Abstract
The hypersensitive response (HR) is a programmed cell death that is commonly associated with plant disease resistance. A novel lesion mimic mutant, vad1 (for vascular associated death1), that exhibits light conditional appearance of propagative HR-like lesions along the vascular system was identified. Lesion formation is associated with expression of defense genes, production of high levels of salicylic acid (SA), and increased resistance to virulent and avirulent strains of Pseudomonas syringae pv tomato. Analyses of the progeny from crosses between vad1 plants and either nahG transgenic plants, sid1, nonexpressor of PR1 (npr1), enhanced disease susceptibility1 (eds1), or non-race specific disease resistance1 (ndr1) mutants, revealed the vad1 cell death phenotype to be dependent on SA biosynthesis but NPR1 independent; in addition, both EDS1 and NDR1 are necessary for the proper timing and amplification of cell death as well as for increased resistance to Pseudomonas strains. VAD1 encodes a novel putative membrane-associated protein containing a GRAM domain, a lipid or protein binding signaling domain, and is expressed in response to pathogen infection at the vicinity of the hypersensitive lesions. VAD1 might thus represent a new potential function in cell death control associated with cells in the vicinity of vascular bundles.
INTRODUCTION
Programmed cell death (PCD) in plants occurs in a variety of cell types during development or in response to environmental stresses. Until now, the hypersensitive response (HR) has been the best studied example of PCD, probably because of its importance in agriculture and because this local and rapid cell death occurs in response to most pathogens and limits growth of the microorganism. Among the different approaches used to identify the molecular players of plant PCD, mutational analyses were particularly fruitful. A large number of mutants exhibiting spontaneous cell death were initially isolated in maize (Zea mays) (Hoisington et al., 1982) and classified as lesion mimics. They were then identified in several plants including rice (Oryza sativa) (Takahashi et al., 1999), barley (Hordeum vulgare) (Wolter et al., 1993), and Arabidopsis thaliana (Greenberg and Ausubel, 1993; Dietrich et al., 1994; Greenberg et al., 1994). Recently, studies have been focused on the characterization of novel lesion mimic mutants; although some of these show similar features as those previously described, others exhibit original traits, such as lesions mimicking disease or HR-like lesions and alteration of the HR. Analyses of these mutants/genes within the context of the signaling defense pathways has revealed cross talk between these pathways (Lorrain et al., 2003). These latter findings raise the question of the implication of such lesion mimic genes in normal response pathways triggered by pathogen infection, some of them probably being not directly associated with defense response but rather caused by physiological alterations. In addition, a dual role in pathogen defense or response to environmental stresses and plant development can be envisaged in some cases, suggesting the recruitment of basic components of plant physiology for response to pathogens.
Plant defenses, and among them the HR, are triggered by specific recognition of pathogen-derived molecules. Arabidopsis R (resistance) genes have been cloned that confer specific recognition to bacterial, viral, and fungal pathogens (Parker et al., 2000). After this recognition event, plant defense is regulated through a complex network of transduction pathways involving several signaling molecules: reactive oxygen species, nitric oxide, salicylic acid (SA), jasmonic acid (JA), and ethylene (Kunkel and Brooks, 2002). In addition to these, mutational analyses in Arabidopsis have uncovered genes acting as positive regulators that are required for resistance conferred by several R genes. Enhanced Disease Susceptibility1 (EDS1) and Non-Race Specific Disease Resistance1 (NDR1) are such regulators and are hypothesized to belong to two downstream pathways triggered by R genes, encoding two kinds of protein structures (Toll Interleukin1 Receptor-Nucleotide Binding-Leucine Rich Repeat or Coiled-Coil-Nucleotide-Binding-Leucine Rich Repeat, respectively) determining which downstream factors are required (Century et al., 1995; Parker et al., 1996; Aarts et al., 1998). Additional independent pathways also have been found (McDowell et al., 2000; Bittner-Eddy and Beynon, 2001). Subsequent recent discovery of two downstream resistance components, Required for Mla Dependent Resistance1 and Suppressor of G2 Allele of SKP1, introduced further complexities in our understanding of HR. These components are required for signaling by sets of resistance genes that overlap the boundaries between the pathways previously defined (Austin et al., 2002; Azevedo et al., 2002; Muskett et al., 2002; Tör et al., 2002; Tornero et al., 2002). In this context, the lesion mimic mutants, altered in disease resistance, regulation of cell death, and defense responses, could be affected in genes of general importance for signaling pathways. This idea is supported by several reports, including crosses between lesion mimic mutants and mutants affected in signaling pathways leading to resistance and/or cell death (Lorrain et al., 2003). For example, the lesion mimic phenotype of suppressor of salicylic acid insensitivity1 (ssi1), accelerated cell death5 (acd5), and acd11 is SA dependent and can be restored by application of SA or its functional analogs 2,6-dochloroisonicotinic acid/benzothiadiazol, suggesting that cell death and SA are involved in a feedback amplification loop and that the mutated genes could be components of such a pathway (Shah et al., 1999; Greenberg et al., 2000; Brodersen et al., 2002). Surprisingly, the nahG transgene, which encodes an SA-degrading salicylate hydroxylase, suppressed the constitutive expression of PDF1.2 in the ssi1 mutant, suggesting an SA-dependent regulation of PDF1.2 (Shah et al., 1999) and that SSI1 (Stokes and Richards, 2002) could be a component of early signaling pathways leading to the activation of defense mechanisms, or at a branch point between SA and JA/ethylene pathways. A new function for EDS1 in cell death amplification emerged from crosses with constitutive PR5 (cpr5) or lsd1 (for lesion simulating disease resistance1) (Clarke et al., 2001; Rusterucci et al., 2001). Indeed, lesions were reduced in a cpr5 eds1 double mutant, and propagative lesions in lsd1 are totally EDS1 dependent. This function is independent of R signaling functions because EDS1-dependent runaway cell death is effective whatever R gene mediated pathway is activated. Phytoalexin Deficient4 (PAD4) is also involved in the amplification of cell death in lsd1, whereas NDR1 contributes to the control of cell death to a less extent than EDS1 and PAD4.
To date, among the 37 lesion mimic mutants that have been identified, only five mutants show propagative lesions: the acd (for accelerated cell death) mutants acd1 and acd2 (Greenberg and Ausubel, 1993; Greenberg et al., 1994) and the lsd1 mutant (Dietrich et al., 1994), showing necrotic, HR-like lesions, and the disease like lesion1 (dll1) and acd11 mutants, exhibiting chlorotic, disease-like lesions (Brodersen et al., 2002; Pilloff et al., 2002). These mutants, unable to control the rate and extent of the lesions, are thought to be affected in genes controlling the suppression/limitation of PCD, whereas the other lesion mimic mutants, the so-called initiation mutants, would be altered in the initiation of the process (Walbot et al., 1983). Here, we report the characterization of a novel propagation mutant, vad1 (for vascular associated death1, previously named svn1; Lorrain et al., 2003), which displays necrotic HR-like lesions propagating along the vascular system and whose appearance is dependent on light intensity. The vad1 lesion phenotype is SA dependent but Nonexpressor of PR1 (NPR1) independent, and both EDS1 and NDR1 are necessary for the proper timing and amplification of cell death; interestingly, defense activation and the resulting enhanced disease resistance to virulent and avirulent bacterial pathogens in the mutant require both NDR1- and EDS1-mediated pathways. VAD1 was found to encode a putative membrane associated protein containing a GRAM domain.
RESULTS
Identification and Genetic Characterization of the vad1 Mutant
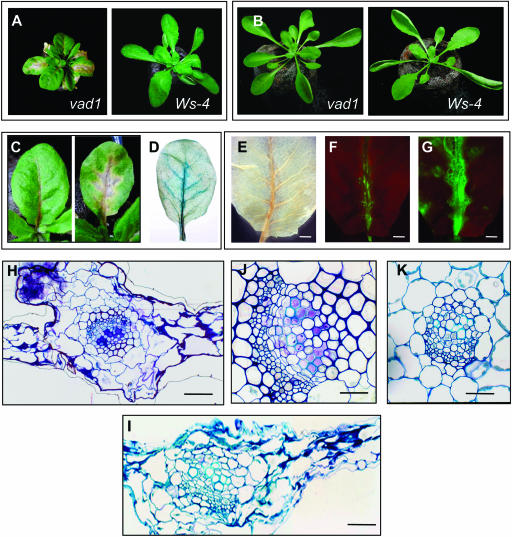
The vad1 mutant was identified by screening a population of Arabidopsis mutagenized by T-DNA insertion (Bechtold et al., 1993) for their ability to display spontaneous lesions on the leaves. Under normal growth conditions, the vad1 mutant displayed necrotic lesions that started at the petiole base and progressed upward along the midvein and then along the secondary veins, resulting in the necrosis of the whole leaf (Figures 1A and 1C). Newly emerging rosette leaves were not affected, only adult leaves formed lesions. The mutant phenotype appeared 19 to 22 d after transfer of seedlings to a growth chamber and was conditional, depending on light intensity. Lesions were suppressed under low light intensity (96 μE/m2/s; lesion− conditions) (Figure 1B). Under lesion-promoting conditions (normal growth conditions, 192 μE/m2/s), the vad1 plants were smaller than those of the wild type, with a more compact stature, and the leaves were smaller with shorter petioles (Figure 1A). These lesions occurred in vad1 plants in the absence of a pathogen and consistently appeared in all mutant progeny plants. In addition, another mutant allele (called vad1-2, the first one being called vad1-1), showed a similar phenotype (data not shown). Similar lesions were formed under sterile culture conditions, demonstrating that lesion formation in this mutant does not require an exogenous biotic trigger. Evans blue staining revealed the presence of areas of dead cells corresponding to the sites where there are macroscopic signs of lesions before staining (Figure 1D). Such staining was not found in wild-type leaves or in the mutant under low light intensity conditions (data not shown).
