Abstract
Tumors utilize aerobic glycolysis to support growth and invasion. However, the molecular mechanisms that link metabolism with invasion are not well understood. The nutrient sensor O-linked-β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) modifies intracellular proteins with N-acetylglucosamine. Cancers display elevated O-GlcNAcylation and suppression of O-GlcNAcylation inhibits cancer invasion and metastasis. Here, we show that the regulation of cancer invasion by OGT is dependent on the NAD+-dependent deacetylase SIRT1. Reducing O-GlcNAcylation elevates SIRT1 levels and activity in an AMPK-dependent manner. Reduced O-GlcNAcylation in cancer cells leads to SIRT1-mediated proteasomal degradation of oncogenic transcription factor FOXM1 in a MEK/ERK-dependent manner. SIRT1 is critical for OGT-mediated regulation of FOXM1 ubiquitination and reducing SIRT1 activity reverses OGT-mediated regulation of FOXM1. Moreover, we show that SIRT1 levels are required for OGT-mediated regulation of invasion and metastasis in breast cancer cells. Thus, O-GlcNAcylation is a central component linking metabolism to invasion and metastasis via a SIRT1/ /FOXM1 axis.
Keywords: O-GlcNAc, OGT, sirtuin, SIRT1, FOXM1, cancer, metabolism, invasion, metastasis, deacetylation, MEK, ERK, AMPK
Cancer cells increase nutrient consumption leading to the altered metabolic state known as the Warburg effect(1) (2). The Hexosamine Biosynthetic Pathway (HBP) utilizes major metabolic pathways including glucose, amino acid, fatty acid and nucleotide metabolism to generate the nucleotide sugar uridine diphosphate N-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is used as a substrate for glycosylation of proteins including the post-translational addition of O-linked- β-N-acetylglucosamine (O- GlcNAc) on nuclear and cytosolic proteins, by the enzyme O-GlcNAc transferase (OGT)(3). This modification can be removed by the glycoside hydrolase O-GlcNAcase (OGA) that catalyzes cleavage of O-GlcNAc from proteins(4). This modification can alter protein function directly or, in some cases, indirectly by competing with phosphorylation sites(5).
More recently, elevated O-GlcNAcylation has been described in various types of cancer(6). Our lab provided the first evidence that breast(7) and prostate(8) cancers increase total O-GlcNAcylation by increasing OGT levels. O-GlcNAcylation has been found to be elevated in a host of other cancers including epithelial tumors of the lung(9), liver(10) and colon(11). We have recently shown that the mTOR/MYC pathway can increase OGT levels and O-GlcNAcylation in breast cancer cells(12) thus providing a potential explanation as to why multiple tumor types contain increased O-GlcNAcylation and OGT levels. We have also demonstrated that suppressing O- GlcNAcylation in breast cancer cells leads to reversal of the Warburg effect via regulation of metabolic regulator HIF-1α (13). Importantly, reducing OGT activity inhibits cancer cell invasion in vitro(7) and metastasis in vivo(14) (8). However, the mechanism by which O-GlcNAcylation regulates invasion and metastasis remains unclear.
The oncogenic transcription factor FOXM1 is increased in invasive cancer cells and tissues and is considered as a signature of invasive tumor behavior and poor clinical outcomes(15). In response to growth factor stimulation, cancer cells stabilize FOXM1 levels to promote cell cycle progression, proliferation and invasion(16) (17). FOXM1 has been shown to be directly phosphorylated, stabilized and activated by ERK(17). We have previously shown that OGT inhibition reduces breast and prostate cancer cell invasion through, in part, inhibition of the oncogenic transcription factor, FOXM1 and two of its transcriptional targets, notably the matrix metalloproteinases 2, and 9 (MMP-2, MMP-9)(7) (8). However, the mechanism by which OGT regulates FoxM1 remains unclear.
The sirtuins or class III NAD+-dependent deacylases, have been implicated in many physiological functions including metabolism and cancer(18) (19). Activation of SIRT1 has been shown to extend the life span and promote health in a variety of organisms including yeast and mammals (20) (21). Emerging evidence also points to SIRT1 as a critical player in tumor suppression (22) (23). Tissue-specific loss of SIRT1 is reported to be associated with the increased growth of tumors of the breast(24), colon(22) and liver(21). Recently, SIRT1 was shown to prevent epithelial-to-mesenchymal transition, invasion, and metastasis of breast cancer cells via inhibition of MMP levels(25). Here, we demonstrate that the nutrient sensing pathway HBP connects with the SIRT1 deacetylase via O-GlcNAcylation to regulate cellular invasion via regulation of FOXM1.
Results
O-GlcNAcylation regulates NAD+ metabolism, SIRT1 levels and SIRT1 activity in breast cancer cells
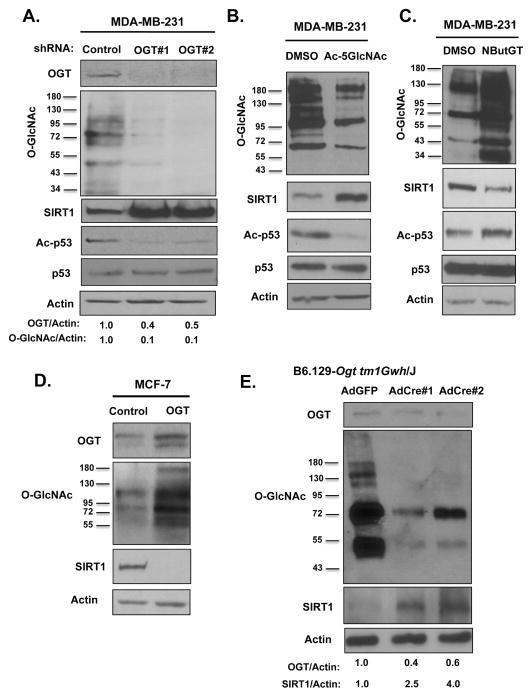
Previously, we have shown that O-GlcNAcylation contributes to metabolic reprogramming in breast cancer cells as reducing O-GlcNAcylation inhibits glycolysis in cancer cells(13). Since reprogramming of cellular metabolism alters intracellular NAD(+) levels(26), we examined the effects on NAD+ metabolites upon alteration of O-GlcNAcylation in cancer cells. MDA-MB-231 breast cancer cells stably expressing OGT shRNA displayed a significant increase in the ratio of NAD+/NADH metabolite levels compared to control cells (Fig. S1A). Since SIRT1 is a NAD+-dependent deacetylase regulated by changes in energy metabolism(18), we hypothesized that OGT could be regulating SIRT1 levels in breast cancer cells. We examined SIRT1 expression in the context of altered OGT/O-GlcNAcylation in a panel of breast cancer cell lines. MDA-MB-231 breast cancer cells stably expressing OGT shRNA (Fig. 1A) contained 2–2.5 fold increase levels of SIRT1 (Fig. S1B), and increased activity as measured by reduced acetylation of p53 on K382 (Fig. 1A), a SIRT1-specific target(27). To ensure effect on SIRT1 levels and activity was dependent on O-GlcNAc cycling, we treated MDA-MB-231 cells with OGT inhibitor (Ac-5SGlcNAc) and also detected a 2.5 fold increase in SIRT1 levels and activity (Fig. 1B, S1C). Similar elevation in SIRT1 levels and activity was observed in other breast cancer cells including MCF-7 (Fig. S1D) and MDA-MB-157 (Fig. S1E) in response to reducing O-GlcNAcylation levels. Conversely, we observed reduced SIRT1 protein levels and activity when we elevated O-GlcNAcylation in MDA-MB-231 cells following treatment with OGA inhibitor NButGT (Fig. 1C, S1F) or in MCF-7 cells stably overexpressing OGT (Fig. 1D, S1G). To test whether OGT regulates SIRT1 levels in vivo in non-tumorigenic tissue, we examined SIRT1 levels in mice containing ogt floxed alleles. We injected adenoviral (Ad)-Cre recombinase or Ad-GFP as control into the mammary fat pads of female mice containing ogt exons flanked by loxP recombination sites(28). To ensure that the majority of each fat pad was infected by adenovirus, we imaged mammary fat pads following Ad-GFP injection and found over 60% of the mammary gland positive for GFP (Fig. S1H). Injecting Ad-Cre to mammary fat pad resulted in reduced OGT and O-GlcNAc levels and elevated SIRT1 levels compared to Ad-GFP treated mice (Fig. 1E). Thus, O-GlcNAcylation regulates SIRT1 levels in mammary tissue in vivo and also SIRT1 levels and activity in cancer cells.
