). Many of the genes in this cluster are closely related, and they include the closest Arabidopsis homologs of the recently cloned CMS restorer genes from petunia, radish, and rice. The radish restorer Rfo (Rfk1) lies in a region that is very clearly syntenic to this Arabidopsis cluster (Desloire et al., 2003).
A noticeable characteristic of PPR genes is that they rarely contain introns within the coding sequence. More than 80% (363/441) of our PPR ORF models contain a single exon, and only 7% (31/441) contain more than one intron. These figures are astonishingly low compared with the average of five introns or more for other Arabidopsis genes (Arabidopsis Genome Initiative, 2000). This characteristic explains why PPR genes are relatively short (on average <2 kb from ATG to stop codon) despite the fact that PPR proteins are comparatively large proteins (642 amino acids on average).
PPR and PPR-Related Motifs in Other Genomes
To characterize the PPR family in a variety of other organisms, we collected data on several fully sequenced and annotated genomes (Caenorhabditis elegans, Drosophila melanogaster, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Cyanidioschyzon merolae, Escherischia coli, Ralstonia solanacearum, Rickettsia prowazekii, and Synechocystis sp 101) or almost completely annotated genomes (human, rice, and Trypanosoma brucei). Using the HMMER matrices defined using Arabidopsis PPR, E, and DYW motifs, we searched for these motifs in the collected genomes (Table 1). The criteria for the motif search were (1) an hmmsearch E-value < 10 and (2) at least one pair of PPR or PPR-like repeats in any protein hit. Using these criteria, no PPR proteins were identified in any of the prokaryotic genomes that we analyzed except for a single protein from R. solanacearum. This PPR-like protein appears to be a fairly recent capture by horizontal transfer (Salanoubat et al., 2002). By contrast, PPR motifs were found in all eukaryotes analyzed but with an extraordinary discrepancy in numbers between plant and nonplant organisms. Thus, whereas Arabidopsis and rice have several hundreds of PPR genes in their genome, the human genome encodes only six putative PPR proteins. Trypanosomes, recently linked to the plant kingdom (Hannaert et al., 2003; Martin and Borst, 2003) and the red alga C. merolae (Matsuzaki et al., 2004), have more PPR proteins than humans but many fewer than the two plant genomes. The list of putative nonplant PPR proteins is given in Supplemental Table S3 online. We also looked for E and DYW motifs (Table 1) and for L and S variants of PPR motifs (data not shown), but we were unable to find conclusive examples of any of them outside the higher plant genomes, indicating that the P-L-S half of the family is strictly restricted to plants.
Table 1.
PPR Proteins Are Specific to Eukaryotes
| Organism | Sequences | PPR Hits | E Hits | DYW Hits |
|---|---|---|---|---|
| Homo sapiens | 37,490 | 6 | 0 | 0 |
| Drosophila melanogaster | 17,087 | 2 | 0 | 0 |
| Caenorhabditis elegans | 20,673 | 2 | 0 | 0 |
| Schizosaccharomyces pombe | 5,010 | 6 | 0 | 0 |
| Saccharomyces cerevisiae | 6,304 | 5 | 0 | 0 |
| Trypanosoma brucei | 16,757 | 19 | 0 | 0 |
| Cyanidioschyzon merolae | 4,772 | 10 | 0 | 0 |
| Arabidopsis thaliana | 28,581 | 470 | 193 | 87 |
| Oryza sativa | 74,385 | 655 | 292 | 104 |
| Ralstonia solanacearum | 5,118 | 1 | 0 | 0 |
| Rickettsia prowazekii | 834 | 0 | 0 | 0 |
| Synechocystis sp | 3,169 | 0 | 0 | 0 |
Searches for PPR and PPR-associated motifs were conducted on complete or quasicomplete predicted proteomes from a range of species using HMMER. The figures indicate the number of proteins found to contain two or more PPR motifs with an E-value of <10 or one or more E or DYW motifs with a score of >0.
Subcellular Localization of the PPR Proteins
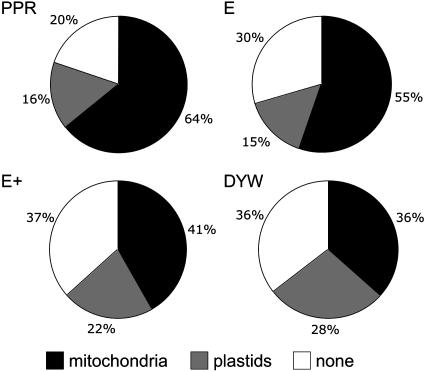
Most of the few known PPR proteins have roles in mitochondria or plastids, and the PPR family was discovered while systematically screening Arabidopsis proteins for those predicted to be targeted to mitochondria or chloroplasts (Small and Peeters, 2000). We therefore checked for predicted targeting signal peptides at the N terminus of PPR proteins using the TargetP (Emanuelsson et al., 2000) and Predotar (Small et al., 2004) prediction programs. Both programs assigned most of the proteins of the family to either mitochondria or plastids (see Supplemental Table S4 online). Using the Predotar program and a threshold probability of 0.25, 54 and 19% of the PPR proteins were predicted to be targeted to mitochondria or plastids, respectively (Figure 4). TargetP gave similar results (see Supplemental Table S4 online). Differences were observed in the proportion of the different targeting predictions between the various subclasses (Figure 4). For example, whereas pure PPR proteins are mostly predicted to be targeted to mitochondria, DYW proteins are almost as likely to be predicted to be targeted to plastids as to mitochondria. Furthermore, the percentage of predicted untargeted proteins in the E, E+, and DYW subclasses is significantly higher than in the pure PPR subfamily (35% versus 20%, P < 0.001, χ2 test). These differences might reflect fundamental differences in the function of these proteins, but they may simply be because of major differences in ORF model quality, members of the pure PPR subfamily being better known and more expressed than the other subclasses (see below). Overall, the percentage of observed false negative results using Predotar or TargetP is ∼20 to 30% (Emanuelsson et al., 2000; Small et al., 2004) (i.e., not very different than the number of apparently untargeted PPR proteins). Furthermore, because less than half of the PPR ORF models could be validated by cDNA sequences (see below) and because small exons were found at the 5′-end of a few cDNA-validated PPR ORF models, some of the models are undoubtedly lacking the correct initiation codon and thus are missing a potential targeting peptide. Although it is not possible to definitively conclude, our targeting prediction results are consistent with the idea that very few, if any, PPR proteins are localized outside the organelles of plant cells.
Figure 4.
Predicted Subcellular Localization of PPR Proteins.
The proportions of each PPR subclass predicted by Predotar to be targeted to mitochondria (black segments) or plastids (gray segments) or to lack targeting signals (white segments) are indicated. PPR and E proteins are mostly predicted to be mitochondrial; E+ and DYW proteins are predicted to be more evenly distributed between the two organelles.
To further characterize the subcellular localization of PPR proteins, we experimentally analyzed the localization of several PPR-green fluorescent protein (GFP) or PPR-DsRed2 fluorescent fusion proteins. A set of 48 PPR proteins representing each subclass of the PPR family was chosen for the targeting experiments. The corresponding full-length ORFs (from start codon to the last codon before the stop codon) and the N-terminal targeting signal coding sequences (arbitrarily fixed as being the first 300 bp of the coding region of the genes) were systematically cloned into a pDONR207 Gateway entry vector. This recombinational cloning system was subsequently used to further subclone PPR ORFs and PPR presequences into several GFP or DsRed2 destination vectors (see Methods for details). pOL-type vectors (Peeters et al., 2000) were used for electroporation of tobacco protoplasts. Agrobacteria carrying binary vectors were used for agroinfiltration of Nicotiana benthamiana leaves. The subcellular localization of 45 PPR proteins was analyzed in these experiments. We were able to observe fluorescence for 26 out of these 45 PPR protein fusions (Table 2). For the remaining 19 PPR proteins, we were unable to detect any interpretable fluorescence signal for any of the constructs tested (data not shown), the fusion proteins either aggregating or not accumulating to detectable levels.
