Abstract
The sensitive to freezing2-1 (sfr2-1) mutation causes freezing sensitivity in Arabidopsis thaliana. By mapping, transgenic complementation, and sequencing, sfr2-1 was revealed to be a mutation in gene At3g06510. A new knockout allele was obtained, and its identical freezing-sensitive phenotype confirmed that the SFR2 gene product is essential for freezing tolerance. Transcription of SFR2 was observed to be constitutive rather than stress inducible and was distributed throughout most aerial tissues. SFR2 encodes a protein homologous to family 1 glycosyl hydrolases (β-glycosidases), but the predicted AtSFR2 protein is divergent from all other family 1 β-glycosidases of Arabidopsis, showing closer homology to the sequences of several β-glycosidases from thermophilic archea and bacteria. After purification from a heterologous expression system, AtSFR2 displayed a specific hydrolytic activity against β-d-glucosides.
INTRODUCTION
What genes are responsible for the freezing tolerance of cold-acclimated plants? The characterization of genes whose expression is induced during cold acclimation has been a powerful approach to this problem (reviewed in Thomashow, 1999). Its success is manifest in the manipulation of freezing and drought tolerance by transgenesis with transcription factors (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000) and in the molecular genetic description of significant portions of the signaling pathway (Stockinger et al., 2001; Thomashow et al., 2001; Fowler and Thomashow, 2002; Seki et al., 2003). However, the contributions of individual genes to freezing tolerance, except via the control of other genes' expression, have not been readily ascertained by this approach (Xin and Browse, 2000).
A complementary approach has been to discover genes by their effects on phenotype. Natural variation has been exploited in some commercially important species to identify quantitative trait loci affecting freezing tolerance (Teutonico et al., 1995; Byrne et al., 1997; Sutka et al., 1997; Lerceteau et al., 2000). However, quantitative trait loci are difficult to map with sufficient accuracy for gene identification and can only reveal genes in which there is natural variation between cultivars. Therefore, some groups have pursued approaches involving the isolation of mutants in the model plant Arabidopsis thaliana.
In one study, mutants were obtained that caused elevated levels of freezing tolerance in the absence of cold acclimation (Xin and Browse, 1998). The mutations may point to components of the signaling pathway(s) that are responsible for cold acclimation or identify genes that can make self-sufficient individual contributions to freeze protection. Another group used a reporter-based screen to identify mutants specifically altered in their signaling response to cold (and also to osmotic stress) (Ishitani et al., 1997), revealing several novel components of the signal transduction pathway (Xiong et al., 2002).
Mutants deficient in freezing tolerance have also been described. The frs1 mutation was found to be an allele of ABA3 (Llorente et al., 2000), confirming the necessity for normal abscisic acid signaling during cold acclimation (Heino et al., 1990). The freezing sensitivity of two sensitive to freezing (sfr) mutants (Warren et al., 1996) has been interpretable in terms of prior knowledge about freezing tolerance. The sfr4-1 mutant fails to elevate sugar levels during cold acclimation (McKown et al., 1996) and consequently fails to prevent the lesion known as loss of osmotic responsiveness (Uemura et al., 2003). The sfr6-1 mutant fails to induce genes with C repeat/dehydration-responsive (CRT/DRE) elements in their promoters (Knight et al., 1999; Boyce et al., 2003) and thus lacks a set of protein and biochemical changes that, collectively, are known to engender freezing tolerance (Gilmour et al., 2000). The sfr2-1 mutation, on the other hand, does not affect any known aspect of the cold response (McKown et al., 1996; Knight et al., 1999). Therefore, characterization of the SFR2 gene should reveal a previously undetected mechanism contributing to freezing tolerance.
No pleiotropic effects of the sfr2-1 mutation have been observed (McKown et al., 1996)—not even minor effects on vigor, fertility, or morphology—either at normal growth temperatures or during cold acclimation. This suggests that the function of SFR2 is likely to be quite specific to the prevention of freezing injury. The sfr2-1 mutant is unusual among the sfr mutants in that its strong phenotype in the whole-plant freezing assay contrasts with a relatively weak phenotype in the electrolyte leakage assay (Warren et al., 1996). This might indicate an unusual mode of protective action for the product of the SFR2 gene. Here, we identify the SFR2 gene, characterize its expression and its homology to proteins of known function, and discuss the implications for its role in freezing tolerance.
RESULTS
Classical Genetic Mapping of the SFR2 Gene
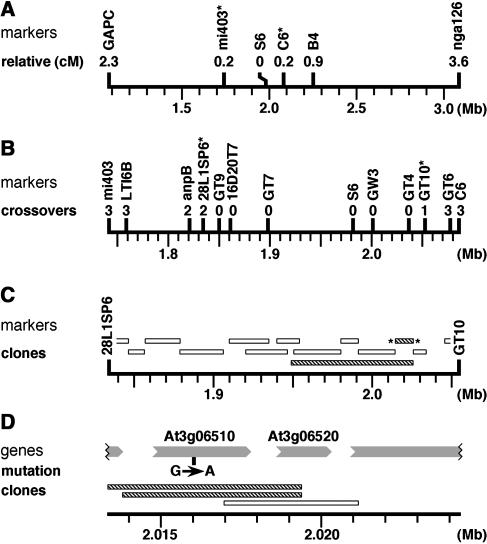
The sfr2-1 mutation was previously mapped to the region between markers GAPC and nga126 on chromosome III (Thorlby et al., 1999). From the mapping cross of sfr2-1/sfr2-1 Columbia (Col) × SFR2+/SFR2+ Landsberg erecta, we screened 690 F2 zygotes for crossovers in this region. Among those zygotes showing crossovers, we determined SFR2 genotypes by freeze testing the F3 progeny of each zygote. By screening these lines with publicly available mapping markers, we obtained a position for sfr2-1 between markers mi403 and C6 (Figure 1A).
Figure 1.
Mapping of the SFR2 Gene.
The positions of all features are shown against the physical map of chromosome III. Features with asterisks flank the interval that is expanded in a succeeding map. Mb, megabase pairs.
(A) Classical genetic mapping of SFR2. Relative distances to SFR2 (cM, centimorgans) are best estimates derived from crossover frequencies.
(B) Fine genetic mapping in a reduced interval. The number of crossovers between each marker and SFR2 is indicated.
(C) Complementation mapping of SFR2. Bars indicate extents of genetic material in the tested clones. Hatched bars denote restoration of freezing tolerance to an sfr2-1 line; open bars denote no effect on freezing tolerance.
(D) Fine complementation mapping and sequence comparison. Predicted genes are shown as shaded arrows pointing in the direction of transcription. The position of the sequence change (G>A) in the sfr2-1 line is indicated.
An assembly of contiguous sequence (contig) was constructed from BAC clones covering this genetic interval. Further mapping markers in this interval were generated as PCR amplicons, using sequence information from members of the contig. (For Arabidopsis, this approach has now been superseded by the creation of an online polymorphism database by Monsanto Company, http://www.arabidopsis.org/Cereon/index.html.) Analysis with the new mapping markers (Figure 1B) allowed us to revise both boundaries of the interval containing sfr2-1. It also showed that the mapping population had been employed to the effective limit of its resolution because markers just inside the new boundaries (by 16 and 17 kb, respectively) had no crossovers with SFR2. Thus, classical genetic mapping placed SFR2 within a 218-kb interval bounded by markers 28L1SP6 and GT10.
