Abstract
Retrograde signaling is a mechanism by which mitochondrial dysfunction is communicated to the nucleus for inducing a metabolic shift essential for cell survival. Previously we showed that partial mtDNA depletion in different cell types induced mitochondrial retrograde signaling pathway (MtRS) involving Ca+2 sensitive Calcineurin (Cn) activation as an immediate upstream event of stress response. In multiple cell types, this stress signaling was shown to induce tumorigenic phenotypes in immortalized cells. In this study we show that MtRS also induces p53 expression which was abrogated by Ca2+ chelators and shRNA mediated knock down of CnAβ mRNA. Mitochondrial dysfunction induced by mitochondrial ionophore, carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and other respiratory inhibitors, which perturb the transmembrane potential, were equally efficient in inducing the expression of p53 and downregulation of MDM2. Stress-induced p53 physically interacted with HIF-1α and attenuated the latter’s binding to promoter DNA motifs. Additionally, p53 promoted ubiquitination and degradation of HIF-1α in partial mtDNA depleted cells. The mtDNA depleted cells, with inhibited HIF-1α, showed upregulation of glycolytic pathway genes, glucose transporter 1–4 (Glut1–4), phosphoglycerate kinase 1 (PGK1) and Glucokinase (GSK) but not of prolyl hydroxylase (PHD) isoforms. For the first time we show that p53 is induced as part of MtRS and it renders HIF-1α inactive by physical interaction. In this respect our results show that MtRS induces tumor growth independent of HIF-1α pathway.
Keywords: Mitochondrial DNA, mitochondrial retrograde signaling, p53, HIF-1α, ubiquitination
INTRODUCTION
The ubiquitous tumor suppressor, p53 is a multifunctional protein, which mediates a number of physiological and pathological processes in addition to being an important determinant of cellular response to stress1. Paradoxically, wild type p53 (wtp53) also plays a role in the regulation of genes involved in tumor invasion (cathepsin D, MMP-2, and MMP-9)2, tumor growth (example, VEGF receptor)3 and decreased expression of E-cadherin4 that are involved in tumor metastasis. The ability to sense DNA damage, transcriptionally modulate the expression of a number of genes involved in DNA repair and cell cycle progression are critically important in its role as a tumor suppressor5. Yet another important function of p53 is to modulate cellular metabolism and oxidative phosphorylation by regulating the transcription of SCO2 gene, which is involved in the assembly of mitochondrial cytochrome c oxidase (CcO) complex6. Multiple types of post-translational modifications including phosphorylation, acetylation and sumoylation regulate the stability of p53 by interfering with its association with MDM2 and subsequent degradation7.
The p53 protein carries out several functions that are not dependent on its DNA binding and transcriptional activity. Channeling of glucose preferentially to glycolysis by direct association with glucose 6-phosphate dehydrogenase (G6PDH), and inhibition of pentose phosphate pathway (PPP) is one example8. Several studies suggest a direct role of p53 in inducing cellular apoptotic pathway by physical interaction with anti-apoptotic BcL2 family proteins as well as pro-apoptotic Bid and Bax proteins9–11. p53 has also been shown to localize inside the mitochondrial inner membrane-matrix compartment where it is proposed to play a protective role on mitochondrial function10.
Mitochondria play a central role in cellular energy generation, metabolic integration and initiation of apoptosis. Mitochondrial dysfunction is detrimental to cellular viability, and has been linked to many neurodegenerative, muscular, myocardial diseases as well as aging and cancer12, 13. Mitochondrial DNA (mtDNA) mutations and reduced mtDNA contents are frequently found in many neuromuscular, cardiac diseases and cancers14. A series of studies from various laboratories have shown that dysfunctional mitochondria induce stress signaling, called retrograde signaling (MtRS)15, which culminates in altered nuclear gene expression, altered metabolism and tumorigenesis in otherwise non-tumor producing cells16–19. Several mechanisms of stress signal propagation including Ca2+/Calcineurin (Cn) activation pathway20–22, mitochondrial ROS induced HIF-1α pathway23, ERK/JNK pathway24, and AMPK pathway activation24, 25, among others, have been described. In the Ca2+/Cn pathway we showed the activation of several Ca2+ sensitive transcription factors including C/EBPδ, NFAT, CREB and IkBβ permissive NFkB (cRel;p50) activation26 and also Akt mediated activation of a common transcription coactivator, hnRNPA227. The signaling cascade induced metabolic shift towards glycolysis, altered cell morphology and resistance to apoptosis17. In previous studies we observed that partial mtDNA depletion which did not alter mitochondrial mass in C2C12 cells induced the level of p53 protein in addition to transcription activation of other ~120 nuclear genes28, although the role of p53 in signal propagation was not investigated.
Altered metabolism and induced glycolysis are also the hallmark of the HIF-1α signaling pathway29, 30. Furthermore, HIF-1α and p53 are reported to play both complementary and contradicting roles in cellular metabolic shift, apoptosis and cancer progression31, 32. In this study, therefore, we investigated the regulation of HIF-1α pathway and its connection to Ca2+/Cn pathway and p53 using partial mtDNA depletion (~70%) as stress initiator. In six different immortalized or transformed cell types, HIF-1α was not activated by partial mtDNA depletion as tested by ChIP analysis, and DNA-protein binding by gel mobility shift analysis. In cells where the steady state level of HIF-1α was increased, its activity was attenuated by physical interaction with p53, which is also induced under mitochondrial stress conditions. This was accompanied by transcription down regulation of MDM2 protein which probably causes the accumulation of p53 in cells containing low mtDNA copy number and p53 interaction induced ubiquitination and degradation of HIF-1α protein. We therefore demonstrate here that the Ca2+/Cn pathway, activated in response to low mtDNA copy number induces the expression of p53 which in turn attenuates HIF-1α activity.
RESULTS
Role of MtRS in cellular HIF-1a and p53 protein levels in C2C12 cells
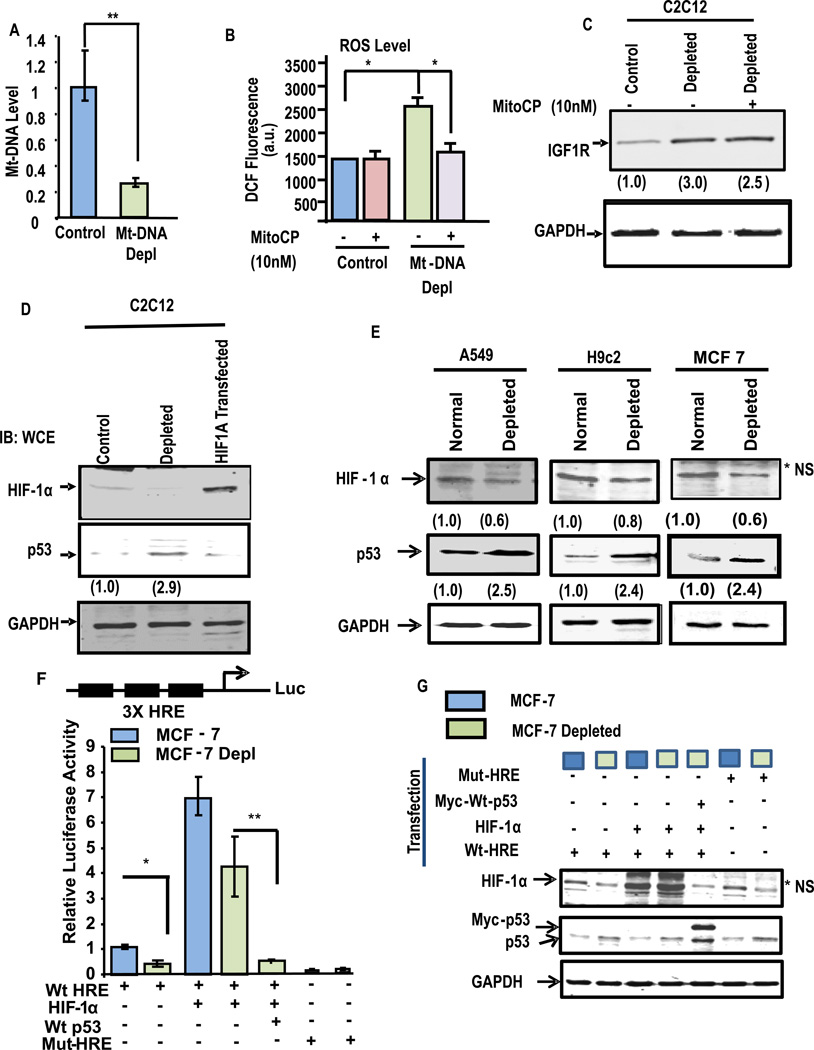
We first investigated if HIF-1α was activated in addition to the Ca2+/Cn pathway in C2C12 and five other cell models by partial mtDNA depletion. Fig. 1A shows that ddC treatment which caused about 70% reduction in mtDNA content in C2C12 cells in about 5 growth cycles (hereafter called depleted cells) generate ~55% higher H2O2 than the control cells (Fig. 1B), which was attenuated by treatment with 10 nM mitochondria-targeted SOD-mimic antioxidant, Mito-CP33. Mito-CP, however, failed to attenuate the increase in steady state level of IGF1R (Fig. 1C), which is an important marker for mitochondrial retrograde signaling34. The results suggest that ROS generated in depleted cells may not have any significant role in propagating MtRS in these cells. Despite showing an increase in ROS production, depleted C2C12 cells did not show any detectable increase in HIF-1α protein (Fig. 1D). The extract from C2C12 cells transfected with HIF-1α expression cDNA was used as a positive control in Fig 1D.
