Abstract
DNA insertional transformants of Chlamydomonas reinhardtii were screened chemochromically for attenuated H2 production. One mutant, displaying low H2 gas photoproduction, has a nonfunctional copy of a gene that shows high homology to the family of isoamylase genes found in several photosynthetic organisms. DNA gel blotting and gene complementation were used to link this isoamylase gene to previously characterized nontagged sta7 mutants. This mutant is therefore denoted sta7-10. In C. reinhardtii, the STA7 isoamylase gene is important for the accumulation of crystalline starch, and the sta7-10 mutant reported here contains <3% of the glucose found in insoluble starch when compared with wild-type control cells. Hydrogen photoproduction rates, induced after several hours of dark, anaerobic treatment, are attenuated in sta7 mutants. RNA gel blot analysis indicates that the mRNA transcripts for both the HydA1 and HydA2 [Fe]-hydrogenase genes are expressed in the sta7-10 mutant at greater than wild-type levels 0.5 h after anaerobic induction. However, after 1.5 h, transcript levels of both HydA1 and HydA2 begin to decline rapidly and reach nearly undetectable levels after 7 h. In wild-type cells, the hydrogenase transcripts accumulate more slowly, reach a plateau after 4 h of anaerobic treatment, and maintain the same level of expression for >7 h under anaerobic incubation. Complementation of mutant cells with genomic DNA corresponding to the STA7 gene restores both the starch accumulation and H2 production phenotypes. The results indicate that STA7 and starch metabolism play an important role in C. reinhardtii H2 photoproduction. Moreover, the results indicate that mere anaerobiosis is not sufficient to maintain hydrogenase gene expression without the underlying physiology, an important aspect of which is starch metabolism.
INTRODUCTION
The search for alternative energy resources is necessitated by the environmental and political concerns associated with fossil fuel consumption and depletion. The benefits of hydrogen as an energy carrier have long been recognized. Hydrogen can be combusted or used with fuel cell technologies to provide power with only water as the end by-product. Several microorganisms, including the green alga Chlamydomonas reinhardtii, hold significant promise as H2 producers (Weaver et al., 1980; Benemann, 1996; Hansel and Lindblad, 1998; Asada and Miyake, 1999; Ghirardi et al., 2000; Melis and Happe, 2001; Boichenko et al., 2004). Biological hydrogen production is mediated either by the hydrogenase enzymes found in several microbes or by the nitrogenase enzymes found in a variety of prokaryotes (Weaver et al., 1980; Vignais et al., 1985, 2001; Boichenko and Hoffmann, 1994; Tamagnini et al., 2002; Boichenko et al., 2004).
Hydrogen production by green algae was first reported using Scenedesmus obliquus in seminal experiments performed by Gaffron and Rubin (1942). Several additional strains of green algae, including C. reinhardtii, have subsequently been shown to produce H2 (Weaver et al., 1980; Brand et al., 1989; Boichenko et al., 2004). The highest rates of H2 production are typically observed in the light after anaerobic induction (Gaffron and Rubin, 1942; Gfeller and Gibbs, 1984). However, H2 photoproduction is not sustainable in the light unless the O2, coproduced by photosynthesis, is continually removed from the medium to prevent it from inactivating the reaction (Greenbaum, 1988; Ghirardi et al., 1997). Electrons for H2 photoproduction are supplied by the photosynthetic electron transport chain, originating either from water oxidation by photosystem II (PSII) and/or from the metabolic oxidation of endogenous substrate in the chloroplast via its attendant electron flow to the plastoquinone (PQ) pool. Hydrogen production is also observed during fermentative algal metabolism in the dark, albeit at much lower rates (Gaffron and Rubin, 1942; Miura et al., 1982; Gfeller and Gibbs, 1984).
In C. reinhardtii, two [Fe]-hydrogenase enzymes, HydA1 and HydA2, have been reported (Happe and Naber, 1993; Happe et al., 2002; Forestier et al., 2003). The HydA1 enzyme has been purified and assayed (Roessler and Lien, 1984; Happe and Naber, 1993; Happe et al., 1994) and is located in the chloroplast stroma. Purification and characterization of the HydA2 enzyme has not been reported. Algal [Fe]-hydrogenases contain only the catalytic H-cluster (Melis and Happe, 2001; Happe and Kaminski, 2002) and are thus distinguished from the corresponding proteins found in certain anaerobic bacteria (e.g., Clostridium pasteurianum [Peters et al., 1998; Nicolet et al., 1999]), which contain accessory iron sulfur clusters.
A new experimental approach to produce and accumulate H2 gas from C. reinhardtii was reported recently (Ghirardi et al., 2000; Melis et al., 2000; Melis and Happe, 2001; Kosourov et al., 2002, 2003; Zhang et al., 2002). When deprived of sulfate nutrients, the activity of PSII in C. reinhardtii declines (Wykoff et al., 1998) to the point where O2 consumption by respiration is greater than the rate of photosynthetic O2 evolution (Melis et al., 2000; Kosourov et al., 2002). Sealed cultures under these conditions become anaerobic in the light and produce H2 gas for several days.
To gain a better fundamental understanding of the underlying biochemical pathways and metabolic conditions that promote H2 production in green algae, we screened a DNA insertional mutagenesis library for strains lacking the ability to produce H2, after anaerobic induction. Screening DNA insertional mutagenesis libraries in C. reinhardtii has become a popular strategy for identifying important genes involved in specific cellular pathways and processes (Debuchy et al., 1989; Rochaix, 1995; Tam and Lefebvre, 1995; Niyogi et al., 1997; Adam and Loppes, 1998; Davies et al., 1999; Moseley et al., 2000; Van et al., 2001; Dame et al., 2002; Polle et al., 2003). Our screening efforts resulted in the identification of several mutants that were compromised in their ability to photoproduce H2. The characterization of one of these mutants, sta7-10, revealed the disruption of a gene with high homology to the isoamylase gene family, a class of enzymes found in a variety of photosynthetic organisms. This enzyme is critical for the formation of insoluble starch in C. reinhardtii (Ball et al., 1996; Mouille et al., 1996; Ball, 1998; Myers et al., 2000; Dauvillee et al., 2001b). The mutant reported here stores <3% of the insoluble starch compared with the wild-type strain. Moreover, the mutant's ability to photoproduce H2 and maintain hydrogenase transcription after anaerobic induction is attenuated. Complementation of the sta7-10 mutant, with genomic DNA corresponding to the wild-type isoamylase gene, leads to the recovery of both starch and H2 production, confirming the linkage between the two phenotypes. We also demonstrate that the isoamylase gene identified from our mutant is allelic to previously characterized sta7 mutants, which were not tagged (Mouille et al., 1996; Dauvillee et al., 2001b). Additionally, we show that another previously characterized starch mutant, sta6 (BAFJ5), also has severely reduced H2 photoproduction rates and decreased hydrogenase gene transcription.
RESULTS
Mutant Isolation and Characterization
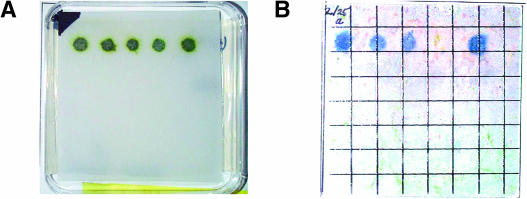
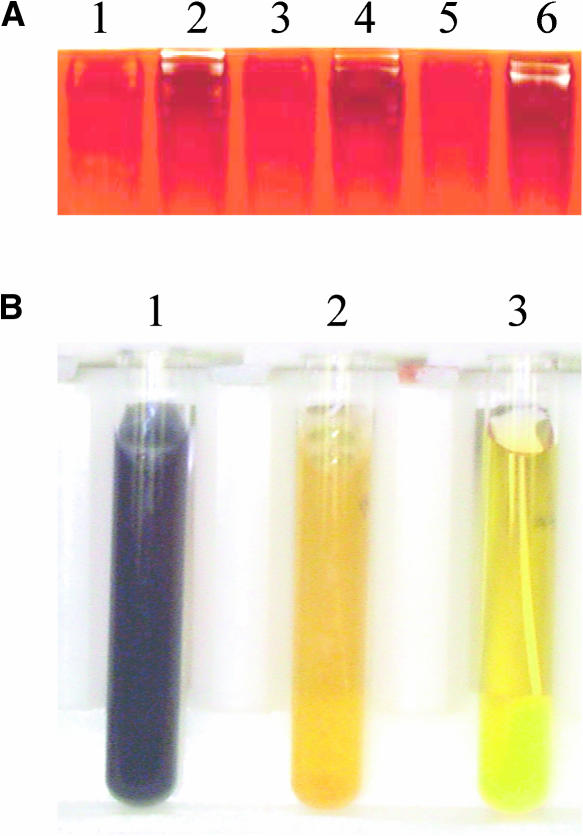
To identify mutants attenuated in the photoproduction of H2, we used a DNA insertional mutagenesis library, generated by transformation of C. reinhardtii strain CC425 with the plasmid pJD67 (Davies et al., 1999). This library was screened for H2 production using chemochromic sensors (Seibert et al., 2001b; Flynn et al., 2002; Posewitz et al., 2004). Figure 1 shows results typical of the screens, in which mutants were identified by their inability to produce sufficient amounts of H2 for detection by the sensor. As shown in Figure 1B, the fourth colony from the left is unable to generate H2 in sufficient quantities for detection. The dark blue spots observed for the other four colonies from the same library are indicative of H2 production.
