Abstract
The Ustilago maydis mrb1 gene specifies a mitochondrial matrix protein with significant similarity to mitochondrial p32 family proteins known from human and many other eukaryotic species. Compatible mrb1 mutant strains were able to mate and form dikaryotic hyphae; however, proliferation within infected tissue and the ability to induce tumor development of infected maize (Zea mays) plants were drastically impaired. Surprisingly, manifestation of the mrb1 mutant phenotype selectively depended on the a2 mating type locus. The a2 locus contains, in addition to pheromone signaling components, the genes lga2 and rga2 of unknown function. Deletion of lga2 in an a2Δmrb1 strain fully restored pathogenicity, whereas pathogenicity was partially regained in an a2Δmrb1Δrga2 strain, implicating a concerted action between Lga2 and Rga2 in compromising pathogenicity in Δmrb1 strains. Lga2 and Rga2 localized to mitochondria and Mrb1 interacted with Rga2 in the yeast two-hybrid system. Conditional expression of lga2 in haploid cells reduced vegetative growth, conferred mitochondrial fragmentation and mitochondrial DNA degradation, and interfered with respiratory activity. The consequences of lga2 overexpression depended on the expression strength and were greatly exacerbated in Δmrb1 mutants. We propose that Lga2 interferes with mitochondrial fusion and that Mrb1 controls this activity, emphasizing a critical link between mitochondrial morphology and pathogenicity.
INTRODUCTION
Ustilago maydis is a member of the order Ustilaginales, causing worldwide smut diseases in more than 75 plant families of the angiosperms. Among the estimated 1200 species of smut fungi, the economically significant pathogens are U. hordei, U. nuda, U. nigra, U. tritici, Urocystis agropyri, U. maydis, U. scitaminea, and species of the genus Sporisorium, causing smut diseases on barley (Hordeum vulgare), oat (Avena sativa), wheat (Triticum aestivum), maize (Zea mays), sugarcane (Saccharum officinarum), and sorghum (Sorghum bicolor), respectively, as well as Tilletia species causing bunt diseases on wheat (Martínez-Espinoza et al., 2002). In particular, it is the ability of smut fungi to develop within grain kernels that can result in drastically lowered crop yields. U. maydis and U. hordei are the most exclusively studied members of the Ustilaginales. U. maydis infects young meristematic tissue above ground and triggers the formation of tumors, most prevalent on infected ears, tassels, stems, and nodal shoots. In these tumors, massive proliferation of the fungus occurs. A different strategy is adopted by U. hordei, which maintains the mycelium state in the apical meristem until flowering and then ramifies ovary tissue, resulting in kernels filled with teliospores (Martínez-Espinoza et al., 2002).
U. maydis is genetically tractable, and a broad spectrum of molecular methods, including an efficient gene knockout system, stage-specific and inducible promoters, reporter genes, and insertional mutagenesis, has been established over the last 15 years. Recently, the complete genome sequence of U. maydis has been released to the public database providing for novel strategies to identify pathogenicity functions based on transcriptome analysis and candidate approaches. This has rendered U. maydis a prime model organism to uncover general strategies elaborated by phytopathogenic fungi (Basse and Steinberg, 2004).