Figure 1. Effects of partial depletion of mtDNA on the HIF-1α and p53 protein levels in different cell lines.
(A) Relative mt-DNA content in control and ddC treated C2C12 cells was measured by q-PCR of total cell DNA using primers specific for the cytochrome oxidase subunit I gene (CcOI). GAPDH and nuclear encoded CcOIVi1 gene were used as internal controls. The values represent mean ±SEAM of four independent assays. P<0.005 (‘**’). (B) The level of ROS in control and mtDNA depleted C2C12 cells were measure by DCF fluorescence method. Mito-CP (10 nM) was added at the start of the measurement. Values represent average of triplicates. Where *, p< 0.05. (C) Immunoblot analysis of cell extracts (50 µg each) for IGF1R and GAPDH as loading control. (D) Immunoblot for HIF-1α and p53 levels in control and mtDNA depleted C2C12 cells using total lysates (40µg protein). Protein extract from C2C12 cells transfected with HIF-1α was used as a positive control. The blot was also probed with GAPDH antibody as loading control. Values in parentheses indicate relative band intensities. (E), Immunoblot analysis of protein extracts (40µg each) of control and mtDNA depleted A549, H9c2, and MCF7 cells. Values in parentheses indicate relative band intensities normalized with GAPDH expression. Iimmunoblots were also probed with GAPDH as a loading control. (F) HRE promoter-reporter assay in control and mtDNA depleted MCF7 cells. A trimeric HRE promoter-reporter DNA construct or a mutant version was transfected. Cells were also cotransfected with Renilla luciferase, with or without pCEP4–HIF-1α or pCDNA-Myc-wtp53 as indicated in figure. After 48hrs dual luciferase activity were measured and the data were normalized to Renila luciferase activity and represent the mean ± S.E. of 3 independent assays. (G) Immunoblot analysis of cellular extracts (50 µg protein each) of control and various transfected cells from figure F was used to ensure protein expression. Antibody to GAPDH was used to assess loading levels.
Notably, we observed about 2.9-fold increased p53 level in depleted C2C12 cells compared to control cells. Induction of p53 by partial mtDNA depletion was also observed in A549 lung carcinoma, H9C2 cardiomyocytes, MCF-7 mammary carcinoma cells (Fig. 1E) and COS-7 cells (Fig. S1A). As shown later, HCT116 colon carcinoma cells also induced p53 expression in response to mtDNA depletion.
Next, we carried out the 3×HRE promoter-reporter assay for assessing HIF-1α activity in control and mtDNA depleted MCF7 cells (Fig. 1F). Nearly 50% decrease in luciferase activity was observed in mtDNA depleted MCF7 cells compared to control cells. The luciferase activity was 7-fold and 4-fold higher when HIF1α cDNA was cotransfected with 3×HRE reporter plasmid in control and depleted cells, respectively. However, luciferase activity in control cell was attenuated by Myc tagged wt-p53 when cotransfected with HIF1α and 3×HRE plasmids. Notably, Mut-HRE DNA did not show any significant HIF-1α activity in both control and depleted cells. A control experiment in Fig. 1G shows that cells cotransfected with HIF-1α and p53 along with the promoter-reporter construct as shown in Fig. 1F indeed expressed the proteins as expected. For example, HIF-1α levels were several-fold higher in cells co-transfected with HIF-1α cDNA and cells co-transfected with MYC tagged p53 showed higher level of p53 which migrated slower than the endogenous p53. However, co-transfection of HIF-1α and MYC-tagged p53 cDNAs caused reduced HIF-1α protein level as explained in the next sections. These results together show that p53 is induced in response to MtRS, which in turn attenuates HIF-1α activity.
HIF-1α levels in WT and p53 deleted human colon and lung carcinoma cells
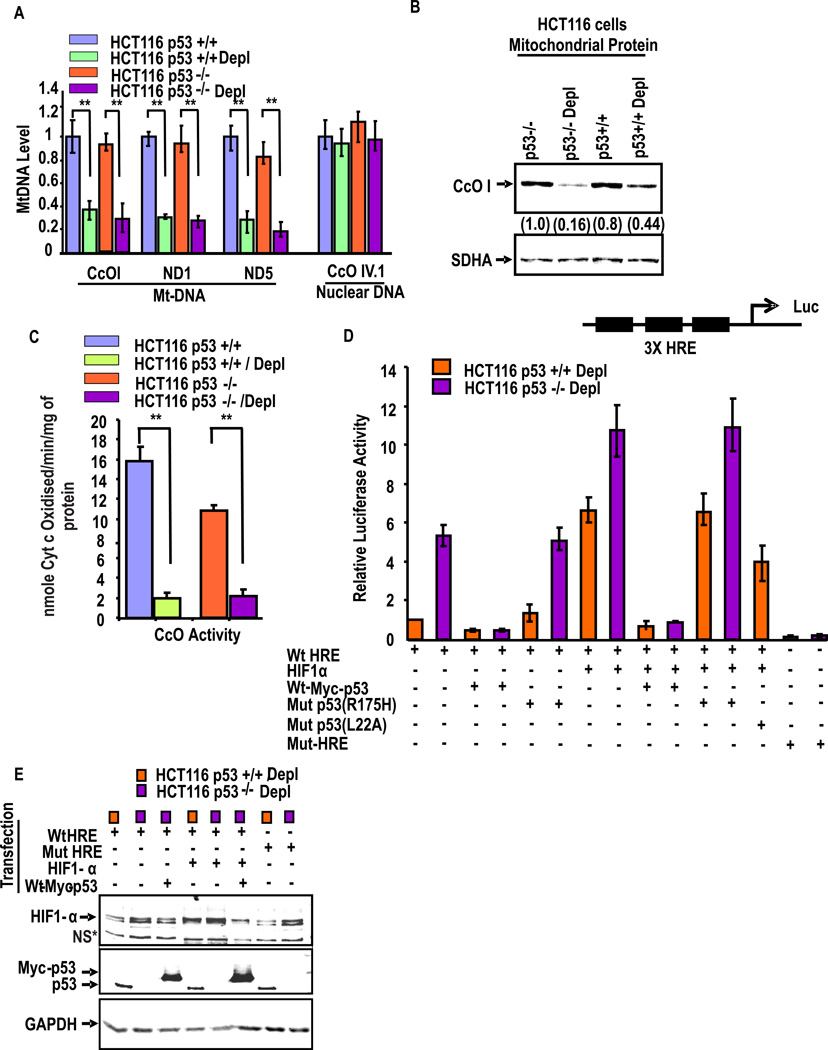
We investigated the roles of p53 and HIF-1α on MtRS in response to mtDNA depletion in human lung carcinoma H1299 cells, which harbor naturally deleted p53 gene35. We also used human colon carcinoma HCT116 cells in which p53 gene was deleted by homologous recombination (p53−/− cells)36. Fig. 2A shows that levels of mtDNA amplified in multiple mitochondrial genomic regions such as CcO I, ND1 and ND5 was reduced by about 60–70% in mtDNA depleted HCT116p53+/+ and p53−/− cells compared with the respective control cells. Results of long stretch PCR presented in Suppl. Fig. S1B also shows a similar reduction of mtDNA in depleted HCT116 cells. As expected the levels of nuclear encoded CcO IVi.1 DNA was not altered in any of the four cell lines tested. Additionally, the level of mtDNA encoded CcO 1 protein was reduced in depleted p53+/+ and p53−/− cells (Fig. 2B). Consistent with reduced mtDNA levels, the CcO activity was diminished by >70% in both of the mtDNA depleted cells in comparison to respective controls (Fig. 2C). Notably, the CcO activity in p53−/− HCT116 cells was significantly lower, possibly because of the predicted role of p53 in CcO assembly or function6, 37. Additionally, MDM2 mRNA levels in both HCT116p53+/+ cells (see Supplemental Fig. S1C) was markedly low suggesting a possible basis for increased p53 protein levels. Although not shown HCT116p53−/− cells as well as other cells used in this study showed a similar down regulation of MDM2 gene expression in partial mtDNA depleted cells.
Figure 2. Retrograde response of p53 and HIF-1α in HCT colon cancer cells.