Figure 1. O-GlcNAcylation Regulates SIRT1 levels in breast cancer cells and mammary epithelial cells in vivo.
(A). Targeting OGT using shRNA results in increased SIRT1 protein levels and activity as measured by increased deacetylation of p53 at lysine-382 in MDA-MB-231 breast cancer cells following immunoblot analysis with indicated antibodies. (B). Treatment with OGT inhibitor Ac-5SGlcNAc (100 μM) for 48 hrs reduces total O-GlcNAc levels and results in elevated SIRT1 protein levels and activity in MDA-MB-231 cells. (C). Treatment with OGA inhibitor NButGT (100 μM) for 48 hours reduces SIRT1 protein levels in breast cancer cells. (D). Stable overexpression of OGT reduces SIRT1 protein levels in MCF7 breast cancer cells. (E). Adenoviral-Cre or control Adenoviral-GFP injections into the mammary fat pad of B6.129-Ogttm1Gwh/J females results in decreased OGT and O- GlcNAc levels and an induction of SIRT1 expression.
O-GlcNAcylation regulation of SIRT1 protein levels requires the metabolic sensor AMPK
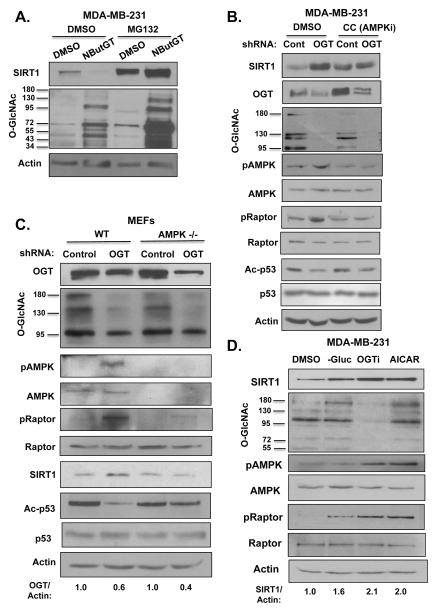
To understand how O-GlcNAcylation regulates SIRT1, we first examined whether O-GlcNAcylation regulates SIRT1 at the level of RNA. Although breast cancer cells expressing OGT RNAi contain a 2–2.5-fold increase in SIRT1 protein levels (Fig. 1A, Fig. S1B), these cells contain decreased levels of SIRT1 RNA as measured by quantitative RT-PCR (qRT-PCR) compared to control cells (Fig. S2A). This indicated that OGT regulation of SIRT1 is independent of RNA and may be at the level of protein. We next examined whether O-GlcNAcylation could influence SIRT1 protein degradation. MDA-MB-231 cells were treated with either DMSO or OGA inhibitor and also treated with the proteasome inhibitor MG132. O-GlcNAc-mediated inhibition of SIRT1 was reversed by treatment with MG132 (Fig. 2A, S2B), suggesting that the change in SIRT1 protein levels was proteasome-dependent. Consistent with this idea, we found that elevating O-GlcNAcylation in MDA-MB-231 cells, by treating with OGA inhibitor, decreased SIRT1 protein stability in cells incubated with cyclohexamide compared to control treated cells (Fig. S2C, S2D). SIRT1 protein does not appear to be directly O-GlcNAcylated, as immunoprecipitatation with RL2 (O-GlcNAc antibody) did not pull down endogenous SIRT1 (Fig. S2E) and exogenous Flag-tagged SIRT1 protein (Fig. S2F) contained no detectable O-GlcNAcylation when compared to the positive control, the transcription factor Sp1 (Fig. S2E). Although this data does not rule out that SIRT1 may be O-GlcNAcylated, it suggests that O-GlcNAcylation regulates SIRT1 protein stability possibly via an indirect mechanism.
Figure 2. O-GlcNAcylation regulates SIRT1 protein stability in an AMPK-dependent manner.
(A). Treatment with proteasome inhibitor reverses OGT-mediated decrease in SIRT1 levels. MDA-MB-231 cells were treated with either DMSO or (100 μM) NButGT for 48 hrs and subjected to treatment with (10 μM) proteasome inhibitor MG132 for 24 hrs following immunoblot analysis with indicated antibodies. (B). OGT regulates SIRT1 levels in an AMPK manner. MDA-MB-231 cells were infected with shControl or shOGT lentivirus for 48 hours, then treated with 10 μM Compound C (AMPKi) for 24 hours following immunoblot analysis using indicated antibodies. (C). Wild type (WT) mouse embryonic fibroblasts (MEFs) and AMPK−/− mouse embryonic fibroblasts were infected with shControl or shOGT lentivirus for 48 hours following immunoblot using indicated antibodies. (D). OGT inhibition mimics increases in SIRT1 induced under low glucose or AICAR treatment. MDA-MB-231 cells were grown in media with DMSO, no glucose, 100 μM OGT inhibitor (Ac-5SGlcNAc), or 1 mM AICAR for 24 hours following immunoblot with indicated antibodies.
