Abstract
Despite its importance in a variety of plant defense responses, our understanding of how jasmonic acid (JA) functions at the biochemical level is limited. Several amino acid conjugates of JA were tested for their ability to complement the JA-insensitive Arabidopsis thaliana mutant jar1-1. Unlike free JA, JA-Ile inhibited root growth in jar1-1 to the same extent as in the wild type, whereas JA-Val, JA-Leu, and JA-Phe were ineffective inhibitors in both genotypes. Thin-layer chromatography and gas chromatography–mass spectrometry (GC-MS) analysis of products produced in vitro by recombinant JAR1 demonstrated that this enzyme forms JA-amido conjugates with several amino acids, including JA-Ile. JA-Val, -Leu, -Ile, and -Phe were each quantified in Arabidopsis seedlings by GC-MS. JA-Ile was found at 29.6 pmole g−1 fresh weight (FW) in the wild type but was more than sevenfold lower in two jar1 alleles. JA-Leu, -Val, and -Phe were present at only low levels in both genotypes. Expression of wild-type JAR1 in transgenic jar1-1 plants restored sensitivity to JA and elevated JA-Ile to the same level as in the wild type. The ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) conjugated to JA was also found in plant tissue at 18.4 pmole g−1 FW. JA-ACC was determined not be an effective jasmonate root inhibitor, and surprisingly, was twofold higher in the mutants than in the wild type. This suggests that another JA-conjugating enzyme(s) is present in Arabidopsis. Synthesis of JA-ACC might provide a mechanism to coregulate the availability of JA and ACC for conversion to the active hormones JA-Ile and ethylene, respectively. We conclude that JAR1 is a JA-amino synthetase that is required to activate JA for optimal signaling in Arabidopsis. Plant hormone activation by conjugation to amino acids and the enzymes involved in their formation were previously unknown.
INTRODUCTION
Plants use chemical signals to regulate growth and development and to control biotic and abiotic stress responses. Among these signals are members of the jasmonate family of oxylipins that are derived from the oxidation of linolenic acid (Vick and Zimmerman, 1987). Jasmonates function in a variety of important roles, including defense against insects and pathogens (Kessler and Baldwin, 2002; Turner et al., 2002; Weber, 2002), protection from abiotic stresses (Conconi et al., 1996; Rao et al., 2000), and in reproductive development (Feys et al., 1994; Sanders et al., 2000; Li et al., 2004). Although jasmonic acid (JA) is the best studied of the jasmonates, its biosynthetic intermediates can also be biologically active. Most notable is 12-oxophytodienoic acid (OPDA), which is involved in plant defense (Weiler et al., 1993; Mueller, 1997; Gundlach and Zenk, 1998; Stintzi et al., 2001) and is considered the primary signal for mechanosensitive tendril coiling in Bryonia dioica (Stelmach et al., 1998). OPDA is also a stronger inducer of phytoalexin production in soybean (Glycine max) cell cultures than is JA (Fliegmann et al., 2003).
JA has been assumed to be the primary signal in most jasmonate-dependent responses. Strong evidence for this has been provided for male fertility in Arabidopsis thaliana (Stintzi and Browse, 2000) and in the induction of alkaloid biosynthesis in Eschscholtzia californica cells (Haider et al., 2000). However, little is known about the signaling role of free JA, as opposed to its many metabolites. The diverse array of jasmonates in plants affords the possibility for a signaling repertoire that is more complex than presently recognized (Weber et al., 1997).
JA occurs in a variety of modified forms, including the methyl ester (MeJA), glycosyl esters, and amide-linked conjugates with various amino acids (for review, see Sembdner and Parthier, 1993). MeJA is a potent signal, functioning as a volatile in the atmosphere in interplant communication (Farmer and Ryan, 1990). Enhanced pathogen resistance in Arabidopsis plants overexpressing a JA methyl transferase (JMT) also supports a signaling role for MeJA (Seo et al., 2001).
Considerable evidence indicates that both glycosyl and amino acid conjugates of indoleacetic acid (IAA) are inactive and help to regulate auxin homeostasis (for review, see Normanly, 1997), but the function of JA conjugates is still unclear. JA–amino acid conjugates accumulate in osmotically stressed barley (Hordeum vulgare) leaves and in arbuscular mycorrhizal barley roots, suggesting possible roles in stress response and in interactions with microorganisms (Kramell et al., 1995; Hause et al., 2002). The level of JA-Ile also varies markedly during tomato (Lycopersicon esculentum) flower development, which might imply a regulatory role (Hause et al., 2000). Exogenous application of JA–amino acid conjugates induces phytoalexin synthesis in rice (Oryza sativa) (Tamogami et al., 1997), and the Ile or Leu conjugates with 1-oxo-indanoyl mimic the JA induction of volatile compounds in Phaseolus lunatasi (Krumm et al., 1995). On the other hand, JA-Ile elevates only a subset of the transcripts that are induced by JA or MeJA in barley leaves (Kramell et al., 2000). Interpretation of experiments with exogenously applied jasmonates is complicated by the potential for the release of JA by hydrolysis of the applied conjugate and by the possible metabolism of JA to other forms that may be either biologically active or inactive.
The bacterial produced phytotoxin coronatine has potent jasmonate-like properties. The coronatine-resistant mutant coi1 is strongly insensitive to JA (Feys et al., 1994; Xie et al., 1998), but the structure of coronatine suggests that it may mimic a JA–amino acid conjugate, rather than JA itself (Krumm et al., 1995). Coronatine is composed of the polyketide coronafacic acid, with weak structural similarity to JA, linked through an amide bond to 1-amino-2-ethylcyclopropane-1-carboxylic acid (AEC), also called coronamic acid. AEC is an ethyl derivative of the ethylene precursor ACC that in Pseudomonas is synthesized from l-Ile (Parry et al., 1994).
With the exception of JMT, the enzymes involved in modifying JA are unknown. This has limited our ability to deduce the function of specific JA derivatives by genetic and transgenic approaches. By contrast, plant enzymes that form glycosyl esters of IAA have been studied (Szerszen et al., 1994), and a bacterial IAA-Lys synthetase has been characterized (Roberto et al., 1990). Expression of the latter in plants was shown to cause auxin-associated phenotypes (Spena et al., 1991), which substantiates other biochemical and physiological evidence that regulating IAA conjugate formation is important for IAA homeostasis. Plant enzymes that hydrolyze glycosyl and amino acid conjugates of IAA have also been identified (Jakubowska et al., 1993; Davies et al., 1999), but the biosynthetic pathway for IAA–amino acid conjugate formation remains unknown.