(A) Mt-DNA contents were measured by qPCR anlysis in control and depleted human colon adenocarcinoma cell lines (HCT116) differing only in their p53 status. Use of the paired Student’s t-test indicated that all mentioned genes were inhibited in mt-DNA depleted cells with a confidence level of P<0.005 (‘**’). (B) Immunoblot analysis of control and mtDNA depleted HCT116 p53+/+ and p53−/− cells using CcOI antibody. The blot was also probed with SDHA antibody for assessing loading levels. (C) The CcO activity was measured with 20µg of freeze-thawed mitochondria as described in the Materials and Methods section. Means ± S.E. were calculated from 3 independent assays. ** indicates p<0.005. (D) HRE promoter-reporter assay in mt DNA depleted p53+/+ and p53−/− HCT116 cells. A trimeric HRE promoter-reporter DNA construct or a mutant version was transfected. Cells were also cotransfected with Renilla luciferase, with or without pCEP4-HIF-1α or pCDNA-Myc-wtp53 or Mut-p53 (R175H, L22A) as indicated. After 48hrs cell extracts were assayed for dual luciferase activity. The data were normalized to Renila luciferase activity and represent the mean ± S.E. of 3 independent assays. (E) Represents an immunoblot of cell extracts from Fig. D for assessing HIF-1α and p53 contents. The blot was also probed with GAPDH antibody for assessing loading levels.
We further tested the relationship between p53 and HIF-1α activity using 3×HRE reporter assay38 and occupancy of the protein on promoter DNA by ChIP analysis. The 3×HRE-reporter activity (Fig. 2D) was very low in HCT116p53+/+ cells but a 6-fold higher activity was seen in depleted HCT116p53−/− cells. Transient expression of WT Myc-tagged p53 attenuated activity in both cell lines, while expression of mut-p53 (R175H) had no effect. Further, transfection with HIF-1α cDNA induced the activity in both p53+/+ and p53−/− cells, while co-transfection with WT-Myc-tagged p53 cDNA markedly inhibited the activity in both cell lines. As expected, however, co-transfection with Mut-p53 (R175H) did not inhibit HIF-1α induced reporter activity. Co-transfection with transcription activation domain mutant of p53 (L22A and W23A) was only marginally effective in reducing the HIF-1α induced reporter activity. An immunoblot was carried out with the luciferase reporter cell lysates for ascertaining the expected levels of HIF-1α and p53 from the transcriptional assays in Fig. 2D. The blot in Fig. 2E shows that the steady state levels of HIF-1α (top panel) are increased in cells co-transfected with HIF-1α cDNA which was attenuated by expression of WT Myc-tagged p53 cDNA. Immunoblot analysis with p53 antibody shows the levels of endogenous p53 (faster migrating band) in p53+/+ cells and slower migrating band in cells transfected with WT Myc-tagged p53. As expected, p53−/− cell extracts did not show any immunoreactive band. These results together show that p53 negatively modulates the activity of HIF-1α.
Role of MtRS in inducing the p53 expression
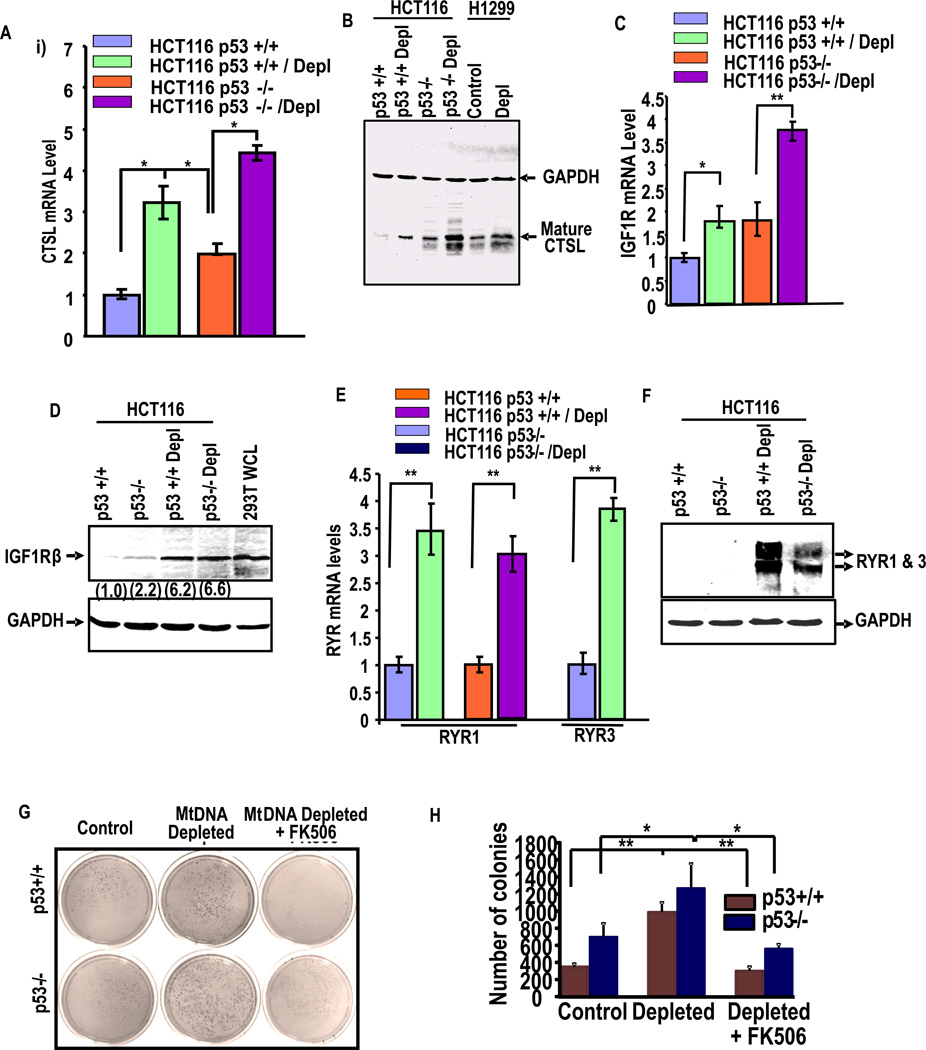
We investigated if expression of p53 is downstream of Cn activation or if it occurs through an alternate pathway in response to stress. Cathepsin-L (CTSL), IGF1Rβ, RYR1/3 are some of the key marker genes induced by Cn mediated MtRS19–21. Fig. 3A shows that CTSL mRNA expression is induced in both p53+/+ and p53−/− cells following mtDNA depletion by about 60–70% of control cells, although the level of expression in p53−/− cells (both control and depleted) were significantly higher. As seen in Fig. 3C, mtDNA depletion in p53+/+ and p53−/− cells increased IGF1Rβ mRNA levels by 1.7-fold and 2-fold compared to their respective controls. About 3.5-fold increase in RYR1 and RYR3 mRNA level was observed in depleted p53+/+ cells while 3-fold higher RYR1 gene expression was observed in depleted p53−/− cells (Fig. 3E). The protein levels (Fig 3B, 3D, and 3F) also showed a similar increase in response to mtDNA depletion. These results demonstrate that the markers of retrograde signaling are induced in HCT116 cells in response to mtDNA depletion.
Figure 3. Activation of Retrograde signaling markers in p53+/+ and p53−/− cells.
(A) Shows the induction of cathepsin L (CTSL) mRNA, (B) CTSL protein; (C) shows the increase of IGFR1 mRNA and (D) IGF1R protein. (E) Shows the mRNA levels of Ryanodine receptor 1 and 3 in indicated cell lines and (F) shows protein levels. Values in parentheses indicate relative band intensities normalized with relative GAPDH levels. (G) Anchorage-independent growth of control and mtDNA depleted p53+/+ and p53−/− cells were analyzed by soft agar colony formation assay as described in Materials and methods. Both sets of mtDNA depleted cells were treated with or without FK506, an inhibitor calcineurin after 3 weeks of growth are shown. (H) The histograms show the average number of colonies of three different plates. Means ± S.E. were calculated from 3 independent assays. ** indicates p<0.005 and *, p< 0.05
We also tested the effects of other known mitochondrial stress inducers including treatment with mitochondrial ionophore, CCCP, and mtDNA depletion by shRNA-mediated silencing of mitochondrial transcription/replication factor, TFAM on p53 induction and induction of MtRS markers. Fig.S2A and S2B show that expression of TFAM shRNA markedly reduced both TFAM mRNA and Protein level. Additionally, TFAM mRNA silencing induced the expression of IGF1R, and pAkt levels that are the markers of MtRS, in addition to p53. Treatment of HCT116 cells with CCCP (25 µM) induced IGF1R protein level (Fig. S2D), p53 mRNA/protein levels, and also RYR1 mRNA (Fig. S2C and S2D). These results show that both mtDNA depletion and membrane damage induce p53 expression
Previously we showed that Cn-initiated retrograde signaling induced anchorage independent growth by colony formation on the soft agar26, 39. Results in Fig 3G and 3H show that mtDNA depletion increased colony formation both in p53+/+ and p53−/− cells though the induction was significantly higher in the latter cell type. Notably, FK506, a known inhibitor of Cn markedly inhibited colony formation in both cell types suggesting the role of Cn in this signaling. These results are consistent with our previous model in which Cn acts as a master regulator of stress signaling which promotes tumor growth.
