Abstract
The mechanism of oxygen evolution by photosystem II (PSII) has remained highly conserved during the course of evolution from ancestral cyanobacteria to green plants. A cluster of manganese, calcium, and chloride ions, whose binding environment is optimized by PSII extrinsic proteins, catalyzes this water-splitting reaction. The accepted view is that in plants and green algae, the three extrinsic proteins are PsbO, PsbP, and PsbQ, whereas in cyanobacteria, they are PsbO, PsbV, and PsbU. Our previous proteomic analysis established the presence of a PsbQ homolog in the cyanobacterium Synechocystis 6803. The current study additionally demonstrates the presence of a PsbP homolog in cyanobacterial PSII. Both psbP and psbQ inactivation mutants exhibited reduced photoautotrophic growth as well as decreased water oxidation activity under CaCl2-depleted conditions. Moreover, purified PSII complexes from each mutant had significantly reduced activity. In cyanobacteria, one PsbQ is present per PSII complex, whereas PsbP is significantly substoichiometric. These findings indicate that both PsbP and PsbQ proteins are regulators that are necessary for the biogenesis of optimally active PSII in Synechocystis 6803. The new picture emerging from these data is that five extrinsic PSII proteins, PsbO, PsbP, PsbQ, PsbU, and PsbV, are present in cyanobacteria, two of which, PsbU and PsbV, have been lost during the evolution of green plants.
INTRODUCTION
Photosynthetic water oxidation is catalyzed by photosystem II (PSII), a multisubunit pigment protein complex in the thylakoid membranes (Hillier and Babcock, 2001). In PSII, a cluster of three inorganic ions, manganese, calcium, and chloride, catalyzes light-induced oxidation of water in cyanobacteria and chloroplasts (Debus, 2000). Studies on PSII from plants, algae, and cyanobacteria have revealed several PSII proteins that collectively regulate the unique redox environment of this inorganic catalytic center (Debus, 2000). The Mn-Ca-Cl cluster is near the surface of the lumenal helix between membrane spanning helices C and D of the D1 subunit, and direct ligation of the manganese ions appears to come from at least five amino acid residues of the D1 protein and one residue of the CP43 subunit (Ferreira et al., 2004). Furthermore, large hydrophilic domains of the core intrinsic membrane proteins, D1, D2, CP47, and CP43, as well as several lumen-localized extrinsic proteins surround the Mn-Ca-Cl cluster and interact with each other to stabilize these ions (Bricker and Ghanotakis, 1996; Ferreira et al., 2004).
Ancestral cyanobacteria are the progenitors of chloroplasts in plants and algae (Goksoyr, 1967), and many PSII subunits are conserved between plants, green algae, and cyanobacteria (Hankamer et al., 2001). Of an ancient lineage, cyanobacteria are thought to be primarily responsible for increasing the oxygen concentration in the biosphere to the current day level, thus transforming the terrestrial atmosphere (Sepkoski, 2001). Present day cyanobacteria are the only prokaryotic oxygenic photosynthetic autotrophs (oxyphototrophs).
The functional features of PSII in all oxygenic photosynthetic organisms are remarkably similar. However, the protein components of the extrinsic domain of this enzyme are significantly different among cyanobacteria, eukaryotic algae, and green plants. The model developed from biochemical analysis of PSII shows that in plants and green algae, the three extrinsic proteins are PsbO (33 kD), PsbP (23 kD), and PsbQ (16 kD), whereas in cyanobacteria, they are PsbO, PsbU, and PsbV (cytochrome c550) (Hankamer et al., 2001; Enami et al., 2003). Therefore, the implication is that the composition of the PSII extrinsic domain has undergone extensive and discontinuous changes, although the mechanism of water oxidation has remained virtually unchanged during the time scale of evolution of green plants from cyanobacteria.
These extrinsic proteins were originally identified during resolution-reconstitution studies of isolated O2-evolving PSII preparations from plants and cyanobacteria (Bricker and Ghanotakis, 1996; Seidler, 1996). For example, treatments with high concentrations of salt (e.g., NaCl and CaCl2) as well as Tris, pH 8, result in decreased O2-evolution activity and release of the PsbO, PsbP, and PsbQ proteins from plant PSII, whereas similar treatments of cyanobacterial PSII release the PsbO, PsbU, and PsbV proteins (Koike and Inoue, 1985; Shen and Inoue, 1993; Kashino et al., 2002). Moreover, upon readdition, such proteins bind to the depleted PSII complexes, with concomitant recoveries of activity (Miyao and Murata, 1985; Shen and Inoue, 1993; Lydakis-Simantiris et al., 1999; Enami et al., 2000). Consistent with such biochemical data, targeted inactivation of the psbO and psbV genes in cyanobacteria results in reduced PSII activity (Debus, 2000). In particular, psbO and psbV mutants exhibit impaired photoautotrophic growth as well as reduced PSII activity in CaCl2-deficient medium (Philbrick et al., 1991; Shen et al., 1998). Furthermore, a Chlamydomonas reinhardtii mutant lacking the PsbP protein has decreased oxygen evolution activity (Mayfield et al., 1987). In such a mutant, normal levels of PSII complexes are produced, but increased concentrations of Cl− are required to restore oxygen evolution activity (Rova et al., 1996).
The cyanobacterial PsbU and PsbV proteins are significantly different from the plant PsbP and PsbQ proteins in many respects, including their primary sequences, heme binding in PsbV, and PSII association characteristics. Binding and reconstitution studies revealed that in plants, PsbP binding is dependent on the presence of only PsbO, whereas PsbQ binding requires both PsbO and PsbP (Miyao and Murata, 1983, 1989; Andersson et al., 1984). In cyanobacteria, PsbV can bind almost independently of the other extrinsic proteins, but binding of PsbU requires the presence of both PsbO and PsbV (Shen and Inoue, 1993). Therefore, it is highly unlikely that PsbP and PsbQ in plants evolved directly from ancestral PsbV and PsbU in cyanobacteria.
Despite biochemical evidence suggesting differences between plant and cyanobacterial extrinsic domains, homologs to plant PsbP and PsbQ genes have been identified in cyanobacteria. During a recent proteomic study of PSII isolated from the cyanobacterium Synechocystis 6803, the presence of a cyanobacterial homolog of the PsbQ protein from plants was revealed (Kashino et al., 2002). Recent genomic studies also predict the presence of PsbP orthologs in cyanobacteria, even though the function of such a protein has remained unexplored (De Las Rivas et al., 2004). Neither PsbP nor PsbQ is present in the current crystallographic model of PSII structure (Ferreira et al., 2004), and it is unknown if they contribute to PSII water oxidation activity in cyanobacteria.
A unique feature of PSII, a multisubunit membrane-bound protein machine, is its high rate of turnover. Under normal photosynthetic conditions, PSII is damaged, and the protein complex is at least partially degraded and then reassembled (Aro et al., 1993; Keren et al., 1997). As a consequence, under steady state conditions, the population of PSII in a cyanobacterial cell or a chloroplast is a heterogeneous mixture of fully active as well as partially assembled complexes. To fully understand the details of the biogenesis and function of the Mn-Ca-Cl cluster, we need to consider this dynamic nature of PSII. This study demonstrates that cyanobacterial PsbP and PsbQ are regulatory proteins that are necessary for the assembly and/or maintenance of optimally active PSII complexes, particularly under calcium and chloride depleted conditions.
RESULTS
Identification of PsbP and PsbQ Proteins in Synechocystis 6803
Our recent proteomics study was designed for global identification of the components of PSII from Synechocystis 6803 (Kashino et al., 2002). For this purpose, highly active oxygen-evolving PSII was isolated from the HT3 mutant, which contains a hexa-histidine tag at the C terminus of the CP47 protein (Bricker et al., 1998). This PSII preparation contained 31 distinct polypeptides, including five novel proteins (Kashino et al., 2002). One of the newly identified PSII components, Sll1638, was homologous to the 16-kD PsbQ protein in Arabidopsis thaliana. Because PsbQ in plants is known to be associated with the 23-kD PsbP protein (Bricker and Ghanotakis, 1996), we searched the Synechocystis 6803 genome using the protein sequence of PsbP from Arabidopsis and identified a predicted homolog, Sll1418.