We recently identified the gene responsible for defective jasmonate response in jar1-1, an Arabidopsis mutant that exhibits decreased sensitivity to exogenous JA (Staswick et al., 1992, 2002). The JAR1 locus is necessary for resistance to the opportunistic soil fungus Pythium irregulare (Staswick et al., 1998), for systemic pathogen resistance induced by nonpathogenic bacteria (van Loon et al., 1998; Clarke et al., 2000), and for protection against ozone damage (Overmyer et al., 2000; Rao et al., 2000). Surprisingly, JAR1 encodes an enzyme that is structurally related to adenylate-forming enzymes of the firefly luciferase family (Conti et al., 1997), and it adenylates JA. Adenylation of substrate carboxyl groups by some of these enzymes is a prerequisite for the synthesis of various peptides and polyketides (Kleinkauf and Von Dohren, 1996). Therefore, one possible function for JAR1 is the biochemical activation of JA for its conjugation to amino acids. To test this, we further examined the biochemical activity of JAR1, the level of JA–amino acid conjugates in jar1 mutants, and the ability of JA conjugates to complement the jar1-1 mutation. We establish that JAR1 is a JA-amino synthetase and that conjugation of JA to an amino acid is necessary for optimal signaling in some jasmonate responses in Arabidopsis.
RESULTS
The JAR1 Enzyme Conjugates JA to Amino Acids
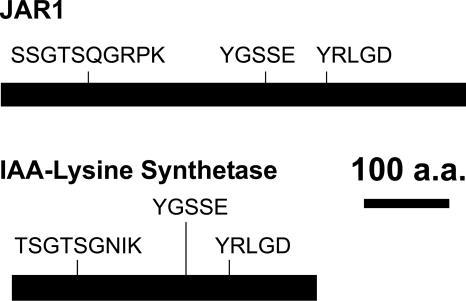
Analysis of (indole-3-acetyl)-l-Lys synthetase, encoded by iaaL in Pseudomonas syringae pv savastanoi, revealed that it contained three short sequence motifs previously identified in JAR1 (Staswick et al., 2002) (Figure 1). IAA-Lys synthetase catalyzes the conjugation of Lys to IAA (Roberto et al., 1990), and although no other similarities between these proteins were found, this prompted us to examine whether JAR1 might also catalyze amide bond formation between JA and amino acids.
Figure 1.
Conserved Motifs and Their Relative Position in JAR1 and Bacterial IAA-Lys Synthetase.
The remainder of the sequence has no significant similarity. a.a., amino acids.
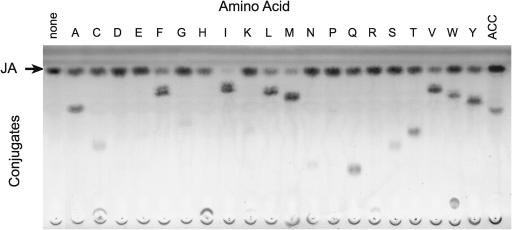
Each of 20 l-amino acids was included along with JA as substrates in independent enzymatic reactions with recombinant glutathione S-transferase (GST)-JAR1. Figure 2 shows that several amino acids yielded products that stained for JA after thin-layer chromatography (TLC) but had lower mobility compared with free JA. Furthermore, there was a consistent correlation between the loss of free JA and the apparent amount of new product formed. The putative JA-Ile, JA-Leu, JA-Phe, and JA-Val conjugates produced by the enzyme reactions also had RF values that were identical to those obtained with the respective chemically synthesized standards (data not shown). To verify their chemical structures, enzymatically produced JA-Ile, -Val, and -Phe were isolated from preparative TLC plates and analyzed by gas chromatography–mass spectrometry (GC-MS). Each yielded products with a mass spectrum consistent with the corresponding standards (data not shown) and in agreement with previously published values for the methyl ester derivatives of these conjugates (Kramell et al., 1988).
Figure 2.
Synthesis of Amino Acid Conjugates of JA by Recombinant GST-JAR1.
Reactions with JA and each amino acid (single letter abbreviations) were analyzed by TLC and stained for JA with vanillin reagent.
To determine whether the enzyme from mutant jar1-1 was defective in conjugating activity, it was compared with JAR1 from the wild type. Each thrombin-cleaved enzyme was used in an assay with JA and Ile as substrates. JAR1 produced JA-Ile at 316 ± 29 pmole min−1 μg−1 of protein. By contrast, the enzyme from mutant jar1-1 did not yield measurable JA-Ile at a detection limit of 2 pmole min−1 μg−1 of protein, <1% of the JAR1 activity. This supports our previous conclusion that jar1-1 is functionally a null allele (Staswick et al., 2002).
JA-ACC Is Present in Arabidopsis Leaves
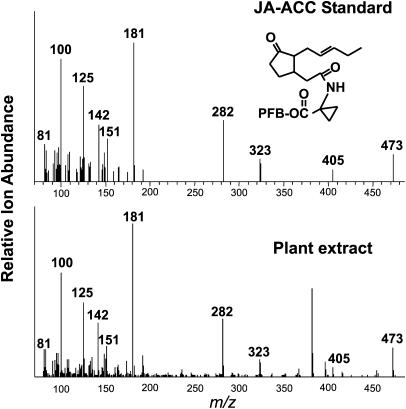
The similarity of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) to the AEC moiety of coronatine prompted us to investigate whether JAR1 could synthesize JA-ACC and whether this compound occurred in plant tissue. JAR1 did indeed produce JA-ACC in vitro (Figure 2), and the product had a mass spectrum identical with chemically synthesized JA-ACC (data not shown). Although several different JA–amino acids have been identified in plants, JA-ACC had not previously been reported. To determine whether this was the case, Arabidopsis leaf extracts were analyzed by GC-MS. Figure 3 shows the spectrum of the peak from the plant sample that eluted at the same time as the pentafluorobenzyl ester derivative of JA-ACC. The predicted molecular ion (473 mass-to-charge ratio [m/z]) and the major fragmentation ions are present in the tissue sample at similar relative proportions to the standard, confirming that Arabidopsis leaves do indeed contain JA-ACC.
Figure 3.
Detection of JA-ACC in Arabidopsis Leaves by GC-MS.
The spectra of JA-ACC, the standard eluting at 18.17 min, and the same peak from 6 g of leaf tissue extract are shown. The molecular ion of pentafluorobenzyl (PFB) ester derivative of JA-ACC (473 m/z) and major fragmentation ions are indicated.
jar1 Alleles Are Deficient in JA-Ile
To determine whether jar1 mutants have reduced JA conjugate levels, five conjugates were purified from Arabidopsis seedling tissue and quantified by GC-MS. The mass spectra of the pentafluorobenzyl ester derivatives of JA, JA-Ile, JA-Leu, JA-Phe, and JA-Val are available in the supplemental material online. The two mutant alleles jar1-1 and jar1-8 were compared with the wild type, and the results are summarized in Table 1. Free JA levels from the wild type and the mutants were not significantly different, ranging from 25.9 to 32.8 pmole g−1 FW. By contrast, the level of JA-Ile was similar to that of JA in the wild type, but was at least sevenfold lower in the mutants. JA-Val, JA-Leu, and JA-Phe were much lower in the wild type compared with JA-Ile, and the first two were not significantly different from the values for jar1-1 and jar1-8. JA-ACC was present at 18.4 pmole g−1 FW in the wild type, and surprisingly, both JA-ACC and JA-Phe were about twofold higher in both jar1 mutants. These results suggest that a critical function for JAR1 might be to synthesize the JA-Ile conjugate in plants, making JA a more active signal.