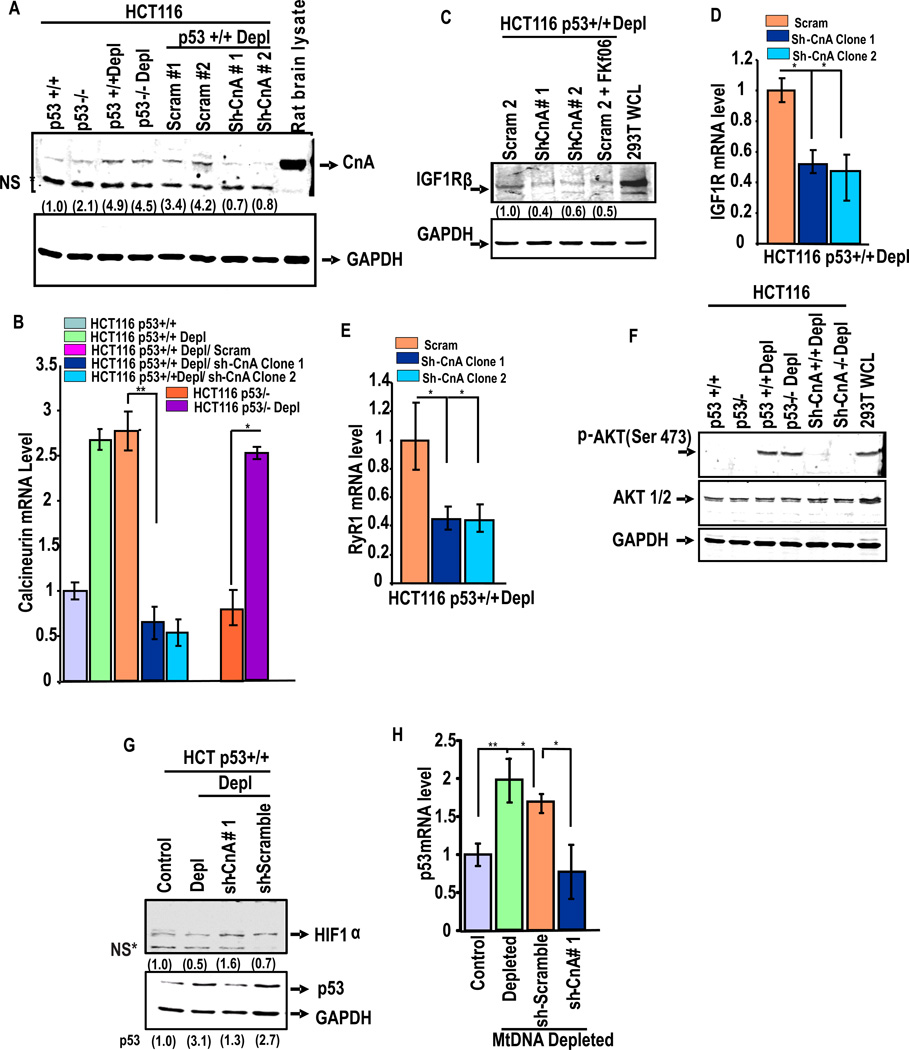
To further evaluate the role of Cn in stress signaling and its role in p53 expression, we generated cell lines stably expressing shRNA targeted to CnAβ subunit (PP2B Aβ) and also scrambled control RNA in depleted HCT116p53+/+ cells. Fig. 4A and B show that two of the shRNA constructs (#1 and #2) were effective in silencing Cn protein and mRNA while scrambled shRNA did not have any significant effect. Although not shown, Cn activity was induced by about 2.5-fold in mtDNA depleted cells which was reduced to near control cell level in shRNA expressing cells. Knock down of CnAβ mRNA also caused a reduction in IGF1R protein (Fig. 4C) and mRNA levels (Fig. 4D) as well as RyR1 mRNA level (Fig. 4E). Immunoblot in Fig. 4F shows that the level of p-AKT (Ser473) is induced by mtDNA depletion in both p53−/− and p53+/+ cells, which was attenuated in cells expressing shRNA targeted to CnAβ mRNA. Most significantly, p53 mRNA (Fig. 4H) and protein (Fig. 4G) levels were markedly reduced in cells expressing shRNA against CnAβ mRNA suggesting that p53 gene expression is also under the regulation of the Ca2+/Cn pathway.
Figure 4. Increased CnAβ protein and mRNA levels in Mt-DNA depleted cells.
(A) Represents the immunoblot analysis of control and mt-DNA depleted HCT116 p53+/+ and p53−/− cells stably expressing shRNA against CnAβ mRNA or scrambled shRNA (Scram 1 and 2). Rat brain protein extract was used as a positive control. (B) CnAβ mRNA level was measured by qPCR. Values represent average of three estimates that were normalized to GAPDH as an internal control. (C) Immunoblot analysis of cell extracts with antibody to IGF1R β. (D) mRNA levels of IGF1R measured by qPCR in mtDNA depleted cells expressing Scrambled and shRNA against CnAβ. (E) RyR1 mRNA levels measured as in Fig D. Values in D and E represent average of triplicates and normalized against GAPDH mRNA level as an internal control. (F) shows the immunoblot analysis of cell extracts using Akt1/2 and Phos-AKT antibodies. (G) Represents the immunoblot analysis of HIF-1α and p53 in HCT116 cells transfected with various shRNA constructs. The blots in A, C, F and G were reprobed with GAPDH antibody as loading controls. (H) p53 mRNA level was measured by qPCR analysis among the indicated cell lines. Means ± S.E. were calculated from 3 independent assays. ** indicates p<0.005 and *, p< 0.05. Values in parentheses of all immunoblots indicate relative band intensities normalized with GAPDH band intensities.
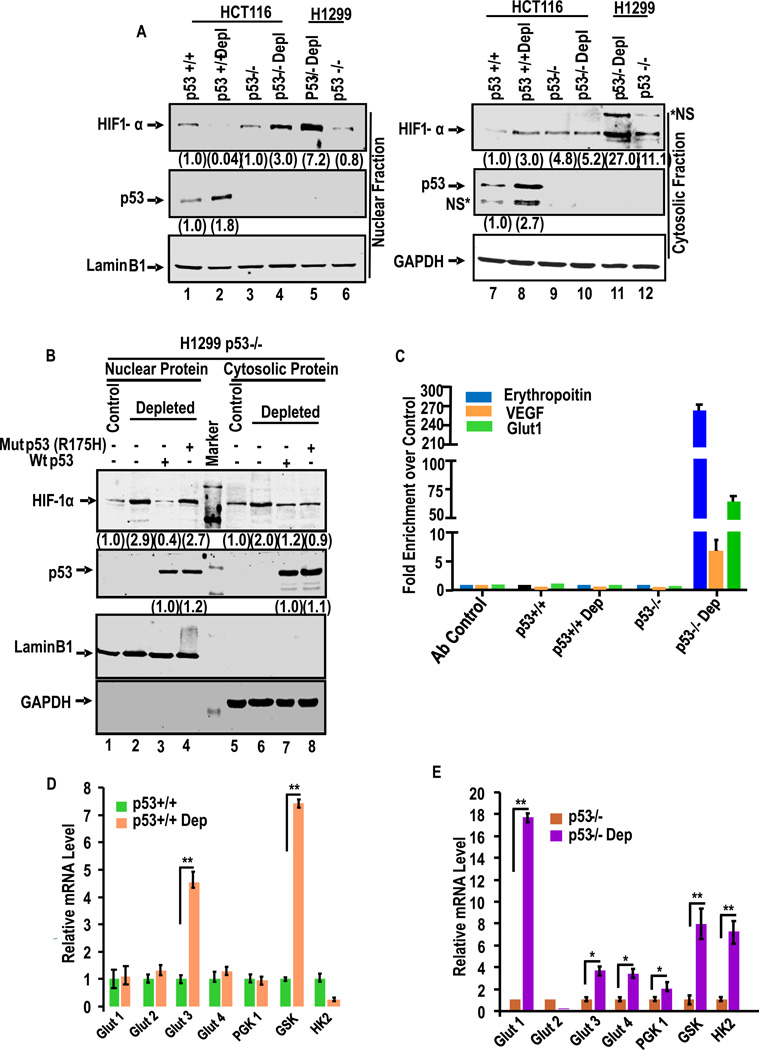
Fig. 5A shows immunoblot analysis of nuclear and cytoplasmic fractions of p53+/+ and p53−/− cells containing normal level of mtDNA and 60–70% depleted mtDNA. It is seen that nuclear HIF-1α was markedly reduced by mtDNA depletion in HCT116 p53+/+ cells. The cytosolic HIF-1α was considerably higher in mtDNA depleted cells than control cells. Interestingly, mtDNA depletion in any of the p53−/− cells caused much higher accumulations of nuclear HIF-1α (lane 4 for HCT116p53−/− and lane 5 for H1299), while the cytosolic levels varied in two different cell types (lanes 10 and 11). The marked difference in HIF-1α levels in these two cell types were contrasted by the levels of p53 or Hif-1α in control and mtDNA depleted cells. The contrasting levels of these two proteins in depleted cells were further tested using H1299 cells which do not express p53, but express relatively high levels of HIF-1α (see lanes 1 and 5, Fig 5A). Fig. 5B shows that mtDNA depletion induced both nuclear (lane 2) and cytosolic (lane 6) HIF-1α levels in H1299 cells. The possible inverse relationship between p53 induction and nuclear HIF-1α was investigated by ectopic expression of WT and R175H mutant p53 in control and mtDNA depleted H1299 cells. MtDNA depletion markedly increased HIF-1α levels in both the nuclear and cytoplasmic fractions (Fig.5B). Expression of WT p53 but not mutant p53 markedly blunted the increase in nuclear HIF-1α (lane 3 compared to lane 4, Fig. 5B) level suggesting the possible role of p53 in regulating the nuclear HIF-1α levels.