Identification of the SFR2 Gene and sfr2-1 Mutation
We generated a series of transformable subclones from BACs in the contig and additionally used one preexisting clone, K1P18 (Liu et al., 1999). Plants of the sfr2-1 mutant line were transformed with the various constructs, and primary transformants from each construct were freeze tested. Two constructs (carrying sequences shown by hatched bars in Figure 1C) restored freezing tolerance in most of their transformants; the level of restored tolerance was indistinguishable from that of the wild type. This appeared to represent complementation of the sfr2-1 mutation by the 11-kb region of Arabidopsis genomic DNA that was common to both clones.
The DNA sequence of this region contained two complete putative genes according to contemporary genome annotations, along with incomplete portions of two other flanking genes (Figure 1D). We sequenced the complete genes in amplicons derived from the sfr2-1 mutant plants and compared them with published genomic data. Gene At3g06510 contained a single base change, a G>A transition consistent with mutagenesis by ethyl methanesulfonate (Figure 1D), whereas gene At3g06520 showed no changes. We noticed that the sequence alteration in gene At3g06510 would create a recognition site for the restriction endonuclease BsrI (ACTGG) and confirmed that this site was present in amplicons obtained from sfr2-1 material and absent in the cognate amplicons from wild-type Arabidopsis.
In parallel with the sequence analysis, we subcloned portions of the 11-kb complementing region that contained At3g06510 and At3g06520 individually; these were transformed into the sfr2-1 mutant line. Two independent clones of gene At3g06510 (identified by hatched bars in Figure 1D) restored freezing tolerance to the sfr2-1 mutant, with freezing sensitivity reappearing and segregating as expected in the next generation. A subclone of the other gene, At3g06520, had no effect on the mutant's freezing sensitivity. SFR2 was thus identified with the putative gene At3g06510 both by mutant sequence analysis and by complementation.
Transcription and Splicing of SFR2
A pair of primers was designed to amplify the whole of the predicted SFR2 coding region. These primers were used in an RT-PCR reaction on mRNA from cold-grown plants, and a product of 1.9 kb (the predicted size) was obtained. This amplicon was cloned and sequenced: it contained a coding region of 623 codons, inclusive of the termination codon. The implied splice sites in pre-mRNA were canonical, and they accord perfectly with the transcript structure predicted for At3g06510 in current annotations of the genome database. An equivalent amplicon was produced from mRNA of the sfr2-1 line, cloned, and partially sequenced. This confirmed the presence of the previously identified G>A mutation in sfr2-1 mRNA.
To determine the transcriptional initiation site of SFR2, we designed a primer complementary to SFR2 mRNA, 182 nucleotides downstream from the presumed initiation codon. Using a PCR protocol to define the 5′ end of the mRNA (Frohman et al., 1988), we obtained an amplicon with an additional 302 nucleotides of homologous sequence; this indicated transcriptional initiation 120 nucleotides upstream of the presumed initiation codon. There were no alternative in-frame initiator codons in this upstream region.
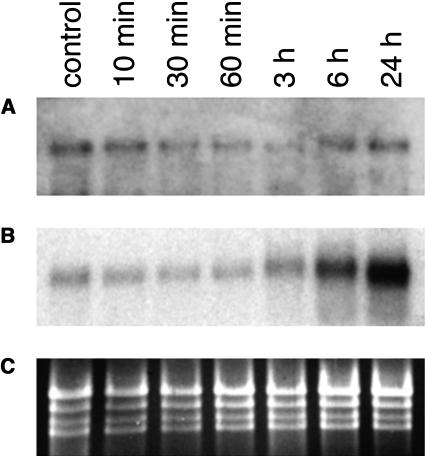
We compared the abundance of SFR2 transcripts in wild-type plants before and during cold acclimation by RNA gel blotting. RNA samples were extracted from plants and tested in three independently replicated experiments. Blots were probed with a transcript region (the 5′ end of the cDNA, amplified by PCR) sufficiently nonhomologous to other Arabidopsis sequences to preclude cross-hybridization. SFR2 transcripts were detected at low levels in all samples: there was no indication of induction by cold, even after 24 h (Figure 2A). As a positive control, the illustrated blot was reprobed with a known cold-inducible gene, KIN1 (Kurkela and Franck, 1990). This showed the expected strong induction over the cold time course (Figure 2B) and confirmed that the sampled plants were undergoing a normal response to low temperature.
Figure 2.
RNA Blot Analysis of Gene Expression after Cold Treatment of Various Durations.
(A) SFR2 expression: heavily exposed autoradiograph of blot probed with SFR2 cDNA. Control: untreated plants.
(B) Control for cold-induced gene expression: normally exposed autoradiograph probed with KIN1 cDNA.
(C) Loading control: the ethidium bromide–stained gel showing rRNA bands.
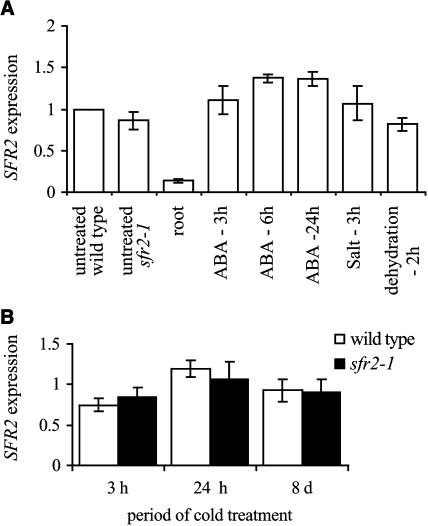
The expression of SFR2 was characterized further using real-time PCR. Wild-type plants were subjected to various dehydrative stress treatments (dehydration, salt, and abscisic acid application) and the level of SFR2 transcript determined (Figure 3A). None of the treatments had other than a small effect on the level of SFR2 transcript. This suggests that the tested stresses have, at most, a limited effect on the transcriptional regulation of SFR2; this contrasts with many stress-induced genes that show a large amplification. Monitoring of a known stress-induced gene, COR47, demonstrated that the experimental plants were undergoing a normal pattern of gene induction in response to these treatments (data not shown). In the same experiment, it was shown that the level of SFR2 transcript was approximately sixfold lower in the root than in the leaf of wild-type plants. A comparison between wild-type and sfr2-1 plants, both untreated, showed that they had very similar levels of SFR2 transcript.
Figure 3.
Analysis of SFR2 Expression Levels Using Real-Time PCR.