Table 1.
Quantitation of JA and Some of Its Conjugates in Arabidopsis Seedlings
| Genotype
|
||||
|---|---|---|---|---|
| Conjugate | Wild Type | jar1-1 | jar1-8 | jar1S2-1 |
| JA | 32.2 (1.8) | 32.8 (3.4) | 25.9 (4.0) | 39.4 (5.1) |
| JA-Ile | 29.6 (7.3)a | 4.2 (0.7)b | 2.5 (0.4)b | 27.4 (8.2)a |
| JA-Val | 3.2 (0.4) | 2.6 (0.7) | 2.3 (0.5) | 1.9 (0.4) |
| JA-Leu | 6.8 (1.6) | 4.5 (3.4) | 3.4 (1.5) | 5.7 (1.3) |
| JA-Phe | 5.9 (1.1)a | 9.6 (0.8)b | 9.3 (1.0)b | 5.8 (0.8)a |
| JA-ACC | 18.4 (4.0)a | 43.5 (2.7)b | 43.8 (1.7)b | 14.3 (1.2)a |
Values are the mean (pmol/g fresh weight [FW]) of three extractions of 2-week-old whole seedling tissue, excluding roots. Values in parentheses are se of the means. Means for conjugates that differ significantly (5% level) between genotypes are indicated by different letters (a and b).
Overexpression of JAR1 Restores JA-Ile Level in jar1-1
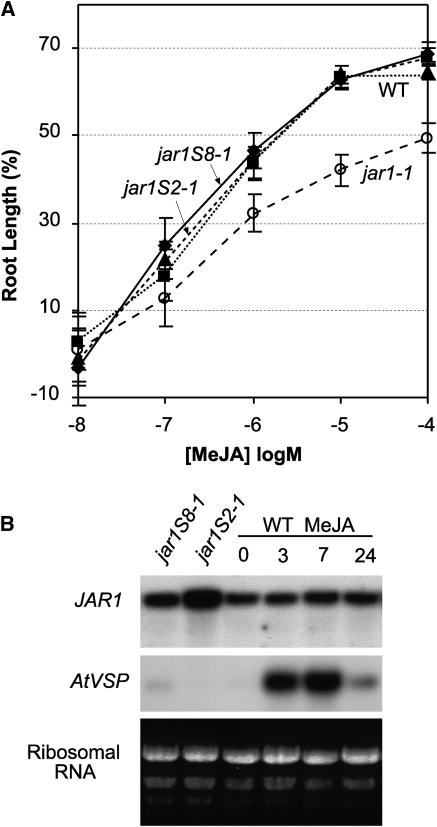
Next we examined whether overexpression of the wild-type gene in jar1-1 plants would complement the insensitivity to JA and increase conjugate levels in this mutant. Figure 4A summarizes the dose response for root growth in the presence of JA for two transformed lines, jar1S2-1 and jar1S8-1, in which JAR1 is expressed under the control of the constitutive 35S promoter of Cauliflower mosaic virus. The level of sensitivity to JA in these transformants was restored to that of the wild type, but there was no evidence for increased sensitivity above that of the wild type in either transformant. Figure 4B shows that the level of mRNA detected with the JAR1 probe in jar1S2-1 was higher than in jar1S8-1, and both were higher than in the wild type (lane 3). There was no detectable difference in JAR1 mRNA level between the wild type and jar1-1 (data not shown). Overexpression of JAR1 in jar1S2-1 restored JA conjugates to essentially the same level as in the wild type (Table 1). In addition to elevating the level of JA-Ile, JA-ACC and JA-Phe were reduced to wild-type levels in this transformant. This result supports the idea that elevated JA-ACC in the two jar1 alleles examined is directly related to the lack of a JAR1 enzyme and not because of a secondary mutation.
Figure 4.
Analysis of jar1-1 Transformants (jar1S2-1 and jar1S8-1) Overexpressing the 35S-JAR1 Construct.
(A) Seedling root growth is expressed as a percentage of the same genotype in the absence of JA. Error bars indicate 95% confidence intervals.
(B) JAR1 transcript level of transformants and the wild type sprayed with 50 μM MeJA. Each lane was loaded with 9 μg of total leaf RNA from overexpression lines or the wild type. The wild type treated with MeJA was subsequently harvested at times indicated. Hybridization probes were full-length cDNA for JAR1 and AtVSP for the positive control, as indicated. Ribosomal RNA bands stained with ethidium bromide in original gel are shown.
The fact that JA is elevated by environmental stresses raised the question of whether JAR1 mRNA might be increased by JA treatment as a means to increase JA conjugate level. However, plants treated with 50 μM MeJA showed no increase in JAR1 mRNA up to 24 h after treatment (Figure 4B), whereas the positive marker (AtVSP) displayed the expected transient rise in mRNA level. Thus, MeJA does not appear to regulate JAR1 expression at the mRNA level.
JA-Ile Complements Jasmonate Insensitivity in jar1-1
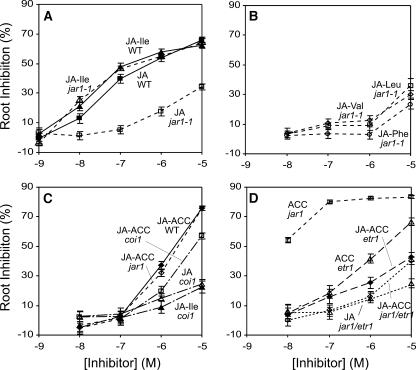
We next tested whether the defect in root growth inhibition seen in jar1-1 could be complemented by specific JA amino acid conjugates. The dose response for root inhibition shown in Figure 5A confirms that jar1-1 is less sensitive to JA than the wild type. JA-Ile inhibited the wild type at least as effectively as did JA, and in contrast with JA, this conjugate strongly inhibited jar1-1, fully complementing the defect in this mutant. By contrast, JA-Leu, JA-Val, and JA-Phe were only weakly effective inhibitors of jar1-1 at the highest concentration tested (Figure 5B). Inhibition of the wild type by these conjugates was similarly ineffective (data not shown). These results confirm the earlier evidence that a deficiency in the ability to produce JA-Ile in jar1-1 might be the cause of its insensitivity to JA.
Figure 5.