Figure 1.
Chemochromic Detection of C. reinhardtii Colonies Deficient in the Photoproduction of H2.
(A) Colonies grown on a TAP agar plate.
(B) Chemochromic sensor after illumination of candidate colonies on the TAP agar plate.
Hydrogenase activity was induced by placing the plates in the dark under a N2 atmosphere overnight. Subsequent actinic illumination resulted in the photoproduction of H2, which is visually detected on the sensor as a dark blue spot. The fourth colony from the left failed to produce H2 and thus was the subject of further investigation.
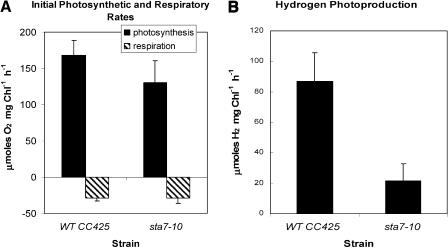
The sta7-10 mutant, identified by chemochromic screening, was successfully grown on minimal media agar plates to verify that the cells were photosynthetically competent. Photosynthetic and respiratory rates of both the parental and mutant strains were measured in liquid media using a Clark-type electrode (Figure 2). Compared with the wild type, the sta7-10 mutant exhibits normal rates of respiration and slightly lower rates of photosynthetic O2 evolution (Figure 2A). Under the assay conditions used (see Methods), the initial rates of H2 photoproduction were attenuated to 20% to 40% of that in the wild type (Figure 2B). Attenuated H2 photoproduction rates were also observed when the sta7-10 mutant was cultured in Tris-acetate-phosphate (TAP) medium containing Arg, eliminating the possibility that this amino acid, which is required to grow strain CC425, is responsible for the observed differences in H2 production.
Figure 2.
Initial Rates of Photosynthesis (O2 Evolution), Respiration (O2 Uptake), and H2 Photoproduction.
(A) Measured rates of photosynthesis and respiration.
(B) Measured rates of H2 production.
Initial rates were measured using a Clark electrode apparatus. Results are shown for both the background strain CC425 and the sta7-10 mutant. The rates of respiration of the two strains are similar, whereas the rate of O2 evolution is slightly lower in the sta7-10 mutant. Hydrogen photoproduction rates are substantially attenuated in the sta7-10 mutant. All samples were induced as described in Methods.
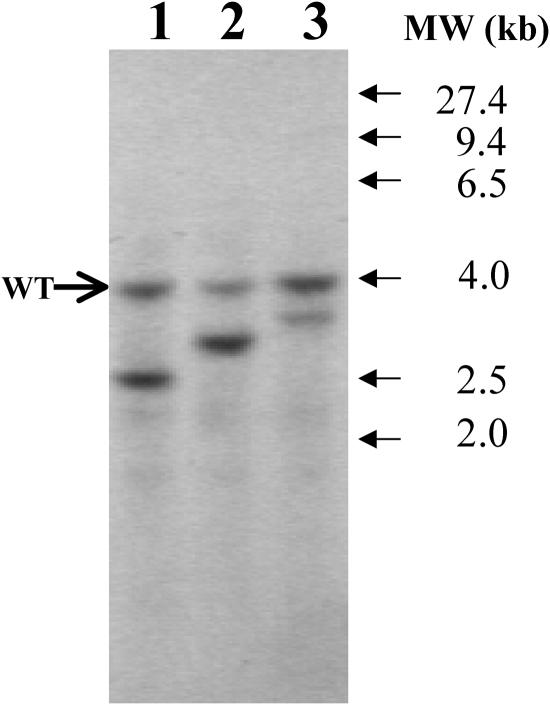
DNA gel blot analysis was performed to determine the number of pJD67 plasmid insertions in this mutant. Genomic DNA was digested with the PstI restriction enzyme and probed using the 130-bp StuI-SphI fragment from the Arg7 gene. This probe contains little redundant genomic sequence and is specific for Arg7 (Debuchy et al., 1989; Adam and Loppes, 1998). As shown in Figure 3, the sta7-10 mutant exhibits one additional site of hybridization in addition to the nonfunctional wild-type copy of the arg7 gene. The other mutants shown in Figure 3 are included to confirm the electrophoretic mobility of the nonfunctional wild-type copy band and to demonstrate that plasmid insertions occur randomly within the genomic DNA, as evidenced by the variable electrophoretic mobility of the different insert bands. These results indicate that the sta7-10 mutant probably contains only one site of pJD67 plasmid insertion, and efforts were undertaken to determine the genomic DNA sequence flanking the pJD67 insertion.
Figure 3.
Genomic DNA Gel Blot for Arg7 Insertion.
Genomic DNA isolated from mutants attenuated in H2 photoproduction was digested with the restriction enzyme PstI. Lane 1 corresponds to DNA isolated from the sta7-10 mutant. The blot was probed using the 130-bp StuI-SphI fragment of the Arg7 gene. The band at 4.0 kb corresponds to the nonfunctional wild-type copy of the arg7 gene, and the band at 2.5 kb corresponds to a single copy of the Arg7 plasmid insert that caused the mutation. Lanes 2 and 3 correspond to other mutants of interest from the same library and demonstrate that, in addition to the wild-type band, one additional site of insertion is observed in these mutants as well.
Gene Identification
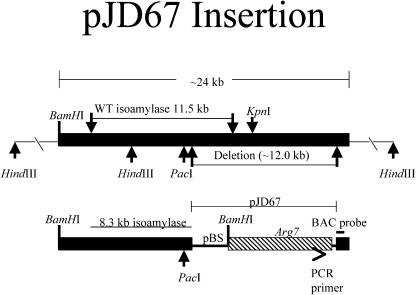
Genome walking PCR and plasmid rescue strategies were used to obtain the genomic DNA sequence flanking the insertion (see Methods). Products from these experiments were sequenced and revealed a disrupted gene with highest homology to isoamylase genes found in several photosynthetic organisms. The integration of pJD67 into the genome of the sta7-10 mutant, the location of the BamHI sites used for plasmid rescue, and the location of the PCR primer used in genome walking are illustrated in Figure 4. To obtain the genomic DNA sequence deleted by the pJD67 insertion, a commercially available C. reinhardtii genomic library was screened, using a probe obtained from the genome walking PCR reaction (Figure 4, BAC probe). The screening revealed 10 positive hybridization clones from a library that is approximately eightfold redundant. Four of the identified BAC clones were obtained, and the presence of a sequence corresponding to the isoamylase gene was confirmed by PCR (data not shown). One of the clones was used to obtain the genomic STA7 sequence deleted in the sta7-10 mutant. Restriction mapping and sequencing indicate that a large fragment of genomic DNA (∼12.0 kb) was deleted upon insertion of the pJD67. The map in Figure 4 illustrates the location of selected restriction enzyme sites and the genomic region deleted by the pJD67 insertion. Most of the pJD67 plasmid remained intact, with the exception of deletions at the ends of the linearized plasmid: ∼200 bp of the pBluescript KS+ plasmid upstream of the Arg7 gene and ∼50 bp at the 3′ end of pJD67 were missing.
Figure 4.
Insertion Map of pJD67 into the C. reinhardtii Genomic DNA of the sta7-10 Mutant.
The plasmid pJD67 was linearized by digestion with the restriction enzyme HindIII and transformed into C. reinhardtii strain CC425 using the glass bead method. Integration of pJD67 resulted in the deletion of ∼12 kb of genomic DNA, of which 3.2 kb was located at the 3′ end of the wild-type STA7 gene. This resulted in a 3′ end-truncated and, therefore, nonfunctional copy of the isoamylase gene. Locations of select restriction enzyme sites, the PCR primer used in genome walking, and the probe used to assay the BAC library are also shown.
The STA7 cDNA coding sequence was obtained in three separate reverse-transcription reactions, using three unique gene-specific primer sets that provided overlapping sequence. These products were PCR-amplified and sequenced, and used to deduce the protein coding sequence of the STA7 gene. The gene encodes an 875–amino acid protein. The genomic sequence is 65% GC rich and contains 21 exons. Figure 5 shows an alignment of the protein sequence translated from the cDNA of STA7 with the three isoamylase isoforms identified from Solanum tuberosum (Hussain et al., 2003).
Figure 5.
Alignment of the C. reinhardtii Isoamylase Amino Acid Sequence with Three Isoamylase Isoforms from S. tuberosum.
