Abstract
Personalized cancer medicine aims to accurately predict the response of individual patients to targeted therapies, including tyrosine kinase inhibitors (TKIs). Clinical implementation of this concept requires a robust selection tool. Here, using both cancer cell lines and tumor tissue from patients, we evaluated a high-throughput tyrosine kinase peptide substrate array to determine its readiness as a selection tool for TKI therapy. We found linearly increasing phosphorylation signal intensities of peptides representing kinase activity along the kinetic curve of the assay with 7.5–10 μg of lysate protein and up to 400 μM adenosine triphosphate (ATP). Basal kinase activity profiles were reproducible with intra- and inter-experiment coefficients of variation of <15% and <20%, respectively. Evaluation of 14 tumor cell lines and tissues showed similar consistently high phosphorylated peptides in their basal profiles. Incubation of four patient-derived tumor lysates with the TKIs dasatinib, sunitinib, sorafenib and erlotinib primarily caused inhibition of substrates that were highly phosphorylated in the basal profile analyses. Using recombinant Src and Axl kinase, relative substrate specificity was demonstrated for a subset of peptides, as their phosphorylation was reverted by co-incubation with a specific inhibitor. In conclusion, we demonstrated robust technical specifications of this high-throughput tyrosine kinase peptide microarray. These features required as little as 5–7 μg of protein per sample, facilitating clinical implementation as a TKI selection tool. However, currently available peptide substrates can benefit from an enhancement of the differential potential for complex samples such as tumor lysates. We propose that mass spectrometry-based phosphoproteomics may provide such an enhancement by identifying more discriminative peptides.
Introduction
Tyrosine kinases are key regulators of normal cellular processes, including differentiation, proliferation, migration and apoptosis.1, 2 Although only 1% of the phosphoproteome results from tyrosine phosphorylation, nearly half of the 90 tyrosine kinases encoded in the human genome have been implicated in cancer, emphasizing their role in this disease.3, 4 When mutated or overexpressed, receptor tyrosine kinases may become oncoproteins, causing and promoting tumor growth by aberrant tyrosine signaling.5 Since the introduction of imatinib in 2003, nearly 20 tyrosine kinase inhibitors (TKIs) that interfere with these proteins have reached clinical approval, while more than 40 targeted therapies have been approved for the treatment of patients with advanced solid and hematological tumors6 (http://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm279174.htm). Apart from looking at the epidermal growth factor receptor (EGFR) mutation status, the anaplastic lymphoma kinase and c-ros oncogene 1 rearrangement, and the breakpoint cluster region-Abelson gene sequence, there are no clinically available tests indicative of response to TKIs.7, 8, 9, 10 Considering the aberrant signaling activities that occur in tumors, it has been hypothesized that kinase activity profiling could be a valuable clinical tool to select TKI treatment for patients with advanced cancer, thereby enhancing efficacy of available drugs and expanding the therapeutic arsenal. Such therapy selection tools should include a robust screening method with a short turnaround time to evaluate available drugs or drug combinations based on tumor biology from an individual patient. We hypothesize that determination of kinase activity in a tumor biopsy may be used in such a screening method.
The PamChip microarray contains 144 tyrosine kinase peptide substrates representing key signal transduction pathways (PamGene, Den Bosch, The Netherlands). Consisting of a porous membrane through which a tumor tissue or cell line lysate is repeatedly transported by a miniature pumping system, this chip (hereafter referred to as PTK (peptide tyrosine kinase) microarray) enables ‘kinetic' measurement of phosphorylation changes over time. Spot intensities on the arrays are derived from the binding of a fluorescently labeled anti-phosphotyrosine antibody to the peptide substrates that become phosphorylated by kinases present in the sample.11, 12, 13 This antibody can recognize most, if not all, phosphotyrosine-containing motifs in proteins and peptides. Several studies have discussed its potential for target identification in clinical samples,14, 15 while others have suggested application of a PTK microarray for the identification of responders versus non-responders.16, 17, 18 Here, we have evaluated the PTK microarray for measurement of kinase activity in cancer cell lines and patient-derived tumor tissues under various experimental conditions to determine optimal test conditions and to evaluate the array's potential for clinical implementation.
Materials and methods
Cell culture and lysis
The cell lines 786-O (renal cell cancer), HCT116 (colorectal cancer) and H460 (non-small cell lung cancer) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 5% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin and were maintained in a humidified incubator containing 5% CO2 at 37 °C. The cell lines were tested for their authenticity by short tandem repeat profiling DNA fingerprinting (Baseclear, Leiden, The Netherlands).
Cells were seeded in 10 cm2 dishes and allowed to attach for 48 h to obtain 70–80% confluence. At t=48 h, cells were lysed as described elsewhere using M-PER (mammalian protein extraction reagent) (M-PER; Thermo Scientific, Rockford, IL, USA) unless stated otherwise. Protein concentrations were determined using the Micro BCATM Protein Assay Kit (Thermo Scientific). Additional buffers were used in one experiment: T-PER (tissue protein extraction reagent) (T-PER; Thermo Scientific) and radioimmunoprecipitation assay (RIPA) buffer (homemade; 30 mM Tris-HCl pH 7.48, 150 mM NaCl, and 1% NP-40). Buffers were supplemented with ethylenediaminetetraacetic acid (EDTA)-free 1% Protease Inhibitor Cocktail and 1% Halt Phosphatase Inhibitor Cocktail (both 100X, Thermo Scientific).