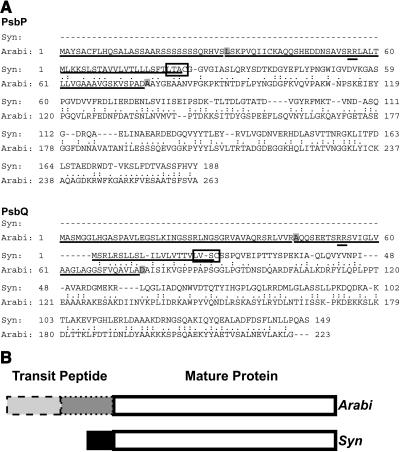
Figure 1A shows the entire PsbP and PsbQ protein sequences from Arabidopsis and Synechocystis 6803 aligned by a global alignment tool (Huang and Miller, 1991), indicating that only the mature proteins were similar. Sequence identities are 27.3 and 32.9% in the mature protein region for PsbP and PsbQ, respectively. The localization sequences are underlined, with both transit peptide cleavage sites highlighted with a gray box for the Arabidopsis proteins. The first part of the bipartite transit peptide is the chloroplast targeting sequence (ChloroP, Emanuelsson et al., 1999), and the second part is the thylakoid lumen-localizing signal (SignalP, Nielsen et al., 1997). Double underlining identifies a twin Arg sequence, important for targeting of these proteins into thylakoid lumen (Settles and Martienssen, 1998). The N terminus of the Arabidopsis protein was recently verified in a proteomics study (Gomez et al., 2003). Figure 1B diagrams the PsbP and PsbQ protein sequences, illustrating the presence of sequence identities primarily in the mature proteins.
Figure 1.
Sequence Comparison of PsbP and PsbQ Proteins from Synechocystis 6803 and Arabidopsis.
(A) Alignments of the predicted amino acid sequences of PsbP and PsbQ proteins from Synechocystis 6803 (Syn) and Arabidopsis (Arabi). Similar residues are indicated by (.) and identical residues by (:). Targeting sequences are underlined. Synechocystis 6803 lipoprotein recognition sites are boxed, with cleavage before the Cys residue. Arabidopsis chloroplast transit peptide cleavage sites (as predicted by ChloroP) are in the first gray box, and thylakoid transit peptide cleavage sites (as predicted by SignalP and determined by N-terminal sequencing by Gomez et al., 2003) are in the second gray box in each sequence. The precursor forms of PsbP and PsbQ proteins in Synechocystis 6803 are 18 and 17.6% identical to their respective homologs in Arabidopsis in this global alignment, respectively.
(B) Diagram depicting sequence alignments of Synechocystis 6803 and Arabidopsis PsbP and PsbQ proteins. Both PsbP and PsbQ have bipartite transit peptides in Arabidopsis, one part for chloroplast envelope translocation (dashed outline) and one for thylakoid lumen localization (dotted outline). These sequences share very little similarity with the much shorter signal peptides on the Synechocystis 6803 proteins (black box). The white boxes represent the mature protein sequences for Arabidopsis and Synechocystis 6803 that are 27.3 and 32.9% identical for PsbP and PsbQ, respectively.
Synechocystis 6803 PsbP and PsbQ have a lipobox (Figure 1A, boxed), suggesting that a type II signal peptidase cleaves their signal peptides (reviewed in Paetzel et al., 2002; predicted by LipoP, Juncker et al., 2003). This enzyme cleaves the signal peptide from an S-glyceride derivative of a prolipoprotein, with the lipid-modified Cys at the signal cleavage site. The presence of an N-terminal modification on PsbQ has some experimental support because the N terminus of the mature protein is blocked (Kashino et al., 2002).
The precursor forms of PsbP and PsbQ from Arabidopsis are translocated into the thylakoid lumen by the ΔpH pathway; similar machinery in prokaryotes is called the twin arginine translocation (Tat) pathway (for review, see Settles and Martienssen, 1998; Dalbey and Robinson, 1999). Even though Synechocystis 6803 has a Tat pathway, PsbP and PsbQ do not appear to be localized to the thylakoid lumen in the same manner as their corresponding plant proteins because they do not contain the twin Arg motif required for ΔpH/Tat mediated localization.
Presence of psbP and psbQ Genes in Many Cyanobacterial Genomes
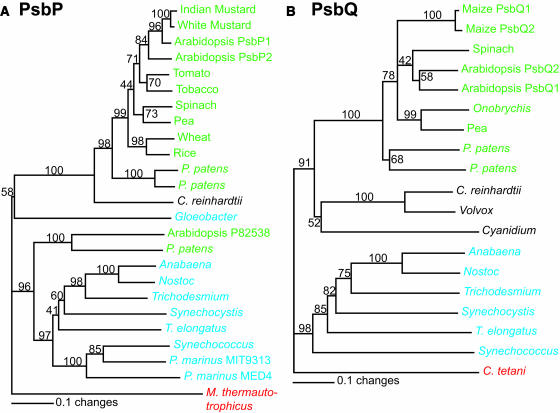
Because PsbP and PsbQ homologs were found in Synechocystis 6803, sequence similarity was compared between these proteins and other PsbP and PsbQ sequences. Figure 2 shows the phylogenetic trees generated from ClustalX alignments of PsbP and PsbQ protein sequences. Most of the plant species group together as one clade (green), separate from the cyanobacterial clade (blue) and the algal clade (black). Outgroups for rooting the trees are shown in red. The observed clade grouping is an expected result based on the evolutionary divergence of plants, algae, and cyanobacteria.
Figure 2.
Phylogenetic Analysis of the PsbP and PsbQ Proteins.
(A) PsbP proteins.
(B) PsbQ proteins.
The numbers at the branch points represent bootstrap values as a percentage from 1000 replications. The trees are rooted with Methanothermobacter thermautotrophicus (for PsbP) and Clostridium tetani (for PsbQ). Green, plant species; blue, cyanobacterial species; black, algal species; red, outgroups. See Methods for further details. Bars = 0.1 mutations/site.
There are several other noteworthy features, including (1) both psbP and psbQ genes are present in at least six different cyanobacterial species, including two filamentous ones. (2) PsbP is found in Gloeobacter violaceus, whereas PsbQ is not. Based on 16S rRNA analysis, G. violaceus is an ancient species (Nelissen et al., 1995). Thus, it is likely that PsbP arose as a component of PSII before PsbQ. Interestingly, several Prochlorococcus species, which are the dominant oxyphototrophs in the open ocean (Ting et al., 2002), also have only psbP but no psbQ. (3) PsbQ is found in the red algae Cyanidium caldarium as well as the recently sequenced Cyanidioschyzon merolae (Matsuzaki et al., 2004), again suggesting that PsbQ was present in their cyanobacterial ancestor. (4) At least 10 copies of psbP, a nuclear gene, are present in the genome sequence of Arabidopsis (Peltier et al., 2000; Schubert et al., 2002). As shown in Figure 2A, only one of these copies (P82538) clusters within the cyanobacterial clade along with one copy of PsbP from the moss Physcomitrella patens. Because the Arabidopsis protein has been identified in a proteomic study of the thylakoid lumen (Schubert et al., 2002), it will be of interest to determine when and where this isoform of PsbP functions in Arabidopsis. It is possible that this isoform of PsbP comes from an ancestral gene still present in both Arabidopsis and P. patens. It is noteworthy that there is no plant PsbQ isoform that groups with the cyanobacterial sequences.
