ABSTRACT
The prolyl-4-hydroxylase domain (PHD) enzymes are regarded as the molecular oxygen sensors. There is an interplay between oxygen availability and cellular metabolism, which in turn has significant effects on the functionality of innate immune cells, such as macrophages. However, if and how PHD enzymes affect macrophage metabolism are enigmatic. We hypothesized that macrophage metabolism and function can be controlled via manipulation of PHD2. We characterized the metabolic phenotypes of PHD2-deficient RAW cells and primary PHD2 knockout bone marrow-derived macrophages (BMDM). Both showed typical features of anaerobic glycolysis, which were paralleled by increased pyruvate dehydrogenase kinase 1 (PDK1) protein levels and a decreased pyruvate dehydrogenase enzyme activity. Metabolic alterations were associated with an impaired cellular functionality. Inhibition of PDK1 or knockout of hypoxia-inducible factor 1α (HIF-1α) reversed the metabolic phenotype and impaired the functionality of the PHD2-deficient RAW cells and BMDM. Taking these results together, we identified a critical role of PHD2 for a reversible glycolytic reprogramming in macrophages with a direct impact on their function. We suggest that PHD2 serves as an adjustable switch to control macrophage behavior.
KEYWORDS: PDK, prolyl-4-hydroxylase domain, dioxygenases, hypoxia, macrophages
INTRODUCTION
Macrophages are an essential component of innate immunity and are well recognized to play critical roles in inflammation, tumor progression, and tissue repair, for example, after an ischemic insult (1). Under aerobic conditions, the oxidative breakdown of pyruvate within the mitochondria is the prevalent source of energy in most cells. Upon a decrease in oxygen availability, cells shift the metabolism toward anaerobic glycolysis. In line with this, macrophages can use aerobic or anaerobic glycolysis for energy production, depending on the context. There is a growing understanding that macrophage function can be altered by cellular metabolism (2). One of the key factors in switching aerobic to anaerobic metabolism at the transcriptional level is the hypoxia-inducible factor (HIF). HIF comprises two subunits: the constitutively regulated HIFβ subunit and one of three oxygen-regulated HIFα subunits (HIF-1α, HIF-2α, or HIF-3α) (3). The protein stability of HIFα is regulated by the three prolyl-4-hydroxylase domain (PHD) enzymes, PHD1, -2, and -3, which hydroxylate HIFα in an oxygen-dependent manner (for a review, see references 4 and 5). The hydroxylated product is recognized by the pVHL protein, which results in ubiquitination and proteasomal degradation of the α-subunit. In hypoxia, the hydroxylation and degradation are inhibited, and thus HIFα is stabilized, which finally results in HIF-dependent transcriptional activation of a repertoire of target genes. Besides many others, genes encoding glycolytic enzymes and pyruvate dehydrogenase kinase 1 (PDK1) belong to the target genes (6, 7). Both determine the glycolytic cellular program. PHD enzymes are of interest for the ongoing development of small-molecule inhibitors which will allow stimulation of HIF-dependent gene expression in normoxia (8).
PHD1 to -3 have common but also nonredundant functions (9). In the case of innate immunity, the role of PHD3 has been analyzed in detail (10–12). The role of PHD2, however, is less well understood. In particular, if and how PHD2 affects macrophage metabolism have not been described before. We therefore analyzed the consequences related to cell metabolism and function for PHD2 knockouts in bone marrow-derived macrophages (BMDM) isolated from LysMCre+/− × Phd2flox/flox mice (PHD2 conditional knockout [PHD2 cKO] mice) and in the monocyte/macrophage cell line RAW264.
RESULTS
PHD2-deficient macrophages induce a hypoxic gene expression pattern in normoxia, including that of PDK1, a central regulator of pyruvate dehydrogenase (PDH).
BMDM isolated from PHD2flox/flox × LysMCre mice (PHD2 cKO) and RAW cells transfected with a constitutively active short hairpin RNA (shRNA) targeting PHD2 (shPHD2 cells) showed an 80% reduction of PHD2 RNA, with a consequential increase of PHD3 RNA expression, compared to that in wild-type (wt) BMDM and wt RAW cells (Fig. 1A). The compensatory increase of the HIF-1 target PHD3 is in line with other cell/tissue-specific PHD2 knockout mouse models (13). Besides PHD3, other metabolism-related HIF gene targets, such as the Glut-1, PFK1, PDK1, COX4-2, LonP, and BNIP3 genes, were upregulated. The gene expression patterns for the PHD2 cKO and shPHD2 cells resembled the pattern of HIF target genes in wt BMDM and wt RAW cells after incubation under hypoxic conditions. Quantitatively, however, the levels of the HIF target genes were lower in the shPHD2 and PHD2 cKO cells in normoxia than in the respective wt cells in hypoxia, which indicates that the reduction of PHD2 induced a partial HIF response, possibly due to the fact that the other PHDs, i.e., PHD1 and PHD3, were still active. This assumption was further supported by the fact that after hypoxic incubation of shPHD2 and PHD2 cKO cells the RNA levels of the HIF target genes were further increased, to an extent similar to that in the respective wt cells in hypoxia. Levels of cell viability/cell death, as determined by the number of annexin V (AV) single-positive cells, were not different in untreated wt BMDM and wt RAW cells compared to PHD2 cKO and shPHD2 cells, respectively, or after treatment with 1 mM dimethyloxalylglycine (DMOG) (Fig. 1B).
FIG 1.
PHD2 knockdown RAW cells and PHD2 knockout (PHD2 cKO) BMDM display increased PDK1 expression and activity. (A) wt RAW and shPHD2 knockdown cells as well as wt BMDM and PHD2 cKO macrophages were incubated for 24 h at 20% or 1% O2. RNA levels of the indicated genes were analyzed by qRT-PCR. RNA levels in wt RAW cells and wt BMDM were set to 1. Fold changes of the RNA levels for the indicated genes in shPHD2 cells, PHD2 cKO BMDM, or wt cells in hypoxia were determined by comparison to the levels in wt cells in normoxia (n = 3 to 6 independent samples per condition). (B) Annexin V (AV) single-positive cells were analyzed in wt BMDM and PHD2 cKO macrophages, with and without treatment with 1 mM DMOG for 24 h. (C) HIF-1α, HIF-2α, PHD2, and β-actin protein levels in wt RAW and shPHD2 cells as well as wt BMDM and PHD2 cKO macrophages in normoxia (20% O2) or hypoxia (1% O2 for 24 h). (D) Phospho-PDH, total PDH, PDK, and β-actin protein levels in wt RAW and shPHD2 cells as well as wt BMDM and PHD2 cKO macrophages in normoxia (20% O2) or hypoxia (1% O2 for 24 h). (E) PDH activities in normoxia or hypoxia (1% O2 for 24 h) for wt RAW and shPHD2 cells and wt RAW cells treated with 1 mM DMOG for 24 h (n = 6 independent samples per condition). Data are means and SEM. *, P < 0.05.