Tumor tissue collection and lysis
Frozen patient-derived tumor tissues were obtained from archival primary resection specimens (breast cancer, melanoma, colorectal cancer), metastasectomy tissue (lung cancer), and a liver metastasis needle biopsy from a patient with a pancreatic neuroendocrine tumor and a patient with lung cancer according to an Institutional Review Board-approved protocol for biobanking of remaining human specimen (VUmc, Amsterdam, The Netherlands). Additionally, two previously harvested and frozen tumors, one from a murine breast cancer xenograft model of the MDA-MB-231 cancer cell line19 and one from a chicken embryo chorioallantoic membrane experiment using the colorectal cancer cell line HT29, were obtained. Approximately 10–20 μm cryosections per sample were prepared using a LKB-Reichert-Jung ultra cryomicrotome (Leica, Rijswijk, The Netherlands) and transferred to 1.5 ml Eppendorf vials while keeping samples frozen at −20 °C in the cryotome. Samples were snap-frozen in liquid nitrogen and stored at −80 °C. Using M-PER and/or T-PER as described in the results section, lysates were prepared as previously reported. 11, 12
Tyrosine kinase activity profiling
Tyrosine kinase peptide microarray (PamGene International, 's-Hertogenbosch, The Netherlands) experiments were performed as previously described.11 Briefly, 40 μl of control sample mix for the kinase activity array was prepared using a reaction buffer containing 10 × Abelson buffer (Westburg, Leusden, The Netherlands), 10 × bovine serum albumin, 400 μM ATP (Sigma-Aldrich, Zwijndrecht, The Netherlands) unless stated otherwise, fluorescein-labeled antibody PY20 (Exalpha, Maynard, MA, USA), dithiotreitol and 7.5 μg of lysate protein unless stated otherwise. For drug inhibition experiments, concentrations of dimethyl sulfoxide were equalized to 2.5% between inhibition and control samples. Applied TKI concentrations were based on IC50s of proliferation assays performed in cancer cell lines (data not shown).20 After blocking the arrays with 2% bovine serum albumin or other buffers as specified and subsequent loading of the sample mix in triplicate onto the arrays, incubation at 30 °C was started for 60 cycles utilizing a PamStation12 instrument. During this time period, the sample mix was transferred through the array once per minute. Repeated fluorescent imaging was performed with a 12-bit CCD camera, monitoring fluorescence intensities resulting from binding of the anti-phosphotyrosine antibody over time.
Data analysis
Spot intensity at each time point was quantified (and corrected for local background), and the resulting time-resolved curves were fit to calculate the initial phosphorylation rate and end levels using specific kinetic algorithms and appropriate statistical methods (Bionavigator software version 5.1, PamGene International, 's-Hertogenbosch, The Netherlands). Unless otherwise stated, data are expressed as the average signal intensity ±s.d. of the 143 peptide spots based on end levels of the phosphorylation curve using ⩾3 technical replicates per sample. Significant differences between two conditions were determined using two-sided Student's t-test using Microsoft Excel, with P<0.05 considered to be statistically significant. One-way ANOVA comparisons were applied when comparing more than two conditions.
Results
Conditions influencing basal profile signal intensity protein and ATP concentration
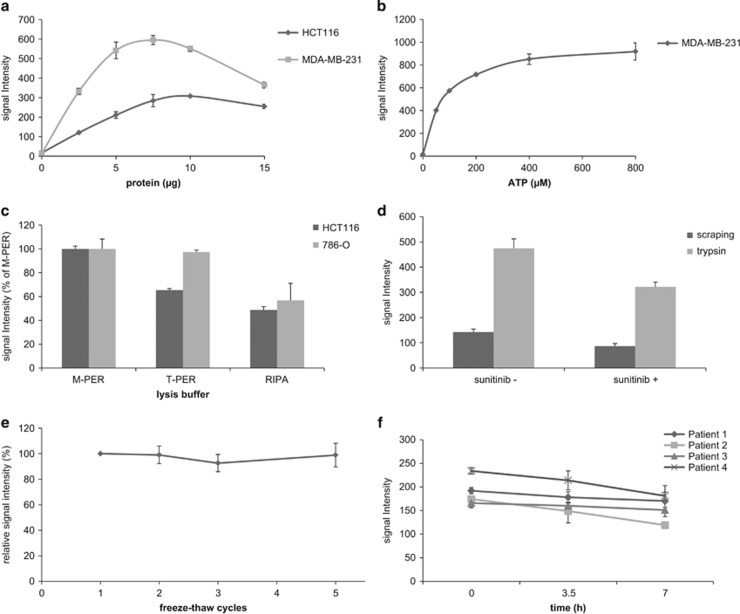
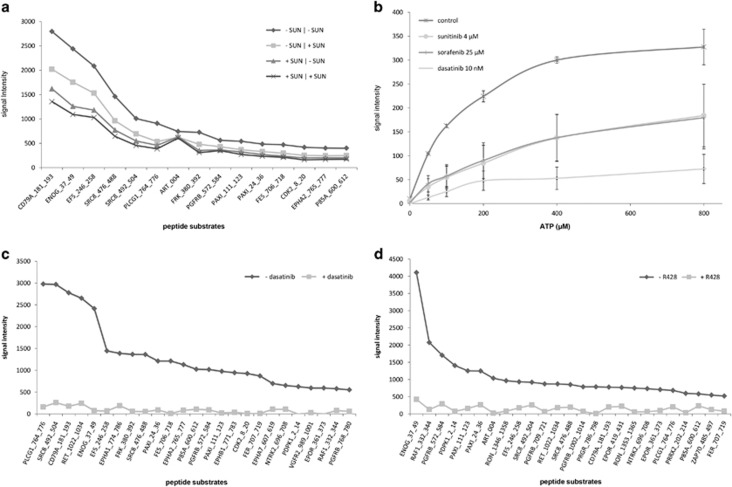
To determine the amount of protein, which reflects kinase abundance, and the amount of ATP that were needed for optimal basal profile signal intensities, arrays were incubated with increasing protein and ATP concentrations using HCT116 cancer cells and/or MDA-MB-231 xenograft tissue lysate. Increasing protein concentrations resulted accordingly in elevation of the signal intensity for both lysates, reaching optimal signal intensity at 10 and 7.5 μg of protein input per array, respectively (Figure 1a). The signal dropped slightly when more protein was added, probably due to clogging of the array membrane. For the xenograft lysate, ATP concentrations up to 400 μM strongly increased signal intensities, while higher concentrations yielded similar signal intensities (Figure 1b).
Figure 1.