Figure 1.
Phenotype of the vad1 Mutant.
(A) and (B) Five-week-old plants of vad1 in comparison with the wild-type plant (Ws-4) (photographed at the same distance) grown under normal light intensity (A) and under low light conditions (B).
(C) Spontaneous lesion formation in vad1 plants. Leaves show lesions propagating along the vascular system starting at the petiole basis.
(D) Evans blue staining reveals regions of intensely stained dead cells along the vascular system of vad1 leaves.
(E) Observation of vad1 leaves after bleaching, under a stereomicroscope, showing the lesions along the vascular system. Bar = 2 mm.
(F) Observation of the same leaf as in (E) under a fluorescence microscope (Leica MZ FLIII) shows autofluorescence associated to the lesions observed in (E) (excitation filter 470/40 nm, barrier filter 515 nm). Bar = 2 mm.
(G) H2O2 staining of the same leaf using H2DCFDA reveals production of this molecule at the site of lesion formation (excitation filter 470/40 nm, barrier filter 515 nm). Bar = 2 mm.
(H) to (K) Microscopic analysis of leaf sections of vad1 under lesion-promoting conditions ([H] and [J]) and of the wild type (Ws-4), healthy (K) and after inoculation with an avirulent strain of Xanthomonas (Xcc147) (I). vad1 lesions (H) resemble Xcc147-induced HR (I). Xylem vessels in vad1 lesions are occluded by pink-stained material (J) not seen in healthy leaves (K). Bars = 12 μm for (J) and (K) and 6 μm for (H) and (I).
The vad1 mutation was isolated in the homozygous state. The original homozygous vad1/vad1 mutant was crossed to Wassilewskija-4 (Ws-4) wild type (VAD1/VAD1). None of the vad1/VAD1 F1 plants developed lesions. These plants were allowed to self-pollinate, and the segregation of the vad1 phenotype was monitored in the F2 generation: 73 out of 312 plants showed lesions. This closely approximates a 3:1 ratio (χ2 = 0.427; P = 0.6), indicating that the mutant phenotype was caused by a recessive mutation at a single locus.
The vad1 Mutant Exhibits Several Defense-Associated Responses
To examine whether vad1 plants, as some other lesion mimic mutants, express defense-associated responses under lesion promoting conditions, several histochemical, biochemical, and molecular markers usually associated with the hypersensitive cell death program were analyzed. Fluorescence microscopy revealed the accumulation of autofluorescent compounds (Figure 1F) in regions corresponding to areas of lesion formation (Figure 1E), a hallmark of plant defense responses that is also observed in some lesion mimic mutants. Because PCD and HR are often preceded by a transient burst of reactive oxygen species, production of H2O2 was visualized by staining with H2DCFDA (2′,7′-dichlorofluorescin diacetate) (Figure 1G) and with diaminobenzidine tetrahydrochloride (data not shown) in vad1 mutants showing early symptoms. The fluorescence produced by the H2DCFDA reaction was clearly visible and distinguishable from the autofluorescence observed in Figure 1F: it coincides with the propagation front of the lesion and is absent in mutant leaves without lesions and in the wild type (data not shown). Similar data were obtained with diaminobenzidine tetrahydrochloride staining (data not shown).
Histology of the lesion-positive leaves revealed that cell death was restricted predominantly to mesophyll cells neighboring the veins (Figure 1H). Dying cells showed a dark blue–stained coagulated cytoplasm, retracted from the cell wall, revealing a cell collapse similar to that observed during Xanthomonas-induced HR cell death in the wild type (Figure 1I). Interestingly, a feature typical of vad1 lesions was observed in certain xylem elements surrounded by the lesions; vessels were seen to be occluded by a pink-stained material (Figure 1J) that was never seen in tissues evolving an HR (Figure 1I) or in healthy wild-type tissues (Figure 1K). Ultrastructural observations of this occluding material revealed a fibrillar-like structure that positively immunoreacted with JIM 5 antipectin monoclonal antibody (data not shown), a phenomenon usually observed in defense responses during certain plant–pathogen interactions.
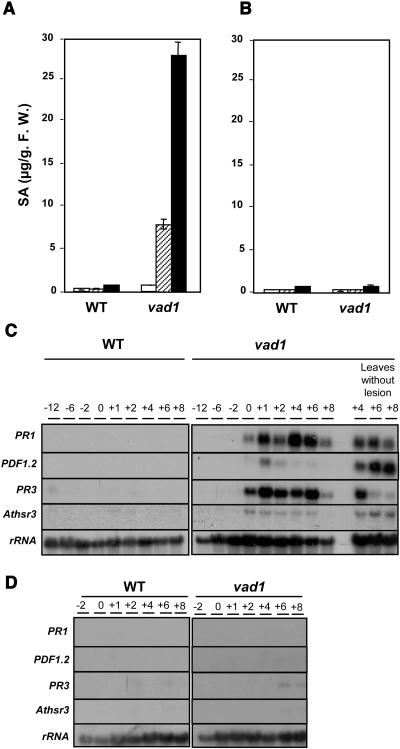
To further analyze the phenotype of the vad1 mutant, defense gene expression and production of SA were analyzed (Figure 2). SA, a key endogenous signaling molecule involved in disease resistance and the HR (Kunkel and Brooks, 2002), accumulates to a high level in several lesion mimic mutants (Lorrain et al., 2003). The levels of total SA were monitored in vad1 plants, in comparison with wild-type plants, at different times in relation to lesion formation. Under lesion-promoting conditions, vad1 plants accumulate 7- to 10-fold higher levels of SA than the wild-type plants when the lesions appeared, this accumulation being even higher (30-fold the wild type) 5 d after lesion appearance (Figure 2A). In the absence of lesions, vad1 showed similar levels of SA to the wild type (Figure 2B). In the same way, RNA was extracted from lesion-negative and lesion-positive plants, and RNA gel blots were analyzed with different defense gene probes: PR1, whose expression is dependent on SA and the SA signaling pathway; PR3 and PDF1-2, whose expression depends on the ethylene/jasmonate signaling pathway; and Athsr3 as a marker of the HR (Lacomme and Roby, 1999). In the mutant under lesion-promoting conditions, expression of these markers was induced, especially for PR1, PDF1-2, and PR3 (Figure 2C), whereas in wild-type plants and in mutant plants under lesion− conditions, the expression of these defense genes remains undetectable (Figure 2D). Athsr3 is expressed at a lower level but is clearly detectable in the mutant, whereas it is absent in the wild type or in the mutant under lesion− conditions. This result suggests that several defense pathways are activated in the mutant, including genes that are not regulated by SA. In addition, all these genes are also expressed in leaves without lesions from mutant plants harboring lesions, indicative of the activation of systemic acquired resistance in these plants.
Figure 2.
SA Levels and Defense Gene Expression in Wild-Type and vad1 Plants.
(A) and (B) Total SA levels in wild-type and vad1 plants. Leaves were harvested from plants grown on soil under lesion-promoting conditions (A) 10 d before lesion appearance (white bars), at the lesion appearance (hatched bars), and 5 d after the lesion appearance (black bars) and from plants grown under lesion− conditions at the same times (B). SA measurements and standard errors are derived from two replicates. F.W., fresh weight.
(C) and (D) Transcript levels of PR1 (Pathogenesis-Related 1), PDF1-2 (Plant Defensin 1-2), PR3, and Athsr3 (Arabidopsis thaliana Hypersensitivity-Related 3) in wild-type and vad1 plants at different times before, during, and after lesion appearance in leaves with or without lesions under lesion-promoting conditions (C). The same analysis was performed in wild-type and vad1 plants under lesion− conditions at the same times (D). Transcript levels were determined by gel blot analysis.
vad1 Displays Enhanced Resistance to Bacterial Pathogens
Because vad1 showed activation of the basal defense responses and of genes known to be involved in the regulation of cell death and defense, the effect of the mutation on resistance to pathogens was determined. Pathological tests were performed not only with virulent bacterial pathogens but also avirulent strains to determine whether the basal resistance and another plant defense response, the HR, were affected. This was of particular interest because vad1 is a propagation mutant, putatively affected in the control of the limitation of cell death (Walbot et al., 1983).
