Abstract
Phytochrome, a major photoreceptor in plants, consists of two domains: the N-terminal photosensory domain and the C-terminal domain. Recently, the 651–amino acid photosensory domain of phytochrome B (phyB) has been shown to act as a functional photoreceptor in the nucleus. The phytochrome (PHY) domain, which is located at the C-terminal end of the photosensory domain, is required for the spectral integrity of phytochrome; however, little is known about the signal transduction activity of this domain. Here, we have established transgenic Arabidopsis thaliana lines expressing an N-terminal 450–amino acid fragment of phyB (N450) lacking the PHY domain on a phyB-deficient background. Analysis of these plants revealed that N450 can act as an active photoreceptor when attached to a short nuclear localization signal and β-glucuronidase. In vitro spectral analysis of reconstituted chromopeptides further indicated that the stability of the N450 Pfr form, an active form of phytochrome, is markedly reduced in comparison with the Pfr form of full-length phyB. Consistent with this, plants expressing N450 failed to respond to intermittent light applied at long intervals, indicating that N450 Pfr is short-lived in vivo. Taken together, our findings show that the PHY domain is dispensable for phyB signal transduction but is required for stabilizing the Pfr form of phyB.
INTRODUCTION
Light acts not only as an energy source for plants but also as an environmental signal that regulates plant development. Plants can sense the intensity, wavelength, direction, and timing of illumination using diverse photoreceptors including red-light photoreceptors, phytochromes, blue-light photoreceptors, cryptochromes, and phototropins (Briggs and Huala, 1999; Neff et al., 2000; Briggs and Christie, 2002). Of these photoreceptors, phytochrome is the best characterized.
Phytochromes are large, soluble proteins that exist as dimers in solution (Furuya, 1993). A wide range of physiological and developmental processes are under the control of phytochrome (Neff et al., 2000). The phytochrome monomer is a polypeptide of ∼120 kD, with a single, covalently attached open tetrapyrrole chromophore responsible for the absorption of visible light. Phytochromes undergo photoreversible absorption changes between two spectrally distinct forms: red light absorbing form of phytochrome (Pr) and far-red light absorbing form of phytochrome (Pfr). Red light activates phytochrome by converting it from the Pr to the Pfr form. Conversely, far-red light inactivates phytochrome by converting Pfr back to Pr. Thus, phytochrome functions as a light-driven biological switch. In addition, in the dark, Pfr is converted back to Pr by a thermally driven process called dark reversion. Dark reversion has been proposed to attenuate phytochrome signaling because it reduces the amount of active Pfr (Mancinelli, 1994).
Higher plants contain ensembles of phytochromes, which differ in amino acid sequence by ∼50%. They are selectively responsible for sensing different light quality. In Arabidopsis thaliana, the phytochrome family comprises five genes, PHYTOCHROME A (PHYA) to PHYE (Mathews and Sharrock, 1997). Analysis of mutants deficient in different phytochromes has defined differential, as well as overlapping, physiological roles for members of this family (Quail et al., 1995; Smith and Whitelam, 1997; Franklin et al., 2003; Monte et al., 2003). The functional distinction is particularly striking between phytochrome A (phyA) and phytochrome B (phyB) in the control of seedling deetiolation, for which the effects of continuous far-red (cFR) light are mediated exclusively by phyA, whereas the effects of continuous red light (cR) are mediated predominantly by phyB.
Phytochromes translocate from the cytoplasm to the nucleus and form nuclear speckles upon light activation (Kircher et al., 1999, 2002; Yamaguchi et al., 1999; Hisada et al., 2000). Consistent with this observation, phytochromes interact with the nuclear-localized basic helix-loop-helix proteins PIF3 and PIF4 in a light-dependent manner (Ni et al., 1998, 1999; Huq and Quail, 2002). These PIFs bind to various G-box sequences found in several light-regulated promoters (Martinez-Garcia et al., 2000; Huq and Quail, 2002) and probably act as negative regulators for the gene expression (Kim et al., 2003). These data suggest that phytochromes transduce the light signal by interacting with transcription factors inside the nucleus.
The phytochrome molecule folds into two major domains separated by a protease-sensitive hinge region: an N-terminal chromophore bearing photosensory domain of ∼70 kD and a C-terminal domain of ∼55 kD that contains two PAS homology domains (Kay, 1997) and the His kinase homology domain (Schneider-Poetsch, 1992). The C-terminal domain contains all of the structures necessary for dimerization (Quail, 1997) and by itself accumulates in the nucleus (Sakamoto and Nagatani, 1996). Although the C-terminal domain had long been assumed to transduce the light signal to downstream components of the pathway, recent structure/function analysis has demonstrated that the N-terminal photosensory domain of phyB alone is sufficient for signaling when dimerized and expressed in the nucleus (Matsushita et al., 2003).
The photosensory domain is highly conserved throughout phytochrome species. Without the C-terminal domain, the photosensory domain exhibits photoreversible spectral changes that are indistinguishable from those observed for full-length phytochrome. This domain is further divided into three regions: a short N-terminal extension (6 to 10 kD), the central chromophore-bearing region (∼40 kD), and a C-terminal remnant of ∼20 kD, which is referred to as the PHY domain (Montgomery and Lagarias, 2002; Figure 1A). Deletion of the N-terminal extension results in a blue shift of the absorption peaks, a decrease in the Pfr absorbance, and an increase in the rate of dark reversion compared with full-length phytochrome (Cherry et al., 1992). A fragment of 39 kD corresponding to the central chromophore-bearing region possesses chromophore lyase activity (Quail, 1997; Wu and Lagarias, 2000). The PHY domain contributes to the integrity of Pfr (Montgomery and Lagarias, 2002), and deletion of this domain results in substantial alterations in the spectral properties of phytochrome (Cherry et al., 1993).
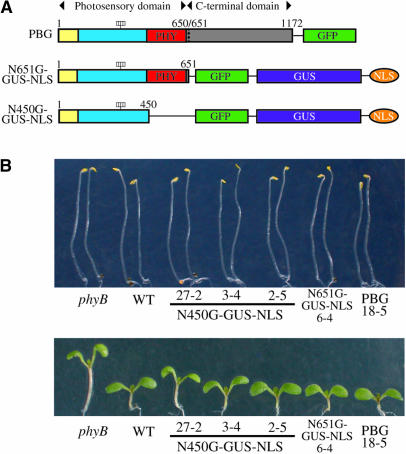
Figure 1.
Morphology of N450G-GUS-NLS, N651G-GUS-NLS, and PBG Lines under cW.
(A) Diagram of phyB derivatives. Dotted line indicates the junction between the photosensory and C-terminal domains. Yellow box, N-terminal extension; blue box, central chromophore–bearing region; red box, PHY domain at amino acid positions 444 to 623. The four rectangles indicate the chromophores.
(B) Morphology of the phyB mutant seedlings expressing N450G-GUS-NLS, N651G-GUS-NLS, or PBG in the dark (top) or under cW (bottom).
Although the photosensory domain has been studied intensively with respect to its spectral nature, the mechanism by which this domain transduces the light signal in the nucleus remains unclear. For example, the short N-terminal extension has been shown to be important for the function of phytochromes (Quail, 1997), and replacing all of the Ser residues in this region with Ala results in enhanced biological activity in rice (Oryza sativa) phyA (Stockhaus et al., 1992). To date, however, there is no experimental evidence to indicate that the PHY domain is involved in the signal transduction process in the nucleus.
In this study, we have examined the functions of the PHY domain with respect to its involvement in light signal transduction. In addition, we have examined how alterations in the spectral properties of phyB caused by removing the PHY domain affect physiological responses. We expressed an N-terminal 450-residue (N450) fragment of phyB fused to green fluorescent protein (GFP), β-glucuronidase (GUS), and nuclear localization signal (N450G-GUS-NLS) in the phyB mutant of Arabidopsis to examine its biological activity in the nucleus. In parallel, we expressed N450 in Escherichia coli and examined its spectral properties in vitro. Our results indicate that the N450 fragment is capable of transducing the signal in response to light in planta. In vitro spectral analysis indicates that the Pfr form of N450 is less stable and rapidly reverts back to the Pr form in the dark. Consistent with this, physiological analysis suggests that the Pfr form of N450G-GUS-NLS is short-lived in planta. Taking these findings together, the PHY domain is not essential for signal transduction in the nucleus but is required for stabilizing the Pfr form of phyB.