For stress treatments, plants were either transferred to a 4°C growth room for cold treatment, sprayed with 100 μM abscisic acid, drenched with 200 mM salt solution, or, for dehydration stress, excised leaves were placed in a flow hood at room temperature. Samples were collected at the indicated times after the stress treatment was initiated. For all RNA isolations, leaves from 3- to 4-week-old soil-grown plants were used except for the leaf/root comparison where plants grown in agar were used. Except where indicated, all analysis was with wild-type plants. In (A), SFR2 expression levels are relative to the level in untreated wild-type plants. In (B), expression levels are relative to untreated plants of the same genotype (either wild type or sfr2-1). Error bars represent ±sd.
In a similar set of experiments, real-time PCR was used to determine the levels of transcript in wild-type and sfr2-1 plants over a prolonged period in the cold. Figure 3B extends the results obtained by RNA gel blotting, demonstrating little variation in the level of SFR2 transcript even after 8 d in the cold. Transcript levels in sfr2-1 were similarly stable, suggesting that transcriptional differences between the two are unlikely to explain the mutant phenotype.
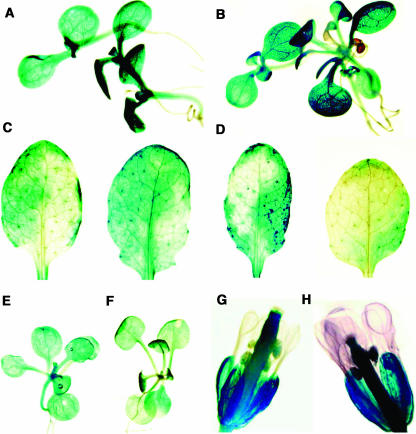
The localization of SFR2 expression was determined using a translational fusion of a β-glucuronidase (GUS) reporter to the 31st codon of the SFR2 gene, which was introduced into the wild-type background. GUS activity was observed in 7 out of 11 primary transformants. Activity was seen in most aerial organ types (hypocotyls, cotyledons, stems, leaves, pedicels, sepals, anthers, and pistils) but not in petals or filaments (Figure 4). In support of the real-time PCR experiments, expression in roots was very limited, with only a few plants showing small and weak patches of expression. The distribution of reporter activity appeared heterogeneous in the leaves of older plants (Figures 4C and 4D). Consistently with the result of RNA gel blotting and real-time PCR, we did not observe any differences in localization between untreated and cold-treated plants (Figure 4, comparing A with B, C with D, E with F, and G with H).
Figure 4.
Tissue Localization of SFR2 Transcription by Visualization of GUS Reporter Gene Expression.
(A) and (B) Seedlings grown in tissue culture.
(C) and (D) Leaves from soil-grown untreated plants of two independent reporter lines.
(E) and (F) Plants of reporter lines grown in tissue culture.
(G) and (H) Flowers from soil-grown plants.
Plants in (A), (C), (E), and (G) were untreated, whereas those in (B), (D), (F), and (H) were treated at 4°C for 24 h.
Protein Sequence Homologies
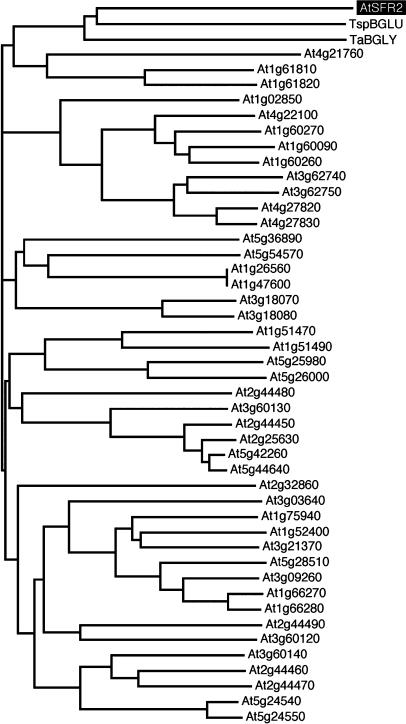
Public databases were searched for protein sequences homologous with the conceptual translation product of SFR2. The highest similarities that we found were to β-glycosidase enzymes from thermophilic and halophilic archea and bacteria (Table 1, Figure 5). These enzymes were members of glycosyl hydrolase family 1 (Henrissat, 1991), and a characteristic family 1 motif was detected in AtSFR2 protein (residues 264 to 268 and 427 to 431). The various activities reported for the prokaryotic homologs included specific cleavage of β-d-glucosidic, β-d-mannosidic, and β-d-galactosidic bonds and more catholic recognition of multiple types of β-glycosidic bonds (Table 1). We examined the relationship between all the family 1 β-glycosidase homologs of Arabidopsis and the two prokaryotic β-glycosidases most closely homologous to AtSFR2. A multiple protein sequence alignment was calculated and used to derive a cladogram relating the proteins by minimum substitution distances (Figure 6). This confirmed that AtSFR2 and the prokaryotic β-glycosidases form a distinct clade. Their grouping highlights the unusual extent of AtSFR2's divergence from every other family 1 β-glycosidase enzyme in Arabidopsis.
Table 1.
Closest Protein Homologs with Defined Function
| Activity | Accession | Source | Similaritya |
|---|---|---|---|
| β-Glucosidase | CAA94187 | Thermococcus sp | 2 × 10−36 |
| β-Glycosidase | AAD43138 | T. aggregans | 3 × 10−36 |
| β-Glucosidase | BAB05642 | Bacillus halodurans | 1 × 10−33 |
| β-Galactosidase | CAA34074 | Sulfolobus solfataricus | 5 × 10−33 |
| β-Glucosidase | AE010133 | Pyrococcus furiosus | 6 × 10−32 |
| β-Glycosidase | AAA79030 | Sulfolobus shibatae | 1 × 10−30 |
| β-Mannosidase | CAB49848 | Pyrococcus abyssi | 2 × 10−30 |
Similarity is represented by the probability of finding at least the observed quality of match in a database containing the same quantity of randomized sequence information (thus, smaller numbers represent greater similarities).
Figure 5.
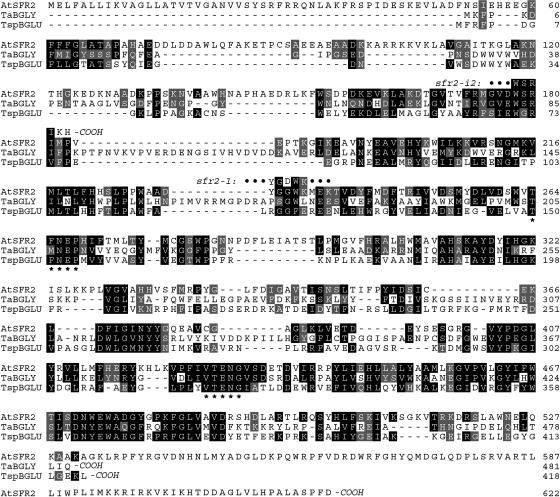
Sequence Comparison of the Proteins AtSFR2, TaBGLY, and TspBGLU.
TaBGLY (Thermococcus sp β-glycosidase) and TspBGLU (Thermosphaera aggregans β-glucosidase) represent the closest protein homologs of defined function available in public databases. The family 1 glycosyl hydrolase motif (TFNEP, I/VTENG) is shown (*). The position of the changed amino acid in sfr2-1 (G to D) and the predicted site of early translation termination in sfr2-i2 are also indicated. Black, identical or gonnet350 substitution values of 15 or more; gray, Gonnet350 substitution values of 5 to 14.