Table 2.
Subcellular Localization of Selected PPR Proteins
| AGI Code | PPR Subclass | Electroporation Experiments | Agroinfiltration Experiments | Organellar Proteomics | TargetP Prediction | Predotar Prediction |
|---|---|---|---|---|---|---|
| At1g11630 | PPR | ND | ND | Mitochondriaa | Mitochondria | Mitochondria |
| At1g26460 | PPR | ND | ND | Mitochondriaa | Mitochondria | Mitochondria |
| At1g55890 | PPR | ND | ND | Mitochondriaa | Mitochondria | Possibly mitochondria |
| At1g59720 | DYW | ND | Mitochondriabc | Plastid | None | |
| At1g60770 | PPR | ND | ND | Mitochondriaa | None | Possibly mitochondria |
| At1g61870 | PPR | Mitochondriacde | Mitochondriac | Mitochondriaa | Mitochondria | Mitochondria |
| At2g01390 | PPR | Mitochondriac | ND | Plastid | Possibly mitochondria | |
| At2g19280 | PPR | Mitochondriab | ND | Mitochondria | Possibly plastid | |
| At2g20540 | E+ | No signald | Mitochondriab | None | None | |
| At2g34370 | DYW | Mitochondriace | ND | Mitochondria | Mitochondria | |
| At3g02010 | DYW | Mitochondriabde | ND | Mitochondria | None | |
| At3g11460 | DYW | Mitochondriade | Mitochondriab | Plastid | Plastid | |
| At3g13160 | PPR | ND | ND | Mitochondriaa | Mitochondria | Mitochondria |
| At3g13770 | DYW | Mitochondriab | No signalc | Mitochondria | Mitochondria | |
| At3g13880 | E+ | Mitochondriab | No signalc | Mitochondria | Possibly mitochondria | |
| At3g24000 | DYW | Mitochondriad | Mitochondriab | Mitochondria | Mitochondria | |
| At3g26780 | DYW | ND | ND | Mitochondriaa | Mitochondria | Mitochondria |
| At3g60960 | PPR | ND | ND | Mitochondriaa | Mitochondria | Possibly mitochondria |
| At4g02750 | DYW | Mitochondriab | ND | Mitochondria | None | |
| At4g20090 | PPR | ND | Mitochondriab | Plastid | Possibly ER | |
| At4g35850 | PPR | ND | ND | Mitochondriaa | Mitochondria | Mitochondria |
| At4g36680 | PPR | ND | ND | Mitochondriaa | Mitochondria | Mitochondria |
| At5g09950 | DYW | ND | Mitochondriac | Mitochondria | Plastid | |
| At5g13230 | DYW | Mitochondriab | ND | Mitochondria | Mitochondria | |
| At5g14770 | PPR | Mitochondriab | No signalc | Mitochondria | Mitochondria | |
| At5g15340 | DYW | Mitochondriab | ND | Mitochondria | Mitochondria | |
| At5g55740 | E+ | Mitochondriab | Mitochondriab | Plastid | Plastid | |
| At1g74850 | PPR | ND | ND | Plastidf | Plastid | Possibly plastid |
| At2g01860 | PPR | Plastidsb | ND | Plastid | Possibly plastid | |
| At2g03880 | DYW | Plastidsb | ND | Mitochondria | Mitochondria | |
| At2g29760 | DYW | Plastidsb | ND | Plastid | Plastid | |
| At3g04760 | PPR | ND | ND | Plastidf | Plastid | Possibly plastid |
| At3g23020 | PPR | ND | ND | Plastidf | None | Possibly plastid |
| At3g42630 | PPR | ND | ND | Plastidf | None | Possibly mitochondria |
| At3g46870 | PPR | ND | ND | Plastidf | Plastid | Possibly mitochondria |
| At4g16390 | PPR | Plastidsde | ND | Plastid | Possibly plastid | |
| At4g31850 | PPR | Plastidsce | ND | Plastid | Plastid | |
| At5g13270 | DYW | Plastidsb | ND | Plastid | Plastid | |
| At5g52630 | DYW | Plastidsce | Plastidsb | None | None |
Organellar proteomics data from Heazlewood et al. (2004).
Localization was studied by fusion of presequences to the following:
DsRed2,
GFP, or
full-length ORFs to DsRed2.
The sequence of the entry clone used for the LR reaction was verified.
Organellar proteomics data from Kleffmann et al. (2004).
Experimentally proven localizations were compared to bioinformatics predictions from TargetP and Predotar. The first and second parts of the table correspond to mitochondrial- and plastid-localized PPR proteins, respectively.
When several different constructs or transformation methods (Table 2) were used for the experimental localization of the same PPR protein, the results were always consistent. In summary, as shown in Table 2, 18 PPR proteins were shown to be targeted to mitochondria, and eight were shown to be targeted to plastids. The results were in general consistent with the bioinformatics predictions for the proteins tested, which implies that the predictions for the family as a whole are likely to be not far wrong. Whereas some PPR proteins were predicted to be nonorganellar, mitochondria and plastids were the only subcellular localization that we observed in our experiments. In addition, no cases of dual targeting to mitochondria and plastids were observed for any of the fluorescent fusions.