RESULTS
PhyB N450 Is Biologically Active
The N-terminal half of phyB alone, consisting of 651 amino acids, is sufficient to transduce the light signal in the nucleus (Matsushita et al., 2003). To examine whether a smaller fragment is also active in the nucleus, we expressed a fragment of the N-terminal 450 amino acids of phyB (N450), which possesses the chromophore lyase activity (Wu and Lagarias, 2000) but lacks the PHY domain, as a fusion protein with GFP, GUS, and NLS (N450G-GUS-NLS) in transgenic Arabidopsis (Figure 1A). The Arabidopsis phyB-deficient mutant was transformed with T-DNA containing N450G-GUS-NLS driven by the Cauliflower mosaic virus 35S promoter by the Agrobacterium tumefaciens–mediated method. As a result, 46 independent transformants were obtained, of which 16 lines exhibited recovery from the phyB phenotype (see below).
The seedlings were grown either in the dark or under continuous white light (cW) to examine the biological activity of the N450G-GUS-NLS protein. A representative result is shown in Figure 1B. In the dark, neither the parental phyB mutant nor the N450G-GUS-NLS lines exhibited a morphological phenotype. Under cW, the long-hypocotyl phenotype of the phyB mutation was complemented in N450G-GUS-NLS lines, as well as in phyB plants expressing either an N-terminal 651–amino acid fragment of phyB (N651) fused to GFP, GUS, and NLS (N651G-GUS-NLS) (Matsushita et al., 2003) or full-length phyB fused to GFP (PBG) (Yamaguchi et al., 1999). This phenotype complementation was further supported by quantitative analysis (Figure 2A). The extent of complementation varied depending on the lines.
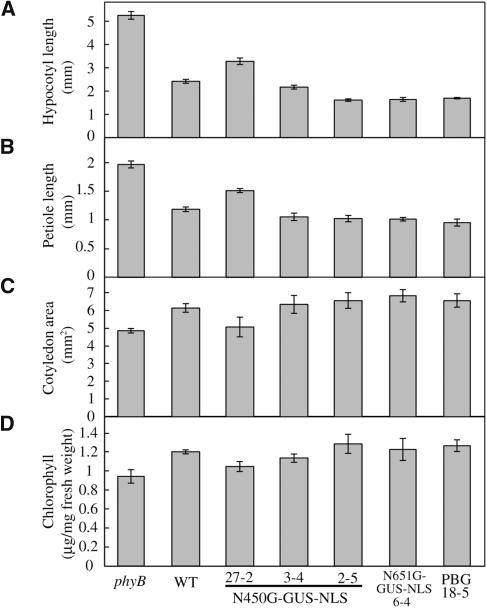
Figure 2.
Physiological Responses of the phyB Mutant Seedlings Expressing N450G-GUS-NLS, N651G-GUS-NLS, and PBG to cW.
(A) Hypocotyl length of seedlings grown under cW (52 μmol m−2 s−1) for 1 week. Data are the mean ± se (n = 25).
(B) Petiole lengths of seedlings grown under cW (52 μmol m−2 s−1) for 1 week. Data are the mean ± se (n = 25).
(C) Cotyledon area of seedlings grown under cW (52 μmol m−2 s−1) for 1 week. Data are the mean ± se (n = 25).
(D) Chlorophyll levels in seedlings grown under cW (52 μmol m−2 s−1) for 1 week. Data are the mean ± se (n = 6).
In addition to the long hypocotyl phenotype, phyB mutant seedlings exhibit features such as long petioles, smaller cotyledons, and low chlorophyll accumulation (Reed et al., 1993). The N450G-GUS-NLS seedlings exhibited complementation of all of these phenotypes (Figures 2B to 2D). Hence, we concluded that the N450G-GUS-NLS protein was physiologically functional. Furthermore, it appeared to be active only when it absorbed light because no morphological phenotypes were observed in the dark.
N450G-GUS-NLS Protein Accumulates in the Nucleus
We examined the integrity of the N450G-GUS-NLS protein expressed in transgenic Arabidopsis by immunoblot analysis (Figure 3A). The monoclonal anti-phyB antibody mBA1, which was raised against the N-terminal 598–amino acid fragment of phyB (Shinomura et al., 1996), did not detect the N450 fusion protein, probably because the N450 fragment lacks the epitope for mBA1. Therefore, an anti-GFP antibody was used to detect the N450G-GUS-NLS protein.
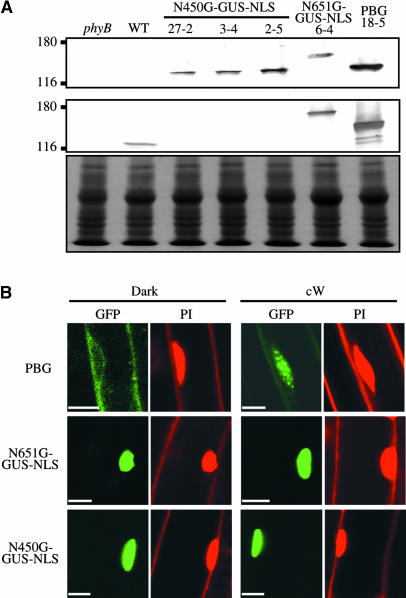
Figure 3.
Detection of the N450G-GUS-NLS Fusion Protein in Transgenic Arabidopsis.
(A) Immunoblot analysis of N450G-GUS-NLS, N651G-GUS-NLS, and PBG proteins in extracts from the respective transgenic lines detected with mouse monoclonal anti-GFP (top panel) and anti-phyB (mBA1) (middle panel) antibodies. Fifty micrograms of total protein was loaded in each lane. To confirm equal protein loading, the same blots were subjected to Coomassie Brilliant Blue (CBB) staining (bottom panel).
(B) Confocal microscopy observation of N450G-GUS-NLS, N651G-GUS-NLS, and PBG proteins in transgenic Arabidopsis seedlings. Seedlings were grown for 5 d in complete darkness (left) or under cW (right). Fluorescence from GFP and propidium iodide (PI) is shown separately in a root epidermal cell. Scale bars = 10 μm.
As shown in Figure 3A, a band at the expected size was detected on the blots of plant extracts. To estimate the expression levels of N450G-GUS-NLS protein in these plants, control lines (N651G-GUS-NLS 6-4, PBG 18-5, and the wild type) were analyzed in parallel. We found that N450G-GUS-NLS was expressed at relatively high levels; in addition, we confirmed that line 27-2, which showed the weakest physiological activity (Figures 1B and 2A to 2D), accumulated less N450G-GUS-NLS protein than did the other lines.
Next, we examined the subcellular localization of N450G-GUS-NLS (Figure 3B). Arabidopsis seedlings grown in the dark or under cW for 7 d were stained with propidium iodide to visualize nuclei and cell walls and were then examined for GFP and propidium iodide fluorescence with a confocal laser scanning microscope. As expected, N450G-GUS-NLS, which contains an NLS, accumulated exclusively in the nucleus regardless of the light conditions. No nuclear speckles were observed even under cW for either N450G-GUS-NLS or N651G-GUS-NLS.
PhyB N450 Mediates the Response to cR
Inhibition of hypocotyl elongation is mediated by phyB and phyA in response to cR and cFR, respectively (Quail et al., 1995). We examined whether N450G-GUS-NLS retained this spectral specificity by comparing the hypocotyl lengths of transgenic Arabidopsis under cR and cFR (Figure 4A). As expected, inhibition of hypocotyl elongation in N450G-GUS-NLS–expressing phyB plants was elicited by monochromatic cR. By contrast, no response to cFR was observed when N450G-GUS-NLS was expressed on the phyA phyB double mutant background. Hence, N450G-GUS-NLS can complement the loss of phyB but not that of phyA. In addition, no interference of N450G-GUS-NLS with endogenous phyA was observed because plants expressing both phyA and N450G-GUS-NLS (on the phyB mutant background) responded to cFR normally. It should be noted that the response of N450G-GUS-NLS, N651G-GUS-NLS, and PBG to cR was independent of the presence of phyA. The responses to cR of lines on the phyB single mutant and the phyA phyB double mutant background were indistinguishable.