Figure 6.
Inferred Cladistic Relationships among β-Glycosidase–Like Proteins.
Horizontal branch lengths are proportional to numbers of residue substitutions.
Enzymatic Activity of AtSFR2 Protein
The full coding sequence from both SFR2 and sfr2-1 cDNAs were expressed in the yeast Pichia pastoris. In the supernatant of induced cultures, a protein was detected whose migration on SDS-PAGE indicated a size (∼70 kD) consistent with AtSFR2 protein. No such protein was detected in control cultures transformed with the empty vector. The secretion of AtSFR2 and AtSFR2.G234D protein into the medium suggests that the N terminus of AtSFR2 functions in yeast as a signal sequence, supporting the prediction of current annotations of At3g06510 by the Munich Information Center for Protein Sequences (München, Germany; http://mips.gsf.de/proj/thal/).
Protein concentrates from culture supernatants were tested for their glycosidic activity against a variety of synthetic glycosides (Table 2). A β-glucosidase purified from almond emulsin (EC 3.2.1.21; Sigma) was included in the tests as a positive control. Protein concentrated from both the SFR2 and sfr2-1 transformants produced significant hydrolysis of o-nitrophenyl-β-d-glucoside and of p-nitrophenyl-β-d-glucoside but not of several other p-nitrophenyl-β-d-glycosides nor of a p-nitrophenyl-α-d-glucoside. No activity against any of these substrates was detected in the preparations from the empty-vector control. This seems to indicate that AtSFR2 protein is an enzymatically active β-d-glucosidase with strong glycone selectivity. The presence of comparable β-d-glucosidase activity in the protein produced from the mutated sfr2-1 transcript suggests that enzymatic activity is maintained, at least in vitro, when the protein is tested against synthetic glycosides.
Table 2.
Specific Activities against Nitrophenyl d-Glycosides
| Glycoside | AtSFR2 | AtSFR2.G234D | Almond β-Glucosidase |
|---|---|---|---|
| ONP-β-d-glucoside | 151.0 | 158 | 17,104 |
| PNP-β-d-glucoside | 273.0 | 302 | 8,484 |
| PNP-β-d-fucoside | nd | nd | 17,523 |
| PNP-β-d-cellobioside | 1.9 | nd | nd |
| PNP-β-d-mannoside | nd | nd | nd |
| PNP-β-d-galactoside | nd | nd | 200 |
| PNP-α-d-glucoside | nd | nd | nd |
Specific activities have units of nmol min−1 mg−1. nd, not detected (detection limit: 1.5). PNP, p-nitrophenyl; ONP, o-nitrophenyl.
Mutant Alleles of SFR2
The sfr2-1 mutation affects codon 234 of SFR2, converting a Gly (GGC) into an Asp (GAC) codon; thus, sfr2-1 is a missense mutation (Figure 5). An automatically generated alignment of At3g06510 with its closest homologs was obtained from the Munich Information Center for Protein Sequences (data not shown). This revealed strong conservation of G234 among a wide variety of β-glycosidases; hence, it is likely that G234 is important for the function of β-glycosides.
The recessive nature of the sfr2-1 allele led Warren et al. (1996) to infer that it represented a loss-of-function mutation. This inference underlay our supposition that the wild-type SFR2 gene encoded an essential freeze-protective activity. If sfr2-1 truly caused a simple loss of function, then severely deficient alleles of SFR2 (e.g., early nonsense or insertional mutations) should display a similar phenotype. We therefore sought such alleles. An insertional mutant allele, which we designate sfr2-i2, was identified in the SIGnAL database of the Salk Institute (La Jolla, CA). We sequenced sfr2-i2 to determine the precise upstream and downstream junctions between host and inserted DNA. The upstream junction predicted termination of translation after two additional amino acids had been translated from the inserted sequence (Figure 5). Thus, sfr2-i2, encoding a protein of only 183 residues and <30% of the molecular mass of wild-type AtSFR2 protein, should be a strongly deficient allele.
We used PCR to distinguish mutant homozygotes from heterozygotes. Homozygous sfr2-i2 mutants were selfed, and their progeny were tested for freezing tolerance. All the tested progeny (33/33) showed freezing sensitivity, and the appearance of freezing damage was indistinguishable from that of sfr2-1 plants frozen in parallel (Figure 7). Further progeny were tested under freezing conditions of varying severity, but in no test did we observe any difference between the phenotypes of the sfr2-1 and sfr2-i2 mutants.
Figure 7.
Comparison of the Postfreezing Phenotypes of the sfr2-1 and sfr2-i2 Mutant Alleles with Wild-Type (SFR2+) Plants.
The photograph was taken after freezing at −6.0°C and 5 d of recovery at 20°C.
Immunoblot Analysis
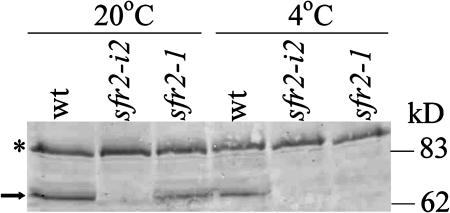
A polyclonal antibody raised against a peptide generated from the C-terminal region of the AtSFR2 protein was used to compare both the abundance of the AtSFR2 protein in different alleles of SFR2 and the effect of cold treatment (Figure 8). The antibody detected a protein with an apparent molecular mass of ∼65 kD in extracts of wild-type and sfr2-1 plants but not in material isolated from sfr2-i2 plants. This is consistent with the expected early termination of translation in sfr2-i2, predicted from DNA sequence analysis, and suggests that the 65-kD protein detected in wild-type and sfr2-1 plants is AtSFR2. A cross-reacting band (∼110 kD) present in all plants tested is unlikely to be AtSFR2 because its molecular weight is much higher than predicted for unprocessed AtSFR2 and because of its presence in sfr2-i2 plants.
Figure 8.
Detection of AtSFR2 Protein.
Total protein was extracted from wild-type, sfr2-1, and sfr2-i2 plants either grown in unstressed conditions or after 24 h at 4°C and used to conduct a protein gel blot with anti-SFR2 serum. The arrow shows the position of the AtSFR2 band. The asterisk indicates a cross-reacting band acting as a loading control.
The abundance of protein detected in plants of sfr2-1 grown at 20°C was slightly reduced compared with that in the wild type, suggesting that the mutation may have some effect on the translation or stability of the protein. After transfer of plants to 4°C for 24 h, there was a reduction in the level of AtSFR2 protein in wild-type plants, but no AtSRF2 protein could be detected in sfr2-1 plants. A time-course experiment (data not shown) showed that AtSFR2 protein was not detected in sfr2-1 plants over a period of at least 5 d after transfer to growth at 4°C. This suggests that the missense mutation in sfr2-1 plants has created a cold-sensitive allele of SFR2. The apparent mobility of the AtSFR2 protein in planta (∼65 kD) was smaller than that produced by expression in P. pastori (∼70 kD). This is most likely the result of posttranslational modification, possibly involving cleavage of the predicted N-terminal signal sequence.