Regions of identical amino acids are shown in black and homology regions are shaded in gray. Included in this figure are the C. reinhardtii STA7 protein (STA7) and three separate isoamylase isoforms from S. tuberosum (Stisa1, Stisa2, and Stisa3). The S. tuberosum isoamylase isoforms are ordered according to homology with the C. reinhardtii STA7 protein. The STA7 protein is 54% identical and 65% similar to Stisa1, 42% identical and 56% similar to Stisa3, and 34% identical and 48% similar to Stisa2.
Starch Characterization
The STA7 enzyme is involved in starch metabolism, and, in C. reinhardtii, it is necessary for the accumulation of insoluble starch (Ball et al., 1996; Mouille et al., 1996; Ball, 1998; Myers et al., 2000; Dauvillee et al., 2001b). To verify that the sta7-10 mutant was compromised in the accumulation of starch, glucose levels were assayed from the insoluble starch pellet (see Methods) and shown to be <3% of those found in the CC425 background strain (data not shown). To further assess the role of this putative isoamylase enzyme in starch metabolism, cells were grown to early stationary phase and used in zymograms to detect isoamylase activity. Figure 6A shows an iodine-stained native protein gel containing glycogen. The top of the gel has several bands, visualized as light bands on the dark background, corresponding to heteromeric protein complexes capable of degrading glycogen. Two prominent bands are seen for CC425 (lane 2) in this zymogram; however, depending on gel-incubation times and digital imaging contrasts, up to five bands may be detected. By contrast, the mutant lane (lane 1) contains only very faint bands. This is in agreement with earlier studies using zymograms to assess the activity of starch enzymes in C. reinhardtii, which demonstrated that isoamylase mutants in C. reinhardtii are compromised in their ability to form these high molecular weight heteromeric complexes (Dauvillee et al., 2001b). The in vitro phenotype of the sta7-10 mutant is therefore the inability to degrade glycogen; however, the in vivo phenotype is attenuated starch formation. These results are consistent with the biochemical characterization of previously isolated C. reinhardtii STA7 mutants (Mouille et al., 1996; Ball, 1998; Myers et al., 2000).
Figure 6.
Differential Starch Metabolism between the sta7-10 Mutant and the CC425 Background Strain.
(A) Zymogram illustrating isoamylase activity.
(B) Color-based assay of the starch levels.
The zymogram in (A) illustrates the activity of heteromeric complexes containing isoamylase that are capable of breaking down glycogen. Crude cellular extracts were centrifuged, and the supernatant was loaded onto a 7.5% acrylamide native gel containing glycogen. An iodine solution was used to stain areas still containing glycogen after incubation. Lane 1 illustrates the breakdown of starch by the sta7-10 mutant and lane 2, the control CC425. The remaining lanes demonstrate identical alternate samples of the sta7-10 mutant (lanes 3 and 5) and CC425 (lanes 4 and 6). Areas of glycogen breakdown are shown as white bands on a dark background. Assay of starch levels contained in cell extracts stained with iodine solution is shown in (B). The CC425 (tube 1) extract stains a dark purple indicative of high starch levels. By contrast, the sta7-10 mutant (tube 2) remains a light yellow similar to the negative control with no cells (tube 3), indicating a substantially attenuated starch concentration.
The same cultures were also used to assess starch levels using iodine staining. The cells were harvested, the pigments extracted twice with ethanol, and the pellet resuspended in water and autoclaved to solubilize the starch. An iodine solution was then added to the solution, and it was visually inspected for staining. As shown in Figure 6B, the CC425 background strain (tube 1) turns a dark purple color indicative of high starch levels. The mutant (tube 2) remains a light yellow color, indicative of low starch levels, similar in intensity to the control in which no cells were added (tube 3).
STA7 Gene Complementation
To link the H2 production phenotype to pJD67 insertion, we used gene complementation. The plasmid pMP25 contains DNA corresponding to the full-length isoamylase gene (Figure 4) and also contains the Ble gene, which confers resistance to the antibiotic zeocin. The sta7-10 mutant was transformed with either pMP25 or pSP124S, which contains only the Ble gene, in separate transformations. Colonies that grew on zeocin-containing TAP plates after transformation with either pMP25 or pSP124S were analyzed for starch levels using an iodine solution. The ratios of colonies with wild-type starch levels versus colonies that grew on zeocin plates were 10:95 and 0:42 for transformation with pMP25 or pSP124S, respectively. The low number of colonies with both wild-type starch levels and zeocin resistance after transformation with pMP25 (10.5%) may be a consequence of the large size of pMP25. The isoamylase gene represents >11 kb of DNA on this plasmid, whereas the Ble portion constitutes only 1.3 kb. It is possible that vortexing with glass beads during the transformation protocol resulted in sheared DNA or that the integration and proper expression of the STA7 gene is not as efficient as the integration and expression of the Ble gene. Elrad et al. (2002) recently reported mutant complementation frequencies between 30% and 40% after transformation of the npq5 mutant using a DNA construct containing both the Ble gene and the C. reinhardtii Lhcbm1 gene, which was 4.4 kb.
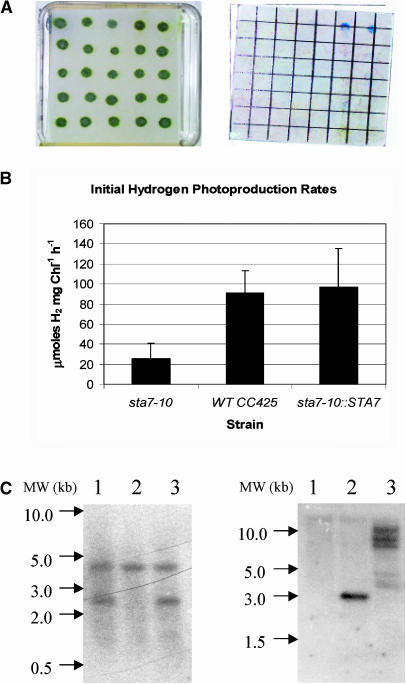
The 10 colonies that showed complementation of the starch defect were grown on TAP plates and screened for H2 production using chemochromic sensors. The original sta7-10 mutant and four colonies, which displayed no starch complementation, were also placed on the plate. All colonies that showed starch complementation exhibited recovered H2 production capacity and were competent to turn the chemochromic sensors blue. The other five colonies displayed no H2 production (data not shown). Additionally, a zeocin-containing plate with 25 potentially complemented colonies was chemochromically screened. Only two colonies were shown to produce H2 using the sensors (Figure 7A). Starch analysis demonstrated that only the two colonies capable of producing H2 contained wild-type starch levels.
Figure 7.
Complementation of the sta7-10 Background with STA7 Genomic DNA.
(A) Algal colonies on TAP plates after complementation with the STA7 gene (left) and chemochromic sensors after illumination (right).
(B) Hydrogen production rates.
(C) DNA gel blot analysis.
The chemochromic sensor (A) indicates that only two colonies at the far right of the first row are able to produce sufficient quantities of H2 for detection. These two clones were also the only colonies with wild-type levels of starch. Hydrogen-production rates (B) in solution measured using a Clark electrode apparatus. The mutant sta7-10 strain shows rates that are 20% to 40% those of the wild-type CC425. Mutant background complemented with STA7 (sta7-10::STA7) shows H2-production rates similar to CC425. DNA gel blot analysis of PstI-digested genomic DNA probed using the 130-bp /StuI-SphI fragment from the Arg7 gene ([C], left). DNA extracted from sta7-10, CC425, and sta7-10::STA7 cultures are shown in lanes 1, 2, and 3, respectively. DNA gel blot analysis of NotI-digested genomic DNA probed with the 114-bp KpnI-SacI fragment from the 3′ end of the genomic STA7 gene ([C], right). DNA samples are as described for the Arg7 blot.
To verify that the H2 photoproduction rates of the complemented clones were similar to wild-type cells in solution, two of the complemented mutants were grown in TAP media, and their initial H2 production rates were assayed using a Clark electrode. The rates for the complemented clones were comparable to the CC425 rates and were 2.5 to 5 times greater than the H2 photoproduction rates observed for the sta7-10 mutant (Figure 7B). One of the putatively complemented clones was probed by DNA gel blotting for the Arg7 insertion to verify that it contained the STA7 disruption. As shown in Figure 7C (left), the complemented clone contains both the endogenous nonfunctional arg7 and the pJD67 insertion found in the sta7-10 mutant, confirming that the mutant background was complemented. A separate blot was probed for the STA7 gene and showed three distinct bands of different electrophoretic mobility, verifying multiple integration of STA7 (Figure 7C, right).
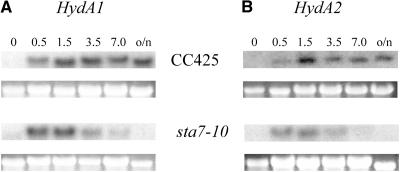
RNA Gel Blot Analysis
RNA gel blot analysis was performed with aliquots obtained at various time points after hydrogenase induction to determine the effect of the isoamylase gene disruption on the levels of HydA1 and HydA2 mRNA accumulation during anaerobic induction. Figure 8A compares the HydA1 expression profiles from CC425 and sta7-10 as a function of anaerobic treatment time, and Figure 8B shows the same comparison for HydA2 expression. In CC425, hydrogenase mRNA reaches maximal levels at ∼1.5 h after dark, anaerobic treatment, and both HydA1 and HydA2 maintain nearly steady state levels after overnight treatment. By contrast, the hydrogenase mRNA levels in the sta7-10 mutant reach maximal levels shortly after induction (∼0.5 h) and then steadily decay to almost undetectable levels by 7 h after anaerobic induction. These data clearly illustrate that the sta7-10 mutant downregulates the expression of both hydrogenase genes upon prolonged anaerobiosis.