Root Growth Inhibition by JA and Its Amino Acid Conjugates.
Inhibition is expressed as percentage untreated for each genotype. Error bars indicate 95% confidence intervals.
JA-ACC Is Not an Effective Root Inhibitor
Although JA-ACC was increased rather than decreased in jar1, we investigated whether this conjugate was able to inhibit root growth. JA-ACC was at least as effective as JA-Ile in both the wild type and jar1-1 at the highest concentration tested (Figure 5C). However, at lower concentrations, JA-ACC was less effective than JA-Ile. This suggested that JA-ACC itself might not be responsible for the observed inhibition. The jasmonate signaling mutant coi1-35 also showed little resistance to JA-ACC, indicating that JA-ACC did not require the jasmonate response pathway defined by COI1. By contrast, coi1-35 was strongly resistant to both JA and JA-Ile, confirming that JA-Ile uses the COI1 jasmonate signaling path.
As the immediate precursor of ethylene, ACC is a strong inhibitor of root growth. To examine whether ACC was responsible for the observed sensitivity to JA-ACC, the ethylene/ACC insensitive mutant etr1 was evaluated alone and as a double mutant with jar1-1. Both etr1 and etr1 jar1-1 exhibited strong resistance to JA-ACC (Figure 5D) compared with the wild type and jar1-1 (Figure 5C). Together, these results indicate that JA-ACC is not an effective jasmonate signal and that ACC is responsible for the majority of the root inhibition observed in the presence of JA-ACC.
JAR1 Is Not Active on JA Biosynthetic Intermediates
Because several JA biosynthetic intermediates are structurally similar to JA and at least one (OPDA) is known to have biological activity, we examined the substrate specificity for JAR1 adenylating activity. Isotope exchange activity was determined in the presence of the JA precursor linolenic acid or the JA biosynthetic intermediates 13-hydroperoxy-linolenic, 12-oxo-phytodienoic, and 3-oxo-cyclopentane octanoic acids. Table 2 shows that compared with the control reaction without substrate, the amount of [32P]PPi exchanged into ATP was negligible for each substrate tested, except for JA. The enzyme also failed to form detectable levels of OPDA conjugates with Ala, Asp, Gly, Phe, Val, or Ile when assayed by TLC and stained with vanillin reagent (data not shown). Thus, although JAR1 appears to have low specificity for the amino donor, it is highly specific for the JA carboxyl group.
Table 2.
Isotope Exchange Activity of JAR1 on JA and Related Substrates
| Substrate | Activity (Percentage of ATP/Total) |
|---|---|
| None | 4.2 ± 0.6 |
| Linolenate | 1.9 ± 0.6 |
| HPOE | 2.6 ± 0.6 |
| OPDA | 2.0 ± 0.7 |
| OCPOA | 2.6 ± 0.6 |
| JA | 42.3 ± 1.8 |
Data shown are means (±se) of five experiments expressed as a percentage of total radioactivity exchanged into ATP. HPOE, 13-hydroperoxy-linolenic acid; OCPOA, 3-oxo-cyclopentane octanoic acid.
DISCUSSION
The Discovery of Hormone Amino Acid Conjugating Enzymes
The jar1-1 mutant has been widely used to study the role of JA in plant biology, but the biological function of the gene identified by this mutant was unknown. We have now established that JAR1 encodes a JA-amino synthetase that activates JA by conjugating it to Ile. Although known to occur in plants for more than two decades, no definitive role for JA–amino acid conjugates had previously been established. The identification of a mutant compromised in its ability to synthesize JA-Ile has allowed us for the first time to demonstrate that an amino acid conjugate of a plant hormone is essential in signaling. Notably, the activation of JA by conjugation contrasts with the general assumption for conjugates of IAA, which appear to be inactive and help to regulate the level of active IAA (for review, see Normanly, 1997).
The identification of an enzyme responsible for synthesis of JA–amino acid conjugates is an important milestone because it also has broader significance. It has been almost 50 years since the discovery of amino acid conjugates of IAA in plants (Andrea and Good, 1955), but the enzyme(s) responsible for their biosynthesis has not been reported. We now know that several JAR1-related enzymes in Arabidopsis have this activity (Staswick et al., 2002; P.E. Staswick, unpublished data). Similar genes occur in several other plant species, including the auxin-responsive GH3 gene of soybean (Hagen et al., 1991), which conjugates amino acids to IAA (P.E. Staswick, unpublished data). This opens the door to solving important questions regarding the regulation and function of hormone conjugation in plants.
Modification of JA for Biological Activity
Although free JA is commonly assumed to be biologically active, our findings, along with results from other laboratories, indicate that for some responses JA must be modified for optimal activity. In Arabidopsis, these include the responses defined by jar1-1, which include pathogen defense and ozone sensitivity (see Introduction). Our attempts to complement the P. irregulare susceptibility of jar1 with JA-Ile were inconsistent (data not shown), but this may reflect difficulty in supplying adequate amounts of JA conjugate to the roots to effect a sustained resistance response. Although the other conjugates used in this study were also ineffective, we cannot rule out the possibility that JAR1 forms other novel conjugates with JA and that these are active in defense. The fact that JAR1 was inactive on other carboxy acids (Staswick et al., 2002), including closely related biosynthetic precursors of JA (Table 1), suggests that JA is the only lipid-derived precursor that would be involved in these defense responses.
The ability of low levels of atmospheric MeJA to stimulate plant responses strongly suggests that the methylated form of JA is also an active signal (Farmer and Ryan, 1990). More recent work showed that overexpression of a JMT in Arabidopsis enhanced jasmonate responses, providing further evidence that MeJA is biologically active (Seo et al., 2001). Surprisingly, however, this contrasts with our finding that MeJA does not complement the defect in root inhibition in jar1-1 (Staswick et al., 1998). It appears that at least for this response, MeJA is first demethylated and then activated by conjugation to Ile. In view of our results, it will be important to determine whether JA-Ile levels are altered in JMT transgenic plants. The elevation of MeJA in these plants did not diminish the amount of free JA, suggesting that JA biosynthesis may have been stimulated in these plants. The quantity of various jasmonates was altered differentially in plants overexpressing an intermediate in JA biosynthesis (Miersch et al., 2004). Thus, it is possible that overaccumulation of MeJA might influence JA-Ile synthesis as well.
The molar quantity of the five conjugates quantified in this study was more than twice that of free JA. Additional conjugates with other amino acids, as well as with sugars, are also likely to occur (Sembdner and Parthier, 1993). This suggests that a relatively small fraction of JA in Arabidopsis occurs as the free acid. MeJA is typically present at a lower level than is JA (Weber et al., 1997; Seo et al., 2001), and if MeJA and JA-Ile are the only active forms of JA, then it appears that much of the JA in Arabidopsis seedlings is inactive. This is also the case for IAA, which occurs predominately in conjugated forms that are inactive in plants (see Normanly, 1997). Unlike JA, however, no IAA conjugates are presently known to have biological activity.