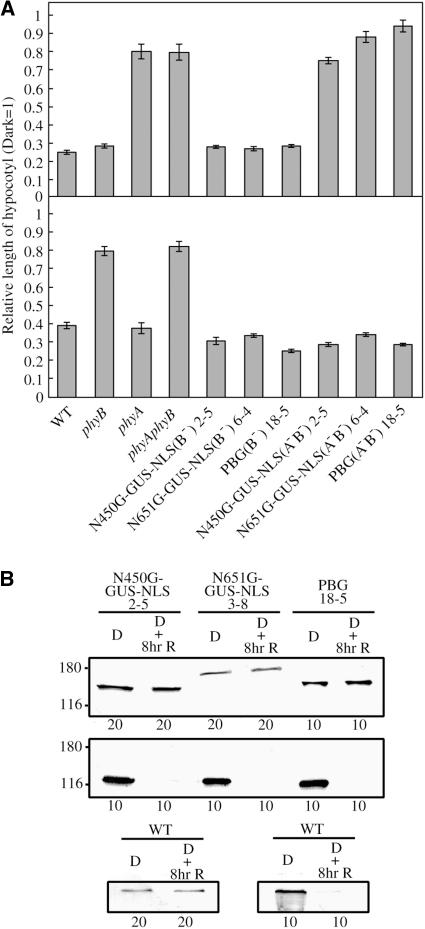
Figure 4.
Spectral Specificity of the N450G-GUS-NLS Responses.
(A) Hypocotyl lengths of seedlings grown for 5 d under cFR (top) or cR (bottom). Values are expressed as a ratio of the respective dark controls. Lines on the phyB mutant background (N450G-GUS-NLS 2-5, N651G-GUS-NLS 6-4, and PBG 18-5) and those on the phyA phyB double mutant background [N450G-GUS-NLS(A−B−) 2-5, N651G-GUS-NLS(A−B−) 6-4, and PBG(A−B−) 18-5] were examined. Data are the mean ± se (n = 25).
(B) Effects of light on the levels of N450G-GUS-NLS, N651G-GUS-NLS, PBG, and endogenous phyA proteins. Seedlings were grown for 5 d in the dark and then subsequently exposed or not to cR (18 μmol m−2 s−1) for 8 h. Extracts were analyzed by immunoblotting with anti-GFP (top), anti-phyA (mAA1) (middle and bottom right), or anti-phyB (mBA1) (bottom left) monoclonal antibodies. The amounts of total protein (in μg) loaded are indicated at the bottom.
Next, we confirmed that the N450G-GUS-NLS protein level was not influenced by light. Extracts from etiolated seedlings exposed or not to cR for 8 h were subjected to immunoblot analysis (Figure 4B). As expected, the levels of the endogenous phyB, N450G-GUS-NLS, N651G-GUS-NLS, and PBG were not influenced by light. In addition, the levels of endogenous phyA were unaffected by the expression of N450G-GUS-NLS, N651G-GUS-NLS, or PBG.
PhyB N450 Is Moderately Hypersensitive to cR
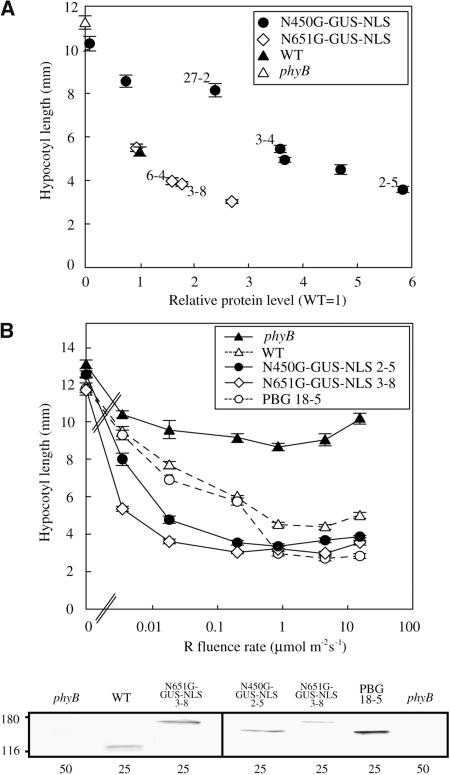
To compare the biological activity of N450G-GUS-NLS with that of N651G-GUS-NLS, several independent lines exhibiting different expression levels of the fusion proteins were analyzed. The hypocotyl lengths of these lines grown under saturating cR were plotted against protein expression levels (Figure 5A). The results showed that ∼3.5 times more N450G-GUS-NLS protein than endogenous phyB was required to obtain a wild-type response. By contrast, N651G-GUS-NLS (Figure 5A) or PBG (Matsushita et al., 2003) expressed at the endogenous phyB level was sufficient to elicit a wild-type response. Hence, N450G-GUS-NLS appears to be less active than N651G-GUS-NLS or PBG under saturating light.
Figure 5.
Quantitative Analysis of the Physiological Activity of N450G-GUS-NLS.
(A) Relationship between the hypocotyl length under cR (ordinate) and the level of protein accumulation (abscissa) in independent transgenic lines expressing N450G-GUS-NLS or N651G-GUS-NLS in the phyB mutant. Protein levels were measured by densitometric analysis of the immunoblots and are expressed as a ratio of the endogenous phyB level in wild-type plants. The symbols with numbers denote lines that were used for other experiments. Data are the mean ± se (n = 25).
(B) Fluence-rate response curves for the inhibition of hypocotyl elongation in the phyB mutant seedlings expressing N450G-GUS-NLS, N651G-GUS-NLS, or PBG. Seedlings were grown for 5 d under cR. The levels of protein expression were analyzed by immunoblotting (bottom panel) with anti-phyB (mBA1) (left) and anti-GFP (right) monoclonal antibodies. The amounts of total protein (in μg) loaded are indicated at the bottom.
Compared with PBG plants, N651G-GUS-NLS plants respond to an extremely low intensity of cR, even when the expression level of N651G-GUS-NLS is not very high (Matsushita et al., 2003). We therefore examined the fluence-rate dependence of the N450G-GUS-NLS response to cR (Figure 5B). For this experiment, we used the following lines, which showed a decreasing level of protein expression: PBG18-5 > N450G-GUS-NLS 2-5 > N651G-GUS-NLS 3-8. Seedlings of these lines were grown under a wide range of intensities of cR, and their hypocotyls lengths were determined. As previously reported (Matsushita et al., 2003), N651G-GUS-NLS 3-8 was ∼100 times more sensitive to cR than PBG 18-5. A similar, albeit smaller (∼30-fold), increase in light sensitivity was observed for N450G-GUS-NLS 2-5 compared with PBG 18-5. As mentioned above, the protein level in N450G-GUS-NLS 2-5 was lower than that in PBG-18-5 but higher than that in N651G-GUS-NLS 3-8. Hence, the difference in apparent sensitivity cannot be attributed to the difference in protein expression. In conclusion, N450G-GUS-NLS plants are moderately hypersensitive to cR compared with those expressing full-length phyB. These observations demonstrate that the PHY domain is not essential for signal transduction activity but may contribute positively to phyB signaling.
Spectral Properties of PhyB N450
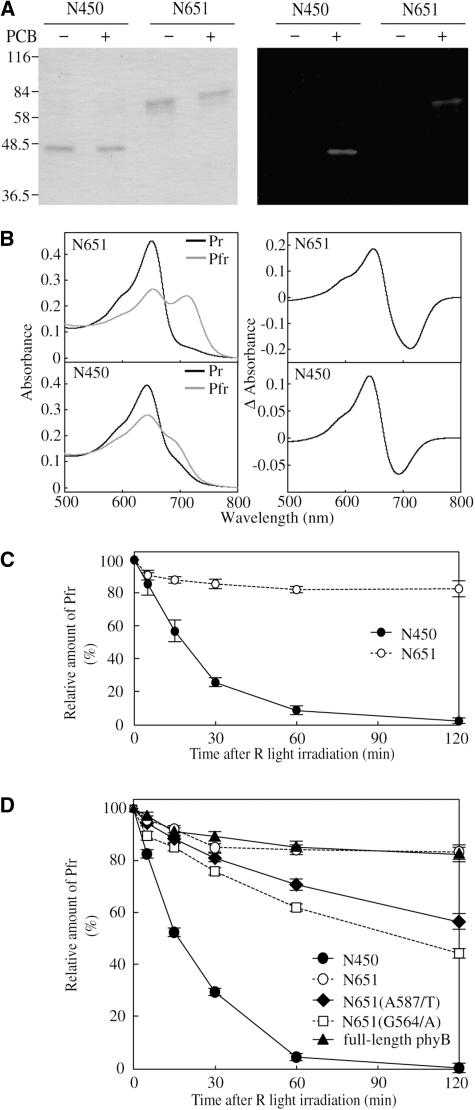
The PHY domain contributes to the integrity of Pfr (Montgomery and Lagarias, 2002). To examine whether deletion of the PHY domain altered the spectral properties of the N450 fragment, we expressed N450 and N651 fused to the intein/chitin binding domain (Intein/CBD) in E. coli. The expressed proteins were bound to a chitin column and self-cleaved on the column according to the manufacturer's instructions (New England Biolabs). This procedure results in proteins that should have only two additional amino acids (Pro and Gly) at the C terminus. We found that both N450 and N651 were expressed at high levels in E. coli. The purified N450 and N651 polypeptides exhibited the expected molecular masses of 48 and 70 kD, respectively, on electrophoresis gels (Figure 6A). N-terminal sequence analysis revealed that the N termini of these polypeptides were intact except for the first Met, which was missing (data not shown).
