ABSTRACT
Inhibition of protein synthesis serves as a general measure of cellular consequences of chemical stress. A few proteins are translated selectively and influence cell fate. How these proteins can bypass the general control of translation remains unknown. We found that low to mild doses of oxidants induce de novo translation of the NRF2 protein. Here we demonstrate the presence of a G-quadruplex structure in the 5′ untranslated region (UTR) of NRF2 mRNA, as measured by circular dichroism, nuclear magnetic resonance, and dimethylsulfate footprinting analyses. Such a structure is important for 5′-UTR activity, since its removal by sequence mutation eliminated H2O2-induced activation of the NRF2 5′ UTR. Liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics revealed elongation factor 1 alpha (EF1a) as a protein binding to the G-quadruplex sequence. Cells responded to H2O2 treatment by increasing the EF1a protein association with NRF2 mRNA, as measured by RNA-protein interaction assays. The EF1a interaction with small and large subunits of ribosomes did not appear to change due to H2O2 treatment, nor did posttranslational modifications, as measured by two-dimensional (2-D) Western blot analysis. Since NRF2 encodes a transcription factor essential for protection against tissue injury, our data have revealed a novel mechanism of cellular defense involving de novo NRF2 protein translation governed by the EF1a interaction with the G-quadruplex in the NRF2 5′ UTR during oxidative stress.
KEYWORDS: RNA binding proteins, RNA structure, antioxidant genes, protein translation, proteomics
INTRODUCTION
Increasing evidence suggests that low to moderate levels of chemical stress activate cellular defense systems. A centerpiece of such defense systems involves the expression of antioxidant and detoxification genes, many of which contain the antioxidant response element in the promoters where the NRF2 transcription factor binds. Examples of such genes include the genes for hemeoxygenase 1, NAD(P)H:quinone oxidoreductase 1, glutamate-cysteine ligase, glutathione S-transferases, thioredoxin, and UDP-glucuronosyltransferase (1–4). Works from our laboratory and others have shown that the induction of the NRF2 protein contributes to elevated expression levels of these genes (5–8). Gene knockout studies have demonstrated that NRF2 functions as a cytoprotective gene in multiple organ systems, including the brain, cardiovascular system, airway, lung, liver, stomach, gastrointestinal tract, kidney, and bladder (3, 9–11). These lines of evidence support the importance of understanding the molecular pathways regulating NRF2 protein expression.
Various chemical stressors, including those inducing oxidative stress, cause an accumulation of the NRF2 protein at the cellular level. The human NRF2 gene encodes an mRNA species of 2,859 nucleotides (nt) (NCBI RefSeq accession number NM_006164.4) and a protein of 605 amino acids (12–15). The N-terminal hydrophilic domain of the NRF2 protein interacts with KEAP1, which regulates the stability of the NRF2 protein by ubiquitination and proteasome-mediated degradation (13, 15). The C-terminal half of the NRF2 protein contains a cap-and-collar domain, a basic amino acid region for DNA binding, and a leucine zipper for heterodimerization with a cotranscription factor, such as the small musculoaponeurotic factor (12–16). Although the NRF2 protein can be stabilized due to chemically induced dissociation from KEAP1, we have found that oxidative stress induces de novo NRF2 protein translation in vitro and in vivo (7, 8, 17).
The process of protein translation is divided into three sequential stages, initiation, elongation, and termination, with initiation being the rate-limiting step. Under normal physiological conditions, translation initiation requires a 7-methyl-guanine (m7G) cap structure at the 5′ end of mRNA for recognition by eukaryotic translation initiation factor 4E (eIF4E) (18–20). eIF4E is a component of the eIF4F complex, which contains the scaffold eIF4G and the ATP-dependent helicase eIF4A. Binding of the eIF4F complex to the 5′ m7G cap is catalyzed by eIF4A in an ATP-dependent manner, triggering the joining of eIF4B, an enhancer of the eIF4A helicase that removes inhibitory RNA structures. The poly(A) binding protein interacts with eIF4G to circularize mRNA and with eIF4B to stabilize the complex. In parallel to the process of mRNA recognition, the 43 Svedberg (43S) preinitiation complex (PIC) is formed, containing eIF1, eIF1A, eIF3, eIF5, the eIF2/GTP/tRNAiMet ternary complex, and the 40S subunit of the ribosome. Binding of the 43S PIC to mRNA prebound with the eIF4F complex results in the 48S initiation complex. During chemical stress, this 5′-m7G-dependent translation initiation is inhibited. It is estimated that 3 to 5% of genes encode mRNA species containing an internal ribosomal entry site (IRES) in the 5′ untranslated region (UTR) and can undergo translation initiation in a 5′-m7G cap-independent manner (21–25). However, previously reported IRESs are diverse in sequence and conformation, adding to the mystery about the identity of IRESs and how they coordinate the process of translation initiation.
How certain proteins such as NRF2 can escape general suppression by translational machinery and be selectively translated under stress conditions remains unknown. Human NRF2 mRNA contains a 555-nucleotide 5′ UTR, with 70% of the sequence being G's or C's. Four guanine bases can form a G-tetrad due to Hoogsteen hydrogen bonds between each guanine base (26–28). Two to four G-tetrads can stack up to form a stable three-dimensional (3-D) structure, i.e., the G-quadruplex. The G-quadruplex is a four-stranded nucleic acid secondary structure typically consisting of three stacks of G-tetrads. A DNA or RNA strand containing 4 or more blocks of 2 to 4 consecutive guanines can form a stable G-quadruplex structure (26, 27, 29). Although the G-quadruplex has been studied mostly in DNA, the structure requires the dissociation of the canonical Watson-Crick base pairing in double-stranded DNA to permit Hoogsteen bond formation. In contrast, given the presence of a consensus G-quadruplex sequence, RNA is prone to the formation of the 3-D structure due to its single-stranded nature (29, 30). In addition to the favorable thermal dynamics, there is evidence that the RNA G-quadruplex is more stable than its DNA counterpart (28, 31). Here we test whether a G-quadruplex is formed in the NRF2 5′ UTR and whether such a structure plays a role in de novo NRF2 protein translation under oxidative stress.
RESULTS
De novo Nrf2 protein translation induced by H2O2 in HEK293 cells.
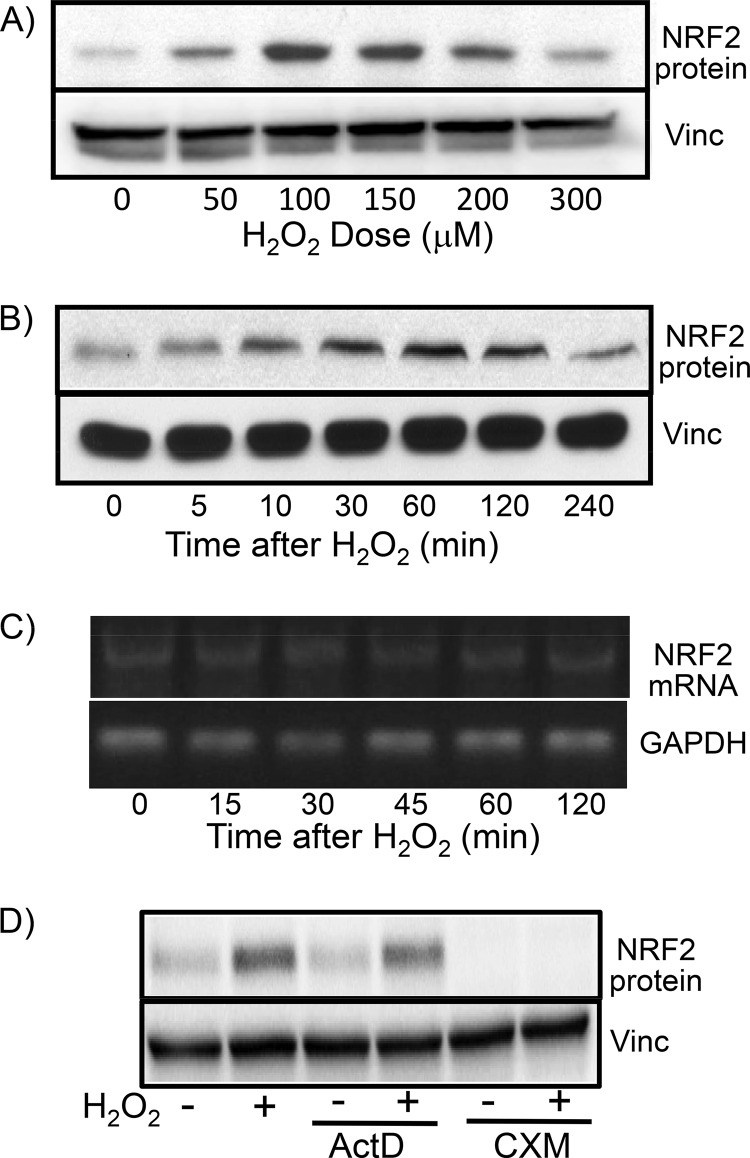
Previous works from our laboratory indicate that low to moderate doses of H2O2 induce de novo NRF2 protein translation in cardiomyocytes and HeLa cells (7, 8, 17). With human embryonic kidney 293 (HEK293) cells, we found that a brief treatment with H2O2 at concentrations of 50 to 200 μM induced an increase in the level of the NRF2 protein (Fig. 1A). The best dose for NRF2 protein induction, i.e., 100 μM, caused the NRF2 protein level to increase within 5 min and to reach a peak at 1 h (Fig. 1B). Cellular levels of NRF2 mRNA did not change due to H2O2 treatment (Fig. 1C). To demonstrate that increased NRF2 protein levels result from posttranscriptional regulation, we used cycloheximide, a protein synthesis inhibitor, and actinomycin D, an RNA synthesis inhibitor. Cycloheximide was able to inhibit the elevation of the NRF2 protein level by H2O2 but not actinomycin D (Fig. 1D), which was effective in blocking the elevation of the level of positive-control cyclooxygenase-2 mRNA by H2O2 treatment (data not shown). These data point to posttranscriptional mechanisms responsible for the rapid induction of the NRF2 protein by H2O2 treatment.
FIG 1.
H2O2 treatment causes increased NRF2 protein levels in the absence of increased NRF2 mRNA levels. HEK293 cells were treated with various doses of H2O2 (A) or 100 μM H2O2 (B to D) for 10 min before harvesting 1 h later (A and D) or at the indicated time points (B and C) for Western blotting to measure the NRF2 protein level with vinculin (Vinc) as a loading control (A, B, and D) or for RT-PCR and agarose gel electrophoresis to measure the NRF2 mRNA level with GAPDH as a loading control (C). Actinomycin D (ActD) (0.5 μM) or cycloheximide (CXM) (0.5 μg/ml) was added to cells 10 min before H2O2 treatment (D). Data are from one experiment representative of three.