Figure 8.
RNA Gel Blot Analysis of the Hydrogenase Transcripts.
(A) HydA1 in CC425 and sta7-10.
(B) HydA2 in CC425 and sta7-10.
Samples were grown in TAP media until late log phase, and RNA was isolated as described in Methods at the indicated times (hours) of anaerobic induction. RNA transcripts for both HydA1 and HydA2 were undetected from samples isolated from the oxygenated growth flasks. CC425 and sta7-10 RNA were run on the same blot and probed together for either HydA1 or HydA2 transcripts. Both hydrogenase transcripts from the sta7-10 mutant are detected shortly after the beginning of anaerobic induction, and their levels peak at ∼0.5 h. Thereafter, a steady decline to undetectable levels is observed. Background CC425 cells showed slower induction of hydrogenase transcript, which peaked at ∼1.5 h and remained at essentially steady state levels for the duration of the experiment. The ribosomal 23S RNA band is shown as a loading control below each RNA gel blot. o/n, overnight.
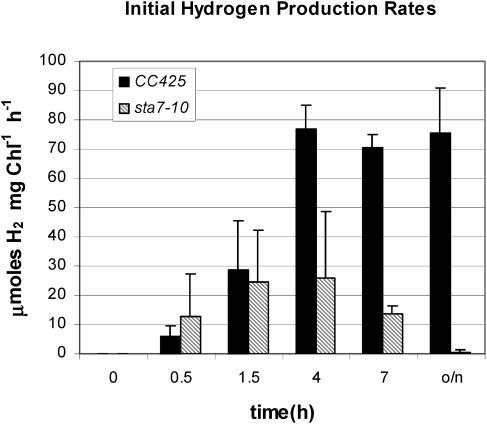
The induction of H2 production activity, monitored over a similar dark, anaerobic incubation period, differs significantly between the wild type and the sta7-10 mutant (Figure 9). At induction times <2 h, CC425 and sta7-10 cultures photoproduce similar amounts of H2. The initial rates of H2 photoproduction increase to a maximum in wild-type cells at the end of 4 h and remain constant, even after overnight incubation at 4°C. However, in the mutant cultures, the rates of H2 photoproduction reach ∼30% of the maximal wild-type rates at the end of 4 h and decline thereafter. No significant H2 photoproduction rates are detected after overnight incubation.
Figure 9.
Hydrogen Evolution Rates as a Function of Anaerobic Induction Time.
Rates of hydrogen evolution measured using a Clark electrode apparatus. Results are shown for the sta7-10 mutant (shaded bars) and the background strain CC425 (closed bars). Hydrogenase activity was induced as described in the Methods section. Samples were withdrawn from anaerobic cultures and assayed at the indicated times.
Additional Starch Mutants
Several additional C. reinhardtii mutants with variable starch content have been characterized previously (Ball, 1998), and these include the sta1, sta6, sta8, and sta7 strains (Mouille et al., 1996; Van den Koornhuyse et al., 1996; Dauvillee et al., 2001a; Zabawinski et al., 2001). These starch mutants were generously provided to us by Steven Ball and were used to determine whether other low-starch mutants have attenuated H2 photoproduction rates. A total lack of starch is observed only for the sta6 and sta7 strains, and both strains have attenuated H2 photoproduction rates compared with their parental strain, 330 (Figure 10). The sta7 mutants, which are isoamylase mutants (Mouille et al., 1996; Dauvillee et al., 2001b), show H2 photoproduction rates similar to those of our sta7-10 mutant (Figure 9). The sta6 mutant has a lesion in the catalytic subunit of ADP-glucose pyrophosphorylase (Zabawinski et al., 2001), and it has H2 photoproduction rates that are severely reduced relative to the wild type at all time points. Interestingly, the sta1 and sta8 mutants have rates of H2 photoproduction that are attenuated (data not shown) but only at later anaerobic induction times (∼7 to 10 h). It is important to note that although these strains have decreased starch levels, they have significantly more starch than the sta6 and sta7 mutants under our growth conditions.
Figure 10.
Initial H2 Photoproduction Rates from Starch Mutants.
Initial H2 photoproduction rates were measured from previously isolated starch mutants using a Clark-type electrode apparatus. Results are shown, respectively, for the parental background strain (330), a sta6 mutant (BAFJ5), two sta7 mutants (BAFJ4 and S), which were combined into a single data set, and a sta7 mutant (BAFJ4) that was complemented with DNA corresponding to the isoamylase gene. Hydrogen photoproduction rates are attenuated in the sta6 and sta7 mutants. Complementation of the sta7 (BAFJ4) mutant with the isoamylase gene restores wild-type H2 photoproduction rates. All samples were induced as described in Methods. The starch mutants were previously isolated in Steven Ball's laboratory.
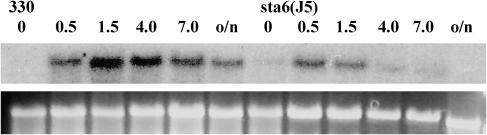
To determine whether hydrogenase gene transcription is also attenuated in sta6, RNA gel blot analyses were performed. As is observed for sta7-10, the levels of HydA1 (Figure 11) and HydA2 (data not shown) transcripts in sta6 are attenuated relative to the parental strain. In contrast with sta7-10, the hydrogenase transcript levels in sta6 are lower relative to the parental strain and become nearly undetectable at earlier times of anaerobic induction. This is consistent with the observed lower rates of H2 photoproduction from sta6 in comparison with sta7 mutants as shown in Figure 10.
Figure 11.
RNA Gel Blot Analysis of the Hydrogenase Transcripts in Parental Wild-Type Strains 330 and sta6 (BAFJ5).
Samples were grown in TAP media until late log phase, and RNA was isolated as described in Methods at the indicated times (hours) of anaerobic induction. RNA transcripts for HydA1 were undetected from samples isolated from the aerobic growth flasks. Wild-type (330) and sta6 (BAFJ5) RNA were run on the same blot and probed together for HydA1 transcripts. HydA1 transcripts from the 330 and sta6 (BAFJ5) strains were detected shortly after the beginning of anaerobic induction (0.5 h). In the sta6 (BAFJ5) mutant, HydA1 transcript levels peak at 0.5 h. Immediately thereafter, a steady decline to nearly undetectable levels is observed by 4.0 h. In the background 330 cells, HydA1 transcript levels peak at 1.5 h. A slow decline in HydA1 transcript levels is then observed during the remainder of the experiment. The ribosomal 23S RNA band is shown as a loading control below each RNA gel blot.
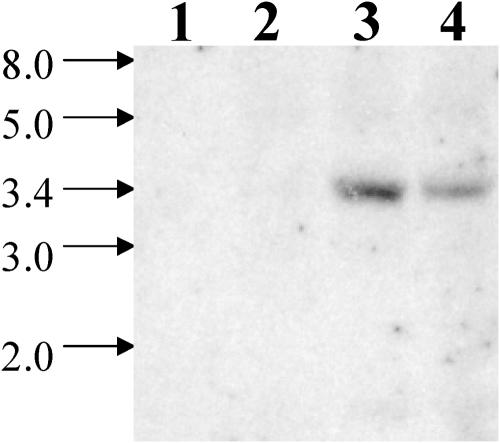
Given the similarities in the phenotypes between sta7-10 and the other sta7 mutants, we obtained DNA gel blots to determine whether the isoamylase gene described in this article is disrupted in these sta7 mutants as well. As shown in Figure 12, DNA isolated from the sta6 and sta8 strains hybridize to an isoamylase-specific probe at the expected electrophoretic mobility. However, this probe does not hybridize to DNA isolated from two sta7 mutants (BAFJ4 and S). Complementation of these sta7 mutants with genomic DNA, containing the wild-type isoamylase gene, restored both starch accumulation (data not shown) and H2 photoproduction (Figure 10). These data provide conclusive evidence that the isoamylase gene described here is allelic to the gene disrupted in the previously reported sta7 mutants.
Figure 12.
Genomic DNA Gel Blot Using Previously Identified Starch Mutants.
Genomic DNA isolated from previously identified starch mutants was digested with the restriction enzyme NcoI. Lanes 1 to 4 correspond, respectively, to DNA isolated from two sta7 mutants (BAFJ4 and S), a sta6 mutant (BAFJ5), and a sta8 strain (BAFV13). The blot was probed using the 690-bp NcoI-SphI DNA fragment from the isoamylase gene. The band at 3.4 kb corresponds to the isoamylase gene. The isoamylase band is absent in DNA isolated from both of the sta7 mutants.