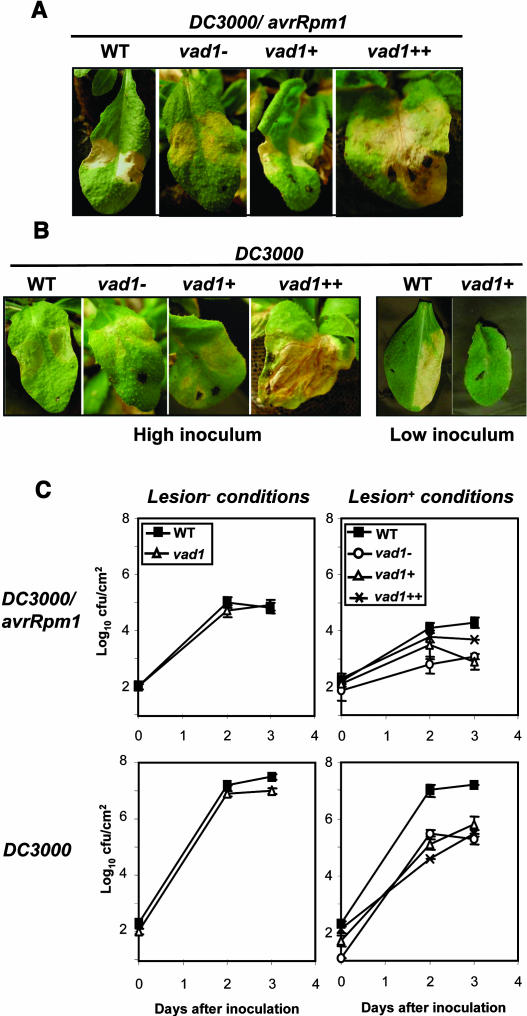
We first examined the responses of wild-type and vad1 plants grown under lesion− conditions. The phenotypes of the mutant plants in response to virulent and avirulent bacterial strains are similar to the wild type (data not shown). In the case of plants grown under lesion promoting conditions, a typical HR although slightly delayed, was observed in the mutant leaves that did not form lesions (vad1−) or develop lesions only on the half of the midvein at the inoculation time (vad1+) (Figure 3A). By contrast, the mutant leaves that display propagative lesions along the whole primary vein showed spreading cell death (runaway cell death [RCD]), as described for the lsd1 mutant (Dietrich et al., 1994) (Figure 3A, vad1++). In response to the virulent strain DC3000, lesion phenotypes of the mutant are a little less strong and slightly delayed at high inoculum, whereas very faint (or no) symptoms were observed in vad1+ and vad1− at low inoculum compared with those observed in the wild type (Figure 3B). Like the response observed to the avirulent strain, the mutant leaves that display propagative lesions showed RCD (Figure 3B, vad1++).
Figure 3.
Lesion Phenotypes and Bacterial Growth in Wild-Type and vad1 Plants after Pathogen Inoculation.
Leaves of 5-week-old wild-type and vad1 mutant plants grown under lesion-promoting conditions ([A] and [B]) were infiltrated on both sides of the leaf with suspensions (2.107 colony-forming units [cfu]/mL) of P. syringae pv tomato strain DC3000 expressing avrRpm1 (A) and DC3000 (B). Leaves were photographed 54 h after inoculation and were classified according to the propagation rate of the lesions before inoculation: no lesion (vad1−), presence of lesion only halfway up the primary vein (vad1+), and lesions propagating along the whole primary vein (vad1++). In the case of DC3000, inoculations with a lower inoculum (2.105 cfu/mL) were performed for a better observation of the disease phenotypes in the wild-type and vad1+ leaves. All treatments were repeated at least three times with similar results.
(C) Growth of P. syringae pv tomato DC3000 and DC3000 expressing avrRpm1 in wild-type and vad1 plants grown under lesion-promoting conditions or lesion− conditions. In the case of mutant plants grown under lesion-promoting conditions, bacterial growth was evaluated in the different leaves classified as previously described. Inoculation was performed with a bacterial suspension of 2.105 cfu/mL, and bacterial growth determinations were performed at the times indicated. Mean bacterial densities are shown (three to five replicates with corresponding standard deviations) for one representative experiment from two or three independent experiments performed for each strain.
These observations were confirmed by evaluating the bacterial growth in the mutant leaves, as compared with the wild-type plants (Figure 3C). No significant differences in resistance could be found under lesion− conditions (Figure 3C). Under lesion-promoting conditions and in response to the avirulent strain, evaluation of bacterial growth showed that resistance is increased (2- to 25-fold) compared with the wild type and that resistance in vad1++ leaves is comparable to that displayed by the other leaves of the mutant, indicating that the spreading lesions observed correspond to RCD and do not result from resistance loss and pathogen invasion (Figure 3C). In response to the virulent strain, in planta pathogen growth is significantly decreased in the mutant compared with the wild type (25- to 250-fold) (Figure 3C). Therefore, the vad1 mutation enhanced host resistance to virulent and avirulent strains of Pseudomonas syringae. This observation could be explained by the constitutive expression of defense genes in the mutant under lesion-promoting conditions; in addition, PR-1 gene expression in the mutant in response to pathogen attack is highly increased over the constitutive expression level, as compared with the wild type (data not shown).
Cell Death and Basal Disease Resistance in vad1 Is Dependent on SA Production
Because cell death in lesion mimic mutants may be SA dependent or SA independent (Lorrain et al., 2003), vad1 was crossed with a transgenic Arabidopsis line harboring the nahG gene, unable to accumulate SA (Ryals et al., 1996), to determine the role of this signaling molecule in vad1 phenotypes. Because nahG plants recently have been shown to display side effects from catechol production (van Wees and Glazebrook, 2003), vad1 sid1 double mutants were also generated. Finally, to understand the role of another component of SA pathway, NPR1, which is involved in SA signaling, we generated vad1 npr1 double mutants. With nahG, sid1, and npr1 mutations being in a Columbia-0 (Col-0) background, a backcross of vad1 (background Ws-4) to the wild-type Col-0 was performed as a control, showing that vad1-conferred cell death phenotype segregated as in a parental backcross and appeared with the same kinetics and intensity under the same conditions (data not shown).
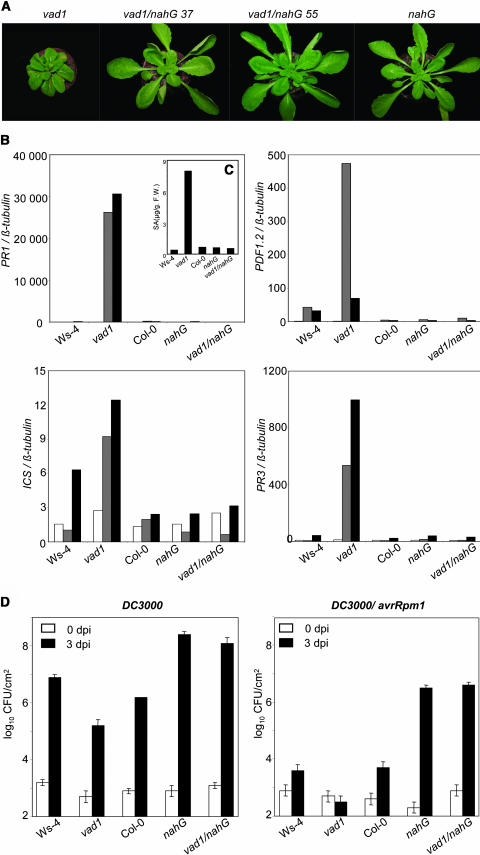
Under conditions where vad1 would normally form lesions, the double mutant vad1 nahG never exhibited lesions, as shown for two independent lines in Figure 4A. To quantify these observations, the expression of several defense genes, previously shown to be expressed in lesion-positive plants, was studied in the double mutant compared with the single ones and the wild type. As shown in Figure 4B, not only the activation of SA-dependent genes, such as PR-1 and Isochorismate Synthase (ICS), but also the activation of SA-independent genes, such as PDF1-2 and PR3, were suppressed in the double mutant. As a control, SA level was determined (Figure 4C) and shown to be abolished in the double mutant, as in the nahG line. So, in the double mutant that did not present SA accumulation, lesions and defense gene expression were abolished. The phenotypes of these different lines were then evaluated after inoculation with virulent (DC3000) and avirulent strains (DC3000 harboring avrRpm1) of P. syringae by measuring in planta bacterial growth (Figure 4D). Resistance was clearly compromised in vad nahG plants, which were as susceptible as nahG plants to both bacterial strains. Thus, the increased basal resistance observed in vad1 in response to both virulent and avirulent pathogens is dependent on SA.
Figure 4.
Lesion Phenotypes, Bacterial Growth, and Defense Gene Expression in Wild-Type, Single Mutant (nahG and vad1), and Double Mutant vad1 nahG Plants.
Two lines, vad1/nahG 37 and vad1/nahG 55, out of three lines are presented as examples.
(A) Five-week-old wild-type, single, or double mutant plants 7 d after lesion formation in vad1.
(B) Defense gene expression in wild-type, single, or double mutant plants. Transcript levels of PR-1, PR-3, and PDF1-2 were determined by quantitative PCR in plants grown under lesion-promoting conditions 8 d before (white bars), at day 0 (gray bars), and 7 d (black bars) after lesion formation in vad1. See Methods for further details. This experiment was repeated twice with different sets of plants, and similar results were obtained. F.W., fresh weight.
(C) Total SA levels in wild-type, single, or double mutant plants 7 d after lesion formation. The plant material used in (B) was also used for SA measurements.
(D) Bacterial growth in wild-type, single, or double mutant plants. Inoculation with P. syringae strain DC3000 and strain DC3000 expressing avrRpm1 was performed with a bacterial suspension of 2.105 cfu/mL, and bacterial growth determinations were performed at the times indicated. Mean bacterial densities are shown (three to five replicates with corresponding standard deviations) for one representative experiment from two or three independent experiments performed for each strain.