The infection process of U. maydis has been documented in previous cytological studies (Snetselaar and Mims, 1994 and references therein; Banuett and Herskowitz, 1996; Kahmann et al., 2000). Dikaryotic hyphae that emerge from fusion of compatible, haploid sporidia are able to penetrate the surface of aerial plant parts by developing appressoria-like structures at their tips. Hyphae initially grow through epidermal cells and proceed with intercellular proliferation in underlying tissue. After karyogamy, diploid spore precursors are released by fragmentation from highly branched, sporogeneous hyphae and further mature to teliospores. During all these stages U. maydis maintains a biotrophic relationship with its host. Early host responses are chlorosis and anthocyanin formation (Banuett and Herskowitz, 1996), with the latter being an indicator that the fungus has entered the plant tissue. Full virulence, however, coincides with the formation of host tumors. Pathogenicity is genetically controlled by the mating type loci a and b. The a locus specifies a pheromone/receptor system that triggers cell fusion in response to pheromone recognition by the receptor of opposite mating type (Bölker et al., 1992; Spellig et al., 1994). This locus exists in two alleles termed a1 and a2. Whereas both loci code for pheromone precursor and receptor genes, the a2 locus comprises two additional genes, lga2 and rga2, of which lga2 has been proposed to encode a putative mitochondrial protein. However, these genes are not critical for mating and their absence is not affecting pathogenic development (Urban, 1995; Urban et al., 1996a). The b loci encode the bE and bW homeodomain proteins, which can dimerize in nonallelic combinations and then form an active transcription factor required for stability and development of the infectious filamentous dikaryon (Gillissen et al., 1992 and references therein; Kämper et al., 1995; Brachmann et al., 2001). All genes residing in the a locus are pheromone induced, and lga2 expression is additionally stimulated in the presence of an active b heterodimer (Urban et al., 1996b). More recent studies have shown that lga2 is a direct target of the b proteins (Romeis et al., 2000). Although dikaryotic hyphae represent the infectious agent in nature, haploid, solopathogenic strains, which bypass the requirement for a mating partner and cause infections when inoculated singly into the maize plant, can be generated in the laboratory. This is achieved by the introduction of b mating-type genes whose products can dimerize with those of resident genes. In particular, it has been demonstrated that an a1b1 strain transformed with a b2 allele is pathogenic (Kronstad and Leong, 1989; Schulz et al., 1990; Bölker et al., 1995).
By differential display analysis, we recently identified the mig2 cluster, which consists of five highly similar genes lacking homologies to database entries. All these genes are extensively upregulated after the fungus has entered the host tissue (Basse et al., 2002). The mig2 locus is flanked by the constitutively expressed U. maydis mrb1 gene (Basse et al., 2002). mrb1 encodes a protein with significant homologies to the so-called p32 family proteins and is predicted to reside in mitochondria. The founding member of this protein family is the human p32 protein, which was originally identified in association with the SR family splicing factor ASF/SF2 (Krainer et al., 1991). p32 family proteins are implicated in diverse regulatory processes, including transcriptional activation by cooperating with viral transcription factors, pre-mRNA splicing, and mitochondrial RNA editing (Krainer et al., 1991; Yu et al., 1995; Petersen-Mahrt et al., 1999; Van den Brulle et al., 1999; Van Scoy et al., 2000; Hayman et al., 2001 and references therein). Because the function of p32 family proteins has not yet been addressed in a pathogenic fungus, we became interested in analyzing a possible regulatory function of Mrb1 connected to pathogenicity.
Here, we describe that Mrb1 functions as a novel, mitochondrial virulence determinant in U. maydis. Our results indicate that Mrb1 is specifically required for pathogenicity in the presence of the a2 locus genes lga2 and rga2. We explicitly show that Lga2 interferes with cell growth, mitochondrial morphology, and fitness and that Mrb1 counteracts these Lga2 effects. Our results suggest the formation of a mitochondrial protein complex containing Mrb1, Lga2, and Rga2 and emphasize a critical connection between mitochondrial morphology and fungal development during the pathogenic growth stage.
RESULTS
mrb1 Encodes an Acidic Protein of the p32 Family
We have previously identified the U. maydis mrb1 gene based on its neighboring position to the mig2 cluster (Basse et al., 2002). The predicted mrb1 open reading frame (ORF) encodes a putative protein of 274 amino acids corresponding to a molecular mass of 30.4 kD. The protein sequence revealed highest similarity to Aspergillus nidulans SuAprgA1 (36.5% identity), Mam33p of Saccharomyces cerevisiae (22.3% identity), as well as the p32 and p22 proteins from human and Trypanosoma brucei, respectively (23.7 to 24.1% identity; Figure 1). These proteins all have a mitochondrial target sequence and are highly acidic with pI values in the range of 4 to 4.5. The predicted pI value of Mrb1 is 4.35. The majority of acidic amino acid positions are conserved among p32 family proteins, and the most pronounced sequence conservation is found in the C-terminal portion between positions 216 and 274 of Mrb1 (Figure 1). The respective region of human p32 comprises two α-helical regions implicated in oligomerization as predicted from the crystal structure (Jiang et al., 1999). In addition, Mrb1 shares several conserved amino acid positions with p32 family proteins that are surface exposed in p32 and have been proposed to participate in protein–protein interactions (Figure 1; Jiang et al., 1999).