The level of JA detected in wild-type Arabidopsis tissue (32 pmole g−1 FW) was somewhat lower than previous reports of ∼170 pmole g−1 FW (Weber et al., 1997). To limit potential variability in conjugate levels, we grew plants aseptically, which might reduce the basal level of jasmonates compared with plants grown in nonsterile conditions. Reported JA levels in plants are known to differ up to approximately three orders of magnitude, depending on tissue type, age, and environmental stress applied (Creelman and Mullet, 1995). JA conjugate levels have not been as well studied, but Hause et al. (2000) reported marked variation in both JA and JA-Ile during tomato flower development. In some tissues JA-Ile was at least 100-fold higher than what we found in Arabidopsis seedlings. Only two of the 18 jar1-1 transformants we recovered were complemented by expression of JAR1 under the 35S promoter, and neither of these had conjugate levels or sensitivity to JA that differed from the wild type. Although we do not know whether the elevated mRNA resulted in increased JAR1 enzyme in jar1S2-1 and jar1S8-1, this result may suggest that the quantity of JA-Ile is tightly controlled at the level of synthesis or by catabolism of conjugates at abnormally high levels.
Other JA Conjugating Enzymes in Arabidopsis
We found that JAR1 forms conjugates with Leu, Val, Phe, and ACC in vitro, but JA-Ile was the only conjugate negatively affected in the jar1 alleles. Several explanations for this discrepancy are possible. First, kinetic analysis might reveal a preference for Ile that was not evident in our endpoint TLC assays. Second, cellular conditions might alter enzyme specificity in a way not evident in vitro. Third, the availability of specific amino acids would affect conjugate formation in vivo. Fourth, conjugate hydrolases might regulate the level of individual conjugates. Finally, the presence of additional JA-amino synthetases might mask the activity of JAR1 for specific conjugates.
All known jar1 alleles are moderately insensitive to JA and fully male fertile, in contrast with the more severe phenotype of signaling mutant coi1 (Figure 5; Staswick et al., 2002). Partial redundancy for JAR1 function might explain the weak phenotype of jar1 alleles. No activity was detected for the mutant enzyme from jar1-1. Based on the sensitivity of this assay, the upper limit of jar1-1 activity was 1% of the wild-type enzyme activity. Furthermore, both jar1-1 and the nonsense mutant jar1-8 exhibited the same level of JA-Ile deficiency, and yet JA-ACC and JA-Phe were elevated in both mutants. This indicates that at least one additional JA conjugating enzyme is present in Arabidopsis. Tissue-specific roles for different enzymes might also account for the requirement of JAR1 in some jasmonate responses, such as root inhibition and pathogen defense, but not in others, such as male fertility. Studies are currently underway to determine the expression pattern for JAR1, its response to various stimuli, as well as the identity of other enzymes active on JA.
JA-ACC Formation and the Coregulation of Jasmonate and Ethylene Synthesis
The discovery that JA-ACC is present in significant quantity in plant tissue is a novel finding that suggests a possible role in coordinating interaction or cross talk between plant hormone signaling pathways (Gazzarrini and McCourt, 2003). Although at the highest concentration tested JA-ACC inhibited jar1-1 root growth to a similar level as JA-Ile, at lower concentrations it was less effective than JA-Ile (Figure 5C). The JA-ACC response also did not require COI1 but was dependent on the ethylene response pathway defined by etr1. This led us to conclude that the effect of JA-ACC was primarily an ethylene response and not a jasmonate signaled response. In our highly purified JA-ACC preparation, ACC could not have been present at >0.03 mol % of the JA-ACC level. However, because of the high sensitivity of Arabidopsis roots to ACC (Figure 5D), even a low concentration of contaminating ACC would have contributed to the inhibition observed at the highest JA-ACC concentration tested. Another possibility is that ACC was released by hydrolysis after assimilation of JA-ACC by roots. IAA–amino acid conjugate hydrolases from Arabidopsis have been characterized (Davies et al., 1999), but similar enzymes with specificity for JA–amino acids have not yet been described.
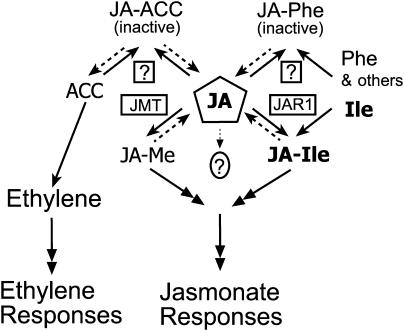
JA and ethylene have been implicated in common environmental stress responses, and synergy between them has been reported (Wang et al., 2002; Lorenzo et al., 2003; Schmelz et al., 2003). As the immediate precursor of ethylene, ACC level is regulated and its synthesis is stimulated by wounding, pathogen infection, and insect feeding (McKeon et al., 1995). These are conditions that also increase JA level in plants. Synthesis of the malonate-ACC conjugate by ACC N-malonyltransferase is thought to help regulate the amount of ACC available for ethylene synthesis (Yang and Hoffman, 1984). In the same way, a JA-ACC conjugating enzyme might regulate the availability of both ACC and JA for conversion to the respective active signals. Conversely, hydrolysis of JA-ACC could supply precursors for both ethylene formation and JA activation. In this context it may be significant that JAR1 appears to negatively regulate JA-ACC accumulation, based on its increased abundance in the jar1 mutants. Some of these possible relationships are illustrated in a model shown in Figure 6.
Figure 6.
Model for the Role of JAR1 and Related Enzymes in Jasmonate and Ethylene Signaling.
The JAR1 (JA-amino synthetase) and JMT are boxed. Other putative JA-amino synthetase(s) are indicated by boxed question marks. Solid and dashed arrows indicate biosynthetic and possible hydrolytic steps, respectively. Double-headed arrows represent signal transduction pathways. The circled question mark indicates uncertainty about the signaling role of free JA. JA-Me, methylester.
Coronatine Is a Structural Mimic of JA-Ile
As for coronatine, JA-Ile uses the response pathway defined by coi1 (Figure 5). Our findings strongly support the idea that coronatine is a structural mimic of JA-Ile rather than JA. It was noted previously that opening of the cyclopropane ring of the AEC moiety of coronatine would convert it to Ile (Krumm et al., 1995). Although it is not known if this occurs in plants, this could form an important basis for the jasmonate-like activity of coronatine. The fact that the Leu isomer of JA-Ile did not complement jar1-1, as well as the ability of P. lunatus to discriminate between the d- and l-isomers of Ile conjugated to 1-oxo-indan-carboxylic acid (Krumm et al., 1995) highlights the specificity of this constituent group. Even so, JA-Ile was no more active than JA in root inhibition, indicating that there are features unique to coronatine that contribute to its stronger biological activity compared with JA (Koda et al., 1996). AEC is not commercially available, but we are currently attempting to synthesize sufficient quantities to test its efficacy as a conjugate with JA.