PHD2 protein levels were likewise decreased in PHD2 cKO and shPHD2 cells (Fig. 1C). Whereas HIF-1α and HIF-2α were detectable in BMDM isolated from wt mice in hypoxia only, PHD2 cKO BMDM revealed high HIF-1α and HIF-2α protein levels in normoxia. In the RAW cells, HIF-2α was not detectable with the antibodies applied. For HIF-1α, a pattern similar to that in the BMDM was observed. Comparable to the HIF target RNA expression levels, HIF-1α and HIF-2α protein levels were further increased after exposure of the PHD2 cKO cells and shPHD2 RAW cells to hypoxia. Taken together, these data demonstrate a biologically relevant reduction of PHD2, with subsequent stabilization of the HIFα proteins and induction of HIF target genes, such as PDK1, in a cell line model and in genetically modified primary macrophages partially mimicking hypoxia. The increased expression of PDK1 was further analyzed at the protein level. PDK1 is a major regulator in central metabolic pathways, including glucose consumption pathways. It acts in part by regulating the activity of the pyruvate dehydrogenase (PDH) by phosphorylation, which results in inactivation of the enzyme. PDH is one part of a mitochondrial multienzyme complex that catalyzes the oxidative decarboxylation of pyruvate and is one of the major enzymes responsible for the regulation of homeostasis of carbohydrate fuels in mammals. The induction of PDK1 in PHD2-deficient cells in normoxia was also detectable at the protein level (Fig. 1D). In line with this, PDH was found to be more phosphorylated in the shPHD2 RAW cells and the PHD2 cKO BMDM than in their wild-type counterparts. In hypoxia, both wt and PHD2-deficient cells showed increased PDK1 protein levels and phosphorylation of PDH. In parallel, we determined the PDH activity in cell extracts isolated from wt cells and PHD2-deficient cells after exposure to normoxic or hypoxic conditions (Fig. 1E). shPHD2 cells exhibited a significantly lower PDH activity in normoxia, which was also observed in wt cells after treatment with the PHD inhibitor DMOG. In hypoxia, a decreased PDH activity was observed in wt and shPHD2 cells, still with significantly lower levels in the knockdown cells. Since PDK1 is a critical regulator of cellular metabolism, we next analyzed the glycolytic capacity of the PHD2-deficient cells.
PHD2-deficient macrophages show a switch to glycolytic metabolism.
The utilization of glycolysis to meet energy demands can be characterized by the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) after stimulation. The basal oxygen consumption rate and maximal respiration after uncoupling of the mitochondria by use of carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) were significantly decreased in the PHD2-deficient cells or after treating wt cells with DMOG, which indicates that as a consequence of inhibiting PHD activity, macrophages shift their source of energy to anaerobic glycolysis (Fig. 2A and B). This was further supported by a significantly increased glycolytic function as determined by the extracellular acidification rate after glucose or oligomycin treatment, which reflects basal glycolysis or the overall glycolytic capacity, respectively (Fig. 2C to F). In line with this, we found increased lactate levels in the supernatants of shPHD2 RAW cells and PHD2 cKO BMDM compared to those in the supernatants of their respective wt cells (Fig. 2G). Lactate levels were likewise increased after DMOG treatment or exposure of the cells to hypoxic conditions. Incubating wt and PHD2 cKO BMDM without glucose at 20% O2 abolished lactate production, demonstrating that the glucose in the culture medium was indeed the major source of lactate production (Fig. 2H).
FIG 2.
Macrophages shift their metabolism toward anaerobic glycolysis as a consequence of a reduction of PHD2 expression. (A) wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells or wt RAW cells treated with 1 mM DMOG for 24 h were tested for their oxygen consumption rate (OCR) after addition of oligomycin, FCCP, and rotenone plus antimycin A (Rot+AA) (n = 6 [RAW cells] or 10 [BMDM] independent samples per condition). (B) Basal respiration and maximum respiration were analyzed in wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells based on the data shown in panel A. The OCR after addition of Rot+AA was subtracted from the OCRs after addition of oligomycin and FCCP to obtain basal respiration and maximum respiration values, respectively. (C and D) The extracellular acidification rate (ECAR) was determined for wt RAW and shPHD2 RAW cells (C) as well as wt BMDM and PHD2 cKO cells (D) or wt RAW cells treated with 1 mM DMOG for 24 h after addition of glucose, oligomycin, and 2-desoxyglucose (2-DG) (n = 7 independent samples per condition). (E and F) Glycolysis and anaerobic glycolytic capacity were analyzed based on the data shown in panels C and D. The ECAR after addition of 2-DG was subtracted from the ECARs after addition of glucose and oligomycin to obtain glycolysis and glycolytic capacity values, respectively. (G) Lactate levels in the supernatants of wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells were determined after incubation of the cells under the indicated conditions (n = 4 independent samples per condition). Data are means and SEM. *, P < 0.05. (H) Lactate levels in the supernatants of wt BMDM and cKO cells were determined after incubation of the cells at 20% O2 or 1% O2, with or without addition of glucose to the cell culture medium. Cells were incubated for 24 h in the respective cell culture medium (n = 4 independent samples per condition).
A fully operative cellular metabolism of macrophages is important to provide the necessary amount of ATP. Aerobic and anaerobic glycolysis pathways differ in the net production of ATP. In line with the less efficient anaerobic glycolysis pathway, levels of ATP were significantly diminished in the PHD2 cKO BMDM and the shPHD2 RAW cells as well as after treating wt RAW cells with DMOG or exposing them to hypoxic conditions (Fig. 3A), which might affect their functionality, namely, polarization, migration, and phagocytosis. Macrophages display remarkable plasticity and can change their phenotype upon stimulation (14). The most prominent macrophage populations are the M1- and M2-polarized macrophages. To analyze if the metabolic alterations in the PHD2-deficient cells affected macrophage polarization, we analyzed RNA levels for the M1 markers tumor necrosis factor alpha (TNF-α), inducible nitric oxide synthase (iNOS), and macrophage chemoattractant protein 1 (MCP-1) as well as the M2 markers Ym1 and Fizz in wt and PHD2 cKO BMDM. Arginase, which is also an M2 marker, was not detectable in nonstimulated wt BMDM. None of the M1 or M2 markers were changed in the PHD2 cKO BMDM (Fig. 3B). Subsequently, we stimulated the cells with lipopolysaccharide (LPS) and the Th1 cytokine gamma interferon (IFN-γ) or with the Th2 cytokine interleukin-4 (IL-4) to functionally characterize M1 and M2 polarization (Fig. 3C). Successful M1 and M2 polarization after stimulation was verified by increased RNA levels for TNF-α, iNOS, and MCP-1 after treatment with LPS as well as those for Ym1, arginase, and Fizz after treatment with IL-4. Expression levels of the M1 and M2 marker RNAs were not different between wild-type and PHD2 cKO BMDM. A lack of macrophage polarization in the PHD2 cKO BMDM was additionally confirmed by a nonbiased RNA microarray gene expression assay (see Tables S1 and S2 in the supplemental material). In total, 42 genes were found to be upregulated significantly in PHD2 cKO BMDM, including the PHD3, BNIP3, PFK1, and PDK1 genes, and 56 were found to be downregulated significantly. Pathway analysis of the RNAs identified to be up- or downregulated in the PHD2 cKO BMDM versus wt BMDM did not reveal any indications of macrophage polarization. Likewise, treatment of wt or PHD2 cKO BMDM with DMOG resulted in altered RNA expression but no clear pattern of M1- or M2-associated genes (Fig. 3D). Whereas DMOG resulted in significantly decreased RNA levels for TNF-α and MCP-1, those for iNOS, Fizz, and arginase were significantly increased in wt as well as cKO BMDM. Taken collectively, the metabolic alterations in response to DMOG treatment resulted in the downregulation of RNAs for some M1-associated genes; however, no stringent impact on the polarization of macrophages was observed in cKO BMDM.