Protein Abundance
The protein encoded by the psbQ (sll1638) gene has been previously detected (Kashino et al., 2002), but there was no experimental evidence that the psbP (sll1418) gene is expressed in Synechocystis 6803. Antibodies were raised against both PsbP and PsbQ from Synechocystis 6803. Immunoblot analysis revealed the presence of PsbP and PsbQ proteins in membrane extracts from wild-type cells. Using the overexpressed PsbP and PsbQ proteins as standards, the amount of each protein present in Synechocystis 6803 membrane extracts was quantified by comparative immunoblotting experiments (see Supplemental Figures S1A and S1B online). This experiment revealed that there are 0.064 and 2.8 pmoles protein per microgram of chlorophyll-containing membranes for PsbP and PsbQ, respectively.
Based on the crystal structure (Zouni et al., 2001; Kamiya and Shen, 2003; Ferreira et al., 2004), each of the core subunits of PSII is present in one copy. Hence, we used CP47 as an indicator to quantitate the number of PSII centers in the membranes. Supplemental Figure S1C online shows the standard curve generated for CP47 in PSII, which was used to determine that 2.25 pmoles of CP47 was present per microgram of chlorophyll-containing membrane. Hence, for every CP47 molecule, there is 0.03 PsbP and 1.2 PsbQ. Therefore, one PsbQ protein is present in every PSII complex, whereas one PsbP protein is present in only 3% of the PSII centers.
Knockouts of psbP and psbQ Genes
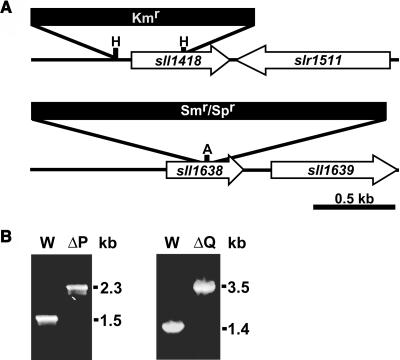
Knockout mutations in the sll1418 and sll1638 genes (ΔpsbP and ΔpsbQ mutants, respectively) were engineered to gain insights into the functions of these cyanobacterial proteins. Strategies for mutagenesis are diagrammed in Figure 3A. Figure 3B shows that complete segregation was obtained for each mutation, which indicates that wild-type copies of the gene are no longer present in the mutant strains. The closest neighboring open reading frames for each of the genes disrupted in this study are shown in Figure 3A. For psbP, the closest open reading frame is oriented in the opposite coding direction, so the expression of it is not expected to be affected by the disruption mutation. There is an open reading frame 147 nucleotides downstream from the psbQ gene, but this is at the outer limit for proximity of genes in the same operon, based on sequence comparisons of known operons (data not shown). Furthermore, this open reading frame downstream of psbQ encodes a urease accessory protein involved in nickel incorporation whose function is not expected to influence PSII activity.
Figure 3.
Inactivation of the psbP and psbQ Genes in Synechocystis 6803.
(A) Restriction maps of the constructs for creating ΔpsbP (sll1418, top) and ΔpsbQ (sll1638, bottom) mutants. Restriction sites used are HincII (H) and AvaI (A). White arrows indicate the coding region for the disrupted gene and its closest neighboring open reading frame. Antibiotic resistance cassettes are for kanamycin (Kmr) and spectinomycin (Smr/Spr).
(B) PCR segregation analysis of the ΔpsbP (left panel) and ΔpsbQ (right panel) mutants. Genomic DNA from wild-type (W), ΔpsbP (ΔP), or ΔpsbQ (ΔQ) strains was the template for amplification with gene-specific primers to confirm segregation of the respective mutations.
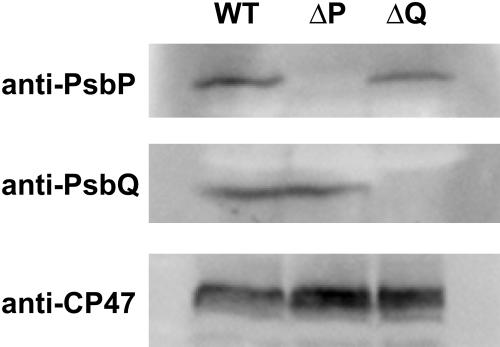
Immunoblot analysis confirmed that the corresponding proteins have been knocked out in both the ΔpsbP and ΔpsbQ mutants (Figure 4). This figure also shows that the abundance of PsbP did not significantly change in membranes isolated from the ΔpsbQ mutants. Similarly, the abundance of PsbQ was not altered in the ΔpsbP mutant. In addition, probing for CP47 in these mutants indicated that there was no significant change in PSII abundance when PsbP or PsbQ was missing (Figure 4, bottom panel).
Figure 4.
Effect of ΔpsbP and ΔpsbQ Mutations on the Accumulation of PsbP, PsbQ, and CP47 Proteins.
Membranes were isolated from wild-type, ΔpsbP (ΔP), and ΔpsbQ (ΔQ) strains and probed using antisera against PsbP (top), PsbQ (middle), or CP47 (bottom). Each lane was loaded with 7 (top) or 5 (middle, bottom) μg chlorophyll-containing membranes.
Photoautotrophic Growth of the Mutant Strains
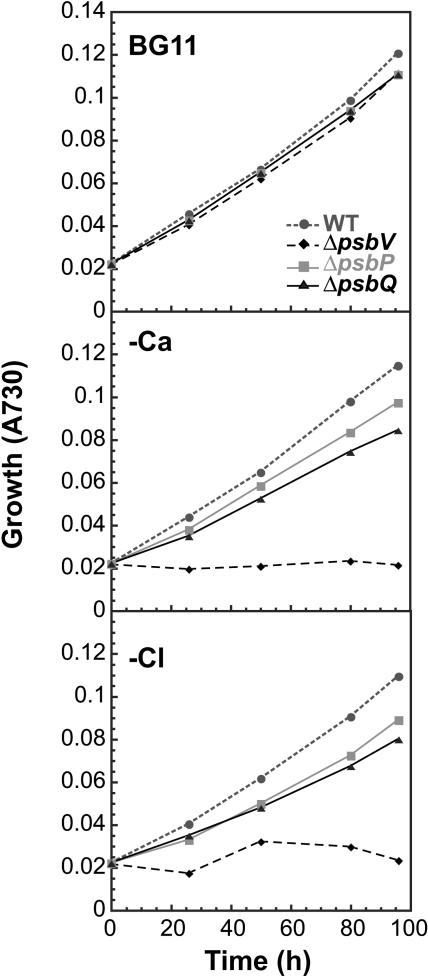
As discussed above, past investigations on the functions of the PsbP and PsbQ proteins in plants typically involved in vitro analysis. During this study, deletion mutations in the psbP and psbQ genes allowed in vivo analysis of the individual functional role of each protein. Figure 5 (top panel) shows that photoautotrophic growth rates of ΔpsbP and ΔpsbQ mutants are similar to wild-type cells under normal growth conditions. By contrast, both mutants showed decreased growth relative to wild-type cells in the absence of calcium (Figure 5, middle panel) or chloride (Figure 5, bottom panel). Calcium and chloride were tested separately to verify that each ion was a requirement in both mutants. These data correlated well with the requirements of PsbP- and PsbQ-depleted plant PSII preparations for high concentrations of CaCl2 for optimal O2-evolving activity (Bricker and Ghanotakis, 1996).
Figure 5.
Photoautotrophic Growth.
Growth of wild-type (circles, small dashed line), ΔpsbP (squares), ΔpsbQ (triangles), and ΔpsbV (diamonds, large dashed line) cells in BG11 medium (top graph), Ca-deficient medium (middle graph), and Cl-deficient medium (bottom graph). A730, absorbance at 730 nm.