OGT inhibition induces phosphorylation and activation of AMP-activated protein kinase α(AMPK)(13). Since SIRT1-deacetylase activity has been shown to be regulated through activation of AMPK(29),(30), we hypothesized that OGT/O-GlcNAcylation may regulate SIRT1 protein stability in an AMPK-dependent manner. To test this, MDA-MB-231 cells stably expressing control or OGT shRNA were treated with the AMPK inhibitor, Compound C. MDA-MB-231 cells treated with Compound C contained decreased AMPK phosphorylation and activation as measured by phosphorylation of its substrate Raptor (Ser792) (Fig. 2B). The increased SIRT1 levels and activity observed upon suppression of OGT were reversed by treatment with Compound C compared to control cells (Fig. 2B, S3A). Similar results were obtained in MCF-7 (Fig. S3B) and MDA-MB-157 (Fig. S3C) breast cancer cells, suggesting that the regulation of SIRT1 by OGT was AMPK-dependent in multiple breast cancer cells. Similar to our observation in cancer cell lines, wild-type mouse embryonic fibroblasts (MEFs) expressing OGT shRNA displayed increased SIRT1 levels compared to control cells (Fig. 2C, S2G). However, OGT depleted MEFs containing deletion of both alpha subunits of the AMPK gene (MEF-AMPKα−/−) did not increase SIRT1 levels or activity upon OGT suppression (Fig. 2C, S2G). To test whether AMPK activation alone was sufficient to increase SIRT1 levels in MDA-MB-231 cells we treated cells with AICAR (5-aminoimidazole-4-carboxamide-ribonucleotide), an AMP analog known to activate AMPK and increase phosphorylation of downstream AMPK targets including Raptor(31). Treatment of cells with AICAR, as well as glucose deprivation increased SIRT1 levels and increased both AMPK activation and Raptor phosphorylation. These changes were similar to those observed with OGT inhibitor treatment (Fig. 2D) and consistent with previous results showing that increase of O-GlcNAcylation, via treatment with OGA inhibitor, inhibits AMPK activity(13). To test whether OGT regulation of AMPK and SIRT1 is dependent on changes in metabolism, we tested whether treating OGT depleted cells with Methyl-pyruvate, a permeable nutrient that supports bioenergetics, would reverse AMPK activation and SIRT1 levels. Reducing OGT levels in MDA-MB-231 cells leads to activation of AMPK as measured by increase in Raptor phosphorylation (Fig. S3D). However, this activation was reduced in OGT-depleted cells treated with Methyl-pyruvate compared to control cells (Fig. S3D). SIRT1 levels were slightly increased in cells treated with Methyl-pyruvate compared to control. However, knocking down OGT did not effect SIRT1 levels in the presence of Methyl-pyruvate suggesting OGT regulation of SIRT1 is, in part, dependent on metabolic changes. Consistent with reversing AMPK-associated signaling and metabolic stress, Methyl-pyruvate treatment also reversed ATP level depletion in OGT knockdown cells (Fig. S3E). Taken together, these results suggest that O-GlcNAcylation regulation of SIRT1 protein stability is dependent on the metabolic sensor AMPK in cancer cells and associated with O-GlcNAcylation regulation of cell metabolism.
O-GlcNAcylation-mediated cancer cell invasion requires SIRT1 regulation of FOXM1 signaling
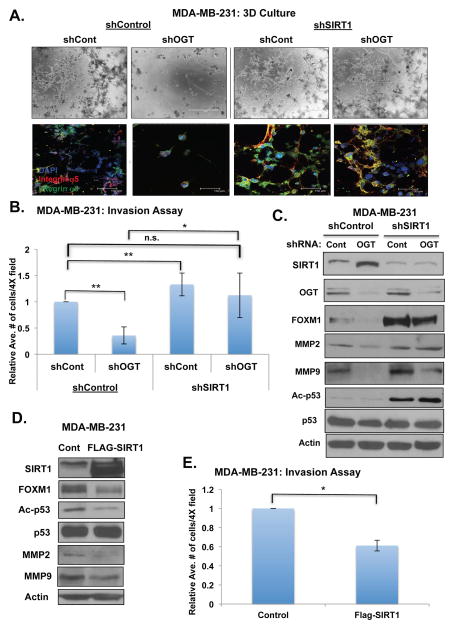
We have previously shown that inhibition of OGT levels or activity reduces breast(7) and prostate(8) cancer cell invasion in vitro. SIRT1 activity has been shown to be critical in the regulation of EMT and metastasis of breast cancer cells in vivo(25). Therefore, we sought to characterize the role of SIRT1 in OGT-mediated breast cancer cell invasion. Reducing OGT expression in MDA-MB-231 cells blocks cellular invasion in a 3-dimensional (3D) culture system (Fig. 3A) and as measured by transwell chamber invasion assays (Fig. 3B). In invasion assay, reducing SIRT1 expression also slightly increased invasion (Fig. 3B) compared to control (shControl, shControl) cells (one-way ANOVA, p<0.05). We were able to rescue invasion defects in 3D culture (Fig. 3A) and invasion assays (Fig. 3B) in OGT knockdown cells to control levels by reducing SIRT1 expression (Fig. 3C). Conversely, overexpression of SIRT1 alone significantly decreased MDA-MB-231 breast cancer cell invasion (Fig. 3D, 3E).
Figure 3. OGT modulates breast cancer cell invasion and FOXM1 levels in a SIRT1-dependent manner.
(A). Stably reducing SIRT1 levels using RNAi in MDA-MB-231 cells rescues OGT- mediated growth/invasion inhibition in 3D culture. Cells were photographed on day 5 using confocal microscopy either in bright field (BF) or upon confocal immunoflourescence staining (IF) using indicated antibodies. (B). Stably reducing SIRT1 levels using RNAi in MDA-MB-231 cells rescues OGT-mediated reduction in invasive potential using Boyden chamber invasion assay (one-way ANOVA used to compare all groups to Control (shCont, shCont), **p<0.05). Student’s two-tailed test was used to compare shCont-shOGT and shSirt1-shOGT. (*p<0.05). Relative average of number of cells in 4 different fields was quantified. (C). Stably reducing SIRT1 levels using RNAi in MDA-MB-231 cells rescues OGT- mediated inhibition of oncogenic transcription factor FOXM1 and targets MMP2 and MMP9. (D). Overexpression of SIRT1 results in reduced FOXM1 levels and MMP2/MMP9 expression in MDA-MB-231 cells. (E). Overexpression of SIRT1 results in inhibition of invasion in MDA-MB-231 cells using Boyden chamber invasion assay (*p<0.05).