Conditions influencing optimal basal profile signal intensity. Average signal intensity ±s.d. of the 143 peptide spots is represented unless stated otherwise. (a) Basal profile signal intensities obtained with increasing lysate protein input. Optimal signal intensity was obtained using 7.5 and 10 μg of lysate protein input per array for HCT116 and MDA-MB-231, respectively. (b) Basal profile signal intensities obtained with different ATP concentrations. ATP concentrations up to 400 μM strongly increased signal intensity, while at higher concentrations the curve deviated from linearity. (c) Comparison of signal intensities with different lysis buffers. Compared to T-PER and RIPA lysis buffers, M-PER resulted in more efficient and consistent lysis of HCT116 (P<0.001 compared to both buffers, Student's t-test) and 786-O cells (P<0.001 relative to RIPA; T-PER P=0.807). Average signal intensity relative to M-PER is shown. (d) Comparison of trypsin-based cell lysis and scraping-based lysis. Trypsin-based lysis of 786-O cells enhanced signal intensity when compared to the standard scraping procedure. Incubation with 2 μM sunitinib prior to trypsin-based lysis and ex-vivo spiking of the same concentration in a scraping-based lysate resulted in an approximately 25% decrease of average signal intensity compared to the control sample (P<0.001 in both comparisons). (e) Freeze-thaw cycles. Relative to the first freeze-thaw cycle after lysate storage, average signal intensity of HCT116 lysate was not affected by additional cycles (P=0.98, ANOVA). (f) Sample conservation on ice. Conservation of sample constituents of four patient-derived tumor lysates on ice for three consecutive measurements resulted in a non-significant decline of average sample signal intensity (P=0.25, ANOVA).
Lysis buffer efficiency
To determine whether signal intensity may be influenced by the type of buffer used for lysis, M-PER, T-PER and RIPA buffer were evaluated. As concluded from the signal intensities, M-PER resulted in more efficient tumor cell lysis when compared to T-PER and RIPA (M-PER versus T-PER: P<0.001 for HCT116, P=0.81 for 786-O; M-PER versus RIPA: P<0.0001 both cell lines, Figure 1c) while protein yield was equal (data not shown). To compare M-PER and T-PER in two colorectal cancer tissues, four samples per tumor consisting of ten consecutive cryosections (A1–4 and B1–4) were alternatingly lysed using buffer such that two samples per tumor were lysed with each. There were no significant differences between the signal intensities obtained using either buffer within the same samples (data not shown), suggesting equal protein extraction efficiency for M-PER compared to T-PER when applied to tissue. The differences in composition between the two buffers is not revealed by the manufacture but probably lies in the detergent added in 25 mM bicine buffer (pH 7.6).
Because time may also be a critical factor in conserving kinase activity during the actual lysis procedure of tumor cells,21 we compared trypsin-based lysis to the recommended method of cell scraping on ice. Significant differences in terms of signal intensity were observed between trypsinized and scraped cells: trypsin resulted in a three to four-fold increase in signal intensity when compared to scraping (Figure 1d). It is not clear whether this is indeed a result of enhanced lysis efficacy, as activation of stress kinases may also play a role. Both incubation with 2 μM sunitinib prior to trypsin-based lysis and ex-vivo spiking of the same concentration in a scraping-based lysate of cell line X resulted in an approximately 25% decrease of average signal intensity compared to the control sample (Figure 1d).
Effects of freeze-thaw cycles
As shown in Figure 1e, average signal intensity was not significantly influenced by freezing and thawing cell lysates up to a practically relevant number of four additional cycles. However, when both the cell lysate and the ATP- and fluorescent antibody-containing sample mix were kept on ice for 7 h to perform three consecutive microarray experiments, a potentially relevant trend of decreasing signal intensity was observed (Figure 1f). The average decrease in signal intensity across four patient tumor tissue-derived lysates, measured three times using triplicate samples, was 18% (range 9–31%). Fresh sample constituents should therefore be prepared for each experiment.
Substrate specificity
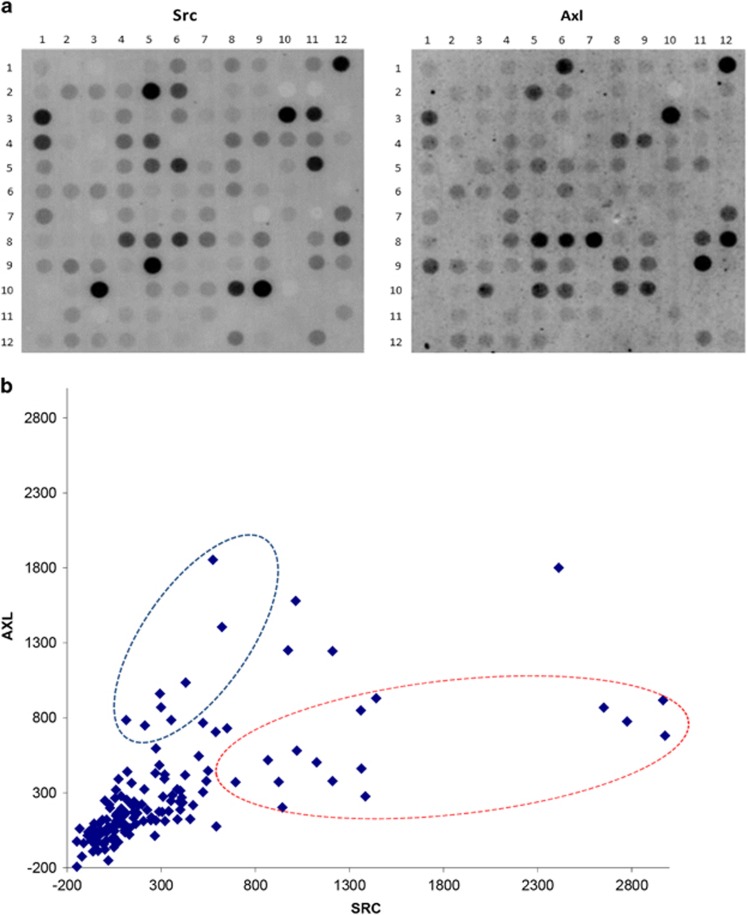
Kinases can phosphorylate multiple peptide substrates, although the number of phosphorylation substrates may differ substantially per kinase.22 As peptide phosphorylation intensity on the PTK microarray represents the cumulative activity of multiple kinases, we studied substrate specificity by incubating separate arrays with the recombinant kinases Src and Axl (Figure 2a). While some peptides were predominantly phosphorylated by either Src (for example, spots on row 3—column 11 and 5–6, corresponding to substrates EPHA1_774_786 (site Y781) and FES_706-718 (Y713), respectively) or Axl (9–11 and 10–5 corresponding to RAF1_332_344 (Y340, 341) and RON_1346_1358 (Y1353), respectively), most were phosphorylated by both (3–1, 3–10). The putative upstream kinases for the four substrates are unknown, FES, Src and RON/MET (http://www.phosphosite.org/siteSearchAction.do).23, 24 A correlation plot also showed substrate preference by these two kinases (R2=0.29, Figure 2b) but otherwise clearly demonstrated the promiscuous nature of phosphorylation for the majority of the peptide substrates in this assay.