Figure 6.
Spectral Properties of N450, N651, and Full-Length phyB.
(A) CBB staining (left) and zinc blot (right) analyses of the samples used for spectral measurements. The indicated proteins were expressed in E. coli. Purified N450 and N651 polypeptides were separated by SDS-PAGE after incubation with (+) or without (−) 5 μM PCB. Each lane contained 250 ng of protein. The molecular masses of the protein standards are indicated on the left.
(B) Absorption (left) and difference spectra (right) of the recombinant N450 (bottom) and N651 (top) holoproteins. The spectra were recorded for the purified recombinant holoproteins shown in (A) using a spectrophotometer.
(C) Dark reversion in purified N450 and N651 holoproteins. Proteins were converted to the Pfr form by irradiation with saturating red light for 10 min, and then Pfr dark reversion was monitored with a spectrophotometer. The level of Pfr at the beginning of the dark incubation was set as 100% Pfr. The incubation temperature was 22°C. Each value represents the mean of at least three measurements with independent protein preparations. Data are the mean ± se (n = 3).
(D) Dark reversion in crude preparations of N450, N651, N651(A587/T), N651(G564/A), and full-length phyB holoprotein. Ammonium sulfate fractions of the recombinant proteins were mixed with PCB and used to measure dark reversion as described in (C). Data are the mean ± se (n = 3).
Spectrally active holoproteins were reconstituted using phycocyanobillin (PCB) as a chromophore (Lagarias and Lagarias, 1989). A zinc blot analysis revealed that chromophore was incorporated efficiently into the recombinant protein (Figure 6A). The N651 fragment exhibited typical phytochrome absorption spectra, with peaks at 650 and 711 nm for Pr and Pfr, respectively (Figure 6B). The difference spectrum for the Pr/Pfr photoconversion exhibited a peak at ∼650 nm and a trough at ∼710 nm (Figure 6B). These values are consistent with those reported for other phytochromes reconstituted with PCB, although authentic phytochromes exhibit peaks at a longer wavelength (Li and Lagarias, 1992). The spectral change ratio (i.e., increase in red light absorbance/decrease in far-red light absorbance) was 1.16.
In contrast with N651, the Pfr form of N450 exhibited a marked reduction in the far-red peak, which is characteristic of phytochrome fragments lacking the PHY domain (Cherry et al., 1993). The Pr form of N450 exhibited a typical Pr absorption spectrum with a peak at 644 nm (Figure 6B), which is slightly blue-shifted compared with N651 Pr. In N450 Pfr, the peak in the far-red region was not clearly recognized. The difference spectrum for the Pr/Pfr photoconversion exhibited a peak at 644 nm and a trough at 693 nm (Figure 6B). Compared with the difference spectrum for N651, the trough shifted toward shorter wavelength by ∼17 nm, and the spectral change ratio increased to 1.83 (Figure 6B).
Because the Pfr form has lower thermodynamic stability, it spontaneously converts back to the Pr form in the dark, a process referred to as Pfr dark reversion (Furuya and Song, 1994). Compared with phyA, phyB exhibits faster dark reversion both in vitro and in vivo (Elich and Chory, 1997; Sweere et al., 2001). Here, we compared the rate of dark reversion for the purified N450 and N651 holoproteins (Figure 6C). N651 showed a relatively fast dark reversion, but only 20% of the Pfr molecules was involved in this reaction. More than 80% of the total phytochrome remained as Pfr even after 2 h in the dark. This kinetics is very similar to the one reported for the full-length phyB (Eichenberg et al., 2000; Sweere et al., 2001). By contrast, almost all the N450 Pfr reverted back to Pr within 2 h. The half-life of the N450 Pfr in the dark was ∼15 min. We confirmed that the total amount of N450 protein did not change during the experiment by converting the Pr back to Pfr after the dark incubation (data not shown).
To compare the spectral properties of N450 and N651 with those of full-length phyB, we expressed full-length phyB, N651, and N450 fused to the c-Myc epitope tag in E. coli. Because the expression of full-length phyB was very low, we used crude ammonium sulfate fractions for these experiments. Immunoblot analysis with anti-Myc antibody detected fragments of the proteins in these preparations, but a zinc blot analysis after the addition of PCB indicated that chromophore was incorporated mostly into a polypeptide of the expected size (data not shown). The kinetics of dark reversion in these samples is shown in Figure 6D. Approximately 20% of the total full-length phyB Pfr reverted back to Pr after 2-h incubation in darkness. The dark reversion rate of N651 was indistinguishable from that of full-length phyB. By contrast, almost all the N450 Pfr reverted back to Pr within 2 h, similar to that of the purified polypeptide (Figure 6C).
The above results imply that the PHY domain is involved in the Pfr stabilization. However, the instability of N450 Pfr could have been because of the inappropriate protein folding caused by the truncation of the PHY domain. Hence, we examined the effects of the missense mutations in the PHY domain (Figure 6D). Because the G564/E mutation in Arabidopsis phyB is known to reduce the dark reversion rate (Kretsch et al., 2000), we examined N651 derivatives carrying G564/A, G564/D, G564/K, or G564/H mutations. Of these, the G564/A mutation increased the dark reversion substantially. We also examined another mutation, A587/T, which causes the mislocation of phyB in the cell (Chen et al., 2003). This mutation resulted in increased dark reversion as well. Hence, two missense mutations that reduced the stability of N651 were identified within the PHY domain.
Instability of N450G-GUS-NLS Pfr in Vivo
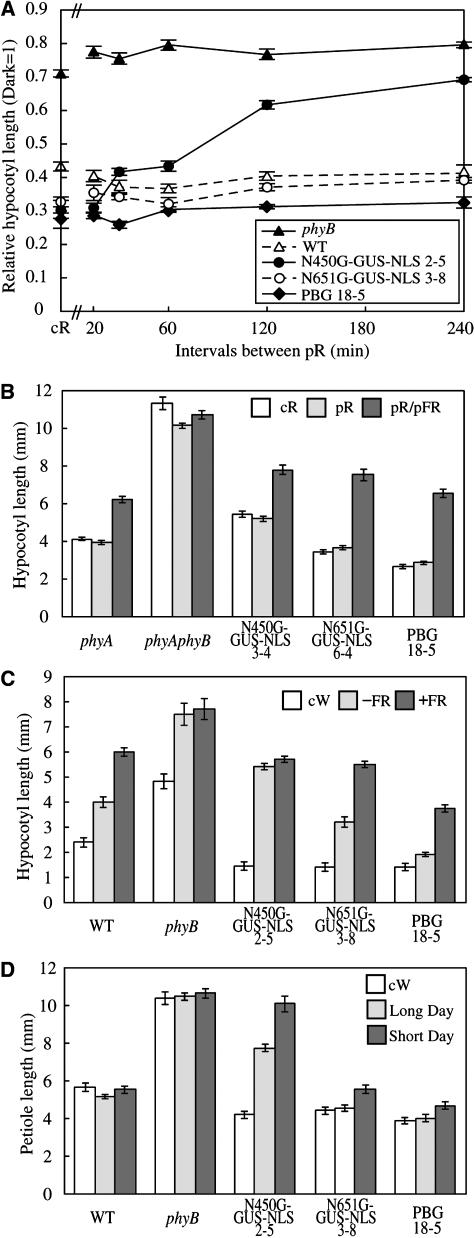
Intermittent irradiation with red light pulses (pR) is often as effective as cR for eliciting phyB responses (Mancinelli, 1994). Indeed, the phyB-mediated inhibition of hypocotyl elongation in Arabidopsis can be induced by intermittent pR (McCormac et al., 1993). This property of the phyB response reflects the stability of the Pfr form of phyB in the dark. We therefore examined the response of N450G-GUS-NLS to intermittent pR light exposure at various intervals. As has been described for wild-type seedlings (McCormac et al., 1993), pR given at intervals shorter than 4 h was as effective as cR in N651G-GUS-NLS and PBG plants (Figure 7A). By striking contrast, N450G-GUS-NLS plants barely responded to pR, even at 2-h intervals. The intervals had to be <20 min to obtain full inhibition. These results suggest that the Pfr form of N450G-GUS-NLS rapidly converts back to the Pr form in the dark in vivo.
