Abstract
The plant cyclotides are a family of 28 to 37 amino acid miniproteins characterized by their head-to-tail cyclized peptide backbone and six absolutely conserved Cys residues arranged in a cystine knot motif: two disulfide bonds and the connecting backbone segments form a loop that is penetrated by the third disulfide bond. This knotted disulfide arrangement, together with the cyclic peptide backbone, renders the cyclotides extremely stable against enzymatic digest as well as thermal degradation, making them interesting targets for both pharmaceutical and agrochemical applications. We have examined the expression patterns of these fascinating peptides in various Viola species (Violaceae). All tissue types examined contained complex mixtures of cyclotides, with individual profiles differing significantly. We provide evidence for at least 57 novel cyclotides present in a single Viola species (Viola hederacea). Furthermore, we have isolated one cyclotide expressed only in underground parts of V. hederacea and characterized its primary and three-dimensional structure. We propose that cyclotides constitute a new family of plant defense peptides, which might constitute an even larger and, in their biological function, more diverse family than the well-known plant defensins.
INTRODUCTION
In the last few years the concept of peptides and proteins being linear chains of amino acids has been challenged by the discovery of various naturally occurring circular proteins isolated from microorganisms, plants, and even a mammal (Trabi and Craik, 2002). In one family of circular plant proteins, called the cyclotides (Craik et al., 1999), the unusual feature of a head-to-tail cyclized peptide backbone is combined with a cystine knot motif, formed by six absolutely conserved Cys residues. In this motif, two disulfide bonds and their connecting backbone segments form a ring that is penetrated by the third disulfide bond. The combined cyclic backbone and cystine knot is referred to as a cyclic cystine knot (CCK; Craik et al., 2001), and it is thought to be responsible for making the cyclotides extremely resistant to endoproteinase and exoproteinase digest and for conferring exceptional thermal stability onto them. As a result of these unusual properties, the cyclotide framework constitutes a promising template for drug design and agrochemical applications (Craik, 2001; Craik et al., 2002) via the exploitation of natural cyclotide activities or potentially via grafting new bioactive functionalities onto the stable framework.
The background to the discovery of cyclotides may be traced back to native medicine applications in Africa. During two Red Cross relief missions to the Congo in the 1960s, a Norwegian physician observed accelerated labor and childbirth, unusually strong uterine contractions, and an abnormally high rate of birth complications and necessary caesarean sections among the women of the local Lulua tribe (Gran, 1970). Gran noted that the women boiled the dry aerial parts of a local weed, Oldenlandia affinis DC (Rubiaceae), and sipped the resulting decoction during labor. Analysis of the plant material revealed the presence of serotonin and a uterotonic peptide named kalata B1 (Sletten and Gran, 1973). At that stage neither kalata B1's primary sequence nor the CCK motif were characterized, and it was more than two decades after the initial report that the circular nature of the peptide backbone of kalata B1 and its disulfide bonding pattern were discovered (Saether et al., 1995).
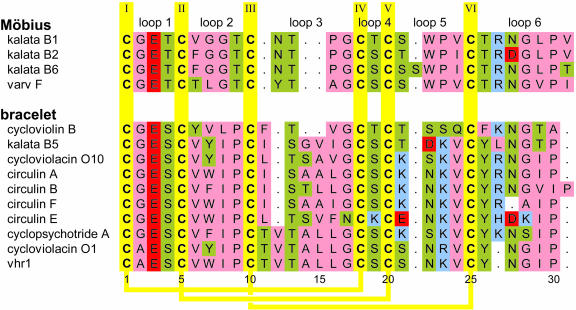
Recent years have seen the discovery of an ever-growing number of cyclotides, mainly found through screening programs for various bioactivities. A summary of selected cyclotides along with their bioactivities and sources is presented in Table 1. Irrespective of these various activities, the natural role of cyclotides is assumed to be a protective one, a theory corroborated by the observation that kalata B1 inhibits the growth and development of Helicoverpa punctigera larvae in feeding trials (Jennings et al., 2001). Figure 1 shows several sequences representative of the cyclotide family. Because of the circular nature of the peptide backbone and the conserved number of Cys residues in the cyclotides, there are nominally six backbone segments (or loops) between successive Cys residues, also shown in Figure 1. Because the site and mechanism(s) of cyclotide precursor processing are not yet known, we have decided to number the sequences starting from the first absolutely conserved residue in the most conserved loop of the mature cyclotide sequence. The core group of cyclotides can be divided into two subfamilies, called Möbius and bracelet, which were originally named based on the presence or absence of a conceptual twist in the peptide backbone associated with a cis-Pro residue in loop 5 of the sequence (Craik et al., 1999).
Table 1.
Bioactivities of Selected Cyclotides, Their Sources, Sizes, and Masses
| Cyclotide | Plant Species | Fa | aab | Mass | Activity | Potency | Reference |
|---|---|---|---|---|---|---|---|
| Circulins A and B | Chassalia parvifolia Schum. | R | 30/31 | 3152/3284 | Anti-HIV | EC50c 40–260 nM | Gustafson et al. (1994) |
| Antimicrobial | MICd 0.19–25.5 μM | Tam et al. (1999) | |||||
| Cycloviolins A–D | Leonia cymosa Mart. | V | 28–31 | 2886–3212 | Anti-HIV | EC50 ∼130 nM | Hallock et al. (2000) |
| Kalata B1 | O. affinis DC | R | 29 | 2892 | Uterotonic | NDe | Gran (1973b) |
| Antimicrobial | MIC 0.26 μM | Tam et al. (1999) | |||||
| Insecticidal | 0.8 μM/g diet | Jennings et al. (2001) | |||||
| Violapeptide I | V. tricolor L. | V | 29 | ND | Hemolytic | ND | Schöpke et al. (1993) |
| Palicourein | Palicourea condensata Standl. | R | 37 | 3905 | Anti-HIV | EC50 100 nM | Bokesch et al. (2001) |
| Cyclopsychotride A | Psychotria longipes Muell. Arg. | R | 30 | 3231 | Neurotensin antagonism | IC50f 3 μM | Witherup et al. (1994) |
| Antimicrobial | MIC 1.55–39.0 μM | Tam et al. (1999) | |||||
| Cycloviolacin O2 | V. odorata L. | V | 30 | 3141 | Antitumor | IC50 0.1–0.3 μM | Lindholm et al. (2002) |
| Vitri A | V. tricolor L. | V | 30 | 3155 | Cytotoxic | IC50 0.6–1 μM | Svangård et al. (2004) |
| MCoTI-I and -II | M. cochinchinensis (Lour.) Spreng. | C | 34 | 3482/3454 | Trypsin inhibition | 0.6–0.7 IUg | Hernandez et al. (2000) |
Plant family (R, Rubiaceae; V, Violaceae; C, Cucurbitaceae).
Size in amino acids (aa).
EC50, effective concentration that provides 50% cytoprotection.
MIC, Minimum inhibitory concentration.
ND, Not determined.
IC50, inhibitory concentration that decreases neurotensin binding to cell membranes by 50%, and in the case of the cytotoxic data, the concentration that yields a survival index of 50%.
IU, inhibitory units (1 IU = the amount of inhibitor that reduces the activity of 2 mg of trypsin by 50%).
Figure 1.
Amino Acid Sequences of Various Cyclotides Representative of the Bracelet and Möbius Subfamilies.