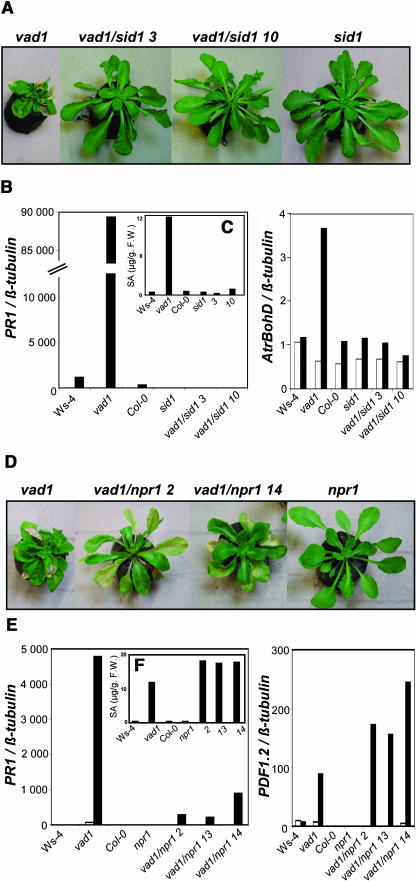
In the vad1 sid1 double mutants, the spontaneous lesions did not appear under conditions where vad1 would normally form lesions (Figure 5A). However, faint lesions that are less pervasive and that affect only one or two leaves per plant were occasionally observed in the double mutants at least 2 weeks later than in vad1. As for vad1 nahG double mutants, the expression of defense genes, such as PR1 (Figure 5B), PDF1-2, or PR-5 (data not shown), was suppressed, and SA levels were found very low (Figure 5C). AtrbohD, a gene encoding a major putative subunit of the NADPH oxidase complex (Keller et al., 1998; Torres et al., 2002), is more expressed in the mutant compared with the wild type, but this elevated expression is reduced to background levels in vad1 sid1 plants. Similar results were obtained for vad1 resistance to Pseudomonas strains (data not shown). These data demonstrate that similar results were obtained using sid1 or nahG plants as mutants impaired in SA biosynthesis and that the vad1 mutant phenotypes are dependent on SA accumulation.
Figure 5.
Lesion Phenotypes and Defense Gene Expression in Wild-Type, Single Mutant (sid1, npr1, and vad1), and Double Mutant vad1 sid1 and vad1 npr1 Plants.
Two lines, vad1 sid1 3 and vad1 sid1 10, out of nine lines, and vad1/npr1 1 and vad1/npr1 14, out of 16 lines, are presented as examples.
(A) and (D) Five-week-old single or double vad1 sid1 (A) and vad1 npr1 (D) mutant plants 10 d after lesion formation in vad1.
(C) and (F) Total SA levels in wild-type, single, or double mutant plants vad1 sid1 (C) and vad1 npr1 (F) after lesion formation in vad1. The plant material used in (B) or (E) was also used for SA measurements.
(B) and (E) Defense gene expression in wild-type, single, or double vad1 sid1 (B) and vad1 npr1 (E) mutant plants. Transcript levels of PR-1 and AtbohD were determined by quantitative PCR in plants grown under lesion-promoting conditions 9 d before (white bars) and 6 d (black bars) after lesion formation in vad1 (B). In (E), transcript levels of PR-1 and PDF1-2 were also determined by quantitative PCR in plants grown under lesion-promoting conditions 7 d before (white bars) and 5 d (black bars) after lesion formation in vad1. See Methods for further details. F.W., fresh weight.
The timing of appearance, frequency, and propagation of the lesions in the vad1 npr1 double mutants was very similar to the lesions observed in vad1 (Figure 5D), but the stature of the double mutants was intermediary between those of the parental lines. Expression of the PR1 gene (Figure 5E) was only partially affected in the double mutants, indicating that this gene in vad1 is regulated by NPR1-dependent and NPR1-independent pathways. PDF1-2 gene expression (Figure 5E) and SA levels (Figure 5F) are slightly enhanced in the double mutants, whereas AtrbohD gene expression is not significantly modified in the double mutant (data not shown). Thus, NPR1 does not seem to be required for the vad1-conferred cell death and is partially required for defense activation in vad1.
The vad1 Cell Death Phenotype Is Partially EDS1 and NDR1 Dependent, whereas the vad1 Resistance Phenotype Requires the Two Genes
Mutational analyses in Arabidopsis have demonstrated the existence of positive regulators required for resistance conferred by several resistance genes (Aarts et al., 1998). EDS1, a lipase-like protein, and NDR1, a putative membrane protein, whose roles already have been investigated in some lesion mimic mutants (Clarke et al., 2001; Rusterucci et al., 2001; Yoshioka et al., 2001), are components of these regulatory pathways. To determine whether these resistance components could influence the phenotypes of vad1, we constructed double mutant lines between eds1, ndr1, and vad1 and assessed first their effects on cell death phenotypes. We observed that vad1-induced lesion formation was delayed by 6 to 7 d in vad1 eds1 double mutant lines and by 3 to 4 d in vad1 ndr1 double mutants lines (Figures 6A and 6C ). As shown in Figure 6B, the delay in lesion formation is clear for the double mutant line vad1 eds1, whereas in the case of ndr1 (Figure 6D), this is a more quantitative effect. These observations were confirmed by a kinetic and quantitative evaluation of lesion-positive plants in the double mutants in comparison with vad1 (Table 1). Thus, EDS1 and NDR1, although not required for lesion initiation in vad1, are involved in cell death amplification and acceleration in the mutant.
Figure 6.
Cell Death Phenotypes in Wild-Type, Single Mutants (eds1, ndr1, and vad1), and Double Mutant vad1 eds1 and vad1 ndr1 Plants.
One line, vad1 eds1 15, out of six lines, and vad1 ndr1 4, out of five lines, are presented as examples.
(A) and (B) Single or double mutant plants (vad1 eds1) 5 d (A) and 8 d (B) after lesion formation in vad1.
(C) and (D) Single or double mutant (vad1 ndr1) plants 3 d (A) and 11 d (B) after lesion formation in vad1.
(E), (H), (G), and (J) Defense gene expression in wild-type, single, or double mutant plants. Transcript levels of PR-1 and PDF1-2 were determined by quantitative PCR in plants grown under lesion-promoting conditions 6 d before (white bars) and 6 d (gray bars) and 12 d (black bars) after lesion formation in vad1 for vad1 eds1 (E) and 6 d before (white bars) and 2 d (gray bars) and 8 d (black bars) after lesion formation in vad1 for vad1 ndr1 lines (F). See Methods for further details. This experiment was repeated twice with different sets of plants, and similar results were obtained. F.W., fresh weight.
(F) and (I) Total SA levels in wild-type, single, or double mutant plants vad1 eds1 (F) and vad1 ndr1 (I) at two time points after lesion formation in vad1. The plant material used in (E) or (H) was also used for SA measurements.
Table 1.
Lesion Formation in vad1 eds1 and vad1 ndr1 Mutant Plants Compared with vad1
| Plants Exhibiting Lesions (Percentage of Total Number of Plants)a
|
||||
|---|---|---|---|---|
| Daysb
|
||||
| Mutants | 5 | 7 | 11 | 14 |
| vad1 | 100 | 100 | 100 | 100 |
| vad1/eds1-15 | 0 | 0 | 41 | 88 |
| vad1/eds1-36 | 0 | 0 | 48 | 83 |
| vad1/ndr1-4 | 12 | 12 | 75 | 91 |
| vad1/ndr1-42 | 11 | 30 | 83 | 100 |
The number of plants used in the experiment varies from 40 to 77 according to the line.
Days after lesion formation in vad1 plants.
The effect of eds1 and ndr1 was then examined on the expression of defense-related genes and SA accumulation in vad1. The elevated PR1 gene expression (Figure 6E) and SA accumulation (Figure 6F) observed in the vad1 mutant was strongly suppressed in the double mutant line vad1 eds1, 6 d after lesion appearance in vad1, and was significantly reduced 12 d after lesion formation in vad1. These results are consistent with the phenotypes previously described (i.e., delayed and reduced lesions on the double mutant line vad1 eds1). PDF1.2 expression was slightly increased in the double mutant lines compared with vad1 (Figure 6G). A similar approach was conducted for gene expression in vad1 ndr1 double mutant lines. In this case, PR1 expression (Figure 6H), SA accumulation (Figure 6I), and PDF1.2 expression (Figure 6J) were suppressed or reduced 2 d after lesion appearance in vad1, when vad1 ndr1 still did not present lesions. However, 8 d after lesion formation in vad1, when vad1 ndr1 presented lesions, the level of PR-1 gene expression and SA in the vad1 ndr1 lines was found to be increased to a level comparable to that observed in vad1 2 d after lesion appearance, suggesting that the expression of this defense marker together with the production of SA is delayed rather than reduced in the double mutant. This concurs with the observed cell death phenotypes, which were found to be affected less by the mutation ndr1 than by eds1.