FIG 3.
Decreased ATP levels and unaltered polarization in PHD2-deficient macrophages. (A) (Left and middle) wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells were incubated at 20% O2 or 1% O2 for 24 h, and subsequently, intracellular ATP levels were determined (n = 6 independent samples per condition). (Right) wt RAW cells were incubated with 1 mM DMOG for the indicated times, and ATP levels were then determined (n = 6 or 7 independent samples per condition). (B and C) RNA levels for M1 and M2 markers in resting wt BMDM and cKO cells (B) or after stimulation with IL-4 (20 nM) or LPS (100 ng/ml) and IFN-γ (20 nM) for 24 h (C). Numbers of copies of the respective RNAs were quantified relative to the number of RNA copies of the housekeeping gene mS12. (D) RNA levels for M1 and M2 markers in resting wt BMDM and PHD2 cKO cells were analyzed after treatment of the cells with 1 mM DMOG for 24 h. Fold changes of the RNA levels for the indicated genes in DMOG-treated wt or cKO BMDM were determined by comparison to the levels in the nontreated cells (n = 3 to 6 independent samples per condition). Data are means and SEM. *, P < 0.05.
Impaired migratory and phagocytic capacities of PHD2 cKO BMDM and shPHD2 RAW cells.
Whereas most cells in the body are fixed in place, macrophages are motile and able to migrate into surrounding tissues, where one of their major tasks is phagocytosis of invading pathogens or cell debris. We analyzed the migration capacities of PHD2 cKO BMDM and shPHD2 RAW cells by confronting the macrophages with conditioned supernatant from MDA-MB231 breast carcinoma cells in a Boyden chamber and in single-cell migration experiments. Significantly fewer PHD2 cKO BMDM and shPHD2 RAW cells than their respective wt cells migrated in the Boyden chamber (Fig. 4A). Additionally, the accumulated distances in single-cell migration experiments were found to be reduced (Fig. 4B). The impaired migration capacity of the PHD2-deficient RAW cells could be mimicked by treating wt RAW cells with the PHD inhibitor DMOG or exposing them to hypoxic conditions (Fig. 4C). In line with the migration deficit, the phagocytosis capacity was disturbed in the shPHD2 RAW cells and the PHD2 cKO BMDM (Fig. 4D). Comparable to the migration capacity, the decreased phagocytosis could be mimicked in wt RAW cells by incubating the cells under hypoxic conditions or by treatment with DMOG.
FIG 4.
A reduction of PHD2 expression in RAW cells or BMDM results in defects in macrophage migration and phagocytosis. (A) wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells were tested for their migration capacity in Boyden chambers by use of FCS or conditioned medium from MDA-MB231 cells as a stimulant (n = 4 independent samples). (B) The accumulated migration distances over 6 h for wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells were tested in single-cell migration experiments using FCS or conditioned medium from MDA-MB231 cells as a stimulant (n = 59 to 64 cells per condition [RAW cells] or 51 to 71 cells per condition [BMDM]). (C) wt RAW cells were incubated at 20% O2, with or without 1 mM DMOG, or at 1% O2 for 6 h. The accumulated migration distance was tested in single-cell migration experiments using FCS or conditioned medium from MDA-MB231 cells as a stimulant. For cells analyzed at 1% O2, the hypoxic conditions were kept during the single-cell migration experiments, without reoxygenation (n = 50 to 53 cells per condition). (D) wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells were incubated at 20% O2, with or without 1 mM DMOG, or at 1% O2 for 20 h in total. Subsequently, the capacity of the cells to phagocytose labeled beads was analyzed. Fluorescent beads were added to the cells for 4 h without reoxygenation. Data are means and SEM (n = 5 independent samples per condition). *, P < 0.05. (E) RNA levels for chemokine receptors in resting wt BMDM and PHD2 cKO cells after incubation under normoxic or hypoxic (1% O2) conditions for 24 h. Fold changes of the RNA levels for the indicated genes in the PHD2 cKO BMDM were determined by comparison to the levels in the wt cells in normoxia (n = 3 independent samples per condition). Data are means and SEM. *, P < 0.05.
Differential migration was not due to modulated chemokine receptor expression, since quantifying CCR2, CCR4, CCR5, CCR7, and CXCR4 RNA levels by reverse transcription-PCR (RT-PCR) after exposure of wild-type and PHD2 cKO BMDM to 20% O2 or 1% O2 did not reveal any difference between the cell types besides an upregulation of CCR1 in the PHD2 cKO cells (Fig. 4E). However, we observed a significant upregulation of CCR1 and CCR5 and a slight upregulation of CXCR4 in hypoxia, which is in line with the literature (15, 16). Since the migration deficit was not due to a striking differential expression of chemokine receptors, we further analyzed the impacts of the metabolic alterations on the migratory and phagocytosis capacities.
Regulation of PDH activity by PDK is critical for macrophage function.