Analysis of PPR Gene Expression
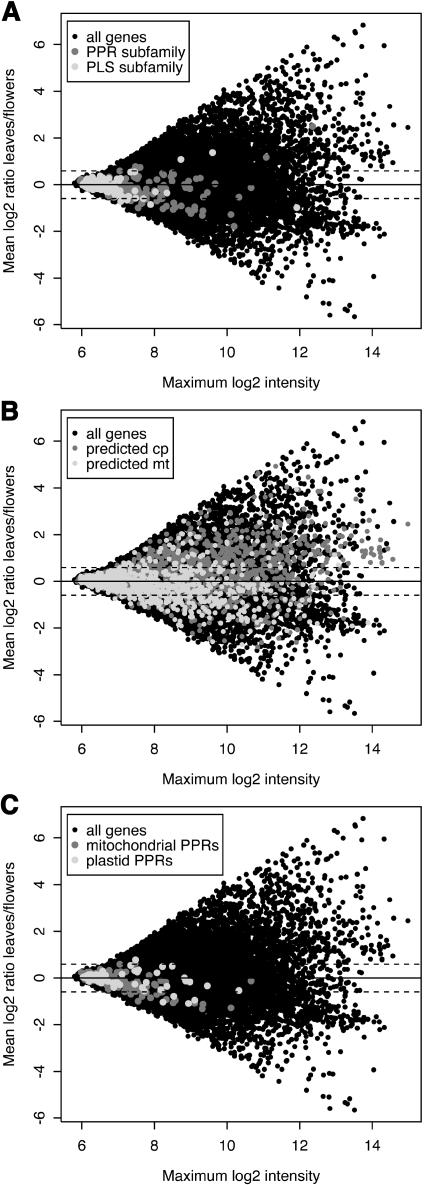
One of the reasons why such a huge family of proteins was discovered only very recently in Arabidopsis is that few ESTs corresponding to PPR genes were previously identified. Among the 210,388 cDNA and EST sequences (including many sequences from normalized and subtracted libraries) available from FLAGdb++ (Samson et al., 2004), only 843 transcripts are from PPR genes, corresponding to only 215 out of the 441 genes (48.7%) described here (Table 3; see Supplemental Table S5 online). Using the same database, 69.1% of all predicted Arabidopsis genes have proof of expression from EST or cDNA data. This implies that PPR genes are expressed at low levels and could even mean that many are not expressed at all. To investigate this further, we analyzed PPR transcripts in RNA from two organs expected to show high expression of genes for mitochondrial and plastid proteins, namely flowers and leaves. This was performed by systematic RT-PCR amplification using 372 primer pairs from the Complete Arabidopsis Transcriptome MicroArray (CATMA) project (Hilson et al., 2004), each corresponding to a different PPR transcript, and also by microarray experiments using the full set of CATMA gene-specific probes. Each technique detected expression of ∼40 to 50% of PPR genes, but taken together, evidence of expression of the vast majority of PPR genes could be obtained (Table 3). Interestingly, all three methods of transcript analysis detected a much higher proportion of transcripts from PPR subfamily genes than from P-L-S subfamily genes. This difference is highly significant (χ2 test, P < 10−11). The same tendency toward higher expression of PPR subfamily genes was found in a quantitative analysis of the microarray data (Figure 5A). We found that members of the pure PPR subclass were in general expressed at higher levels than the other members of the family (Wilcoxon rank sum test, P < 10−15). Although hybridization signals between different genes on microarrays are not strictly comparable because of variations in probe efficiency, the large number of genes in the family allows broad conclusions to be drawn. Taken together, these results confirm the low overall level of expression of the PPR gene family and the extremely low expression levels of the P-L-S subfamily. We looked at various experiments comparing different plant organs, and we never observed any obvious deviation of the PPR cloud from the central diagonal, indicating that these genes are expressed generally constitutively. This is illustrated in Figures 5B and 5C, where the genes encoding predicted plastid-targeted PPR proteins fail to show the bias toward higher expression in leaves exhibited by the bulk of genes encoding predicted chloroplast proteins and in fact behave indistinguishably from genes encoding predicted mitochondrial PPR proteins.
Table 3.
Expression of PPR Genes Detected by Different Methods
| Detection of Gene Expression
|
||||
|---|---|---|---|---|
| Subfamily | ESTs | RT-PCR | Microarrays | By at Least One Technique |
| PPR | 148 (61.4%) | 93 (50.3%) | 128 (69.2%) | 228 (94.6%) |
| P-L-S | 67 (33.5%) | 63 (35.0%) | 58 (32.2%) | 139 (69.5%) |
| Total | 48.8% | 41.9% | 50.0% | 83.2% |
The number of genes whose expression could be detected by different techniques is given for both subfamilies (the percentage detected in the subfamily is given in parentheses). EST data is from FLAGdb++; RT-PCR experiments were conducted on Arabidopsis leaf and flower mRNA with 372 gene-specific primer pairs from the CATMA collection; microarray hybridization results are from the data in Figure 5, covering 384 PPR genes.
Figure 5.
Expression of Sets of Genes as Measured by Microarray Hybridization Data.
RNA was extracted from Arabidopsis rosette leaves or flowers and hybridized to CATMA arrays containing 24,576 gene-specific probes. Data from four independent two-color hybridizations (comprising two dye swaps) were corrected and averaged as described in Methods. The scales are logarithmic (log2), representing the mean signal ratio (leaves/flowers) against the maximum mean signal intensity (leaves or flowers). The dotted lines indicate the ratios (0.46) above or below that which the statistical analysis indicates the genes to be differentially expressed in this set of experiments.
(A) Expression of PPR genes. Data points corresponding to the PPR and P-L-S subfamilies are depicted in dark gray or light gray, respectively. In general, the PPR subfamily is more highly expressed (Wilcoxon rank sum test, P < 10−15).
(B) Expression of genes predicted to encode plastid or mitochondrial proteins. Data points corresponding to genes encoding Predotar-predicted plastid or mitochondrial proteins (cutoff 0.5) are depicted in dark gray or light gray, respectively. The predicted plastid set shows a strong bias toward higher expression in leaves (Wilcoxon rank sum test, P < 10−15). cp, chloroplast; mt, mitochondria.
(C) Expression of genes predicted to encode plastid or mitochondrial PPR proteins. Data points corresponding to genes encoding Predotar-predicted mitochondrial or plastid PPR proteins (cutoff 0.25) are depicted in dark gray or light gray, respectively. The two sets do not show significantly different distributions of leaf/flower expression ratios (Wilcoxon rank sum test, P > 0.75) and are much less biased toward expression in leaves than the complete predicted plastid set shown in (B) (Wilcoxon rank sum test, P < 10−15) while being slightly less biased toward expression in flowers than the complete mitochondrial set (Wilcoxon rank sum test, P < 0.04).
Analysis of the Mutant Collections
Reverse genetics and especially sequenced insertion mutant collections are valuable resources for obtaining information about the function of unknown genes. The collections from Versailles (Samson et al., 2002), from GABI (Li et al., 2003), and from the Salk Institute (Alonso et al., 2003) were screened to look for insertions in Arabidopsis PPR genes. Not surprisingly, given the large size of the family, many putative insertion mutants are listed in these databases. We further characterized some of these mutants corresponding to different subclasses of the PPR family. Among the 25 mutants (corresponding to 21 genes) we have genetically characterized so far, we were unable to find homozygous plants for six mutants (five genes) when we looked at progeny from heterozygous plants (Table 4). Dissection of the siliques of heterozygous mutant plants showed that about one-fourth of the embryos aborted at an early stage (data not shown), indicating that the homozygous mutation was lethal in early stages of embryo development. Furthermore, in direct genetic screens for embryo-lethal mutations, the Arabidopsis SeedGenes project (Tzafrir et al., 2003) has identified many PPR mutants (included in Table 4). Taken together, these results indicated that many PPR genes have an essential function in plant embryos that leads to the abortion of the embryos if the genes are mutated. For the 19 remaining PPR mutants, homozygous plants were found. Most of them did not have any obvious macroscopic phenotype (when looking at growth and development of plants in soil or in vitro). Five mutant lines were found with a clear visible phenotype on homozygous plants: three of these present short siliques containing many deformed embryos. Two of these lines carry an insertion in the same gene (At3g25970). A fourth line with a clear slow growth phenotype carries an insertion in a gene (At1g20230) that when knocked out by an independent insertion gives rise to an embryo-lethal phenotype. Twelve out of fourteen genes encoding proteins predicted to be targeted to plastids give rise to a phenotype when the mutation is homozygous (usually embryo lethality) but 7 of the 12 mutations in genes encoding predicted mitochondrial proteins generate no observable macroscopic phenotype.
Table 4.