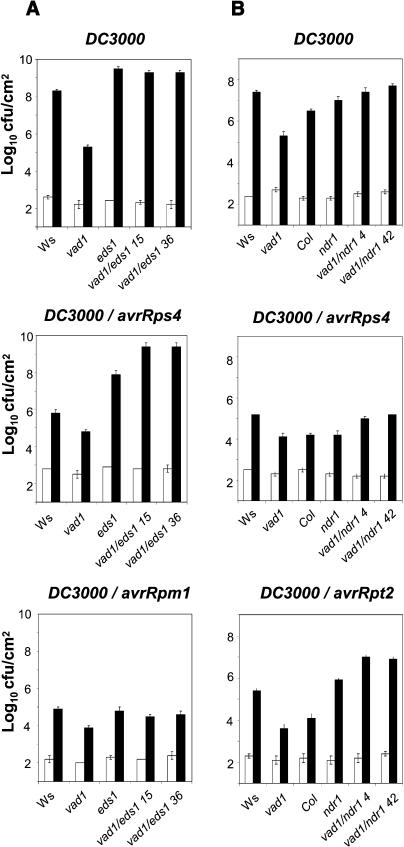
In response to inoculation with virulent and avirulent strains of P. syringae, resistance was clearly compromised in vad1 eds1 and vad1 ndr1 plants, which were as susceptible as eds1 and ndr1 plants, respectively, to all bacterial strains (Figures 7A and 7B). This is also true for RPM1 resistance, which was diminished only partially by the ndr1 mutation in the Col-0 background, as already reported (Tornero et al., 2002). Thus, the increased basal resistance observed in vad1 is dependent on EDS1 and NDR1.
Figure 7.
Resistance Phenotypes in Wild-Type, Single Mutant (eds1, ndr1, and vad1), and Double Mutant vad1 eds1 and vad1 ndr1 Plants.
Bacterial growth in wild-type, single, or double mutant plants: vad1 eds1 (A) and vad1 ndr1 (B). Inoculation with P. syringae strain DC3000 and strain DC3000 expressing avrRpm1, avrRpt2, and avrRps4 was performed with a bacterial suspension of 2.105 cfu/mL, and bacterial growth determinations were performed at the times indicated (day 0, white bars; day 3, black bars). Mean bacterial densities are shown (three to five replicates with corresponding standard deviations) for one representative experiment from two or three independent experiments performed for each strain.
Cloning and Expression of VAD1
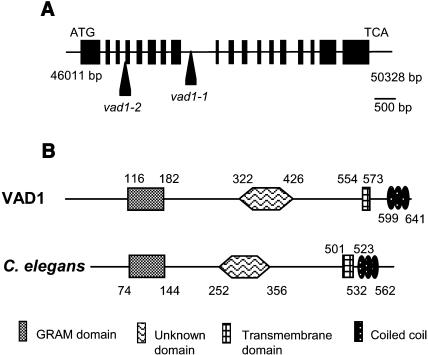
As mentionned before, the vad1 mutant was identified by screening a population of Arabidopsis mutagenized by T-DNA insertion, and the mutant phenotype was caused by a recessive mutation at a single locus. The T-DNA, containing genes that conferred dominant traits for resistance to kanamycin (Bouchez et al., 1993), segregated in a 3:1 ratio in the F2 population, indicating a single integration locus in the mutant line. Because the T-DNA cosegregated with the mutant phenotype, the mutation was probably tagged. DNA gel blot analysis confirmed the presence of a single copy of the T-DNA (data not shown). The sequence of the plant DNA flanking the T-DNA borders was determined and used in a BLAST search of the Arabidopsis genome database, revealing that the T-DNA was inserted into the eighth intron of a gene (At1g02120) (Figure 8A) located on the T7I23 BAC (www.arabidopsis.org). The VAD1 genomic sequence is 4317 bp long and has 18 exons. The predicted VAD1 protein is composed of 644 amino acids, contains a GRAM domain (Doerks et al., 2000), a transmembrane domain, and a coil domain (Figure 8B), and shows a structural organization very similar to a Caenorhabditis elegans protein (ZC328.3, http://elegans.swmed.edu), whose function is unknown. The two proteins present a domain of unknown function that is found in other Arabidopsis proteins that also possess a GRAM domain associated with a C2 domain (Rizo and Südhof, 1998) (At1g03370, At3g59660, and At5g50170). To test genetic complementation of vad1-1 with a wild-type copy of the VAD1 gene, a 6765-bp DNA fragment containing the VAD1 gene coding regions plus 1774 bp of upstream and 894 bp of downstream DNA was used for Agrobacterium-mediated transformation. vad1-1 plants transformed with the genomic clone clearly had a wild-type phenotype (data not shown), demonstrating that the disruption in VAD1 was responsible for the mutant phenotype in vad1-1 mutant plants.
Figure 8.
Molecular Identification of the VAD1 Gene.
(A) Genomic organization of vad1. The arrows indicate the insertion sites of the T-DNA in the mutants vad1-1 and vad1-2 within the VAD1 gene sequence. Gene organization in exons (boxes) and introns (black line) is presented.
(B) Comparison of the predicted VAD1 protein with a protein from C. elegans (ZC328.3, http://elegans.swmed.edu). The regions delimiting the different domains were deduced from ProDom and SMART analysis.
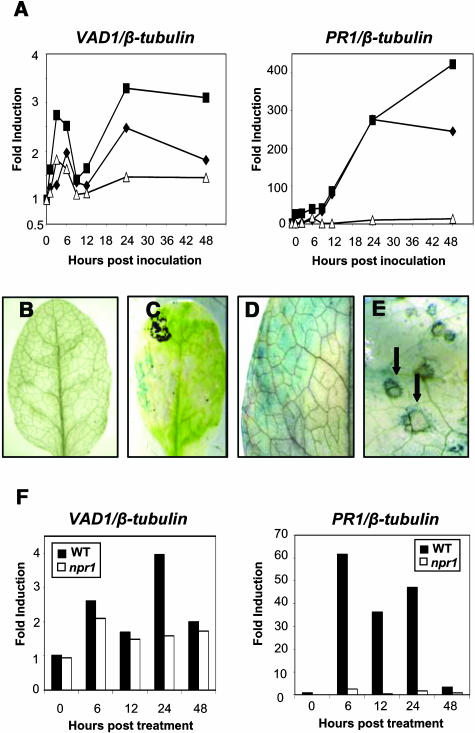
Expression of the VAD1 gene was assessed during plant–pathogen interactions, and interestingly, whereas the gene is constitutively expressed at very low levels in healthy plants, it was shown to be induced upon inoculation with avirulent strains of Xanthomonas campestris pv campestris (Xcc147) and P. syringae pv tomato (DC3000/avrRpm1) (Figure 9A). Transient expression of VAD1 was observed with a maximum between 3 and 6 h postinoculation; this expression decreased during the next few hours, to reach the basal level of expression observed in noninoculated plants. Twenty-four hours postinoculation, activation of the gene was again observed and maintained until 48 h postinoculation. Localization of VAD1 gene expression was performed using either transgenic lines containing a fusion between the VAD1 gene promoter and the β-glucuronidase (GUS) coding region or vad1 lines where the GUS gene contained in the T-DNA was in frame with VAD1. GUS staining of these plants during development did not reveal any GUS activity (Figure 9B). In response to an avirulent bacterial pathogen, GUS activity was found transiently in the cells evolving an HR (Figure 9D) and then (48 h and 72 h postinoculation) at the vicinity of the hypersensitive lesions (Figures 9C and 9E). It hardly could be detected in the case of a compatible interaction (data not shown). Because the vad1 phenotypes were found SA dependent, VAD1 expression was assessed in response to SA, as compared with PR1 expression under the same conditions (Figure 9F). VAD1 was found to be induced also, and this induction was shown to be NPR1 independent at early time points but NPR1 dependent 24 h after treatment.
Figure 9.
VAD1 Gene Expression after Pathogen Inoculation and in Response to SA Treatment.
(A) VAD1 and PR1 transcript accumulation in wild-type plants (Col-0) at different times after inoculation with an avirulent (Xcc147) strain of X. campestris pv campestris (squares), an avirulent (DC3000/avrRpm1) strain of P. syringae pv tomato (diamonds), or after treatment with water (triangles). Transcript levels of VAD1 and PR-1 were determined by quantitative PCR as described in Methods. Results are expressed as fold induction compared with the noninoculated wild type.
(B) to (E) Histochemical localization of GUS activity in leaves from vad1 plants or plants containing a VAD1 promoter-GUS fusion, both healthy (B) and after inoculation with an avirulent (Xcc147) of X. campestris pv campestris ([C], [D], and [E]). Undetached leaves were infiltrated in a small region (1 cm2) with the bacterial strain at 108 cfu/mL and observed 48 h postinoculation (C) or sprayed with the bacterial suspension at 108 cfu/mL 6 h postinoculation (D) or 72 h postinoculation (E).
(F) VAD1 and PR1 transcript accumulation in wild-type (Ws-4) and npr1 mutant plants at different times after treatment with SA (1 mM). One representative experiment is shown from two independent experiments.