As mentioned earlier, JA is not required for some jasmonate responses that are apparently mediated by OPDA. We have found no evidence that JAR1 (Table 2) or any of its homologs in Arabidopsis (Staswick et al., 2002) are active on OPDA. OPDA contains an eight-carbon carboxyl side chain at the C-1 position of the cyclopentanone ring. This side chain is reduced to two carbons by β-oxidations to form JA. It may be that the extended side chain of OPDA is functionally analogous to Ile in the conjugate with JA. However, discrimination between these probably occurs based on the different specificities of JA and OPDA in signaling.
In summary, we have demonstrated that JAR1 is a JA-amino synthetase, and its activity in conjugating Ile to JA is required for optimal jasmonate signaling in at least some Arabidopsis responses. Although the activity defined by the JAR1 locus acts downstream of JA, contrary to earlier assumptions, it is upstream of the jasmonate signal transduction pathway. JA-Ile may play roles that are distinct from other active jasmonates and thereby contribute to the diversity and specificity of jasmonate signaling.
METHODS
Plant Materials and Chemicals
Methyl jasmonate, linolenic acid, and the (±) JA used for enzymatic and root inhibition studies were from Sigma-Aldrich (St. Louis, MO). Methyl dihydrojasmonate was obtained from Bedoukian Research (Danburry, CT). JA and dihydrojasmonic acid (DHJA) used for synthesis of amino acid conjugates was produced by saponification from MeJA or methyl dihydrojasmonate according to Dathe et al. (1981). B. Vick and F. Schaller provided 12-oxo-phytodienoic acid and 3-oxo-cyclopentane octanoic acid, respectively, and 13-hydroperoxy-linolenic acid was purchased from Cayman Chemical (Ann Arbor, MI). Arabidopsis thaliana (Columbia) and genotypes jar1-1, jar1-8, and coi1-35 were previously described (Staswick et al., 2002). The ethylene insensitive mutant etr1-1 (Bleecker et al., 1988) was obtained from the ABRC (Columbus, OH). The double mutant jar1-1 etr1-1 was recovered from a cross using independent assays for resistance to both JA and to ACC.
Seedling Growth
Seedlings were grown on agar media containing the inhibitors indicated using surface sterilized seeds as previously described (Staswick et al., 2002). Plates containing seeds were incubated at 4°C for 4 d and then placed vertically in a culture chamber and grown 6 d at 23°C under 16-h fluorescent light/8-h dark cycles. Root lengths were measured, and inhibitor treatments were expressed as percentage of inhibition relative to the untreated control for each genotype. Confidence intervals (95%) were calculated using the delta method (n = 25 seedlings). Seedlings for extraction of jasmonates were grown similarly for 12 d and tissue excised from roots was frozen, ground to a powder, and stored at −80°C.
Synthesis of JA–Amino Acid Conjugates
Amino acid conjugates of JA were prepared by condensation reactions as outlined by Kramell et al. (1988, 1999). Reaction products were purified by silica gel chromatography (1.5 × 50 cm) in chloroform, ethyl acetate, acetic acid (14:6:1 [v/v]). Organic solvents were evaporated from the appropriate column fractions under a stream of nitrogen, and the conjugates were purified on RP18 SPE columns (Burdick and Jackson, Muskegon, MI; 500 mg/8 mL). JA-ACC was further purified by HPLC on a Luna 5-μm C18 reverse phase column (Phenomenex, Torrance, CA) with isocratic 80% MeOH and 0.2% acetic acid (1 mL/min). Residual ACC in JA-ACC was assayed by the method of Lanneluc-Sanson et al. (1986). Purity and structure of each conjugate was verified by GC-MS after derivatization with diazomethane. The mass spectra were consistent with values previously reported (Kramell et al., 1988), and JA comprised <0.25 mol % of each conjugate. JA-ACC had not previously been described, and the methyl derivative yielded a chromatographic peak between JA-Val and JA-Leu. The mass spectrum included the expected molecular ion of 307 m/z and major fragmentation ions of 83, 116, 125, 151, and 157 for both the chemically synthesized standard and the enzymatically produced conjugate. Dihydro-JA conjugates for internal standards were synthesized similarly by substituting DHJA for JA. JA conjugates used as standards for quantitation in plant extracts were derivatized with 2,3,4,5,6-pentafluorobenzyl bromide as described below, purified by HPLC, measured by weight after solvent removal, and then dissolved in isooctane to appropriate concentrations for GC-MS.
Isolation and Quantitation of Jasmonates from Tissue
Solvents were HPLC grade, and chloroform contained 1% ethanol as stabilizer. The general methods of Kramell et al. (2000) were followed with additions and modifications as described. Weighed frozen tissue (∼0.5 g) was added to 7 mL of 80% MeOH with DHJA, and its amino acid conjugates added as internal controls. Tissue was ground 1 min on high speed with a Tissue Tearor homogenizer (Biospec Products, Bartlesville, OK) and then centrifuged 5 min at 6000 rpm. DEAE A-25 Sephadex (Sigma-Aldrich) was equilibrated in 0.5 M sodium acetate and then washed thoroughly with 80% methanol. Ion exchange was performed with a 4-mL bed volume of DEAE resin in a 10-mL disposable syringe with a filter paper frit to retain the resin. After sample loading, columns were washed with 14 mL of 100% MeOH and then eluted with 14 mL of 1 M acetic acid in MeOH. The eluate was dried in a stream of nitrogen at 40°C. Residue was dissolved in 1.2 mL of MeOH 0.2% acetic acid to which 4.8 mL of water was then added. The sample was loaded onto an SPE RP C18 column (Burdick and Jackson; 500 mg/8 mL) equilibrated in 20% MeOH and 0.2% acetic acid. The column was washed with 6 mL of 20% MeOH and 0.2% acetic acid and then eluted with 6 mL of 80% MeOH and 0.2% acetic acid.
The sample was dried and then derivatized in 50 μL of acetone with 1 μL of 1-ethylpiperidine and 5 μL of 2,3,4,5,6-pentafluorobenzyl bromide for 45 min at 65°C as described (Epstein and Cohen, 1981). This derivatization was found to enhance sensitivity for GC-MS analysis compared with the more standard derivatization with diazomethane. After drying, the residue was applied to a silica column (Burdick and Jackson; 300 mg/2.5 mL) equilibrated in 0.2% acetic acid in chloroform. The column was eluted with 0.2% acetic acid in ethyl acetate. The dried residue was dissolved in MeOH, and fractions were separated by HPLC as described earlier. Fractions containing conjugates and internal standards were dried and resuspended in isooctane for analysis by GC-MS. Analysis was done on an HP6890 gas chromatograph with a DB-5ht column (15 m × 0.25 mm, 0.1 μ) from J.W. Scientific (Folsom, CA). The injector port was 280°C, and the column gradient was 60 to 300°C in 25 min. Detection was with a HP5973 EI mass selective detector operated in SIM mode. Quantitative data was obtained from integrated peak areas for the respective molecular ion of each compound and comparison with a standard curve. Recovery of DHJA conjugate internal standards was typically 50 to 80%.