Figure 1.
Multiple Alignment of Mrb1 with Related Sequences from A. nidulans (SuAprgA1), S. cerevisiae (Mam33p), T. brucei (p22), and Human (p32).
Identical (≥60%) positions are boxed and shaded. Predicted N-terminal amino acids generated after cleaving the presequence are encircled. Conserved acidic positions in the Mrb1 sequence are marked by dots. Asterisks mark highly conserved amino acids of p32 proteins (Glu89, Arg118, Arg242, Gly243, Glu263, and Tyr264) assigned to the protein surface in p32 (Jiang et al., 1999).
Localization and Expression of Mrb1
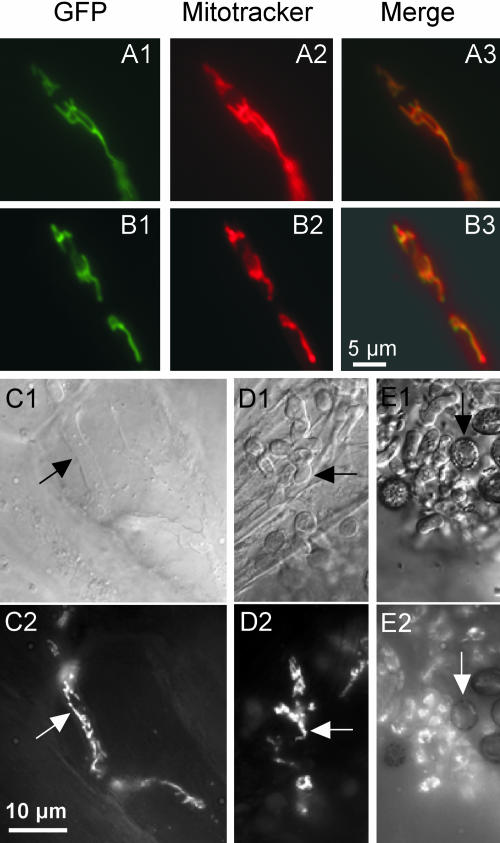
For Mam33p and p32, it has been documented that these proteins are predominantly localized in the mitochondrial matrix (Muta et al., 1997; Seytter et al., 1998). The existence of a mitochondrial signal sequence was also predicted for Mrb1 (Figure 1). To assess whether the potential sorting signal is functional, we generated plasmids pMB2-1 and pMB2-2, which allow constitutive expression of enhanced green flourescent protein (eGFP) fused to the regions coding for the N-terminal 46 and 58 amino acids of Mrb1, respectively. These plasmids were inserted ectopically into U. maydis strain FB1. As control, plasmid pMGH1 was generated, which contained the complete mrb1 ORF translationally fused to eGFP under control of the native mrb1 promoter, and inserted by homologous integration into strain FB1. In all cases, fluorescence microscopy of transformants revealed a thread-like mitochondrial pattern (Figures 2A and 2B; data not shown). This indicated that mitochondrial localization was conferred by all three constructs and that the N-terminal 46 amino acids of Mrb1 were sufficient for targeting. To determine the spatial and temporal control of mrb1 expression during pathogenic development, we used compatible FB1mrb1:eGFP and FB2mrb1:eGFP strains, which expressed the mrb1-eGFP fusion construct under the mrb1 promoter. Expression of mrb1 remained high in the initial phase of pathogenic development as reflected from mitochondrial GFP fluorescence in appressoria on the leaf surface (data not shown) as well as during biotrophic growth of hyphae in planta until the stage of hyphal fragmentation (Figures 2C and 2D). By contrast, mature teliospores emitted only barely detectable fluorescence (Figure 2E).
Figure 2.
Localization and Expression of Mrb1.