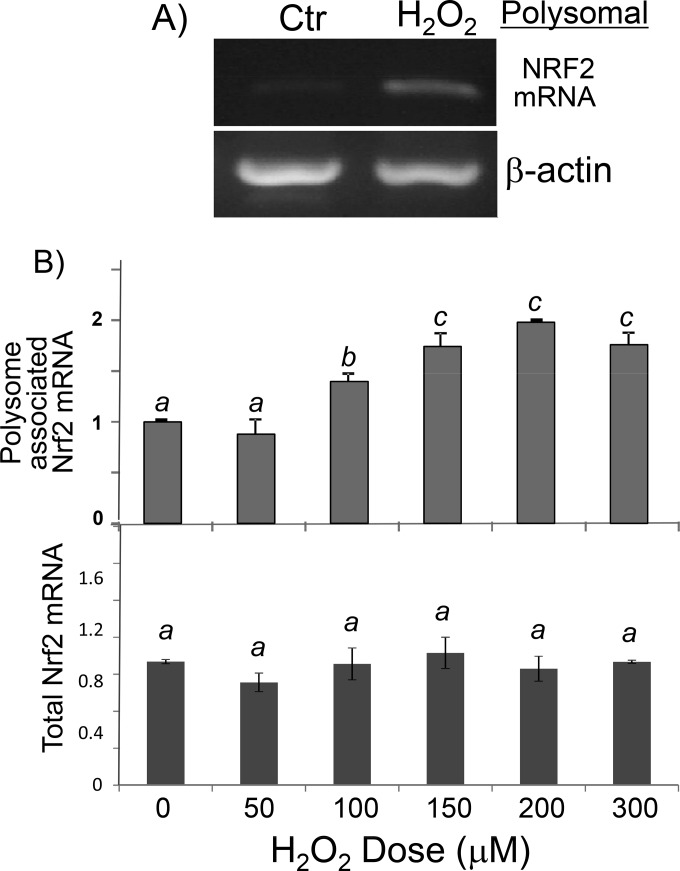
To demonstrate that the NRF2 protein undergoes de novo translation, we isolated polysomes from control and H2O2-treated cells for measurement of NRF2 mRNA levels, since an mRNA species in the process of being translated into a protein is bound with multiple ribosomes. An increased abundance of NRF2 mRNA was detected in polysomes from cells treated with H2O2 (Fig. 2C and D). In contrast, β-actin mRNA did not increase its association with polysomes in H2O2-treated cells, serving as a negative control (Fig. 2C). Since the cellular NRF2 mRNA level does not change with H2O2 treatment (Fig. 1C and 2B, bottom), the observed increases of NRF2 mRNA associations with polysomes along with elevated levels of the NRF2 protein in the absence of increases in total cellular mRNA support that H2O2 treatment induces de novo Nrf2 protein translation in HEK293 cells.
FIG 2.
H2O2 causes an increased association of NRF2 mRNA with ribosomes. Shown are data for HEK293 cells after treatment with 100 μM H2O2 (A) or various doses of H2O2 (B) for 10 min and harvesting 1 h later. RNA was isolated from polysomes (A and B) or cell lysates (B) for RT-PCR (A) or real-time RT-PCR (B). The RT-PCR output of NRF2 mRNA in relative fluorescent units was normalized to that of the reference gene 18S rRNA (B). Data are from one representative experiment (A) or averages ± standard deviations of triplicates from one experiment representative of three (B). A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). Therefore, the means labeled “a” are significantly different from those labeled “b” or “c.” Ctr, control.
The NRF2 5′ UTR contains a G-quadruplex structure.
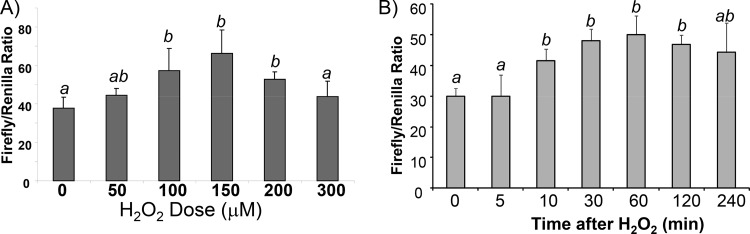
To demonstrate that the 5′ UTR of Nrf2 drives protein translation under oxidative stress, we cloned the human Nrf2 5′ UTR into a dicistronic reporter vector, pRF (32). The resulting pRL-NRF2 5′UTR-FL reporter construct contains a simian virus 40 (SV40) promoter and expression cassettes for two luciferases: the 5′-m7G-driven renilla luciferase in front of the Nrf2 5′-UTR-regulated firefly luciferase. Whereas the SV40 promoter drives the transcription of both renilla and firefly luciferase genes, renilla luciferase corrects for the transcription rate and 5′-m7G cap-dependent general translation, while firefly luciferase activity reflects Nrf2 5′-UTR-driven translation. An increase in the ratio of firefly over renilla luciferase in the absence of a reduction of renilla luciferase activity indicates NRF2 5′-UTR-driven translation. When the construct was transfected into HEK293 cells and after treatment of cells with H2O2 at concentrations varying from 50 to 300 μM, the highest ratio of firefly over renilla luciferase was observed with treatment with 100 to 150 μM H2O2 (Fig. 3A). The activity of the Nrf2 5′ UTR increased within 30 min and reached a maximum ∼60 min after 100 μM H2O2 treatment (Fig. 3B). These dose and time course studies indicate that H2O2 activated the Nrf2 5′ UTR.
FIG 3.
NRF2 5′-UTR activation by H2O2 treatment. The dicistronic pRL-Nrf2 5′UTR-FL reporter construct was transfected into HEK293 cells. Transfected cells were treated with various doses of H2O2 (A) or 100 μM H2O2 (B) before harvesting 1 h later (A) or at the indicated times (B) for dual-luciferase assays. Data indicate averages ± standard deviations for triplicate ratios of firefly over renilla luciferase from one experiment representative of three. A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). Therefore, the means labeled “a” are significantly different from those labeled “b,” whereas the label “ab” indicates means with no significant difference from those labeled “a” or “b.”
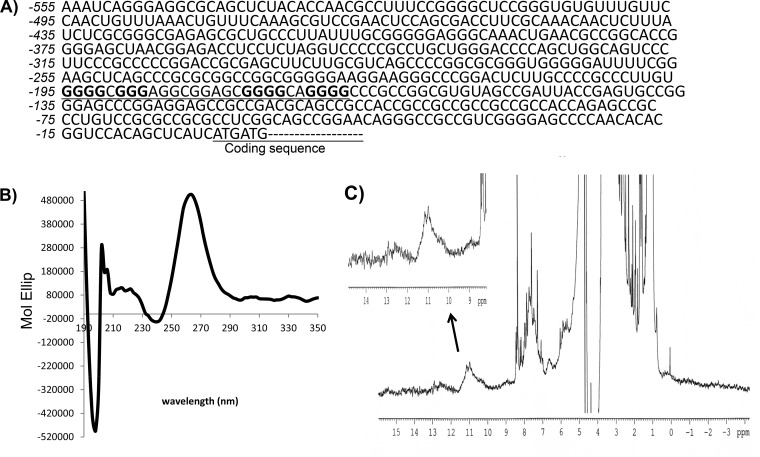
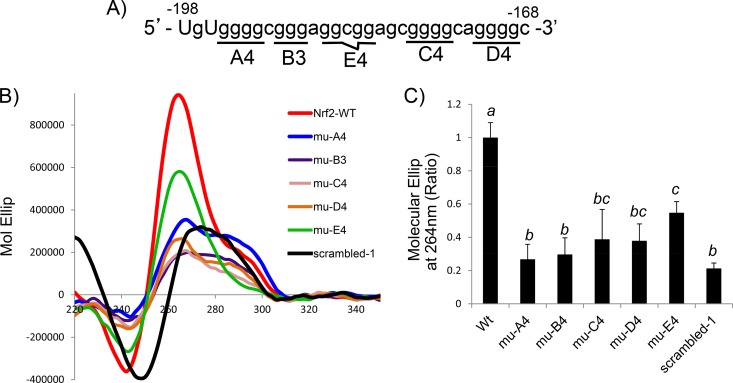
When examining the sequence of the NRF2 5′ UTR, we found that the region spanning nt −195 to −169, i.e., GGGGCGGGAGGCGGAGCGGGGCAGGGG, resembles the consensus G-quadruplex sequence G3+N1–7G3+N1–7G3+N1–7G3+ (Fig. 4A). A 31-mer RNA fragment containing the sequence spanning nt −198 to −168 from the NRF2 5′ UTR was generated to determine the presence of the G-quadruplex structure using circular dichroism (CD) spectroscopy and nuclear magnetic resonance (NMR). CD spectroscopy revealed a peak of molar ellipticity at a 264-nm wavelength and a downward peak at 240 nm, which is typical of a parallel G-quadruplex structural fold (Fig. 4B). The NMR results showed imino proton peaks at 10 to 12 ppm, which is characteristic of the G-quadruplex structure (Fig. 4C).
FIG 4.
The NRF2 5′ UTR contains the G-quadruplex sequence and structure. (A) The NRF2 5′-UTR sequence is shown, with the putative G-quadruplex consensus sequence underlined. (B) A 31-mer RNA oligonucleotide (5 μM) synthesized from the DNA template containing the sequence spanning nt −198 to −168 of the NRF2 5′ UTR was used for CD spectroscopy. (C) 1H NMR was performed by using the same RNA sequence. Data are from one experiment representative of three.
There are several possible conformations of the G-quadruplex depending upon which set of G's is in play for the stacks of the G-tetrad. Blocks of G's were replaced with T's to determine which set of G's is essential for forming the G-quadruplex structure (Fig. 5A). The basal level of molar ellipticity was documented with a sequence in which most G's are replaced with A's or T's, 5′-TGTATTACTATAGGCGGAGCATTACATATAC-3′ (scrambled-1). Replacement of G's with T's in the first (A4), second (B3), third (C4), or fourth (D4) block reduced molar ellipticity to a level similar to that of scrambled-1 (Fig. 5B and C), indicating that these four blocks of G's are the most likely basis for G-quadruplex formation.
FIG 5.
Measurement of the G-quadruplex structure using mutant sequences. (A and B) The 31-mer RNA oligonucleotides containing the wild type or the mutant with G's replaced with T's, as indicated (A), were used for CD spectroscopy (B). (C) The molar ellipses (Mol Ellip) at 264 nm from mutated (mu) sequences were compared to that of the wild-type (WT) sequence, with the value for the signal of the wild-type sequence being set at 1. Data are from one representative experiment (B) or averages ± standard deviations from three separate experiments (C). A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). Therefore, the means labeled “a” are significantly different from those labeled “b,” “bc,” or “c,” whereas the label “bc” indicates means with no significant difference from those labeled “b” or “c.”