The amino acids are colored according to their properties (hydrophobic, pink; hydrophilic, green; acidic, red; basic, blue). Cys residues are given in yellow, and intercysteine loops are numbered at top of figure. The yellow lines on the bottom of the figure indicate the disulfide connectivities. The subfamilies can be distinguished by the presence (Möbius) or absence (bracelet) of a cis-Pro residue in loop 5. Note also the loop sizes (loop 3) and loop sequences characteristic for each subfamily. As a result of the cyclic nature of the peptide backbone in cyclotides and some ambiguity about precursor processing sites and mechanism(s), the numbering of residues is arbitrary, starting with the first absolutely conserved residue in the mature cyclotide sequence (Cys1) in the loop with highest conservation within and across the two subfamilies. The sequences have been aligned using MultAlin (Corpet, 1988) with Blosum62, a gap opening penalty of 10 and a gap extension penalty of 2, followed by a manual alignment of the structurally crucial hydroxyl bearing residue in loop 3 (Rosengren et al., 2003) The amino acid sequence and residue numbering of vhr1, determined in this study, are given at bottom of figure. Original citations to the various cyclotides are as follows: kalata B1, Saether et al. (1995); kalata B2, kalata B5, cycloviolacin O1, and cycloviolacin O10, Craik et al. (1999); kalata B6, Jennings et al. (2001); varv F, Göransson et al. (1999); cyclopsychotride A, Witherup et al. (1994); circulin A and circulin B, Gustafson et al. (1994); circulin E and circulin F, Gustafson et al. (2000); cycloviolin B, Hallock et al. (2000); and vhr1, this study. A complete list of cyclotide sequences and database access IDs is given as supplemental material (see Supplemental Table S1 online).
Recently, two structurally related circular trypsin inhibitors have been reported from the seeds of Momordica cochinchinensis, a Cucurbitaceae plant (Hernandez et al., 2000; Felizmenio-Quimio et al., 2001; Heitz et al., 2001). However, because their amino acid sequences and biological activities differ significantly from those of Möbius and bracelet cyclotides, there is still debate whether these trypsin inhibitors should be included in the cyclotide family (Craik et al., 2004). To date, no cyclotides have been found in the common model plant Arabidopsis thaliana, and genes encoding for cyclotides are not apparent in a search of the Arabidopsis genome.
Although ∼50 cyclotides from 11 different Violaceae and Rubiaceae species have been described to date, previous studies of the discovery of cyclotides have typically reported only a small number of peptides for a given species. Here, we demonstrate that it is not atypical for one plant species to contain dozens of different cyclotides. Specifically, we report the presence of at least 57 new cyclotides in the native Australian violet, Viola hederacea.
In his early work, Gran states that for the uterotonic decoction used by the Lulua aerial parts of O. affinis were collected during the rainy season and dried for later use (Gran, 1970; Gran, 1973a). This might be an indication of variation in cyclotide expression with both time and the plant tissue used (or O. affinis, an herbaceous Rubiaceae, simply dies off during the dry season). In this study, we investigate the cyclotide profiles of different Viola species (Violaceae) with regard to variations in cyclotide expression, putting particular emphasis on the differences in the cyclotide profiles of several plant parts. A recent report mentions the presence of cyclotides in the underground parts of Viola odorata (Göransson et al., 2003), but no cyclotides from root tissue have been characterized to date, and, therefore, our specific interest was in this plant part because roots are subject to a vastly different array of pests compared with aerial plant parts. Here, we show that the roots of various Viola species, like their aerial counterparts, contain a large number of different cyclotides. We also present the amino acid sequence and solution structure of vhr1 (V. hederacea root cyclotide1), the first cyclotide isolated from roots, and compare its three-dimensional fold to that of other cyclotides.
RESULTS
The focus of this study was to ascertain the extent and nature of the cyclotide distribution in different plant parts. HPLC and liquid chromatography–mass spectrometry (LC-MS) techniques were used to separate and characterize cyclotides from different plant parts of several Viola species (Violaceae). We compared the masses of these cyclotides, derived from LC-MS studies, with the masses of known cyclotides to gauge the overall number of new peptides.
Cyclotide Profiles
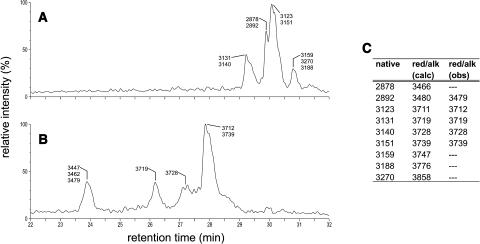
Cyclotides typically elute between 45% and 65% acetonitrile on C18 reverse-phase (RP) HPLC and have masses between ∼2800 and 3900 D (Table 1). In addition to making use of these characteristics, we used chemical modification of the disulfide bonds as well as MS experiments to establish that the late eluting peaks seen in the LC-MS traces are indeed cyclotides. The peptide fraction isolated from underground runners was subjected to reduction of the disulfide bonds with Tris (2-carboxyethyl) phosphine hydrochloride and alkylation with maleimide. LC-MS analysis of the reaction mixture after reduction and alkylation showed significantly shortened elution times and masses that differed by 588 D from those of the native peptides (Figures 2A and 2B). This is consistent with the reduction of three disulfide bonds and the subsequent addition of one maleimide moiety to each of the six resulting half-cystines. Even under harsh MS conditions, there was no evidence for fragmentation of the reduced and alkylated peptides, which strongly suggests the presence of a head-to-tail cyclized backbone.
Figure 2.
Effects of Disulfide Bond Modification on the LC-MS Profile of Cyclotides.
LC-MS profile of a native cyclotide fraction (A). After reduction and alkylation of the disulfide bonds (B), the modified cyclotides elute significantly earlier and show masses that correspond to six alkylated half-cystines, providing proof that three disulfide bonds have been broken. (C) provides a tabular listing of the masses observed in (A) (column native) together with the calculated [red/alk (calc)] and observed [red/alk (obs)] masses of the reduced and alkylated peptides. Note that reduction and alkylation of such a complex reaction mixture can lead to incomplete reaction of some species; therefore, masses corresponding to some native cyclotides found in (A) are missing in (B) and (C).
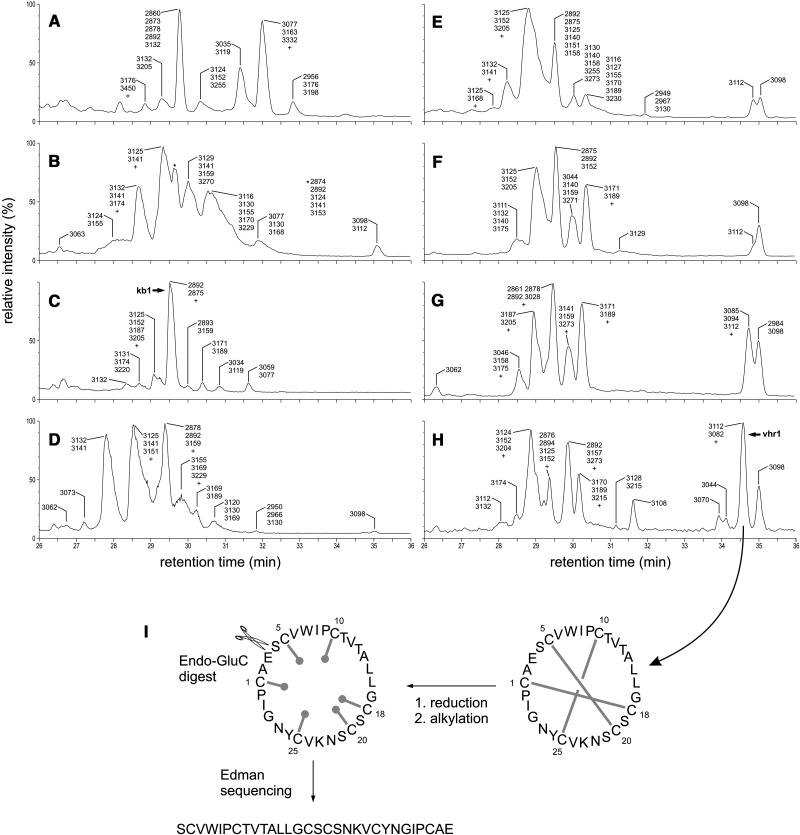
Having established this identification procedure for cyclotides, we proceeded to investigate the cyclotide content of a range of crude extracts. Analysis of crude extracts derived from various plant parts (leaves, petioles, flowers, pedicels, roots, bulbs, as well as aboveground and belowground runners) of V. hederacea showed markedly different cyclotide profiles for each plant part (Figures 3A to 3H), indicating tissue-specific expression of certain cyclotides. Kalata B1 (2892 D at retention time ∼29.5 min) was found in all tissue types tested and was generally accompanied by the coeluting varv peptide A (2878 D; Claeson et al., 1998), also called kalata S (Craik et al., 1999).