List of Arabidopsis T-DNA Mutants with Insertions in PPR Genes
| Mutant ID | AGI Code | Subclass | Targeting | Homozygotes | Phenotype of Homozygotes |
|---|---|---|---|---|---|
| DNR2 | At1g14470 | PLS | ? | Yes | Dwarf, reduced fertility |
| EAN42 | At1g20230 | DYW | ? | No | –a |
| Salk_033409 | At1g20230 | DYW | ? | Yes | Dwarf, late flowering |
| DVM10 | At2g40720 | E+ | ? | Yes | – |
| DDU146 | At3g25970 | E+ | ? | Yes | Dwarf, reduced fertility |
| Salk_127777 | At3g25970 | E+ | ? | Yes | Dwarf, reduced fertility |
| EMB 2744 | At5g39680 | DYW | ? | No | Embryo defective |
| DJU9 | At1g06710 | PPR | Mitochondria | Yes | – |
| T243 | At1g22830 | E | Mitochondria | Yes | – |
| DYK12 | At1g51965 | PPR | Mitochondria | Yes | – |
| ABP1 | At1g80550 | PPR | Mitochondria | No | – |
| Salk_024306 | At1g80550 | PPR | Mitochondria | No | – |
| DNP1 | At2g20540 | E+ | Mitochondria | Yes | – |
| DXZ45 | At3g13150 | PPR | Mitochondria | Yes | – |
| Salk_015469 | At3g49140 | DYW | Mitochondria | Yes | – |
| EMB 1796 | At3g49240 | PPR | Mitochondria | No | Embryo defective |
| EMB 1025 | At4g20090 | PPR | Mitochondria | No | Embryo defective |
| EMB 2745 | At5g39710 | PPR | Mitochondria | No | Embryo defective |
| Salk_033891 | At5g46460 | DYW | Mitochondria | Yes | – |
| DSN8 | At5g62370 | PPR | Mitochondria | No | – |
| DZY11 | At5g65570 | DYW | Mitochondria | Yes | – |
| EMB 1444 | At1g06150 | E+ | Plastids | No | Embryo defective |
| Salk_067029 | At1g09410 | DYW | Plastids | Yes | – |
| Gabi_104E05 | At1g09410 | DYW | Plastids | Yes | – |
| EMB 2279 | At1g30610 | PPR | Plastids | No | Embryo defective |
| CYK8 | At2g01860 | PPR | Plastids | No | Embryo defective |
| hcf152 | At3g09650 | PPR | Plastids | Yes | Defective processing of petB |
| EMB 1270 | At3g18110 | PPR | Plastids | No | Embryo defective |
| Salk_040629 | At3g22150 | E+ | Plastids | Yes | – |
| crr2 | At3g46790 | DYW | Plastids | Yes | Defective processing of ndhB |
| Salk_046131 | At3g46790 | DYW | Plastids | Yes | Defective processing of ndhB |
| Gabi_425F06 | At3g46790 | DYW | Plastids | Yes | Defective processing of ndhB |
| EMB 2261 | At3g49170 | DYW | Plastids | No | Embryo defective |
| pgr3 | At4g31850 | PPR | Plastids | Yes | Cytochrome b6f and/or NDH-deficient |
| CRI6 | At4g31850 | PPR | Plastids | Yes | Yellow leaves |
| EMB 2453 | At4g39620 | PPR | Plastids | No | Embryo defective |
| EMB 1899 | At5g03800 | DYW | Plastids | No | Embryo defective |
| DYH216 | At5g27270 | PPR | Plastids | No | Embryo defective |
| EMB 1006 | At5g50280 | PPR | Plastids | No | Embryo defective |
Not determined.
The mutants are named according to the nomenclature of their collections of origin: Salk_xxx mutants are from the Salk collection (Alonso et al., 2003), Gabi_xxx mutants are from the GABI-Kat collection (Li et al., 2003), EMB xxx mutants are from the SeedGenes project (Tzafrir et al., 2003), and the other mutants are from the Versailles collection (Samson et al., 2002). The targeting column indicates predictions based on Predotar and TargetP scores (see Supplemental Table S3 online), or if the protein localization has been experimentally determined (Table 2), the organelle is indicated in bold. For each mutant, we indicate whether or not homozygotes could be obtained, and if so, the phenotype of the homozygous plants. When homozygotes could not be obtained, in some cases the siliques of heterozygous plants were examined for evidence of embryo abortion, and the phenotype is indicated as embryo defective. Several alleles of the mutants hcf152, crr2, and pgr3 have been described (Hashimoto et al., 2003; Meierhoff et al., 2003; Yamazaki et al., 2004). Unlike the other mutants listed here, these are mostly point mutations and are included for comparison.
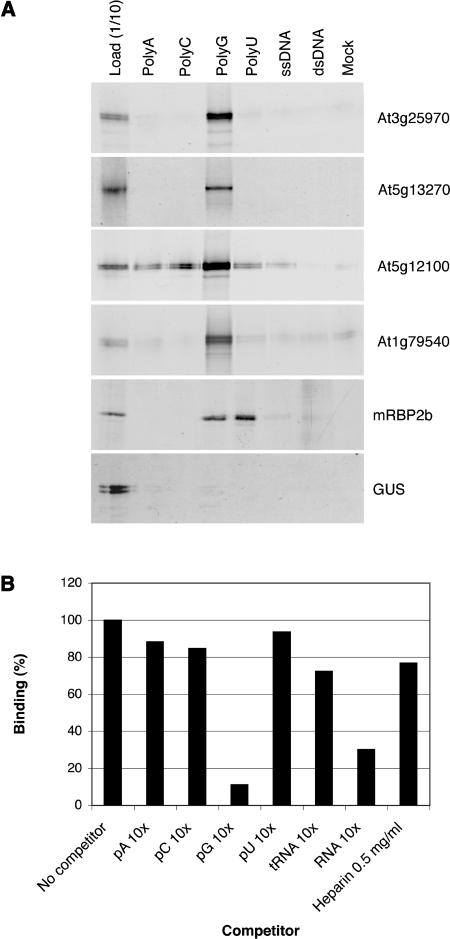
RNA Binding Activity of Some PPR Proteins
As a preliminary study of the RNA binding capacity of the PPR protein family, the proteins At3g25970, At5g13270, At5g12100, and At1g79540, representing different subclasses of the family, were synthesized in vitro and incubated with homoribopolymers, ssDNA, and dsDNA. All four PPR proteins preferentially bound poly(G) homoribopolymers (Figure 6A), whereas the previously characterized RNA recognition motif–containing mRBP2b protein (Vermel et al., 2002), which was used as a positive control in this study, bound poly(U) and poly(G) RNA. Consistent with these data, competition experiments indicated that for each of the four PPR proteins analyzed, the addition of 50 μg of either poly(G) RNA or total RNA, competed for their poly(G) binding capacity (Figure 6B; data not shown). In conclusion, these results demonstrate that the four PPR proteins tested show a much higher affinity to poly(G) RNA than to the other three homoribopolymers and DNA. The biological significance of these findings remains to be elucidated in future experiments, hopefully via the identification of physiological RNA targets.
Figure 6.
RNA Binding Assay for Four Representative Arabidopsis PPR Proteins.
Two of these proteins (At1g79540 and At5g12100) are P subfamily members. At3g25970 is an E+ protein, and At5g13270 is a DYW protein.
(A) Radioactively labeled protein retained on Sepharose columns carrying various polyribonuculeotides, single-stranded DNA (ssDNA), or double-stranded DNA (dsDNA). The left lane was loaded with one-tenth of the protein loaded on the columns. The far right lane shows binding to a Sepharose column lacking added nucleotides. mRBP2b is a previously characterized RNA binding protein of the RNA recognition motif family, used here as a positive control. The bottom panel shows binding of β-glucuronidase (GUS) as a negative control.
(B) Competition assays for At5g12100. Labeled proteins were preincubated with competitor RNA or heparin before being loaded on a poly(G) Sepharose column and binding quantified by a phosphoimager, using binding in absence of competitor as 100%.