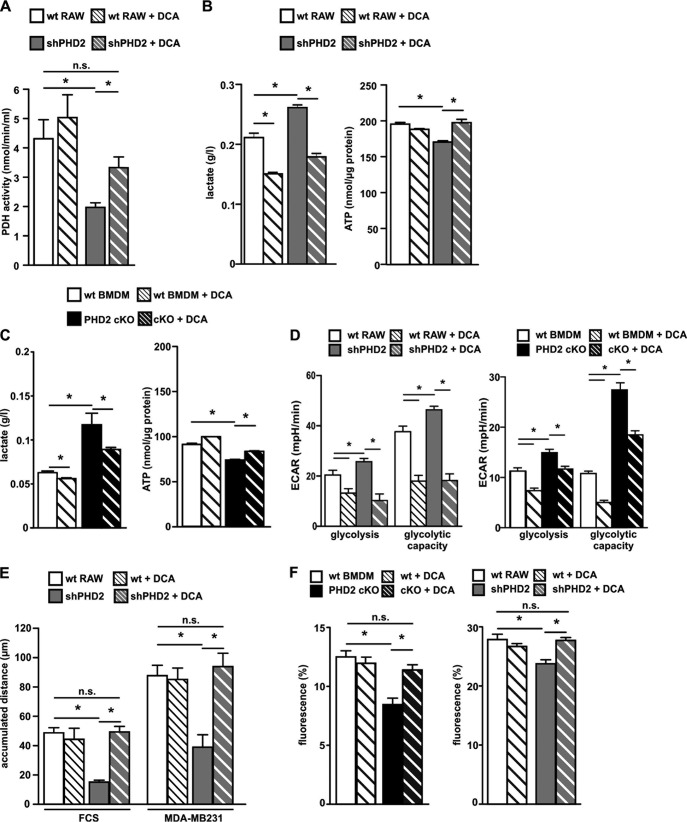
To obtain insight into the importance of altered PDK1 levels and PDH activity for the functional impairment of the PHD2-deficient macrophages, we treated cells with the PDK inhibitor dichloroacetate (DCA). DCA binds to PDK1 near the helix bundle in the N-terminal part of the protein. Bound DCA promotes local conformational changes that are communicated to both nucleotide-binding and lipoyl-binding pockets of PDK1, leading to the inactivation of kinase activity (17). DCA treatment reestablished the decreased PDH activity in the shPHD2 RAW cells (Fig. 5A). In line with this, shPHD2 cells and PHD2 cKO BMDM produced less lactate and demonstrated significantly increased ATP levels after DCA treatment (Fig. 5B and C). Reversal of the metabolic reprogramming after DCA treatment was also observed by analyzing the ECAR. Whereas the nontreated shPHD2 cells and PHD2 cKO BMDM had significantly increased glycolysis and glycolytic capacity, these were significantly decreased in the DCA-treated cells (Fig. 5D). Finally, we tested if DCA treatment was able to normalize the migration and phagocytosis deficits (Fig. 5E and F). Most interestingly, DCA treatment was indeed able to increase the impaired migration and phagocytosis capacities of the shPHD2 RAW and PHD2 cKO cells, to levels comparable to those of the wild-type cells, strongly indicating that the metabolic programming via PDK1 per se was responsible for the functional alterations observed as a consequence of downregulating PHD2 expression.
FIG 5.
Inhibition of PDK1 by dichloroacetate (DCA) reverses the metabolic phenotype and the migration defect in PHD2-deficient macrophages. (A) PDH activity was determined in lysates of wt RAW and shPHD2 RAW cells after incubation of the cells with or without 5 mM DCA for 24 h (n = 6 independent samples per condition). (B and C) Lactate levels in the supernatants and intracellular ATP levels of wt RAW and shPHD2 RAW cells (B) as well as wt BMDM and cKO macrophages (C) after incubation of the cells with or without 5 mM DCA for 24 h. (D) Glycolysis and glycolytic capacity were analyzed in wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells after incubating the cells with or without 5 mM DCA for 24 h (n = 6 to 10 independent samples per condition). (E) Accumulated migration distances over 6 h for wt RAW and shPHD2 RAW cells after incubation of the cells with or without 5 mM DCA for 24 h were determined in single-cell migration experiments using FCS or conditioned medium from MDA-MB231 cells as a stimulant (n = at least 20 cells per condition). (F) wt RAW cells, shPHD2 RAW cells, wt BMDM, and PHD2 cKO cells were incubated with or without 5 mM DCA for 24 h. Subsequently, the capacity of the cells to phagocytose labeled beads was analyzed. Data are means and SEM (n = 5 to 8 independent samples per condition). *, P < 0.05; n.s., not significant.
PDK1 has been described as a HIF target gene (7). In a previous report, we demonstrated that HIF-1α in PHD3 cKO BMDM is not stabilized in normoxia (12), which is in sharp contrast to the effect seen in the PHD2 cKO BMDM described here. This was further supported by the fact that HIF target genes, including the PDK1 gene, were not significantly increased in PHD3 cKO BMDM, in contrast to PHD2 cKO and wt BMDM in normoxia and hypoxia, respectively (Fig. 6A). The genes identified in the gene array analysis (PHD2 cKO versus wt BMDM) were additionally compared to PHD3-regulated genes described earlier (12). Most interestingly, just 13 genes were found to be regulated in the PHD2 cKO as well as the PHD3 cKO BMDM, indicating PHD isoform-specific effects (Table S3). In line with this, ATP and lactate levels as well as the accumulated migration distance were not altered in PHD3 cKO versus wt BMDM (Fig. 6B). To further analyze if PHD2 mediates its effects via HIF-1α, we generated double cKO (dcKO) mice (PHD2flox/flox × HIF-1αflox/flox × LysMcre+/− mice). dcKO BMDM had a blunted response in RNA levels for the HIF target genes encoding PHD3, Glut-1, PFK1, PDK1, Cox4.2, and LonP in hypoxia (Fig. 7A). The metabolic phenotypes of the PHD2 cKO BMDM, i.e., decreased ATP levels, increased lactate levels, decreased OCR, and decreased migration, were rescued as a consequence of the HIF-1α knockout, demonstrating that HIF-1 and PDK1 are the two main mediators of the PHD2-induced metabolic alterations (Fig. 7B to D).
FIG 6.
No metabolic phenotype in PHD3-deficient macrophages. (A) wt BMDM and PHD3 cKO macrophages were incubated for 24 h at 20% or 1% O2. RNA levels for the indicated genes were analyzed by qRT-PCR. RNA levels in wt BMDM were set to 1. Fold changes of the RNA levels for the indicated genes in PHD3 cKO BMDM or wt cells in hypoxia were determined by comparison to the levels in the wt cells in normoxia (n = 3 independent samples per condition). (B) ATP (n = 5 independent samples) and lactate (n = 4 independent samples) levels as well as accumulated migration distances were determined for wt BMDM and PHD3 cKO macrophages.
FIG 7.
HIF-1α mediates the metabolic alterations in PHD2-deficient macrophages. (A) wt BMDM and dcKO macrophages were incubated for 24 h at 20% or 1% O2. RNA levels for the indicated genes were analyzed by qRT-PCR. RNA levels in wt BMDM were set to 1. Fold changes of the RNA levels for the indicated genes in dcKO BMDM or wt cells in hypoxia were determined by comparison to the levels in the wt cells in normoxia (n = 3 independent samples per condition). Data are means and SEM. *, P < 0.05 compared to wt cells at 20% O2; #, P < 0.05 compared to wt cells at 1% O2. (B) Intracellular ATP levels were determined for wt BMDM, PHD2 cKO cells, and dcKO cells after incubation for 24 h in 20% O2 or 1% O2. (C) OCRs and lactate levels in the supernatants of wt BMDM, PHD2 cKO cells, and dcKO cells. (D) The accumulated migration distances over 6 h for wt BMDM, PHD2 cKO cells, and dcKO cells were determined in single-cell migration experiments using FCS or conditioned medium from MDA-MB231 cells as a stimulant (n = at least 20 cells per condition). Data are means and SEM. *, P < 0.05.