Figure 7.
Responses of N450G-GUS-NLS Plants to Various Light Treatments.
(A) Responses of wild-type, phyB, and the phyB mutant expressing N450G-GUS-NLS, N651G-GUS-NLS, or PBG to intermittent pR exposure at variable intervals. Plants were grown under intermittent pR (55.2 μmol m−2 s−1 for 5 min) or under cR (2.3 μmol m−2 s−1) for 5 d, and then hypocotyl lengths were determined. Values are expressed as a ratio of the respective dark controls. For comparison, hypocotyl lengths under cR (2.3 μmol m−2 s−1) are also shown. Data are the mean ± se (n = 25).
(B) Effect of pFR exposure immediately after intermittent pR exposure in the wild type, phyB, and the phyB mutant expressing N450G-GUS-NLS, N651G-GUS-NLS, or PBG. Plants were grown under cR (2.3 μmol m−2 s−1), intermittent pR (55.2 μmol m−2 s−1 for 5 min) at intervals of 20 min, or intermittent pR (55.2 μmol m−2 s−1 for 5 min) followed by pFR (24.6 μmol m−2 s−1 for 5 min) at intervals of 20 min for 5 d, and then hypocotyl lengths were determined. Values are expressed as a ratio of the respective dark values. Data are the mean ± se (n = 25).
(C) Responses of the wild type, phyB, and the phyB mutant expressing N450G-GUS-NLS, N651G-G-NLS, or PBG to EOD-FR. Seedlings were grown for 7 d under 10-h-light/14-h-dark cycles with or without 20 min of pFR exposure at the beginning of the dark period. Data are the mean ± se (n = 25).
(D) Petiole lengths in plants grown under cW (45.6 μmol m−2 s−1), the long-day condition (16 h light/8 h dark), or the short-day condition (8 h light/16 h dark) for 3 weeks. Data are the mean ± se (n = 25).
The effect of intermittent pR on hypocotyl elongation of Arabidopsis can be cancelled by far-red light pulses (pFR) (Kretsch et al., 2000). We examined whether this reversibility would occur in plants expressing N450G-GUS-NLS. To eliminate interference from the phyA response elicited by multiple pFR (Shinomura et al., 2001), plants in the phyA phyB double mutant background were used for this experiment. Seedlings were treated intermittently with pR for 5 min, followed by pFR for 5 min, and then placed in the dark for 10 min. As expected, N450G-GUS-NLS was sensitive to pFR if it was given immediately after pR (Figure 7B), as is the case with N651G-GUS-NLS and PBG. This suggests that the Pfr form of N450 is the physiologically active form in vivo even though it is less stable and short-lived in the dark.
The end-of-day far-red light (EOD-FR) effect on hypocotyl elongation in Arabidopsis is mediated predominantly by phyB (Robson et al., 1993). The EOD-FR response requires a stable pool of Pfr. We therefore examined the response to EOD-FR in N450G-GUS-NLS seedlings (Figure 7C). Consistent with the previous study, EOD-FR treatment resulted in a substantial increase in the hypocotyl length in wild-type seedlings, whereas the phyB mutant exhibited long hypocotyls regardless of EOD-FR treatment. Like the wild type, N651G-GUS-NLS and PBG plants responded to EOD-FR treatment. By contrast, N450G-GUS-NLS seedlings exhibited long hypocotyls regardless of EOD-FR treatment, indicating that the Pfr form of N450G-GUS-NLS disappears or becomes inactive rapidly in the dark, even without pFR treatment. It should be noted here that N450G-GUS-NLS plants responded to cW normally (Figure 7A).
Figure 7D shows the petiole length of 3-week-old wild-type, phyB, and transgenic plants expressing N450G-GUS-NLS grown under cW, the long-day condition (16 h light/8 h dark) or the short-day condition (8 h light/16 h dark). Under cW, the morphology of N450G-GUS-NLS plants was indistinguishable from that of the wild type. By contrast, N450G-GUS-NLS plants grown in the short-day condition resembled the phyB mutant rather than the wild type. The plants exhibited intermediate phenotype under the long-day condition. Hence, the N450G-GUS-NLS phenotype correlated well with the daylength. In addition, as has been observed for the phyB mutant, N450G-GUS-NLS exhibited early flowering under the short-day condition (data not shown). These observations are consistent with our finding that the N450G-GUS-NLS Pfr form is short-lived in the dark.
DISCUSSION
Physiological Activity of N450
The C-terminal half of phyB is known to be dispensable for signal transduction in the nucleus (Matsushita et al., 2003), and this study has further expanded this view. We have demonstrated that the PHY domain of ∼180 amino acids, which is located in the N-terminal half of phyB, is dispensable for signaling activity (Figures 1, 2, 4, and 5). Full complementation of the phyB-deficient mutant was observed in lines expressing N450G-GUS-NLS, which lacks the PHY domain.
The physiological activity of N450G-GUS-NLS was observed only under light and not in the dark (Figures 1B and 5B) as is the case with N651G-GUS-NLS (Matsushita et al., 2003). This indicates that the N450G-GUS-NLS protein is active only when it absorbs red light. Although it remains possible that a photoreceptor other than N450G-GUS-NLS triggered the response, it is not likely because N651G-GUS-NLS with a chromophore attachment site mutation does not have physiological activity (Matsushita et al., 2003).
In theory, N450G-GUS-NLS may transduce the signal in combination with endogenous phytochromes using their C-terminal domains. This data demonstrated that phyA was not required for the function of N450G-GUS-NLS (Figure 4A). Because the contribution of phyC, phyD, and phyE to the phytochrome responses is relatively small (Aukerman et al., 1997; Devlin et al., 1998; Franklin et al., 2003; Monte et al., 2003), it is less likely that N450G-GUS-NLS requires these phytochromes for its function. Nevertheless, this should be tested using the multiple phytochrome mutant in the future.
Taken together, we conclude that the N450G-GUS-NLS fusion protein can transduce the light signal to downstream components almost normally in response to light stimulus. N450 is the shortest fragment of phytochrome so far that has been shown to possess such activity. It should be noted here that this study focused on the phyB activity in the nucleus. It remains possible that phyB may transduce the signal in the cytoplasm as well (Schafer and Bowler, 2002). The C-terminal and/or the PHY domains may be required for such activity.
Quantitative analysis of lines expressing N450G-GUS-NLS at various levels demonstrated that higher expression of the protein is required to obtain a wild-type response under saturating light (Figure 5A). This might be because of reduced interaction of the Pfr form of N450G-GUS-NLS with downstream components, compared with full-length or N651 phyB Pfr (see below). Interestingly, a lower fluence rate of light was needed to elicit the response of N450G-GUS-NLS than to elicit the response of phyB (Figure 5B). As previously discussed (Matsushita et al., 2003), this increased sensitivity to light could be because of removal of the C-terminal domain. The sensitivity of N450G-GUS-NLS to low fluence rates of light was lower than that of N651G-GUS-NLS (Figure 5B), which might be because of the instability of N450G-GUS-NLS Pfr (Figures 6C and 7; see below).
The mode of phyB action is very different from that of phyA (Quail et al., 1995; Shinomura et al., 2001). PhyA responds to cFR as well as to very low fluences of light, whereas phyB acts mainly under cR. A previous domain-swap analysis (Wagner et al., 1996b) has shown that determinants for the wavelength specificity of phyA and phyB are located in the photosensory domain. Consistent with this view, both N450G-GUS-NLS and N651G-GUS-NLS responded to cR but not to cFR (Figure 4A). Furthermore, the response of N450G-GUS-NLS to intermittent pR was far-red reversible (Figure 7B), which is a characteristic of the phyB response (McCormac et al., 1993; Shinomura et al., 2001).
A dominant-negative effect of phyB on phyA activity has been reported. Plants overexpressing the photosensory domain of Arabidopsis phyB (Wagner et al., 1996a) or oat (Avena sativa) phyA (Boylan et al., 1994) exhibit a reduction in phyA response. However, we did not observe such an effect for either N450G-GUS-NLS or N651G-GUS-NLS. This was probably because of the lower expression of these proteins in the present lines, compared with the overexpression used in the previous studies.