Our lab has previously demonstrated that OGT-mediated invasion in breast and prostate cancer cells was associated with regulation of FOXM1 protein stability(7) (8) and its transcriptional targets the MMP2 and MMP9. Therefore, we investigated whether SIRT1 played a role in the regulation of FOXM1 levels and activity by OGT. We compared the effects of OGT knockdown on levels of FOXM1 in control MDA-MB-231 cells and those stably expressing SIRT1 RNAi. Reducing OGT expression in breast cancer cells reduced levels of FOXM1 (Fig. 3C, S4A). Interestingly, reducing SIRT1 levels in MDA-MB-231 cells increased not only the basal expression of FOXM1 protein but also reversed OGT-mediated inhibition of FOXM1 levels (Fig. 3C, S4A). Similar reversal of OGT-mediated regulation of FOXM1 was seen when SIRT1 levels were reduced in two additional breast cancer cell lines MDA-MB-157 (Fig. S4B) and MDA-MB-468 (Fig. S4C) and similar reversal of invasive phenotype was seen in MDA-MB-157 cells in 3D culture when SIRT1 levels were reduced (Fig. S4D). Consistent with SIRT1 reversing FOXM1 protein levels, we also observed rescue of FOXM1 transcriptional targets MMP2 (Fig. 3C) and MMP9 (Fig. 3C, S4B, S4C) at both the protein and mRNA levels of MMP9 (Fig. S4E) in OGT depleted MDA-MB-231 cells. To ensure that the changes in MMP2 and MMP9 were dependent on FOXM1, we examined effect on these targets in FOXM1-depleted cells. Increased levels of MMP2 and MMP9 in MDA-MB-231 cells expressing SIRT1 shRNA was reversed in cells co-expressing FOXM1 RNAi (Fig. S5A) thus establishing that the effect on MMP2 and MMP9 by SIRT1 was indeed FOXM1-dependent. We were also able to reverse OGT-depletion-mediated inhibition of FOXM1 and MMP9 levels (Fig. S5B) and the invasive phenotype (Fig. S5C) in MDA-MB-231 cells after treatment with the SIRT1-specific inhibitor EX-527. Treatment of MDA-MB-231 cells with EX-527 had similar effects on p53 acetylation (Fig. S5B) compared to cells containing SIRT1 knockdown (Fig. 3C). Conversely, overexpression of SIRT1 in MDA-MB-231 breast cancer cells resulted in reduced expression of FOXM1 as well as that of its transcriptional targets MMP2 and MMP9 (Fig. 3D) and was also consistent with concomitant decreases in cellular invasion (Fig. 3E). Thus, these data suggest that OGT regulation of the key cell invasion protein FOXM1 and its targets MMP2 and MMP9 in breast cancer cells are dependent on O-GlcNAc suppression of SIRT1.
SIRT1 regulates FOXM1 ubiquitination and proteasomal degradation via D and KEN-boxes and E3-ubiquitin ligase Cdh1
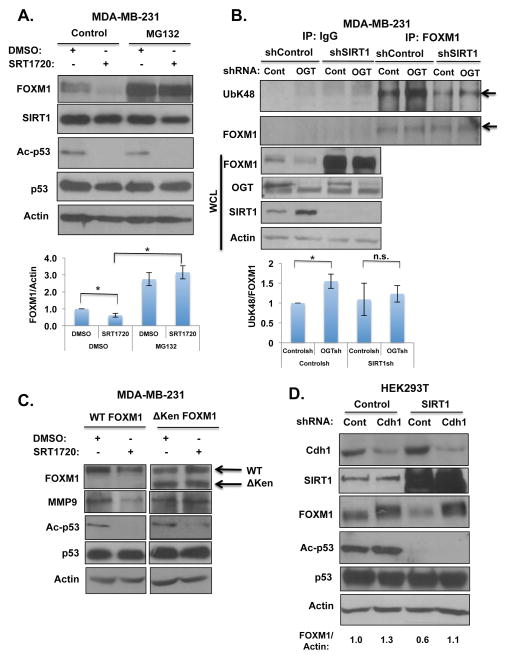
Since we were unable to detect SIRT1-dependent acetylation changes on FOXM1 (data not shown), we hypothesized that SIRT1 may be regulating FOXM1 activity via an alternate mechanism. To test whether SIRT1 regulates FOXM1 degradation, MDA-MB-231 cells treated with the SIRT1-specific activator, SRT1720(32), were subjected to proteasome inhibition using MG132. Downregulation of FOXM1 protein levels upon SIRT1 activation could be reversed by treatment with MG132 (Fig. 4A), suggesting that SIRT1 regulation of FOXM1 was proteasome-dependent. We also detected a 1.5-fold increase in FOXM1 ubiquitination under conditions of decreased OGT levels in MDA-MB-231 cells compared to controls (Fig. 4B). However, this OGT-mediated increase in FOXM1 ubiquitination was diminished in the presence of SIRT1 shRNA (Fig. 4B). The N-terminus of FOXM1 contains degron destruction D-box and KEN-box sequences that are known to be essential for FOXM1 degradation(33). Since OGT regulates FOXM1 proteasome-mediated degradation via the D-box and KEN-box sequences(7), we investigated whether SIRT1 activation reduces FOXM1 levels in MDA-MB-231 cells stably expressing either wild-type or a degradation-resistant FoxM1 mutant. Treatment with SIRT1 activator SRT1720 resulted in decreased levels of exogenous wildtype FOXM1 and its target MMP9 in MDA-MB-231 cells (Fig. 4C, S5D). However, in MDA-MB-231 cells overexpressing the non-degradable FoxM1 mutant lacking the N-terminus, SIRT1 activation did not alter levels of FOXM1 mutant or MMP9 levels compared to wildtype FOXM1 expressing cells (Fig. 4C). Since the E3-ubiquitin ligase Cdh1 is known to regulate the FOXM1 degradation pathway(34), we examined whether SIRT1 effects on FOXM1 were dependent on Cdh1-mediated degradation. We compared the effects of SIRT1 overexpression on FOXM1 levels in HEK293T cells stably expressing control or Cdh1 RNAi. Reducing levels of Cdh1 increased endogenous expression of FOXM1 levels (Fig. 4D). Overexpression of SIRT1 in breast cancer cells (Fig. 3D) or in HEK293T cells reduced levels of FOXM1 (Fig. 4D). This inhibition was lost in HEK293T cells containing Cdh1 stable RNAi compared to control cells (Fig. 4D). Taken together, these data suggest that the regulation of FOXM1 by OGT requires SIRT1-mediated ubiquitination and proteasomal degradation of FOXM1 which is dependent on its D-box/KEN-box degron domains and its E3-ligase Cdh1.
Figure 4. OGT regulates FOXM1 stability through SIRT1.
(A). Proteasome inhibition reverses SIRT1-mediated decreases in FOXM1 protein. MDA-MB-231 cells were treated for 24 hrs with either DMSO or (1μM, SIRT1 activator) SRT1720 and subjected to treatment with (10 μM) proteasome inhibitor MG132 following immunoblot analysis with indicated antibodies. (B). Immunoprecipitation of endogenous FOXM1 in the presence OGT shRNA results in elevated FOXM1 ubiquitination which is reversed by SIRT1 depletion. Arrows indicate bands quantified. Graph represents quantified UbK48/FOXM1 levels in indicated treatments (*p<0.05). (C). Treatment with SIRT1 activator SRT1720 (1μM) in MDA-MB-231 breast cancer cells stably expressing either WT or degradation deficient (ΔKen) HA-tagged FOXM1. SIRT1 activator, SRT1720 results in reduced levels of WT FOXM1 but not degradation deficient (ΔKen) FOXM1. (D). HEK293T cells infected with either Cdh1 shRNA or Control (pBabe) and transiently transfected with either control or SIRT1 plasmid and immunoblot analysis with indicated antibodies.