Figure 2.
Recombinant Src and Axl kinase substrate specificity. Microarrays were incubated with 125 ng ml−1 of recombinant Src or 500 ng ml−1 Axl kinase. (a) Raw images of array spot phosphorylation after 1 h demonstrate significant overlap in peptide phosphorylation. Some peptides were predominantly phosphorylated by either Src (row 3-column 11 and 5–6) or Axl (for example, spots 9–11 and 10–5). (b) Correlation plot of 143 peptide signal intensities showing relative substrate preference for Src and Axl kinase, indicated by the red and blue circle, respectively.
Profile reproducibility
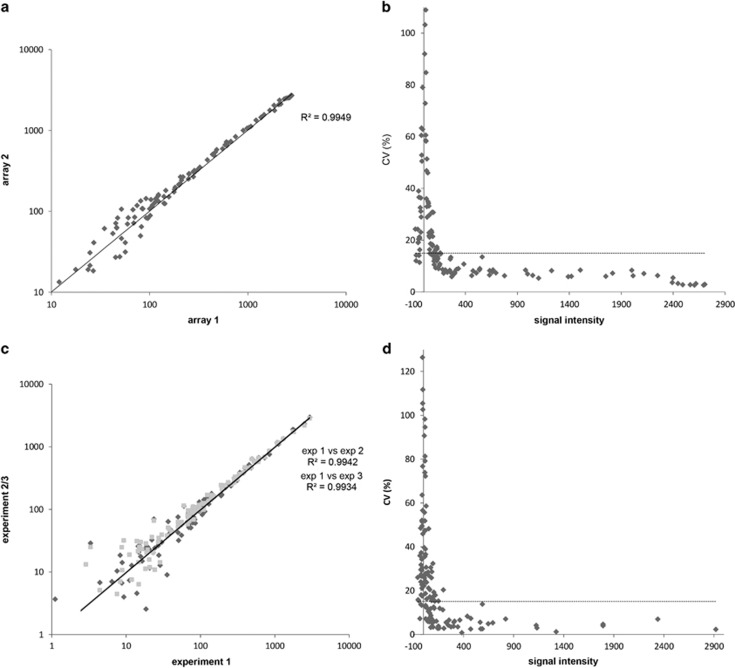
We determined intra-experimental technical reproducibility using HCT116 lysate replicates on 3 chips, comprising 12 arrays. Intra- and inter-chip correlations between peptide phosphorylation levels as assessed using Pearson's correlations (r) ranged from 0.9954 to 0.9993. As an example, a regression analysis of two replicates showed that they differed from each other by 0.87% as revealed using the linearity equation with a coefficient of determination (r2) of 0.9949 (Figure 3a). High technical reproducibility of the microarray was further demonstrated by the obtained total median CV of <15% (Figure 3b), while this was <20% between experiments using MDA-MB-231 xenograft lysate (data not shown). The median CV for well-expressed signals was much lower, at 7% for peptides with signal intensity of ⩾200 in the aforementioned experiment, but these represented only 35% of available substrates (Figure 3b).
Figure 3.
Microarray reproducibility and linearity. Inter- and intra-chip correlations between peptide phosphorylation levels as assessed by Pearson's correlations (r) using 12 technical replicates of HCT116 lysate (7.5 μg) measured on 3 chips in a single run. (a) Linearity plot of two representative technical replicates. (b) CV plot of the technical replicas shown in (a). The threshold was set at 15% as recommended by the microarray manufacturer. (c) Linearity plot of biological replicates of 786-O cells in three independent experiments. (d) CV plot of the biological replicas shown in (c). Threshold at CV 15%. CV, coefficient of variation as determined by the ratio of the standard deviation to the mean signal intensity.
To determine reproducibility of biological replicates, 786-O cells were lysed at three different time points. A correlation plot of three independent measurements yielded r2 values of 0.9942 and 0.9934, respectively, while the median CV was <20% (Figures 3c and d).
Differences between samples of different biological origin
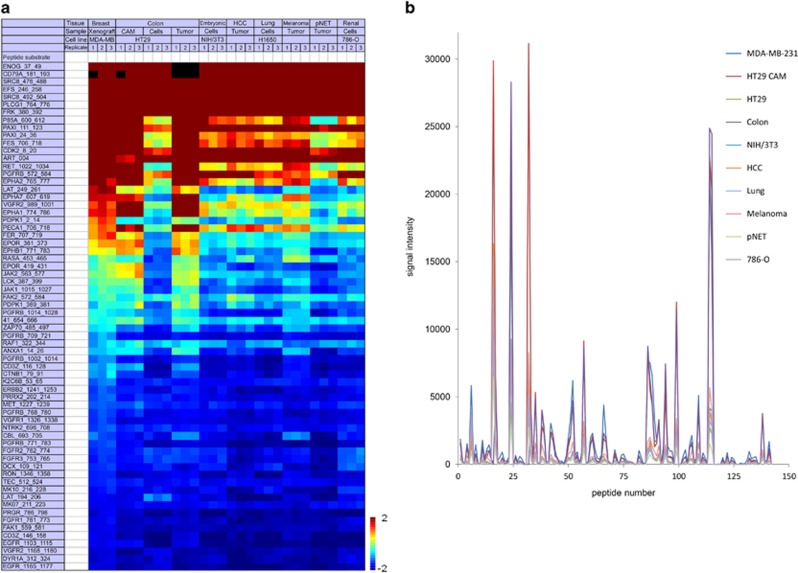
To investigate whether PTK microarray analysis can differentiate between samples of different biological origin based on the presence of sample-specific profiles, we first measured four tumor tissues, two from patients with metastatic non-small cell lung cancer (pt1 harboring KRAS exon 1 mutation, pt2 EGFR exon 20 mutation) and two with primary breast cancer (pt2 human epidermal growth factor receptor 2 amplification). The kinase activity profiles of these tumors were largely similar as determined by the presence and absence of phosphorylation of similar peptides (Supplementary Figure 1; top 65 peptides). An additional 10 samples, including cancer cell lines, xenograft and patient-derived tumor tissues, were compared. A heatmap of obtained basal profiles demonstrated phosphorylation of similar peptides across the samples with modest variability of signal intensity, but no apparent ‘on-off' differences in terms of peptide phosphorylation (Figure 4a). These inter-sample profile similarities are even more notable when the absolute signal intensities are represented in a ‘landscape view' (Figure 4b). Moreover, these analyses revealed that in these samples approximately 2/3 of available peptide substrates were being phosphorylated. The top 15 peptide substrates with highest phosphorylation intensity in both data sets included ENOG, CD79A, EFS, SRC8, PLCG1, FRK, PAXI, CDK2, FES and RET. These substrates were shown to be highly phosphorylated in other data sets as well.11, 25, 26
Figure 4.