(A) and (B) U. maydis strains FB1mrb1:eGFP (A) and FB1/pMB2-1 (B) were grown in yeast extract peptone sucrose medium and assayed for fluorescence of GFP (left) and the mitochondrial marker dye CM-H2Xros (middle). The yellow color (right) indicates coincidence. The bar refers to all panels.
(C) to (E) Spatial mrb1 expression during the life cycle of U. maydis. Plants were inoculated with mixtures of compatible wild-type reporter strains FB1mrb1:eGFP (a1b1) and FB2mrb1:eGFP (a2b2), which express mrb1 under its own promoter. Maize tissue samples were assayed by differential interference contrast (DIC) ([C1] to [E1]) and fluorescence microscopy ([C2] to [E2]). GFP fluorescence in intracellularly growing hyphae 5 DAI (C) and in sporogenic hyphae 10 and 12 DAI (arrow in [D] and [E]), respectively, are shown. The arrow in (E) points to a mature teliospore. The bar refers to all panels.
Deletion of mrb1 and Complementation of mrb1 Null Mutant Strains
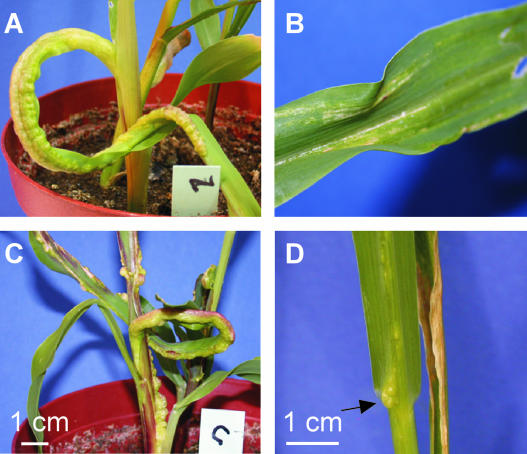
To assess the function of mrb1, the gene was replaced by the carboxin (cbx) resistance cassette in compatible, haploid U. maydis strains FB1 (a1b1) and FB2 (a2b2). The resulting Δmrb1 strains were unaffected in growth in either yeast extract peptone sucrose or minimal medium, and when combined, they readily formed dikaryotic filaments on solid charcoal complete medium (CM). A previous study has shown that growth of yeast mam33 mutants was retarded on solid YPG medium with glycerol as nonfermentable carbon source at 18°C (Muta et al., 1997). However, under these conditions, mrb1 mutant strains were not affected in growth (data not shown). Remarkably, none of the six combinations of compatible FB1Δmrb1 and FB2Δmrb1 strains tested was able to trigger tumor development within 9 d after inoculation (DAI), whereas plants infected with isogenic wild-type strains produced tumors with a frequency of 92% within that period (Table 1, Figures 3A and 3B). Furthermore, the induction of anthocyanin formation was drastically reduced in infections with Δmrb1 mutants (data not shown). After a period of 10 to 12 DAI, only 5% of infected plants produced tumors, and these were significantly reduced in size with diameters of ∼2 mm. Combinations of compatible Δmrb1 and wild-type strains were able to mate and cause disease symptoms in all cases; however, incidence of tumor formation was markedly diminished in all combinations with FB2Δmrb1 strains (Table 1). To unequivocally demonstrate that the absence of mrb1 accounted for the loss of pathogenicity, plasmid pMGH1 (see above) was introduced into strains FB1Δmrb1 and FB2Δmrb1. Combinations of these transformants induced wild-type disease symptoms (Figure 3C, Table 1). This shows that the pathogenicity defect of Δmrb1 strains can be complemented and illustrates further that the mrb1-eGFP fusion protein is functional.
Table 1.