PHY Domain Function in Relation to the Spectral Properties of Phytochrome
The PHY domain is required for the integrity of the Pfr form of phytochrome (Montgomery and Lagarias, 2002). Proteolytically prepared 39-kD fragments of pea (Pisum sativum) and oat phyA exhibit blue shifts in the Pr absorbance peak and bleached spectra for Pfr (Yamamoto and Furuya, 1983; Reiff et al., 1985). More recently, a 398–amino acid fragment of oat phyA, which roughly corresponds to the N450 fragment of phyB, was expressed in tobacco (Nicotiana tabacum) and found to exhibit a blue shift in the absorption peaks for both Pr and Pfr as well as a substantial decrease in the absorption peak for Pfr (Cherry et al., 1993). Our reconstitution experiments demonstrated similar effects for phyB (Figure 6B).
Dark reversion reflects the thermal stability of Pfr and is thought to be an effective mechanism for signal attenuation (Mancinelli, 1994). Phytochromes exhibit diverse rates of dark reversion depending on their type (Eichenberg et al., 2000). Furthermore, dark reversion rates can differ substantially between in vivo (expressed in plant tissues) and in vitro (purified or reconstituted holoproteins in solutions) studies (Sweere et al., 2001). We observed clear difference between N450 and N651 or the full-length phyB with respect to the dark reversion kinetics (Figures 6C and 6D). The results indicated that N450 Pfr was much less stable compared with N651 and the full-length phyB Pfr, suggesting that the PHY domain stabilizes Pfr. It should be noted here that the dark reversion kinetics in vitro may not match exactly with that in planta. A large part of recombinant phyB Pfr remains as Pfr after prolonged dark incubation, whereas the Pfr disappears gradually during the 2-h dark incubation in planta (Sweere et al., 2001).
Little is known about the structural molecular basis of dark reversion. The Pfr chromophore of phytochrome is in a distorted, high-energy C15-E, anticonfiguration, which is presumably maintained through chromophore–protein interactions (Andel et al., 1996). We speculate that the PHY domain contributes to the stability of the Pfr chromophore through such interactions. The involvement of the PHY domain in Pfr stabilization is further supported by two missense mutations, A587/T and G564/A, within the PHY domain (Figure 6D). In contrast with the G564/E mutation, which results in extremely slow dark reversion (Kretsch et al., 2000), these mutations increased the dark reversion.
It is well established that truncation of the N-terminal extension results in an increase in the rate of dark reversion (Cherry et al., 1992). Furthermore, much evidence suggests that there is a physical interaction between the Pfr chromophore and the N-terminal extension (Furuya and Song, 1994). Interestingly, both ends of the photosensory domain (the N-terminal extension and the PHY domain) stabilize the Pfr chromophore, which may give some clues to the molecular topography of these subdomains in the photosensory domain.
In Vivo Stability of the N450 Pfr and Its Physiological Implication
Our in vitro measurements demonstrated that almost all the N450 Pfr reverted back to Pr within 2 h in darkness (Figures 6C and 6D). Physiological analysis of the N450G-GUS-NLS plants further suggested that the Pfr form of N450 is unstable in vivo. Intermittent pR given at 4-h intervals was as effective as cR in wild-type, PBG, and N651G-GUS-NLS lines (Figure 7A). By contrast, intervals had to be <20 min to obtain full inhibition in N450G-GUS-NLS lines. This duration of 20 min fits well with our in vitro observation that the half-life of N450 Pfr is ∼15 min (Figure 7A).
The EOD-FR effect on hypocotyl elongation is mediated by phyB in Arabidopsis seedlings (Robson et al., 1993). The effect depends on the presence of stable Pfr during the dark period (Casal, 1996). We demonstrated that the N450G-GUS-NLS seedlings were constitutively tall, regardless of the EOD-FR treatment, as is the case for the parental phyB mutant (Figure 7C). This observation indicates that N450G-GUS-NLS Pfr disappears or rapidly becomes inactive in the dark, even though N450G-GUS-NLS functions normally under continuous irradiation. Hence, rapid dark reversion of N450 Pfr appears to occur not only in vitro but also in vivo. Indeed, phyB-101, whose holoprotein exhibits extremely rapid dark reversion in vitro, is null for the EOD-FR response (Elich and Chory, 1997). The importance of the Pfr stability in natural conditions was further demonstrated by the short-day experiment (Figure 7D). Under the short-day condition, N450G-GUS-NLS plants looked almost like the phyB-deficient mutant, although they appeared normal under cW. By virtue of the long life of Pfr in the dark, plants can perceive the light quality (red/far-red ratio) independently of the daylength.
In addition, the reduced stability of Pfr might affect its light sensitivity. The ratio of Pfr to Pr should depend on the rates of both photochemical and nonphotochemical conversion under the condition that the photochemical reaction is relatively slow under a low fluence rate of light. In fact, the sensitivity to cR of N450G-GUS-NLS was significantly lower than that of N651G-GUS-NLS (Figure 5B).
The Core Signaling Domain of phyB
Various deletion derivatives of phyA and phyB have been tested in transgenic plants with respect to their biological activity (Boylan and Quail, 1991; Cherry et al., 1992, 1993; Boylan et al., 1994; Wagner et al., 1996a). Interestingly, the first 103 residues of Arabidopsis phyB are not critical for phyB signaling activity under cR (Wagner et al., 1996a). Taken together with these results, the core region for the phyB signaling can be narrowed to a region as small as ∼350 amino acids encompassing positions 103 to 450.
Although the PHY domain may not be essential for phyB signaling, it contributes to the normal function of phyB. Under saturating light, the physiological response of N450G-GUS-NLS was weaker than that of N651G-GUS-NLS (Figure 5A). Because dark reversion is much slower than photoconversion under such conditions (Eichenberg et al., 1999), this difference in physiological response cannot be because of differences in the rate of dark reversion. A possible explanation is that removal of the PHY domain may result in a reduction in the interaction of phyB with downstream components. Furthermore, the PHY domain may be directly involved in interactions with signaling components and/or in the light-induced nuclear localization. The PHY domain is located at the end of the photosensory domain close to the flexible hinge region, which is highly exposed and alters its structure during photoconversion (Furuya and Song, 1994).
N651G-GUS-NLS has been proposed to transduce the light signal through interaction with transcription factors, such as PIF3 (Zhu et al., 2000; Shimizu-Sato et al., 2002; Matsushita et al., 2003). PIF3 has been recently shown to negatively regulate the photomorphogenesis (Kim et al., 2003) and to be degraded by the action of photoreceptors, including phyB (Bauer et al., 2004). It remains less clear how the N450 fragment of phyB transduces the signal in the nucleus. We speculate that the N450 fragment retains the binding site for these factors, although this should be examined experimentally in future work.
It is interesting here that six missense mutations, G118/R, S134/G, I208/T, H283/T, C327/Y, and A372/T, have been described in this region (Reed et al., 1993; Krall and Reed, 2000; Chen et al., 2003). Of these, the G118/R, C327/Y, and A372/T mutations result in mislocation of phyB in the nucleus, and the H283/T mutation results in slightly rapid dark reversion. However, the molecular lesions in these mutants with respect to their signaling activity remain unclear. Some of these residues might be directly involved in interaction with downstream components of the pathway.
Although several factors have been shown to interact with phytochrome, most of these, with the exception of PIF3 (Zhu et al., 2000; Shimizu-Sato et al., 2002) and ARR4 (Sweere et al., 2001), recognize the C-terminal but not the photosensory domain of phytochrome (Fankhauser et al., 1999; Jarillo et al., 2001; Liu et al., 2001; Yang et al., 2001). Among them, PIF3 is particularly interesting because both the N-terminal chromophoric and the C-terminal domains of phyB interact with PIF3 (Zhu et al., 2000). Our previous work (Matsushita et al., 2003) and this work have suggested that the C-terminal domain attenuates the signaling activity of phyB. Complex interactions between these two domains of phyB and PIF3 may play a key role in the regulation of the phyB sensitivity. In any case, efforts should be made to search for components that interact with the core region of phyB defined by our study using techniques such as yeast two-hybrid screening (Shimizu-Sato et al., 2002).
METHODS
Plant Materials, Growth Conditions for Seedlings, and Growth Measurements
The Arabidopsis thaliana mutants (phyB-5, Reed et al., 1993; phyA-201, Nagatani et al., 1993) were null alleles on the Landsberg erecta background. The PBG (Yamaguchi et al., 1999) and N651G-GUS-NLS (originally NG-GUS-NLS) (Matsushita et al., 2003) lines on the phyB-5 background have been described elsewhere. Preparation of the N450G-GUS-NLS line is described below. We established lines expressing N450G-GUS-NLS, N651G-GUS-NLS, or PBG on the phyA phyB double mutant background by crossing.