Figure 3.
LC-MS Profiles of Cyclotides in Various V. hederacea Plant Parts and Sequencing Strategy for vhr1.
(A) to (H) LC-MS profiles of various parts of V. hederacea. Leaves (A); petioles (B); flowers (C); pedicels (D); aboveground runners (E); bulbs (F); belowground runners (G); and roots (H). The peaks are labeled with the masses of the cyclotides they contain. The presence of one or more minor components with a signal intensity of <30% of that of the strongest corresponding signal is indicated by a plus symbol (+); the masses of these components are not given in the figure. For peaks that are identified by a symbol, the masses are given elsewhere in the figure panel, preceded by the corresponding symbol. The peaks containing kalata B1 (kb1) and vhr1 are labeled ([C] and [H], respectively).
(I) Characterization of vhr1. The disulfide bonds of the native, circular peptide are reduced and alkylated to destabilize the fold. Enzymatic digest with endoproteinase GluC yields a linear peptide whose amino acid sequence can be determined by Edman degradation.
Altogether 66 different masses were seen in the various extracts of V. hederacea, 57 of which did not match any known cyclotide mass and can therefore be regarded as unique and belonging to an as yet uncharacterized cyclotide. Only nine masses, or <15%, corresponded to those of known cyclotides. These previously reported masses match those of kalata B4 (2894), kalata B2 (2956), vodo M (3077), cycloviolacin O1 (3116), cyclopsychotride A (3129), cycloviolacin H1 (3131), cycloviolacin O2 and O9 (3140), circulin A (3151), and vico A (3270). However, despite their matching masses, there is the possibility that the molecules found in V. hederacea are other new cyclotides that just happen to have the same masses.
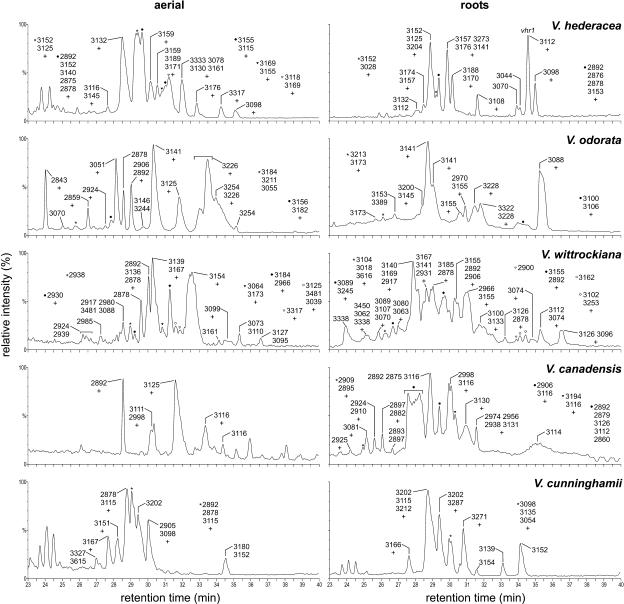
Comparison of aboveground and belowground plant parts for other Viola species also showed marked differences in the cyclotide profiles (Figure 4). The overall levels and numbers of individual cyclotides were generally comparable between the different plant parts of one species. One remarkable exception was Viola canadensis, whose roots contained ∼5 times the number of cyclotides found in aerial parts. The most complex cyclotide pattern was found in the roots of Viola wittrockiana, the humble garden pansy.
Figure 4.
LC-MS Profiles of Aerial and Underground Parts of Various Viola Species.
For labeling of the peaks, see Figure 3. The aerial parts of V. odorata contain a further cyclotide peak not shown in the profile (elution time 22.3 min, mass of the main compound 2748 D).
As indicated by the rather complex LC-MS traces in Figure 4, all species and plant parts examined in this study contained a plethora of different cyclotides. Some of these cyclotides were found in various species and plant parts, although often in varying amounts. Kalata B1 (2892 D at retention time ∼29.5 min), for example, constituted the major component of its respective peak in each of the aerial samples but was often virtually absent in roots, like in Viola cunninghamii roots, where it represented <5% of the main peak component. Of the species screened in this study, V. wittrockiana produced the largest cyclotide with a mass of 3481 D. The latest eluting peak (elution time 36.6 min) containing the most hydrophobic cyclotides was also found in V. wittrockiana, specifically in the roots. The main component of this peak showed a mass of 3126 D and was accompanied by second peptide (mass 3096 D) with a signal intensity of ∼35% and half a dozen of compounds with signal intensities between 3% and 10% of the main compound. The most hydrophilic cyclotide with an elution time of 22.3 min was found in V. odorata. With a mass of 2748 D, it also is the smallest cyclotide reported to date.
Amino Acid Sequence and Disulfide Pattern of vhr1
We isolated the cyclotide most abundant in the roots of V. hederacea, vhr1, to determine its primary and solution structure. Mass spectrometric analysis of native and reduced vhr1 confirmed the presence of three disulfide bonds. All except for three cyclotides (circulins D and E as well as varv peptide E) contain one and only one Glu residue; therefore, Endo GluC was chosen to linearize the continuous peptide backbone of vhr1 (Figure 3I). As is frequently seen with cyclotides, native vhr1 proved resistant to enzymatic cleavage, emphasizing the role of the cystine knot motif in stabilizing the peptide's fold. After reduction and alkylation of the disulfide bonds, the peptide backbone was amendable to enzymatic cleavage. Digest of the Cys-reduced and Cys-alkylated peptide increased its molecular weight by 18 D, consistent with the presence of a head-to-tail cyclized peptide backbone and one Glu residue. Automatic Edman degradation of the reduced, alkylated, and linearized peptide revealed an amino acid sequence with conserved residues and loop sizes characteristic of a bracelet cyclotide (Figure 1).
Solution Structure of vhr1
The solution structure of vhr1 was determined using homonuclear two-dimensional NMR methods combined with simulated annealing calculations. Sequence specific resonance assignments of vhr1 were obtained from total correlation spectroscopy (TOCSY) and double quantum filtered correlation spectroscopy (COSY) spectra, to identify the spin systems, and nuclear Overhauser enhancement spectroscopy (NOESY) spectra, to establish the connections between the spin systems and identify their position in the primary structure, as described previously for other cyclotides (Rosengren et al., 2003). The two-dimensional NMR spectra showed good chemical shift dispersion in the fingerprint region, making resonance assignment relatively straightforward. Any ambiguities in assignment were resolved by comparison of the respective spectra at 298K and 310K. Sample spectra and chemical shift values of all assigned protons are given as supplemental information.