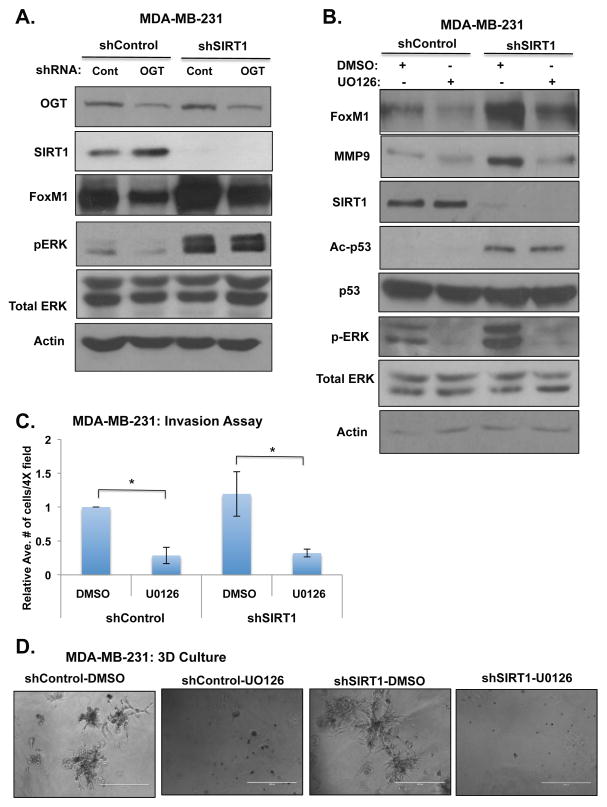
O-GlcNAc/SIRT1 regulation of FOXM1 requires the MEK/ERK pathway in breast cancer cells
Growth factor-dependent stabilization of FOXM1 protein can be mediated through ERK phosphorylation to promote an invasive signature in various cancer types(16) (17). Recent reports have demonstrated that SIRT1 can inhibit MAP-Kinase pathway activity through direct deacetylation of MEK, thus affecting it’s ability to phosphorylate and activate ERK(35). Therefore, we sought to investigate whether O-GlcNAc and SIRT1 could promote FOXM1 stability via the MEK-ERK pathway. To address this question, we first measured levels of ERK phosphorylation (Thr202/Tyr204) in OGT-depleted cells with and without stable expression of SIRT1 RNAi. Depletion of OGT levels reduced ERK phosphorylation in MDA-MB-231 (Fig. 5A, S6A), MDA-MB-468 (Fig. S6B), and MDA-MB-157 (Fig. S6C) breast cancer cells. Changes in ERK phosphorylation upon OGT depletion were reversed in SIRT1 depleted cells in all three breast cancer cell lines, MDA-MB-231 (Fig. 5A, S6A), MDA-MB-468 (Fig. S6B) and MDA-MB-157 (Fig. S6C), indicating that OGT-mediated regulation of ERK is SIRT1-dependent. Consistent with this data, treating MDA-MB-231 cells with SIRT1 activator EX-527 also increased basal ERK activation but also reversed OGT-depleted inhibition of ERK activity (Fig. S6D). Conversely, MDA-MB-231 cells treated with SIRT1 activator SRT1720 reduced ERK activity (Fig. S6E), FOXM1 levels (Fig. 4C) and invasion in 3D culture (Fig. S6F). To test whether SIRT1 regulation of FOXM1 stability requires activation of MEK/ERK pathway, MDA-MB-231 cells were treated with the MEK inhibitor, U0126. As previously shown, cells depleted of SIRT1 display elevated FOXM1 levels and ERK activity compared to control cells (Fig. 5B). However, treatment of SIRT1 depleted cells with U0126 abrogated changes in FOXM1 levels (Fig. 5B, S6G) and decreased levels of the FOXM1 transcriptional target MMP-9 (Fig. 5B). Consistent with reduced FOXM1 and MMP9 levels, SIRT1-depleted MDA-MB-231 cells treated with MEK inhibitor mitigated cellular invasion (Fig. 5C) and invasion in 3D culture (Fig. 5D). Lastly, to test whether SIRT1 regulation of ERK/FOXM1 pathway occurs in non-cancer cells, we examined FOXM1 levels and ERK activation in SIRT1 deficient (SIRT1 −/−) mouse embryonic fibroblasts (MEFs) compared to wild-type MEFs. SIRT1 deleted MEFs contained higher FOXM1 levels and ERK activation compared to control cells (Fig. S6H). These data indicate that OGT-mediated increase in SIRT1 levels and activity can suppress ERK signaling leading to reduction of FOXM1 activity and invasion in aggressive breast cancer cells.
Figure 5. OGT Regulates FOXM1 levels via SIRT1-ERK dependent mechanism.
(A). Stably reducing SIRT1 levels using RNAi in MDA-MB-231 cells reverses OGT-mediated inhibition of oncogenic transcription factor FOXM1 and ERK activity. (B). SIRT1 regulation of FOXM1 is MEK-dependent. Treatment with MEK inhibitor (U0126, 10 μM) for 24 hrs reverses SIRT1-mediated increases in FOXM1 and target gene MMP9. (C). Treatment with MEK inhibitor (U0126, 10 μM) for 16 hrs blocks SIRT1-knockdown invasion using Boyden chamber invasion assay (*p<0.05). Relative average of number of cells in 4 different fields was quantified (n=3). (D). Treatment with MEK inhibitor (U0126, 10 μM) blocks growth and invasion in SIRT1-depleted MDA-MB-231 cells in 3-dimensional culture (day 5). Scale bar: 400μ.
SIRT1 is critical for OGT-mediated regulation of breast cancer metastasis in vivo
Based on our in vitro studies above, we hypothesized that OGTs regulation of metastasis may involve modulating SIRT1 levels in breast cancer cells. To test directly whether OGT regulation of SIRT1 plays a critical role in breast cancer metastasis, we compared the effects of OGT knockdown in an intracardiac model of metastasis in MDA-MB-231 cells stably expressing control or SIRT1 RNAi. MDA-MB-231 cells expressing luciferase and either control shRNA or OGT shRNA in the presence or absence of SIRT1 shRNA (Fig. 6A) were injected into the left ventricle of female NSG mice. The progression of systemic metastasis of tumor cells following intracardiac injection was monitored by bioluminescence imaging using a stably expressed firefly luciferase reporter. Reducing OGT in MDA-MB-231 cells caused a decrease in metastatic dissemination in vivo (Fig. 6B) and decreased establishment of liver macrometastasis (Fig. 6C) and micrometastasis (Fig. 6D). However, cells depleted of SIRT1 were able to rescue the metastatic effect of OGT RNAi in vivo (Fig. 6B), since SIRT1 RNAi in the context of OGT depletion reversed liver macrometastasis (Fig. 6C) and significantly reversed micrometastasis (Fig. 6D) compared to control OGT-depleted cells. Consistent with bioluminescence imaging, histological sections from the control shRNA group exhibited massive liver metastatic tumor (Fig 6Ea). By comparison, a striking decrease in the size and intra-hepatic spread of metastatic cancer deposits was demonstrated in OGT depleted MDA-MB-231 cells (Fig. 6Eb). In contrast, widespread metastatic involvement accompanied by tumor necrosis, comparable in severity to the control group, was detected in the SIRT1 shRNA group (Fig. 6Ec), as well as in the OGT shRNA group albeit in the presence of SIRT1 shRNA (Fig. 6Ed). Thus, at the histological level, inhibition of hepatic metastasis by OGT depletion could be reversed in the presence of SIRT1 shRNA. Similar results were seen in histological analysis of bone metastasis to vertebral column (Fig. S7). Thus, OGT regulation of cancer metastasis in vivo is partly dependent on SIRT1 expression.