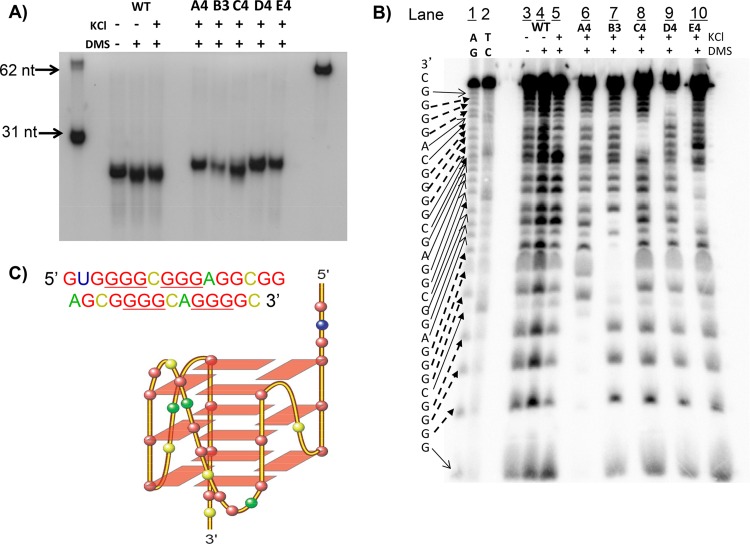
G-quadruplexes can form intramolecularly from a single strand or intermolecularly from two or more strands of DNA or RNA. We measured the size of the oligomers using polyacrylamide gel electrophoresis to confirm that the 31-mer sequence from the NRF2 5′ UTR indeed forms an intramolecular G-quadruplex (Fig. 6A). To address which GGG in the sequence forms the foundation for the G-quadruplex, we analyzed the 31-mer NRF2 5′-UTR sequence by a dimethylsulfate (DMS) footprint, which defines the position of G's for forming the G-quadruplex since these G's are protected from methylation by DMS and subsequent cleavage by piperidine (33). Whereas KCl facilitates G-quadruplex formation, the addition of DMS under conditions not favoring G-quadruplex formation sets the baseline methylation of nucleotides (Fig. 6B, lane 4). In comparison, those G's protected from methylation indicate that they are the component of the G-quadruplex structure (Fig. 6B, lane 5). The mutant sequences were included to confirm the location of G's in the G-quadruplex (Fig. 6B, lanes 6 to 10). The results revealed that the GGG at positions −194 to −192, −190 to −188, −177 to −175, and −172 to −170 were protected from piperidine cleavage, indicating their involvement in G-quadruplex structural folding. Taken together, the G-quadruplex formed in the 31-mer NRF2 5′-UTR sequence appears to be a parallel-stranded G-quadruplex, as shown in Fig. 6C.
FIG 6.
Methylation and footprinting to define G-quadruplex structure. (A) The 31-mer fragments corresponding to the region spanning nt −198 to −168 of the wild-type or mutant NRF2 5′ UTR were treated with DMS to determine the monomeric or dimeric G-quadruplex. (B) Oligonucleotides were treated with piperidine for sequencing by polyacrylamide gel electrophoresis. (C) Predicted G-quadruplex conformation. Data are from one experiment representative of three.
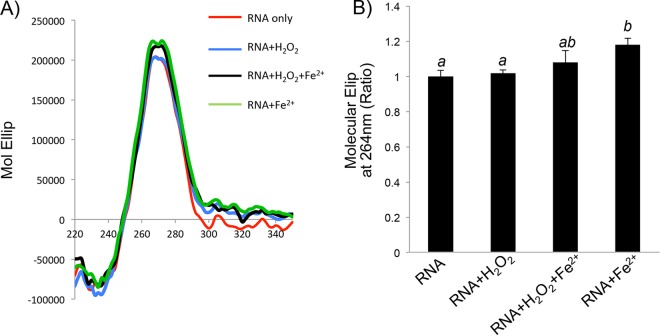
G-quadruplex structural folding is usually measured by using the molecule of interest in solution. Since H2O2 treatment caused an increase in NRF2 protein translation in cells, the presence of the G-quadruplex consensus sequence in the 5′ UTR led us to question whether H2O2 enhances or decreases G-quadruplex formation in solution. Inside cells, H2O2 is converted to reactive hydroxyl radicals via the Fenton reaction. Ferrous iron (Fe2+) was added to the solution for the Fenton reaction to determine the effect on G-quadruplex formation. Although Fe2+ alone can cause a minor increase in G-quadruplex formation, as measured by the molar ellipticity at 264 nm, H2O2 alone or together with Fe2+ did not enhance G-quadruplex formation (Fig. 7).
FIG 7.
G-quadruplex structure in solution containing H2O2 and Fe2+. A 31-mer RNA oligonucleotide synthesized from the DNA template containing the sequence spanning nt −198 to −168 of the NRF2 5′ UTR was diluted in a solution containing 5 μM RNA, with or without 100 μM H2O2 and/or 1 μM FeSO4. Data are from one representative experiment (A) or averages ± standard deviations from three independent experiments (B).
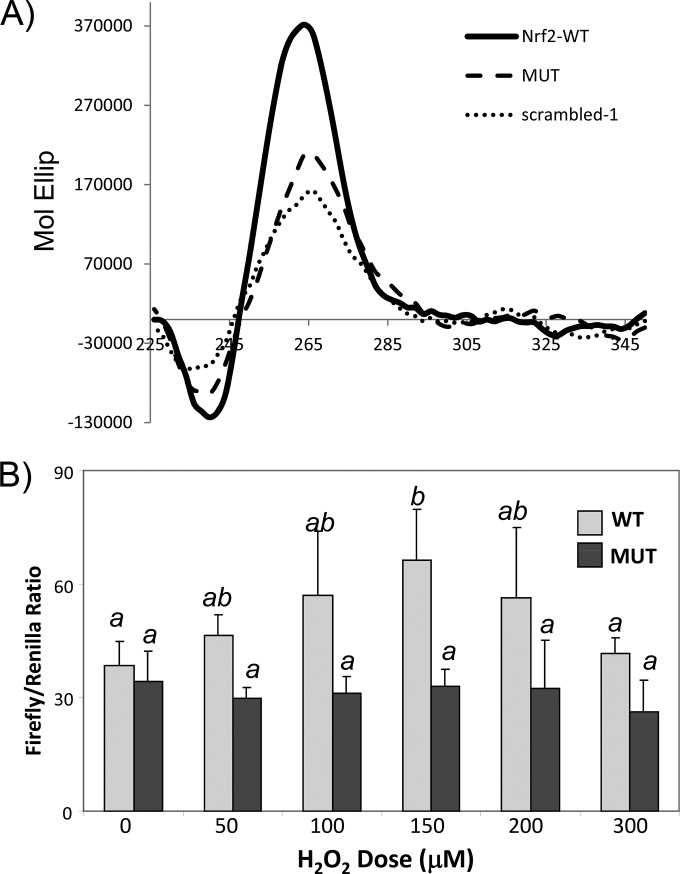
The functional impact of the NRF2 5′-UTR G-quadruplex at the cellular level was determined by using the dicistronic reporter construct as described above. Unlike 5′-m7G cap-mediated translation, IRES-mediated protein translation does not occur with in vitro translation systems, such as with rabbit reticulocyte lysates (22, 34). Therefore, such a reporter assay becomes important for testing the role of the G-quadruplex in NRF2 5′-UTR-mediated protein translation inside cells. Replacement of GGGG at nt −195 to −192 of the NRF2 5′ UTR with TTTT, i.e., the A4 mutant, inhibited G-quadruplex formation in solution (Fig. 8A). A full-length NRF2 5′-UTR reporter construct containing A4 mutations was transfected into HEK293 cells for measurements of H2O2-induced activation. Compared to the wild type, the G-quadruplex-eliminated mutant NRF2 5′ UTR was no longer responsive to H2O2-induced activation (Fig. 8B). This suggests that the presence of the G-quadruplex is important for NRF2 5′-UTR activation by H2O2 at the cellular level.
FIG 8.
G-quadruplex sequence-dependent NRF2 5′-UTR activation by H2O2 in cells. A dicistronic luciferase reporter construct of the wild-type human NRF2 5′ UTR (555 nt) or a mutant NRF2 5′ UTR with GGGG at nt −195 to −192 being replaced with TTTT (A4 mutant) was transfected into HEK293 cells. (A) The A4 mutant in a 31-mer RNA oligonucleotide containing the sequence spanning nt −198 to −168 from the NRF2 5′ UTR was used for confirmation of the lack of a G-quadruplex structural fold by CD spectroscopy. (B) Transfected cells were treated with various doses of H2O2 for 10 min and harvested 1 h later for measurements of the ratio of firefly versus renilla luciferase. Data indicate averages ± standard deviations of triplicates from one experiment representative of three. A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). Therefore, the means labeled “a” are significantly different from those labeled “b,” whereas the label “ab” indicates means with no significant difference from those labeled “a” or “b.”
EF1a binds to the NRF2 5′-UTR G-quadruplex during H2O2-induced NRF2 protein translation.
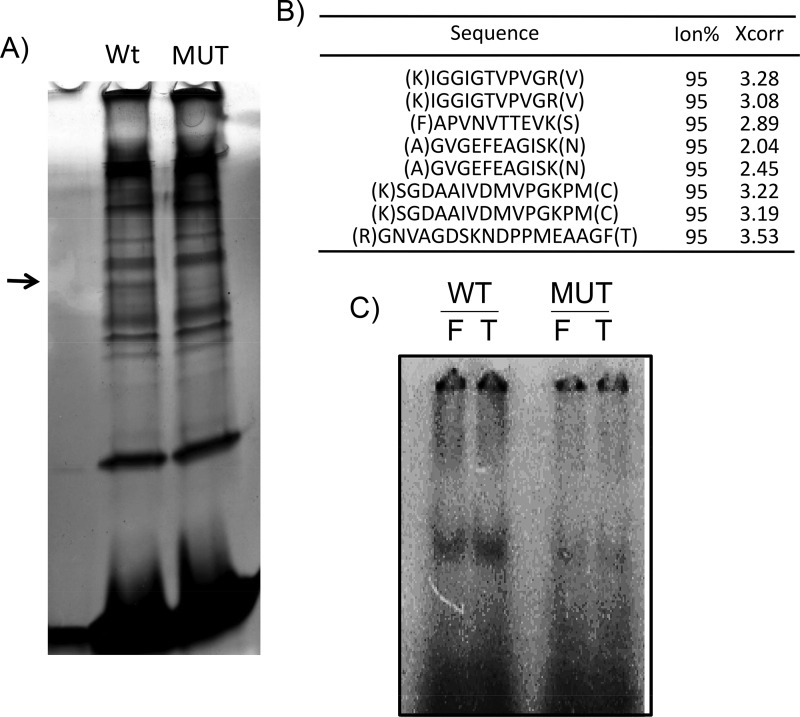
An RNA molecule is rarely free of protein binding in a cell (35–37). To understand how the 5′-UTR G-quadruplex affects NRF2 protein translation at the cellular level, we identified proteins capable of binding to the NRF2 5′-UTR G-quadruplex using liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics. A biotinylated RNA bait was generated from the 31-mer oligonucleotide of the wild-type or A4 mutant NRF2 5′-UTR sequence for the isolation of binding proteins. The bound proteins were resolved by SDS-PAGE for silver staining to identify the difference (Fig. 9A). A band present in the wild type but not the G-quadruplex-eliminated mutant was excised for LC-MS/MS analyses. A sequence database search indicates that this protein is eukaryotic elongation factor 1 alpha (EF1a; IPI00025447) (Fig. 9B).