DISCUSSION
The isolation of several lesion mimic mutants that show aberrant regulation of cell death constitutes a powerful approach for the identification of genes involved in the regulation and/or execution of PCD, and more specifically the hypersensitive cell death. Although some of them might represent perturbations of cell homeostasis, unrelated to disease defense responses, several of them have been shown to constitutively express markers associated with pathogen infection, and more convincingly, to be affected in resistance to pathogens. More recently, crosses of these mutants with mutants affected in signaling pathways leading to resistance and/or cell death revealed either the implication of the corresponding genes in these pathways or, more indirectly, the existence of novel functions for known genes belonging to these pathways or cross talks between them. Thus, at least some of them might be used as keys to decipher cell death and defense pathways in plants. We identified a novel lesion mimic mutant, vad1, which exhibits propagative HR-like lesions along the vascular system. To our knowledge, only five other propagation mutants have been isolated to date, and among them two exhibit disease-like (not HR-like) lesions (Brodersen et al., 2002; Pilloff et al., 2002); therefore, vad1 may be a new function involved in cell death control. The observations that (1) the mutant constitutively expresses defense genes, accumulates elevated levels of SA, and exhibits enhanced resistance to P. syringae and (2) that its phenotypes are dependent on SA accumulation, and partially or completely on the resistance components EDS1 and NDR1, are in favor of its involvement in cell death/defense pathways. In addition, the vad1 mutation is unique because of the tissue specificity of the lesions.
vad1 Exhibits Propagative and Tissue-Specific Lesions
Histology of mutant leaves harboring lesions revealed that lesions were predominantly restricted to parenchyma cells neighboring veins, showing cytological features of cell death very similar to those described for cells undergoing the HR: increase in vacuolization, condensed cytoplasm more or less retracted from the cell wall, alteration of membranes, collapse of organelles, and increase in nucleus size. In addition to these characteristic changes of cells undergoing PCD in vad1 lesions, xylem vessels were occluded by accumulation of a pectin-containing fibrillar material, originated from vessel-associated parenchyma (Clérivet et al., 2000) and which was not observed in HR cells responding to X. campestris pv campestris. This defense reaction was previously described as a typical response of vascular tissues to vascular pathogens, such as bacteria (Kpémoua et al., 1996) and fungi (Roussel et al., 1999). This tissue specificity of vad1 lesions is of particular interest, considering that these cells have been shown to be more prone to undergo PCD than others (Hammond-Kosack and Jones, 1996; Alvarez et al., 1998), and the increased sensitivity of these cells might contribute to reducing long distance movement of viral (Carrington et al., 1996) or other pathogens. Our observations suggest the existence of a highly regulated system controlling predisposition of specific cells to undergo PCD, in which VAD1 could be an important component. The fact that VAD1 is not specifically expressed in these cells suggests that VAD1 would not exert an executive function in these cells but might rather control the production of a specific signal(s), which could be perceived differently by each cell/tissue leading to its own response: execution of cell death in accordance with its specific requirement. Alternatively, it might affect the perception of these signals in these specific cells. The signals that trigger PCD in the mutant and their relationship to those involved in initiation of hypersensitive cell death are clearly of great interest: JA (Reymond and Farmer, 1998; Hilpert et al., 2001; Stenzel et al., 2003), H2O2 (Dat et al., 2003; Overmyer et al., 2003), and SA (Mur et al., 1997, 2000; Aviv et al., 2002) could represent key regulators of this potentiation system.
VAD1 as a Regulator of Defense and Resistance
The induced expression of all defense genes tested, the accumulation of SA, and the enhanced disease resistance phenotypes of the mutant imply that VAD1 may play a role in regulating defense response, as some other propagation mutants (Lorrain et al., 2003). The increased resistance to P. syringae found for vad1 is only observed under lesion-promoting conditions, the mutant exhibiting a wild-type phenotype under lesion− conditions. Therefore, the enhanced resistance phenotype is closely related to the cell death phenotype and can be explained at least partially by the constitutive expression of defense genes under lesion promoting conditions and their accelerated and enhanced expression above that of the constitutive expression level seen in the lesion+ mutant in response to pathogen inoculation.
Placement of vad1 in the Resistance/Defense Signaling Pathways
The comprehensive epistasis study between the vad1 mutant showing deregulation of cell death and increase of general resistance and the mutants blocking the SA-, EDS1-, and NDR1-mediated resistance generated a large amount of data; only some of them have been presented and only the main important conclusions are drawn here. Through this study, we found that the disease resistance induced in the vad1 mutant follows biologically relevant signaling pathways.
As in several other lesion mimic mutants or in response to different pathogens, SA is a requisite modulator of cell death and resistance pathways activated in vad1. However, unlike lsd1, whose cell death phenotype but not basal resistance is SA dependent, and unlike lsd2, lsd4, lsd5, cet2, cet4.1, or agd2, whose basal resistance but not lesion formation are suppressed in the nahG background, all the phenotypes of vad1 are abolished in the double mutant vad1 nahG and most of them in vad1 sid1. This implies that VAD1 operates upstream of SA and that basal resistance is intimately related to cell death in vad1. Surprisingly, blocking the SA pathway in vad1 either by sid1 or nahG results in a dramatic decrease not only in PR1 expression but also in PDF1-2 and PR5 genes, suggesting that antagonism between SA and JA/ethylene defense pathways is no longer operating in vad1 or that vad1 acts upstream in the regulatory pathway.
Because the production of reactive oxygen species (ROS) has been involved in the HR cell death and SA, in combination with an avirulent pathogen, has been shown to potentiate a sustained H2O2 burst and cell death (Shirasu et al., 1997), we studied the expression of the AtbrohD gene, a gene encoding a major subunit of NADPH oxidase and supposed to participate in superoxide generation (Keller et al., 1998; Torres et al., 1998). The vad1 mutant accumulates elevated levels of AtrbohD transcripts, in accordance with the microscopic observation of H2O2 production at the lesion sites. Interestingly, depletion of SA in vad1 sid1 plants reduces AtrbohD expression to the background level, suggesting that SA (or SA-regulated signals) may potentiate the accumulation of ROS by positively regulating the transcription of AtrbohD and possibly other related genes.
NPR1 is required for some aspects of SA signaling (Cao et al., 1994). Cell death in vad1 is not significantly affected in absence of NPR1, indicating that cell death is essentially regulated in vad1 by SA-dependent, NPR1-independent pathways. Regulation of defense activation seems to be more complex, a reduction of PR1 expression being observed, suggesting the involvement of NPR1-dependent and NPR1-independent pathways. Several precedents, including the cpr and ssi mutants, have shown the existence of an SA-dependent, NPR1-independent pathway (Clarke et al., 1998, 2000; Shah et al., 1999, 2001; Aviv et al., 2002; Devadas et al., 2002; Shirano et al., 2002). This NPR1-independent resistance pathway, which has been suggested to resemble the local response induced during the HR (Clarke et al., 2000), is clearly functioning in vad1. Positive regulation of VAD1 gene expression by SA in an NPR1-independent manner, as recently shown for early SA-responsive genes (Uquillas et al., 2004), suggests also the existence of a feedback loop controlling VAD1 expression.
Epistasy analysis of the vad1 mutant was also performed with mutants affected in the resistance/defense regulators EDS1 and NDR1, showing that the resistance phenotype of vad1 required these two components, whatever the pathogen strain used. EDS1 is a component of a basal resistance pathway that limits growth of virulent pathogens in the absence of activation of a specific resistance pathway; besides, EDS1 and NDR1 have recently been found to be important in regulating the local ROS status (Rusterucci et al., 2001). Our observations are in agreement with these novel functions proposed for EDS1 and NDR1 in plant defense potentiation. In addition, the fact that the cell death phenotype of vad1 was differentially affected by the two regulators also could be related, at least in part, through ROS generation or independently of them, to their ability to differentially modulate SA levels (Shapiro and Zhang, 2001; our work). Consistent with these results, PR1 gene expression is clearly affected in the double mutants. However, PDF1-2 appeared to be overexpressed (although at a low extent) in the vad1 eds1 double mutant, which is reminiscent of observations with the cpr6 eds1 mutant (Clarke et al., 2001) and mutants disrupting SA-mediated responses that become sensitized for activation of JA/ethylene pathways (Bowling et al., 1997).
Based on our results, we propose that VAD1 acts as an important element in a plant defense potentiation system controlling predisposition of specific cells to undergo PCD and highly regulated by the ROS and SA-dependent defense signal amplification loop.
The Function of VAD1
Further insights into the roles of VAD1 within the complex interplay of plant defense signaling networks and its functions in cell death control should be gained from examination of its biochemical function. VAD1 encodes a novel plant membrane protein containing a GRAM domain (Doerks et al., 2000), which has never been identified in plants through a mutational analysis. The GRAM domain has been reported to be an intracellular protein binding or lipid binding signaling domain, which has an important function in membrane-associated processes. It is present in a variety of species and organisms and appeared to be ubiquitous in putative Rab-like GTPase activators, myotubularins, Sbf1 proteins, and other hypothetical proteins (Doerks et al., 2000). On the basis of the results presented here and the putative function of this domain, it can be speculated that VAD1 plays a role in defense and cell death signaling associated with the cell membrane.
Its proposed function as a negative regulator of cell death is also supported by its expression pattern and timing during plant–pathogen interactions. Its transcription is induced rapidly by avirulent pathogen inoculation at the inoculation site. VAD1 transcript appeared as early as 6 h postinoculation with avirulent strains of Xanthomonas and Pseudomonas; this gene activation can be interpreted as a part of the highly regulated process of PCD associated with HR—positive and negative regulators should be first present to control cell death kinetics and intensity. Then, the gene is rapidly repressed to reach the background level observed in healthy plants: this might allow the cell death process to develop. Finally, VAD1 started again to be expressed 24 h postinoculation, at the borders of the HR lesions, probably in relation to cell death limitation.