Pathogenicity of Δmrb1 Strains and Complementation by pMGH1
| Inoculum | Inoculated Plants | Plants with Tumors/Percentagea |
|---|---|---|
| FB1Δmrb1 × FB2Δmrb1 (4/6)bc | 199 | 0/0 |
| FB1 × FB2 (4/1)b | 101 | 93/92 |
| FB1Δmrb1#12 × FB2 (1/1) | 20 | 20/100 |
| FB1Δmrb1#15 × FB2 (1/1) | 20 | 19/95 |
| FB1 × FB2Δmrb1#4 (1/1) | 15 | 7/47 |
| FB1 × FB2Δmrb1#9 (1/1) | 19 | 6/32 |
| FB1Δmrb1#15 × FB2Δmrb1#9 (1/1) | 15 | 0/0 |
| FB1Δmrb1#12 × FB2Δmrb1#4 (1/1) | 16 | 0/0 |
| FB1 × FB2 (1/1) | 20 | 19/95 |
| FB1Δmrb1/pMGH1 × FB2Δmrb1/pMGH1 (1/4) | 84 | 59/70 |
| FB1mrb1:eGFP × FB2mrb1:eGFP (1/1) | 14 | 8/57 |
| SG200Δmrb1 (2/5) | 138 | 82/59 |
| SG200 (2/1) | 65 | 43/66 |
Comparable results were obtained with Δmrb1 strains, in which the ip cassette was substituted by the hph cassette.
Number of plants possessing at least one tumor and indication of the respective percentage.
(x/y): x is the number of independent pathogenicity tests performed; y is the number of different strains or strain combinations.
Plants were inspected 6 to 9 DAI. Tumors of wild-type infected plants were already present 6 DAI.
Figure 3.
Complementation of Δmrb1 Strains with pMGH1.
(A) to (C) Infection symptoms of maize plants 6 DAI with mixtures of wild-type FB1 (a1b1)/FB2 (a2b2) strains (A), FB1Δmrb1/FB2Δmrb1 strains (B), and FB1Δmrb1/pMGH1/FB2Δmrb1/pMGH1 strains (C).
(D) Leaf tumor (arrow) 11 DAI with strains FB1Δmrb1/FB2Δmrb1.
The Role of Mrb1 during the Infection Cycle
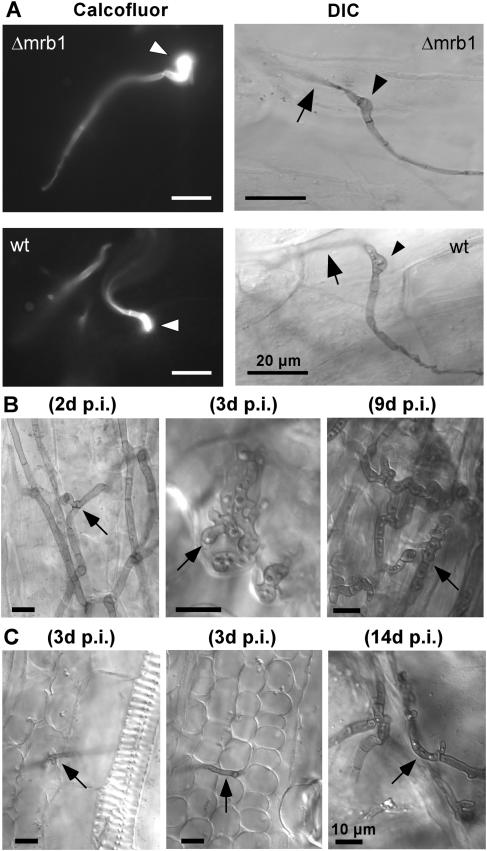
To investigate at which stage during pathogenic development Mrb1 is required, we followed the fate of dikaryotic hyphae after inoculation. Mutant Δmrb1 hyphae were able to grow on the leaf surface and differentiated appressoria that penetrated into leaf tissue (Figure 4A). However, a significant difference in proliferation between mutant and wild-type hyphae became apparent during subsequent biotrophic growth. Already 2 DAI wild-type hyphae had ramified through epidermal cells followed by proliferation in underlying tissue 3 DAI, and 9 DAI fragmenting sporogeneous hyphae were detected (Figure 4B). By contrast, only few Δmrb1 hyphae were detected in infected tissue and these grew predominantly unbranched (Figure 4C). In one of the small tumors induced by Δmrb1 hyphae, moderate accumulation of branched hyphae was detected 14 DAI (Figure 4C). Moreover, teliospores had developed in only one of the seven small tumors that resulted from infection with FB1Δmrb1 and FB2Δmrb1 strains after a prolonged period (data not shown). In conclusion, the pathogenicity defect of Δmrb1 mutants results from severely attenuated hyphal development in epidermal and underlying cells.