We also noted that the growth dependence on calcium and chloride was less severe in the ΔpsbP and ΔpsbQ mutants than in the ΔpsbV mutant lacking the cytochrome c550 protein (Figure 5; Shen et al., 1998). A deletion mutation in the psbO gene also prevents photoautotrophic growth of cyanobacteria when CaCl2 is absent (Philbrick et al., 1991). Therefore, it appears that PsbP and PsbQ are necessary for optimal photoautotrophic growth in the absence of calcium and chloride. However, they do not share the critical roles that PsbO and PsbV play in cyanobacterial PSII-dependent growth under these same conditions.
PSII Activities in the ΔpsbP and ΔpsbQ Mutants
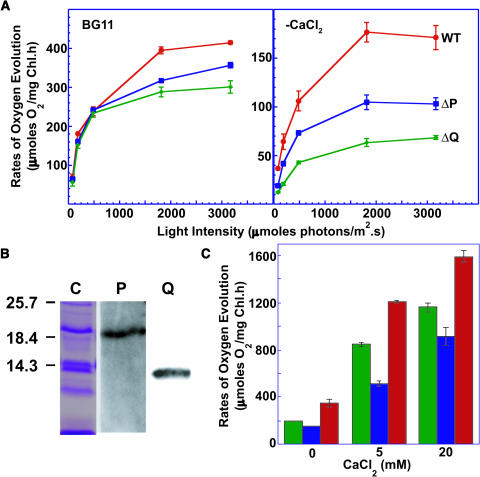
When grown in normal medium, the light-saturated rates of PSII-mediated steady state oxygen evolution from the ΔpsbP and ΔpsbQ mutant cells were between 75 and 85% of the wild-type activity (left panel, Figure 6A, Table 1). However, when grown in the absence of CaCl2, such rates were 63 and 40% from the ΔpsbP and ΔpsbQ mutant cells, respectively, compared with that from the wild-type cells (right panel, Figure 6A, Table 1). As shown in Table 1, the Vmax for the light intensity dependence of this activity in cells grown without CaCl2 was significantly different in the mutants. By contrast, the Km values were not significantly different from that of the wild-type cells, implying that the active PSII complexes in the mutant cells function as well as those in the wild-type cells. However, there were not as many active complexes in the mutant cells as in the wild-type cells. Immunoblot analysis indicated that the abundance of core PSII proteins was not altered in the mutants (Figure 4). Therefore, of the assembled complexes in the ΔpsbP and ΔpsbQ mutants, fewer had water oxidation activity.
Figure 6.
PsbP and PsbQ in PSII.
(A) Dependence of PSII-mediated oxygen evolution activities of wild-type (red circles), ΔpsbP (blue squares), and ΔpsbQ (green diamonds) cells on light intensity. Measurements were in BG11 at 30°C after growth in BG11 (left panel) or in BG11 without CaCl2 (right panel). Error bars represent standard deviation among three replicates.
(B) Immunoblot analysis of purified PSII preparations from the HT3 strain of Synechocystis 6803. Protein samples were fractionated on denaturing SDS-PAGE, transferred to nitrocellulose filter, and immunostained using antibodies against PsbP (P) and PsbQ (Q). Five micrograms of chlorophyll-containing sample was loaded in each lane. Lane C shows the Coomassie-stained profile of the fractionated proteins. Positions of molecular mass markers (kD) are shown on the left.
(C) Rates of oxygen evolution from purified PSII preparations from HT3 (red bar), HT3/ΔpsbP (blue bar), and HT3/ΔpsbQ (green bar) mutant cells in the presence of different concentrations of CaCl2. Error bars represent standard deviations among three replicates.
Table 1.
Kinetic Parameters for Light Intensity Dependence of PSII Activities
| BG11
|
BG11-CaCl2
|
|||
|---|---|---|---|---|
| Km (μmoles photons/m2·s)a | Vmax (μmoles O2/mg Chl·h) | KM (μmoles photons/m2·s)a | Vmax (μmoles O2/mg Chl·h) | |
| Wild type | 362.0 ± 95b | 450.9 ± 59 | 373.7 ± 38 | 197.6 ± 11 |
| ΔpsbP | 324.3 ± 74 | 385.6 ± 43 | 397.8 ± 59 | 123.7 ± 10 |
| ΔpsbQ | 280.5 ± 95 | 331.6 ± 50 | 457.9 ± 48 | 78.6 ± 4.5 |
Calculated from an Eadie-Hofstee plot of data presented in Figure 6A.
Standard errors of three replicates.
As described above, only 3% of the PSII centers had a copy of PsbP (see Supplemental Figure S1 online). By contrast, Figure 6A shows that the absence of PsbP resulted in almost 40% decrease in water oxidation activity. Hence, PsbP must be a regulator of PSII activity even though it is not a part of every functional complex.
To further address the roles of PsbP and PsbQ in PSII, purified PSII complexes from wild-type, ΔpsbP, and ΔpsbQ strains were isolated. Figure 6B shows that isolated PSII complexes from wild-type cells contained both the PsbP and the PsbQ proteins. This confirms the presence of PsbQ in PSII as reported previously (Kashino et al., 2002). Figure 6B also establishes that PsbP was also associated with the PSII complex under these isolation conditions.
We examined the dependence of the O2-evolving activities of these isolated complexes on the concentration of CaCl2 (Figure 6C). In the presence of 20 mM CaCl2, the activity of the wild-type sample was high (1590 μmoles O2 mg Chl−1 h−1), whereas the ΔpsbP sample had 60% and the ΔpsbQ sample had 70% of this activity. When only 5 mM CaCl2 was added, the activity of the ΔpsbP sample decreased to 40% of the wild type, whereas the ΔpsbQ sample still had ∼70% activity. Finally, when no CaCl2 was added, the ΔpsbP sample retained 40% of the wild-type activity, but that of the ΔpsbQ sample decreased to <60%. Even though both the wild type and the mutants were affected by decreasing amounts of CaCl2, the mutants were affected more severely than the wild-type cells. Similar findings have been reported for biochemically resolved spinach (Spinacia oleracea) PSII preparations lacking PsbP and PsbQ (Ghanotakis et al., 1984; Miyao and Murata, 1984). We conclude that the PsbP and PsbQ proteins in cyanobacterial PSII are required for optimal O2-evolution activity, both in vivo and in vitro, and they are involved in modulating the CaCl2 requirement for PSII activity.
DISCUSSION
The data presented in this article demonstrate that homologs of the plant PsbP and PsbQ proteins (1) are present in biochemically purified PSII from the cyanobacterium Synechocystis 6803 and (2) function in optimizing PSII water oxidation activity. These two proteins were previously thought to be restricted only to eukaryotic PSII. Thus, our findings raise intriguing new questions about the evolution of the water oxidation machinery in oxyphototrophs.
Presence of PsbP and PsbQ in Synechocystis 6803 PSII
We have shown that the PsbP and PsbQ proteins are present and functional in cyanobacteria. Significant inferences about the role of these proteins relative to their plant homologs could be made from examining their amino acid sequence. Sequence similarity for Synechocystis 6803 PsbP and PsbQ to their corresponding plant proteins is only maintained in the mature protein sequences (Figure 1). The targeting sequences in the Synechocystis proteins are distinctly different from those for the corresponding plant proteins. During the evolutionary time scale, the genes for the plant PsbP and PsbQ proteins were obtained when chloroplasts were acquired, but they have since been transferred to the nucleus. Many genes have been transferred from the chloroplast genome to the nuclear genome, and the transit peptides needed to get the proteins back to their sites of action in the chloroplast have been genetically fused to the mature protein.