Basal profile comparison of 10 tumor-related samples. Samples were measured using triplicate samples of 7.5 μg of lysate protein per array. (a) Heatmap of log2-transformed normalized signal intensities of the top 65 phosphorylated peptides in 10 different tumor cell or tissue-derived samples, sorted from high (red) to low (blue) signal intensity. The top 10 substrates included CD79A, ENOG, EFS, SRC8, PLCG1, FRK, P85A and PAXI. (b) ‘Phosphorylation landscape' indicating marginal absolute signal intensity differences between the tumor tissues (colorectal cancer, HCC, melanoma, lung cancer, pNET, MDA-MB-231 human breast cancer xenograft, HT29 chorioallantoic membrane model) and cell lines (HT29 colorectal cancer, NIH/3T3 mouse fibroblast, 786-O renal cell cancer). CAM chick chorioallantoic membrane; HCC hepatocellular carcinoma, pNET pancreatic neuroendocrine tumor.
Inhibition profiles
‘Off-chip' versus ‘on-chip' TKI treatment
The results from tumor tissue or cancer cell line inhibition profiles using several protein kinase inhibitors have previously been published.11, 18, 26, 27 To compare drug incubation of cancer cells in vitro (off-chip treatment) with ‘on-chip' spiking of the corresponding control lysate, we incubated HCT116 cells prior to lysis for 1 h using dimethyl sulfoxide control or 2 μM sunitinib, a concentration shown to inhibit phosphorylation of key downstream signaling proteins.20 For subsequent PTK microarray analysis, lysates from control and sunitinib incubated cells were spiked with dimethyl sulfoxide control (SUN −/−, SUN +/−, respectively) or 2 μM sunitinib (SUN −/+, SUN +/+). Although the resulting inhibition patterns were similar, inhibition of phosphorylation was more pronounced in sunitinib-incubated (SUN +/−) than in spiked (SUN −/+) samples (Figure 5a). This can be explained by high uptake of sunitinib in cancer cells within 1 h,28 which may contribute to prolonged inhibition ‘on-chip' by sunitinib present in the lysate, in the absence of ‘spiked' sunitinib. This should be taken into account in future drug screens using this array, but these data support the use of drug spiking in tumor lysates derived from patients for evaluating inhibition profiles.
Figure 5.
Inhibition of kinase activity. (a) TKI incubation versus spiking. Prior to lysis, HCT116 cells were incubated in vitro ±2 μM sunitinib, and subsequent lysates were spiked ±2 μM sunitinib (drug incubation/drug spiking). Signal intensity of the top 15 substrates is represented from high to low. Although inhibition patterns were similar, inhibition potency increased for sunitinib-incubated versus spiked samples, while additional spiking of previously incubated samples did not increase the degree of inhibition. (b) Competition with ATP. The HCT116 lysate was spiked with 4 μM sunitinib, 25 μM sorafenib or 10 nM dasatinib in the presence of increasing ATP concentrations. Despite average phosphorylation inhibition of 65–85% at 100 μM ATP, increasing ATP concentrations induced signal intensity and attenuated the inhibitory effects of sunitinib and sorafenib. With dasatinib, signal intensity increased to a lesser extent, suggesting partial ATP-independent inhibition of kinase activity. (c and d) Substrate-specific inhibition. Microarrays were incubated with 125 ng ml of recombinant Src or 500 ng ml Axl kinase±85 nM and 2 μM of the Src-inhibitor dasatinib and Axl-inhibitor R428, respectively. Both drugs resulted in near-complete inhibition of phosphorylation. The results of the top 25 phosphorylated substrates are shown.
Competition with ATP
The majority of TKIs binds the active conformation of the target kinase, competing with ATP binding.29, 30 These class I inhibitors include the clinically approved, multitargeted TKIs sunitinib, sorafenib and dasatinib. As shown above, the addition of ATP increases signal intensity of peptides spotted on the PTK microarrays. Therefore, we spiked HCT116 lysate with a fixed concentration of sunitinib (4 μM), sorafenib (25 μM) or dasatinib (10 nM), and investigated whether increasing ATP concentrations could partially neutralize their inhibitory effect. While inhibition of phosphorylation was observed at a mean of 66% for all peptide substrates at 100 μM ATP for sunitinib, 65% for sorafenib and 85% for dasatinib, increasing ATP concentrations induced signal intensity and attenuated the inhibitory effects of sunitinib and sorafenib (Figure 5b). For dasatinib, a smaller increase in signal intensity was observed, suggesting partial ATP-independent inhibition of kinase activity by additional allosteric binding of the target kinase. While allosteric binding sites may be obscure, their presence has been previously suggested for dasatinib.31, 32 This, as well as its high affinity for off-targets, may also explain the approximately 1000-fold lower concentration of dasatinib needed for inhibition.