DISCUSSION
A Model for the Action of PPR Proteins
At first sight, the genetic analysis of the PPR mutants is quite surprising. How can a mutation in a single gene of a 441-gene family lead to a phenotype as dramatic as the death of the embryo? How can a mutation in another very similar gene of the same family give no obvious phenotype?
The data accumulated so far concerning PPR proteins are mostly consistent with the view that they are RNA binding proteins involved in posttranscriptional processes (RNA processing and translation) in mitochondria and chloroplasts. Posttranscriptional processes in organelles are incredibly complex, involving many highly specific processing events (trimming of 5′ and 3′ ends, cis- and in some cases trans-splicing, stabilization or destabilization of transcripts, etc.) and a relatively poorly understood control of translation initiation at internal AUG codons (Barkan and Goldschmidt-Clermont, 2000; Giege and Brennicke, 2001). The PPR family could potentially be involved in many of these functions. Plant cells are not viable without mitochondrial respiration, and in addition, mutations in genes encoding essential plastid proteins lead to an embryo-lethal phenotype in Arabidopsis (Budziszewski et al., 2001). The most frequent phenotype observed for T-DNA insertions in PPR genes (i.e., a lack of homozygous mutants) is therefore consistent with a role for PPR proteins in organelle biogenesis. The genetic results also imply that there is little or no redundancy of function between many PPR proteins, despite the huge size of the family. Furthermore, the very limited primary defects observed in the few mutants that have been studied, the specificity of action of the cloned CMS restorer genes, and the low expression levels of the majority of the genes in the family all imply that these proteins are for the most part targeted to specific transcripts rather than being general unspecific RNA binding factors. The large majority of these proteins lack any obvious domains likely to have catalytic activity; thus, we imagine their role as adaptors for directing the action of other factors (Figure 7). In this case, the phenotype of the mutants will be entirely dependent on the identity of the target transcripts; for example, the macroscopic phenotypes of the mutants crr2, hcf152, and crp1 range from wild type to lethal despite the fact that the molecular functions of these three PPR proteins are apparently very similar.
Figure 7.
A Model for PPR Protein Action.
We assume that the putative superhelix formed by tandemly repeated PPR motifs forms a sequence-specific RNA binding surface either alone (A) or in the presence of an additional factor (B). The resulting protein-RNA complex recruits one or more other transfactors to a specific site on the RNA target (in this case an endonuclease). We assume that in most cases the catalytic site is in the partner protein; for the DYW class of PPR proteins, it may lie in the C-terminal domain itself.
This model is probably basically correct in its broad details, but several points still need to be investigated. The RNA binding activity of PPR proteins now seems to be beyond doubt (Lahmy et al., 2000; Mancebo et al., 2001; Meierhoff et al., 2003; Mili and Pinol-Roma, 2003; Nakamura et al., 2003), but there is very little evidence showing binding specificity, in particular to known target transcripts. This may be attributable to technical difficulties (PPR proteins are difficult to produce in soluble form; C. Lurin and H. Mireau, unpublished data) but may also be explained if, in vivo, PPR proteins bind in concert with one or more other RNA binding proteins (Figure 7B). Such complexes are relatively common, one of the best-studied examples being the binding of Pumilio, Nanos, and Brat to the 3′ UTR of hunchback mRNA (Sonoda and Wharton, 1999; Edwards et al., 2003). This brings in the more general question of which other proteins are associated with PPR proteins. Several PPR proteins have been shown to be in protein complexes in vivo (Fisk et al., 1999; Liu et al., 2002; Liu and McKeehan, 2002; Tsuchiya et al., 2002; Williams and Barkan, 2003), but in very few cases to date have any of the partners been identified.
Evolution of the Family and Putative Functions
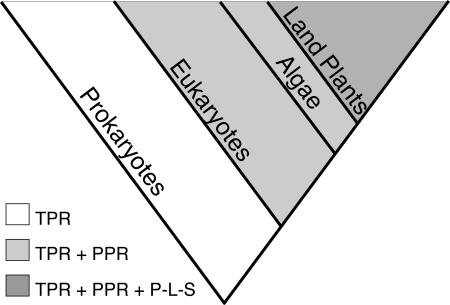
Probably the most striking feature of the PPR family is its spectacular expansion in plants (Table 1). The Arabidopsis genome codes for 70 times more PPR proteins than the human genome, and all the evidence so far from genome sequencing and large-scale EST projects suggests that other plant genomes encode even more. The P-L-S subfamily is entirely specific to plants but accounts for only about half of the extra plant proteins. From EST data, this subfamily can be traced as far as bryophytes (e.g., Physcomitrella patens ESTs AW599911 and AW561409) but is apparently absent from Chlamydomonas. This unusual distribution poses at least three major questions. (1) Where did the plant-specific proteins originate? (2) How did the family grow at least 70-fold in the plant lineage? (3) Why do plants need so many PPR proteins?
It seems more than probable that the pure PPR subfamily in plants has arisen by expansion of the small family of pure PPR genes in all eukaryotes, given the similarities in structure and function (Figure 8). The PPR-like repeats of the plant-specific P-L-S subfamily probably arose by mutation of classical PPR repeats, but the origins of the characteristic C-terminal E, E+, and DYW motifs are mysterious. Many plant-specific genes can be traced to the endosymbiotic ancestor of plastids, but in the case of PPR genes there is no evidence for this whatsoever. As far as we can tell, PPR proteins are absent from almost all prokaryotes that we have looked at, including the closest living relatives of the organellar endosymbionts. What is more, the lack of these plant-specific proteins in trypanosomes and Chlamydomonas suggests that they were acquired long after the acquisition of plastids.
Figure 8.
Order of Appearance and Likely Evolutionary Relationships between PPR Families Based on Phylogenetic Distribution.
TPR proteins are ubiquitous, whereas PPR proteins are only found in eukaryotes and the P-L-S subfamily only in land plants.
If the origin of many of these proteins remains obscure, the expansion of the family may be easier to explain. PPR genes stand out from the vast majority of other plant genes in their paucity of introns. When introns are present, they are generally outside the zone of the repeats or in proteins rather divergent from the bulk of the family. This is characteristic of other large gene families with repeated motifs, such as Leu-rich repeat proteins that can evolve rapidly via unequal crossing-over events (Michelmore and Meyers, 1998; Meyers et al., 2003). Such events can fuse, split, duplicate, or delete all or part of a gene (Jelesko et al., 1999). However, a general consequence of this type of evolution is the generation of clusters of related genes, again as typified by disease resistance genes. Apart from the restorer gene–related cluster on chromosome 1, PPR genes do not show such obvious clustering in Arabidopsis. An alternative hypothesis would be that PPR genes have been amplified by one or more bursts of reverse transcription/integration (Lecharny et al., 2003), perhaps by fortuitous association with a retrotransposon. This could also explain the lack of introns while accounting for a wide chromosomal distribution. However, if this is the true explanation, no obvious trace of these events persists.