DISCUSSION
Our identification of the SFR2 gene was undertaken because SFR2 was expected to encode an essential freeze-protective function in the cold-acclimated plant. This was based on an inference that the sfr2-1 mutant phenotype reflected a loss (rather than a gain or change) of function. However, sfr2-1 was found to be a missense mutation. By isolating a second mutant allele of SFR2, of a type that would be expected a priori to destroy the gene's function absolutely, the rationale for gene identification has been tested. Because the second mutant sfr2 allele had a similarly freezing-sensitive phenotype, we can be confident that SFR2 is indeed essential for freezing tolerance.
The predicted AtSFR2 protein is homologous to enzymes with glycosyl hydrolase (glycosidase) activity, in particular the group of β-glycosidases designated family 1 (Henrissat, 1991). This group includes β-glucosidases (EC 3.2.1.21), 6-phospho-β-glucosidases (EC 3.2.1.86), β-galactosidases (EC 3.2.1.23), 6-phospho-β-galactosidases (EC 3.2.1.23), β-mannosidases (EC 3.2.1.25), lactases/phlorizin hydrolases (EC 3.2.1.62/108), and myrosinases (EC 3.2.3.1) (Henrissat, 1991). Among family 1 β-glycosidases with empirically defined functions, the least distant homologs of AtSFR2 were archeal and bacterial enzymes with several different glycone specificities. It is not possible, on the basis of this pattern of homologies, to infer the specificity of AtSFR2 protein for either the glycone or aglycone portion of its substrate.
When produced by heterologous expression, AtSFR2 protein displayed hydrolytic activity against two nitrophenyl β-d-glucosides. Activity was not detected when the glucose moiety of the substrate was replaced by any of several other glycones nor when the β linkage to glucose was replaced by an α linkage: this indicates that its specificity for β-linked glucosides is sufficient for AtSFR2 to be considered a β-glucosidase. The specific activity of AtSFR2 against both nitrophenyl β-glucosides is nevertheless low in comparison with that of the almond β-glucosidase with which it was compared. This might be because of lower catalytic capacity or to lack of appropriate modification in the heterologous host but could also be because of greater discrimination against the artificial aglycones (the p-nitrophenyl and o-nitrophenyl groups) used in these assays; it would be expected that AtSFR2 should have strong specificity for the aglycone of its natural substrate.
The β-d-glucosidase activity present in the mutated AtSFR2.G234D protein was surprising. It seemed likely that the mutant protein would lack activity because it has the same freezing phenotype as the sfr2-i2 insertional mutant that is expected to be totally deficient in activity. Indeed, it cannot be ruled out that the protein produced in the sfr2-1 mutant, although active against synthetic substrates in vitro, fails to cleave its natural substrate. The results obtained from protein blotting, however, suggest another possible explanation for the freezing sensitivity of the sfr2-1 allele.
Given the minor differences in the level of SFR2 RNA transcript between the wild-type and sfr2-1 plants and the conservation of the in vitro β-d-glucosidase activity of the AtSFR2.G234D mutant protein, the elimination of AtSFR2 protein during the cold acclimation period may explain the freezing sensitivity of the sfr2-1 plants. If sfr2-1 plants do contain biologically active protein before cold acclimation, this is not sufficient to protect the plant from freezing. This suggests a specific requirement for SFR2 during the cold acclimation process rather than the manifestation of a latent lesion that is undetected in the warm. Evidence only exists, however, for β-glucosidase activity in vitro and not for the appropriate biological activity of the mutant protein in planta.
β-Glucosidases are ubiquitous. Glucosylation (reversible by the appropriate glucosidase) can affect various characteristics of the glucosylated moiety (the aglycone), including reactivity, solubility, and transport (Li et al., 2001). Many roles for glucosidases in plants have been postulated (reviewed in Esen, 1993). Some β-glucosidases are capable of affecting the properties of the cell wall (Gerardi et al., 2001; Li et al., 2001), which might be a crucial function in preparing cells for the physical deformations associated with freezing. In stress responses, β-glucosidases commonly release active molecules from inert precursors: the various released molecules include a variety of antimicrobials (Cicek and Esen, 1998; Sue et al., 2000), phytohormones (Brzobohaty et al., 1993), and at least one antioxidant (Chong et al., 2002). Stress-related roles have also been suggested for several β-glucosidases of unknown function on the basis of their stress-responsive expression (Fujiki et al., 2001; Seki et al., 2001; Chen et al., 2002).
Both bioinformatic analysis and its secretion from yeast cells indicate that AtSFR2 has a signal peptide at its N terminus. If AtSFR2 is indeed secreted from the protoplast in planta, the range of possible substrates for the AtSFR2 β-glucosidase would be limited to apoplastic metabolites, membrane components, and structural components of the cell wall.
The constitutive expression of SFR2, both in response to cold and to several dehydrative stresses, indicates that this gene would not have been detected by the approach of characterizing cold-inducible genes. Constitutive expression of the SFR2 gene does not necessarily equate to constitutive activity of AtSFR2 protein; in particular, posttranslational control is conceivable because most plant β-glucosidases are themselves glycosylated, and glycosylation has been implicated in their stabilization (Cicek and Esen, 1999). Transcription of SFR2 was detected in all green tissues and at a much lower level in roots; this implies that AtSFR2 plays a role in freeze protection of photosynthetic tissues or possibly a more specific role in plastid protection.
METHODS
Freeze Testing
To screen individuals for freezing tolerance, seedlings were grown for ∼5 weeks at 18 to 20°C with a 9-h photoperiod at 250 μmol m−2 s−1, and then subjected to 11 d of cold acclimation at 4°C, with an 8-h photoperiod at 220 μmol m−2 s−1. They were placed in a freezer with air temperature at a minimum of −6.0°C for 16 h and then returned to their preacclimation growth conditions. Injury was assessed after 5 d.
Generation and Use of Physical Markers in Mapping
Contemporary sequence information was available from the ends of many BAC clones in the contig (see http://www.tigr.org/tdb/e2k1/ath1/abe/bac_end_search.shtml). PCR amplicons were designed from these using the Primer3 program (S. Rozen and H. Skaletsky, unpublished data; http://www-genome.wi.mit.edu/genome_software/other/primer3.html). Primers were synthesized by MWG Biotech (Ebersberg, Germany; http://www.mwg-biotech.com). Amplicons separately produced from Col and Landsberg erecta DNA were screened for restriction endonuclease site polymorphisms (Konieczny and Ausubel, 1993). The cleaved amplified polymorphic sequence marker GAPC (Konieczny and Ausubel, 1993) and simple sequence length polymorphism marker nga126 (Bell and Ecker, 1994) have previously been described. For other markers, primer sequences and cognate restriction endonucleases are shown in Table 3.
Table 3.