DISCUSSION
Macrophages are critical effector cells for innate immunity but also for adaptive immune function, by recruiting further cells to the inflamed tissue (18). To fulfill their functions, macrophages need to migrate efficiently into the affected tissue and phagocytose invading pathogens or cell debris. Both characteristic features were severely impaired in the PHD2-deficient RAW cell model as well as the primary BMDM used in our study. Impaired migration is in line with two previous studies analyzing the migratory capacity of PHD2-deficient peritoneal macrophages as well as shPHD2 RAW cells toward MCP-1 as a stimulus (19, 20). Infiltrating inflammatory cells, including macrophages, play an important role in tissue remodeling after an insult. In line with this, LysMCre PHD2flox/flox animals showed less macrophage infiltration in the aorta during hypertensive cardiovascular remodeling (19). This was associated with protection from hypertension-induced left ventricular hypertrophy and reduced ejection fraction. The basis for impaired macrophage migration, however, has not been analyzed in more depth so far.
The functionality of macrophages is significantly affected by their polarization as well as their metabolic phenotype. Macrophages can generally be classified into two major groups, i.e., M1 (classically activated) and M2 (alternatively activated) macrophages (21). Both subgroups have specific functions in the inflammatory clearance of pathogens and in tissue repair (22). HIF-1α and HIF-2α are known to affect macrophage polarization, with predominant roles of HIF-1α for M1 macrophages and HIF-2α for M2 macrophages (23). Since the PHD enzymes regulate HIFα stability, it is tempting to speculate that inhibition of their enzymatic activity will also affect macrophage polarization. Characterization of resting as well as stimulated wt and PHD2 cKO macrophages, however, did not reveal any striking pattern which would indicate a clear polarization. It is important that this observation does not contrast with a study by Takeda et al. in which M2 polarization as a consequence of PHD2 knockout was described (24). They observed M2 polarization in heterozygous but not homozygous PHD2-deficient macrophages, which matches our observations with homozygous PHD2-deficient macrophages.
In stark contrast to the unaltered polarization, we found an influence of PHD2 on reprogramming of mitochondrial metabolism. This effect was mediated via PHD2-dependent regulation of PDK1 expression. This is in accordance with a recent report showing that PHD2 can regulate PDK1 in livers/hepatocytes (25). In line with higher levels of PDK1 and more phosphorylation of its target, PDH, the basal and stimulated oxygen consumption rates were significantly lower in the PHD2-deficient cells, accompanied by an increased glycolytic capacity. PDK1 is a key regulatory enzyme in glucose metabolism. The PDH complex, which is regulated by PDK1, converts pyruvate produced from the glycolytic flux to acetyl coenzyme A (acetyl-CoA). Pyruvate-derived acetyl-CoA then enters the tricarboxylic acid (TCA) cycle, which generates the NADH that fuels the electron transport chain for oxidative phosphorylation. In hypoxia, PDH activity is inhibited via PDK1-mediated phosphorylation, which induces anaerobic glucose metabolic homeostasis under conditions of limited oxygen availability. This mechanism is widely used by tumor cells and is part of the so-called Warburg effect. Warburg described that unlike most normal tissues, cancer cells tend to “ferment” glucose into lactate even in the presence of sufficient oxygen to support mitochondrial oxidative phosphorylation (26). In the case of immune cells, however, the metabolic adaptation is part of their physiological response. Innate immune cells, such as neutrophils, likewise depend on anaerobic glycolysis for ATP production, which is also suggested by the fact that they harbor only a few mitochondria (27). In contrast, macrophages have numbers of mitochondria comparable to those in other body cells, and thus have a higher metabolic flexibility, which allows a quick metabolic switch from aerobic to anaerobic glycolysis. The hallmarks of anaerobic glycolysis are reduced ATP production, increased lactate levels, and decreased oxygen consumption (28). All three features were significantly altered in the PHD2-deficient macrophages, indicating an anaerobic metabolic shift. Neutrophils and monocytes/macrophages fulfill their physiological function in severely hypoxic areas, such as those with inflammation or ischemia. Unlike short-lived neutrophils, macrophages survive longer in the body, up to a maximum of several months. Compared to that in the oxygenated blood, the partial O2 pressure (pO2) in most tissues is significantly lower. Thus, compared to monocytes, macrophages need to be able to adapt to the hypoxic conditions which reflect their physiological environment. In contrast, short-lived neutrophils enter inflamed hypoxic tissue, where they die quickly to fulfill their function. This is also reflected by the fact that in contrast to other cells, they die upon exposure to hypoxia. Most interestingly, the metabolic phenotypes observed for the shPHD2 RAW cells and cKO BMDM mimicked the effects seen in wt cells in hypoxia. The metabolic adaptations thus might ensure function and viability of the cells as long as possible under hypoxic conditions. A lower migration rate as a consequence helps to keep the cells in place until they are stimulated during the course of acute inflammation.
Recent evidence suggests an intricate link between metabolism and macrophage activation (29). Thus, it becomes important to know what kind of metabolic changes occur after immune cell activation and if the altered metabolism per se can serve as a controller of the immunomodulatory functionality. Analyzing metabolic aspects as a consequence of blocking PHD2 thus might answer the question of what changes in the regulation of energy metabolism are necessary for macrophages and if these can be targeted to control innate immune function. In this regard, our data clearly indicate that deleting PHD2 in macrophages is sufficient to drive an anaerobic glycolytic phenotype in normoxia and interferes with migration and phagocytosis. PHD enzyme activity can be inhibited by use of competitive oxoglutarate analogues. Respective inhibitors are being developed to specifically interfere with the PHD/HIF signaling pathway (30). Short-term treatment of wt macrophages with the PHD inhibitor DMOG or exposure of the cells to hypoxia was indeed sufficient to mimic the metabolic features of the genetically modified macrophages. Moreover, the metabolic switch in the PHD2-deficient macrophages was readily reversible by inhibition of PDK1, demonstrating that PHD2-mediated metabolic changes are not decretory. DCA treatment rescued all hallmarks of anaerobic glycolysis in the PHD2-deficient cells, including the impaired functionality. A critical role of PDK1 in macrophage metabolism and function has been described earlier and is in line with our findings (31). Via alteration of PDK1 activity, the cellular ATP levels as well as extracellular lactate levels are modulated. ATP as well as a lactate-enriched environment has been demonstrated to add to immunomodulatory functions by altering the migratory activity of defense cells (32, 33). The use of PDK1 as a molecule to target deregulated energy metabolism is an emerging strategy for cancer therapy (34). Redirection of glucose metabolism from glycolysis to oxidation, which reverses the Warburg effect, leads to inhibition of proliferation and induction of caspase-mediated apoptosis in tumor cells. Thus far, DCA is the most extensively studied PDK1 inhibitor; however, it has limited use for therapeutic purposes because of its low potency and high toxicity. PHD inhibitors, on the other hand, have entered preclinical models and clinical trials, so interfering with PHD2 activity might serve as a better strategy to influence immune functions via metabolic reprogramming (35).