RNA Analysis
Total RNA was isolated from leaf tissue of 5-week-old plants grown under a 12-h/12-h (light/dark) cycle. Procedures outlined in the instructions supplied with the Qiagen (Valencia, CA) RNeasy plant mini kit were followed, the resulting RNA was electrophoresed, and RNA gel blot hybridizations were performed as previously described (Tiryaki and Staswick, 2002). Although Arabidopsis contains up to 18 other genes encoding related proteins, the nucleic acid identity of the JAR1 coding sequence with the related transcripts was ≤55%. Thus, the full-length JAR1 cDNA probe is not expected to detect other transcripts under the hybridization stringency employed here. The AtVSP cDNA probe was obtained as described previously (Tiryaki and Staswick, 2002). MeJA treatment was by spraying a 50-μM aqueous solution on plants. Leaf tissue was harvested at indicated times and immediately frozen.
Plant Transformation
The full-length cDNA corresponding to JAR1 was obtained by RT-PCR and inserted in frame with the tobacco etch virus translational leader into the Cauliflower mosaic virus 35S-driven cassette in pRTL2. The 35S-JAR1 construct was then introduced into pZP212 and transformed into jar1-1 as described previously (Clough and Bent, 1998; Staswick et al., 2002). Transformants were screened for their conversion to the wild-root phenotype when assayed on media containing 50-μM MeJA, and homozygous lines were subsequently identified similarly. The genotype of these transformants was confirmed with a cleaved-amplified polymorphic sequence marker for the jar1-1 mutation.
Enzyme Assays
The wild-type GST-JAR1 fusion protein was produced as described previously (Staswick et al., 2002). The gene coding sequence for the mutant allele jar1-1 was recovered by RT-PCR from mRNA obtained from 3-week-old seedlings using the same primers as for the wild-type gene. The fusion protein was expressed from the pGEX4-T-1 vector in Escherichia coli (BL21). Cell sonication and protein purification on glutathione agarose were done in 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, pH 7.3, according to the manufacturer's protocols (Amersham Pharmacia, Uppsala, Sweden). Fusion proteins recovered in the supernatant were analyzed by SDS-PAGE as described previously (Staswick et al., 2002).
Isotope exchange assays were performed as previously described (Staswick et al., 2002) using glutathione agarose-bound enzyme at ∼0.17 μg/μL. Radioactivity was determined by cutting the appropriate area from the TLC sheet and counting by liquid scintillation. Assays for conjugate formation were done in 50 mM Tris-HCl, pH 8.6, 3 mM MgCl2, 3 mM ATP, 1 mM DTT, 1 mM JA, and 1 mM amino acid. Reaction products were analyzed on silica gel 60 F260 TLC plates (EM Sciences, Gibbstown, NJ) developed in 35% chloroform, 55% ethyl acetate, and 10% formic acid and then stained with vanillin reagent (6% vanillin [w/v] and 1% sulfuric acid [v/v] in ethanol). For comparison of the activity of mutant and wild-type JAR1, each enzyme was proteolytically cleaved from GST and used in soluble form (64 to 82 μg/mL) in the same assay as above with JA and Ile as substrates. The mean of three assays is reported with standard errors. Products were extracted with chloroform, derivatized with diazomethane, and then quantified by GC-MS.
Supplementary Material
Acknowledgments
The generous advice of J. Cohen regarding jasmonate quantitation is gratefully acknowledged. D. Stanley and S. Putnam are also thanked for providing the GC-MS and helpful advice. We thank B. Vick and F. Schaller for their gift of 12-oxo-phytodienoic acid and 3-oxo-cyclopentane octanoic acid, respectively. This is Journal Series Paper number 14288, a contribution of the University of Nebraska Agricultural Research Division, Lincoln, NE 68583. Funding was provided by the Nebraska Research Initiative and the National Science Foundation (Award MCB-0130868).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Paul E. Staswick (pstaswick1@unl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023549.
References
- Andrea, W.A., and Good, N.E. (1955). The formation of indoleacetyl aspartic acid in pea seedlings. Plant Physiol. 30, 380–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A.B., Estelle, M.A., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene is conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conconi, A., Smerdon, M.J., Howe, G.A., and Ryan, C.A. (1996). The octadecanoid signaling pathway in plants mediates a response to ultraviolet radiation. Nature 383, 826–829. [DOI] [PubMed] [Google Scholar]
- Conti, E., Stachelhaus, T., Marahiel, M.A., and Brick, P. (1997). Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16, 4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1995). Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 92, 4114–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe, W., Ronsch, H., Preiss, A., Schade, W., Sembdner, G., and Schreiber, K. (1981). Endogenous plant hormones of the broad bean, Vicia faba L. (-)- jasmonic acid, a plant growth inhibitor in pericarp. Planta 153, 530–535. [DOI] [PubMed] [Google Scholar]
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, E., and Cohen, J.D. (1981). Microscale preparation of pentafluorbenzyl esters. Electron-capture gas chromatographic detection of indole-3-acetic acid from plants. J. Chromatogr. 209, 413–420. [Google Scholar]
- Farmer, E.E., and Ryan, C.A. (1990). Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 87, 7713–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann, J., Schuler, G., Boland, W., Ebel, J., and Mithofer, A. (2003). The role of octadecanoids and functional mimics in soybean defense responses. Biol. Chem. 384, 437–446. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2003). Cross-talk in plant hormone signaling: What Arabidopsis mutants are telling us. Ann. Bot. 91, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach, H., and Zenk, M.H. (1998). Biological activity and biosynthesis of pentacyclic oxylipins: The linoleic acid pathway. Phytochemistry 47, 527–537. [Google Scholar]
- Hagen, G., Martin, G., Li, Y., and Guilfoyle, T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. [DOI] [PubMed] [Google Scholar]
- Haider, G., von Schrader, T., Fusslein, M., Blechert, S., and Kutchan, T.M. (2000). Structure-activity relationships of synthetic analogs of jasmonic acid and coronatine on induction of benzo[c]phenanthridine alkaloid accumulation in Eschscholzia californica cell cultures. Biol. Chem. 381, 741–748. [DOI] [PubMed] [Google Scholar]
- Hause, B., Maier, W., Miersch, O., Kramell, R., and Strack, D. (2002). Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol. 130, 1213–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: Allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24, 113–126. [DOI] [PubMed] [Google Scholar]