DISCUSSION
Isoamylase
To elucidate the genes and enzymes involved in H2 photoproduction by C. reinhardtii, we screened an insertional mutagenesis library for mutants attenuated in H2 photoproduction, after dark, anaerobic induction. One mutant isolated from the screen contains a disrupted gene with high homology to the isoamylase gene family. This mutant contains significantly depressed levels of insoluble starch in comparison with wild-type strains and exhibits H2 photoproduction rates that are ∼20% to 40% of those found in wild-type cells after 4 to 5 h of anaerobic induction.
Previous biochemical studies of sta7 mutants have demonstrated the vital role that this isoamylase plays in C. reinhardtii starch accumulation in vivo (Mouille et al., 1996). Although the precise role of isoamylase in starch synthesis is not completely understood, three models have been proposed (Zeeman et al., 1998; Smith, 2001; Burton et al., 2002; Ball and Morell, 2003; Bustos et al., 2004). The first suggests that isoamylase is required to trim excess branching sites in preamylopectin to yield a properly ordered structure capable of forming the crystalline starch of amylopectin. The second suggests that isoamylase rapidly degrades soluble glucans. In the absence of isoamylase, these glucans are not degraded but rather accumulate as phytoglycogen, which competes with the formation of insoluble starch (Zeeman et al., 1998). The first model was proposed to explain the role of isoamylase in C. reinhardtii (Ball et al., 1996; Mouille et al., 1996; Ball, 1998; Myers et al., 2000). However, recent isoamylase mutant studies in barley (Hordeum vulgare; Burton et al., 2002) and isoamylase antisense RNA studies in potato tubers (Bustos et al., 2004) indicate that heteromultimeric isoamylases control the frequency of starch granule initiation. This third model, first proposed by Burton et al. (2002), is based on the observation that an abnormally large number of smaller starch granules are formed in both potato and barley when isoamylase activity is disrupted (Burton et al., 2002; Bustos et al., 2004). Despite a clear understanding of the mechanism by which isoamylase influences starch synthesis in vivo, the results presented here confirm that disruption of the isoamylase STA7 gene in C. reinhardtii drastically reduces the accumulation of starch in this alga.
Three isoamylase isoforms from potato are known (Hussain et al., 2003). Two of the isoforms, Stisa1 and Stisa3, have hydrolytic activity, whereas the third isoform, Stisa2, forms a complex with Stisa1 and potentially regulates the activity and/or specificity of Stisa1 (Hussain et al., 2003). As shown in Figure 5, the STA7 isoamylase has the highest homology to Stisa1 (65% similarity and 54% identity), which forms a multimeric enzyme complex. This complex is responsible for the majority of the observed isoamylase activity in native gels containing S. tuberosum protein extracts (Hussain et al., 2003). The STA7 enzyme is therefore likely to represent a major hydrolytic rather than a regulatory isoamylase isoform.
Starch and Hydrogen Production
Starch mobilization has been implicated in H2 production from green algae (Healey, 1970; Bamberger et al., 1982; Gfeller and Gibbs, 1984; Ohta et al., 1987; Kosourov et al., 2001; Melis and Happe, 2001); however, our work directly links the disruption of a specific enzyme involved in starch metabolism to attenuated H2 photoproduction. Two different metabolic pathways provide reductant used in the photoproduction of H2 by algal cells anaerobically induced in the dark. One of the pathways originates in PSII-catalyzed water oxidation (Bamberger et al., 1982; Greenbaum et al., 1983; Gfeller and Gibbs, 1984), and it is sensitive to the PSII inhibitor DCMU. This pathway involves the full photosynthetic electron transport chain. The second pathway is DCMU insensitive and depends on the transfer of reductant from endogenous substrate degradation, particularly starch, to the photosynthetic electron transport chain. Starch breakdown is proposed to provide electrons in the dark to the PQ pool through the putative NAD(P)H-PQ oxidoreductase (Godde and Trebst, 1980; Gfeller and Gibbs, 1984; Bennoun, 1998). These electrons are subsequently transferred to hydrogenase via photosystem I and ferredoxin after subsequent illumination.
It is tempting to attribute the lower levels of H2 photoproduction observed in the sta7-10 mutant solely to an absence of reducing equivalents provided by starch catabolism. However, the RNA gel blot data (Figure 8) demonstrate that both putative C. reinhardtii hydrogenase genes are downregulated in the sta7-10 mutant after extended periods of anaerobic induction, strongly implying that the lower rates are the result of a transcriptional component. Interestingly, the sta7-10 mutant contains more hydrogenase transcript than the wild type after 0.5 h of induction (Figure 8), which further suggests that H2 production and hydrogenase transcription in C. reinhardtii are regulated by factors other than merely the absence of O2.
The linkage between starch content and anaerobically induced H2 production activity is further strengthened by the observed decrease in H2 photoproduction rates in the sta6 mutant. Although both sta6 and sta7 mutants contain drastically depressed starch levels, H2 photoproduction and hydrogenase transcription are more seriously attenuated in the sta6 mutant. This could be because of the presence of small amounts of phytoglycogen, which is found in sta7 mutants (Mouille et al., 1996; Dauvillee et al., 2001a) but not sta6. Glucose derived from phytoglycogen may be able to sustain the brief H2 photoproduction activity observed during the early anaerobic induction period in sta7 strains. These results illustrate the importance of starch for obtaining maximal rates of H2 production from C. reinhardtii cultures anaerobically induced in the dark. The sta1 and sta8 strains were less affected in total starch accumulation and demonstrate a much weaker H2 photoproduction phenotype. This implies that a significant reduction in cellular starch content is required to adversely affect H2 photoproduction at earlier time points.
It is evident that the catabolism of starch in C. reinhardtii is necessary to maintain hydrogenase transcription and H2 photoproduction after several hours of dark, anaerobic induction. Hydrogen production from a variety of nonphotosynthetic organisms is mediated by the breakdown of carbon substrates and the resultant reduced redox carriers formed during fermentation (Adams, 1990; Hackstein et al., 1999; Vignais et al., 2001). Although green algae obtain significant amounts of reductant for H2 production from PSII after illumination, it is likely that under dark, anaerobic conditions, hydrogenase transcription and subsequent H2 photoproduction also depend on reducing equivalents provided by starch degradation.
The precise mechanism by which starch metabolism influences hydrogenase transcription is presently unclear; however, it is possible that the degradation of starch affects intracellular levels of NAD(P)H and/or the oxidation state of the PQ pool, both of which have been demonstrated to regulate gene transcription either directly or through signal transduction (Escoubas et al., 1995; Pfannschmidt et al., 2001a, 2001b; Rutter et al., 2001). Further experimentation is under way to more clearly investigate this hypothesis.
METHODS
Strains
An insertional mutagenesis library of ∼6000 colonies was generated by transforming Chlamydomonas reinhardtii strain CC425 (cw15, sr-u-60, arg7-8, mt+) with the pJD67 plasmid (Davies et al., 1999). This plasmid contains a functional copy of the genomic argininosuccinate lyase (Arg7) gene (Debuchy et al., 1989) subcloned into the plasmid pBluescript KS+ (Statagene, La Jolla, CA). The pJD67 plasmid was linearized by digestion with the restriction enzyme HindIII and transformed into C. reinhardtii strain CC425 using the glass bead transformation method of Kindle (1990). Colonies were maintained on TAP plates without Arg under low light (∼14 μE m−2 s−1) at 25°C. The sta6, sta7, and sta8 starch mutant strains, as well as the parental strain (330), were obtained from Steven Ball (Université des Sciences et Technologies de Lille, France). The CC425 and sta1 strains were obtained from the Duke University Genetics Center (Durham, NC).
Hydrogen and Oxygen Assays and Cell Growth Conditions
Chemochromic screening was performed using colonies growing on TAP agar plates. The sensors were prepared as described previously (Seibert et al., 2001a; Flynn et al., 2002). Colonies were replica plated onto TAP plates and grown for ∼1 to 2 weeks at a light intensity of ∼14 μE m−2 s−1 PAR (25°C). Hydrogenase activity was induced anaerobically in the dark. Briefly, the agar plates were placed in a glove bag, which was filled and evacuated four times with nitrogen, and left under a nitrogen atmosphere overnight in the dark. Photoproduction of H2 was monitored the following day using chemochromic sensors placed over the agar plates, and the reaction was started by illumination at ∼380 μE m−2 s−1 PAR of light. Normally transparent, the sensors turn blue in the presence of H2 gas and were used to visualize colonies deficient in H2 production (those that did not elicit a colorimetric response). For each assay, every plate containing potential mutants was analyzed in duplicate.