Seeds were surface-sterilized and sown on 0.6% agar plates containing MS medium with or without 2% sucrose. The plates were kept in the dark at 4°C for 72 h and then irradiated with cW for 3 h at 22°C to induce seed germination. The plates were then placed under various light conditions, as specified in the figure legends. For hypocotyl length measurements, the seedlings were grown on MS agar plates without sucrose for 5 d at 22°C and then pressed gently onto the surface of agar medium before photographs were taken. Hypocotyl lengths, cotyledon area, and petiole lengths were determined by NIH image software (Bethesda, MD). The chlorophyll assay was performed as described (Mochizuki et al., 2001). For immunoblot analysis, the seedlings were grown on MS agar plates with 2% (w/v) sucrose for 1 week at 22°C in cW (45 μmol m−2 s−1).
Plasmid Construction and Plant Transformation
To generate the N450G-GUS-NLS construct, the N651 fragment in N651G-GUS-NLS (Matsushita et al., 2003) was replaced with a PCR-amplified N450 fragment at the 5′ XbaI and 3′ ClaI restriction sites. N450G-GUS-NLS was inserted between the constitutive Cauliflower mosaic virus 35S promoter and the nopaline synthase terminator of pPZP211/35S-nosT, which is derived from pPZP211 (Hajdukiewicz et al., 1994).
The phyB-5 mutant was used as the host for the N450G-GUS-NLS construct and was transformed by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). Transformed plants were selected on MS medium containing 25 mg/mL of kanamycin and 166 mg/mL of claforan (Hoechst, Frankfurt, Germany). For N450G-GUS-NLS, 46 independent lines were generated, and 16 independent lines that segregated to give 75% kanamycin-resistant plants in the T2 generation were selected. Next, T3 self-progeny of homozygous T2 plants were identified, and the level of overexpression was determined immunochemically in each line. Two of the lines, N450G-GUS-NLS 2-5 and N450G-GUS-NLS 3-4, were crossed with phyA-201 to obtain the lines homozygous for the N450G-GUS-NLS gene on the phyA phyB mutant background.
Immunochemical Experiments
Protein extraction, SDS-PAGE, protein blotting, and immunodetection were performed as described by Yamaguchi et al. (1999). The monoclonal antibodies were mAA1 and mBA1, which are specific to phyA and phyB, respectively (Shinomura et al., 1996). The anti-GFP monoclonal antibody was from Sigma (St. Louis, MO).
Light Sources
The white light was obtained from fluorescent tubes (FLR40SW/M-B; Hitachi, Tokyo, Japan). The red light was obtained by a combination of red fluorescent tubes (FL20S/R-F; National, Tokyo, Japan) and a 3-mm thick, red plastic plate (Shinkolite A102; Mitsubishi, Tokyo, Japan). The far-red light was obtained by a combination of fluorescent tubes (FL20S FR-74; TOSHIBA, Tokyo, Japan) and a 3-mm methacrylic plate (Dalaglass A-900; Asahi Chemical Industry, Tokyo, Japan). The fluence rates and spectral qualities were measured by an optical power meter (Model LI-1000; Li-Cor, Lincoln, NE) and a spectroradiometer (USR-40V; USHIO, Tokyo, Japan), respectively.
Analysis of Subcellular Localization in Transgenic Arabidopsis Seedlings
Transgenic seedlings were grown on MS agar plates without sucrose for 1 week at 22°C either in the dark or under cW of 44 μmol m−2 s−1 and stained with 20 μg/mL of propidium iodide (Molecular Probes, Eugene, OR) to visualize nuclei and cell walls. The subcellular localization of the GFP and propidium iodide fluorescence was visualized by a confocal laser scanning microscope (Zeiss LSM510; Jena, Germany) with the fluorescein isothiocyanate channel (green: GFP) and the tetramethylrhodamine isothiocyanate channel (red: propidium iodide). For etiolated seedlings, a dim green safelight was used in every step before microscopy, and images were recorded during the first 1 min of microscopic observation. The observations were done on various parts of the seedlings, and essentially the same results were obtained. Hence, the results shown in root cells are representative of the whole seedlings.
Escherichia coli Expression and Purification of Recombinant Phytochrome Fragments
The N450, N651, and full-length PHYB gene sequences were amplified from PBG (Yamaguchi et al., 1999) by PCR. These fragments, to which 5′ NdeI and 3′ SmaI sites were introduced, were inserted into the pTYB2 vector (New England Biolabs, Beverly, MA) at the NdeI/SmaI sites to make an in-frame fusion with the Intein/CBD sequence, as previously described (Hanzawa et al., 2001). Consequently, vectors expressing N450-Intein/CBD, N651-Intein/CBD, and full-length phyB-Intein/CBD were obtained. To make an in-frame fusion with the c-Myc epitope tag sequence, the Intein/CBD sequence in the above constructs was replaced with the PCR-amplified c-Myc epitope tag sequence at the 5′ SmaI and 3′ PstI sites. Missense mutations (G564/A, G564/D, G564/K, G564/H, and A587/T) were generated using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
The plasmids were transformed into E. coli strain ER2566 (New England Biolabs). Single colonies containing plasmid were inoculated into 5 mL of LB medium containing 30 μg mL−1 of carbenicillin and grown for 16 h at 37°C. The cultures were then added to 1 liter of LB medium containing 30 μg mL−1 of carbenicillin and shaken vigorously at 37°C until the cell density reached A600 = 0.5. Isopropyl-thio-β-d-galactoside was then added at 0.1 mM to induce expression of the construct. The cells were cultured at 15°C for 16 h, collected by centrifugation, and washed with 50 mL of extraction buffer (20 mM Tris-HCl, 500 mM NaCl, and 0.1 mM EDTA, pH 7.8). Washed cell pellets were frozen in liquid nitrogen and stored at −80°C until use. Frozen cells were resuspended in 1:4 (w/v) extraction buffer and broken by sonication. The homogenate was clarified by centrifugation at 12,000g for 30 min.
The Intein/CBD fusion proteins were purified with the IMPACT system kit using a chitin affinity column according to the manufacturer's instruction (New England Biolabs). The extract was washed onto the column, and bound protein was self-cleaved in the presence of 40 mM 1,4-dithiothreitol for 2 d at 4°C. The N-terminal amino acid sequences of the proteins obtained were determined with a Procise 494 HT sequencer (Applied Biosystems, Foster City, CA) at APRO Life Science Institute (Tokushima, Japan).
To prepare crude ammonium sulfate fractions for the c-Myc epitope-tagged proteins, ammonium sulfate was added at a ratio of 0.25 g mL−1 to extracts to precipitate proteins. After incubation for 1 h, the precipitate was collected by centrifugation for 30 min at 12,000g. The resulting pellet was dissolved in 2 mL of the extraction buffer and clarified by centrifugation at 12,000g for 30 min.
The holoproteins were reconstructed using PCB as a chromophore (Lagarias and Lagarias, 1989). PCB was obtained from Spirulina platensis cells (a kind gift from Dainippon Ink and Chemicals, Tokyo, Japan), essentially as described previously (Fu et al., 1979), and stocked in methanol. For reconstitution, PCB was added at 5 μM to samples of purified recombinant protein or the ammonium sulfate fractions and kept in the dark for 1 h at 4°C. The resultant holoproteins were subjected to further analysis. For the measurement of absorption spectra, excess PCB was removed by dialysis.
Detection of E. coli–Derived Recombinant Protein and Zinc Blot Analysis
For CBB staining and zinc blot analysis, reconstituted samples were separated by SDS-PAGE on 10% gels. Zn2+-induced fluorescence of the holoprotein was visualized under UV irradiation after incubating the gel in a buffer containing 20 mM zinc acetate and 150 mM Tris-HCl, pH 7.0 (Berkelman and Lagarias, 1986). Immunodetection of c-Myc fusion proteins was performed using anti c-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Spectrophotometric Assays
Absorption and difference spectra of reconstituted samples were obtained with a UV-1600PC spectrophotometer (Shimadzu, Kyoto, Japan). A jacketed cuvette holder was maintained at 22°C by a circulating water bath. Actinic irradiation was from a light emission diode (red, peak at 655.3 nm, half-width = 66 nm; far-red, peak at 755.2 nm, half-width = 30 nm). The fluence rates of the red and far-red light irradiation at the top of the sample cuvette were 89 μmol m−2 s−1 and 93 μmol m−2 s−1, respectively. For standard assays, 10-min irradiation periods of red and far-red light were used to ensure saturation. For the measurement of dark reversion, the sample in the cuvette was irradiated with saturating red light, and the baseline was recorded immediately. After an appropriate period in the dark, the sample was scanned to measure the absorbance changes in the red and far-red light regions, from which the loss of Pfr was estimated.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number X17342.