Figure 6. OGT regulates breast cancer metastasis in a SIRT1-dependent manner.
(A). MDA-MB-231-Luciferase cells stably expressing either control or SIRT1 shRNA and either control or OGT shRNA were lysed and immunoblotted with the indicated antibodies before being injected intracardially into 6-week-old female NSG mice. (B). SIRT1 knockdown prevents OGT-mediated inhibition of breast cancer metastatic lesions to bone, brain and liver. Animals were imaged using the IVIS Lumina system at 5 weeks post-injection with control or SIRT1 shRNA expressing and control and OGT shRNA-expressing MDA-MB-231 cells. (C). Representative ex vivo liver images of mice from each group using bioluminescence imaging. (D). Average number of metastatic foci detected using bioluminescence imaging were counted for control (n = 5), OGT (n = 5), SIRT1 (n = 5) and SIRT1/OGT shRNA (n = 6) mice and graphed. S.E. *, < 0.05. (E). Representative images (magnification ×4) from immunohistochemistry analysis (hematoxylin/eosin) of liver from mice from all four groups. Asterisks indicate metastatic tumor cell infiltration. Scale bars: 50μ.
Discussion
Taken together, our findings have demonstrated a novel mechanism by which the nutrient sensor, O-GlcNAcylation contributes to the acquisition of invasive and metastatic phenotypes in breast cancer cells through regulation of SIRT1/FOXM1 axis. For the first time, we demonstrate that SIRT1 inhibits stability of the oncogenic transcription factor FOXM1 effectively suppressing tumor cell invasion and provide a mechanism by which reduced SIRT1 levels may contribute to metastasis. Recently, we have demonstrated that O-GlcNAcylation promotes a Warburg phenotype to alter metabolic signaling and energy status in cancer cells (13). Our data strongly suggest a model (Fig. S8) in which OGT-mediated alterations in the energy metabolism can regulate AMPK signaling which regulates SIRT1 levels and activity in breast cancer cells. O-GlcNAcylation regulation of SIRT1 contributes to the stabilization of the oncogenic transcription factor FOXM1 via an ERK-dependent mechanism to regulate MMP2 and MMP9 levels and thus breast cancer invasion and metastasis. Collectively, these results establish a novel molecular pathway linking the metabolic regulator O-GlcNAcylation to invasion and metastasis via a SIRT1/ERK/FOXM1 axis.
Cross-talk between O-GlcNAcylation and the AMPK is emerging as important node in regulating cell metabolism, growth and signaling in both normal physiology and diseased states
Previous studies have shown that AMPK can phosphorylate OGT and regulate its substrate specificity and that AMPKα and γ subunits can be O-GlcNAcylated in skeletal muscle cells(36). In cancer cells, our data suggests O-GlcNAcylation regulates AMPK indirectly via regulation of metabolism. Reducing O-GlcNAcylation in cancer cells leads to robust increase in AMPK activity that can be reversed by addition of a permeable nutrient metabolite to support bioenergetics such as Methyl-pyruvate or overexpressing glucose transporter GLUT1(13). Interestingly, OGT encodes three splice variants with the longest OGT isoform nucloecytoplasmic OGT being localized in the nucleus and cytoplasm(37, 38). This isoform has been linked to transcriptional regulation, proteasomal regulation, and stress response. A mitochondrial isoform is thought to be proapoptotic. Future studies will determine which OGT isoform is a key regulator of AMPK, SIRT1 and metabolic pathways in cancer cells.
SIRT1 has a predominately tumor suppressing function in various tumor mouse models, including mice with mutant p53(39) or BRCA1(24). It is possible that the role of SIRT1 in breast cancer may be dependent on genetic make-up or breast cancer subtype, as our data shows that SIRT1 functions as tumor suppressor in basal/triple-negative breast cancers. A decrease of SIRT1 levels caused reversal of the mitigating effect on metastatic potential of OGT-depleted cells thus suggesting that reduced SIRT1 levels and activity is beneficial to metabolically stressed cancer cells and contributes to metastatic dissemination. SIRT1 also directly regulates transcription and chromatin silencing through deacetylation of histone substrates(40), as well as a variety of non-histone targets involved in cancer including p53 and PGC-1α to combat cellular stress and promote oxidative metabolism(41). Interestingly, O-GlcNAcylation is also known to regulate transcription and epigenetic regulators(42). It will of interest to determine whether an inverse relationship exists between genes regulated by O-GlcNAcylation and SIRT1. It was recently demonstrated that in response to growth factor stimulation, SIRT1 can directly deacetylate MEK (K175) to inhibit downstream ERK activation(35). In addition, it was recently shown that SIRT2 depletion in colon and lung cancer cells increases activation of ERK signaling and leads to resistance to EGFR inhibitors(43). Our studies are consistent with this data and extend a role of SIRT1/ERK signaling to regulate oncogenic transcription factor FOXM1 which is critical for invasion and metastasis in aggressive breast cancer cells.
Complete knockout of OGT in MEFs lead to defects in cell growth(28) and other studies have shown that O-GlcNAcylation is important in cell cycle progression(38). Our studies and others suggest that knockdown of OGT or pharmacological inhibition of O-GlcNAcylation impairs growth and survival of cancer cells but has no effect on non-transformed epithelial cells of the prostate(8), mammary(7) (12) and pancreas(44). This data suggest that cancer cells may be more dependent on O-GlcNAcylation for growth and survival compared to normal counterparts. However, developing specific OGT inhibitors may help address this important question.
Activity of sirtuins, including SIRT1, have been reported to decrease during aging and are associated with altering longevity(18) (45). Interestingly, recent reports have shown that various tissues derived from aging mice contain elevated levels of O-GlcNAcylation(46) (47). Since we have shown that reducing O-GlcNAcylation in normal mammary tissue increases SIRT1 levels in vivo, it will be interesting to further understand the inverse relationship between O-GlcNAcylation and SIRT1 as it relates to aging and longevity in future studies. Thus, aberrant O-GlcNAcylation contributes to cancer progression via regulation of SIRT1 and may also connect to pathways associated with aging and longevity.