Furthermore, the type of signal peptide on the cyanobacterial proteins is quite interesting. Based on the sequence of their genes, PsbP and PsbQ each have a single N-terminal cleavable signal peptide recognized by bacterial signal peptidase II (Figure 1A). Several prolipoproteins in Synechocystis 6803 are modified in this manner (Maeda and Omata, 1997). We postulate that the presence of a lipid moiety at the N termini of the mature forms of both PsbP and PsbQ anchors them to the cyanobacterial thylakoid membrane.
The presence of a lipid anchor on PsbP and PsbQ explains why these proteins were not previously detected in PSII from cyanobacteria. Traditional resolution and reconstitution experiments do not release the PsbP and PsbQ proteins from the PSII complex of Synechocystis 6803 (Kashino et al., 2002) or thermophilic cyanobacteria (Shen and Inoue, 1993; Enami et al., 2003). Because of the lipid anchor on PsbP and PsbQ, high-salt treatment could not release them, even though these proteins are largely hydrophilic. Therefore, these proteins would not have been previously characterized as extrinsic PSII proteins.
Additionally, PsbP was not identified in our previous proteomics study (Kashino et al., 2002). PsbP migrates at the same position as PsbV in SDS-PAGE, so a unique band was not seen. Furthermore, PsbP is present in isolated membranes at 3% of the abundance of PSII (see Supplemental Figure S1 online), so its abundance in isolated PSII may have been too low to detect in the proteomics study. Even though PsbP is substoichiometric, its absence creates up to 40% decrease in activity (Table 1). The implication is that PsbP in cyanobacteria is a regulatory protein and is not a structural component of every functional PSII complex.
Functional Role of PsbP and PsbQ in Synechocystis 6803 PSII
We have presented evidence that the PsbP and PsbQ proteins could have a similar function in cyanobacteria as in plants, based on their role in modifying the CaCl2 requirements for PSII activity. Figure 6C shows that increasing CaCl2 availability to the complex increased the capacity for oxygen evolution in the absence of PsbP or PsbQ more than in wild-type PSII. The in vivo data from the ΔpsbP and ΔpsbQ mutants show that the absence of CaCl2 inhibits optimal photoautotrophic growth and water oxidation activity (Figures 5 and 6A). This finding is similar to results for some of the extrinsic loop mutants of CP47 (Clarke and Eaton-Rye, 1999) as well as the ΔpsbO (Philbrick et al., 1991) and ΔpsbV mutants (Shen et al., 1998) of Synechocystis 6803. The intermediate phenotype of the ΔpsbP and ΔpsbQ mutants as opposed to the large decrease in activity in the ΔpsbV mutant upon CaCl2 removal suggests that PsbP and PsbQ are more peripheral in the extrinsic domain of cyanobacterial PSII. Such a peripheral location may explain why these two proteins have not yet been seen in the current crystallographic models of cyanobacterial PSII (Zouni et al., 2001; Kamiya and Shen, 2003; Ferreira et al., 2004). Furthermore, the lipid anchor of PsbP and PsbQ increases their affinity for the lipid bilayer. Therefore, the highly purified PSII used for crystallographic modeling may not contain either of these proteins.
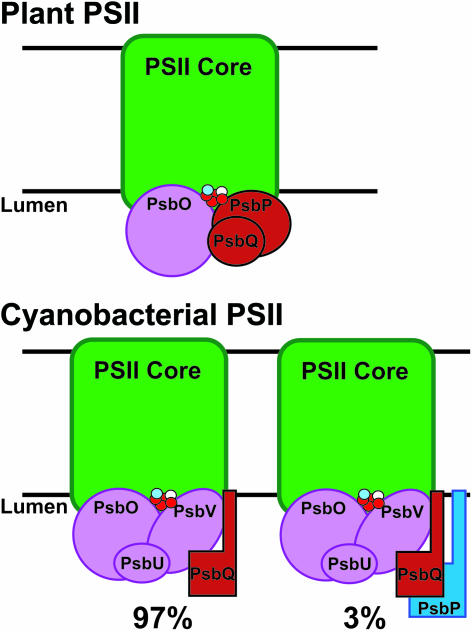
We suggest that PsbP and PsbQ have regulatory roles in aiding calcium and chloride association with the PSII oxygen evolution domain, which is not dependent on their tight association with the complex. PsbP, especially, may be required for assembly of PSII and therefore would not remain tightly bound to the highly resolved complex. Figure 7 is a model depicting the water oxidation domain in plants and cyanobacteria. In plants, there is most likely a homogenous mix of PSII complexes, each containing PsbO, PsbP, and PsbQ in the extrinsic domain. The model shows that cyanobacteria have PsbQ associated with the other extrinsic proteins and attached to the membrane bilayer. PsbP also has a membrane anchor, but it is only associated with a small percentage of the PSII complexes in the population. This model reflects the notion that PSII is a dynamic complex, and at any given time, the population of PSII centers is a heterogeneous mixture of partially assembled and fully active complexes. Further studies are needed to better understand the individual contributions of PsbP and PsbQ during the life cycle of PSII.
Figure 7.
Schematic Models of PSII in Cyanobacteria and Plants.
The core PSII subunits (D1, D2, CP43, and CP47, shown as green boxes) provide the primary binding sites for the Mn-Ca-Cl cluster (red, blue, and white dots, respectively) and extrinsic proteins (shown in red, blue, or purple). Based on the suggested stoichiometry for plant PSII, there should be one PsbP and PsbQ molecule for every PSII core (top). Cyanobacterial PSII has five extrinsic subunits in the thylakoid lumen, with PsbP ant55d PsbQ anchored to the membrane. PsbP (blue) is substoichiometric, so not every PSII core has an associated PsbP protein.
PsbP and PsbQ Homologs in Other Cyanobacteria
Many recently sequenced cyanobacterial genomes have homologs of the genes encoding PsbP and PsbQ, which suggests that these proteins were present in the ancestral PSII complex before the split dividing the chloroplast and cyanobacterial lineages. This finding answers the long-standing question as to how plants and green algae acquired PsbP and PsbQ. Figure 2 shows that the homologs separate into well-supported plant and cyanobacterial groups, with the exception of PsbP isoforms that fall into the cyanobacterial group.
It is now generally accepted that the reaction centers in anoxygenic and oxygenic photosynthetic organisms have a common origin (Raymond et al., 2002). However, little is known about the way in which the reaction center complex in PSII acquired the ability to oxidize water to form molecular oxygen, a reaction that eventually transformed the redox environment of the biosphere. The mechanisms of the fairly involved reactions that result in the production of O2 have remained unaltered during billions of years of evolution from ancestral cyanobacteria to modern land plants. To date, a disturbing gap in our understanding of the composition of PSII has been the apparently discontinuous evolution of the lumenal extrinsic domain, which directly influences the binding environment of the catalytic Mn-Ca-Cl cluster for water oxidation.