Specificity of phosphorylation inhibition
Peptide substrates on the array can be phosphorylated by multiple kinases present in the lysate, while the relative contribution of these kinases to the signal intensity of a given peptide substrate is not known. Above, we showed kinase-specific activity of Src and Axl-kinase, while here we studied the potency and specificity of their inhibitors. We incubated separate microarrays with 125 ng/ml recombinant Src- and 500 ng/ml Axl-kinase +/− Src-inhibitor dasatinib (85 nM) and Axl-inhibitor R428 (2 μM). Both drugs caused almost complete inhibition of phosphorylation of the Src- and Axl-phosphorylated peptides (Figures 5c and d). In addition, we studied potential substrate-specific inhibition in tumor lysates. We assessed phosphorylation of the seven EGFR-substrates on the PTK microarray, revealing consistent phosphorylation of three substrates (EGFR_1103_1115, 1165_1177, 1190_1202) in all replicates for the tumor lysates from the four patients reported above. However, EGFR_908_920 was not phosphorylated in any of the lysates nor in any of the previously discussed experiments (data not shown). For the three remaining substrates, we did not observe specific inhibition upon spiking with 20 μM of the EGFR-specific inhibitor erlotinib. For the aforementioned EGFR_1103_1115, significant inhibition of this substrate was observed using dasatinib (85 nM), sunitinib (4 μM) and erlotinib in 4/4 patients and using sorafenib (25 μM) in 2/4 patients, indicating that this peptide is phosphorylated by kinases other than EGFR. Incubation with recombinant Src and, to a lesser extent with Axl kinase, indeed caused phosphorylation of this substrate, while dasatinib and RF428 each resulted in complete inhibition of activity (data not shown). The phosphorylation site of EGFR_1103_1115, Y1110, is known to be an EGFR autophosphorylation site (http://www.phosphosite.org/siteSearchAction.do) and has been previously shown to be inhibited by similar dasatinib concentrations.33
TKI treatment selection
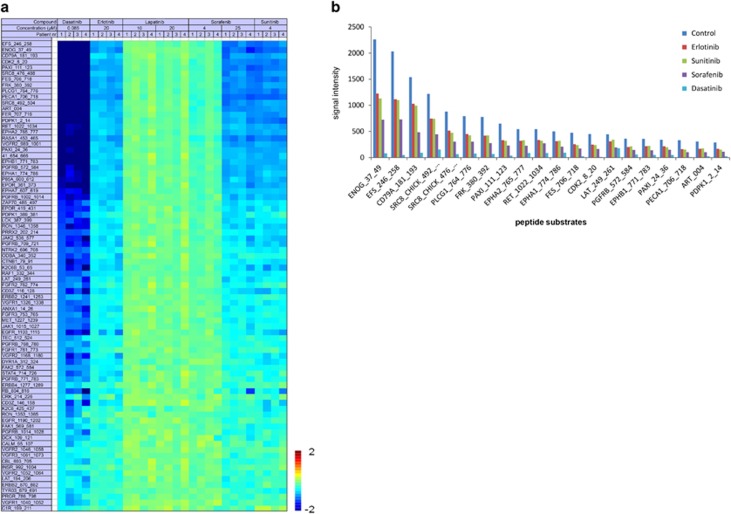
As a therapy selection pilot, we compared phosphorylation inhibition by five TKIs, including two concentrations of lapatinib and sorafenib, in tumor tissue lysates from four patients. As there are no algorithms available to evaluate inhibition profiles for potential efficacy in vivo and activated pathways cannot be inferred from basal microarray profiles, the inhibition potency for a given drug may be assessed by the number of significantly inhibited peptides as well as their inhibition/control ratios. Additionally, the presence of a ‘drug response' relationship may also indicate sensitivity. The inhibition ratios in Figure 6a suggest that inhibition occurred in similar peptide substrates and to the same extent. Dasatinib exhibited the highest inhibition potency, followed by sunitinib and sorafenib 25 μM, while no inhibition for lapatinib and a clear dose-response relationship for sorafenib was observed. Figure 6b further highlights that the inhibition potency of these drugs varies rather than the differential inhibition of highest intensity substrates is observed.
Figure 6.
TKI inhibition profiles of four patient samples. (a) Inhibition heatmap of four patient-derived tumor lysates (two breast, two lung cancer) demonstrating inhibition ratios of the top 80 peptides achieved by spiking with five TKIs. Peptides are sorted from high inhibition ratio (dark blue) to low, and green indicates absence of inhibition. (b) Representative patient example of top 30 highest intensity substrates sorted from high to low based on control samples. The control samples revealed the absolute phosphorylation intensities without inhibitor for each sample. Subsequently, absolute phosphorylation inhibition achieved by spiking with sorafenib, erlotinib, sunitinib and dasatinib are shown. Inhibition potency varies between the drugs, while no differential inhibition of individual peptides is observed.
Discussion
In this study, we evaluated kinase activity profiling of cancer cell lines and patient-derived tumor tissue using a commercially available array (PamChip) consisting of 144 PTK substrates. The goal was to evaluate the performance of the array under several experimental conditions for future TKI therapy selection in patients with cancer. We determined linearity and reproducibility, as well as optimal lysis and experimental running conditions. As a positive control for readout of kinase activity, we demonstrated phosphorylation activity of recombinant Axl and Src kinase, which could be reverted by co-inhibition with a specific inhibitor. Ideally, when aiming to predict TKI-sensitivity in individual patients, peptide phosphorylation on chip should be exclusively related to a single kinase and/or pathway, such that inhibition may be directly related to clinical sensitivity.13 The currently included peptide substrates can be phosphorylated by multiple kinases present in the lysate, while the relative contribution of these kinases to the signal intensity of a given peptide substrate is not known. This promiscuity may be enhanced in vitro by insufficient specificity of the kinases for the applied short peptides containing the tyrosine residue in this assay because both the amino acid sequence context of the phosphorylation site and the three-dimensional structure of the substrate protein are known contributing factors to this specificity.34, 35 Furthermore, cellular compartmentalization has been lost in lysates. Therefore, ‘hyper-reproducible' phosphorylation of several (Src-) substrates, including ENOG, CD79A, EFS and SRC8, which has been shown in several of our experiments (Figures 4a and 6a, Supplementary Figure 1), may be the result of low kinase specificity for these substrates rather than a mere reflection of higher abundant upstream kinases.36 Together, these factors increase the complexity of properly analyzing and comparing biological samples, which is illustrated by the apparent absence of sample-specific basal profiles of cancer cells and tissues (Figure 4a, Supplementary Figure 1). The observed profile similarities between samples may be the result of ‘household' kinase activity essential for proliferation and survival, while differential kinases contributing to cell- or patient-specific biologic behavior may be present at low or lower abundance and therefore not reproducibly detected using this array. The above is equally relevant for the evaluation of specific inhibitory drug profiles. As shown in Figures 5a and 6a, inhibition is most prominent in peptides shown to be highly phosphorylated in basal profile experiments, while the (off-) target affinity of the applied TKIs, rather than their supposed differential targets, seems to define the inhibition profiles obtained with this assay.