Materials and Methods
Reagents
NButGT, Ac-5SGlcNAc were provided by D. Vocadlo (Simon Fraser University)(48). SRT1720, EX-527, Compound C purchased from Selleck-Chemicals (Houston, TX). MG132 purchased from Sigma-Aldrich (St. Louis, MO). pcDNA3.1-SIRT1-Flag was gift from E. Verdin (Addgene plasmid#: 13812). pLenti4-HA-OGT (provided by K. Vosseller, Drexel University). Antibodies used: anti-actin, anti-FOXM1 and anti-RL2 from Santa Cruz Biotechnology; pERK1/2, anti-OGT and anti-O-GlcNAc from Sigma-Aldrich; anti-acetylated p53(K382), anti-AMPK, anti-pAMPK(S172), anti-ERK1/2, anti-MMP2, anti-pRaptor(792), anti-Raptor, anti-SIRT1 and anti-Ubiquitin(K48) from Cell Signaling (Danvers, MA); anti-Cdh1 and anti-p53 from Neobiolabs; anti-MMP9 from Novus-Biologicals (Littleton, CO); integrin α5 and α6 are from BD-Pharmingen (San Jose, CA).
Cell Culture
MDA-MB-231, MDA-MB-157, MCF-7 and MDA-MB-468 cells were obtained from ATCC (Manassas, VA) and cultured according to ATCC. SIRT1+/+ and SIRT1−/− MEFs were cultured as previously described(49). MEFs and AMPK null (AMPKα1/α2 KO) MEFs were kindly provided by B. Viollet (INSERM) and cultured as previously described(50).
Metabolic Assays
NAD+/NADH levels were measured with the NAD/NADH Cell Based Assay Kit (Cayman Chemical, Ann Arbor, MI) and ATP levels were measured with the ATP Assay Kit (Calbiochem, San Diego, CA) according to the manufacturer’s protocol.
Immunoblotting
Immunoblotting protocols have been previously described(51).
3D assay and indirect immunofluorescence
Immunoprecipitation
Assays were performed as described previously(13).
Transwell Invasion Assays
Invasion assays were performed as previously described(8). Assays utilizing chemical inhibitors were performed by adding inhibitors at indicated concentrations prior to seeding.
shRNA Generation
shRNA sequences and lentiviral particles were generated as previously described(7, 13). Sequences used for SIRT1 were MissionshRNA clones; SIRT1-1: (TRCN0000229630, Sigma-Aldrich) and SIRT1-2 (TRCN0000218734, Sigma-Aldrich, St. Louis, MO).
Animal Studies
Female NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory) (6–8 weeks old) were randomized and inoculated via intracardiac injection into the left ventricle with 2.0×105 MDA-MB-231 cells as previously described (8). Mice were injected intraperitoneally with 200 ul of D-luciferin solution (9 mg/ml; Caliper-Life-Sciences, Hopkinton, MA), and bioluminescence imaging was performed 4 hours after intracardiac injection to detect the distribution of breast cancer cells followed by weekly imaging using the IVIS 200. Only animals that showed successful injections by bioluminescence were included in analysis. Sample size was determined via previous analysis. B6.129-Ogttm1Gwh/J females used for fat pad injections have been previously described(28) and were obtained as a kind gift from S. Jones (University of Louisville). Mice were injected directly into the mammary fat pad with Ad5-CMV-GFP or Ad5-CMV-Cre (2.7×107 PFU, from the Baylor College of Medicine). Animals groups were not blinded. Histology was performed as previously described(8). All animal protocols were approved by the Institutional Animal Care and Use Committee.
qRT-PCR
QRT-PCR was performed as previously described(13). TaqMan gene expression assay primer/probes were purchased from Applied Biosystems (Foster City, CA) for cyclophilin A (Hs99999904_m1), OGT (Hs00914634_g1), SIRT1 (Hs01009005_m1), MMP-9 (Hs00234579_m1).
Statistics
All results shown are results of at least three independent experiments and shown as averages and presented as mean ± SE. P-values were calculated using a Student’s two-tailed test: (* represents at least p-value <0.05). Where indicated, one-way analysis of variance (ANOVA) was used to compare differences between multiple groups (** represents at least p-value <0.05). Appropriate sample size was chosen to achieve significance using this analysis.
Supplementary Material
Acknowledgments
We thank Valerie Sodi for critical reading of this manuscript. This work is supported by NCI grants CA183574 (to C.M.F.), CA155413 (to M.J.R.) and NIH/NIA grant AG028730 (to D.A.S).
Footnotes
Conflict of interest
D.A.S. is a consultant to GlaxoSmithKline, Segterra, Ovascience, and MetroBiotech. The authors declare no conflict of interest.
References
- 1.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19345–50. doi: 10.1073/pnas.0709747104. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008 Jan;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PubMed] [Google Scholar]
- 3.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007 Apr 26;446(7139):1017–22. doi: 10.1038/nature05815. Research Support, N.I.H., Extramural Review. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001 Mar 30;276(13):9838–45. doi: 10.1074/jbc.M010420200. Research Support, U.S. Gov’t, P.H.S. [DOI] [PubMed] [Google Scholar]
- 5.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010 Feb;1800(2):96–106. doi: 10.1016/j.bbagen.2009.07.018. Research Support, N.I.H., Extramural Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch TP, Reginato MJ. O-GlcNAc transferase: a sweet new cancer target. Cell Cycle. 2011 Jun 1;10(11):1712–3. doi: 10.4161/cc.10.11.15561. Comment Editorial. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010 May 13;29(19):2831–42. doi: 10.1038/onc.2010.41. Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S. [DOI] [PubMed] [Google Scholar]
- 8.Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012 Mar 30;287(14):11070–81. doi: 10.1074/jbc.M111.302547. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, et al. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011 Apr;1812(4):514–9. doi: 10.1016/j.bbadis.2011.01.009. Research Support, Non-U.S. Gov’t. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Q, Zhou L, Yang Z, Lai M, Xie H, Wu L, et al. O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med Oncol. 2012 Jun;29(2):985–93. doi: 10.1007/s12032-011-9912-1. Comparative Study Research Support, Non-U.S. Gov’t. [DOI] [PubMed] [Google Scholar]
- 11.Yang YR, Kim DH, Seo YK, Park D, Jang HJ, Choi SY, et al. Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-kappaB signaling. Oncotarget. 2015 May 20;6(14):12529–42. doi: 10.18632/oncotarget.3725. Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sodi VL, Khaku S, Krutilina R, Schwab LP, Vocadlo DJ, Seagroves TN, et al. mTOR/MYC Axis Regulates O-GlcNAc Transferase Expression and O-GlcNAcylation in Breast Cancer. Mol Cancer Res. 2015 May;13(5):923–33. doi: 10.1158/1541-7786.MCR-14-0536. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell. 2014 Jun 5;54(5):820–31. doi: 10.1016/j.molcel.2014.04.026. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y, Mi W, Ge Y, Liu H, Fan Q, Han C, et al. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010 Aug 1;70(15):6344–51. doi: 10.1158/0008-5472.CAN-09-1887. Research Support, Non-U.S. Gov’t. [DOI] [PubMed] [Google Scholar]