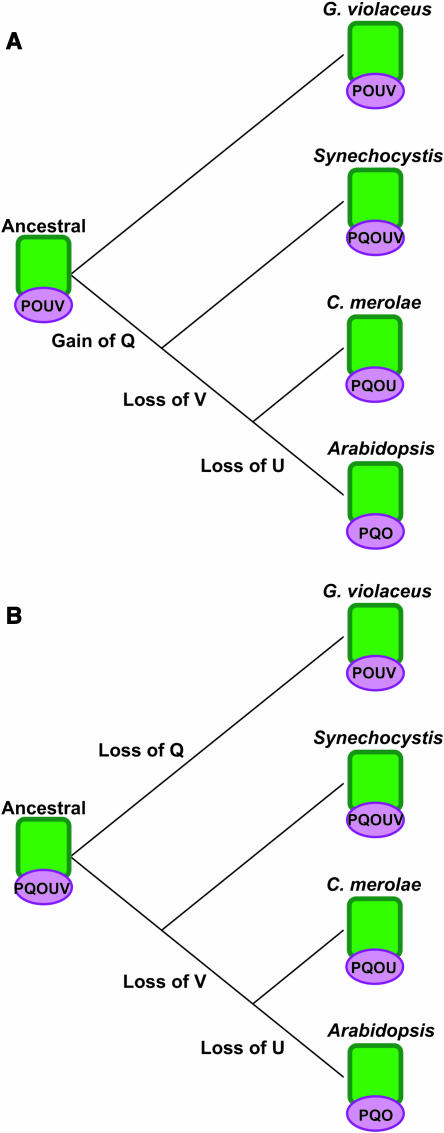
According to the currently accepted model, PsbO is the only extrinsic protein conserved between cyanobacteria and plants, and both the PsbP and PsbQ proteins are new inventions in green plants and algae (Hankamer et al., 2001; Enami et al., 2003). It has also been suggested that PsbO was the only extrinsic protein in ancestral PSII (De Las Rivas et al., 2004). Based on our findings and the phylogenetic relationships within the PsbP and PsbQ families, we propose a new model for the evolution of the PSII extrinsic domain in which the ancestral PSII had multiple extrinsic subunits. As shown in Figure 8A, it is possible that the ancestral PSII had PsbO, PsbP, PsbU, and PsbV in the extrinsic domain. This ancestor is presently represented by G. violaceus. Support for this model is in the primitive structure of the photosynthetic membranes in G. violaceus, which are not developed into thylakoid structures as in the other oxyphototrophs. The model in Figure 8A suggests that PsbQ was gained as an extrinsic protein before the branch point for any of the thylakoid membrane containing organisms. After the evolutionary separation of the cyanobacterial lineage, PsbV was lost. Here, C. merolae represents the red algae because its entire genome was recently sequenced (Matsuzaki et al., 2004). PsbU was lost in the green photosynthetic eukaryote lineage, and PsbP and PsbQ acquired a more central role. Figure 8B is an alternate model for PSII evolution. In this model, the ancestral PSII had all five extrinsic proteins contributing to PSII activity, which is represented by the Synechocystis 6803 lineage. In the G. violaceus lineage, PsbQ was lost after this line separated from what eventually led to other oxyphototrophs. In the red algal lineage, PsbV was lost to result in what has presently been seen in C. merolae. As mentioned above, PsbU was lost after the red algae lineage separated from the plant lineage. It is important to note that biochemical data suggest that the red alga C. caldarium has PsbV and not PsbP (Ohta et al., 2003), suggesting that the evolutionary history of the water oxidation domain in the red algae may be more complex.
Figure 8.
Model of Evolution of the PSII Extrinsic Protein Domain.
(A) Ancestral PSII with four subunits in the extrinsic domain. In this model, G. violaceus is the modern representation of such an ancestral organism. PsbQ was acquired before the split separating the cyanobacteria (Synechocystis) from the eukaryotic photosynthetic organisms. PsbV was lost before the separation of red algae (C. merolae) and green photosynthetic eukaryotes (Arabidopsis), and PsbU was lost only in the plant lineage. O, PsbO; P, PsbP; Q, PsbQ; U, PsbU; V, PsbV.
(B) Ancestral PSII with five subunits in the extrinsic domain. In this model, the nonthylakoid cyanobacterial lineage lost PsbQ (G. violaceus), and other cyanobacteria (Synechocystis) represent the ancestral organism. As described above, PsbV was lost before the red algae (C. merolae) separation, and PsbU was lost after this separation from green photosynthetic eukaryotes (Arabidopsis).
Both models in Figure 8 reflect that the presence of PsbU and PsbV is sufficient for PSII activity without both PsbP and PsbQ. When PsbU and PsbV are lost, PsbP is recruited to have a more central role in water oxidation activity than it does in the presence of PsbU and PsbV. Replacing PsbV with PsbP in plant PSII is supported by a recent PsbP structure analysis, in which the authors suggested that the electrostatic surface potential of PsbP is similar to PsbV (Ifuku et al., 2004). Ifuku and colleagues suggested that the binding potential, and possibly the functional contribution, of PsbP in plants is not much different than PsbV in cyanobacteria. Furthermore, PsbV is a heme binding protein, but there is no known contribution to PSII redox activities. If the redox contributions of PsbV are no longer needed in plant PSII activity, it follows that evolution would select for a simpler protein to take its place.
These discussions lead to three major conclusions about the PSII water oxidation domain, in general. (1) It is apparent that PsbO must be present for oxygenic photosynthesis. (2) Two or more other extrinsic subunits must also be present to optimize water oxidation activity. (3) There is flexibility as to which of the known extrinsic subunits (PsbP, PsbQ, PsbV, or PsbU) are used to optimize PSII activity. It appears that the known oxyphototrophs have evolved to represent different combinations of extrinsic subunits (Figure 8); however, there are anomalies within each group. For example, some of the cyanobacteria do not have PsbQ (Figure 2), and some red algae have PsbV (Ohta et al., 2003). Such variability in the composition of the PSII water oxidation domain may be a direct consequence of the need for optimization of oxygen evolution activities in various oxyphototrophs that occupy diverse ecological niches.
METHODS
Bacterial Strains and Culture Conditions
Synechocystis 6803 cells were grown at 30°C under 30 μmoles photons m−2 s−1 of white light in the BG11 medium (Allen, 1968). Growth media for ΔpsbP, ΔpsbQ, and ΔpsbV mutant strains was supplemented with 10 μg/mL of kanamycin, 10 μg/mL of spectinomycin, and 2 μg/mL of gentamycin, respectively. CaCl2-depleted BG11 was prepared in glassware, which was rinsed with HCl to remove any contaminating ions. When needed, Ca2+ or Cl− was added back individually as Ca(NO3)2 or NaCl salt. Growth of cyanobacteria was monitored by measuring OD730nm on a DW2000 spectrophotometer (SLM-Aminco, Urbana, IL) or a μQuant plate reader (Bio-Tek Instruments, Winooski, VT).
Gene Cloning and Isolation of Mutants
The sll1418 (psbP) and sll1638 (psbQ) genes of Synechocystis 6803 were amplified by PCR using the following oligonucleotide primers: 5′-GGAAAAAGAGCCAAGCGTGGGAAG-3′ and 5′-CCTCCGACTTGGACTGGGTAATTC-3′ for psbP; 5′-CCTAAACTCTGTTGCACA-3′ and 5′-CTGGAGCTTAATGCAGAA-3′ for psbQ. These PCR products were cloned into the TA cloning vector (Invitrogen, Carlsbad, CA). The ΔpsbP mutant was generated by replacing a HincII fragment in psbP with a kanamycin resistance (Kmr) cartridge. To introduce the same mutation into the HT3 (Kmr) strain (see below), the ΔpsbP mutant was also generated using a spectinomycin resistance (Smr/Spr) cartridge in a similar manner. The ΔpsbQ mutant was generated by inserting an Smr/Spr cartridge into an AvaI site in psbQ. The ΔpsbV mutant was engineered by replacing Synechocystis 6803 nucleotides 21322151 to 21322577 (Cyanobase, http://www.kazusa.or.jp/cyanobase/, 11-5-03) in the psbV (sll0258) gene with a gentamycin resistance cartridge.
Protein Overexpression and Immunodetection
The coding regions of Synechocystis 6803 psbP and psbQ genes were engineered to overexpress PsbP and PsbQ proteins in Escherichia coli using the pET-41b(+) system (Novagen, Darmstadt, Germany). Antibodies against each of the purified proteins were raised in rabbits (Cocalico Biologicals, Reamstown, PA). Membrane extracts were fractionated by SDS-PAGE (16% acrylamide and 6 M urea); the fractionated proteins were blotted onto nitrocellulose membranes and reacted with individual antisera. The hybridization signals were developed using enhanced chemiluminescence reagents (Pierce Biotechnology, Rockford, IL) and then visualized on a Fujifilm LAS-1000 plus imager (Fujifilm, Stamford, CT). Negative background (thick white bands) appeared on the blots from phycobiliproteins in the membrane preparation. For protein quantitation by comparative immunoreaction, increasing concentrations of antigen were fractionated on the same gel that also had various concentrations of membrane extracts. The immunoblot signal intensities were quantified using ImageJ 1.31 (National Institutes of Health, http://rsb.info.nih.gov/ij/). Using the equation of the curve fits (R > 0.94) shown in Supplemental Figure S1 online, protein abundance on a per chlorophyll basis was determined.