Rates of photosynthesis, respiration, and H2 production in liquid cultures were determined using a water-jacketed chamber (2.5 mL volume held at 25°C) that was equipped with two Clark electrodes (YSI 5331; Yellow Springs, OH), one poised for the measurement of H2 and the other for O2 (Ghirardi et al., 1997). Unless otherwise noted, algal cells used for H2 photoproduction measurements were grown in TAP liquid medium (Arg was added to the CC425 cultures at 150 to 200 μg/mL) with constant stirring at room temperature, bubbled with 2% CO2 in air, and continuously illuminated with cool-white fluorescent light at a light intensity of 120 μE m−2 s−1 PAR. Cultures were inoculated with <1.0 μg/mL total chlorophyll and grown to a density of between 15 and 30 μg/mL of total chlorophyll. Cells were centrifuged at 2500g for 5 min, and the pellet was resuspended to 200 μg/mL of total chlorophyll in anaerobic induction buffer consisting of 50 mM potassium phosphate, pH 7.0, and 3 mM magnesium chloride (Ghirardi et al., 1997). Hydrogen production activity was then induced by placing 5.0 mL of concentrated cells into a vial wrapped with aluminum foil to exclude light. The vial was sealed with a rubber septum and flushed with argon. An enzymatic O2-scrubbing system was also included to ensure complete anaerobiosis for hydrogenase induction (McTavish et al., 1989). After 4 h of induction, the cells were stored anaerobically at 4°C overnight. For each measurement, the following day 250 μL of induced cells were placed in the Clark electrode assay chamber, which was maintained at 25°C and contained 2.25 mL of Mops buffer (50 mM, pH 6.8) deoxygenated with argon. A shutter was used to control illumination of the assay chamber. The algae were adapted for 2 min in the dark and then exposed for 3 min to ∼700 μE m−2 s−1 PAR of actinic light filtered through a solution of 1% CuSO4. Concentrations of H2 and O2 were monitored simultaneously as a function of time using the two electrodes, and initial H2 production rates were calculated.
Hydrogen photoproduction rates for the complemented clones (Figure 7B) were measured as above, except no O2-scrubbing enzymes were added, and the H2 production rates were measured 5 h after induction at room temperature. Anaerobic induction for the H2 photoproduction time courses (Figures 9 and 10) were also performed without the O2-scrubbing enzymes, and samples were analyzed at various times for a total period of 7 h, after which the samples were stored at 4°C. The overnight sample was assayed the following morning. The O2-scrubbing enzymes were omitted from these latter assays because argon purging and cellular respiration were subsequently determined to be sufficient for inducing hydrogenase activity.
Photosynthesis (O2 evolution) and respiration (O2 uptake) rates were measured using early log phase cells, cultured in TAP media as described above using the same Clark electrode apparatus.
DNA and RNA Gel Blot Analysis
DNA gel blotting experiments were performed using standard methodology (Southern, 1975). Briefly, 2.0 μg of genomic DNA was digested with PstI, NotI, or NcoI, as indicated, and the fragments were separated on a 0.8% agarose Tris/acetate/EDTA gel. Genomic DNA was extracted and purified using the DNeasy plant mini kit from Qiagen (Valencia, CA) and after digestion with the appropriate restriction enzyme was transferred to positively charged nylon membranes (Amersham Pharmacia Biotech, Uppsala, Sweden). Indicated probes were labeled using [α-32P]dCTP (ICN, Costa Mesa, CA) and the rediprime II DNA random prime labeling system from Amersham Pharmacia Biotech. The blots were washed twice using 2× SSC and 0.1% SDS at 65°C for 10 min each, twice using 0.5× SSC and 0.5% SDS at 65°C for 10 min each, and once using 0.25× SSC and 0.2% SDS at 65°C for 10 min.
RNA gel blot analysis was performed as described previously using 10 μg of total RNA for each sample (Forestier et al., 2003). Total RNA was isolated from anaerobically induced samples using a SNAP RNA isolation kit from Invitrogen (Carlsbad, CA). DNA was removed by treatment with RNase-free DNaseI. RNA was separated by electrophoresis on denaturing 1.0% agarose, 0.22 M formaldehyde gels and then blotted onto a Nytran N+ nylon membrane (Schleicher and Schuell, Keene, NH) using 10× SSC as the transfer buffer. Transfer of RNA to the nylon membrane was confirmed by UV illumination of the ethidium bromide–stained RNA, and it was determined to be essentially quantitative as a result of the complete lack of fluorescence remaining in the gel and the intense fluorescence of the membrane. Hydrogenase probes specific for HydA1 and HydA2 contained the 5′ untranslated region and ∼150 bp of coding sequence for each gene. The probes were labeled as described for DNA gel blotting, and the blots were prehybridized and hybridized using 6× SSC, 5× Denhardt's solution, 1.0% SDS, and 50 μg/mL of salmon sperm DNA. The blots were washed twice using 2× SSC and 0.2% SDS at 65°C for 5 min each, twice using 2× SSC and 0.1% SDS at 65°C for 10 min each, and twice using 0.5× SSC and 0.5% SDS at 65°C for 10 min each. Samples were induced as described above for the anaerobic induction used in the H2 photoproduction time course study.
Gene Identification
DNA regions flanking the insertion site of pJD67 were determined using genome walking and plasmid rescue techniques. DNA downstream of the insertion site was amplified using the PCR methods outlined in the Universal GenomeWalker kit and the Advantage-GC Genomic PCR mix, both from Clontech (Palo Alto, CA). We followed the manufacturer's instructions for the procedure and used the gene-specific primer 5′-GGGACGGGTGTGACAGAGTTACGG-3′, corresponding to a region at the 3′ end of the Arg7 gene. DNA flanking the 5′ site of insertion was cloned using plasmid rescue, where ∼5.0 μg of genomic DNA from the sta7-10 mutant was digested with the restriction enzymes BamHI or StuI. After digestion, the DNA was extracted once with phenol:chloroform: isoamyl alcohol (25:24:1) and once with chloroform:isoamyl alcohol (24:1), precipitated with ethanol, resuspended in 0.5 mL of 1× ligation buffer (New England Biolabs, Beverly, MA), and then ligated overnight at room temperature. The DNA was then recovered by organic extraction and ethanol precipitation as described above, resuspended in 20 μL of 10.0 mM Tris, pH 8.0, and transformed into Escherichia coli DH5α cells using electroporation (Dower, 1990). Reverse transcription was used to obtain the STA7 cDNA coding sequence, and it was performed using gene-specific primers and the Improm-II reverse transcriptase from Promega (Madison, WI). The reactions were performed using three sequentially stepped temperatures starting at 48°C for 45 min, followed by 53°C and 58°C for 45 min each. The cDNA products were then amplified using standard PCR protocols. RNA was isolated as described for the RNA gel blot experiments. All DNA products were sequenced by the University of California, Davis sequencing facility (http://davissequencing.com).
BAC Library Screening
A BAC clone containing the isoamylase gene was identified using the C. reinhardtii BAC library filter from Incyte Genomics (Palo Alto, CA), following the manufacturer's instructions. The BAC filter was prepared by Incyte Genomics in collaboration with Peter Lefebvre (University of Minnesota, St. Paul) and consists of a partial HindIII digest of genomic DNA (Lefebvre and Silflow, 1999). The filter contains DNA from 15,000 BAC clones, with an average insert size of 70 kb. The library was probed using a restriction fragment from the PCR product obtained from genome walking, downstream from the site of pJD67 insertion. Positive BAC clones were obtained from Incyte Genomics.
Zymograms and Starch Analysis
Zymograms were performed as described by Dauvillee et al. (2001b). Briefly, C. reinhardtii cultures were grown in TAP media to early stationary phase under the conditions described above. Cultures were centrifuged at 3000g for 5 min, and the cell pellet was stored frozen at −80°C. Samples were resuspended in fresh lysis buffer (50 mM Tricine, 10 mM NaCl, 5 mM MgCl2, 1 mM aminocaproic acid, pH 7.8, 1.0 mM phenylmethylsulfonyl fluoride, and 0.1% β-mercaptoethanol) at a 1:50 concentration of the initial volume, placed in a sonication bath for 5 min, and then centrifuged to remove cellular debris. One hundred micrograms of crude protein extract were loaded onto a 29:1 acrylamide:bisacrylamide, 7.5% (w/v), 1.5-mm-thick native polyacrylamide gel containing 0.6% (w/v) rabbit liver glycogen. Gels were run at 30 mA for 4 h at 4°C in 25 mM Tris Gly buffer, pH 8.0, and 0.1% β-mercaptoethanol, incubated overnight in running buffer containing 0.1% β-mercaptoethanol, and stained with a solution containing 0.025% (w/v) I2 and 3.1% (w/v) KI.
Glucose was assayed using the Glucose (HK) Assay kit from Sigma (St. Louis, MO). Cells were spun down, and the pigments were extracted twice with 95% ethanol. The cell pellet was washed once with 100 mM sodium acetate, pH 4.5, resuspended in 100 mM sodium acetate, pH 4.5, and then disrupted by sonication for 10 min. The sample was autoclaved to convert starch granules into colloidal solution and then digested with 4 units of amyloglucosidase (Sigma) overnight at 55°C. The resultant glucose was subsequently measured according to the manufacturer's instructions.