Figure 4.
Development of Δmrb1 Strains.
(A) Appressoria development of hyphae emerging from a cross of compatible wild-type and Δmrb1 strains 26 h after inoculation. Left panels, Calcofluor staining of appressoria (arrowheads) from a cross of FB1/FB2 (wt) or FB1Δmrb1/FB2Δmrb1 (Δmrb1) strains on the leaf surface before penetration. Right panels, appressoria (arrowheads) on the leaf surface from combinations of FB1/pMB2-2/FB2/pMB2-2 strains (wt) and mutant FB1Δmrb1/pMB2-2/FB2Δmrb1/pMB2-2 strains (Δmrb1), from which filaments penetrate into leaf tissue (arrows). Filaments and appressoria were stained with Chlorazol Black E (CBE). Samples were assayed by DIC light microscopy. Bars in all panels = 20 μm.
(B) and (C) Biotrophic growth of wild-type and Δmrb1 hyphae. Maize seedlings were inoculated with either mixtures of FB1/FB2 (B) or FB1Δmrb1/FB2Δmrb1 (C) strains. Tissue samples were collected as indicated. In all tissue samples, fungal hyphae (arrows) were stained with CBE and assayed by DIC light microscopy. Bars in all panels = 10 μm. The arrows point to branched hyphae growing through epidermal cells (left panel in [B]), lobed hyphal branches growing beneath the epidermal layer (middle panel in [B]), and a fragmenting sporogeneous hyphae (right panel in [B]). The arrows point to a short hyphal branch in epidermal cells (left panel in [C]) and a hyphae growing in the mesophyll layer (middle panel in [C]). Hyphal structures in a small leaf tumor that resulted from infection of a mixture of FB1Δmrb1/FB2Δmrb1 strains are shown (right panel in [C]). d p.i., days after inoculation.
mrb1 Is Dispensable for Pathogenicity of Solopathogenic Strains Carrying the a1 Locus
To assess whether mrb1 was also necessary for pathogenicity of strain SG200 (a1mfa2bE1bW2), which is derived from strain FB1 and is solopathogenic because of the presence of the bE1 and bW2 genes (Bölker et al., 1995), SG200Δmrb1 strains were generated. Surprisingly, these strains were unaffected in pathogenicity (Table 1), suggesting a specific mrb1 requirement for pathogenicity of dikaryotic hyphae. Therefore, we expected to render FB1Δmrb1 and FB2Δmrb1 strains solopathogenic by introducing either a bE2 gene (pUb2-2) or a bE1 gene (pUb1). Whereas independent FB1Δmrb1/pUb2-2 transformants acquired the ability to induce tumor and teliospore development (Table 2), none of the six tested FB2Δmrb1/pUb1 strains was able to trigger tumor formation in infected plants. To demonstrate functionality, pUb1 was introduced into wild-type FB2, and here all three transformants tested had become pathogenic (Table 2). These results suggested the existence of a FB2-specific factor that interfered with pathogenic development in the absence of mrb1.
Table 2.
Pathogenicity of Haploid Wild-Type and Δmrb1 Strains Harboring Compatible b Alleles
| Inoculum | Inoculated Plants | Plants with Tumors/Percentagea |
|---|---|---|
| FB1Δmrb1/pUb2-2 (3/2)b | 62 | 29/47 |
| FB1/pUb2-2 (2/2) | 47 | 21/45 |
| FB2Δmrb1/pUb1 (2/6) | 121 | 1c/<1 |
| FB2/pUb1 (1/3) | 48 | 29/60 |
Plants were inspected 6 to 7 DAI.
Number of plants possessing at least one tumor and indication of the respective percentage.
(x/y): x is the number of independent pathogenicity tests performed; y is the number of different strains.