FIG 9.
NRF2 5′-UTR G-quadruplex binds to EF1a protein. A biotinylated 31-mer RNA fragment from the region spanning nt −198 to −168 of the NRF2 5′ UTR (WT) or the A4 mutant (MUT) was used as a bait for isolation of binding proteins from cytoplasmic HeLa cell lysates. (A) The bound proteins were resolved by SDS-PAGE for detection by silver staining. The band indicated by an arrow was excised for LC-MS/MS analyses. (B) Peptides of EF1a detected by mass spectrometry. (C) The same probes, wild type (WT) or A4 mutant (MUT), but labeled with [32P]ATP were used for in vitro binding with the purified recombinant full-length (F) or C-terminally truncated (T) EF1a protein, and the binding complex was detected by autoradiography following EMSAs.
The interaction of the EF1a protein with the 31-mer G-quadruplex-containing sequence from the NRF2 5′ UTR was measured by electrophoretic mobility shift assays (EMSAs). Using purified EF1a protein and the 31-mer wild type or A4 mutant as the probe, we found an interaction of the EF1a protein with the G-quadruplex-containing sequence but not the mutant (Fig. 9C). The EF1a protein has three functional domains: domain I binds GTP, domain II binds to aminoacyl-tRNA, and domain III in the C terminus together with domain II interacts with actin (38). When cloning the EF1a expression vector, an internal HindIII site enabled us to obtain a truncated version of EF1a without the C terminus. We found that the truncated EF1a protein was capable of binding to the Nrf2 5′-UTR G-quadruplex sequence like full-length EF1a (Fig. 9C).
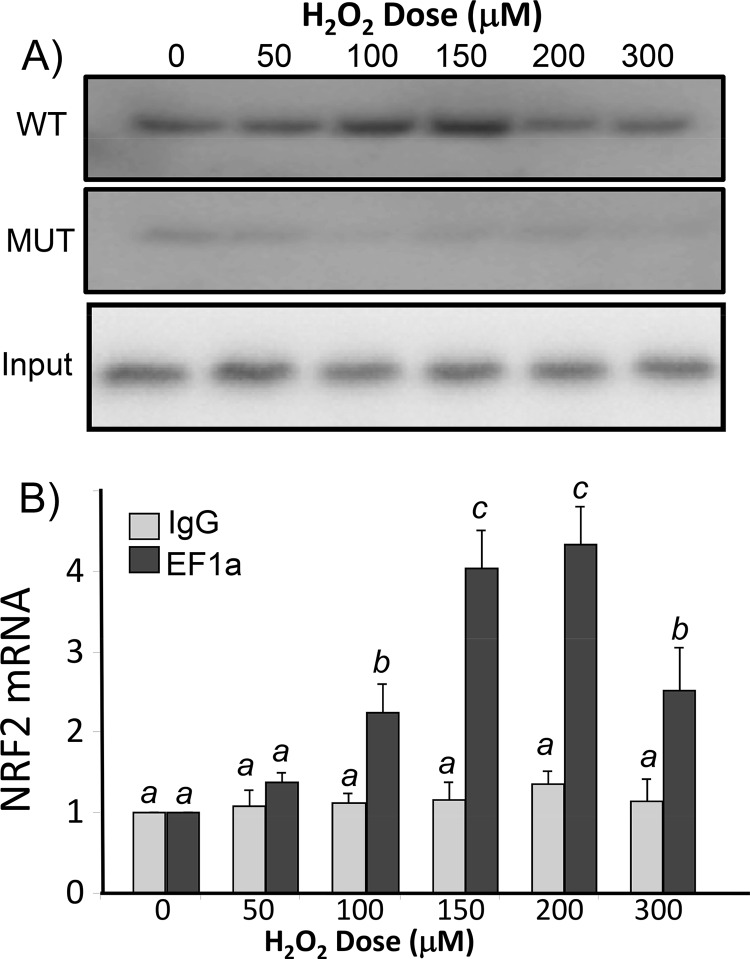
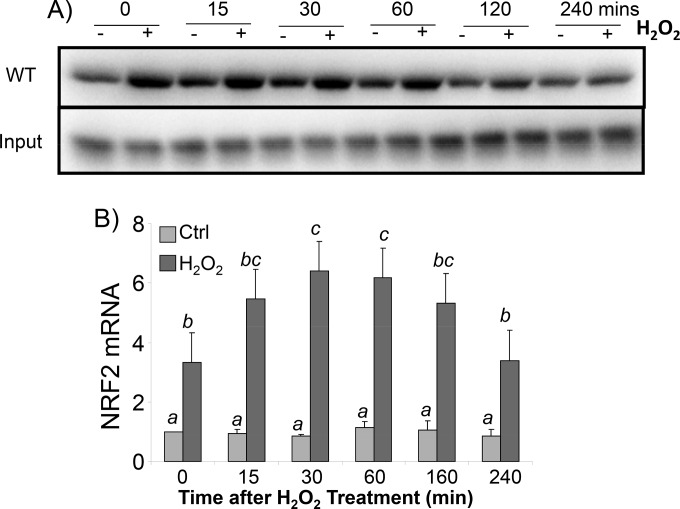
To demonstrate that EF1a inside cells can interact with the NRF2 5′-UTR G-quadruplex under conditions of oxidative stress, we generated a biotinylated 31-mer oligonucleotide of the NRF2 5′-UTR RNA for isolating binding proteins from cell lysates. Western blotting for EF1a using RNA bait-associated proteins revealed that H2O2 treatment induced a dose- and time-dependent increase of the EF1a association (Fig. 10A and 11A). This interaction is specific to the G-quadruplex since the A4 mutant was not able to pull down EF1a (Fig. 10A). To confirm that EF1a indeed binds to NRF2 mRNA inside cells, we performed immunoprecipitation analysis of the EF1a protein from cell lysates for quantification of NRF2 mRNA by reverse transcription-PCR (RT-PCR). This RNA binding protein immunoprecipitation assay indicated that H2O2 caused a dose- and time-dependent increase in the association of EF1a with NRF2 mRNA (Fig. 10B and 11B). These data support that H2O2 treatment results in an increased EF1a interaction with NRF2 mRNA at the cellular level.
FIG 10.
H2O2 dose-dependent increase of EF1a interaction with NRF2 mRNA in HEK293 cells. HEK293 cells were treated with various doses of H2O2 for 10 min and harvested 1 h later. (A) Cytoplasmic extracts were incubated with biotinylated 31-mer wild-type or A4 mutant probes to isolate binding proteins for Western blot analysis to detect EF1a (top), with EF1a from total cell lysates serving as the input (bottom). (B) In parallel, cytoplasmic extracts were used for immunoprecipitation with EF1a antibody or IgG. The complex from EF1a antibody immunoprecipitation was used for RT-PCR to determine the level of NRF2 mRNA, with that from the immunoprecipitates of IgG serving as a negative control. The levels of NRF2 mRNA are expressed as fold increases in comparison to the control sample without H2O2 treatment, with the RT-PCR output, expressed as relative fluorescent units, being set to 1. The data are from one representative experiment (A) or averages ± standard deviations from triplicates of one experiment representative of three (B). A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). Therefore, the means labeled “a” are significantly different from those labeled “b” or “c.”
FIG 11.
H2O2 time-dependent increase of EF1a interactions with the NRF2 5′ UTR in cells. HEK293 cells were treated with 100 μM H2O2 for 10 min and harvested at the indicated time points. (A) Cytoplasmic extracts were incubated with biotinylated 31-mer wild-type probes to isolate binding proteins for Western blot analysis to detect EF1a (top), with the EF1a protein from the total cell lysate serving as the input (bottom). (B) Cytoplasmic lysates were used for immunoprecipitation to isolate EF1a for RT-PCR to detect NRF2 mRNA. The levels of NRF2 mRNA are expressed as fold increases in comparison to the control sample without H2O2 treatment; both groups were corrected by RT-PCR signals from negative-control IgG immunoprecipitates. Data are from one representative experiment (A) or averages ± standard deviations from triplicates of one experiment representative of three (B). A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). Therefore, the means labeled “a” are significantly different from those labeled “b,” “c,” or “bc,” whereas “bc” indicates means with no significant difference from those labeled “b” or “c.”
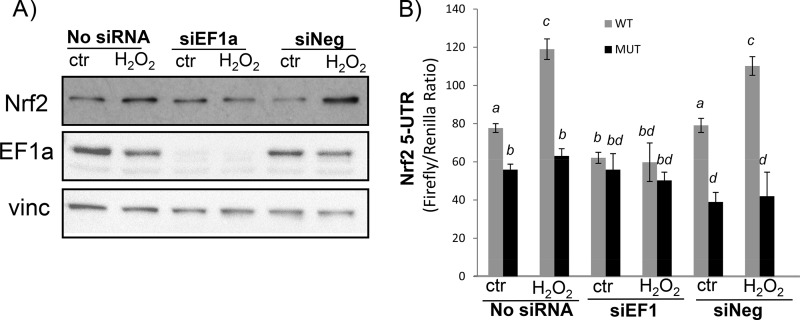
To address whether the binding of EF1a to the NRF2 5′-UTR G-quadruplex mediates protein translation, we used small interfering RNA (siRNA) to knock down EF1a by transfecting HEK293 cells (Fig. 12A). The results showed that EF1a siRNA was able to inhibit NRF2 protein induction by H2O2 treatment (Fig. 12A). The NRF2 5′-UTR dicistronic reporter assay confirmed an inhibitory effect of EF1a siRNA against H2O2-induced increases in the ratio of firefly over renilla luciferase in cells transfected with the wild-type Nrf2 5′-UTR reporter (Fig. 12B). Firefly luciferase activities without correction for renilla luciferase are consistent with data for the ratio showing an inhibitory effect of EF1a siRNA (data not shown). EF1a siRNA or H2O2 treatment does not cause changes in renilla luciferase activity (data not shown). The mutant NRF2 5′-UTR reporter with the G-quadruplex eliminated was not affected by H2O2 treatment or EF1a siRNA (Fig. 12B). These data demonstrate a role of EF1a in mediating H2O2-induced NRF2 protein translation.
FIG 12.
EF1a-dependent Nrf2 protein translation. HEK293 cells were transfected with either EF1a siRNA (siEF1) or negative-control siRNA (siNeg) without (A) or with (B) the pRL-Nrf25′UTR-FL reporter construct. Forty-eight hours after transfection, cells were placed into 0.5% FBS–DMEM for 16 h before treatment with 100 μM H2O2 for 10 min and harvesting 1 h later for Western blot analyses (A) or dual-luciferase assays (B). Data are from one representative experiment (A) or triplicates of one experiment representative of three (B). A mean that is significantly different from another is labeled with a different letter symbol based on ANOVA (P < 0.05). The means labeled “a” are significantly different from those labeled “b,” “c,” or “d,” whereas “bd” indicates means with no significant difference from those labeled “b” or “d.”