Primer Pairs for PCR
| Marker | Enzyme | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| ANP3 | MvaI | AAGGGGAATTAATCGGTTGC | ATAAGGTTCCTGGCGCTTTC |
| B4a | TaqI | TGGTAGAAAATTTGGGATTG | GTCTTTTGTCATGCGCCTCC |
| C6a | DdeI | GCCTACCATCAATAAACC | ATGAAAGACATCACAGATCC |
| F28L1SP6 | DdeI | TACTCCATGATCCCCGAATG | GCAATTTTCAACTCTTGTTTGG |
| GT10 | (None) | GACGTTTGCTTATTAATTGTGTG | AGCCATAGGGTTTAGGGTTC |
| GT4 | HinfI | GGCCGATTCACACTTACCTC | CCCAAGTGCGTTAGAAAACC |
| GT6 | NdeII | GCGTTTGCGTTTCCATTATC | CTGGAGCATGTCAGCAGAAG |
| GT7 | MvaI | TGCAAGAAACCATCATGAAAAC | CCTCCTTGAAAATGACCGATAC |
| GT9 | BsuRI | GAGGCCTCACCTGTTTCTTG | TGGCCCAGTGACCATATACC |
| GW3 | DdeI | TTGGCAGCAGGAACTGTATG | ATAGGTGTCGTGCCTCTTGG |
| LTI6B | AluI | ATACCGCATAATGCCCAAAG | GGAGAAGAGCACGACGAAAC |
| S6a | (None) | TTTTTCAACCTTTCCCCC | ACCACCAACAACAGATTTC |
| T16D20T7 | AluI | AATTGAACCGACCCGAATC | TTTCCAAATCTTTGTGGATCTTC |
M. Grant, unpublished data (http://www.arabidopsis.org/maps/CAPS_Chr3.html).
DNA samples were isolated from F2 plants (Thorlby et al., 1999) and analyzed for cleaved amplified polymorphic sequence marker genotypes (Konieczny and Ausubel, 1993) and simple sequence length polymorphism marker genotypes (Bell and Ecker, 1994) as described. One restriction fragment length polymorphism marker, mi403, was also used. Map positions refer to the May 2001 release by the Nottingham Arabidopsis Stock Centre (Nottingham, UK; http://nasc.nott.ac.uk/) of the recombinant inbred map (Lister and Dean, 1993).
BAC Identification and Alignment
Filters carrying the TAMU (Texas A&M University) library of Arabidopsis (Col) BAC clones were obtained from ABRC (Columbus, Ohio; http://godot.ncgr.org/abrc) and probed with markers S6, C6, and B4. This identified several BAC clones, including T5D11 and T8E24. BAC clones were oriented relative to one another by querying the Washington University BAC fingerprinting database (http://genome.wustl.edu/gsc/arab/arabidopsis.html), which also allowed the identification of several overlapping BAC clones. Subsequent completion of the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000) has provided precise positional data for a subset of these BAC clones. Here, we present physical maps with the retrospective accuracy that this allows.
Subcloning for Complementation
A restriction map of the region of interest was constructed by digesting the overlapping BAC clones F28L1, F8L1, and T5D11 with endonucleases HindIII, BamHI, and BglII. The binary vector pSLJ755I6 (Jones et al., 1992) was then used for subcloning. BamHI subclones were constructed for all BamHI fragments >5 kb and <25 kb. Gaps in the subclone coverage were filled by subcloning from BamHI-partial and BglII-total and -partial digests. For single-gene transformations, genomic fragments were cloned into the vector pCAMBIA3300 (CAMBIA, Canberra, Australia; http://www.cambia.org). At3g06520 was inserted as an XbaI fragment and At3g06510 as SacI/BglII and SacI/BamHI fragments.
Plant Transformation and Selection
Plasmids were transferred to Agrobacterium tumefaciens GV3101/pM90 (Koncz and Schell, 1986) by triparental mating for plasmids derived from pSLJ755I6 (Jones et al., 1992) or by electroporation (Cangelosi et al., 1991) for plasmids derived from the pCAMBIA series vectors. Plants of the homozygous sfr2-1 mutant line were transformed by the floral dip method (Clough and Bent, 1998). Primary transformants were selected for Basta resistance (the marker in pSLJ75516 and the pCAMBIA vectors used here) by repeated spraying of the seedlings with a 250-mg/L solution of the herbicide Challenge 60 (AgrEvo, King's Lynn, UK) until the growth differential was clear.
Sequencing of SFR2
The genomic sequences of both At3g06520 and At3g06510 were obtained from homozygous sfr2-1 mutant plant DNA: overlapping PCR products were generated with a proofreading thermostable polymerase (ProofStart; Qiagen, Crawley, UK; http://www.qiagen.com), cloned into pGEM-T-Easy (Promega, Madison, WI; http://www.promega.com), and sequenced using the vector primers SP6 and T7. The sfr2-1 sequence has accession number AJ491321. The cDNA sequence of SFR2 was generated from cold-acclimated wild-type plant mRNA: cDNA was obtained using Omniscript reverse transcriptase (Qiagen), and a 1924-bp fragment was amplified by primers flanking the coding sequence of At3g06510. The fragment was cloned into pGEM-T-Easy and sequenced with SP6, T7, and internal primers. All sequencing was performed by MWG Biotech.
5′ Rapid amplification of cDNA ends was performed using the 5′/3′ RACE kit (Roche Molecular Biochemicals, Lewes, UK; http://www.biochem.roche.com), using a gene-specific primer (5′-TGCAGTAGCTACCCAAAGAAG-3′) positioned 182 nucleotides downstream of the initiation codon. PCR products were cloned into pGEM-T-Easy. A plasmid clone whose insert matched the size of the PCR amplicon was selected for sequence analysis (accession number AJ491320).
RNA Gel Blotting
Samples of total RNA were isolated using the RNeasy plant mini kit (Qiagen). Approximately 6 micrograms of RNA per sample was separated by formaldehyde gel electrophoresis and blotted onto nylon membrane (Roche). Membranes were hybridized with 32P-labeled probes generated using Ready-To-Go DNA labeling beads (Amersham-Pharmacia, Little Chalfont, UK; www.amershambiosciences.com/uk). Quik-hyb hybridization solution (Stratagene, La Jolla, CA; http://www.stratagene.com) was used according to the manufacturer's instructions. The SFR2 probe was a 610-bp PCR fragment amplified from the 5′ end of the SFR2 cDNA. The KIN1 probe was a 342-bp PCR fragment amplified from the coding sequence of KIN1 cDNA.
Real-Time RT-PCR
For quantitative real-time PCR, the Smartcycler (Cepheid, Sunnyvale, CA) was used according to the manufacturer's instructions. RNA was extracted using the RNeasy kit (Qiagen) including the optional, on column, DNase I digestion. Aliquots of RNA (2 μg) were reverse transcribed using SuperScript III (Invitrogen, Paisley, UK) as described in the manufacturer's instructions with 40 units of RNase inhibitor (Promega) and 2.5 μM oligo(dT)20 added to the reaction. A 1:100 (w/v) dilution of the first-strand cDNA was used in PCR reactions.