Our major goals, however, are to discover the functions of PPR proteins in plants to understand why the family has grown so large. Genetic data from yeast and Chlamydomonas strongly suggest that many specific trans-acting factors are required for proper expression of each and every transcript in mitochondria and plastids, leading to estimates that several hundred of these factors must exist (Barkan and Goldschmidt-Clermont, 2000; Costanzo et al., 2000). A fair number of these factors have been identified from these single-celled organisms, but only a few are PPR proteins. Clearly hundreds of PPR proteins are not a prerequisite for organelle biogenesis outside the plant kingdom. This leaves two explanations for the huge increase in the PPR family in plants: either PPR proteins play roles in plants that are performed by other proteins in other organisms or PPR proteins perform novel functions in plants absent from animals, fungi, and algae. The first explanation is likely to be correct to some extent because Arabidopsis lacks recognized homologs to most of the transcript-specific mitochondrial factors identified in yeast. The second explanation is very tempting, especially for the plant-specific half of the family. However, the gene content of land plant organelles is not significantly higher than that of other organisms, and their posttranscriptional processes are not notably more complex, with one exception, RNA editing. The posttranscriptional conversion of specific cytosines to uracil in mitochondrial and plastid transcripts is unique to land plants (Steinhauser et al., 1999). Several hundred editing sites in Arabidopsis mitochondria (Giege and Brennicke, 1999) and 30 or so in plastids (Tsudzuki et al., 2001) have been cataloged. These sites appear to require many specific trans factors to provide the observed editing specificity (Chateigner-Boutin and Hanson, 2002, 2003). It now seems accepted that these factors are probably proteins (Hirose et al., 1999; Miyamoto et al., 2002, 2004), but their identity remains unknown. The evolutionary expansion of the PPR family and the appearance of the plant-specific P-L-S subfamily correlates with the appearance of C-to-U RNA editing within the coarse resolution permitted by the EST data. The only mutants in this subfamily so far described are the Arabidopsis crr2 mutants defective in processing of the plastid ndhB transcript (Hashimoto et al., 2003). However, editing of ndhB transcripts is not obviously affected in these mutants (data not shown), and it should be stressed that there is as yet no physical evidence for an involvement of PPR proteins in RNA editing.
Conclusion
We have performed an exhaustive, systematic bioinformatics analysis of Arabidopsis PPR genes and proteins. The resulting descriptions of the family and its members should provide a solid and unified platform on which future genetic and functional studies can be based. The preliminary functional characterization of a range of family members, together with the sparse data on these proteins published, allows us to propose a plausible model (Figure 7) in which PPR proteins play the role of sequence-specific adaptors for a variety of other RNA-associated proteins. This model makes two testable predictions: PPR proteins should (directly or indirectly) associate with specific RNA sequences and with defined effector proteins. Future work needs to be directed toward the identification of these factors to elucidate the precise functions of one of the largest and least understood protein families in plants.
METHODS
Bioinformatics
Arabidopsis thaliana genomic sequences, predicted protein sequences, and functional annotations were obtained from TIGR (ftp://ftp.tigr.org/pub/data/a_thaliana/). ClustalW version 1.81 (Thompson et al., 1994) was used for aligning multiple peptide sequences to construct alignments as input to the HMMER package versions 2.2g (Eddy, 1998), which was used to construct models of different PPR motifs and then to search peptide sequences for these motifs. The programs hmmbuild and hmmcalibrate were used to construct matrices describing the motifs, and hmmsearch was used for discovering motifs within protein sequences. The default parameters were used in all cases.
A modified version of the FLAGdb++ database and interface (Samson et al., 2004) was used for much of the visualization needed for manual annotation, including display of PPR motifs, AGI ORF models, known mRNAs, and predicted mitochondrial and plastid targeting sequences. Predotar version 1.03 (Small et al., 2004) and TargetP version 1.01 (Emanuelsson et al., 2000) were used for prediction of organelle targeting from protein sequences. A modified version of Predotar was used to predict regions encoding putative targeting sequences in genomic DNA. Secondary structure predictions of PPR motifs were performed with the PredictProtein server (Rost and Liu, 2003) using the alignments of motifs described previously.
The complete predicted proteomes of various organisms used to create Table 1 were obtained from the National Center for Biotechnology Information RefSeq database, apart from Trypanosoma brucei sequences (from ftp://ftp.sanger.ac.uk/pub/databases/T.brucei_sequences/) and Cyanidiodschyzon merolae sequences (from http://merolae.biol.s.u-tokyo.ac.jp/).
The alignment files, hmm matrices, and search results are available in the supplemental data online.
Transcriptome Studies
The microarray analysis has been performed with the CATMA array containing 24,576 gene-specific tags (GSTs) from Arabidopsis (Hilson et al., 2004). The GST amplicons were purified on Multiscreen plates (Millipore, Bedford, MA) and resuspended in TE-DMSO at 100 ng/μL. The purified probes were transferred to 1536-well plates with a Genesis workstation (TECAN, Männedorf, Sweden) and spotted on UltraGAPS slides (Corning, Corning, NY) using a Microgrid II (Genomic Solution, Huntingdon, UK). The CATMA array printed at the Unité de Recherche en Génomique Végétale consists of three metablocks, each composed of 64 blocks of 144 spots. A block is a set of spots printed with the same print tip. In these arrays, a print tip is used three times to print a block in each metablock. For the transcriptome studies, eight sets of 25 Arabidopsis Columbia-0 plants were grown on horticultural potting soil (Tref substrate with NFU 44-571 fertilizer; BAAN, Vulaines, France) under cool white light at 100 μmol m−2 s−1 with a 16-h photoperiod at 22°C, 50% humidity. Four pooled samples of young leaves were harvested at stage 3.9, and four pooled samples of flower buds harvested at stage 6.0, the material being collected 5 to 7 h into the light period. RNA was extracted from these samples using TRIzol extraction (Invitrogen, Carlsbad, CA) followed by two ethanol precipitations and then checked for RNA integrity with the Bioanalyzer from Agilent (Waldbroon, Germany). cRNAs were produced from 2 μg of total RNA from each sample with the Message Amp aRNA kit (Ambion, Austin, TX). Then 5 μg of cRNAs were reverse transcribed in the presence of 300 units of SuperScript II (Invitrogen), cy3-dUTP, and cy5-dUTP (NEN, Boston, MA) according to Puskas et al. (2002) for each slide. Samples were combined, purified, and concentrated with YM30 Microcon columns (Millipore). Slides were prehybridized for 1 h and hybridized overnight at 42°C in 25% formamide. Slides were washed in 2× SSC + 0.1% SDS for 4 min, 1× SSC for 4 min, 0.2× SSC for 4 min, and 0.05× SSC for 1 min and dried by centrifugation. Four hybridizations (two dye swaps) were performed. The arrays were scanned on a GenePix 4000A scanner (Axon Instruments, Foster City, CA), and images were analyzed by GenePix Pro 3.0 (Axon Instruments).
For RT-PCR validation, mRNAs were purified from the extracts described above with Dynabeads (Dynal, Oslo, Norway) according to the manufacturer's specifications. Fifty nanograms of mRNA per 20-μL reaction were reverse transcribed in the presence of 10 units of Superscript II (Invitrogen). PCR reactions for each sample were performed in 96-well plates each with 0.5 μL of the RT reaction and the corresponding set of specific CATMA primers for each of 384 PPR genes in presence of 2.5 units of Platinum Taq DNA polymerase (Invitrogen). The PCR conditions were 94°C for 1 min, followed by 10 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s (with a 1°C decrease of the annealing temperature at each cycle), followed by 25 cycles of 94°C for 30 s, 68°C for 30 s, 72°C for 30 s, and finally 72°C for 7 min. Additional PCR reactions were performed on RT minus samples as negative controls. The RT-PCR products were validated by size verification after electrophoresis on agarose gels.