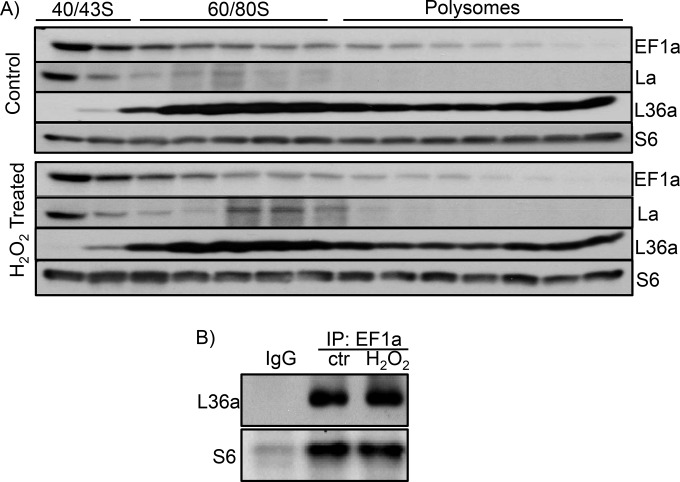
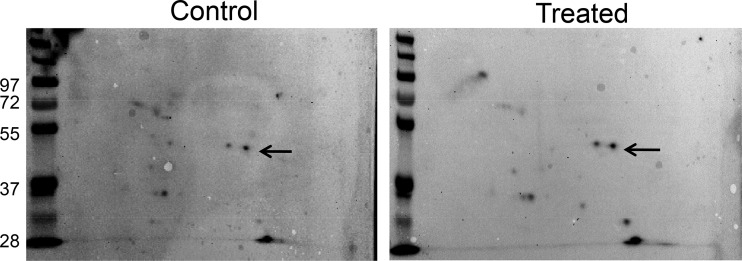
In an effort to understand how EF1a increases its contact with the NRF2 5′ UTR during oxidative stress, we tested whether the level of the EF1a protein or the subcellular localization of EF1a changed due to H2O2 treatment. Neither assay showed positive data (data not shown). When ribosomes were collected for measurements of EF1a, we found EF1a in the 40/43S small-subunit fraction predominantly regardless of H2O2 treatment (Fig. 13A). This differs from La autoantigen, an RNA binding protein capable of binding to NRF2 mRNA and increasing the association with 60/80S ribosomal fractions during oxidative stress (17). The association of EF1a with ribosomal small or large subunits did not appear to change due to H2O2 treatment, as measured by coimmunoprecipitation of the large- or small-subunit protein L36a or S6, respectively (Fig. 13B). Two-dimensional (2-D) gel electrophoresis and Western blotting failed to detect a shift in the molecular weight or isoelectric focus point of the EF1a protein due to H2O2 treatment (Fig. 14). Therefore, how oxidative stress causes an increased association of EF1a with the G-quadruplex in the 5′ UTR of NRF2 remains unknown.
FIG 13.
EF1a association with 40/43S ribosomes. (A) HEK293 cells were treated with 100 μM H2O2 for 10 min and harvested 1 h later for sucrose gradient ultracentrifugation and collection of ribosomal fractions. The fractions were used for Western blotting to detect the distribution of EF1a. La was included as a positive control for H2O2-induced increases in associations with ribosomes. S6 and L36a are used to indicate the fractions containing small or large subunits of ribosomes. (B) Immunoprecipitation was performed by using cytoplasmic cell lysates from control or H2O2-treated cells with IgG or EF1a antibody for detection of the L36a or S6 protein by Western blotting. The data are from one experiment representative of three.
FIG 14.
Lack of detectable posttranslational modification of the EF1a protein due to H2O2 treatment. HEK293 cells were treated with 100 μM H2O2 for 10 min and harvested 1 h later for 2-D Western blotting. Arrows indicate the signal corresponding to the EF1a protein. The data are from one experiment representative of three.
DISCUSSION
This study reports that de novo NRF2 protein translation contributes to the elevation of NRF2 protein levels in HEK293 cells during oxidative stress. The G-quadruplex structure has been detected in solution by using RNA oligonucleotides from the region spanning nt −198 to −168 of the NRF2 5′ UTR. Although the presence of H2O2 plus Fe2+ did not affect the formation of the G-quadruplex structure in solution from the naked 31-mer RNA fragment, the sequence corresponding to the G-quadruplex structure has been found to bind to the EF1a protein in vitro and in vivo. At the cellular level, H2O2 treatment caused a dose- and time-dependent increase in EF1a protein binding to Nrf2 mRNA. Knocking down of the EF1a protein using siRNA blocked the induction of the NRF2 protein or the activation of the NRF2 5′ UTR by H2O2 treatment. EF1a was detected in 40/43S ribosomal fractions, supporting its role in translation initiation. However, H2O2 treatment did not alter the EF1a distribution among different fractions of ribosomes or its association with the small or large subunit of ribosomes. 2-D Western blotting failed to detect a gain or loss of posttranslational modifications. Our data indicate the importance of the NRF2 5′-UTR G-quadruplex and its interaction with EF1a for de novo NRF2 protein translation under conditions of oxidative stress.
The G-quadruplex formed from single-stranded DNA has been postulated to exist in many biologically active regions of the human genome (39). Computational analyses of the human genome predict the presence of such a noncanonical DNA structure upstream of transcription start sites for about half of known genes (27). In fact, the promoter region of the NRF2 gene has been found to contain a G-quadruplex structure (40). Such a structure affects the unwinding of DNA double strands, protein binding, and, consequently, transcription initiation.
The RNA G-quadruplex has been discovered in association with a variety of functional regulations (41). In mitochondria, the G-quadruplex structure in RNA facilitates the termination of DNA transcription (42). The RNA G-quadruplex affects alternative splicing, ribosomal frameshifts, and stop codon readthrough (43–46). In the 3′ UTR of mRNAs, the G-quadruplex structure drives subcellular relocalization and increases the efficiency of alternative polyadenylation and therefore the expression of shortened transcripts (47, 48). An additional noteworthy function of the 3′-UTR G-quadruplex is its interaction with microRNAs, serving as a mechanism for controlling protein translation (49, 50). For p53 mRNA, the 3′-UTR G-quadruplex can interact with RNA binding proteins and release translation repression during stress, contributing to the elevation of the level of the p53 protein and its function in apoptosis (51). Therefore, the RNA G-quadruplex plays an important role in regulating protein expression.
Many genes contain a GC-rich sequence in the 5′ UTR and therefore may form G-quadruplex structures in the 5′ UTR. Computational analyses predict that ∼3,000 genes encode mRNA species containing a G-quadruplex in the 5′ UTR (52). The G-quadruplex structure has been studied in 5′ UTRs of mRNA species such as the Zic-1 zinc finger protein, bcl-2, N-Ras, the estrogen receptor gene, cyclin D3, AKT-interacting protein, cathepsin B, MT3-matrix metalloproteinase, and the transcription repressor YY1 (52–60). In these mRNA species, the formation of the 3-D G-quadruplex structure appears to repress translation initiation. Contrary to this negative mode of regulation, the presence of a G-quadruplex in the 5′ UTRs of some mRNA species enhances translation, consistent with our findings. A previous study detected an IRES-containing G-quadruplex structure in fibroblast growth factor 2, and such a structure determines IRES-mediated translation from alternative translation initiation sites (61). The G-quadruplex is a part of the IRES for cap-independent translation of the vascular endothelial growth factor (VEGF) protein in HeLa cells (62). However, such a G-quadruplex is dispensable for the stress-induced activation of VEGF translation, whereas stabilization of the VEGF G-quadruplex by ligands and increasing G-stretches results in the inhibition of the IRES-mediated translation of VEGF (63), suggesting the complexity of how the G-quadruplex controls for protein translation under stress conditions. Nevertheless, 5′-UTR G-quadruplex-dependent protein translation has been found with transforming growth factor β-2 (TGF-β-2) and foxhead box E3 (58, 64). Such 5′-UTR G-quadruplex-mediated protein translation can be cap independent as well as cap dependent (29). In fact, RNA domains folding into the G-quadruplex can recruit the 40S ribosomal subunit directly (65), pointing to a possible role of the 5′-UTR G-quadruplex in the assembly of the 48S initiation complex. Therefore, the functional outcomes of the 5′-UTR G-quadruplex can vary depending on genes or conditions and can be inhibitory or stimulatory for cap-dependent or -independent translation.
How the EF1a interaction with the Nrf2 G-quadruplex leads to the translation of the Nrf2 protein remains to be elucidated. With cap-dependent translation, the interaction of the eIF4F complex with the 5′-m7G group is the first step for translation initiation. Although the interplay between the IRES and the eIF4F complex is unclear, it is believed that eIFs in cap-dependent translation also participate in cap-independent translation. A recent report employing a transcriptome-scale ribosome footprinting technique led to the discovery of G-quadruplex structures in RNA conferring eIF4A-dependent translation (66). Many oncogenes and transcription factors undergo eIF4A-dependent translation in association with the G-quadruplex in the 5′ UTR (66). Whether eIF4A interacts with EF1a and mediates G-quadruplex-dependent Nrf2 protein translation remains to be investigated.
EF1a is a 462-amino-acid protein best known for GTP binding and amino acid chain elongation during protein synthesis (38, 67). EF1a controls the hydrolysis of GTP by carrying GTP and aminoacyl-tRNA to the A site of ribosomes, allowing the addition of the amino acid by transferring the peptide chain from the P site of ribosomes. In addition to catalyzing chain elongation, EF1a proofreads codon-anticodon interactions during translation elongation. However, binding of EF1a to an IRES in the 5′ UTR and to a pseudoknot in the 3′ UTR of viral RNAs has been reported (68, 69). EF1a also exhibits functions independent of protein translation, from regulating cytoskeletal actin organization to regulating protein degradation (70, 71). Our data show that the C-terminal deletion mutant of EF1a remains capable of NRF2 G-quadruplex binding (Fig. 9C), suggesting the importance of the N-terminal and middle domains. The middle domain of EF1a alone interacts with 19 different proteins, including eIF4G (72). Phan et al. (73) demonstrated that the G-quadruplex structure fills in the arginine-glycine-rich RGG motif in a binding protein. The importance of the RGG motif for G-quadruplex binding has also been demonstrated for a number of RNA binding proteins (74–76). The EF1a protein sequence shows several putative RGG motifs in the N-terminal or middle domain (77), providing the structural feature for G-quadruplex binding.
Guanine in DNA or RNA is a commonly studied target of oxidative stress, since hydroxyl radicals attack the eighth position of the purine ring, resulting in 8-oxoguanine formation (78). Guanine oxidation does not appear to alter the rate of G-quadruplex formation (79). Consistent with this finding, our studies using the naked RNA oligonucleotide did not reveal an effect of H2O2 with or without Fe2+ on G-quadruplex formation in solution. On the other hand, G residues in a G-quadruplex appear to be more likely oxidized than G residues in duplex DNA (80, 81). 8-Oxoguanine in telomeric DNA influences the binding and activity of telomerase (82, 83). In addition, 8-oxoguanine enhances the binding of helicase and DNA repair enzymes to telomeric DNA (84, 85). This points to the possibility that the oxidation of the NRF2 5′-UTR G-quadruplex facilitates its binding to EF1a.