Sequential connectivities in the NOESY spectra confirmed both the circular nature of the peptide backbone and the trans-conformation of both Pro residues. In particular, sequential Hαi-HNi+1 nuclear Overhauser effect (NOE) connectivities were seen in a continuous circle around the whole backbone except at Pro residues, which lack the amide proton and, therefore, intrinsically do not have such connections. However, the presence of Hαi−1-Hδi cross-peaks for both Pro residues not only completed the backbone circle but also confirmed, when considered together with the lack of corresponding Hαi−1-Hαi connectivities, that both Pro residues were in a trans-conformation. Recently there has been debate about whether the disulfide bonding pattern in kalata B1, the prototypic cyclotide, was indeed knotted as originally proposed (Saether et al., 1995) or in a ladder-like arrangement (Skjeldal et al., 2002). The issue was resolved when it was shown that the use of χ1 angles confirmed the originally proposed connectivity (Rosengren et al., 2003). The question of the disulfide pattern arose because in both knotted and laddered arrangements, Hβ protons of nonbonded Cys residues can be closer in space than those of bonded Cys residues, and it is therefore impossible to deduce the disulfide pattern exclusively from NOE connectivities (Rosengren et al., 2003). Consequently, it is crucial to incorporate Cys χ1 angles in the pattern analysis. In vhr1, the χ1 side chain dihedral angles could be determined for C10 and C18 based on 3JHα−Hβ coupling constants and NOE patterns. For these two residues, dihedral angle constraints of −60° ± 30° were used in structure calculations. These values are consistent with those reported for the knotted disulfide arrangement of kalata B1 (Rosengren et al., 2003), whereas in the laddered arrangement (Skjeldal et al., 2002), the majority of structures show positive χ1 angles for these two residues. Furthermore, for vhr1 the observed S-S distances in initial structure calculations without explicitly set disulfide bonds were again consistent with the knotted but not laddered disulfide arrangement. For the final round of structure calculations, the disulfide bonding pattern was therefore explicitly set to I-IV, II-V, and III-VI (i.e., a knotted disulfide arrangement). This disulfide pattern has recently been further confirmed in kalata B1 using a chemical approach for elucidation of Cys-Cys connectivities (Göransson and Craik, 2003).
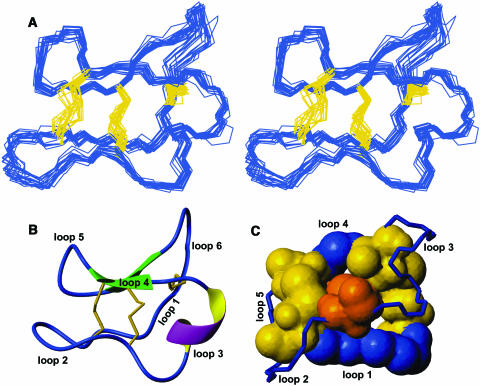
The final family of 20 structures is of high precision, with a root mean square deviation (RMSD) of 0.50 Å when superimposed over the backbone atoms of all 30 residues or 1.04 Å when superimposed over all heavy atoms (Figure 5A). As in other cyclotides, the peptide backbone of vhr1 is folded back onto itself and cross-braced by the three disulfide bonds, forming the CCK motif. Two short β-strands can be found in loops 4 and 5/6, which are connected by a turn region centered on residues 22 to 24, thus forming a β-hairpin motif. A third strand, incorporating residues 2 to 5 of loop 1, is somewhat distorted and is not recognized as part of the β-sheet by the secondary structure checking programs PROMOTIF (Hutchinson and Thornton, 1996) or Procheck-NMR (Laskowski et al., 1996). In 90% of the structures, residues 14 to 16 in loop 3 form a short 310 helix, as indicated in Figure 5B.
Figure 5.
Structural Characteristics of vhr1.
(A) Structural family of vhr1. Stereo view of the 20 lowest energy structures of vhr1 superimposed over the N, C, and Cα atoms of all 30 residues. The peptide backbone is given in blue, and the disulfide bonds are in yellow.
(B) Summary of the structural features present in vhr1. The disulfide bonds are given in yellow, and the intercysteine loops are labeled.
(C) The cystine knot motif in vhr1. The atoms of the cystine knot are scaled according to their van der Waals radii. The two disulfide bonds and the backbone segments forming the ring are shown in blue and yellow, respectively. The third disulfide bond, penetrating this disulfide/backbone ring, is shown in orange. Backbone segments not directly involved in the cystine knot motif are shown as lines with labeled loops.
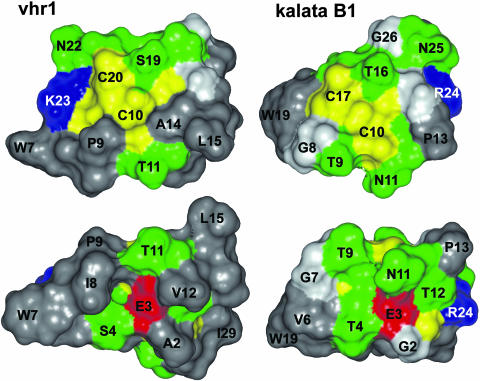
An analysis of the surface-exposed residues showed that hydrophobic residues make up 57% of the surface of vhr1, compared with only 39% of the surface of kalata B1. Figure 6 illustrates that in vhr1 hydrophobic and Cys residues form a continuous hydrophobic surface on one face of the molecule that is only disturbed by T11 and S19, whereas the hydrophobic surface on the opposite face of the molecule is only disturbed by E3, T4, and T11. By contrast, in kalata B1 the hydrophobic patches are smaller and more interspersed with various hydrophilic residues.
Figure 6.
Surface Representation of the Lowest Energy Structure of vhr1 in Comparison with Kalata B1.
The amino acids are colored according to their properties (red, acidic; blue, basic; green, hydrophilic; gray, hydrophobic). Cys residues are given in yellow, and Gly residues are in light gray. Selected residues are labeled with the one letter code and the residue number, following the numbering of residues in the respective PDB entries.
DISCUSSION
Cyclotide Expression and Biosynthesis
Recent reports have shown that cyclotides act as insecticidal (Jennings et al., 2001) and antimicrobial agents (Tam et al., 1999), implying a role in the plant's defense system. Craik et al. (1999) also list the large quantities of cyclotides found in plant material, up to 1 g per kg of leaves, and their Cys-rich nature, not dissimilar to thionins, as indications for the involvement of cyclotides in host defense. However, it is still unclear whether this is their primary natural function in plants. Gran reports that in the Congo the aerial parts of O. affinis are used to prepare the uterotonic decoction kalata-kalata (Gran, 1973a), possibly implying a difference in cyclotide content between the various plant parts. To learn more about the expression of cyclotides and their natural function, we investigated the expression patterns of these unusual proteins in different plant parts of V. hederacea, the native Australian violet, and various other Viola species (Violaceae).
This study reveals clear variations in the cyclotide profiles of different parts of V. hederacea. The differences are most obvious between aerial and belowground parts: in addition to a number of cyclotides found in the aerial parts of V. hederacea, all plant parts in contact with soil (i.e., roots, runners, and bulbs) produce further, late eluting and, therefore, more hydrophobic cyclotides. In roots, these peptides are expressed in approximately the same amounts as the earlier eluting aboveground group. With nine different cyclotides (as assessed by their masses), the roots also contain the largest variety of late-eluting, tissue-specific peptides. These particular cyclotides are missing completely in leaves and flowers and are only present at very low levels in petioles and pedicles (whose lower parts are in contact with soil, too).
Overall, it can be estimated from this study that V. hederacea expresses at least 66 different cyclotides. The masses of only nine of these peptides correspond to masses of cyclotides with known amino acid sequence, leaving at least 57 cyclotides in V. hederacea uncharacterized at this point in time. However, one mass can correspond to different cyclotides, as can be seen for example from varv peptides A, C, and D (all 2874 D), and, therefore, even some of these known masses could correspond to as yet uncharacterized cyclotides. Because all cyclotides sequenced to date were isolated from aerial plant parts, it is not surprising that peptides in belowground parts (i.e., roots, runners, and bulbs) of V. hederacea represent the majority of cyclotides with previously unseen masses.