- Jakubowska, A., Kowalczyk, S., and Leznicki, A. (1993). Enzymatic hydrolysis of 4-O- and 6-O-indol-3-ylacetyl-β-D-glucose in plant tissues. J. Plant Physiol. 142, 61–66. [Google Scholar]
- Kessler, A., and Baldwin, I.T. (2002). Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328. [DOI] [PubMed] [Google Scholar]
- Kleinkauf, H., and Von Dohren, H. (1996). A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236, 335–351. [DOI] [PubMed] [Google Scholar]
- Koda, Y., Takahashi, K., Kikuta, Y., Greulich, F., Toshima, H., and Ichihara, A. (1996). Similarities of the biological activities of coronatine and coronafacic acid to those of jasmonic acid. Phytochemistry 41, 93–96. [Google Scholar]
- Kramell, R., Atzorn, R., Schneider, G., Miersch, O., Bruckner, C., Schmidt, G., and Parthier, B. (1995). Occurrence and identification of jasmonic acid and its amino acid conjugates induced by osmotic stress in barley leaf tissue. J. Plant Growth Regul. 14, 29–36. [Google Scholar]
- Kramell, R., Miersch, O., Atzorn, R., Parthier, B., and Wasternack, C. (2000). Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves. Implications for different signaling pathways. Plant Physiol. 123, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramell, R., Miersch, O., Schneider, G., and Wasternack, C. (1999). Liquid chromatography of jasmonic acid amine conjugates. Chromatographia 49, 42–46. [Google Scholar]
- Kramell, R., Schmidt, J., Schneider, G., Sembdner, G., and Schreiber, K. (1988). Synthesis of N-(jasmonyl)amino acid conjugates. Tetrahedron 44, 5791–5807. [Google Scholar]
- Krumm, T., Bandemer, K., and Boland, W. (1995). Induction of volatile biosynthesis in the lima bean (Phaseolus lunatus) by leucine- and isoleucine conjugates of 1-oxo- and 1-hydroxyindan-4-carboxylic acid: Evidence for amino acid conjugates of jasmonic acid as intermediates in the octadecanoid signaling pathway. FEBS Lett. 377, 523–529. [DOI] [PubMed] [Google Scholar]
- Lanneluc-Sanson, D., Phan, C.T., and Granger, R.L. (1986). Analysis by reverse-phase high-pressure liquid chromatography of phenylisothiocyanate-derivatized 1-aminocyclopropane-1-carboxylic acid in apple extracts. Anal. Biochem. 155, 322–327. [DOI] [PubMed] [Google Scholar]
- Li, L., Zhao, Y., McCaig, B.C., Wingerd, B.A., Wang, J., Whalon, M.E., Pichersky, E., and Howe, G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon, T.A., Fernandez-Maculet, J.C., and Yang, S. (1995). Biosynthesis and metabolism of ethylene. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 118–139.
- Miersch, O., Weichert, H., Stenzel, I., Hause, B., Maucher, H., Feussner, I., and Wasternack, C. (2004). Constitutive overexpression of allene oxide cyclase in tomato (Lycopersicon esculentum cv. Lukullus) elevates levels of some jasmonates and octadecanoids in flower organs but not in leaves. Phytochemistry 65, 847–856. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J. (1997). Enzymes involved in jasmonic acid biosynthesis. Physiol. Plant 100, 653–663. [Google Scholar]
- Normanly, J. (1997). Auxin metabolism. Physiol. Plant 100, 431–442. [Google Scholar]
- Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H., and Kangasjärvi, J. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12, 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, R.J., Mhaskar, S.V., Lin, M.-T., Walker, A.E., and Mafoti, R. (1994). Investigations of the biosynthesis of the phytotoxin coronatine. Can. J. Chem. 72, 86–99. [Google Scholar]
- Rao, M.V., Lee, H., Creelman, R.A., Mullet, J.E., and Davis, K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto, F.F., Klee, H., White, F., Nordeen, R., and Kosuge, T. (1990). Expression and fine structure of the gene encoding N epsilon-(indole-3-acetyl)-L-lysine synthetase from Pseudomonas savastanoi. Proc. Natl. Acad. Sci. USA 87, 5797–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz, E.A., Alborn, H.T., and Tumlinson, J.H. (2003). Synergistic interactions between volicitin, jasmonic acid and ethylene mediate insect-induced volatile emission in Zea mays. Physiol. Plant 117, 403–412. [DOI] [PubMed] [Google Scholar]
- Sembdner, G., and Parthier, B. (1993). The biochemistry and the physiological molecular actions of jasmonates. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 569–589. [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98, 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena, A., Prinsen, E., Fladung, M., Schulze, S.C., and Van Onckelen, H. (1991). The indoleacetic acid-lysine synthetase gene of Pseudomonas syringae subsp. savastanoi induces developmental alterations in transgenic tobacco and potato plants. Mol. Gen. Genet. 227, 205–212. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Yuen, G.Y., and Lehman, C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 16, 747–754. [DOI] [PubMed] [Google Scholar]
- Stelmach, B.A., Muller, A., Hennig, P., Laudert, D., Andert, L., and Weiler, E.W. (1998). Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry 47, 539–546. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen, J.B., Szczyglowski, K., and Bandurski, R.S. (1994). iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265, 1699–1701. [DOI] [PubMed] [Google Scholar]
- Tamogami, S., Rakwal, R., and Kodama, O. (1997). Phytoalexin production by amino acid conjugates of jasmonic acid through induction of naringenin-7-O-methyltransferase, a key enzyme on phytoalexin biosynthesis in rice (Oryza sativa L.). FEBS Lett. 401, 239–242. [DOI] [PubMed] [Google Scholar]
- Tiryaki, I., and Staswick, P.E. (2002). An Arabidopsis thaliana mutant defective in jasmonate response is allelic to the auxin signaling mutant axr1. Plant Physiol. 130, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.), S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C., Bakker, P.A.H.M., and Pieterse, C.M.J. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Plant Physiol. Plant Mol. Biol. 36, 453–483. [DOI] [PubMed] [Google Scholar]
- Vick, B.A., and Zimmerman, D.C. (1987). Oxidative systems for the modification of fatty acids. In The Biochemistry of Plants: Lipids. P. Stumph and E. Conn, eds (New York: Academic Press), pp. 53–90.
- Wang, K.L., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl), S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- Weber, H., Vick, B.A., and Farmer, E.E. (1997). Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 94, 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, E.W., Albrecht, T., Groth, B., Xia, Z.-Q., Luxem, M., Li, H., Andert, L., and Spengler, P. (1993). Evidence for the involvement of jasmonates and their octadecanoid precursors in the tendril coiling response of Bryonia doica. Phytochemistry 32, 591–600. [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yang, S.F., and Hoffman, N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.