In summary, we have found a novel mechanism of NRF2 induction via de novo protein translation. The NRF2 5′ UTR contains a G-quadruplex structure important for EF1a binding and NRF2 protein translation initiation under conditions of oxidative stress. What signals the increased binding of EF1a to the NRF2 5′-UTR G-quadruplex under conditions of oxidative stress and how the EF1a interaction with the G-quadruplex triggers the recruitment of translational machinery remain to be determined.
MATERIALS AND METHODS
Cell culture and H2O2 treatment.
HEK293 cells or HeLa cells were obtained from the American Type Culture Collection and were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 10 U/ml penicillin, and 10 μg/ml streptomycin. Cells were subcultured weekly and seeded into 100-mm dishes or 6-well plates for treatment with H2O2 upon reaching 80% confluence. Prior to H2O2 treatment, cells were placed into DMEM with 0.5% FBS for 16 to 24 h for serum starvation. Cells were typically treated with H2O2 for 10 min, followed by replacement with fresh DMEM containing 0.5% FBS before harvesting at 1 h or the indicated times.
Western blotting.
Cells were washed with ice-cold phosphate-buffered saline (PBS) and subsequently lysed in radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 140 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 10 mM Tris [pH 8.0]) containing freshly added protease inhibitors (Sigma-Aldrich, St. Louis, MO). Following protein concentration measurements by a bicinchoninic acid assay (Pierce, Thermo Scientific, Carlsbad, CA), an equal amount of proteins between samples was loaded onto 10% SDS-PAGE gels for electrophoresis using a Bio-Rad minigel system (Bio-Rad, Hercules, CA). Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane for blotting with antibodies against Nrf2 (EP1808Y monoclonal [Abcam, Cambridge, MA] and H-300 polyclonal [Santa Cruz Biotechnology, CA]), vinculin (ab11194 monoclonal; Abcam, Cambridge, MA), or EF1a (H-300, catalog number sc-28578; Santa Cruz Biotechnology, CA). Secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology, CA) were used for enhanced chemiluminescence reactions.
RT-PCR.
Total RNAs were collected from cells by using TRIzol and ethanol precipitation. Following reverse transcription using a commercial cDNA synthesis kit (Fermentas) with random hexamers and 1 μg of total RNA, PCR was carried out by using the oligonucleotide primer pairs for Nrf2 (forward primer 5′-CAGGTTGCCCACATTCCCAAATCA-3′ and reverse primer 5′-AGCAATGAAGACTGGGCTCTCGAT-3′), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward primer 5′-CGTCTTCACCATGGAGA-3′ and reverse primer 5′-CGGCCATCACGCCACAGTTT-3′), β-actin (forward primer 5′-AATGTGGCCGAGGACTTTGAT-3′ and reverse primer 5′-AGGATGGCAAGGGACTTCCTG-3′), and 18S rRNA (forward primer 5′-TCAACTTTCGATGGTAGTCGCCGT-3′ and reverse primer 5′-TCCTTGGATGTGGTAGCCGTTTCT-3′). The primer sets were designed with an online tool provided by IDTDNA and synthesized by Signosis. Real-time RT-PCR was performed by using a CFX-96 thermal cycler (Bio-Rad, Hercules, CA) and SYBR green dye (TaKaRa Bio USA, Mountain View, CA) with initial denaturation at 95°C for 10 min and 40 cycles of denaturation at 95°C for 15 s, primer annealing at 60°C for 30 s, and extension at 72°C for 30 s. Melting-curve analysis was performed at the end of PCR to verify the specificity of the product. Bio-Rad CFX Manager software was used for data analyses.
Isolation of polysomal RNA and ribosomal fractionation.
Cells were treated with 5 μg/ml cycloheximide for 10 min before harvesting in ribosomal lysis buffer (20 mM Tris-HCl [pH 7.4], 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.1 mg/ml cycloheximide, 0.1 M sucrose, 0.6% Triton X-100). Cell lysates were centrifuged at 400 × g for 30 min to remove the nuclei and cell debris. The supernatant from cell lysates was transferred to a clean Eppendorf tube and centrifuged at 18,000 × g for 10 min to remove mitochondria. The postmitochondrial supernatant was loaded onto 3 ml 10% sucrose layered over 6 ml 35% sucrose in ribosomal lysis buffer for 4 h of centrifugation using an SW41 rotor (Beckman) at 4°C and at 240,000 × g to pellet polysomes according to methods described previously by Hresko and Mueckler (86). This method of polysome preparation was validated by examining the pellets using electron microscopy. The polysomal pellets were resuspended in TRIzol (Invitrogen) for RNA isolation. An equal amount of RNA (100 ng) from each sample was used for reverse transcription and real-time PCR.
For fractionation of ribosomes, the postmitochondrial supernatant was loaded onto a linear sucrose gradient (15 to 50%, wt/vol) cushion and centrifuged at 200,000 × g with an SW40 rotor for 1.5 h at 4°C. The gradient was displaced with 60% sucrose for fractionation with monitoring of the absorbance at 254 nm with a BioLogic LC system (Bio-Rad, Hercules, CA) at a flow rate of 0.5 ml/min. The proteins in each fraction were precipitated by trichloroacetic acid and acetone and resuspended in SDS loading buffer for Western blot analyses.
Cloning and transfection of NRF2 5′-UTR reporter plasmids.
The 5′ UTR of the human NRF2 gene (NCBI RefSeq accession number NM_006164.4) (5′ UTR of 555 nt) was cloned by PCR using cDNAs from HeLa cells as a template. The sequence was inserted between SpeI and NcoI restriction sites in the pRF dicistronic vector upstream of firefly luciferase (32) to generate the pRL-NRF2 5′UTR-FL reporter construct. The mutant NRF2 5′-UTR sequence (pNRF2-5′UTR-A4) was generated by using the full-length wild-type 5′ UTR as a template with a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). Forward primer 5′-GACTCTTGCCCCGCCCTTGTTTTTCGGGAGGCGGAGCGGG and reverse primer 5′-CCCGCTCCGCCTCCCGAAAAACAAGGGCGGGGCAAGAGTC (125 ng) were used for PCR, followed by digestion with the DpnI restriction enzyme to eliminate nonmutated double-strand DNA before transformation in XL10 competent cells and selection for positive clones. Mutations were confirmed by DNA sequencing.
The pRL-NRF2 5′UTR-FL reporter construct (300 ng) was transfected into 30% confluent cells seeded into 24-well plates (Greiner Bio-One, Monroe, NC) by using Fugene 6 (Roche Applied Science) in serum-free medium for 5 h. The medium was changed to fresh DMEM with 10% FBS for 16 h before 18 to 24 h of serum starvation with 0.5% FBS. Approximately 48 h after transfection, cells were treated with H2O2 for 10 min and harvested 1 h later for measurements of dual luciferase activity.
In vitro transcription.
A DNA oligonucleotide with the complementary sequence from nt −198 to −168 of the NRF2 5′ UTR containing an upstream T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′) was synthesized. This DNA fragment was used as a template for 16 h of in vitro transcription at 37°C using a MEGAscript T7 kit (Ambion, San Diego, CA). Newly transcribed RNA was separated from the template oligonucleotide by 16% native polyacrylamide gel electrophoresis, excised, and eluted at 37°C for 1 h before ethanol precipitation and resuspension in nuclease-free water.
CD spectroscopy.
Newly synthesized 31-mer RNA oligonucleotides were diluted to 5 μM in 50 mM Tris-HCl (pH 7.4). After heating at 95°C for 10 min, the solution was cooled gradually to room temperature. CD spectra were recorded on a Jasco (Easton, MD) 810 spectropolarimeter at room temperature, using a quartz cell with a 1-mm optical path and a scanning speed of 100 nm/min with a response time of 1 s. The spectral contribution of buffers was subtracted where appropriate by using the software installed in the spectrometer.
Nuclear magnetic resonance analysis.
NMR experiments were performed on a Bruker DRX-600 spectrometer as described previously (87). Samples in water were prepared in a solution containing 10% D2O–90% H2O. The final NMR samples contained 0.2 mM RNA with the sequence of the NRF2 5′ UTR from the region spanning nt −198 to −168 in 25 mM K-phosphate buffer (pH 7.0) and 70 mM KCl. Before loading onto the instrument, samples were annealed by heating to 95°C for 10 min and then cooled to room temperature. The one-dimensional (1-D) 1H NMR spectra were recorded at 25°C with a Watergate pulse sequence to suppress the water signal. A spectral width of 25 ppm and 512 scans were collected. In each scan, 16,000 data points were used.
Dimethylsulfate footprinting.
DMS footprinting was performed with a 31-mer single-stranded DNA oligonucleotide corresponding to nt −198 to −168 of the NRF2 5′ UTR according to methods described previously by Sun and Hurley (33), with minor modifications. Briefly, after heating in a buffer (10 mM Tris [pH 7.5], 50 mM sodium chloride, and 1.0 mM EDTA) at 95°C for 10 min then cooling down to room temperature, the oligonucleotide was labeled with [γ-32P]ATP at the 5′ end and purified by alcohol precipitation. The DNA was treated with DMS (1%, vol/vol) for 2 min, and the methylation reaction was terminated by the addition of gel loading buffer. After electrophoresis on a 16% native polyacrylamide gel for 3.5 h, the DNA band was visualized via autoradiography and excised for elution. After elution in a buffer (200 mM NaCl, 2 mM KCl) for 1 h at 65°C, the DNA was precipitated, washed with 75% ethanol, and resuspended in 20 μl of nuclease-free water for subsequent treatment with 19% DMS for 1 min, before methylation was stopped by using a solution containing 30% (vol/vol) β-mercaptoethanol and 0.3 M sodium acetate and incubation at −20°C for 2 h. The DMS-treated DNA was precipitated, washed with 75% ethanol, air dried, treated with 10% piperidine for 5 min at 85°C, and vacuum dried. The pellets were then subjected to denaturation at 90°C for 5 min before 3 to 4 h of electrophoresis on a sequencing gel.
Isolation of RNA binding proteins.
The DNA template with the sequence complementary to nt −198 to −168 of the NRF2 5′ UTR containing an upstream T7 promoter was used as a template for 16 h of in vitro transcription in the presence of biotin-11-UTP (Invitrogen). For the A4 mutant sequence, the GGGG sequence in the region spanning nt −195 to −191 was replaced with TTTT in the template. RNA affinity chromatography was performed to pull down proteins, as described previously (17, 88). Briefly, HeLa cells were harvested in nucleic acid binding (NAB) buffer (10 mM HEPES [pH 7.6], 5 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol, 5% glycerol, 5 mg/ml heparin, 1 U/ml RNasin), followed by 3 brief sonications (5 s each) on ice. The cell debris and nuclei were then removed by centrifugation at 14,000 rpm at 4°C, and the supernatant (cytoplasmic extracts) was collected and quantified for subsequent RNA binding. Biotinylated RNA probes (5 μg) were incubated with cytoplasmic proteins (500 μg) for 1 h on ice, followed by incubation with 0.2 ml of streptavidin-Sepharose beads (GE Healthcare) at 4°C overnight with gentle shaking. The beads were then loaded onto a 2-ml centrifugation column (Pierce) and washed three times with 2 ml of 1 M NaCl in NAB buffer to remove nonspecific binding. The RNA binding proteins were released by boiling in SDS-PAGE loading buffer, separated by SDS-PAGE, and identified by mass spectrometry-compatible silver staining (Bio-Rad). Protein bands showing differential binding between the control and H2O2 treatments were excised for LC-MS/MS analyses as described previously (17).