Figure 5. Mitochondrial stress-induced HIF-1α activation in p53+/+ and p53−/− HCT116 cells.
(A) Immunoblot analysis of nuclear and cytosolic fractions with antibodies to HIF-1α and p53 in indicated cells. (B) The effects of ectopically expressed WT and R175H mutant p53 in H1299 prostate cancer cells which lack p53 expression. Details were as in Fig. 2D. Antibody to Laminin B1 used as a nuclear marker and antibody to GAPDH was used as cytoplasmic marker. The numbers in parentheses in A and B represent band intensities (average of two separate estimates). (C) ChIP analysis of EPO, VEGF and Glut-1 promoter regions for HIF-1α binding. Data represent Means ± S.E of three independent assay points. (D and E) mRNA levels for different glycolytic pathway genes in control and depleted p53+/+ and p53−/− cells (Fig. E) by realtime PCR. Means ± S.E. were calculated from 3 independent assays. ** indicates p<0.005 and *, p< 0.05. Values in parentheses of all immunoblots (average of two separate estimates) indicate relative band intensities normalized with relative GAPDH levels.
HIF-1α responsive genes such as erythropoietin (EPO)40, the vascular endothelial growth factor (VEGF)41 and Glucose transporter 142, 43 contain a HRE motif in their promoter regions. ChIP analysis (Fig. 5C) shows occupancy of the HRE motifs of EPO, VEGF and Glut1 genes by by HIF-1α by 262-fold, 7-fold and 73-fold higher only in mtDNA depleted p53 null cells over control cells (Fig. 5C). The mtDNA depleted p53+/+ cells, on the other hand, showed no significant HIF-1α binding to promoter DNA. The results of ChIP analysis fully support the EMSA data in Fig. S3A.
Since HIF-1α is known to induce many glycolytic pathway genes, we tested the levels of glucose transporter 1–4 (Glut1, Glut2, Glut3 and Glut4), phosphoglycerate kinase 1 (PGK1), Glucokinase (GSK), and hexokinase (HK) mRNA levels by qPCR in response to mtDNA depletion and p53 inactivation (Fig. 5D). No significant changes in Glut1, Glut2, Glut4 and PGK1 mRNAs were observed in control and mtDNA depleted depleted p53+/+ cells. However, a 4.5 fold increase in Glut3 and 7 fold increase of GSK mRNA were observed in depleted p53+/+ cells. There was also a marked reduction in the level of HK mRNA in depleted p53+/+ cells (Fig. 5D). However, as seen from Fig 5E, a 17 fold increase in Glut1, 3.6 fold in Glut3, 3.3 fold in Glut4, 2 fold of PGK1, 7.9-fold of GSK and 7-fold of hexokinase mRNA were observed in mtDNA depleted p53−/− cells compared to the control p53−/− cells. We also observed a 90% reduction in Glut2 mRNA by mtDNA depletion in p53−/− cells. Our results clearly show that most of the glycolysis pathway genes are upregulated in p53−/− cells in response to mitochondrial stress, confirming the possible role of p53 in regulating HIF-1α activity.
The generality of p53 regulation of HIF-1α factor was investigated in breast epithelial carcinoma MCF-7 cells. Fig. S3B shows gel shift analysis with nuclear extracts from control and depleted MCF-7 (80% less mtDNA content, data not shown) cells with EPO-HRE DNA probe. Results show that nuclear extract from CoCl2 treated Cos-7 cells yielded a prominent band which was inhibited by excess of unlabeled double stranded DNA. Additionally, nuclear extract from control MCF-7 cells yielded a minor band which was not seen with nuclear extract from mtDNA depleted MCF-7 cells (Fig. S3B). Co-transfection with HIF-1α cDNA markedly increased DNA binding which was inhibited by 50 molar excess of unlabeled DNA probe and also co-expression with WT p53 cDNA. Co-expression with mut-p53 cDNA did not diminish the band intensity. These results confirm that p53 regulation of HIF-1α activity is not cell specific and likely a general mechanism.
Physical association of p53 with HIF-1α protein
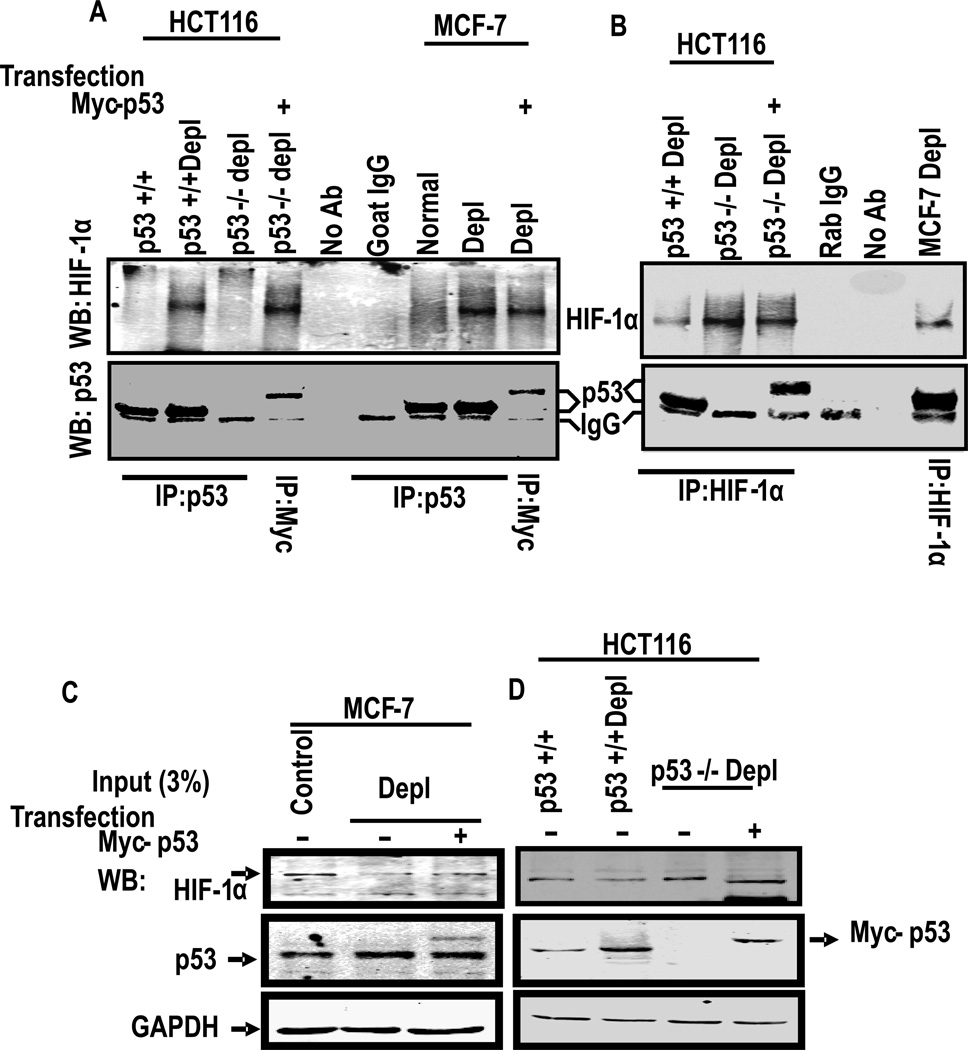
Since induced expression of p53 reduced the nuclear HIF-1α level, we wondered if p53 physically binds to HIF-1α and attenuates its nuclear entry. This was tested by co-immunoprecipitation of the two factors. Immunoblot in Fig. 6A shows that the p53 antibody pulled down negligible HIF-1α protein from HCT116p53+/+ cells, but a substantial amount was pulled down from depleted p53+/+ cells. Furthermore, no detectable HIF-1α protein was pulled down from p53−/− cells, although protein extract from cells transfected with Myc-tagged WT53 showed high level of HIF-1α pull down by Myc-antibody. Immunoprecipitation with goat IgG used as control yielded an IgG specific band which migrated faster than the p53 band. Immunoprecipitation of MCF-7 cell extract yielded essentially similar results in that p53 antibody pulled down higher level of HIF-1α protein in extracts from depleted cells than from control cells. As with HCT116 cells, Myc-antibody pulled down a slower migrating Myc-tagged p53 as well as HIF-1α protein from depleted MCF-7 cells (Fig. 6A). The specificity of p53-HIF-1α interaction was tested by reverse pull down experiments in which immunoprecipitates using HIF-1α antibody were tested for the presence of HIF-1α and also p53 by immunoblot analysis (Fig. 6B). HIF-1α was immunoprecipitated from both HCT116 and MCF-7 cell extracts, though the amount was considerably lower in depleted p53+/+ cells than corresponding p53−/− cells. Also, endogenously expressed p53 was detected only in mtDNA depleted HCT116p53+/+ and MCF-7 cell extracts, but not in p53−/− cell extracts. A slower migrating p53 band was seen in cells transfected with Myc-tagged p53 cDNA. In Fig. 6C and D, about 3% of the lysates used for immunoprecipitation above in Fig. 6A and B were subjected to immunoblot analysis and probed with antibodies to HIF-1α, p53 or GAPDH as loading control. It is seen that the extracts used for immunoprecipitation contained expected levels of HIF-1α, p53 or Myc-p53 protein. These results provide direct evidence for physical association between p53 and HIF-1α most likely in the cytoplasmic compartment of cells.