Primers were designed such that one of the pair spanned the position of an exon/intron border (At1g67090 and At3g06510) and did not amplify genomic DNA, or the amplicon contained an intron and amplified a different sized genomic DNA and cDNA fragments (At1g49240). Fragments were amplified from the 3′ region of the gene transcript and were between 100 and 150 bp in length.
The following primers were used for real-time PCR experiments: SFR2/At3g06510 (5′-GCAATGCTAAAGGGTGTTCC-3′ and 5′-AAGATCATGGGATCGGTCAA-3′), ribulose bisphosphate/At1g67090 (5′-TTCCTGACCTTACCGATTCC-3′ and 5′-ACAAATCCGTGCTCCAACTC-3′), and actin2/At1g49240 (5′-CTTCCCTCAGCACATTCCAG-3′ and 5′-CCCAGCTTTTTAAGCCTTTG-3′).
The Quantitect SYBR Green PCR system (Qiagen) was used according to the manufacturer's recommendations. Diluted cDNA (2 μL) was added to a reaction mix containing primers (0.3 μM) and 1× master mix in a total volume of 25 μL. The following Smartcycler program was used in all experiments. An initial 15 min at 95°C for Taq activation followed by 45 cycles consisting of 15 s at 94°C, 30 s at 56°C, and 30 s at 72°C. At the end of the PCR, a melt curve analysis (60 to 95°C) was performed to verify the fidelity of the amplification. For each test condition, cDNA derived from three independent experiments was used, and each reaction was run in duplicate.
For relative quantification the method of Pfaffl (2001) was used to determine the relative expression ratio. This determines the expression of the target gene (SFR2/At3g06510) relative to a reference gene (actin2/At1g49240 or ribulose bisphosphate/At1g67090) in a test sample compared with a control sample. In all experiments, the control sample was leaf material from warm-grown (untreated) plants. By definition, the expression ratio in the control sample is 1. At1g67090 was used as the reference gene in all experiments except for those comparing root and leaf tissue where At1g49240 was used.
Reporter Gene Analysis
An SFR2 promoter fragment was amplified using the primers 5′-GAAGCTTGTTTGCCTTTTCCTTCTTG-3′ and 5′-GAGATCTACGCGACGGAAACGAGAGTAG-3′. The product, comprising 93 bp of the first exon and 828 bp of contiguous upstream region, was digested with HindIII and BglII and cloned into the GUS expression vectors pCAMBIA1302 and pCAMBIA1391 (CAMBIA). In both constructs, the first 31 codons of SFR2 were fused in frame to a GUS reporter. Both constructs were transformed into the wild type, selecting transformants on MS agar supplemented with 15 mg/L of hygromycin. Their progeny (the T2 generation) were grown under normal conditions and, where indicated, transferred to 4°C for 24 h. Tissue samples (whole seedlings, leaves, or inflorescences) were collected and stained overnight by incubation with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Gallagher, 1992). Tissues were washed with 70% (v/v) ethanol until cleared of chlorophyll and then examined by light microscopy.
Heterologous Expression and Enzymatic Assay of AtSFR2
The coding sequences from SFR2 and sfr2-1 cDNAs were cloned into the vector pPicZB (Invitrogen) downstream of the AOX1 promoter and translational initiation sequences. The resulting constructs were linearized by SacI digestion and introduced into Pichia pastoris X33 by electroporation. Transformants were selected by growth at 28°C on yeast extract peptone dextrose sorbitol medium containing zeocin at 100 μg/mL. The presence of the AOX1-SFR2 fusion and of the zeocin-R gene in the transformed colonies was confirmed by PCR using primer pairs AOSF (5′-GACTTCGTGGAGGACGACTT-3′ and 5′-CAAATTAAAGCCTTCGAGCG-3′) and ZE1 (5′-GACTGGTTCCAATTGACAAGC-3′ and 5′-GCCCACCAAAGGAATTAAGGAA-3′), respectively. A parallel transformation with pPicZB resulted in transformed colonies positive for the zeocin-R gene but negative for the AOX1-SFR2 fusion (empty-vector transformants).
Pichia transformants were cultured for 48 h in 200 mL of buffered methanol complex medium at 28°C. Culture supernatants were collected after 10 min centrifugation at 3000g. Protein was precipitated with 60% saturated ammonium sulfate, redissolved in 20 mL of 20 mM sodium phosphate, pH 7.0, 1 M ammonium sulfate, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride, and loaded onto an octylsepharose hydrophobic interaction column. Protein was eluted by a three-step gradient from 0.75 to 0.0 M ammonium sulfate in the same buffer. Eluted fractions were analyzed by SDS-PAGE, pooled, concentrated by size exclusion chromatography (Vivaspin column, 50,000 kD mass cutoff; Vivascience, Hannover, Germany) and diluted in 100 mM citrate/50 mM phosphate, pH 6.0.
Enzymatic assays were performed in triplicate. Twenty-five microliters of purified protein was mixed with 25 μL of the appropriate chromogenic substrate in 100 mM citrate/50 mM phosphate, pH 6.0, and 250 μL of 100 mM citrate/50 mM phosphate buffer, pH 7.0. Reactions were stopped by addition of 700 μL of 0.4 M sodium carbonate after incubation at 37°C for 0 or 15 min. Release of p-nitrophenol or o-nitrophenol was measured by optical absorbance at 410 nm, assuming extinction coefficients of 18,400 M−1 cm−1 and 3500 M−1 cm−1, respectively.
Isolation of the sfr2-i2 Allele
The SFR2 sequence was used to search the SIGnAL database of sequences flanking T-DNA insertions (Salk Institute; http://signal.salk.edu/cgi-bin/tdnaexpress). This identified a line (SALK 000,226) in which a T-DNA insertion was present in SFR2, creating allele sfr2-i2. Seeds were obtained from the ABRC. Amplicons were generated from the left and right T-DNA borders, using one gene-specific and one border-specific primer in each case, and were sequenced to determine the sfr2-i2 coding sequence (accession number AJ491322). Plants producing such amplicons (i.e., plants containing at least one sfr2-i2 allele) were subjected to a second PCR screening using primers homologous to SFR2 sequences to either side of the insertion. Production of the PCR band typical of wild-type Arabidopsis indicated heterozygosity; absence of the wild-type band indicated that sfr2-i2 was homozygous.
Protein Immunoblot Analysis
Total protein from 5-week-old plants was isolated using the method described by Martinez-Garcia et al. (1999). Protein concentrations were measured using the Bio-Rad protein assay kit (Bio-Rad, Hemel Hempstead, UK). Equal amounts (20 μg) of protein were loaded per lane and separated by SDS-PAGE before transfer onto polyvinylidene difluoride membrane by electro-blot transfer.
For the production of polyclonal antibody against AtSFR2, a cDNA fragment, encoding 109 amino acids at the C terminus of AtSFR2 protein, was cloned by PCR into the expression vector pET28a (Merck Biosciences, Nottingham, UK) in frame with a 6× His tag. The protein was expressed in Escherichia coli strain BL21 and purified using a nickel affinity column (BD Talon; Clontech, BD Biosciences, Cowley, UK). Polyclonal antibodies, raised in rabbit, against the recombinant peptide were used at a dilution of 1:200. Secondary goat anti-rabbit horseradish peroxidase–conjugated antibody (Promega) was used at a 1:5000 dilution and detection performed using 3,3′-diaminobenzidine peroxidase substrate (Sigma-Aldrich, Dorset, UK).