EF1a protein expression and purification.
Human cDNA prepared from HeLa cells was used as the template for generating EF1a cDNA using PCR. The forward primer 5′-TCTGTCGACGGAAAGGAAAAGACTCATATC contained a SalI restriction site (underlined) and intentionally removed the ATG codon of the gene. The reverse primer 5′-GCAAGCTTTCATTTAGCCTTCTGAGCTTTCT had a HindIII site (underlined) and did not include the original stop codon. The PCR products were digested with SalI and HindIII, followed by gel purification. An internal HindIII site in EF1a cDNA produced a truncated version of EF1a without the C terminus. The purified PCR products were ligated with the SalI/HindIII-linearized pEcoli Nterm 6XHN vector (Clontech Laboratories, Mountain View, CA). The plasmid was introduced into Escherichia coli BL21(DE3) cells (Invitrogen, Thermo Fisher, Carlsbad, CA). To express the recombinant protein, 100 ml of LB medium containing ampicillin was inoculated with transformed BL21(DE3) cells and cultured with shaking at 37°C. When the culture reached an optical density at 600 nm (OD600) of 0.6, isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to the culture to a final concentration of 1 mM. The culture remained at 37°C for 6 h with shaking before harvesting.
To purify the recombinant EF1a protein, bacterial cells were pelleted and resuspended in 10 ml PBS containing 2 mM EDTA before the addition of lysozyme and DNase I (1 mg/ml each). The cells were lysed at 37°C for 30 min before sonication 3 times for 10 s each on ice. Soluble proteins were obtained by centrifugation at 20,000 × g and were incubated with 1 ml of PBS-washed Ni-nitrilotriacetic acid (Ni-NTA)–agarose (Qiagen) at room temperature for 2 h in the presence of 20 mM imidazole. After incubation, the agarose was loaded onto a minicolumn (10-ml bed volume) and washed with 20 ml PBS containing 20 mM imidazole. The protein was then eluted with 1 ml of PBS containing 200 mM imidazole. The purity of the protein was analyzed by 10% SDS-PAGE followed by BioSafe Coomassie blue staining (Bio-Rad, Hercules, CA).
Electrophoretic mobility shift assay.
Purified full-length or C-terminally truncated EF1a proteins (100 ng) were incubated with 5′-γ-32P-labeled 31-mer RNA containing the wild type or the G-quadruplex-eliminated mutant for in vitro binding. The RNA probe (1 × 106 cpm) was generated by in vitro transcription using DNA oligonucleotides containing the T7 promoter sequence in the presence of [γ-32P]ATP and was incubated with full-length or truncated EF1a in NAB buffer for 30 min on ice. The bound probe was resolved by 5% polyacrylamide gel electrophoresis at 60 V for 1 h for autoradiography.
Far-Western blotting.
Biotinylated 31-mer RNA probes were generated by in vitro transcription in the presence of biotin-11-UTP. After washing of streptavidin-Sepharose beads (50 μl per reaction; GE Healthcare Life Sciences, Marlborough, MA) with NAB buffer containing freshly added 2 mM DTT, the beads were resuspended in 0.2 ml of NAB buffer for incubation with the biotinylated RNA probe (1 μg/reaction) in the presence of 1 μl RNase inhibitors (RNase inhibitor mixture; Sigma-Aldrich, St. Louis, MO) at 4°C for 4 h. Aliquots of the mixture were incubated with cytoplasmic extracts containing 500 μg of proteins at 4°C for 4 h. After washing 2 times with NAB buffer and an additional 2 times with NAB buffer containing 1 M NaCl, the beads were resuspended in 50 μl of NAB buffer before the addition of SDS-PAGE buffer for boiling and Western blotting using an EF1a antibody (H-300; Santa Cruz Biotechnology, CA). The input as the loading control was EF1a protein detected by Western blotting using total cell lysates.
Immunoprecipitation of the RNA binding protein complex.
Endogenous RNA-protein interactions were measured and presented according to methods described previously by Lal et al. (89), with modifications. To isolate RNA bound with EF1a protein, 2 μg of anti-EF1a antibody (H-300; Santa Cruz Biotechnology, CA) or rabbit IgG (catalog number sc-2027; Santa Cruz Biotechnology, CA) was first incubated with 100 μl of protein A/G Plus beads (catalog number sc-2003; Santa Cruz Biotechnology, CA) for 1 h at 25°C (17). The unbound antibodies were removed by washing the beads 3 times with NAB buffer, followed by incubation with 500 μg HEK293 cytoplasmic extracts at 4°C for 4 h. The unbound proteins were removed by 5 washes with NAB buffer. TRIzol (Invitrogen, Carlsbad, CA) was added to the washed beads for extraction of RNA, which was precipitated in the presence of glycogen (0.5 μg/ml) before being converted to cDNA with Moloney murine leukemia virus (MMLV) reverse transcriptase (Fermentas, Hanover, MD) and an oligo(dT) primer. For quantitative measurements of NRF2 mRNA, the cDNA was used as a template for quantitative PCR (qPCR) using a Bio-Rad CFX96 thermocycler with the primer pair 5′-CAGGTTGCCCACATTCCCAAATCA-3′ and 5′-AGCAATGAAGACTGGGCTCTCGAT-3′ for human NRF2 (NCBI RefSeq accession number NM_006164.4). The primer set had been validated by examination of the melting curve of the product and linearity within a certain dynamic range. The abundance of NRF2 cDNA was analyzed with iQ SYBR green supermix (Bio-Rad) and Bio-Rad CFX Manager software, and the signal from the EF1a immunocomplex was normalized to that of IgG immunoprecipitates.
siRNA transfection.
siRNA against EF1a or negative-control siRNA was obtained from Santa Cruz Biotechnology (catalog numbers sc-77231 and sc-37007, respectively; Santa Cruz, CA). Transfection of siRNA was performed by using Xtreme siRNA transfection reagent (Sigma-Aldrich, MO) according to the manufacturer's protocol in 70% confluent HEK293 cells. The transfection reagent and siRNA at a 1:1 ratio were diluted in serum-free medium and incubated at room temperature for 20 min before they were added to cells. Sixteen hours after transfection, the medium was changed to regular 10% FBS–DMEM. Forty-eight hours after transfection with 24 h of serum starvation in 0.5% FBS–DMEM, cells were treated with H2O2 and harvested for Western blot or luciferase activity assays.
Two-dimensional SDS-PAGE for Western blotting.
Two-dimensional SDS-PAGE was performed by using the Zoom IPGRunner system according to the manufacturer's instructions (Invitrogen). Briefly, 100 μg of cytoplasmic proteins was solubilized in 150 μl 2-D buffer {8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.5% vol/vol Zoom carrier ampholytes, 20 mM DTT, 0.002% bromophenol blue}. Each protein sample (150 μl) was added to one lane of the Zoom IPGRunner cassette with a pH3 to 10 nonlinear (pH3-10NL) strip inserted into each well. The strips were focused according to the manufacturer's recommendations (Invitrogen). The strips were equilibrated in buffer A (106 mM Tris HCl, 141 mM Tris, 2% SDS, 10% glycerol, 0.51 mM EDTA, 50 mM DTT, 0.22 mM bromophenol blue) for 15 min, transferred to buffer B (same as buffer A except that 50 mM DTT was replaced with 125 mM iodoacetamide), and then washed once with ultrapure water. The strips were sealed onto a 10% SDS-polyacrylamide gel with 0.5% agarose. The proteins were separated at 60 V for 150 min for Western blotting as described above.
Statistics.
Means were compared by 2-tailed Student's t test when two samples were compared or by one-way analysis of variance (ANOVA) when multiple groups of data were compared.
ACKNOWLEDGMENTS
The pRL construct containing both renilla and firefly luciferase reporter genes was a gift from Jian-Ting Zhang, Indiana University. We appreciate the technical assistance of Elena Sheveleva for establishing the protocol for the isolation of ribosomes. We acknowledge George Tsaprailis at the Proteomic Core facility of the University of Arizona for proteomics work.
This work was supported by National Institutes of Health grants T32 ES007091 (K.K. and J.S.) and R01 HL089958, R21ES017473, and R01 GM 111337 (Q.M.C.).