Acknowledgments
We thank Hiroko Hanzawa for technical advice on the preparation of recombinant phytochromes. We are grateful to Dainippon Ink and Chemicals for supplying the S. platensis cells. This work was supported, in part, by Grants-in-Aid for Scientific Research (B) (13440239 and 15370020), a Grant-in-Aid for Scientific Research on Priority Areas (2) “Studies on Photoperception and Signal Transduction Pathways of Blue Light Receptor, PHOT, in plants” (13139201), and a Grant-in-Aid for 21st Century COE Research, Kyoto University (A14) from the Ministry of Education, Sports, Culture, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Akira Nagatani (nagatani@physiol.bot.kyoto-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022350.
References
- Andel, F., 3rd, Lagarias, J.C., and Mathies, R.A. (1996). Resonance Raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome. Biochemistry 35, 15997–16008. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., Hirschfeld, M., Wester, L., Weaver, M., Clack, T., Amasino, R.M., and Sharrock, R.A. (1997). A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C.S., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelman, T.R., and Lagarias, J.C. (1986). Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal. Biochem. 156, 194–201. [DOI] [PubMed] [Google Scholar]
- Boylan, M., Douglas, N., and Quail, P.H. (1994). Dominant negative suppression of Arabidopsis photoresponses by mutant phytochrome A sequences identifies spatially discrete regulatory domains in the photoreceptor. Plant Cell 6, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan, M.T., and Quail, P.H. (1991). Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88, 10806–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Huala, E. (1999). Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol. 15, 33–62. [DOI] [PubMed] [Google Scholar]
- Casal, J.J. (1996). Phytochrome A enhances the promotion of hypocotyls growth caused by reductions in levels of phytochrome B in its FR-light-absorbing form in light-grown Arabidopsis thaliana. Plant Physiol. 112, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Schwab, R., and Chory, J. (2003). Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. USA 100, 14493–14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.R., Hondred, D., Walker, J.M., Keller, J.M., Hershey, H.P., and Vierstra, R.D. (1993). Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell 5, 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.R., Hondred, D., Walker, J.M., and Vierstra, R.D. (1992). Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc. Natl. Acad. Sci. USA 89, 5039–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., Patel, S.R., and Whitelam, G.C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberg, K., Baurle, I., Paulo, N., Sharrock, R.A., Rudiger, W., and Schafer, E. (2000). Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 470, 107–112. [DOI] [PubMed] [Google Scholar]
- Eichenberg, K., Kunkel, T., Kretsch, T., Speth, V., and Schafer, E. (1999). In vivo characterization of chimeric phytochromes in yeast. J. Biol. Chem. 274, 354–359. [DOI] [PubMed] [Google Scholar]
- Elich, T.D., and Chory, J. (1997). Biochemical characterization of Arabidopsis wild type and mutant phytochrome B holoproteins. Plant Cell 9, 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochromes that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., Davis, S.J., Stoddart, W.M., Vierstra, R.D., and Whitelam, G.C. (2003). Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15, 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, E., Friedman, L., and Siegelman, H.W. (1979). Mass-spectral identification and purification of phycoerythrobilin and phycocyanobilin. Biochem. J. 179, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya, M. (1993). Phytochromes: Their molecular species, gene families, and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 617–645. [Google Scholar]
- Furuya, M., and Song, P.S. (1994). Assembly and properties of holophytochrome. In Photomorphogenesis in Higher Plants, 2nd ed., R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 105–140.
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hanzawa, H., Inomata, K., Kinoshita, H., Kakiuchi, T., Jayasundera, K.P., Sawamoto, D., Ohta, A., Uchida, K., Wada, K., and Furuya, M. (2001). In vitro assembly of phytochrome B apoprotein with synthetic analogs of the phytochrome chromophore. Proc. Natl. Acad. Sci. USA 98, 3612–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisada, A., Hanzawa, H., Weller, J.L., Nagatani, A., Reid, J.B., and Furuya, M. (2000). Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo, J.A., Capel, J., Tang, R.H., Yang, H.Q., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487–490. [DOI] [PubMed] [Google Scholar]
- Kay, S. (1997). PAS, present and future: Clues to the origin of circadian clocks. Science 276, 753–754. [DOI] [PubMed] [Google Scholar]
- Kim, J., Yi, H., Choi, G., Shin, B., Song, P.S., and Choi, G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Kim, L., Adam, E., Schafer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall, L., and Reed, J.W. (2000). The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl. Acad. Sci. USA 97, 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsch, T., Poppe, C., and Schafer, E. (2000). A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J. 22, 177–186. [DOI] [PubMed] [Google Scholar]
- Lagarias, J.C., and Lagarias, D.M. (1989). Self-assembly of synthetic phytochrome holoprotein in vitro. Proc. Natl. Acad. Sci. USA 86, 5778–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Lagarias, J.C. (1992). Phytochrome assembly. Defining chromophore structural requirements for covalent attachment and photoreversibility. J. Biol. Chem. 260, 2415–2423. [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli, A.L. (1994). The physiology of phytochrome action. In Photomorphogenesis in Higher Plants, 2nd ed., R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 211–269.
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and Sharrock, R.A. (1997). Phytochrome gene diversity. Plant Cell Environ. 20, 666–671. [Google Scholar]
- Matsushita, T., Mochizuki, N., and Nagatani, A. (2003). Dimers of the photosensory domain of phytochrome B are functional in the nucleus. Nature 424, 571–574. [DOI] [PubMed] [Google Scholar]
- McCormac, A.C., Wagner, D., Boylan, M.T., Quail, P.H., Smith, H., and Whitelam, G.C. (1993). Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome B-encoding cDNAs: Evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. Plant J. 4, 19–27. [Google Scholar]
- Mochizuki, N., Brusslan, J.A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98, 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, E., Alonso, J.M., Ecker, J.R., Zhang, Y., Li, X., Young, J., Austin-Phillips, S., and Quail, P.H. (2003). Isolation and characterization of phyC mutants in Arabidopsis reveals complex cross talk between phytochrome signaling pathways. Plant Cell 15, 1962–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, B.L., and Lagarias, J.C. (2002). Phytochrome ancestry: Sensors of bilins and light. Trends Plant Sci. 7, 357–366. [DOI] [PubMed] [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (1997). An emerging molecular map of the phytochromes. Plant Cell Environ. 20, 657–665. [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff, U., Eilfeld, O., and Rudiger, W. (1985). A photoreversible 39-kDalton fragment from the Pfr form of 124-kDalton phytochrome. Z. Naturforsch. 40, 693–698. [Google Scholar]
- Robson, P.R.H., Whitelam, G.C., and Smith, H. (1993). Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica napa deficient in phytochrome B. Plant Physiol. 102, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10, 859–868. [DOI] [PubMed] [Google Scholar]
- Schafer, E., and Bowler, C. (2002). Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep. 3, 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch, H.A.W. (1992). Signal transduction by phytochrome: Phytochromes have a module related to the transmitter modules of bacterial sensor proteins. Photochem. Photobiol. 56, 839–846. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato, S., Huq, E., Tepperman, J.M., and Quail, P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 10, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Uchida, K., and Furuya, M. (2001). Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 122, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H., and Whitelam, G.C. (1997). The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 20, 840–844. [Google Scholar]
- Stockhaus, J., Nagatani, A., Halfter, U., Kay, S., Furuya, M., and Chua, N.H. (1992). Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 6, 2364–2372. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Baurle, I., Kudla, J., Nagy, F., Dchafer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Fairchild, C.D., Kuhn, R.M., and Quail, P.H. (1996. b). Chromophore-bearing NH2-terminal domains of phytochromes A and B determine their photosensory specificity and differential light lability. Proc. Natl. Acad. Sci. USA 93, 4011–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Koloszvari, M., and Quail, P.H. (1996. a). Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell 8, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.-H., and Lagarias, J.C. (2000). Defining the bilin lyase domain: Lessons from the extended phytochrome superfamily. Biochemistry 39, 13487–13495. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B:GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K.T., and Furuya, M. (1983). Spectral properties of chromophore-containing fragments prepared from pea phytochrome by limited proteolysis. Plant Cell Physiol. 24, 713–718. [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97, 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]