Bioinformatics
The BLAST2 algorithm (Altschul et al., 1997) was used to search public databases from the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/BLAST/). EST homologs were obtained from libraries derived from Arabidopsis roots (AV548508), inflorescences (AI996051), seedling hypocotyls (N96111), and whole seedlings (AV785662, AV794172, and AV791716). Other software tools were used at Web sites of the San Diego Supercomputer Center (http://workbench.sdsc.edu/) and European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/). For construction of sequence alignments and cladograms, the Clustal multiple alignment program (Thompson et al., 1994) and the TREEVIEW cladogram drawing program (Page, 1996) were downloaded from European Bioinformatics Institute (ftp://ftp.ebi.ac.uk/pub/software/) and University of Glasgow (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) servers, respectively, and run locally.
Sequence data from his article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ491320, AJ491321, and AJ491322.
Acknowledgments
This research was supported by the Biotechnology and Biological Science Research Council (UK) under Grants A05555 and P10187. Emma Veale made technical contributions. David Bouchez provided physical map information before publication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Glenn Thorlby (g.thorlby@rhul.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024018.
References
- Altschul, F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Boyce, J.M., Knight, H., Deyholos, M., Openshaw, M.R., Galbraith, D.W., Warren, G., and Knight, M.R. (2003). The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 34, 395–406. [DOI] [PubMed] [Google Scholar]
- Brzobohaty, B., Moore, I., Kristoffersen, P., Bako, L., Campos, N., Schell, J., and Palme, K. (1993). Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Byrne, M., Murrell, J.C., Owen, J.V., Williams, E.R., and Moran, G.F. (1997). Mapping of quantitative trait loci influencing frost tolerance in Eucalyptus nitens. Theor. Appl. Genet. 95, 975–979. [Google Scholar]
- Cangelosi, G.A., Best, E.A., Martinetti, G., and Nester, E.W. (1991). Genetic analysis of Agrobacterium. Methods Enzymol. 204, 384–397. [DOI] [PubMed] [Google Scholar]
- Chen, W.Q., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J., Baltz, R., Schmitt, C., Beffa, R., Fritig, B., and Saindrenan, P. (2002). Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek, M., and Esen, A. (1998). Structure and expression of a dhurrinase (β-glucosidase) from sorghum. Plant Physiol. 116, 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek, M., and Esen, A. (1999). Expression of soluble and catalytically active plant (monocot) β-glucosidases in E. coli. Biotechnol. Bioeng. 63, 392–400. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Esen, A., ed (1993). β-Glucosidases: Biochemistry and Molecular Biology. (Oxford: Oxford University Press).
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide orimer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Yoshikawa, Y., Sato, T., Inada, N., Ito, M., Nishida, I., and Watanabe, A. (2001). Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant 111, 345–352. [DOI] [PubMed] [Google Scholar]
- Gallagher, S.R., ed (1992). GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. (San Diego: Academic Press).
- Gerardi, C., Blando, F., Santino, A., and Zacheo, G. (2001). Purification and characterisation of a beta-glucosidase abundantly expressed in ripe sweet cherry (Prunus avium L.) fruit. Plant Sci. 160, 795–805. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Sebolt, A.M., Salazar, M.P., Everard, J.D., and Thomashow, M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino, P., Sandman, G., Lang, V., Nordin, K., and Palva, E.T. (1990). Abscisic acid deficiency prevents development of freezing tolerance in Arabidopsis thaliana L. Heynh. Theor. Appl. Genet. 79, 801–806. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J.K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9, 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Shlumukov, L., Carland, F., English, J., Schofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi, S.K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Knight, H., Veale, E.L., Warren, G.J., and Knight, M.R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TI-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kurkela, S., and Franck, M. (1990). Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol. Biol. 15, 137–144. [DOI] [PubMed] [Google Scholar]
- Lerceteau, E., Plomion, C., and Andersson, B. (2000). AFLP mapping and detection of quantitative trait loci (QTLs) for economically important traits in Pinus sylvestris: A preliminary study. Mol. Breed. 6, 451–458. [Google Scholar]
- Li, S.C., Han, J.W., Chen, K.C., and Chen, C.S. (2001). Purification and characterization of isoforms of β-galactosidases in mung bean seedlings. Phytochemistry 57, 349–359. [DOI] [PubMed] [Google Scholar]
- Lister, C., and Dean, C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4, 745–750. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., and Shibata, D. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente, F., Oliveros, J.C., Martinez-Zapater, J.M., and Salinas, J. (2000). A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta 211, 648–655. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257. [DOI] [PubMed] [Google Scholar]
- McKown, R., Kuroki, G., and Warren, G. (1996). Cold responses of Arabidopsis mutants impaired in freezing tolerance. J. Exp. Bot. 47, 1919–1925. [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Kamei, A., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr. Opin. Biotechnol. 14, 194–199. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Mao, Y.P., Regier, M.K., Triezenberg, S.J., and Thomashow, M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue, M., Ishihara, A., and Iwamura, H. (2000). Purification and characterization of a hydroxamic acid glucoside β-glucosidase from wheat (Triticum aestivum L.) seedlings. Planta 210, 432–438. [DOI] [PubMed] [Google Scholar]
- Sutka, J., Galiba, G., Veisz, O., and Snape, J.W. (1997). Genetic analysis of frost resistance and its contribution to development of frost resistant cereal varieties—A review. Plant Breeding Seed Sci. 41, 39–50. [Google Scholar]
- Teutonico, R.A., Yandell, B., Satagopan, J.M., Ferreira, M.E., Palta, J.P., and Osborn, T.C. (1995). Genetic analysis and mapping of genes controlling freezing tolerance in oilseed Brassica. Mol. Breed. 1, 329–339. [Google Scholar]
- Thomashow, M. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F., Gilmour, S.J., Stockinger, E.J., Jaglo-Ottosen, K.R., and Zarka, D.G. (2001). Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol. Plant 112, 171–175. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlby, G., Veale, E., Butcher, K., and Warren, G. (1999). Map positions of SFR genes in relation to other freezing-related genes of Arabidopsis thaliana. Plant J. 17, 445–452. [DOI] [PubMed] [Google Scholar]
- Uemura, M., Warren, G., and Steponkus, P.L. (2003). Freezing sensitivity in the sfr4 mutant of Arabidopsis is due to low sugar content and is manifested by loss of osmotic responsiveness. Plant Physiol. 131, 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, G., McKown, R., Marin, A., and Teutonico, R. (1996). Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z., and Browse, J. (1998). eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95, 7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z., and Browse, J. (2000). Cold comfort farm: The acclimation of plants to freezing temperatures. Plant Cell Environ. 23, 893–902. [Google Scholar]
- Xiong, L.M., Schumaker, K.S., and Zhu, J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.), S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]