Taken collectively, the data in our study show that while PHD2 is not required for macrophage polarization, it controls macrophage metabolism and function. Mechanistically, the balance between aerobic and anaerobic glycolysis is affected by PHD2 via the expression and activity of PDK1. This adds to our understanding of the functionality of macrophages in normoxia and hypoxia. In addition, our findings might point to the possibility to specifically interfere with the inflammatory function of macrophages by inhibiting PHD2 activity.
MATERIALS AND METHODS
Chemicals.
The oxoglutarate analogue dimethyloxalylglycine (DMOG; Enzo Life Sciences, Lörrach, Germany) and the PDK inhibitor dichloroacetate (DCA; Sigma) were used at final concentrations of 1 mM and 5 mM, respectively. LPS (Enzo Life Sciences, Lörrach, Germany), IFN-γ, and IL-4 (Peprotech, Hamburg, Germany) were applied at concentrations of 100 ng/ml, 20 nM, and 20 nM, respectively, or as indicated.
Myeloid lineage-specific conditional knockout mice.
All animals in this study were backcrossed to C57BL/6 mice at least five times. Phd2flox/flox × LysMcre+/− mice were crossed with Phd2flox/flox mice to obtain PHD2 cKO mice (Phd2flox/flox × LysMcre+/−) and littermate control wild-type mice (Phd2flox/flox). Generation of Phd3flox/flox × LysMcre+/− (PHD3 cKO) mice was described previously (12). To obtain PHD2/HIF-1α double-knockout macrophages, Phd2flox/flox × LysMcre+/− mice were crossed with Hif-1αflox/flox mice (B6.129-Hif1atm3Rsjo/J; Jackson Laboratories) to obtain PHD2flox/flox × HIF-1αflox/flox × LysMcre+/− mice (dcKO mice). All animal work conformed to institutional guidelines and was approved by the Niedersächsische Landesamt für Verbraucherschutz und Lebensmittelsicherheit (approval number 33.9-42502-04-11/0413).
Isolation and differentiation of BMDM.
Bone marrow cells were isolated from the femur. After 24 h of cell culture, nonadherent monocytes were harvested and seeded in Pluznik's medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 0.2 mM l-glutamine, 0.1 mM sodium pyruvate, 50 U/ml penicillin G, 50 μg/ml streptomycin, 10% heat-inactivated fetal calf serum [FCS], 5% heat-inactivated horse serum [Pan Biotech, Aidenbach, Germany], 0.05% 1:1,000-diluted β-mercaptoethanol [Carl Roth GmbH, Karlsruhe, Germany], and 15% L929 cell-conditioned medium [36]) and then allowed to differentiate for 7 days. Adherent bone marrow-derived macrophages (BMDM) were detached with 3.5 ml Accutase (PAA Laboratories, Cölbe, Germany) and resuspended in culture medium (DMEM supplemented with 0.2 mM l-glutamine, 0.1 mM sodium pyruvate, 1 mM HEPES, 50 U/ml penicillin G, 50 μg/ml streptomycin, and 10% heat-inactivated fetal calf serum). For analysis of lactate production without addition of glucose, glucose-free medium (Pan Biotech) was used as indicated. Successful differentiation of the BMDM was controlled by fluorescence-activated cell sorter (FACS) analysis of the macrophage marker F4/80.
Generation of shPHD2 RAW cells.
The mouse macrophage cell line RAW 264.7 was infected with a lentivirus encoding an shRNA targeting mPHD2 (5′-ATTCGAAGAATACCTCCAC-3′) and cotransfected with enhanced green fluorescent protein (EGFP) as described earlier (37). Cells were used as a pool of sorted EGFP+ cells, and knockdown efficiency was tested via quantitative PCR (qPCR).
Preparation of MDA-MB231 cell-conditioned medium.
Five flasks each with 1 × 106 MDA-MB231 cells were cultivated in 15 ml culture medium for 4 days. The medium was transferred to a reaction tube and centrifuged at 4,000 × g for 20 min at 4°C. The supernatant was pooled and frozen at −80°C.
Hypoxic incubation.
For cell culture under defined hypoxic conditions (1% O2), an in vivo hypoxia workstation was used (Ruskinn Technologies, Bridgend, South Wales, United Kingdom).
Phagocytosis.
Fluorescent beads (Protonex Red 600 latex beads; AAT Bioquest) were administered to cells for 4 h. After washing, uptake of fluorescent beads was analyzed by FACS analysis (BD FACSCanto II flow cytometer; BD Biosciences).
Quantification of apoptotic cells.
Supernatant and detached cells were collected. Samples were centrifuged and washed in phosphate-buffered saline (PBS). Subsequently, cells were stained for 30 min at 4°C with Pacific Blue annexin V (70 μg/ml; diluted 1:150) (Biolegend, San Diego, CA) in the dark. Samples were washed with annexin V binding buffer (Biolegend) and analyzed by FACS analysis (BD FACSCanto II; BD Biosciences).
Single-cell migration.
RAW cells (2.5 × 104) or BMDM (5 × 104) were seeded into a 6-well plate. The next day, the medium was replaced with either normal medium or MDA-MB231 cell-conditioned medium. The migration of the cells was investigated using a T1-5M Nikon microscope inside a Sci-tive workstation (Ruskinn Technologies, Bridgend, South Wales, United Kingdom) under normoxic (20% O2) or hypoxic (1% O2) conditions. An image was taken every 10 min for a total of 6 h. Migration was analyzed using the chemotaxis plug-in installed in ImageJ.
Boyden chamber assay.
BMDM or RAW cells (0.7 × 105) were seeded in 500 μl culture medium into inserts (BD Biosciences, Heidelberg, Germany) containing 3-μm pores. The inserts were placed in 24-well plates containing either 500 μl MDA-MB231 cell-conditioned medium or 500 μl cell culture medium (as a control) per well. Eighteen hours later, the inserts were placed in medium with 5 μM calcein. Cells that had not migrated were removed by scraping of the upper side of the insert after 1 h, while migrated macrophages on the lower side were analyzed by fluorescence microscopy (Axio Observer D1 microscope; Carl Zeiss, Göttingen, Germany).
RNA isolation and qRT-PCR.