All Viola species examined in this study contained large numbers of cyclotides. Although these species only constitute a comparatively small part of the genus Viola, expression of cyclotides can probably be regarded as a common theme in the genus. LC-MS analysis of crude plant extracts revealed plant part–specific differences in cyclotide expression for all Viola species. The data seem to indicate that if Violaceae plants express cyclotides, they do so in all tissues, possibly with the exception of Leonia cymosa—the literature only mentions the bark as source for cycloviolins A to D (Hallock et al., 2000). Cyclotides can also be found in plants of a second family, namely the Rubiaceae. However, for this family the literature seems to suggest that in some species cyclotides are only expressed in woody parts, such as the bark (Gustafson et al., 1994; Bokesch et al., 2001).
Apart from cyclization of the peptide backbone, to date there is no evidence of posttranslational modifications of cyclotides, which raises the question of how cyclotide-bearing plants generate such a high number of related proteins. In O. affinis (Rubiaceae), the cyclotides are encoded by a multigene family with up to 12 related genes (Jennings et al., 2001). They are synthesized as prepropeptides consisting of a signal sequence, a proregion, an N-terminal repeat, and between one and three mature cyclotide domains, separated again by N-terminal repeats. In these prepropeptides, cyclotide sequences are generally flanked by a GLP triad, creating four possible processing sites: the mature circular peptide retains one copy of the GLP sequence, which, depending on the initial processing site, can be derived entirely from one of the flanking triads or partially from both. However, in one of the clones isolated, the cyclotide sequences are flanked N terminally by a GLP triad but C terminally by an SLP triad, suggesting processing N terminally of the GLP and SLP elements (Jennings et al., 2001). The mechanism by which this occurs, as well as the intracellular location and timing of the processing, folding, and cyclization events necessary to produce a mature, circular cyclotide, are still not understood. The sequences of the array of new cyclotides reported in this study may provide further clues as to how this processing occurs, and efforts to determine the amino acid sequences of these cyclotides are underway. As an aside, we note that in the absence of definitive knowledge of the processing site in all cyclotides, the circular backbone of the mature peptide makes the choice of numbering scheme for amino acids somewhat arbitrary. We have chosen to number cyclotide sequences starting at the first absolutely conserved residue (Cys), following the loop in which processing occurs.
Comparison with Plant Defensins
Living organisms have evolved various mechanisms to defend themselves against pathogens. Innate immunity is often seen as the oldest and most basic defense strategy, involving, among other responses, the production of antimicrobial peptides (Boman, 1995). One group of defense peptides, called the plant defensins, is apparently ubiquitous throughout the plant kingdom (Thomma et al., 2002). To date, sequences of >80 different (putative) plant defensin genes are known (Thomma et al., 2002), derived from a number of different plant species. In this study, we have shown that on one hand a single Viola species expresses a plethora of cyclotides and that on the other hand there are species and tissue-specific differences in cyclotide expression. We therefore propose that cyclotides can be regarded as a new family of plant defense peptides, complementary to the classic plant defensins. It seems likely at this stage that the cyclotides, although less widespread throughout the plant kingdom, might constitute an even larger and, in their biological function, more diverse family than the plant defensins.
There is evidence for tissue-specific expression of plant defensins: Terras et al. (1995) found that Rs-AFP1, an antifungal defensin isolated from radish (Raphanus sativus), is present in the seeds but barely detectable in healthy leaves. In fungus-infected leaves, the authors identified two other defensins with ∼90% sequence similarity to seed-derived Rs-AFPs. Eight to 10 genes hybridized with an Rs-AFP1 cDNA probe. Comparing these findings to cyclotides, the parallels seem obvious: various cyclotides exhibit antifungal activity (Tam et al., 1999); it has been shown that the cyclotides are derived from a multigene family (Jennings et al., 2001); some cyclotides seem to be pathogen induced (M. Trabi, unpublished data); and, as we have shown here, certain cyclotides are expressed in a tissue-specific manner.
Solution Structure of vhr1 and Comparison with Other Cyclotides
The previously unreported cyclotide vhr1 was chosen for further investigation because it was the most abundant cyclotide expressed only in underground parts of V. hederacea, but also because of its, even for cyclotides, highly hydrophobic character. The primary structure of vhr1 shows high sequence homology to cycloviolacin O1 (Craik et al., 1999), isolated from the aerial parts of V. odorata, including a rather unusual AES triad in loop 1. Only five cyclotides (vhr1, cycloviolacin O1 [Craik et al., 1999], vico A and B [Göransson et al., 2003], and hypa A [Broussalis et al., 2001]) have been reported to contain an Ala residue in position 2 instead of the more common Gly residue. Interestingly, all AES mutants have been isolated from Violaceae plants, raising the question of whether this mutation is a Violaceae-specific phenomenon. Also, no corresponding Möbius cyclotide with A in position 2 has been found to date; the GET triad in loop 1 is absolutely conserved throughout the Möbius subfamily.
Like other cyclotides, vhr1 exhibits a very compact overall fold, with the disulfide bonds occupying the core of the structure. The circular peptide backbone is cross-braced by the three disulfide bonds in their characteristic knotted arrangement. The main features of the fold are two strands of a β-sheet and several turn regions, as well as a short segment of 310 helix in loop 3, stabilized by hydrogen bonds between T13-L16 and A14-G17. The side chain of E3 bridges the gap between loop 1 and the start of this helical region, forming hydrogen bonds with the amide protons of T11, V12, and T13, and with the hydroxyl proton of T13. The Glu residue at position 3 is absolutely conserved across all known cyclotide sequences, and it can be assumed that the stabilizing role of the hydrogen bond network involving its carboxyl group is crucial for the structural integrity not only of the cystine knot framework but also the overall fold in general. Even a highly homologous Asp residue in the same position appears not to be tolerated: with one methylene group less, its side chain would be too short to reach across to loop 3 and provide the acceptor atoms necessary for creating four hydrogen bonds.
The solution structures of one Möbius cyclotide (kalata B1, Protein Data Bank [PDB] IDs 1kal, 1nb1, 1jjz, and 1k48), two bracelet cyclotides (circulin A, PDB ID 1bh4, and cycloviolacin O1, PDB IDs 1df6 and 1nbj), and the rather unusual palicourein (PDB ID 1r1f) have been determined to date. While all these cyclotides were isolated from the aerial parts of the respective plants, vhr1 is the first cyclotide from roots to have its three-dimensional structure determined. In the following discussion, we focus on what we call the core group of cyclotides, i.e., members of the Möbius and bracelet subfamilies, without the structurally related trypsin inhibitor MCoTI-II from M. cochinchinensis (PDB IDs 1ha9 and 1ib9).
The overall structure of the backbone folded back onto itself and surrounding a tightly packed cystine core is very similar in all five cyclotides. In kalata B1, loop 3 contains only four residues, three of which are involved in a type II turn (Rosengren et al., 2003). In the three bracelet cyclotides, however, loop 3 comprises between six and seven residues, which are folded into a short helical segment. Approximately 40% of models in the structure family of circulin A show α-helical geometry in parts of loop 3 (Daly et al., 1999). The structures are slightly disordered in this region, which may reflect conversion between α-helical and 310-helical forms. Indeed, in both cycloviolacin O1 (Rosengren et al., 2003) and vhr1, the respective segment adopts a 310-helical conformation.
All cyclotides contain a highly conserved Pro residue in loop 6 and, within each subfamily, another highly conserved Pro residue in loop 2 (bracelet subfamily) and loop 3 (Möbius subfamily), respectively. In all solution structures determined to date, these Pro residues are in a trans-conformation. An additional absolutely conserved Pro residue can be found in loop 5 of Möbius cyclotides. In kalata B1, this Pro residue was found to be in a cis-conformation (Saether et al., 1995), resulting in a conceptual twist and giving the stylized circular peptide backbone the topology of a Möbius strip, which ultimately gave this subfamily its name.