PSII Purification
The HT3 strain of Synechocystis 6803, which has a hexa-histidine tag on CP47 (Bricker et al., 1998), was a generous gift from T. Bricker. The constructs for the knockout mutants (see above) were used to transform the HT3 strain to generate the HT3/ΔpsbP and HT3/ΔpsbQ mutants. PSII preparations from the three strains were isolated according to Kashino et al. (2002).
Oxygen Evolution
Cells were harvested after 6 d of growth in BG11 or BG11-CaCl2 and resuspended in fresh BG11 at 5 μg Chl/mL. Isolated PSII samples were suspended at 1 μg Chl/mL in a buffer containing 50 mM Mes-NaOH, pH 6.0, and 0.5 M sucrose, with different concentrations of CaCl2 added from a 2 M stock. This assay buffer differed from our previous study (Kashino et al., 2002) because we removed the chloride-containing salts. PSII-mediated oxygen evolution was measured on a Clark-type electrode in the presence of 0.5 mM 2,6-dichloro-p-benzoquinone and 1 mM potassium ferricyanide. Light intensity was determined using a quantum photometer (Licor, Lincoln, NE).
Sequence Alignments
PsbP and PsbQ proteins from Synechocystis 6803 (Sll1418 and Sll1638, respectively) and Arabidopsis thaliana (At1g06680 and At4g21280, respectively) were aligned using the LALIGN program (Huang and Miller, 1991). The sequences were aligned using the global parameters with the Blosum35 matrix and gap penalties set at −10 and −4 for opening and extension, respectively. Similar residues are indicated by (.) and identical residues by (:).
Phylogenetic Analysis
Multiple sequence alignments were performed using the ClustalX program (Thompson et al., 1997). Manual adjustments were made to the alignments before building phylogenetic trees. Resulting alignments were used to generate neighbor-joining trees using the PAUP* 4.0b10 software (Swofford, 2002). Outgroups, M. thermoautotrophicus (for PsbP) and C. tetani (for PsbQ), were chosen after Psi-BLAST analysis of GenBank sequences. These sequences were the closest match for either PsbP or PsbQ from a nonphotosynthetic organism and are not expected to be functional homologs.
Complete Nomenclature and Sequence Accession Numbers
The locus numbers for Arabidopsis sequences referenced are At1g06680 (Q42029), At2g30790 (O49344), At4g15510 (O23403), and At3g55330 (P82538) for PsbP and At4g05180 (Q41932) and At4g21280 (Q9XFT3) for PsbQ, with the Swiss-Prot identifier in parentheses. Sequence data from this article were previously deposited with the EMBL/GenBank data libraries under the following accession numbers: pea (Pisum sativum) (PsbQ: AAP43511), rice (Oryza sativa) (AAC98778), M. thermautotrophicus (NP_275651), C. caldarium (BAD01024), and C. tetani (NP_783832). The Swiss-Prot database numbers (http://us.expasy.org/sprot/, 9-5-03) are Chlamydomonas reinhardtii (P11471 and P12852), Indian mustard (Brassica juncea) (Q96334), maize (Zea mays) (Q41048 and Q41806), Onobrychis (O22591), pea (P. sativum) (PsbP: P16059), spinach (S. oleracea) (P12302 and P12301), tobacco (Nicotiana tabacum) (P18212), tomato (Lycopersicon esculentum) (P29795), Volvox carteri (Q41643), wheat (Triticum aestivum) (Q00434), and white mustard (Sinapis alba) (P11594). Protein sequences for Physcomitrella patens were translated from DNA contigs (10052 and 11296 for PsbP; 1327 and 1042 for PsbQ) in PhyscoBase (http://moss.nibb.ac.jp/, 3-24-04). Protein sequences from Anabaena sp PCC 7120 (All3076 and All1355), G. violaceus PCC 7421 (Gll1440), Synechocystis sp PCC 6803 (Sll1418 and Sll1638), Synechococcus sp WH8102 (SYNW0927 and SYNW2505), P. marinus MIT9313 (PMT1078), Prochlorococcus marinus MED4 (PMM1098), and Thermosynechococcus elongatus (Tlr2075 and Tll2057) were obtained from Cyanobase (http://www.kazusa.or.jp/ cyanobase/, 12-15-03). Nostoc punctiforme ATCC 29133 and Trichodesmium erythraeum sequences were downloaded from the Joint Genome Institute database (http://www.jgi.doe.gov/JGI_microbial/html/index.html, 9-5-03). Where two sequence numbers are listed for any organism, the first one corresponds to the PsbP homolog and the second one corresponds to the PsbQ homolog. The unicellular red alga discussed in Figure 8 is Cyanidioschyzon merolae 10D.
Supplementary Material
Acknowledgments
We thank Terry M. Bricker for the HT3 strain, Victor Bartsevich for creating the ΔpsbV mutant strain in our laboratory, and other members of the Pakrasi laboratory for collegial discussions. This work was supported by funding from the National Science Foundation and the Department of Energy (H.B.P.). L.E.T. and J.L.R. were partially supported by individual graduate research fellowships from the National Science Foundation, and N.K. was partially supported by a postdoctoral fellowship from the International Human Frontier Science Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Himadri B. Pakrasi (pakrasi@wustl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023515.
References
- Allen, M.M. (1968). Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4. [DOI] [PubMed] [Google Scholar]
- Andersson, B., Critchley, C., Ryrie, I.J., Jansson, C., Larsson, C., and Andersson, J.M. (1984). Modification of the chloride requirement for photosynthetic O2 evolution. FEBS Lett. 168, 113–117. [Google Scholar]
- Aro, E.M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosytem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Bricker, T.M., and Ghanotakis, D.M. (1996). Introduction to oxygen evolution. In Oxygenic Photosynthesis: The Light Reactions, D.R. Ort and C.F. Yocum, eds (Dordrecht: Kluwer Academic Publishers), pp. 137–164.