Cells were washed once with PBS and harvested in TRIzol (Invitrogen, Darmstadt, Germany). RNA was isolated according to the manufacturer's instructions, and 1 μg was transcribed using a First Strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany). Transcript levels were analyzed by quantitative reverse transcription-PCR (qRT-PCR) by amplifying 1 μl of cDNA with Brilliant II SYBR green qPCR master mix in an MX3005Pro light cycler (Agilent, Böblingen, Germany). Applied primer sequences were as follows: ms12 for, 5′-GAAGCTGCCAAGGCCTTAGA-3′; ms12 rev, 5′-AACTGCAACCAACCACCTTC-3′; phd2 for, 5′-TTGCTGACATTGAACCCAAA-3′; phd2 rev, 5′-GGCAACTGAGAGGCTGTAGG-3′; phd3 for, 5′-GGCCGCTGTATCACCTGTAT-3′; phd3 rev, 5′-TTCTGCCCTTTCTTCAGCAT-3′; glut1 for, 5′-TGGCCTTGCTGGAACGGCTG-3′; glut1 rev, 5′-TCCTTGGGCTGCAGGGAGCA-3′; pfk1 for, 5′-ACGAGGCCATCCAGCTCCGT-3′; pfk1 rev, 5′-TGGGGCTTGGGCAGTGTCCT-3′; pdk1 for, 5′-TTCACGTCACGCTGGGCGAG-3′; pdk1 rev, 5′-GGCTGGGCACACACCAGTCG-3′; cox4.2 for, 5′-CAGAGAAGGTGGCCTTGTACC-3′; cox4.2 rev, 5′-AGAAGAAGACGCAGCCCATC-3′; LonP for, 5′-CATCGCCTTGAACCCTCTGT-3′; LonP rev, 5′-AGCCGCTTAAGGATGTTGGT-3′; BNIP3 for, 5′-GTCCAGTGTCGCCTGGCCTC-3′; BNIP3 rev, 5′-TGGGAGCGAGGTGGGCTGTC-3′; mCCR2 for, 5′-CCACACCCTGTTTCGCTG-3′; mCCR2 rev, 5′-ACCTCTTCAGCACTTGC-3′; mCCR4 for, 5′-GCCTCTTGTTCAGCACTTGC-3′; mCCR4 rev, 5′-ATAAGCAGCCCCAGGACG-3′; mCCR5 for, 5′-CCAGAGGAGGTGAGACATCCGTTC-3′; mCCR5 rev, 5′-GGCAGGAGCTGAGCCGCAATTT-3′; mCCR7 for, 5′-ATGGACCCAGGGAAACCCAGGAA-3′; mCCR7 rev, 5′-GCACACCGACTCGTACAGGG-3′; mCXCR4 for, 5′-GCTCCGGTAACCACCACGGC-3′; and mCXCR4 rev, 5′-GCGAGGTACCGGTCCAGGCT-3′.
Microarray-based gene expression.
Microarray-based gene expression was analyzed as described before (12).
Western blotting.
Cells were lysed with 50 mM Tris, 150 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 100 mM MgCl2, and 1% NP-40 supplemented with protease inhibitors (Roche). Protein samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Biosciences). Primary antibodies used were as follows: anti-HIF-1α (NB-100-479; Novus), anti-HIF-2α (AF2997; R&D Systems), anti-PHD2 (NB-100-2219; Novus), anti-phospho-pyruvate dehydrogenase (ABS204; Merck), anti-pyruvate dehydrogenase (3205; Cell Signaling), anti-PDK1 (ADI-KAP-PK112-D; Stressgene), and anti-β-actin (A5441; Sigma). For detection of immunocomplexes, horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Santa Cruz Biotechnology) were used, and membranes were incubated with a chemiluminescent HRP substrate (Millipore).
ATP and lactate measurements.
The supernatant of 0.75 × 105 BMDM or RAW cells cultivated for 24 h in a 24-well plate was analyzed for lactate production by use of an l-lactate kit according to the manufacturer's instructions (R-Biopharm, Darmstadt, Germany). For determination of ATP levels, 0.2 × 105 BMDM or RAW cells were seeded. ATP levels were determined using a Cell Titer-Glo ATP kit (Promega, Madison, WI).
PDH activity assay.
For determinations of PDH activity, a MAK183 kit (Sigma, St. Louis, MO) was used.
OCR and ECAR.
The oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) were analyzed in a Seahorse XF96 extracellular flux analyzer (Seahorse Bioscience, Billerica, MA). For OCR analysis, 2.5 × 104 RAW cells or 4.0 × 104 BMDM were seeded per well. The medium was replaced with XF assay medium supplemented with 4.5 g/liter glucose and 1 mM sodium pyruvate and incubated without CO2 for 30 min. After measuring basal respiration, the oxygen consumption was analyzed after sequential addition of 1.5 μM oligomycin, 1 μM FCCP, 2 μM rotenone, and 1 μM antimycin A.
For ECAR determination, 2.5 × 104 RAW cells or 8 × 104 BMDM were seeded per well. Cells were washed with XF Glycostress medium (D5030 DMEM, 134 mM NaCl, 3 mg phenol red, 2 mM l-glutamine, pH 7.35). The cells were incubated for 15 min at 37°C without CO2. ECAR was analyzed after sequential addition of 10 mM glucose, 1.5 to 3 μM oligomycin, and 100 mM 2-deoxyglucose (2-DG).
Statistical analyses.
Statistical analyses were performed using Student's two-tailed t test. Data shown are means ± standard errors of the means (SEM). P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Annette Hillemann for expert technical support.
A.G., A.B., L.S., K.F., S.N., B.W., A.Z., A.J., and M.C. designed and performed research, analyzed data, and wrote the manuscript. J.D. performed research and analyzed data. K.R. analyzed data. P.R., C.X.C.S., and A.M.S. evaluated the data and corrected the paper. D.M.K. designed research and wrote the manuscript. All authors read and edited the manuscript.
We declare that we have no competing financial interests.
This study was supported by research funding from the Deutsche Forschungsgemeinschaft (grant IRTG1816) to A.G. and A.B. P.R. is supported by the SFB1002. M.C., C.X.C.S., and A.M.S. are supported by the British Heart Foundation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00236-16.