Statistical Analysis of Microarray Data
The statistical analysis was based on two dye swaps (i.e., four arrays each containing 24,576 GSTs and 384 controls). The controls were used for assessing the quality of the hybridizations but were not included in the statistical tests or the graphical representation of the results shown in the Results. For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). No background was subtracted. In the following description, log ratio refers to the differential expression between leaves and flowers. It is either log2(red/green) or log2(green/red) according to the experiment design. An array-by-array normalization was performed to remove systematic biases. First, we excluded spots that were considered badly formed features. Then we performed a global intensity-dependent normalization using the loess procedure (see Yang et al., 2002) to correct the dye bias. Finally, for each block, the log-ratio median calculated over the values for the entire block was subtracted from each individual log-ratio value to correct print tip effects on each metablock.
To study the expression of PPR genes, we first tested if the distribution of the maximum signal intensity of the leaf and the flower samples differed between PPR subfamilies. For each array, and for each gene, we calculated according to the method of Yang and Thorne (2003) a normalized logarithm of median feature pixel intensity. We kept the maximum of these normalized intensities for each gene. The statistical test used was a one-tailed Wilcoxon rank sum test.
To determine differentially expressed genes, we performed a paired t test on the log ratios. The number of observations per spot varies between two and four and is inadequate for calculating a gene-specific variance. For this reason, we assume that the variance of the log-ratios is the same for all genes, and we excluded 256 spots displaying extremes of variance (too small or too large). The raw P values were adjusted by the Bonferroni method, which controls the Family Wise Error Rate (FWER) (Ge et al., 2003). A total of 6566 genes (26.7%) were found to be differentially expressed between flowers and leaves taking a FWER of 5%. Amongst these, only 45 are PPR genes, out of the 384 represented by GSTs in the array (i.e., only 11.7% of the PPR genes are differentially expressed in these experiments). The relative stability of PPR gene expression was confirmed by applying Wilcoxon rank sum tests on the normalized log-ratios (Figure 5C) to determine whether the expression differs or not between leaves and flowers.
After the removal of badly formed features, we retained four replications of the normalized log-ratios for 24,391 GSTs, three observations for 67 GSTs, and two observations for 118 GSTs. The absolute value of log-ratio threshold corresponding to a FWER of 5% is 0.46 when four observations are available. It increases respectively to 0.54 and 0.66 when three and two observations are available. In Figure 5, we indicate the thresholds of the log-ratios corresponding to a FWER of 5% when four observations are available.
Construction of a Collection of PPR ORFs
The ORFs of 48 intronless PPR genes were PCR amplified from Arabidopsis (ecotype Columbia-0) genomic DNA using Pfx DNA polymerase and primers flanked with attB1 and attB2 sites and cloned into the entry vector pDONR207 by Gateway recombination (Invitrogen). For targeting experiments, either the whole ORF or just the first 100 codons were cloned in this way. For the nucleic acid binding assays, the ORFs were cloned starting from the predicted start of the mature protein to remove most or all of the putative targeting sequences. TargetP was used for predicting the extent of the targeting sequences. In all cases, the 5′ primer included Kozak and Shine-Dalgarno consensus sequences to maximize translation in eukaryotic or prokaryotic systems. Details on the cloning procedures, including the primer sequences, are available in the supplemental data online.
Subcellular Localization of Proteins
Two types of experiments were performed. The sequences coding for precursor PPR proteins or putative targeting sequences were shuttled into Greengate or Redgate vectors (http://www.evry.inra.fr/public/projects/ppr/greengate.html) from the corresponding pDONR207 vector using Gateway recombination (Invitrogen). These constructs were used to express GFP or DsRed2 fusion proteins in electroporated tobacco protoplasts using previously described protocols (Akashi et al., 1998; Peeters et al., 2000).
Binary destination vectors (p0229-GFP and p0229-RFP) were created by transferring the GFP or DsRed2 cassettes, respectively, from Greengate or Redgate into pGreenII 0229 (Hellens et al., 2000), and the sequences coding for precursor PPR proteins or putative targeting sequences were shuttled into these vectors from the corresponding pDONR207 as before. In this case GFP- and RFP-fusion proteins were transiently expressed in Nicotiana benthamiana leaves via agroinfiltration. Agrobacterium tumefaciens strain GV3101 harboring the transformation helper pSoup (Hellens et al., 2000) and the GFP or DsRed2 constructs were grown overnight and resuspended in infiltration media (10 mM Mes, pH 5.3, 5 mM MgCl2, and 150 mM acetosyringone) at an OD of 0.5. Protoplasts were prepared from leaf material (harvested 48 h after infiltration) cut into small pieces and incubated in enzyme solution (Chupeau et al., 1974) at 30°C for 2 to 3 h. Fluorescent proteins were visualized using an Eclipse TE2000S inverted microscope (Nikon, Kanagawa, Japan), a GFP-B filter block (Nikon), and a custom filter block for detection of DsRed2 (exciter HQ546/12, emitter HQ605/75, beamsplitter Q560lp; Chroma Technology, Rockingham, VT).
Nucleic Acid Binding Assay
The sequences coding for the mature PPR proteins and β-glucuronidase were shuttled into pDEST17 from the corresponding pDONR207 vector using Gateway recombination (Invitrogen). Proteins were synthesized by coupled transcription-translation in the presence of [35S]Met (TNT coupled reticulocyte lysate; Promega, Madison, WI). Homoribopolymers [poly(A), poly(U), poly(G), and poly(C)] cross-linked to agarose beads were obtained from Sigma-Aldrich (St. Louis, MO). Concentrations were equilibrated to 0.5 mg per mL packed gel by dilution with the appropriate volume of Sepharose 4B (Amersham Biosciences, Little Chalfont, UK). Single- and double-stranded DNA cellulose were obtained from Sigma-Aldrich. The nucleic acid binding assay was performed as described by Vermel et al. (2002). A 10-fold excess of competitor RNA was used for competition experiments. The bound fraction of proteins was analyzed by SDS-PAGE, and the relative protein amounts were calculated by a phosphor imager (Fujix Bas 1000; Fuji Photo Film, Tokyo, Japan).
Mutant Analyses
Arabidopsis plants were grown on soil in a growth chamber under the following conditions: photoperiod 16 h day (100 μmol m−2 s−1)/8 h night; temperature 22°C day/16°C night; humidity 80%.
Plants were genotyped by PCR. Two specific primers from each side of the insertion site were used to detect wild-type loci. One primer specific to the genomic sequence near the insertion site and one primer from the T-DNA border were used to amplify a fragment flanking each insertion. These fragments were sequenced for precise localization of the insert. In all mutants described here, the insertion is within the coding sequence. Plants from at least 10 seeds of each initial stock were genotyped to obtain at least one heterozygous plant. Twenty-four seeds from each heterozygote were grown for genotyping to detect plants homozygous for the T-DNA insertion.
The complete set of microarray data from these experiments has been submitted to ArrayExpress (accession numbers A-MEXP-60 and E-MEXP-94).
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ian Small (small@evry.inra.fr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022236.