- Bricker, T.M., Morvant, J., Masri, N., Sutton, H.M., and Frankel, L.K. (1998). Isolation of a highly active photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP 47. Biochim. Biophys. Acta 1409, 50–57. [DOI] [PubMed] [Google Scholar]
- Clarke, S.M., and Eaton-Rye, J.J. (1999). Mutation of Phe-363 in the photosystem II protein CP47 impairs photoautotrophic growth, alters the chloride requirement, and prevents photosynthesis in the absence of either PSII-O or PSII-V in Synechocystis sp. PCC 6803. Biochemistry 38, 2707–2715. [DOI] [PubMed] [Google Scholar]
- Dalbey, R.E., and Robinson, C. (1999). Protein translocation into and across the bacterial plasma membrane and the plant thylakoid membrane. Trends Biochem. Sci. 24, 17–22. [DOI] [PubMed] [Google Scholar]
- De Las Rivas, J., Balsera, M., and Barber, J. (2004). Evolution of oxygenic photosynthesis: Genome-wide analysis of the OEC extrinsic proteins. Trends Plant Sci. 9, 18–25. [DOI] [PubMed] [Google Scholar]
- Debus, R.J. (2000). The polypeptides of photosystem II and their influence on manganotyrosyl-based oxygen evolution. Met. Ions Biol. Syst. 37, 657–711. [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami, I., Iwai, M., Akiyama, A., Suzuki, T., Okumura, A., Katoh, T., Tada, O., Ohta, H., and Shen, J.R. (2003). Comparison of binding and functional properties of two extrinsic components, Cyt c550 and a 12 kDa protein, in cyanobacterial PSII with those in red algal PSII. Plant Cell Physiol. 44, 820–827. [DOI] [PubMed] [Google Scholar]
- Enami, I., Yoshihara, S., Tohri, A., Okumura, A., Ohta, H., and Shen, J.R. (2000). Cross-reconstitution of various extrinsic proteins and photosystem II complexes from cyanobacteria, red algae and higher plants. Plant Cell Physiol. 41, 1354–1364. [DOI] [PubMed] [Google Scholar]
- Ferreira, K.N., Iverson, T.M., Maghlaoui, K., Barber, J., and Iwata, S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838. [DOI] [PubMed] [Google Scholar]
- Ghanotakis, D.F., Topper, J.N., Babcock, G.T., and Yocum, C.F. (1984). Water-soluble 17 and 23 kDa polypeptides restore oxygen evolution activity by creating a high-affinity binding site for Ca2+ on the oxidizing side of photosystem II. FEBS Lett. 170, 169–173. [Google Scholar]
- Goksoyr, J. (1967). Evolution of eucaryotic cells. Nature 214, 1161. [DOI] [PubMed] [Google Scholar]
- Gomez, S.M., Bil, K.Y., Aguilera, R., Nishio, J.N., Faull, K.F., and Whitelegge, J.P. (2003). Transit peptide cleavage sites of integral thylakoid membrane proteins. Mol. Cell Proteomics 2, 1068–1085. [DOI] [PubMed] [Google Scholar]
- Hankamer, B., Morris, E., Nield, J., Carne, A., and Barber, J. (2001). Subunit positioning and transmembrane helix organisation in the core dimer of photosystem II. FEBS Lett. 504, 142–151. [DOI] [PubMed] [Google Scholar]
- Hillier, W., and Babcock, G.T. (2001). Photosynthetic reaction centers. Plant Physiol. 125, 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.Q., and Miller, W. (1991). A time-efficient, linear-space local similarity algorithm. Adv. App. Math. 12, 337–357. [Google Scholar]
- Ifuku, K., Nakatsu, T., Kato, H., and Sato, F. (2004). Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep. 5, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker, A.S., Willenbrock, H., Von Heijne, G., Brunak, S., Nielsen, H., and Krogh, A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, N., and Shen, J.-R. (2003). Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc. Natl. Acad. Sci. USA 100, 98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashino, Y., Lauber, W.M., Carroll, J.A., Wang, Q., Whitmarsh, J., Satoh, K., and Pakrasi, H.B. (2002). Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012. [DOI] [PubMed] [Google Scholar]
- Keren, N., Berg, A., van Kan, P.J., Levanon, H., and Ohad, I.I. (1997). Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: The role of back electron flow. Proc. Natl. Acad. Sci. USA 94, 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike, H., and Inoue, Y. (1985). Properties of a peripheral 34 kDa protein in Synechococcus vulcanus phototsystem II particles. Its exchangeability with spinach 33 kDa protein in reconstitution of O2 evolution. Biochim. Biophys. Acta 807, 64–73. [Google Scholar]
- Lydakis-Simantiris, N., Hutchison, R.S., Betts, S.D., Barry, B.A., and Yocum, C.F. (1999). Manganese stabilizing protein of photosystem II is a thermostable, natively unfolded polypeptide. Biochemistry 38, 404–414. [DOI] [PubMed] [Google Scholar]
- Maeda, S., and Omata, T. (1997). Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 272, 3036–3041. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, M., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. [DOI] [PubMed] [Google Scholar]
- Mayfield, S.P., Rahire, M., Frank, G., Zuber, H., and Rochaix, J.D. (1987). Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 84, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao, M., and Murata, N. (1983). Partial disintegration and reconstitution of the photosynthetic oxygen evolution system: Binding of 24 kilodalton and 18 kilodalton polypeptides. Biochim. Biophys. Acta 725, 87–93. [Google Scholar]
- Miyao, M., and Murata, N. (1984). Calcium ions can be substituted for the 24-kDa polypeptide in photosynthetic oxygen evolution. FEBS Lett. 168, 118–120. [Google Scholar]
- Miyao, M., and Murata, N. (1985). The Cl− effect on photosynthetic oxygen evolution: Interaction of Cl− with 18-kDa, 24-kDa and 33-kDa proteins. FEBS Lett. 180, 303–308. [Google Scholar]
- Miyao, M., and Murata, N. (1989). The mode of binding of three extrinsic proteins of 33 kDa, 23 kDa and 18 kDa in the photosystem II complex of spinach. Biochim. Biophys. Acta 977, 315–321. [DOI] [PubMed] [Google Scholar]
- Nelissen, B., Van de Peer, Y., Wilmotte, A., and De Wachter, R. (1995). An early origin of plastids within the cyanobacterial divergence is suggested by evolutionary trees based on complete 16S rRNA sequences. Mol. Biol. Evol. 12, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Ohta, H., Suzuki, T., Ueno, M., Okumura, A., Yoshihara, S., Shen, J.R., and Enami, I. (2003). Extrinsic proteins of photosystem II: An intermediate member of PsbQ protein family in red algal PS II. Eur. J. Biochem. 270, 4156–4163. [DOI] [PubMed] [Google Scholar]
- Paetzel, M., Karla, A., Strynadka, N.C., and Dalbey, R.E. (2002). Signal peptidases. Chem. Rev. 102, 4549–4580. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Friso, G., Kalume, D.E., Roepstorff, P., Nilsson, F., Adamska, I., and van Wijk, K.J. (2000). Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12, 319–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbrick, J.B., Diner, B.A., and Zilinskas, B.A. (1991). Construction and characterization of cyanobacterial mutants lacking the manganese-stabilizing polypeptide of photosystem II. J. Biol. Chem. 266, 13370–13376. [PubMed] [Google Scholar]
- Raymond, J., Zhaxybayeva, O., Gogarten, J.P., Gerdes, S.Y., and Blankenship, R.E. (2002). Whole-genome analysis of photosynthetic prokaryotes. Science 298, 1616–1620. [DOI] [PubMed] [Google Scholar]
- Rova, E.M., Mc Ewen, B., Fredriksson, P.O., and Styring, S. (1996). Photoactivation and photoinhibition are competing in a mutant of Chlamydomonas reinhardtii lacking the 23-kDa extrinsic subunit of photosystem II. J. Biol. Chem. 271, 28918–28924. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schroder, W.P., and Kieselbach, T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277, 8354–8365. [DOI] [PubMed] [Google Scholar]
- Seidler, A. (1996). The extrinsic polypeptides of photosystem II. Biochim. Biophys. Acta 1277, 35–60. [DOI] [PubMed] [Google Scholar]
- Sepkoski, J.J. (2001). Foundations: Life in the oceans. In The Book of Life, S.J. Gould, ed (New York: W. W. Norton & Co.), pp. 37–44.
- Settles, A.M., and Martienssen, R. (1998). Old and new pathways of protein export in chloroplasts and bacteria. Trends Cell Biol. 8, 494–501. [DOI] [PubMed] [Google Scholar]
- Shen, J.R., and Inoue, Y. (1993). Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32, 1825–1832. [DOI] [PubMed] [Google Scholar]
- Shen, J.R., Qian, M., Inoue, Y., and Burnap, R.L. (1998). Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37, 1551–1558. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2002). PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods). (Sunderland, MA: Sinauer Associates).
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C.S., Rocap, G., King, J., and Chisholm, S.W. (2002). Cyanobacterial photosynthesis in the oceans: The origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142. [DOI] [PubMed] [Google Scholar]
- Zouni, A., Witt, H.T., Kern, J., Fromme, P., Krauss, N., Saenger, W., and Orth, P. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409, 739–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.