Starch was visually assayed using an iodine solution. Cell pigments were removed with 95% ethanol; the sample centrifuged; and the pellet resuspended in 600 μL of water and 400 μL of a 0.75% KI and 0.15% I2 solution. Starch-rich pellets stained a dark brown color, and starch-deficient pellets remained lightly colored. Where noted, the starch pellet was resuspended in water and autoclaved before the addition of the iodine solution.
Complementation
The 3′-half of the isoamylase gene was obtained from the BAC clone that contained the isoamylase gene. The BAC clone was digested with PacI, which cleaves near the middle of the isoamylase gene, and KpnI, which digests past the putative stop codon and polyadenylation site. The 5′-half of the gene was obtained using the BamHI fragment obtained from plasmid rescue. The two portions of the isoamylase gene contain ∼740 bp of overlap, from the PacI restriction site to the site of pJD67 integration. To assemble the full-length isoamylase genomic DNA, the PacI-KpnI fragment containing the 3′ end of the isoamylase gene was subcloned into pNEB193 (New England Biolabs). The 5′ portion of the STA7 gene, obtained from plasmid rescue, was digested with BamHI-PacI. The two fragments were sequentially cloned into pSP124S (Lumbreras et al., 1998; a gift from Saul Purton, University College London, UK) to correctly order the two fragments. The whole gene was excised using BamHI-KpnI digestion and cloned into pUC19. The Ble gene was isolated from plasmid SP124S by digesting with KpnI and EcoRI and subcloned into the plasmid containing the full-length isoamylase gene, resulting in plasmid MP25. This plasmid contains both the full-length isoamylase gene from the BamHI site to the KpnI site and the Ble gene in the same pUC19-based plasmid. Full-length plasmid was linearized by digestion with SwaI and purified using a Qiagen DNA spin column. Transformation into sta7 mutants was done using the glass bead method of Kindle (Kindle, 1990) and 5.0 μg of linearized plasmid. Controls, using cells only or 1 μg of linearized SP124S, were also used. Transformed cells were allowed to recover overnight under low light with gentle shaking. The cells were then plated using 0.8% top agar onto TAP plates containing 1 or 10 μg/mL of zeocin (Invitrogen, San Diego, CA) following the instructions given on Saul Purton's Web site (http://www.ucl.ac.uk/biology/prg/ble1.htm).
The cDNA and genomic sequences of the C. reinhardtii isoamylase have been submitted to the National Center for Biotechnology Information under accession numbers AY324649 and AY323823, respectively.
Acknowledgments
We thank Steven Ball (Université des Sciences et Technologies de Lille) for generously providing us with his sta6, sta7, and sta8 mutants. We also thank Dafna Elrad in Arthur Grossman's laboratory (Stanford University, Stanford, CA) for several helpful discussions and experimental insights; Roland Pitts and Ping Liu (National Renewable Energy Laboratory [NREL]) for fabricating the hydrogen sensors; and Saul Purton (University College London) for generously providing the pSP124S plasmid. We also appreciate the input and advice from the entire NREL Biohydrogen group. This work was funded by the Division of Energy Biosciences, Basic Energy Sciences, Office of Science, U.S. Department of Energy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Matthew Posewitz (matttew_posewitz@nrel.gov).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.021972.
References
- Adam, M., and Loppes, R. (1998). Use of the ARG7 gene as an insertional mutagen to clone PHON24, a gene required for derepressible neutral phosphatase activity in Chlamydomonas reinhardtii. Mol. Gen. Genet. 258, 123–132. [DOI] [PubMed] [Google Scholar]
- Adams, M.W.W. (1990). The structure and mechanism of iron-hydrogenases. Biochim. Biophys. Acta 1020, 115–145. [DOI] [PubMed] [Google Scholar]
- Asada, Y., and Miyake, J. (1999). Photobiological hydrogen production. J. Biosci. Bioeng. 88, 1–6. [DOI] [PubMed] [Google Scholar]
- Ball, S. (1998). Regulation of starch biosynthesis. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Vol. 7, J.D. Rochaix, M. Goldschmiidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 549–567.
- Ball, S., Guan, H.P., James, M., Myers, A., Keeling, P., Mouille, G., Buleon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. [DOI] [PubMed] [Google Scholar]
- Ball, S.G., and Morell, M.K. (2003). From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. [DOI] [PubMed] [Google Scholar]
- Bamberger, E.S., King, D., Erbes, D.L., and Gibbs, M. (1982). H2 and CO2 evolution by anaerobically adapted Chlamydomonas reinhardtii F-60. Plant Physiol. 69, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benemann, J.R. (1996). Hydrogen biotechnology: Progress and prospects. Nat. Biotechnol. 14, 1101–1103. [DOI] [PubMed] [Google Scholar]
- Bennoun, P. (1998). Chlororespiration, sixteen years later. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, J.D. Rochaix, M. Goldschmiidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 675–683.
- Boichenko, V.A., Greenbaum, E., and Seibert, M. (2004). Hydrogen production by photosynthetic microorganisms. In Photoconversion of Solar Energy: Molecular to Global Photosynthesis, Vol. 2, M.D. Archer and J. Barber, eds (London: Imperial College Press), pp. 397–452.
- Boichenko, V.A., and Hoffmann, P. (1994). Photosynthetic hydrogen-production in prokaryotes and eukaryotes: Occurrence, mechanism, and functions. Photosynthetica 30, 527–552. [Google Scholar]
- Brand, J.J., Wright, J.N., and Lein, S. (1989). Hydrogen production by eukaryotic algae. Biotechnol. Bioeng. 33, 1482–1488. [DOI] [PubMed] [Google Scholar]
- Burton, R.A., Jenner, H., Carrangis, L., Fahy, B., Fincher, G.B., Hylton, C., Laurie, D.A., Parker, M., Waite, D., van Wegen, S., Verhoeven, T., and Denyer, K. (2002). Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 31, 97–112. [DOI] [PubMed] [Google Scholar]
- Bustos, R., Fahy, B., Hylton, C.M., Seale, R., Nebane, N.M., Edwards, A., Martin, C., and Smith, A.M. (2004). Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers. Proc. Natl. Acad. Sci. USA 101, 2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame, G., Gloeckner, G., and Beck, C.F. (2002). Knock-out of a putative transporter results in altered blue-light signalling in Chlamydomonas. Plant J. 31, 577–587. [DOI] [PubMed] [Google Scholar]
- Dauvillee, D., Colleoni, C., Mouille, G., Buleon, A., Gallant, D.J., Bouchet, B., Morell, M.K., d'Hulst, C., Myers, A.M., and Ball, S.G. (2001. a). Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 125, 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillee, D., Colleoni, C., Mouille, G., Morell, M.K., d'Hulst, C., Wattebled, F., Lienard, L., Delvalle, D., Ral, J.P., Myers, A.M., and Ball, S.G. (2001. b). Biochemical characterization of wild-type and mutant isoamylases of Chlamydomonas reinhardtii supports a function of the multimeric enzyme organization in amylopectin maturation. Plant Physiol. 125, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F.H., and Grossman, A.R. (1999). Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11, 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy, R., Purton, S., and Rochaix, J.D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: An important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8, 2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower, W.J. (1990). Electroporation of bacteria: A general approach to genetic transformation. Genet. Eng. (N.Y.) 12, 275–295. [DOI] [PubMed] [Google Scholar]
- Elrad, D., Niyogi, K.K., and Grossman, A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas, J.M., Lomas, M., LaRoche, J., and Falkowski, P.G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92, 10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, T., Ghirardi, M.L., and Seibert, M. (2002). Accumulation of O2-tolerant phenotypes in H2-producing strains of Chlamydomonas reinhardtii by sequential applications of chemical mutagenesis and selection. Int. J. Hydrogen Energy 27, 1421–1430. [Google Scholar]
- Forestier, M., King, P., Zhang, L., Posewitz, M., Schwarzer, S., Happe, T., Ghirardi, M.L., and Seibert, M. (2003). Expression of two [Fe]-hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Eur. J. Biochem. 270, 2750–2758. [DOI] [PubMed] [Google Scholar]
- Gaffron, H., and Rubin, J. (1942). Fermentative and photochemical production of hydrogen in algae. J. Gen. Physiol. 26, 219–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller, R.P., and Gibbs, M. (1984). Fermentative metabolism of Chlamydomonas reinhardtii. I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 75, 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi, M.L., Togasaki, R.K., and Seibert, M. (1997). Oxygen sensitivity of algal H2-production. Appl. Biochem. Biotechnol. 63, 141–151. [DOI] [PubMed] [Google Scholar]
- Ghirardi, M.L., Zhang, L., Lee, J.W., Flynn, T., Seibert, M., Greenbaum, E., and Melis, A. (2000). Microalgae: A green source of renewable H(2). Trends Biotechnol. 18, 506–511. [DOI] [PubMed] [Google Scholar]
- Godde, D., and Trebst, A. (1980). NADH as electron donor for the photosynthetic membrane of Chlamydomonas reinhardtii. Arch. Microbiol. 127, 245–252. [Google Scholar]
- Greenbaum, E. (1988). Energetic efficiency of hydrogen photoevolution by algal water splitting. Biophys. J. 54, 365–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum, E., Guillard, R.R.L., and Sunda, W.G. (1983). Hydrogen and oxygen photoproduction by marine algae. Photochem. Photobiol. 37, 649–655. [Google Scholar]
- Hackstein, J.H., Akhmanova, A., Boxma, B., Harhangi, H.R., and Voncken, F.G. (1999). Hydrogenosomes: Eukaryotic adaptations to anaerobic environments. Trends Microbiol. 7, 441–447. [DOI] [PubMed] [Google Scholar]
- Hansel, A., and Lindblad, P. (1998). Towards optimization of cyanobacteria as biotechnologically relevant producers of molecular hydrogen, a clean and renewable energy source. Appl. Microbiol. Biotechnol. 50, 153–160. [Google Scholar]
- Happe, T., Hemschemeier, A., Winkler, M., and Kaminski, A. (2002). Hydrogenases in green algae: Do they save the algae's life and solve our energy problems? Trends Plant Sci. 7, 246–250. [DOI] [PubMed] [Google Scholar]
- Happe, T., and Kaminski, A. (2002). Differential regulation of the Fe-hydrogenase during anaerobic adaptation in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 269, 1022–1032. [DOI] [PubMed] [Google Scholar]
- Happe, T., Mosler, B., and Naber, J.D. (1994). Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222, 769–774. [DOI] [PubMed] [Google Scholar]
- Happe, T., and Naber, J.D. (1993). Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214, 475–481. [DOI] [PubMed] [Google Scholar]
- Healey, F.P. (1970). The mechanism of hydrogen evolution by Chlamydomonas moewusii. Plant Physiol. 45, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, H., Mant, A., Seale, R., Zeeman, S., Hinchliffe, E., Edwards, A., Hylton, C., Bornemann, S., Smith, A.M., Martin, C., and Bustos, R. (2003). Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15, 133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle, K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosourov, S., Seibert, M., and Ghirardi, M.L. (2003). Effects of extracellular pH on the metabolic pathways in sulfur-deprived, H(2)-producing Chlamydomonas reinhardtii cultures. Plant Cell Physiol. 44, 146–155. [DOI] [PubMed] [Google Scholar]
- Kosourov, S., Tsygankov, A., Ghirardi, M.L., and Seibert, M. (2001). The cloning of two hydrogenase genes from the green alga Chlamydomonas reinhardtii. In Proceedings of the 12th International Congress on Photosynthesis. (Melbourne, Australia: CSIRO Publishing).