REFERENCES
- 1.Wynn TA, Vannella KM. 2016. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goda N, Kanai M. 2012. Hypoxia-inducible factors and their roles in energy metabolism. Int J Hematol 95:457–463. doi: 10.1007/s12185-012-1069-y. [DOI] [PubMed] [Google Scholar]
- 3.Bishop T, Ratcliffe PJ. 2015. HIF hydroxylase pathways in cardiovascular physiology and medicine. Circ Res 117:65–79. doi: 10.1161/CIRCRESAHA.117.305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelin WG., Jr 2005. Proline hydroxylation and gene expression. Annu Rev Biochem 74:115–128. [DOI] [PubMed] [Google Scholar]
- 5.Schofield CJ, Ratcliffe PJ. 2004. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol 5:343–354. [DOI] [PubMed] [Google Scholar]
- 6.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. 2006. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Katschinski DM. 2009. In vivo functions of the prolyl-4-hydroxylase domain oxygen sensors: direct route to the treatment of anaemia and the protection of ischaemic tissues. Acta Physiol (Oxf) 195:407–414. doi: 10.1111/j.1748-1716.2008.01952.x. [DOI] [PubMed] [Google Scholar]
- 9.Myllyharju J. 2013. Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol (Oxf) 208:148–165. doi: 10.1111/apha.12096. [DOI] [PubMed] [Google Scholar]
- 10.Walmsley SR, Chilvers ER, Thompson AA, Vaughan K, Marriott HM, Parker LC, Shaw G, Parmar S, Schneider M, Sabroe I, Dockrell DH, Milo M, Taylor CT, Johnson RS, Pugh CW, Ratcliffe PJ, Maxwell PH, Carmeliet P, Whyte MK. 2011. Prolyl hydroxylase 3 (PHD3) is essential for hypoxic regulation of neutrophilic inflammation in humans and mice. J Clin Invest 121:1053–1063. doi: 10.1172/JCI43273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiss J, Mollenhauer M, Walmsley SR, Kirchberg J, Radhakrishnan P, Niemietz T, Dudda J, Steinert G, Whyte MK, Carmeliet P, Mazzone M, Weitz J, Schneider M. 2012. Loss of the oxygen sensor PHD3 enhances the innate immune response to abdominal sepsis. J Immunol 189:1955–1965. doi: 10.4049/jimmunol.1103471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swain L, Wottawa M, Hillemann A, Beneke A, Odagiri H, Terada K, Endo M, Oike Y, Farhat K, Katschinski DM. 2014. Prolyl-4-hydroxylase domain 3 (PHD3) is a critical terminator for cell survival of macrophages under stress conditions. J Leukoc Biol 96:365–375. doi: 10.1189/jlb.2HI1013-533R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hölscher M, Silter M, Krull S, von Ahlen M, Hesse A, Schwartz P, Wielockx B, Breier G, Katschinski DM, Zieseniss A. 2011. Cardiomyocyte-specific prolyl-4-hydroxylase domain 2 knock out protects from acute myocardial ischemic injury. J Biol Chem 286:11185–11194. doi: 10.1074/jbc.M110.186809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosser DM, Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosco MC, Puppo M, Santangelo C, Anfosso L, Pfeffer U, Fardin P, Battaglia F, Varesio L. 2006. Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol 177:1941–1955. doi: 10.4049/jimmunol.177.3.1941. [DOI] [PubMed] [Google Scholar]
- 16.Dong F, Khalil M, Kiedrowski M, O'Connor C, Petrovic E, Zhou X, Penn MS. 2010. Critical role for leukocyte hypoxia inducible factor-1alpha expression in post-myocardial infarction left ventricular remodeling. Circ Res 106:601–610. doi: 10.1161/CIRCRESAHA.109.208967. [DOI] [PubMed] [Google Scholar]
- 17.Kato M, Li J, Chuang JL, Chuang DT. 2007. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure 15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi C, Pamer EG. 2011. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda J, Ichiki T, Matsuura H, Inoue E, Kishimoto J, Watanabe A, Sankoda C, Kitamoto S, Tokunou T, Takeda K, Fong GH, Sunagawa K. 2013. Deletion of phd2 in myeloid lineage attenuates hypertensive cardiovascular remodeling. J Am Heart Assoc 2:e000178. doi: 10.1161/JAHA.113.000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamlouk S, Kalucka J, Singh RP, Franke K, Muschter A, Langer A, Jakob C, Gassmann M, Baretton GB, Wielockx B. 2014. Loss of prolyl hydroxylase-2 in myeloid cells and T-lymphocytes impairs tumor development. Int J Cancer 134:849–858. doi: 10.1002/ijc.28409. [DOI] [PubMed] [Google Scholar]
- 21.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence T, Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 23.Takeda N, O'Dea EL, Doedens A, Kim JW, Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A, Johnson RS. 2010. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev 24:491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda Y, Costa S, Delamarre E, Roncal C, Leite de Oliveira R, Squadrito ML, Finisguerra V, Deschoemaeker S, Bruyere F, Wenes M, Hamm A, Serneels J, Magat J, Bhattacharyya T, Anisimov A, Jordan BF, Alitalo K, Maxwell P, Gallez B, Zhuang ZW, Saito Y, Simons M, De Palma M, Mazzone M. 2011. Macrophage skewing by Phd2 haplodeficiency prevents ischaemia by inducing arteriogenesis. Nature 479:122–126. doi: 10.1038/nature10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhara T, Hishiki T, Kasahara M, Hayakawa N, Oyaizu T, Nakanishi T, Kubo A, Morisaki H, Kaelin WG Jr, Suematsu M, Minamishima YA. 2015. Inhibition of the oxygen sensor PHD2 in the liver improves survival in lactic acidosis by activating the Cori cycle. Proc Natl Acad Sci U S A 112:11642–11647. doi: 10.1073/pnas.1515872112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warburg O. 1956. On respiratory impairment in cancer cells. Science 124:269–270. [PubMed] [Google Scholar]
- 27.Kelly B, O'Neill LA. 2015. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res 25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlova NN, Thompson CB. 2016. The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills EL, O'Neill LA. 2016. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur J Immunol 46:13–21. doi: 10.1002/eji.201445427. [DOI] [PubMed] [Google Scholar]
- 30.Myllyharju J. 2009. HIF prolyl 4-hydroxylases and their potential as drug targets. Curr Pharm Des 15:3878–3885. doi: 10.2174/138161209789649457. [DOI] [PubMed] [Google Scholar]
- 31.Tan Z, Xie N, Cui H, Moellering DR, Abraham E, Thannickal VJ, Liu G. 2015. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol 194:6082–6089. doi: 10.4049/jimmunol.1402469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas R, Smith J, Rocher-Ros V, Nadkarni S, Montero-Melendez T, D'Acquisto F, Bland EJ, Bombardieri M, Pitzalis C, Perretti M, Marelli-Berg FM, Mauro C. 2015. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol 13:e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottfried E, Kunz-Schughart LA, Ebner S, Mueller-Klieser W, Hoves S, Andreesen R, Mackensen A, Kreutz M. 2006. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107:2013–2021. doi: 10.1182/blood-2005-05-1795. [DOI] [PubMed] [Google Scholar]
- 34.Barile E, De SK, Pellecchia M. 2012. PDK1 inhibitors. Pharm Pat Anal 1:145–163. doi: 10.4155/ppa.12.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan MC, Holt-Martyn JP, Schofield CJ, Ratcliffe PJ. 2016. Pharmacological targeting of the HIF hydroxylases—a new field in medicine development. Mol Aspects Med 47–48:54–75. doi: 10.1016/j.mam.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Burgess AW, Metcalf D, Kozka IJ, Simpson RJ, Vairo G, Hamilton JA, Nice EC. 1985. Purification of two forms of colony-stimulating factor from mouse L-cell-conditioned medium. J Biol Chem 260:16004–16011. [PubMed] [Google Scholar]
- 37.Klotzsche-von Ameln A, Muschter A, Mamlouk S, Kalucka J, Prade I, Franke K, Rezaei M, Poitz DM, Breier G, Wielockx B. 2011. Inhibition of HIF prolyl hydroxylase-2 blocks tumor growth in mice through the antiproliferative activity of TGFbeta. Cancer Res 71:3306–3316. doi: 10.1158/0008-5472.CAN-10-3838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.