In all five cyclotides the cystine knot core shows very similar geometry, underlining the high level of conservation in this motif and the structural similarity between members of the Möbius and bracelet subfamilies. Even in palicourein, a rather unusual cyclotide because of its size and amino acid sequence, the cystine knot region superimposes over that of the other cyclotides with an RMSD of no more than 0.91 Å (Barry et al., 2004). It has been suggested that a small residue in position 2 is necessary to enable the conserved cystine/backbone ring to be formed (Rosengren et al., 2003). In kalata B1, G2 adopts a positive Φ angle, whereas both G2 in circulin A and A2 in cycloviolacin O1 and vhr1 show a negative Φ angle.
Because the core of the cyclotides is occupied by the disulfide bonds, the side chains of all other residues, including hydrophobic ones which would normally reside in the center of peptides, are forced to the outside. This inside-out arrangement leads to a number of cyclotide characteristics, such as extended hydrophobic surface patches, late elution time on RP HPLC, and, in some cases, weak self association (Nourse et al., 2004).
As can be seen in Table 1, the amino acid sequence of vhr1 is closely related to that of cycloviolacin O1, a cyclotide isolated from the aerial parts of V. odorata (Craik et al., 1999). Comparing the three-dimensional structures of these two cyclotides, it becomes apparent that the two mutations, Y7W in loop 2 and R23K in loop 5, are located on the same face of the molecule, literally stacked on top of each other. The overall fold of the two cyclotides is practically identical: the backbone atoms of the best 20 models for each of them can be superimposed with an RMSD of 0.44 Å. The most notable differences in the backbone dihedral angles occur in loop 5, around residues 21 and 22. Although the ψ angles of S21 in cycloviolacin O1 cluster around 120°, they are split in two families in vhr1, with the majority around −60°. Consequently, the Φ angles of N22 in vhr1 also fall into two families, clustered around −120° and 60°. These differences result in a slight displacement of loop 5 in vhr1 compared with cycloviolacin O1.
The Cystine Knot Motif
The original publication on the solution structure of kalata B1 predicted the disulfide connectivity to be I-IV, II-V, and III-VI (i.e., knotted; Saether et al., 1995), whereas a later publication claimed it to be I-VI, II-V, and III-IV (i.e., ladder-like; Skjeldal et al., 2002). In the disulfide core of kalata B1, the Cys residues are packed so tightly that some Hβ protons of Cys residues belonging to two different disulfide bonds are closer in space than Hβ protons of covalently bound Cys residues, making it impossible to deduce the correct disulfide connectivity solely from NOE data. However, it was shown that once dihedral angle data are taken into account, only a knotted disulfide arrangement agrees with the experimental data (Rosengren et al., 2003). The structure of vhr1 reported here is consistent with this cystine knot arrangement. As far as we are aware, only one naturally occurring tri-disulfide laddered structure within a circular peptide backbone is known, namely that seen in the antimicrobial peptide RTD-1 (Trabi et al., 2001).
The cystine knot motif is found not only in cyclotides and two circular trypsin inhibitors from the seeds of bitter melon, M. cochinchinensis (Hernandez et al., 2000), but is also present in a carboxypeptidase A inhibitor from potato (Solanum tuberosum; Rees and Lipscomb, 1980), squash trypsin inhibitors (Bode et al., 1989; Heitz et al., 1989; Le Nguyen et al., 1990), as well as growth factors (McDonald and Hendrickson, 1993) and miscellaneous inhibitor molecules (Pallaghy et al., 1994). All cystine knot proteins share the general disulfide connectivity pattern of I-IV, II-V, and III-VI, with numbers following the N-to-C chemical direction. However, whereas in growth factor cystine knots Cys(I-IV) penetrates the ring, the threading disulfide bond in inhibitor cystine knots is Cys(III-VI) (Isaacs, 1995), resulting in two topologically and geometrically different families that cannot be superimposed when backbone orientation is taken into consideration. The cyclotides and MCoTIs constitute an additional family of cystine knot proteins, combining the inhibitor cystine knot motif with a circular peptide backbone; the term CCK was proposed for the combination of these two features (Craik et al., 2001). Although the ring size in cystine knot proteins can be as large as 15 residues, all cyclotides discovered to date, including vhr1, contain the smallest ring (i.e., eight residues) that can still be penetrated by the third disulfide bond, taking the van der Waals radii of all atoms involved into account. In cyclotides the exceptional stability demonstrated by cystine knot proteins, especially those with a tight knot, is further enhanced by the circular peptide backbone, which makes degradation by exopeptidases impossible and seems to confer onto the peptides an outstanding stability against endopeptidase digest.
In conclusion, as a result of their exceptional stability and diverse bioactivities, cyclotides represent an exciting opportunity for both pharmaceutical and agrochemical applications, either using the cyclotides' natural activities or lending the CCK framework to functional but otherwise less stable proteins. In this study we have demonstrated that in various Viola species (Violaceae), some cyclotides are expressed in a plant part–dependent manner, increasing the overall number of exploitable cyclotides and offering the opportunity to target different pathogens.
METHODS
Plant material was collected at the University of Queensland (Viola hederacea) or at Christchurch Botanic Gardens (Viola cunninghamii), or bought at various nurseries (Viola odorata, Viola canadensis, Viola tricolor, and Viola wittrockiana). Cyclotides were extracted according to a procedure developed in our laboratories (Craik et al., 1999) and purified by RP HPLC on an Agilent 1100 series system (Palo Alto, CA) with degasser, quad pump, variable wavelength detector, and Phenomenex Jupiter column (Torrance, CA; 250 × 22 mm, 5 μm, 300 Å). Peptides were eluted with 0.05% trifluoroacetic acid (TFA) in water (v/v) and 90% acetonitrile and 0.05% TFA in water (v/v) at a flow rate of 8 mL/min and a gradient of 1% per minute. LC-MS studies were performed on a Hewlett-Packard HPLC system with Phenomenex Jupiter column (150 × 2 mm, 5 μm, equipped with a SecurityGuard guard column; Phenomenex) run at 200 μL per minute. Because TFA interferes with ionization of the peptides in the ion source and therefore decreases the sensitivity of the mass spectrometric analysis, the solvents used for LC-MS were 0.05% formic acid in water (v/v; solvent A) and 90% acetonitrile and 0.05% formic acid in water (v/v; solvent B). Peptides were eluted with a 2%/min gradient from 0% to 80% B. The eluent was fed directly (i.e., without a split system) into the ionspray source of a Micromass time of flight mass spectrometer controlled by a PC running MassLynx version 3.5 (Micromass, Manchester, UK). Mass spectra were obtained in positive ion mode over a range mass-to-charge ratio 700 to 2000.
Cyclotides are not directly amendable to sequencing by Edman degradation because of their circular peptide backbone and the knotted disulfide pattern. Therefore, the disulfide bonds were reduced and alkylated and the peptide backbone linearized before the amino acid sequence could be determined. For the first step, vhr1 was dissolved in ammonium acetate buffer (0.1 M, pH 7.8) at a concentration of ∼1 mg/mL. Tris (2-carboxyethyl) phosphine hydrochloride solution (0.5 M in water) was added and the reaction mixture incubated at 50°C for 1 h. After adding a 10-fold excess of maleimide solution (0.5 M in water) and incubating at 50°C for another 30 min, the reaction mixture was purified by HPLC and the fractions lyophilized. The reduced and alkylated peptide sample was dissolved in ammonium acetate buffer (0.1 M, pH 7.7) at a concentration of ∼10 mg/mL and incubated with reconstituted endoproteinase GluC solution (10−4 g/L) at 37°C for 1 to 2 h. Progress of the reaction was monitored by mass spectrometry. The cleaved peptide was purified by HPLC and subjected to sequencing by conventional Edman degradation.