REFERENCES
- 1.Nguyen T, Sherratt PJ, Pickett CB. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol 43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 2.Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. 2005. Nrf2, a multi-organ protector? FASEB J 19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 3.Kensler TW, Wakabayashi N, Biswal S. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 4.Ma Q, He X. 2012. Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol Rev 64:1055–1081. doi: 10.1124/pr.110.004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. 2003. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem 278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 6.Purdom-Dickinson S, Lin Y, Dedek M, Johnson J, Chen Q. 2007. Induction of antioxidant and detoxification response by oxidants in cardiomyocytes: evidence from gene expression profiling and activation of the Nrf2 transcription factor. J Mol Cell Cardiol 42:159–176. doi: 10.1016/j.yjmcc.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. 2007. Translational control of Nrf2 protein in activation of antioxidant response element by oxidants. Mol Pharmacol 72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 8.Xu B, Zhang J, Strom J, Lee S, Chen QM. 2014. Myocardial ischemic reperfusion induces de novo Nrf2 protein translation. Biochim Biophys Acta 1842:1638–1647. doi: 10.1016/j.bbadis.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho HY, Kleeberger SR. 2010. Nrf2 protects against airway disorders. Toxicol Appl Pharmacol 244:43–56. doi: 10.1016/j.taap.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Klaassen CD, Reisman SA. 2010. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol 244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Ichikawa T, Janicki JS, Cui T. 2009. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin Ther Targets 13:785–794. doi: 10.1517/14728220903025762. [DOI] [PubMed] [Google Scholar]
- 12.Hayes JD, McMahon M. 2009. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Mitsuishi Y, Motohashi H, Yamamoto M. 2012. The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol 2:200. doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canning P, Sorrell FJ, Bullock AN. 2015. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med 88:101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harder B, Jiang T, Wu T, Tao S, Rojo de la Vega M, Tian W, Chapman E, Zhang DD. 2015. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem Soc Trans 43:680–686. doi: 10.1042/BST20150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal AK. 2004. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med 36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Dinh T, Kapperler K, Chen Q. 2012. La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol Cell Proteomics 11:M111.015032. doi: 10.1074/mcp.M111.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preiss T, Hentze MW. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 19.Babitzke P, Baker CS, Romeo T. 2009. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol 63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holcik M, Sonenberg N. 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 22.Spriggs KA, Bushell M, Mitchell SA, Willis AE. 2005. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ 12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki S, Anderson P. 2008. Reprogramming mRNA translation during stress. Curr Opin Cell Biol 20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komar AA, Mazumder B, Merrick WC. 2012. A new framework for understanding IRES-mediated translation. Gene 502:75–86. doi: 10.1016/j.gene.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faye MD, Holcik M. 2015. The role of IRES trans-acting factors in carcinogenesis. Biochim Biophys Acta 1849:887–897. doi: 10.1016/j.bbagrm.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Bochman ML, Paeschke K, Zakian VA. 2012. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genetics 13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huppert JL. 2010. Structure, location and interactions of G-quadruplexes. FEBS J 277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 28.Lane AN, Chaires JB, Gray RD, Trent JO. 2008. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res 36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bugaut A, Balasubramanian S. 2012. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res 40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanian S, Hurley LH, Neidle S. 2011. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang DH, Fujimoto T, Saxena S, Yu HQ, Miyoshi D, Sugimoto N. 2010. Monomorphic RNA G-quadruplex and polymorphic DNA G-quadruplex structures responding to cellular environmental factors. Biochemistry 49:4554–4563. doi: 10.1021/bi1002822. [DOI] [PubMed] [Google Scholar]
- 32.Han B, Zhang JT. 2002. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol Cell Biol 22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D, Hurley LH. 2010. Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods Mol Biol 608:65–79. doi: 10.1007/978-1-59745-363-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holcik M, Sonenberg N, Korneluk RG. 2000. Internal ribosome initiation of translation and the control of cell death. Trends Genet 16:469–473. doi: 10.1016/S0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- 35.Dreyfuss G, Kim VN, Kataoka N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 36.Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konig J, Zarnack K, Luscombe NM, Ule J. 2011. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet 13:77–83. doi: 10.1038/ni.2154. [DOI] [PubMed] [Google Scholar]
- 38.Mateyak MK, Kinzy TG. 2010. eEF1A: thinking outside the ribosome. J Biol Chem 285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan G, Zhang Q, Zhou J, Li H. 2011. Mass spectrometry of G-quadruplex DNA: formation, recognition, property, conversion, and conformation. Mass Spectrom Rev 30:1121–1142. doi: 10.1002/mas.20315. [DOI] [PubMed] [Google Scholar]
- 40.Waller ZA, Howell LA, Macdonald CJ, O'Connell MA, Searcey M. 2014. Identification and characterisation of a G-quadruplex forming sequence in the promoter region of nuclear factor (erythroid-derived 2)-like 2 (Nrf2). Biochem Biophys Res Commun 447:128–132. doi: 10.1016/j.bbrc.2014.03.117. [DOI] [PubMed] [Google Scholar]
- 41.Wang SK, Wu Y, Ou TM. 2015. RNA G-quadruplex: the new potential targets for therapy. Curr Top Med Chem 15:1947–1956. doi: 10.2174/1568026615666150515145733. [DOI] [PubMed] [Google Scholar]
- 42.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. 2010. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc Natl Acad Sci U S A 107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melko M, Bardoni B. 2010. The role of G-quadruplex in RNA metabolism: involvement of FMRP and FMR2P. Biochimie 92:919–926. doi: 10.1016/j.biochi.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 44.Samatanga B, Dominguez C, Jelesarov I, Allain FH. 2013. The high kinetic stability of a G-quadruplex limits hnRNP F qRRM3 binding to G-tract RNA. Nucleic Acids Res 41:2505–2516. doi: 10.1093/nar/gks1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endoh T, Sugimoto N. 2013. Unusual −1 ribosomal frameshift caused by stable RNA G-quadruplex in open reading frame. Anal Chem 85:11435–11439. doi: 10.1021/ac402497x. [DOI] [PubMed] [Google Scholar]
- 46.Yu CH, Teulade-Fichou MP, Olsthoorn RC. 2014. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res 42:1887–1892. doi: 10.1093/nar/gkt1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. 2011. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep 12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaudoin JD, Perreault JP. 2013. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res 41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefanovic S, Bassell GJ, Mihailescu MR. 2015. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding site accessibility. RNA 21:48–60. doi: 10.1261/rna.046722.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arora A, Suess B. 2011. An RNA G-quadruplex in the 3′ UTR of the proto-oncogene PIM1 represses translation. RNA Biol 8:802–805. doi: 10.4161/rna.8.5.16038. [DOI] [PubMed] [Google Scholar]
- 51.Decorsiere A, Cayrel A, Vagner S, Millevoi S. 2011. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev 25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. 2007. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol 3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. 2008. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA 14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahid R, Bugaut A, Balasubramanian S. 2010. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry 49:8300–8306. doi: 10.1021/bi100957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balkwill GD, Derecka K, Garner TP, Hodgman C, Flint AP, Searle MS. 2009. Repression of translation of human estrogen receptor alpha by G-quadruplex formation. Biochemistry 48:11487–11495. doi: 10.1021/bi901420k. [DOI] [PubMed] [Google Scholar]
- 56.Weng HY, Huang HL, Zhao PP, Zhou H, Qu LH. 2012. Translational repression of cyclin D3 by a stable G-quadruplex in its 5′ UTR: implications for cell cycle regulation. RNA Biol 9:1099–1109. doi: 10.4161/rna.21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris MJ, Basu S. 2009. An unusually stable G-quadruplex within the 5′-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry 48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 58.Agarwala P, Pandey S, Maiti S. 2014. Role of G-quadruplex located at 5′ end of mRNAs. Biochim Biophys Acta 1840:3503–3510. doi: 10.1016/j.bbagen.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 59.Beaudoin JD, Perreault JP. 2010. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res 38:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, Wan M, Giri B, Lei M, Nagamine Y, Vaughn JP, Akman SA, Sui G. 2012. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res 40:1033–1049. doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. 2003. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem 278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. 2010. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc 132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 63.Cammas A, Dubrac A, Morel B, Lamaa A, Touriol C, Teulade-Fichou MP, Prats H, Millevoi S. 2015. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biol 12:320–329. doi: 10.1080/15476286.2015.1017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agarwala P, Pandey S, Mapa K, Maiti S. 2013. The G-quadruplex augments translation in the 5′ untranslated region of transforming growth factor 2. Biochemistry 52:1528–1538. doi: 10.1021/bi301365g. [DOI] [PubMed] [Google Scholar]
- 65.Bhattacharyya D, Diamond P, Basu S. 2015. An Independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry 54:1879–1885. doi: 10.1021/acs.biochem.5b00091. [DOI] [PubMed] [Google Scholar]
- 66.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, Schatz JH, Rodrigo CM, Zhao C, Rondou P, de Stanchina E, Teruya-Feldstein J, Kelliher MA, Speleman F, Porco JA Jr, Pelletier J, Ratsch G, Wendel HG. 2014. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Condeelis J. 1995. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem Sci 20:169–170. doi: 10.1016/S0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- 68.Kou YH, Chou SM, Wang YM, Chang YT, Huang SY, Jung MY, Huang YH, Chen MR, Chang MF, Chang SC. 2006. Hepatitis C virus NS4A inhibits cap-dependent and the viral IRES-mediated translation through interacting with eukaryotic elongation factor 1A. J Biomed Sci 13:861–874. doi: 10.1007/s11373-006-9104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeenko VV, Ryabova LA, Spirin AS, Rothnie HM, Hess D, Browning KS, Hohn T. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J Virol 76:5678–5691. doi: 10.1128/JVI.76.11.5678-5691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gross SR, Kinzy TG. 2005. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol 12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 71.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. 2005. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol 25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamberti A, Sanges C, Chambery A, Migliaccio N, Rosso F, Di Maro A, Papale F, Marra M, Parente A, Caraglia M, Abbruzzese A, Arcari P. 2011. Analysis of interaction partners for eukaryotic translation elongation factor 1A M-domain by functional proteomics. Biochimie 93:1738–1746. doi: 10.1016/j.biochi.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB, Patel DJ. 2011. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol 18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahama K, Oyoshi T. 2013. Specific binding of modified RGG domain in TLS/FUS to G-quadruplex RNA: tyrosines in RGG domain recognize 2′-OH of the riboses of loops in G-quadruplex. J Am Chem Soc 135:18016–18019. doi: 10.1021/ja4086929. [DOI] [PubMed] [Google Scholar]
- 75.Vasilyev N, Polonskaia A, Darnell JC, Darnell RB, Patel DJ, Serganov A. 2015. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc Natl Acad Sci U S A 112:E5391–E5400. doi: 10.1073/pnas.1515737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thandapani P, Song J, Gandin V, Cai Y, Rouleau SG, Garant JM, Boisvert FM, Yu Z, Perreault JP, Topisirovic I, Richard S. 2015. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. eLife 4:e06234. doi: 10.7554/eLife.06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uetsuki T, Naito A, Nagata S, Kaziro Y. 1989. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J Biol Chem 264:5791–5798. [PubMed] [Google Scholar]
- 78.David SS, O'Shea VL, Kundu S. 2007. Base-excision repair of oxidative DNA damage. Nature 447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vorlickova M, Tomasko M, Sagi AJ, Bednarova K, Sagi J. 2012. 8-Oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J 279:29–39. doi: 10.1111/j.1742-4658.2011.08396.x. [DOI] [PubMed] [Google Scholar]
- 80.Delaney S, Barton JK. 2003. Charge transport in DNA duplex/quadruplex conjugates. Biochemistry 42:14159–14165. doi: 10.1021/bi0351965. [DOI] [PubMed] [Google Scholar]
- 81.Huang YC, Cheng AK, Yu HZ, Sen D. 2009. Charge conduction properties of a parallel-stranded DNA G-quadruplex: implications for chromosomal oxidative damage. Biochemistry 48:6794–6804. doi: 10.1021/bi9007484. [DOI] [PubMed] [Google Scholar]
- 82.Szalai VA, Singer MJ, Thorp HH. 2002. Site-specific probing of oxidative reactivity and telomerase function using 7,8-dihydro-8-oxoguanine in telomeric DNA. J Am Chem Soc 124:1625–1631. doi: 10.1021/ja0119651. [DOI] [PubMed] [Google Scholar]
- 83.Vialas C, Pratviel G, Meunier B. 2000. Oxidative damage generated by an oxo-metalloporphyrin onto the human telomeric sequence. Biochemistry 39:9514–9522. doi: 10.1021/bi000743x. [DOI] [PubMed] [Google Scholar]
- 84.Ghosh A, Rossi ML, Aulds J, Croteau D, Bohr VA. 2009. Telomeric D-loops containing 8-oxo-2′-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J Biol Chem 284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. 7 August 2013. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hresko RC, Mueckler M. 2002. Identification of pp68 as the tyrosine-phosphorylated form of SYNCRIP/NSAP1. A cytoplasmic RNA-binding protein. J Biol Chem 277:25233–25238. doi: 10.1074/jbc.M202556200. [DOI] [PubMed] [Google Scholar]
- 87.Dai J, Chen D, Jones RA, Hurley LH, Yang D. 2006. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res 34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A 101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]