Figure 6. HIF-1α directly associates with p53.
(A) Total lysates of indicated cells (1 mg each) were immunoprecipitated (IP) by anti-p53 antibody (goat) or anti-Myc antibody and immunoblotted with anti-HIF-1α and anti-p53 antibody (mouse) as indicated. Lysates without antibody and goat IgG were used as a negative and positive controls, respectively. (B) Association of p53 and HIF-1α proteins were further confirmed by immunoprecipitation with antibody against HIF-1α and immunobloted with indicated antibodies. Rabbit IgG and lysates with no antibody were used as controls. (C–D) 3% input of total cell lysates of indicated cell lines were subject to immunoblot using HIF-1α, p53 and GAPDH antibodies as indicated.
Stress induced p53 expression promotes ubiquitination and degradation of HIF-1α
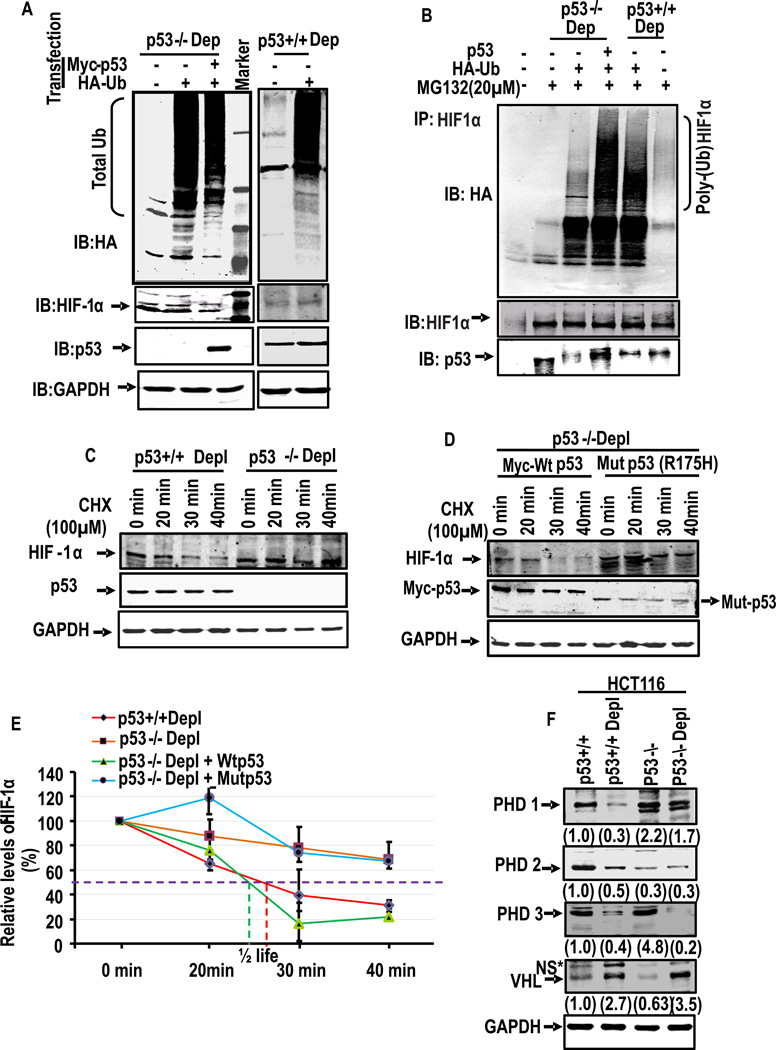
Our results showed significantly lower levels of HIF-1α in depleted cells which express higher levels of p53. Similarly, ectopic expression of p53 caused lowering of HIF-1α protein. We therefore investigated the ubiquitination status of HIF-1α in depleted p53−/− and p53+/+ HCT116 cells. Cells were transfected with cDNA encoding HA-tagged Ubiquitin or Myc-tagged p53 and incubated with proteasome inhibitor MG132 for 6h. The cell extracts were either probed directly by immunoblotting with HA antibody (Fig. 7A) or immunoprecipitated with HIF-1α antibody and probed with HA antibody (Fig. 7B). The immunoblot in Fig. 7A shows that transiently transfected HA-tagged ubiquitin forms a ladder like profile in depleted p53−/− cells even when the cells were co-transfected with p53 cDNA. Similar is the case with depleted p53+/+ cells transfected with HA-tagged ubiquitin cDNA. The immunoblot in the second panel of Fig. 7A (from top), probed with HIF-1α antibody shows a marginal reduction in HIF-1α protein when cells were co-transfected with p53 cDNA. The immunoblot in the third panel, probed with p53 antibody shows that p53 protein was not detected in p53−/− cells excepting when cells were transfected with p53 cDNA. As expected, p53 protein was seen in depleted p53+/+ cells. These results show that treatment with MG132 caused accumulation of ubiquitinated proteins in treated cells.
Figure 7. p53 mediated poly-ubiquitination of HIF-1α and its turnover.
(A–B) depleted HCT116 p53−/− and p53+/+ cells were transiently transfected with indicated expression plasmids. Cells were harvested at 36h after transfections, and total protein was immunoprecipitated (IP) with HIF-1α antibody and immunoblotted with anti-HA to evaluate the level of ubiquitination. The blots were also reprobed with HIF-1α and p53 antibodies. (C–E) Rates of turnover of HIF-1α in p53+/+ and p53−/− cells. Cells were incubated with 100µM cyclohexamide (CHX) to suppress the synthesis of new protein. The cell lysates (40 µg each) at indicated time were subjected to immunoblot analysis with p53 antibody. In (D) cells were transfected with either WT or R175H mutant p53 in HCT116 p53−/− cells. The relative p53 band intensities were calculated using the intensity of zero time of exposure with CHX as 100 percent. Means and standard deviations were calculated from three independent experiments. (F) Total cell lysates (40 µg protein each) of indicated cell line were subjected to immunoblot analysis with antibodies against PHD1, PHD2, PHD3 and VHL antibody. GAPDH was used for the loading control. The values in parentheses indicate relative band intensities normalized to relative levels of GAPDH protein.
To evaluate the level of HIF-1α ubiquitination, depleted p53−/− and p53+/+ cells were transfected with HA-ubiquitin cDNA with or without p53 cDNA. The cell extracts were immunoprecipitated with HIF-1α antibody and probed with HA-antibody, HIF-1α antibody, or p53 antibody. The immunoblot in Fig.7B (upper panel) shows that HIF-1α protein was marginally ubiquitinated in depleted p53−/− cells but profusely unbiquitinated in depleted p53+/+ cells or cells co-tansfected with p53 cDNA. The middle panel from top shows that HIF-1α level was nearly similar in all treated cell extracts. These results suggested the possible role of p53 in the ubiquitination of HIF-1α.
We further investigated this possibility by assessing the turnover rates of HIF-1α in p53+/+ and p53−/− cells when translation of new protein was arrested by the addition of 100 µM cycloheximide. The level of HIF-1α was quantified at different time points from 0 to 40min in both depleted p53+/+ and p53−/− cells by immunoblot analysis (Fig. 7C and D). It is seen that the t1/2 for HIF-1α was considerably longer in p53−/− cells as compared to p53+/+ cells incubated in presence of cycloheximide (Fig. 7E). Similarly, in p53−/− cells, transfection with WT Myc-p53 reduced the t1/2 while transfection with mut p53 did not alter the turnover rate (Fig. 7D and E). These results provide direct proof for the role of p53 in the degradation of HIF-1α.
In oxygenated environment, post-translational modification of HIF-1α at Proline (Pro-402 and Pro-564) residues by prolyl-hydroxylases (PDH 1, 2 and 3) induces ubquitination of HIF-1α44, 45. The tumor suppressor protein VHL46–48 recognizes the hydroxylated moiety and induces the ubiquitination of HIF-1α. Results in Fig. 7F show that there was a significant reduction in the levels of all three PHD isoforms (PHD1, 2 and 3) in response to mtDNA depletion in HCT116p53+/+ cells. Of the three PHDs, PHD2 is thought to be the key oxygen sensor in regulating HIF-1α49. Some studies show that PHD2 and PHD3 are HIF target genes that are part of the negative feedback loop. The lower steady state levels of these proteins in partially mtDNA depleted cells are in keeping with our results of p53 mediated downregulation of HIF-1α. PHD2 is a potent mediator angiogenesis pathway. Dang et. al. showed in xenografts model that tumors that lack of HIF-1α and PHD2 grow faster than control tumors that only lack HIF-1α50 which suggest that PHD2 has additional function independent of HIF-1α. In depleted p53−/− cells, however, the level of PHD1 and 2 were marginally reduced, while PHD 3 was markedly reduced. Because of reported role of PHD3 in the regulation of HIF-1α in hypoxia51, near complete loss of PHD3 in depleted HCT116 suggests that this protein is unlikely involved in the turnover of HIF-1α in HCT116 cells. Fig. 7F also shows that there was a marked increase in the VHL levels in mtDNA depleted cells as compared to control cells. It is therefore likely that VHL acts as an ubiquitin ligase in the degradation of HIF-1α in mtDNA depleted cells and this process is supported by p53. These results suggest that HIF-1α ubiquitination in depleted cells occurs by a mechanism different from the one known for hypoxic condition.