In summary, this work reports the identification of a novel death regulator in plants, putatively involved in a system highly regulated by the ROS and SA-dependent defense signal amplification loop. The role of ethylene/JA-dependent pathways, the biochemical characterization of this protein, and the analysis of the corresponding mutant C. elegans lines should shed some light on this novel lesion mimic mutant gene.
METHODS
Plant Growth Conditions
Arabidopsis thaliana plants, accessions Col and Ws, were used in these experiments. The mutants vad1-1 and vad1-2 were isolated from a Ws-4 ecotype population mutagenized with T-DNA (Bechtold et al., 1993). The mutant allele vad1-2 was identified by comparing the vad1-1 sequence with the flanking insertion site database (Genoplante, Evry, France). npr1-1 (Col-0) plants were obtained from the Nottingham Arabidopsis Stock Centre. nahG (Col-0), eds1-1 (Ws-0), ndr1-1 (Col-0), and sid1-1 (Col-0) mutants were provided respectively by J. Ryals, J. Parker, B. Staskawicz, C. Nawrath, and J.P. Métraux.
For all experiments, mutant and wild-type seeds were sterilized and sown as described previously (Balagué et al., 2003). Seedlings were further transplanted in Jiffy pots and grown in a culture chamber under a light period of 9 h (192 μE/m2/s) at 21°C and 40 to 70% relative humidity (lesions+ conditions). For low light intensity conditions (96 μE/m2/s), plants were covered with a wire mesh 14 d after transplantation and grown under these conditions. Most experiments were performed with 4- to 5-week-old plants when lesions were developing in vad1.
Genetic Analysis
Double mutants were constructed by crossing vad1-1 plants and mutant plants, selfing the F1 plants, and genotyping the segregating F2 plants for the mutations tested. vad1-1 mutation was selected using the kanamycin resistance conferred by the T-DNA and by PCR using primers in the gene (5′-TGATGGATGGTGGGAATATGG-3′) and in the T-DNA right border (5′-CCAGACTGAATGCCCACAGGCCGTC-3′), whereas wild-type plants were selected using primers located on both sides of the T-DNA insertion site (5′-GCAACTTGTGAAGTAGCACC-3′ and 5′-TGATGGATGGTGGGAATATGG-3′). nahG mutants were PCR screened with the primer set 5′-CTGCCGCTACTCCCATATCC-3′ and 5′-CCGATAGGCTTCTCGCAGATGCA-3′. The cleaved amplified polymorphic sequence markers used for eds1-1 were described by Rusterucci et al. (2001), using the primer set 5′-CGCAGAGGAGAATGCGATTTGTGAT-3′ and 5′-GGATAGAAGATGAATACAAGCCAAAGT-3′. The sid1-1 and the npr1-1 mutations respectively created a Tsp509I restriction site and destroyed a NlaIII restriction site that were used in cleaved amplified polymorphic sequence analysis to detect the homozygous plants using the primer set 5′-GGTCGCAGAATCGGTGATAACT-3′ and 5′-GCGTACCAAGAGCAGCGAGTT-3′ for sid1-1 and 5′-GAGGACACATTGGTTATACTC-3′ and 5′-CAAGATCGAGCAGCGTCATCTTC-3′ for npr1-1. The ndr1-1 mutation was detected in vad1 ndr1-1 plants with the primer set 5′-CGGATTGCTCATTGCCATTGGT-3′, 5′-GGGACGGTTTCAATTCTGTGATA-3′, and 5′-AGGTGGTCGAAACTGTTGTACTT-3′. Using an elongation time of 1 min, NDR1 plants give a PCR product of 1108 bp, whereas the ndr1-1 mutation gives a 817-bp PCR product. F2 double mutants were selfed, and experiments were performed with F3 or F4 populations derived from each double mutant.
VAD1 Cloning and Complementation of the Mutant
Using a PCR walking strategy, 514 bp of DNA flanking the T-DNA right border and ∼1.1 kb of DNA flanking the left border were sequenced. Using these sequences as a query, ∼100% homology was found with the T7I23 BAC located at the top of chromosome I. Sequence analysis of the T-DNA-plant DNA junction revealed a deletion of 51 bp and an insertion of a 12-bp filler sequence at the end of the right border.
The T7I23 BAC containing the VAD1 gene was obtained from the TAMU bank (Texas A&M University). The T7I23 DNA was digested with BglII and subcloned into the BamHI site of the binary vector pCambia1302 (www.cambia.org.au). The resulting plasmids were PCR selected using primers in the VAD1 gene (5′-TGACACGAGCTATGACTCTCAA-3′ and 5′-TGATTGTCACCACCACGACTG-3′) to find plasmids with the 8.4-kb fragment containing the VAD1 gene and verified with digestion profiling. A second subcloning was done by digestion of those plasmids with SacI and ligation of the resultant vector, leading to the generation of a 17.3-bp vector (pS1) that had been used for transformation into vad1-1 plants via Agrobacterium tumefaciens strain GV3101 as described by Clough and Bent (1998). A promoter fragment of 194 bp upstream the ATG codon was subcloned in the SphI and BamHI site of the binary vector pBI101 and used to transform plants via A. tumefaciens strain GV3101. Transformants were selected on MS medium supplemented with 30 μg/mL of hygromycin B (H7772; Sigma, St. Louis, MO).
Histochemistry
Cell death localization was observed by Evans blue staining (Balagué et al., 2003). In vivo determination of ROS release was performed using H2DCFDA (Molecular Probes, Leiden, The Netherlands), which was dissolved in DMSO to produce 100 mM stock. Leaves were placed into a small Petri dish containing 10 mL of loading buffer (Tris KCl at 10 and 50 mM, respectively, pH 7.2) and 5 μL of H2DCFDA stock solution, infiltrated for 3 min, and maintained in the dark for 20 min to obtain basal levels of ROS. Leaves were then removed and placed in fresh loading buffer to wash off excess dye. Examination was performed in bright field (or fluorescence) using a Leica MZ FLIII fluorescence stereomicroscope (Leica, Rueil Malmaison, France) (excitation filter 470/40 nm, barrier filter 515 nm). It should be noted that H2DCFDA exhibits selectivity for H2O2 over free radicals (Allan and Fluhr, 1997); nevertheless, this assay provides an integral measurement for several ROS because it is likely that in vivo, other radical species are quickly converted to the more stable H2O2 (Rodriguez et al., 2002).
For microscopy analysis, fragments (2.0 mm2) were sampled on each side from the main vein of vad1 leaves showing necrotic lesions or from HR sites on leaves of the Ws-4 ecotype inoculated with Xcc147. Samples were fixed for 2 h at 4°C in 0.1 M cacodylate buffer, pH 7.2, containing 2% glutaraldehyde, washed in the buffer, and then pots fixed in 1% osmium tetroxide for 1 h. After dehydration in a graded series of ethanol at 4°C and one bath in propylene oxide, the samples were embedded in an epoxy resin (Epon 812; TAAB, Aldermaston, UK) according to the company's recommendation. The resin was polymerized for 24 h at 56°C, and samples were semithin (2 to 2.5 μm) or ultrathin (80 to 90 nm) sectioned with a diamond knife on a Reichert Ultracut E microtome (Leica, Nussloch, Germany).
Histological observations were made using a Leitz-Diaplan light microscope (Leica, Wetzlar, Germany) after staining of semithin sections with toluidine blue (0.05% w/v) in 0.24 M sodium carbonate, pH 9 to 11. Cells showing HR-like cell death displayed a dark-blue, collapsed cytoplasm retracted from the cell wall.
For histochemical GUS assays, inoculated leaves were collected at different times after inoculation and infiltrated under vacuum with GUS staining buffer (Jefferson et al., 1987). Samples were incubated overnight at 37°C in the staining buffer, and leaves were fixed in paraformaldehyde and cleared in 70% ethanol.
Bacterial Strains and Plant Inoculation Procedures
The virulent and avirulent Pseudomonas syringae pv tomato strains were grown at 29°C on King B's medium supplemented with the appropriate antibiotics: 50 μg/mL of rifampicin (DC3000), 50 μg/mL of rifampicin, 20 μg/mL of kanamycin (avrRps4), 50 μg/mL of rifampicin, and 10 μg/mL of tetracycline (avrRpm1). Xanthomonas campestris pv campestris were grown as previously described (Lummerzheim et al., 1993).
Plants used for bacterial inoculations were kept at high humidity 12 h before inoculation. They were injected with a bacterial suspension of 2.107 to 108 cfu/mL (phenotype test) or 2.105 cfu/mL (bacterial growth determination) using a blunt syringe on the abaxial side of the leaves. For some experiments, the plants were inoculated by spraying the plants with the bacterial suspension, as described previously (Lummerzheim et al., 1993). For determination of in planta bacterial growth at 0, 2, and 3 d postinoculation, leaves were harvested on five plants per genotype as distinct replicates for each genotype. Four discs from each plant were ground in 1 mL 10 mM MgCl2 and a succession of 10-fold dilution was made in 10 mM MgCl2. A predetermined range of dilutions for each sample was plated on King's B medium and incubated at 28°C for 2 d.