Samples for NMR spectroscopy contained ∼1 mM vhr1 in either 70% water/10% D2O/20% acetonitrile-d3 or 80% D2O/20% acetonitrile-d3. Preliminary experiments in water yielded poor spectra. The addition of a low percentage of organic solvent was necessary to achieve higher protein concentrations in solution and therefore improve the quality of the spectra. Two sets of spectra. including one-dimensional, TOCSY (Braunschweiler and Ernst, 1983), NOESY (Kumar et al., 1980), double quantum filtered–COSY (Rance et al., 1983), and triple quantum filtered–COSY (Shaka and Freeman, 1983) spectra, were recorded at 298K and 310K on a Bruker DMX750 spectrometer (Karlsruhe, Germany) with a triple-resonance self-shielded z-gradient 5-mm probe. The spectral width was 12 ppm, and the carrier was positioned on the water resonance. Water suppression in COSY experiments was achieved using selective low-power irradiation of the water resonance during the relaxation delay of 1.3 s. For NOESY and TOCSY experiments, a 3-9-19 WATERGATE (Sklenar et al., 1993) scheme was used, employing gradient pulses of ∼6 G cm−1 either side of a 10 kHz 3-9-19 binomial pulse. TOCSY experiments used the mLEV17 sequence (Bax and Davies, 1985) for isotropic mixing; the spin-lock period was set to 80 ms. NOESY spectra were recorded with mixing times of 150 and 250 ms. To determine the temperature dependence of the HN shifts, chemical shift data were extracted from the TOCSY spectra recorded at 298K and 310K and additional one-dimensional spectra recorded at 280K and 300K.
Distance constraints for structure calculations, derived from cross-peak intensities in NOESY spectra recorded at 310K with a mixing time of 250 ms, were grouped into five categories. Appropriate pseudoatom corrections were applied to nonstereospecifically assigned protons. Dihedral angle constraints were generated after stereospecific assignment of side chain protons or deduced from 3J(HN−Hα) coupling constants. Ambiguities were resolved in an iterative process, analyzing preliminary structures generated with X-PLOR 3.851 (Brünger, 1996). In addition to the distance restraints derived from NOE intensities, 23 dihedral angle and 4 J-coupling restraints were used to calculate a final set of 50 structures in CNS (Brünger et al., 1998). Whenever J-coupling constants could be unambiguously translated to dihedral angles, dihedral angle constraints were used. In four cases where the J-coupling could not be directly related to the dihedral angle, the J-coupling itself was entered as a restraint. After final energy minimization in water, 20 models with the lowest overall energies that had no violations of distance restraints >0.2 Å or dihedral angle restraints >2° were retained for analysis. Structures were visualized using the programs InsightII (Biosym Technologies, San Diego, CA) and MOLMOL (Koradi et al., 1996) and analyzed with PROMOTIF (Hutchinson and Thornton, 1996) and PROCHECK_NMR (Laskowski et al., 1996).
The amino acid sequence of vhr1 has been deposited in SwissProt (VHR1_VIOHE; accession number P83937); the structural coordinates have been deposited in the PDB (PDB ID 1vb8).
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the Australian Research Council (D.J.C.). D.J.C. is an ARC Professional Fellow. We thank Silicon Graphics International for providing advanced computing power, Jason Mulvenna for performing a sequence search for cyclotide sequences in the Arabidopsis genome database, and Mark Wellard for critical comments on the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Manuela Trabi (m.trabi@imb.uq.edu.au) and David J. Craik (d.craik@imb.uq.edu.au).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.021790.
References
- Barry, D.G., Daly, N.L., Bokesch, H.R., Gustafson, K.R., and Craik, D.J. (2004). Solution structure of the cyclotide palicourein. Implications for the development of a pharmaceutical framework. Structure 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Bax, A., and Davies, D.G. (1985). MLEV-17 based two dimensional homonuclear magnetisation transfer spectroscopy. J. Magn. Reson. 65, 355–360. [Google Scholar]
- Bode, W., Greyling, H.J., Huber, R., Otlewski, J., and Wilusz, T. (1989). The refined 2.0 A X-ray crystal structure of the complex formed between bovine beta-trypsin and CMTI-I, a trypsin inhibitor from squash seeds (Cucurbita maxima). Topological similarity of the squash seed inhibitors with the carboxypeptidase A inhibitor from potatoes. FEBS Lett. 242, 285–292. [DOI] [PubMed] [Google Scholar]
- Bokesch, H.R., Pannell, L.K., Cochran, P.K., Sowder, R.C.I., McKee, T.C., and Boyd, M.R. (2001). A novel anti-HIV macrocyclic peptide from Palicourea condensata. J. Nat. Prod. 64, 249–250. [DOI] [PubMed] [Google Scholar]
- Boman, H.G. (1995). Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13, 61–92. [DOI] [PubMed] [Google Scholar]
- Braunschweiler, L., and Ernst, R.R. (1983). Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reson. 65, 521–528. [Google Scholar]
- Broussalis, A.M., Göransson, U., Coussio, J.D., Ferraro, G., Martino, V., and Claeson, P. (2001). First cyclotide from Hybanthus (Violaceae). Phytochemistry 58, 47–51. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. (1996). X-PLOR Manual Version 3.851. (New Haven, CT: Yale University Press).