References
- Akashi, K., Grandjean, O., and Small, I. (1998). Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 431, 39–44. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Boudet, N., Kreis, M., and Lecharny, A. (2000). In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol. Biol. 42, 603–613. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Bentolila, S., Alfonso, A.A., and Hanson, M.R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.G., Formanova, N., Jin, H., Wargachuk, R., Dendy, C., Patil, P., Laforest, M., Zhang, J., Cheung, W.Y., and Landry, B.S. (2003). The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35, 262–272. [DOI] [PubMed] [Google Scholar]
- Budziszewski, G.J., et al. (2001). Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 159, 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., and Hanson, M.R. (2002). Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol. 22, 8448–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., and Hanson, M.R. (2003). Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar cis-elements. Nucleic Acids Res. 31, 2586–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupeau, Y., Bourgin, J.P., Missonier, C., Dorion, N., and Morel, G. (1974). Préparation et culture de protoplastes de divers Nicotiana. C. R. Acad. Sci. Paris 278, 1565–1568. [Google Scholar]
- Coffin, J.W., Dhillon, R., Ritzel, R.G., and Nargang, F.E. (1997). The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32, 273–280. [DOI] [PubMed] [Google Scholar]
- Costanzo, M.C., Bonnefoy, N., Williams, E.H., Clark-Walker, G.D., and Fox, T.D. (2000). Highly diverged homologs of Saccharomyces cerevisiae mitochondrial mRNA-specific translational activators have orthologous functions in other budding yeasts. Genetics 154, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desloire, S., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4, 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S.R. (1998). Profile hidden Markov models. Bioinformatics 14, 755–763. [DOI] [PubMed] [Google Scholar]
- Edwards, T.A., Wilkinson, B.D., Wharton, R.P., and Aggarwal, A.K. (2003). Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 17, 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Y., Dudoit, S., and Speed, T.P. (2003). Resampling-based multiple testing for microarray data analysis. TEST 12, 1–44. [Google Scholar]
- Giege, P., and Brennicke, A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege, P., and Brennicke, A. (2001). From gene to protein in higher plant mitochondria. C. R. Acad. Sci. III 324, 209–217. [DOI] [PubMed] [Google Scholar]
- Hannaert, V., Saavedra, E., Duffieux, F., Szikora, J.P., Rigden, D.J., Michels, P.A., and Opperdoes, F.R. (2003). Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl. Acad. Sci. USA 100, 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Tsuyoshi, E., Peltier, G., Tasaka, M., and Shikanai, T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36, 541–549. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Tonti-Filippini, J.S., Gout, A.M., Day, D.A., Whelan, J., and Millar, A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hilson, P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- Hirose, T., Kusumegi, T., Tsudzuki, T., and Sugiura, M. (1999). RNA editing sites in tobacco chloroplast transcripts: Editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet. 262, 462–467. [DOI] [PubMed] [Google Scholar]
- Ikeda, T.M., and Gray, M.W. (1999). Characterization of a DNA-binding protein implicated in transcription in wheat mitochondria. Mol. Cell. Biol. 19, 8113–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelesko, J.G., Harper, R., Furuya, M., and Gruissem, W. (1999). Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama, T., and Toriyama, K. (2003). A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544, 99–102. [DOI] [PubMed] [Google Scholar]
- Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., and Baginsky, S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362. [DOI] [PubMed] [Google Scholar]
- Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., Sakai, T., Kawasaki, S., and Imamura, J. (2003). Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34, 407–415. [DOI] [PubMed] [Google Scholar]
- Komori, T., Ohta, S., Murai, N., Takakura, Y., Kuraya, Y., Suzuki, S., Hiei, Y., Imaseki, H., and Nitta, N. (2004). Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 37, 315–325. [DOI] [PubMed] [Google Scholar]
- Lahmy, S., Barneche, F., Derancourt, J., Filipowicz, W., Delseny, M., and Echeverria, M. (2000). A chloroplastic RNA-binding protein is a new member of the PPR family. FEBS Lett. 480, 255–260. [DOI] [PubMed] [Google Scholar]
- Lecharny, A., Boudet, N., Gy, I., Aubourg, S., and Kreis, M. (2003). Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genomics 3, 111–116. [PubMed] [Google Scholar]
- Li, Y., Rosso, M.G., Strizhov, N., Viehoever, P., and Weisshaar, B. (2003). GABI-Kat SimpleSearch: A flanking sequence tag (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics 19, 1441–1442. [DOI] [PubMed] [Google Scholar]
- Liu, L., Amy, V., Liu, G., and McKeehan, W.L. (2002). Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In Vitro Cell. Dev. Biol. Anim. 38, 582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., and McKeehan, W.L. (2002). Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics 79, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lown, F.J., Watson, A.T., and Purton, S. (2001). Chlamydomonas nuclear mutants that fail to assemble respiratory or photosynthetic electron transfer complexes. Biochem. Soc. Trans. 29, 452–455. [DOI] [PubMed] [Google Scholar]
- Mancebo, R., Zhou, X., Shillinglaw, W., Henzel, W., and Macdonald, P.M. (2001). BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol. Cell. Biol. 21, 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey, G.M., and McEwen, J.E. (1995). The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14, 4031–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey, G.M., Przybyla-Zawislak, B.D., and McEwen, J.E. (1998). The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 255, 156–161. [DOI] [PubMed] [Google Scholar]
- Martin, W., and Borst, P. (2003). Secondary loss of chloroplasts in trypanosomes. Proc. Natl. Acad. Sci. USA 100, 765–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. [DOI] [PubMed] [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15, 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mili, S., and Pinol-Roma, S. (2003). LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 23, 4972–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Obokata, J., and Sugiura, M. (2002). Recognition of RNA editing sites is directed by unique proteins in chloroplasts: Biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell. Biol. 22, 6726–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Obokata, J., and Sugiura, M. (2004). A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts. Proc. Natl. Acad. Sci. USA 101, 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha, V.K., et al. (2003). Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. USA 100, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., Meierhoff, K., Westhoff, P., and Schuster, G. (2003). RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270, 4070–4081. [DOI] [PubMed] [Google Scholar]
- Peeters, N.M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A., and Small, I. (2000). Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 50, 413–423. [DOI] [PubMed] [Google Scholar]
- Puskas, L.G., Zvara, A., Hackler, L., Jr., and Van Hummelen, P. (2002). RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques 32, 1330–1334, 1336, 1338, 1340.. [DOI] [PubMed] [Google Scholar]
- Rost, B., and Liu, J. (2003). The PredictProtein server. Nucleic Acids Res. 31, 3300–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Duchene, S., De Oliveira, Y., Caboche, M., Lecharny, A., and Aubourg, S. (2004). FLAGdb++: A database for the functional analysis of the Arabidopsis genome. Nucleic Acids Res. 32 (Database issue), D347–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590. [DOI] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Sonoda, J., and Wharton, R.P. (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser, S., Beckert, S., Capesius, I., Malek, O., and Knoop, V. (1999). Plant mitochondrial RNA editing. J. Mol. Evol. 48, 303–312. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, N., Fukuda, H., Sugimura, T., Nagao, M., and Nakagama, H. (2002). LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur. J. Biochem. 269, 2927–2933. [DOI] [PubMed] [Google Scholar]
- Tsudzuki, T., Wakasugi, T., and Sugiura, M. (2001). Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 53, 327–332. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., Dickerman, A., Brazhnik, O., Nguyen, Q., McElver, J., Frye, C., Patton, D., and Meinke, D. (2003). The Arabidopsis SeedGenes project. Nucleic Acids Res. 31, 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermel, M., Guermann, B., Delage, L., Grienenberger, J.M., Marechal-Drouard, L., and Gualberto, J.M. (2002). A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA 99, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P.M., and Barkan, A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36, 675–686. [DOI] [PubMed] [Google Scholar]
- Yamazaki, H., Tasaka, M., and Shikanai, T. (2004). PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38, 152–163. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.H., and Thorne, N.P. (2003). Normalization for two-colour cDNA microarray data. In Science and Statistics: A Festschrift for Terry Speed, D.R. Goldstein, ed (Bethesda, MD: IMS), pp. 403–418.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.