RNA Isolation and Quantitative PCR
RNA gel blot experiments were conducted as described by Balagué et al. (2003). For quantitative RT-PCR, RNA was extracted using Macherey-Nagel Nucleospin RNA plant kit (Macherey-Nagel, Hoerdt, France) according to the manufacturer's recommendations. RT-PCR was performed using 2.5 μg of RNA using the superscript reverse transcriptase II (Invitrogen, Carlsbad, CA) with introduction of 5.10−3 ng/μL per sample of RNA of human nebulin. β-Tubulin was used as an internal standard. Quantitative PCR was run on a Roche lightcycler system (Roche Diagnostics, Meylan, France) according to the manufacturer's recommendations. The primer sets used in the different experiments are 5′-GTCCCGAAGCTTACACATGA-3′ and 5′-GCCATACATCCAGCCTTCATCA-3′ (nebulin), 5′-GAGGGAGCCATTGACAACATCTT-3′ and 5′-GCGAACAGTTCACAGCTATGTTCA-3′ (β-tubulin-4), 5′-GGAGCTACGCAGAACAACTAAGA-3′ and 5′-CCCACGAGGATCATAGTTGCAACTGA-3′ (PR1), 5′-TCATGGCTAAGTTTGCTTCC-3′ and 5′-AATACACACGATTTAGCACC-3′ (PDF1.2), 5′-GCCGTCTCTGAACTCAAATCTCAA-3′ and 5′-GTTACGAGCAAGAACAACCTTGTT-3′ (ICS1), 5′-CGCTTGTCCTGCTAGAGGTT-3′ and 5′-GCTCGGTTCACAGTAGTCTGA-3′ (PR3), 5′-CGATGAAAATGAGACGAGGCAA-3′ and 5′-TCGTCGGCGAATCTTGCGTT-3′ (AtrbohD), 5′-GGTTGGAATATGGTAGTGCTGT-3′ and 5′-CTGAACGGATGAAGGTGGAA-3′, or 5′-AGACTCGGTAGAAGGTTGTA-3′ and 5′-CTCCTCGTCACATTCAGATA-3′ (VAD1).
Measurement of and Treatment by SA
Total SA (free SA plus SA conjugate) concentration was measured as described by Chong et al. (2001). For treatment with SA, 4-week-old plants were sprayed with 1 mM solution and maintained at high humidity for 24 h and then under normal growth conditions.
Acknowledgments
S.L. was supported by a grant from the French Ministry of National Education and Research. We thank Olivier Bouchez for technical assistance.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022038.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, A.C., and Fluhr, R. (1997). Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9, 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, M.E., Pennell, R.I., Meijer, P.-J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.F., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Aviv, D.H., Rusterucci, C., Iii, B.F., Dietrich, R.A., Parker, J.E., and Dangl, J.L. (2002). Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 29, 381–391. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Balagué, C., Lin, B., Alcon, C., Flottes, G., Malmstrom, S., Kohler, C., Neuhaus, G., Pelletier, G., Gaymard, F., and Roby, D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bittner-Eddy, P.D., and Beynon, J.L. (2001). The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant-Microbe Interact. 14, 416–421. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expressed both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, S.A., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell to cell and long distance transport of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J., Pierrel, M.A., Atanassova, R., Werck-Reichhart, D., Fritig, B., and Saindrenan., P. (2001). Free and conjugated benzoic acid in tobacco plants and cell cultures. Induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol. 125, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Aarts, N., Feys, B.J., Dong, X., and Parker, J.E. (2001). Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 26, 409–420. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Role of salicilic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clérivet, A., Déon, V., Alami, I., Lopez, F., Geiger, J.P., and Nicole, M. (2000). Tyloses and gels associated with cellulose accumulation in vessels are responses of plane tree seedlings (Platanus acerrifolia) to Ceratocystis fimbriata f. sp. platani. Trees: Structure and Function 15, 25–31.
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dat, J.F., Pellinen, R., Beeckman, T., Van De Cotte, B., Langebartels, C., Kangasjarvi, J., Inze, D., and Van Breusegem, F. (2003). Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 33, 621–632. [DOI] [PubMed] [Google Scholar]
- Devadas, S.K., Enyedi, A., and Raina, R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Doerks, T., Strauss, M., Brendel, M., and Bork, P. (2000). GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem. Sci. 25, 483–485. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Ausubel, F.M. (1993). Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4, 327–341. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Silverman, F.P., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.J. (1996). Resistance gene-dependant plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilpert, B., Bohlmann, H., op den Camp, R.O., Przybyla, D., Miersch, O., Buchala, A., and Apel, K. (2001). Isolation and characterization of signal transduction mutants of Arabidopsis thaliana that constitutively activate the octadecanoid pathway and form necrotic microlesions. Plant J. 26, 435–446. [DOI] [PubMed] [Google Scholar]
- Hoisington, D., Neuffer, M.G., and Walbot, V. (1982). Disease lesion mimics in maize. Dev. Biol. 93, 381–388. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS gene fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, T., Damude, H.G., Werner, D., Doerner, P., Dixon, R.A., and Lamb, C. (1998). A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kpémoua, K., Boher, B., Nicole, M., Calatayud, P., and Geiger, J.P. (1996). Cytochemistry of defense responses of cassava to Xanthomonas campestris pv. manihotis. Can. J. Microbiol. 42, 1131–1149. [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lacomme, C., and Roby, D. (1999). Identification of new early markers of the hypersensitive response in Arabidopsis thaliana. FEBS Lett. 459, 149–153. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., Vailleau, F., Balagué, C., and Roby, D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. [DOI] [PubMed] [Google Scholar]
- Lummerzheim, M., de Olivera, D., Castresana, C., Miguens, F.C., Louzada, E., Roby, D., Van Montagu, M., and Timmerman, B. (1993). Identification of compatible and incompatible interactions between Arabidopsis thaliana and Xanthomonas campestris pv. campestris and characterization of the hypersensitive response. Mol. Plant-Microbe Interact. 6, 532–544. [Google Scholar]
- McDowell, J.M., Cusick, A., Can, C., Beynon, J., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529. [DOI] [PubMed] [Google Scholar]
- Mur, L.A., Brown, I.R., Darby, R.M., Bestwick, C.S., Bi, Y.M., Mansfield, J.W., and Draper, J. (2000). A loss of resistance to avirulent bacterial pathogens in tobacco is associated with the attenuation of a salicylic acid-potentiated oxidative burst. Plant J. 23, 609–621. [DOI] [PubMed] [Google Scholar]
- Mur, L.A.J., Bi, Y.M., Darby, R.M., Firek, S., and Draper, J. (1997). Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersion during lesion establishment in TMV-infected tobacco. Plant J. 12, 1113–1126. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer, K., Brosche, M., and Kangasjarvi, J. (2003). Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 8, 335–342. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Feys, B.J., Van Der Biezen, E.A., Noël, L., Aarts, N., Austin, M.J., Botella, M.A., Frost, L.N., Daniels, M.J., and Jones, J.D.G. (2000). Unravelling R gene-mediated disease resistance pathways in Arabidopsis. Mol. Plant Pathol. 1, 17–24. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilloff, R.K., Devadas, S.K., Enyedi, A., and Raina, R. (2002). The Arabidopsis gain-of-function mutant dll1 spontaneously develops lesions mimicking cell death associated with disease. Plant J. 30, 61–70. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Rizo, J., and Südhof, T.C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879–15882. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A.A., Grunberg, K.A., and Taleisnik, E.L. (2002). Reactive oxygen species in the elongation zone of maize leaves are necessary for leaf extension. Plant Physiol. 129, 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, S., Nicole, M., Lopez, F., Ricci, P., Geiger, J.-P., Renard, M., and Brun, H. (1999). Leptosphaeria maculans and cryptogein induce similar responses in tissues undergoing the hypersensitive reaction in Brassica napus. Plant Sci. 144, 17–28. [Google Scholar]
- Rusterucci, C., Aviv, D.H., Holt III, B.F., Dangl, J.L., and Parker, J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, A.J., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., Nandi, A., and Klessig, D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. [DOI] [PubMed] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C. (1997). Salicylic-acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, I., Hause, B., Maucher, H., Pitzschke, A., Miersch, O., Ziegler, J., Ryan, C.A., and Wasternack, C. (2003). Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato—Amplification in wound signalling. Plant J. 33, 577–589. [DOI] [PubMed] [Google Scholar]
- Stokes, T.L., and Richards, E.J. (2002). Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc. Natl. Acad. Sci. USA 99, 7792–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Kawasaki, T., Henmi, K., Shii, K., Kodama, O., Satoh, H., and Shimamoto, K. (1999). Lesion mimics mutants of rice with alterations in early signalings events of defense. Plant J. 11, 993–1005. [DOI] [PubMed] [Google Scholar]
- Tör, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J.L., and Holub, E.B. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K.E., and Jones, J.D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. [DOI] [PubMed] [Google Scholar]
- Uquillas, C., Letelier, I., Blanco, F., Jordana, X., and Holuigue, L. (2004). NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol. Plant-Microbe Interact. 17, 34–42. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33, 733–742. [DOI] [PubMed] [Google Scholar]
- Walbot, V., Hoisington, D.A., and Neuffer, M.G. (1983). Disease lesion mimics in maize. In Genetic Engeneering of Plants, T. Kosuge and C. Meridith, eds (New York: Plenum Publishing Company), pp. 431–442.
- Wolter, M., Hollricher, K., and Schulze-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]