DISCUSSION
MtRS is an adaptive response to mitochondrial metabolic, genetic, and bioenergetic defects which alters the expression of a large number of nuclear genes involved in myriad of cellular functions including oncogenic progression52. MtRS is also reported to play roles in myocardial disease, familial inherited deafness and aging53. In this paper using five different cell lines and multiple approaches to partially deplete mtDNA or disrupt Δψm we present evidence that p53 gene expression is also induced as part of MtRS, which in turn attenuates HIF-1α activity. Interestingly, MDM2 mRNA levels are reduced in response to partial mtDNA depletion, which is likely the reason for the observed increase in p53 protein levels. Previously we showed that MtRS induces a shift in metabolism, altered cell morphology, altered growth characteristics17, 20, 21, 34. This study therefore shows that the MtRS-induced changes occur independent of HIF-1α activity in immortalized cells.
p53 is an important tumor suppressor protein, which is considered as the guardian of the genome because it renders protection against DNA damage5 Consequently, its expression is induced under chemical or radiation exposure and other stress conditions54. For the first time, we show that mitochondrial chemical and metabolic inhibitors and mtDNA damaging agents induce the expression of p53 and the increase appears to be due to increased transcription in stressed cells. It should be noted that ddC and ethidium bromide used for depleting mtDNA are highly selective for mtDNA depletion and known to cause minimal or no detectable damage to nuclear DNA21, 55. Furthermore, stable expression of shRNA against TFAM which is involved in mtDNA maintenance affects mitochondrial function without any direct effects on nuclear DNA. Using these multiple approaches we show that mitochondrial functional defects induce p53 gene expression.
In addition to the Ca2+/Cn mechanism which was described from our laboratory21, reports suggest the occurrence of other pathways including mitochondrial ROS induced HIF-1α activation which is suggested to alter nuclear gene expression and induce metabolic shift23, 30. We therefore investigated if the 2–4 fold increase in p53 level was part of the Ca2+/Cn pathway or other signaling pathways. Our results suggest that p53 expression is downstream of Cn activation since Cn inhibitor, FK506 and also CnAβ knockdown by shRNA expression markedly attenuated the expression of p53 in mtDNA reduced cells. The pro-apoptotic function of p53 by inducing the expression of Bax and Bid is well established1. mtDNA depleted cells, on the other hand, are resistant to etoposide- and thapsigargin-induced apoptosis by way of inducing several antiapoptotic proteins including BCL2, Akt-Pi3-K, etc.27, 28. In this regard, induced expression of p53 in cells with altered mitochondrial function is somewhat intriguing. However, p53 is known to play yet unknown roles in modulating mitochondrial respiration and oxidative phosphorylation56, 57. It is therefore likely that induced expression of p53 in cells with mitochondrial lesions could be a compensatory response. A notable finding of this study is that p53 induced in response to mtDNA and membrane lesions renders HIF-1α inactive, and that metabolic shift induced in these cells is irrespective of HIF-1α activity.
Some studies suggest that activation of p53 in bone marrow stromal cells reduced the transcription of HIF-1α possibly through binding to transcription co-activator p300, which is also required for transcription of HIF-1α gene58–60. Other reports suggest regulation of p53 degradation by HIF-1α60, 61. Our results demonstrate that HIF-1α activity is not only downregulated by physical association with p53 but also the ubiquitin mediated degradation of the former. We have not observed any degradation of p53 in mtDNA reduced cells. Support for this observation comes from experiments using two different p53 null cells in which mtDNA depletion induced HIF-1α activity and induced expression of its target genes. Furthermore, expression of WT p53, but not mutant p53 in p53−/− cells attenuated HIF-1α activity. These results provide a rigorous proof for a novel pathway of negative modulatory role of p53 on HIF-1α activity and stability. Our results showing reduced nuclear levels of HIF-1α in p53 overexpressing cells suggest that the physical interaction between these two factors may occur in the cytosol. However, interaction between these two proteins within the nuclear compartment cannot be ruled out.
Previous studies showed that in some hypoxic tumors HIF-1α negatively modulated p53 levels by inducing the latter’s degradation62. By contrast, our results with mitochondrial stress show that physical association of p53 induces the degradation of HIF-1α through ubiquitiation, while the level of p53 is not affected. At the present time the molecular bases for this contrasting difference remains unclear.
Under normoxic conditions O2 activates PHD isoenzymes which modify the Pro residues of HIF-1α protein, promoting the VHL mediated ubiquitination. Paradoxically, all three PHD enzymes are downregulated in mtDNA depleted cells while the level of VHL protein is markedly increased (Fig. 7). Results of co-immunoprecipitation in p53−/− and p53+/+ cells demonstrate that p53 markedly increases ubiquitination of HIF-1α. Additionally, pulse chase experiments in presence of added cycloheximede show a more rapid HIF-1α degradation kinetic in p53+/+ cells in comparison to p53−/− cells. These results clearly establish a role for p53 in induced degradation of HIF-1α. We propose that p53 association with HIF-1α makes it more amenable for interaction with VHL ubiquitin ligase thus inducing its degradation. The precise mechanism by which p53 induces ubiquitination of HIF-1α presently remains unclear. Interaction of p53 with HIF-1α is facilitated by downregulation of MDM2 in these stressed cells.
It is well established that HIF-1α is involved in metabolic shift as well as angiogenesis in hypoxic tumors. We propose that in tumors expressing WT p53, MtRS induced hnRNPA2 mechanism plays an important role in metabolic reprogramming of solid tumors, under hypoxic conditions. It is likely that incipient angiogenesis which has been reported to occur in the absence of HIF-1α activity plays a role in tumor progression under these conditions63. In tumors expressing mutant p53, HIF-1α plays a predominant role in metabolic reprogramming, angiogenesis and tumor progression.
Materials and Methods
Cell Lines, Culture Conditions and Transfection
Murine C2C12 skeletal myoblasts (ATCC CRL1772), COS-7, rat myoblast H9c2, human lung carcinoma A549, human non-small cell lung carcinoma cell H1299, colorectal carcinoma cell line HCT116, p53+/+ and its isogenic p53 deficient HCT116 p53−/− were grown in Dulbecco’s modified Eagle’s medium (DMEM, (Gibco, Life Technologies)) as described before21. Human breast adenocarcinoma cell line MCF7 were grown in DMEM/F12 (1:1) medium supplemented with 10% FBS and 1% Pen Strep. MtDNA depletion was carried out by the administration of 10µM 2'–3'-dideoxycytidine (ddC) until the mtDNA content decreased to 70–80% of control cells. mtDNA depleted cells were supplemented with 1mM Sodium pyruvate and 50µg/ml uridine. Chemical hypoxia in COS-7 cells was induced by treatment with 150 µM CoCl2 for 4h.
Electrophoresis Mobility Shift Assay
The HIF-1α binding to an hypoxia responsive element DNA by gel mobility shift assay using 32P end labeled double-stranded DNA probe (5'-GCCCTACGTGCTGTCTCACACAGC-3') as described previously64. For supershift, 2µg of polyclonal antibody to HIF1α (H-206×, Santa Cruz Biotechnology, Inc) was added to the reaction mixture before the addition of labeled oligonucleotides. Excess unlabeled (50–200 fold molar excess) DNA was used for competition. DNA-protein complexes were resolved on a 5% polyacrylamide gel with 0.5 × Tris-borate-EDTA at 100 V and the gel was dried and exposed to X-ray film.
Chromatin immunoprecipitation Assay
ChIP assays were carried out with cells fixed with cells fixed with 1% formaldehyde using a standard protocol, Immunoprecipitation with anti HIF1α antibody (Santacruz) overnight at 4°C. One % of the sample was taken for input of each reaction and preimmune IgG was used as a negative control. Antibody and Chromatin complex were immunoprecipitated by protein A/G agarose beads with salmon sperm DNA (50% Slurry). Eluted DNA-Protein complexes were reversed with 5M NaCl at 65°C overnight. Immunoprecipitated DNA was purified and amplified by PCR of HRE binding regions of human EPO, VEGF and Glut1 promoters (see the primer sequences in Table S1). Data were presented as fold enrichment by subtracting the signal with no antibody and expressed as fold increase over control sample.
HIF1α Stability Assay
MtDNA depleted HCT116 p53+/+ and H1299 were incubated with 100µM Cyclohexamide (Sigma, C7698) for 0 to 40 min and then harvested in RIPA buffer. Equal amounts of total protein from each treatment were used for quantifying HIF-1α protein by immunoblot analysis.