The application of the PTK microarray for kinase substrate optimization in drug development has been recently described,37, 38 while another study confirmed its potential for diagnostic biomarker discovery. In this study, vemurafenib inhibition profiles of 10 BRAF mutant and 16 BRAF wildtype metastatic melanoma patient tissues resulted in a mutation classification accuracy of 77% with a sensitivity (Se) of 90% and specificity (Sp) of 75%. However, predictive information could not be obtained because BRAF inhibitor response data were not available.26 Folkvord et al. analyzed pre-treatment tumor tissues in relation to pathological response (tumor regression grade, TRG) to neoadjuvant chemotherapy followed by neoadjuvant chemoradiation in 66 patients with locally advanced rectal cancer. An 86-peptide substrate-based classification model indicating good versus poor response, constructed from 12 good- (TRG 1–2) and 7 poor-responders (TRG 4), performed with 95% accuracy, 100% Se and 86% Sp in these patients. Classification accuracy in the remaining 37 good- and 10 intermediate-responders (TRG 3) was 85% for the good-responders with 86% Se, while 80% of intermediate responders were predicted to have good response.17 These results are promising and need further evaluation in an independent group of patients. Moreover, clinical implementation may be favored by a high negative predictive value to prevent toxicities in patients who are unlikely to benefit from treatment.
Thus far, predictive tests for clinical use are available as single mutation or amplification tests, but none of the available multiplex peptide assays to determine tyrosine kinase activity have reached the required level of validation for treatment selection tools.39, 40, 41, 42 Only prognostic or diagnostic in vitro diagnostic multivariate index assays have been cleared or approved by the US Food and Drug Agency for use in oncology, including Mammaprint (2007), OVA1 (2009) and Prosigna (2013) (http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm071455.pdf). OVA1, a test to assess likelihood of malignancy in women presenting with an ovarian mass, was the first Food and Drug Agency-cleared in vitro diagnostic multivariate index assays consisting of protein biomarkers.43 Aside from appropriate analytical test performance, critical factors to bringing biomarker discovery to approved clinical diagnostics include clearly defined intended use within a target population and utilization of appropriate specimens in preliminary validation studies to obtain sufficient evidence for well-designed validation studies at multiple clinical sites.43, 44, 45 In conclusion, specific activity of recombinant kinases can be adequately measured on the PTK microarray. For its use with more complex samples containing multiple kinases, the array peptides need further optimization for specificity. The array has clear benefits that can facilitate its clinical implementation as a TKI selection tool, including its reproducibility and short turnaround time, as well as required protein amounts of as little as 5–7 μg per sample. An alternative method to obtain insight into tumor kinase activity is provided using mass spectrometry-based phosphoproteomics.46 This method provides information on thousands of phosphopeptides per sample but in contrast requires sub- or low milligram-range amounts of protein47, which is at the high end of the amount that can be obtained in daily clinical practice from a tumor biopsy (Labots et al., Cancer Res 2015;75(15 Suppl):abstract nr 200745). Taken together, we aim to enhance the differential potential of the PTK array by using mass spectrometry-based phosphoproteomics to select more discriminative peptides and to enable clinical validation of this assay.
Acknowledgments
This study was financially supported by the AEGON International Scholarship in Oncology (M. Labots) and by a research grant from Vitromics Healthcare Services, The Netherlands to VUmc, Department of Medical Oncology.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
This research was supported by a research grant from Vitromics Healthcare Services, The Netherlands to VUmc, Department of Medical Oncology. The authors declare no conflict of interest.
Supplementary Material
References
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2000; 103: 211–225. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 2002; 298: 1912–1934. [DOI] [PubMed] [Google Scholar]
- Gotink KJ, Verheul HM. Anti-angiogenic tyrosine kinase inhibitors: what is their mechanism of action? Angiogenesis 2010; 13: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet 2006; 22: 545–554. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature 2001; 411: 355–365. [DOI] [PubMed] [Google Scholar]
- Keefe DM, Bateman EH. Tumor control versus adverse events with targeted anticancer therapies. Nat Rev Clin Oncol 2012; 9: 98–109. [DOI] [PubMed] [Google Scholar]
- Mazieres J, Zalcman G, Crino L, Biondani P, Barlesi F, Filleron T et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 2015; 33: 992–999. [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010; 363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med 2005; 353: 133–144. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 2001; 344: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Giovannetti E, Labots M, Dekker H, Galvani E, Lind JS, Sciarrillo R et al. Molecular mechanisms and modulation of key pathways underlying the synergistic interaction of sorafenib with erlotinib in non-small-cell-lung cancer (NSCLC) cells. Curr Pharm Des 2013; 19: 927–939. [PubMed] [Google Scholar]
- Hilhorst R, Houkes L, Mommersteeg M, Musch J, van den Berg A, Ruijtenbeek R. Peptide microarrays for profiling of serine/threonine kinase activity of recombinant kinases and lysates of cells and tissue samples. Methods Mol Biol 2013; 977: 259–271. [DOI] [PubMed] [Google Scholar]
- Piersma SR, Labots M, Verheul HM, Jimenez CR. Strategies for kinome profiling in cancer and potential clinical applications: chemical proteomics and array-based methods. Anal Bioanal Chem 2010; 397: 3163–3171. [DOI] [PubMed] [Google Scholar]
- Ter Elst A, Diks SH, Kampen KR, Hoogerbrugge PM, Ruijtenbeek R, Boender PJ et al. Identification of new possible targets for leukemia treatment by kinase activity profiling. Leuk Lymphoma 2011; 52: 122–130. [DOI] [PubMed] [Google Scholar]