- Kosourov, S., Tsygankov, A., Seibert, M., and Ghirardi, M.L. (2002). Sustained hydrogen photoproduction by Chlamydomonas reinhardtii: Effects of culture parameters. Biotechnol. Bioeng. 78, 731–740. [DOI] [PubMed] [Google Scholar]
- Lefebvre, P.A., and Silflow, C.D. (1999). Chlamydomonas: The cell and its genomes. Genetics 151, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras, V., Stevens, D.R., and Purton, S. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14, 441–448. [Google Scholar]
- McTavish, H., Picorel, R., and Seibert, M. (1989). Stabilization of isolated photosystem-II reaction center complex in the dark and in the light using polyethylene-glycol and an oxygen-scrubbing system. Plant Physiol. 89, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis, A., and Happe, T. (2001). Hydrogen production. Green algae as a source of energy. Plant Physiol. 127, 740–748. [PMC free article] [PubMed] [Google Scholar]
- Melis, A., Zhang, L., Forestier, M., Ghirardi, M.L., and Seibert, M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, Y., Yagi, K., Shoga, M., and Miyamoto, K. (1982). Hydrogen production by a green alga, Chlamydomonas reinhardtii, in an alternating light/dark cycle. Biotechnol. Bioeng. 24, 1555–1563. [DOI] [PubMed] [Google Scholar]
- Moseley, J., Quinn, J., Eriksson, M., and Merchant, S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille, G., Maddelein, M.L., Libessart, N., Talaga, P., Decq, A., Delrue, B., and Ball, S. (1996). Preamylopectin processing: A mandatory step for starch biosynthesis in plants. Plant Cell 8, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, A.M., Morell, M.K., James, M.G., and Ball, S.G. (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122, 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet, Y., Piras, C., Legrand, P., Hatchikian, C.E., and Fontecilla-Camps, J.C. (1999). Desulfovibrio desulfuricans iron hydrogenase: The structure shows unusual coordination to an active site Fe binuclear center. Struct. Fold. Des. 7, 13–23. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Bjorkman, O., and Grossman, A.R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, S., Miyamoto, K., and Miura, Y. (1987). Hydrogen evolution as a consumption mode of reducing equivalents in green algal fermentation. Plant Physiol. 83, 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J.W., Lanzilotta, W.N., Lemon, B.J., and Seefeldt, L.C. (1998). X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282, 1853–1858. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Allen, J.F., and Oelmueller, R. (2001. a). Principles of redox control in photosynthesis gene expression. Physiol. Plant 112, 1–9. [Google Scholar]
- Pfannschmidt, T., Schutze, K., Brost, M., and Oelmuller, R. (2001. b). A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem. 276, 36125–36130. [DOI] [PubMed] [Google Scholar]
- Polle, J.E., Kanakagiri, S.D., and Melis, A. (2003). tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217, 49–59. [DOI] [PubMed] [Google Scholar]
- Posewitz, M.C., King, P.W., Smolinski, S.L., Zhang, L., Seibert, M., and Ghirardi, M.L. (2004). Discovery of two novel radical SAM proteins required for the assembly of an active [Fe]-hydrogenase. J. Biol. Chem. 279, 25711–25720. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.D. (1995). Chlamydomonas reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 29, 209–230. [DOI] [PubMed] [Google Scholar]
- Roessler, P.G., and Lien, S. (1984). Purification of hydrogenase from Chlamydomonas reinhardtii. Plant Physiol. 75, 705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, J., Reick, M., Wu, L.C., and McKnight, S.L. (2001). Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293, 510–514. [DOI] [PubMed] [Google Scholar]
- Seibert, M., Benson, D.K., and Flynn, T. (2001. a). Method and Apparatus for Rapid Biohydrogen Phenotypic Screening of Microorganisms Using a Chemochromic Sensor. U.S. patent 6,277,589 B1. (Kansas City, MO: Midwest Research Institute).
- Seibert, M., Flynn, T., and Ghirardi, M.L. (2001. b). Strategies for improving oxygen tolerance of algal hydrogen production. In Biohydrogen II, J. Miyake, T. Matsunaga, and A. San Pietro, eds (Amsterdam, The Netherlands: Elsevier), pp. 67–77.
- Smith, A.M. (2001). The biosynthesis of starch granules. Biomacromolecules 2, 335–341. [DOI] [PubMed] [Google Scholar]
- Southern, E.M. (1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517. [DOI] [PubMed] [Google Scholar]
- Tam, L.W., and Lefebvre, P.A. (1995). Insertional mutagenesis and isolation of tagged genes in Chlamydomonas. Methods Cell Biol. 47, 519–523. [DOI] [PubMed] [Google Scholar]
- Tamagnini, P., Axelsson, R., Lindberg, P., Oxelfelt, F., Wunschiers, R., and Lindblad, P. (2002). Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol. Mol. Biol. Rev. 66, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Koornhuyse, N., Libessart, N., Delrue, B., Zabawinski, C., Decq, A., Iglesias, A., Carton, A., Preiss, J., and Ball, S. (1996). Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J. Biol. Chem. 271, 16281–16287. [DOI] [PubMed] [Google Scholar]
- Van, K., Wang, Y., Nakamura, Y., and Spalding, M.H. (2001). Insertional mutants of Chlamydomonas reinhardtii that require elevated CO(2) for survival. Plant Physiol. 127, 607–614. [PMC free article] [PubMed] [Google Scholar]
- Vignais, P.M., Billoud, B., and Meyer, J. (2001). Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25, 455–501. [DOI] [PubMed] [Google Scholar]
- Vignais, P.M., Colbeau, A., Willison, J.C., and Jouanneau, Y. (1985). Hydrogenase, nitrogenase, and hydrogen metabolism in the photosynthetic bacteria. Adv. Microb. Physiol. 26, 155–234. [DOI] [PubMed] [Google Scholar]
- Weaver, P.F., Lien, S., and Seibert, M. (1980). Photobiological production of hydrogen. Sol. Energy 24, 3–45. [Google Scholar]
- Wykoff, D.D., Davies, J.P., Melis, A., and Grossman, A.R. (1998). The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabawinski, C., Van Den Koornhuyse, N., D'Hulst, C., Schlichting, R., Giersch, C., Delrue, B., Lacroix, J.M., Preiss, J., and Ball, S. (2001). Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183, 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., Umemoto, T., Lue, W.L., Au-Yeung, P., Martin, C., Smith, A.M., and Chen, J. (1998). A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Happe, T., and Melis, A. (2002). Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214, 552–561. [DOI] [PubMed] [Google Scholar]