In Vivo Poly-ubiquitination Assay
About 10 × 106 cells were transfected by electroporation (Bio Rad) with Ha-Ub and indicated plasmid DNAs. Cells were incubated for 36 h and pretreated for an additional 6 h with 20µM MG132 (Cayman chemical company, Michigan, USA) before harvesting. The cells were then lysed in 1% SDS. For denaturing immunoprecipitation, cell lysates were incubated at 95°C for 5 min and sonicated. After removing the debris, one aliquot was saved for Western blot analysis and the rest of the lysates were diluted 10-fold in lysis buffer (supplemented with protease inhibitors and 10mM NEM) to reduce the SDS concentration. Proteins were immunoprecipitated with specific antibodies and resolved by SDS-PAGE. The extent of ubiquitination of immunoprecipitated complexes were detected by HA-specific antibody (12CA5) against HA-Ub tagged proteins.
Statistical analysis
Statistical significance was determined by an unpaired two-tailed Student’s t test and paired wherever needed. All results for the in vitro experiments are presented as means ± S.D. of at least three data points from three different experiments. p values ≤0.05 were considered statistically significant, and p values ≤0.001 were considered highly significant.
Supplementary Material
Acknowledgments
We thank Drs. Craig Thompson, Gregg Semenza, and Bert Vogelstein for generously providing the promoter constructs and cell lines used in this study. This work was supported by NIH grant CA-22762, AR-067066 and an endowment from the Harriet Ellison Woodward trust. We also acknowledge the help of the Imaging Core facility at the School of Veterinary Medicine and the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30DK050306) and its Cell collection core.
Footnotes
Conflict of Interest
The authors declare no conflict of the interest.
Author Contributions
ARC, NGA, SF and AR conceived the experiments. ARC carried out experiments, AL generated mutant cDNA constructs and ARC and NGA wrote the paper.
References
- 1.Amundson SA, Myers TG, Fornace AJ., Jr Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287–3299. doi: 10.1038/sj.onc.1202576. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Wicha M, Leopold WR. Regulation of metastasis-related gene expression by p53: a potential clinical implication. Mol Carcinog. 1999;24:25–28. [PubMed] [Google Scholar]
- 3.Menendez D, Inga A, Snipe J, Krysiak O, Schonfelder G, Resnick MA. A single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol Cell Biol. 2007;27:2590–2600. doi: 10.1128/MCB.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roger L, Jullien L, Gire V, Roux P. Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J Cell Sci. 2010;123:1295–1305. doi: 10.1242/jcs.061002. [DOI] [PubMed] [Google Scholar]
- 5.Liu G, Chen X. Regulation of the p53 transcriptional activity. J Cell Biochem. 2006;97:448–458. doi: 10.1002/jcb.20700. [DOI] [PubMed] [Google Scholar]
- 6.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 7.Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 8.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/ Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, Tai J, Ye H, Kang CB, Yoon HS. The N-terminal domain of tumor suppressor p53 is involved in the molecular interaction with the anti-apoptotic protein Bcl-Xl. Biochem Bio phys Res Commun. 2006;341:938–944. doi: 10.1016/j.bbrc.2005.12.227. [DOI] [PubMed] [Google Scholar]
- 12.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 13.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 15.Boland ML, Chourasia AH, Macleod KF. Mitochondrial dysfunction in cancer. Front Oncol. 2013;3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brar SS, Meyer JN, Bortner CD, Van HB, Martin WJ. Mitochondrial DNA-depleted A549 cells are resistant to bleomycin. Am J Physiol Lung Cell Mol Physiol. 2012;303:L413–L424. doi: 10.1152/ajplung.00343.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 18.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, et al. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J Cell Biol. 2003;161:507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GR, et al. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, et al. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci Signal. 2013;6:rs4. doi: 10.1126/scisignal.2003266. [DOI] [PubMed] [Google Scholar]
- 25.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Tang W, Chowdhury AR, Guha M, Huang L, Van WT, Rustgi AK, et al. Silencing of IkBbeta mRNA causes disruption of mitochondrial retrograde signaling and suppression of tumor growth in vivo. Carcinogenesis. 2012;33:1762–1768. doi: 10.1093/carcin/bgs190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion. 2013;13:577–591. doi: 10.1016/j.mito.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas G, Anandatheerthavarada HK, Avadhani NG. Mechanism of mitochondrial stress-induced resistance to apoptosis in mitochondrial DNA-depleted C2C12 myocytes. Cell Death Differ. 2005;12:266–278. doi: 10.1038/sj.cdd.4401553. [DOI] [PubMed] [Google Scholar]
- 29.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iommarini L, Kurelac I, Capristo M, Calvaruso MA, Giorgio V, Bergamini C, et al. Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum Mol Genet. 2014;23:1453–1466. doi: 10.1093/hmg/ddt533. [DOI] [PubMed] [Google Scholar]
- 31.Obacz J, Pastorekova S, Vojtesek B, Hrstka R. Cross-talk between HIF and p53 as mediators of molecular responses to physiological and genotoxic stresses. Mol Cancer. 2013;12:93. doi: 10.1186/1476-4598-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sermeus A, Michiels C. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2011;2:e164. doi: 10.1038/cddis.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhanasekaran A, Kotamraju S, Karunakaran C, Kalivendi SV, Thomas S, Joseph J, et al. Mitochondria superoxide dismutase mimetic inhibits peroxide-induced oxidative damage and apoptosis: role of mitochondrial superoxide. Free Radic Biol Med. 2005;39:567–583. doi: 10.1016/j.freeradbiomed.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 34.Guha M, Srinivasan S, Biswas G, Avadhani NG. Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J Biol Chem. 2007;282:14536–14546. doi: 10.1074/jbc.M611693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berglind H, Pawitan Y, Kato S, Ishioka C, Soussi T. Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol Ther. 2008;7:699–708. doi: 10.4161/cbt.7.5.5712. [DOI] [PubMed] [Google Scholar]
- 36.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 37.Wanka C, Brucker DP, Bahr O, Ronellenfitsch M, Weller M, Steinbach JP, et al. Synthesis of cytochrome C oxidase 2: a p53-dependent metabolic regulator that promotes respiratory function and protects glioma and colon cancer cells from hypoxia-induced cell death. Oncogene. 2012;31:3764–3776. doi: 10.1038/onc.2011.530. [DOI] [PubMed] [Google Scholar]
- 38.Minet E, Ernest I, Michel G, Roland I, Remacle J, Raes M, et al. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5'UTR. Biochemical and biophysical research communications. 1999;261:534–540. doi: 10.1006/bbrc.1999.0995. [DOI] [PubMed] [Google Scholar]
- 39.Guha M, Srinivasan S, Ruthel G, Kashina AK, Carstens RP, Mendoza A, et al. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene. 2013 doi: 10.1038/onc.2013.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang GL, Semenza GL. Molecular basis of hypoxia-induced erythropoietin expression. Curr Opin Hematol. 1996;3:156–162. doi: 10.1097/00062752-199603020-00009. [DOI] [PubMed] [Google Scholar]
- 41.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Molecular and cellular biology. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barliya T, Mandel M, Livnat T, Weinberger D, Lavie G. Degradation of HIF-1alpha under hypoxia combined with induction of Hsp90 polyubiquitination in cancer cells by hypericin: a unique cancer therapy. PLoS One. 2011;6:e22849. doi: 10.1371/journal.pone.0022849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. The Journal of biological chemistry. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 44.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 45.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, et al. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 47.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 48.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang DT, Chen F, Gardner LB, Cummins JM, Rago C, Bunz F, et al. Hypoxia-inducible factor-1alpha promotes nonhypoxia-mediated proliferation in colon cancer cells and xenografts. Cancer Res. 2006;66:1684–1936. doi: 10.1158/0008-5472.CAN-05-2887. [DOI] [PubMed] [Google Scholar]
- 51.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. The Journal of biological chemistry. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 52.Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene. 2002;21:7839–7849. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- 53.Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 55.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 56.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Puzio-Kuter AM. The Role of p53 in Metabolic Regulation. Genes Cancer. 2011;2:385–391. doi: 10.1177/1947601911409738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu J, Milligan J, Huang LE. Molecular mechanism of hypoxia-inducible factor 1alpha - p300 interaction. A leucine-rich interface regulated by a single cysteine. J Biol Chem. 2001;276:3550–3554. doi: 10.1074/jbc.M009522200. [DOI] [PubMed] [Google Scholar]
- 59.Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995–11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- 60.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 61.Kojima K, McQueen T, Chen Y, Jacamo R, Konopleva M, Shinojima N, et al. p53 activation of mesenchymal stromal cells partially abrogates microenvironment-mediated resistance to FLT3 inhibition in AML through HIF-1alpha-mediated down-regulation of CXCL12. Blood. 2011;118:4431–4439. doi: 10.1182/blood-2011-02-334136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 63.Cao Y, Li CY, Moeller BJ, Yu D, Zhao Y, Dreher MR, et al. Observation of incipient tumor angiogenesis that is independent of hypoxia and hypoxia inducible factor-1 activation. Cancer Res. 2005;65:5498–5505. doi: 10.1158/0008-5472.CAN-04-4553. [DOI] [PubMed] [Google Scholar]
- 64.Camenisch G, Wenger RH, Gassmann M. DNA-binding activity of hypoxia-inducible factors (HIFs) Methods Mol Biol. 2002;196:117–129. doi: 10.1385/1-59259-274-0:117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.