- 15.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012 Jan;1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004. Research Support, Non-U.S. Gov’t Review. [DOI] [PubMed] [Google Scholar]
- 16.Lok GT, Chan DW, Liu VW, Hui WW, Leung TH, Yao KM, et al. Aberrant activation of ERK/FOXM1 signaling cascade triggers the cell migration/invasion in ovarian cancer cells. PLoS One. 2011;6(8):e23790. doi: 10.1371/journal.pone.0023790. Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005 Feb 15;118(Pt 4):795–806. doi: 10.1242/jcs.01657. Research Support, Non-U.S. Gov’t. [DOI] [PubMed] [Google Scholar]
- 18.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014 Aug;24(8):464–71. doi: 10.1016/j.tcb.2014.04.002. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JE, Mostoslavsky R. Sirtuins, metabolism, and DNA repair. Curr Opin Genet Dev. 2014 Jun;26:24–32. doi: 10.1016/j.gde.2014.05.005. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3(4):e2020. doi: 10.1371/journal.pone.0002020. Research Support, N.I.H., Extramural Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2014 Mar 27;33(13):1609–20. doi: 10.1038/onc.2013.120. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008 Oct 10;32(1):11–20. doi: 10.1016/j.molcel.2008.09.011. Research Support, N.I.H., Intramural. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simic P, Williams EO, Bell EL, Gong JJ, Bonkowski M, Guarente L. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Rep. 2013 Apr 25;3(4):1175–86. doi: 10.1016/j.celrep.2013.03.019. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV. Links between metabolism and cancer. Genes Dev. 2012 May 1;26(9):877–90. doi: 10.1101/gad.189365.112. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001 Oct 19;107(2):149–59. doi: 10.1016/s0092-8674(01)00527-x. Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. [DOI] [PubMed] [Google Scholar]
- 28.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004 Feb;24(4):1680–90. doi: 10.1128/MCB.24.4.1680-1690.2004. Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009 Apr 23;458(7241):1056–60. doi: 10.1038/nature07813. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau AW, Liu P, Inuzuka H, Gao D. SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. Am J Cancer Res. 2014;4(3):245–55. [PMC free article] [PubMed] [Google Scholar]
- 31.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008 Apr 25;30(2):214–26. doi: 10.1016/j.molcel.2008.03.003. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013 Mar 8;339(6124):1216–9. doi: 10.1126/science.1231097. Research Support, N.I.H., Extramural Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park HJ, Wang Z, Costa RH, Tyner A, Lau LF, Raychaudhuri P. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene. 2008 Mar 13;27(12):1696–704. doi: 10.1038/sj.onc.1210814. Research Support, N.I.H., Extramural. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol. 2008 Sep;28(17):5162–71. doi: 10.1128/MCB.00387-08. Research Support, N.I.H., Extramural. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeung F, Ramsey CS, Popko-Scibor AE, Allison DF, Gray LG, Shin M, et al. Regulation of the mitogen-activated protein kinase kinase (MEK)-1 by NAD(+)-dependent deacetylases. Oncogene. 2015 Feb 5;34(6):798–804. doi: 10.1038/onc.2014.39. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Hunt DF, Neumann D, et al. Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK) J Biol Chem. 2014 Apr 11;289(15):10592–606. doi: 10.1074/jbc.M113.523068. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997 Apr 4;272(14):9308–15. doi: 10.1074/jbc.272.14.9308. Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. [DOI] [PubMed] [Google Scholar]
- 38.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015 Mar 30;208(7):869–80. doi: 10.1083/jcb.201501101. Research Support, N.I.H., Intramural Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008 Oct 7;14(4):312–23. doi: 10.1016/j.ccr.2008.09.001. Research Support, N.I.H., Intramural. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim Biophys Acta. 2010 Aug;1804(8):1666–75. doi: 10.1016/j.bbapap.2009.10.022. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014 Mar;25(3):138–45. doi: 10.1016/j.tem.2013.12.001. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BA, Hanover JA. O-GlcNAc and the epigenetic regulation of gene expression. J Biol Chem. 2014 Dec 12;289(50):34440–8. doi: 10.1074/jbc.R114.595439. Research Support, N.I.H., Extramural Research Support, N.I.H., Intramural Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajpe PK, Prahallad A, Horlings H, Nagtegaal I, Beijersbergen R, Bernards R. A chromatin modifier genetic screen identifies SIRT2 as a modulator of response to targeted therapies through the regulation of MEK kinase activity. Oncogene. 2015 Jan 22;34(4):531–6. doi: 10.1038/onc.2013.588. Research Support, Non-U.S. Gov’t. [DOI] [PubMed] [Google Scholar]
- 44.Ma Z, Vocadlo DJ, Vosseller K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-kappaB activity in pancreatic cancer cells. J Biol Chem. 2013 May 24;288(21):15121–30. doi: 10.1074/jbc.M113.470047. Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review. [DOI] [PubMed] [Google Scholar]
- 46.Fulop N, Feng W, Xing D, He K, Not LG, Brocks CA, et al. Aging leads to increased levels of protein O-linked N-acetylglucosamine in heart, aorta, brain and skeletal muscle in Brown-Norway rats. Biogerontology. 2008 Jun;9(3):139–51. doi: 10.1007/s10522-007-9123-5. Research Support, N.I.H., Extramural. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang YR, Song M, Lee H, Jeon Y, Choi EJ, Jang HJ, et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell. 2012 Jun;11(3):439–48. doi: 10.1111/j.1474-9726.2012.00801.x. Research Support, Non-U.S. Gov’t. [DOI] [PubMed] [Google Scholar]
- 48.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011 Mar;7(3):174–81. doi: 10.1038/nchembio.520. Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012 May 2;15(5):675–90. doi: 10.1016/j.cmet.2012.04.003. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laderoute KR, Amin K, Calaoagan JM, Knapp M, Le T, Orduna J, et al. 5’-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol. 2006 Jul;26(14):5336–47. doi: 10.1128/MCB.00166-06. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrer CM, Reginato MJ. Cancer metabolism: cross talk between signaling and O-GlcNAcylation. Methods Mol Biol. 2014;1176:73–88. doi: 10.1007/978-1-4939-0992-6_7. Research Support, N.I.H., Extramural. [DOI] [PubMed] [Google Scholar]
- 52.Haenssen KK, Caldwell SA, Shahriari KS, Jackson SR, Whelan KA, Klein-Szanto AJ, et al. ErbB2 requires integrin alpha5 for anoikis resistance via Src regulation of receptor activity in human mammary epithelial cells. J Cell Sci. 2010 Apr 15;123(Pt 8):1373–82. doi: 10.1242/jcs.050906. Research Support, U.S. Gov’t, Non-P.H.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.