- Sikkema AH, Diks SH, den Dunnen WF, ter Elst A, Scherpen FJ, Hoving EW et al. Kinome profiling in pediatric brain tumors as a new approach for target discovery. Cancer Res 2009; 69: 5987–5995. [DOI] [PubMed] [Google Scholar]
- Saelen MG, Flatmark K, Folkvord S, de Wijn R, Rasmussen H, Fodstad O et al. Tumor kinase activity in locally advanced rectal cancer: angiogenic signaling and early systemic dissemination. Angiogenesis 2011; 14: 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkvord S, Flatmark K, Dueland S, de WR, Groholt KK, Hole KH et al. Prediction of response to preoperative chemoradiotherapy in rectal cancer by multiplex kinase activity profiling. Int J Radiat Oncol Biol Phys 2010; 78: 555–562. [DOI] [PubMed] [Google Scholar]
- Versele M, Talloen W, Rockx C, Geerts T, Janssen B, Lavrijssen T et al. Response prediction to a multitargeted kinase inhibitor in cancer cell lines and xenograft tumors using high-content tyrosine peptide arrays with a kinetic readout. Mol Cancer Ther 2009; 8: 1846–1855. [DOI] [PubMed] [Google Scholar]
- Korangath P, Teo WW, Sadik H, Han L, Mori N, Huijts CM et al. Targeting glutamine metabolism in breast cancer with aminooxyacetate. Clin Cancer Res 2015; 21: 3263–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotink KJ, Rovithi M, de Haas RR, Honeywell RJ, Dekker H, Poel D et al. Cross-resistance to clinically used tyrosine kinase inhibitors sunitinib, sorafenib and pazopanib. Cell Oncol 2015; 38: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RJ, Boyce T, Fennell M, Jacobs V, Pinto F, Duffield E et al. The impact of delay in cryo-fixation on biomarkers of Src tyrosine kinase activity in human breast and bladder cancers. Cancer Chemother Pharmacol 2008; 61: 23–32. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE Jr. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 2007; 8: 530–541. [DOI] [PubMed] [Google Scholar]
- Follenzi A, Bakovic S, Gual P, Stella MC, Longati P, Comoglio PM. Cross-talk between the proto-oncogenes Met and Ron. Oncogene 2000; 19: 3041–3049. [DOI] [PubMed] [Google Scholar]
- Mason CS, Springer CJ, Cooper RG, Superti-Furga G, Marshall CJ, Marais R. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J 1999; 18: 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat W, van der Slik AR, Verhoeven DH, Alizadeh BZ, Ly LV, Verduijn W et al. Evidence for natural killer cell-mediated protection from metastasis formation in uveal melanoma patients. Invest Ophthalmol Vis Sci 2009; 50: 2888–2895. [DOI] [PubMed] [Google Scholar]
- Tahiri A, Roe K, Ree AH, de Wijn R, Risberg K, Busch C et al. Differential inhibition of ex-vivo tumor kinase activity by vemurafenib in BRAF(V600E) and BRAF wild-type metastatic malignant melanoma. PLoS ONE 2013; 8: e72692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada I, Hasina R, Arif Q, Mueller J, Smithberger E, Husain AN et al. Dramatic antitumor effects of the dual MET/RON small-molecule inhibitor LY2801653 in non-small cell lung cancer. Cancer Res 2014; 74: 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotink KJ, Broxterman HJ, Labots M, de Haas RR, Dekker H, Honeywell RJ et al. Lysosomal sequestration of sunitinib: a novel mechanism of drug resistance. Clin Cancer Res 2011; 17: 7337–7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009; 9: 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RA, Toader D, Ferguson AD. Structural approaches to obtain kinase selectivity. Trends Pharmacol Sci 2012; 33: 273–278. [DOI] [PubMed] [Google Scholar]
- Shan Y, Kim ET, Eastwood MP, Dror RO, Seeliger MA, Shaw DE. How does a drug molecule find its target binding site? J Am Chem Soc 2011; 133: 9181–9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 2008; 26: 127–132. [DOI] [PubMed] [Google Scholar]
- Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol 2010; 6: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat Biotechnol 2007; 25: 285–286. [DOI] [PubMed] [Google Scholar]
- Beltran L, Casado P, Rodriguez-Prados JC, Cutillas PR. Global profiling of protein kinase activities in cancer cells by mass spectrometry. J Proteomics 2012; 77: 492–503. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhang XC, Chen ZH, Yin XL, Yang JJ, Xu CR et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011; 29: 3316–3321. [DOI] [PubMed] [Google Scholar]
- Sanz A, Ungureanu D, Pekkala T, Ruijtenbeek R, Touw IP, Hilhorst R et al. Analysis of Jak2 catalytic function by peptide microarrays: the role of the JH2 domain and V617F mutation. PLoS ONE 2011; 6: e18522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz SA, Niranjan Y, Hammaren H, Ungureanu D, Ruijtenbeek R, Touw IP et al. The JH2 domain and SH2-JH2 linker regulate JAK2 activity: a detailed kinetic analysis of wild type and V617F mutant kinase domains. Biochim Biophys Acta 2014; 1844: 1835–1841. [DOI] [PubMed] [Google Scholar]
- Nesterov A, Dorsam E, Cheng YC, Schirwitz C, Markle F, Loffler F et al. Peptide arrays with a chip. Methods Mol Biol 2010; 669: 109–124. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Fearns C, Eliceiri B, Yang Y, Lee JD. Big mitogen-activated protein kinase 1/extracellular signal-regulated kinase 5 signaling pathway is essential for tumor-associated angiogenesis. Cancer Res 2005; 65: 7699–7706. [DOI] [PubMed] [Google Scholar]
- Thiele A, Zerweck J, Weiwad M, Fischer G, Schutkowski M. High-density peptide microarrays for reliable identification of phosphorylation sites and upstream kinases. Methods Mol Biol 2009; 570: 203–219. [DOI] [PubMed] [Google Scholar]
- Thiele A, Zerweck J, Schutkowski M. Peptide arrays for enzyme profiling. Methods Mol Biol 2009; 570: 19–65. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev 2010; 19: 2995–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Chan DW. Proteomic cancer biomarkers from discovery to approval: it's worth the effort. Expert Rev Proteomics 2014; 11: 135–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labots M, Schutte LM, van der Mijn JC, Pham TV, Jimenez CR, Verheul HM. Mass spectrometry-based serum and plasma peptidome profiling for prediction of treatment outcome in patients with solid malignancies. Oncologist 2014; 19: 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutillas PR. Role of phosphoproteomics in the development of personalized cancer therapies. Proteomics Clin Appl 2015; 9: 383–395. [DOI] [PubMed] [Google Scholar]
- Piersma SR, Knol JC, de Reus I, Labots M, Sampadi BK, Pham TV et al. Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines. J Proteomics 2015; 127: 247–258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.