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Claeson, P., Göransson, U., Johansson, S., Luijendijk, T., and Bohlin, L. (1998). Fractionation protocol for the isolation of polypeptides from plant biomass. J. Nat. Prod. 61, 77–81. [DOI] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik, D.J. (2001). Plant cyclotides: Circular, knotted peptide toxins. Toxicon 39, 1809–1813. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Bond, T., and Waine, C. (1999). Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Mulvenna, J., Plan, M.R., and Trabi, M. (2004). Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 5, in press. [DOI] [PubMed]
- Craik, D.J., Daly, N.L., and Waine, C. (2001). The cystine knot motif in toxins and applications for drug design. Toxicon 39, 43–60. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Simonsen, S., and Daly, N.L. (2002). The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 5, 251–260. [PubMed] [Google Scholar]
- Daly, N.L., Koltay, A., Gustafson, K.R., Boyd, M.R., Casas-Finet, J.R., and Craik, D.J. (1999). Solution structure by NMR of Circulin A: A macrocyclic knotted peptide having anti-HIV activity. J. Mol. Biol. 285, 333–345. [DOI] [PubMed] [Google Scholar]
- Felizmenio-Quimio, M.E., Daly, N.L., and Craik, D.J. (2001). Circular proteins in plants: Solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J. Biol. Chem. 276, 22875–22882. [DOI] [PubMed] [Google Scholar]
- Göransson, U., Broussalis, A.M., and Claeson, P. (2003). Expression of Viola cyclotides by liquid chromatography-mass spectrometry and tandem mass spectrometry sequencing of intercysteine loops after introduction of charges and cleavage sites by aminoethylation. Anal. Biochem. 318, 107–117. [DOI] [PubMed] [Google Scholar]
- Göransson, U., and Craik, D.J. (2003). Disulfide mapping of the cyclotide kalata B1: Chemical proof of the cystine knot motif. J. Biol. Chem. 278, 48188–48196. [DOI] [PubMed] [Google Scholar]
- Göransson, U., Luijendijk, T., Johansson, S., Bohlin, L., and Claeson, P. (1999). Seven novel macrocyclic polypeptides from Viola arvensis. J. Nat. Prod. 62, 283–286. [DOI] [PubMed] [Google Scholar]
- Gran, L. (1970). An oxytocic principle found in Oldenlandia affinis DC. An indigenous, congolese drug “Kalata-Kalata” used to accelerate delivery. Medd. Nor. Farm. Selsk. 32, 173–180. [Google Scholar]
- Gran, L. (1973. a). Oxytocic principles of Oldenlandia affinis. Lloydia 36, 174–178. [PubMed] [Google Scholar]
- Gran, L. (1973. b). On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol. Toxicol. (Copenh.) 33, 400–408. [DOI] [PubMed] [Google Scholar]
- Gustafson, K.R., Sowder II, R.C., Henderson, L.E., Parsons, I.C., Kashman, Y., Cardellina II, J.H., McMahon, J.B., Buckheit, R.W., Jr., Pannell, L.K., and Boyd, M.R. (1994). Circulins A and B: Novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 116, 9337–9338. [Google Scholar]
- Gustafson, K.R., Walton, L.K., Sowder II, R.C., Johnson, D.G., Pannell, L.K., Cardellina II, J.H., and Boyd, M.R. (2000). New circulin macrocyclic polypeptides from Chassalia parvifolia. J. Nat. Prod. 63, 176–178. [DOI] [PubMed] [Google Scholar]
- Hallock, Y.F., Sowder II, R.C., Pannell, L.K., Hughes, C.B., Johnson, D.G., Gulakowski, R., Cardellina II, J.H., and Boyd, M.R. (2000). Cycloviolins A-D, anti-HIV macrocyclic peptides from Leonia cymosa. J. Org. Chem. 65, 124–128. [DOI] [PubMed] [Google Scholar]
- Heitz, A., Chiche, L., Le-Nguyen, D., and Castro, B. (1989). 1H 2D NMR and distance geometry study of the folding of Ecballium elaterium trypsin inhibitor, a member of the squash inhibitors family. Biochemistry 28, 2392–2398. [DOI] [PubMed] [Google Scholar]
- Heitz, A., Hernandez, J.-F., Gagnon, J., Hong, T.T., Pham, T.T.C., Nguyen, T.M., Le-Nguyen, D., and Chiche, L. (2001). Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry 40, 7973–7983. [DOI] [PubMed] [Google Scholar]
- Hernandez, J.-F., Gagnon, J., Chiche, L., Nguyen, T.M., Andrieu, J.-P., Heitz, A., Hong, T.T., Pham, T.T.C., and Nguyen, D.L. (2000). Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39, 5722–5730. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.G., and Thornton, J.M. (1996). PROMOTIF – A program to identify and analyze structural motifs in proteins. Protein Sci. 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, N.W. (1995). Cystine knots. Curr. Opin. Struct. Biol. 5, 391–395. [DOI] [PubMed] [Google Scholar]
- Jennings, C., West, J., Waine, C., Craik, D.J., and Anderson, M. (2001). Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 98, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. (1996). MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Ernst, R.R., and Wüthrich, K. (1980). A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95, 1–6. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmann, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. (1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Le Nguyen, D., Heitz, A., Chiche, L., Castro, B., Boigegrain, R.A., Favel, A., and Coletti-Previero, M.A. (1990). Molecular recognition between serine proteases and new bioactive microproteins with a knotted structure. Biochimie 72, 431–435. [DOI] [PubMed] [Google Scholar]
- Lindholm, P., Göransson, U., Johansson, S., Claeson, P., Gullbo, J., Larsson, R., Bohlin, L., and Backlund, A. (2002). Cyclotides: A novel type of cytotoxic agents. Mol. Cancer Ther. 1, 365–369. [PubMed] [Google Scholar]
- McDonald, N.Q., and Hendrickson, W.A. (1993). A structural superfamily of growth factors containing a cystine knot motif. Cell 73, 421–424. [DOI] [PubMed] [Google Scholar]
- Nourse, A., Trabi, M., Daly, N.L., and Craik, D.J. (2004). A comparison of the aggregation behaviour of the plant cyclotides kalata B1 and kalata B2 via analytical ultracentrifugation. J. Biol. Chem. 279, 562–570. [DOI] [PubMed] [Google Scholar]
- Pallaghy, P.K., Nielsen, K.J., Craik, D.J., and Norton, R.S. (1994). A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 3, 1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance, M., Sørensen, O.W., Bodenhausen, G., Wagner, G., Ernst, R.R., and Wüthrich, K. (1983). Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117, 479–485. [DOI] [PubMed] [Google Scholar]
- Rees, D.C., and Lipscomb, W.N. (1980). Structure of the potato inhibitor complex of carboxypeptidase A at 2.5-Å resolution. Proc. Natl. Acad. Sci. USA 77, 4633–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren, K.J., Daly, N.L., Plan, M.R., Waine, C., and Craik, D.J. (2003). Twists, knots, and rings in proteins: structural definition of the cyclotide framework. J. Biol. Chem. 278, 8606–8616. [DOI] [PubMed] [Google Scholar]
- Saether, O., Craik, D.J., Campbell, I.D., Sletten, K., Juul, J., and Norman, D.G. (1995). Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry 34, 4147–4158. [DOI] [PubMed] [Google Scholar]
- Schöpke, T., Hasan Agha, M.I., Kraft, R., Otto, A., and Hiller, K. (1993). Hämolytisch aktive Komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci. Pharm. 61, 145–153. [Google Scholar]
- Shaka, A.J., and Freeman, R. (1983). Simplification of NMR spectra by filtration through multiple-quantum coherence. J. Magn. Reson. 51, 169–173. [Google Scholar]
- Skjeldal, L., Gran, L., Sletten, K., and Volkman, B.F. (2002). Refined structure and metal binding site of the Kalata B1 peptide. Arch. Biochem. Biophys. 399, 142–148. [DOI] [PubMed] [Google Scholar]
- Sklenar, V., Piotto, M., Leppik, R., and Saudek, V. (1993). Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. A 102, 241–245. [Google Scholar]
- Sletten, K., and Gran, L. (1973). Some molecular properties of kalatapeptide B-1. A uterotonic polypeptide isolated from Oldenlandia affinis DC. Medd. Nor. Farm. Selsk. 7–8, 69–82. [Google Scholar]
- Svangård, E., Göransson, U., Hocaoglu, Z., Gullbo, J., Larsson, R., Claeson, P., and Bohlin, L. (2004). Cytotoxic Cyclotides from Viola tricolor. J. Nat. Prod. 67, 144–147. [DOI] [PubMed] [Google Scholar]
- Tam, J.P., Lu, Y.-A., Yang, J.-L., and Chiu, K.-W. (1999). An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 96, 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras, F.R.G., Eggermont, K., Kovaleva, V., Raikhel, N.V., Osborn, R.W., Kester, A., Rees, S.B., Torrekens, S., Van Leuven, F., Vanderleyden, J., Cammue, B.P.A., and Broekaert, W.F. (1995). Small cysteine-rich antifungal proteins from radish: Their role in host defense. Plant Cell 7, 573–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Cammue, B.P.A., and Thevissen, K. (2002). Plant defensins. Planta 216, 193–202. [DOI] [PubMed] [Google Scholar]
- Trabi, M., and Craik, D.J. (2002). Circular proteins – No end in sight. Trends Biochem. Sci. 27, 132–138. [DOI] [PubMed] [Google Scholar]
- Trabi, M., Schirra, H.J., and Craik, D.J. (2001). Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from Rhesus macaque leukocytes. Biochemistry 40, 4211–4221. [DOI] [PubMed] [Google Scholar]
- Witherup, K.M., Bogusky, M.J., Anderson, P.S., Ramjit, H., Ransom, R.W., Wood, T., and Sardana, M. (1994). Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57, 1619–1625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.