Tumor with a diameter <1 mm.
The Influence of the lga2 and rga2 Genes on Pathogenic Development in the Absence of mrb1
Prompted by the observation that Mrb1 was specifically required for pathogenicity of FB2 strains, we asked whether the a2-specific lga2 and rga2 genes interfered with pathogenic development in the absence of mrb1. The lga2 gene encodes a putative mitochondrial matrix protein (Urban et al., 1996a) that has been shown to contain a functional N-terminal mitochondrial signal peptide (P. Müller, M. Bölker, and R. Kahmann, unpublished data). By contrast, a low probability (0.06) for a mitochondrial target sequence was predicted for Rga2 (see below). The region comprising lga2 or rga2 was replaced by a hygromycin resistance cassette in the FB2Δmrb1 strain. As expected from previous experiments (Urban et al., 1996a), deleting lga2 and/or rga2 in a wild-type background did not affect pathogenic development (Table 3). Amazingly, mixtures of FB1Δmrb1 and FB2Δmrb1Δlga2Δrga2 strains were as pathogenic as a mixture of compatible wild-type strains (Table 3). Pathogenicity assays with Δmrb1 mutants deleted in either lga2 or rga2 alone revealed that a mixture of FB1Δmrb1 and FB2Δmrb1Δlga2 strains was fully pathogenic, whereas mixtures of FB1Δmrb1 and FB2Δmrb1Δrga2 strains caused only weak disease symptoms (Table 3). Nevertheless, these significantly exceeded disease symptoms induced by a mixture of FB1Δmrb1 and FB2Δmrb1 strains (Table 3). Together, these results reveal that lga2 and rga2 negatively contribute to pathogenic development in the absence of mrb1, although to different extents.
Table 3.
Influence of lga2 and rga2 on Pathogenicity of mrb1 Mutants
| Inoculum | Inoculated Plants | Plants with Tumors/Percentagea |
|---|---|---|
| FB1Δmrb1 × FB2Δmrb1 (2/1)bc | 68 | 0/0 |
| FB1Δmrb1 × FB2Δmrb1Δlga2Δrga2 (2/4)c | 114 | 91/80 |
| FB1 × FB2Δlga2Δrga2 (1/2)c | 31 | 26/84 |
| FB1 × FB2 (2/1)c | 62 | 46/74 |
| FB1Δmrb1 × FB2Δmrb1 (2/2)d | 67 | 1e/1.5 |
| FB1Δmrb1 × FB2Δmrb1Δlga2 (2/2)d | 72 | 69/96 |
| FB1Δmrb1 × FB2Δmrb1Δrga2 (2/2)d | 75 | 19f/25 |
| FB1 × FB2Δlga2 (2/2)d | 50 | 43/86 |
| FB1 × FB2Δrga2 (1/2)d | 23 | 19/83 |
| FB1 × FB2 (3/1)d | 52 | 48/92 |
Number of plants possessing at least one tumor and indication of the respective percentage.
(x/y): x is the number of independent pathogenicity tests performed; y is the number of different strains or strain combinations.
Plants were inspected 6 to 7 DAI.
Plants were inspected 9 to 10 DAI.
Tumor with a diameter <1 mm.
The majority of tumors (68%) developed after 7 DAI and had diameters of 1 to 3 mm.
Interaction of Mrb1 with Lga2 and Rga2
To explore whether Mrb1, Lga2, and Rga2 proteins interact, we conducted yeast two-hybrid assays. For this purpose and to avoid possible interference with subcellular localization, N-terminal deletion derivatives of Mrb1, Lga2, and Rga2 lacking putative mitochondrial targeting sequences were fused to either the LexA DNA binding domain or the B42 activation domain specified by the vectors pLexA and pB42AD, respectively. In the resulting constructs (Table 4), the lower case number reflects the first amino acid position of the respective U. maydis protein that is present in the fusion. Yeast transformants harboring the pLexA constructs alone displayed no LacZ activity, whereas pB42AD constructs as well as the combination of empty pLexA/pB42AD vectors conferred slightly enhanced expression levels (Table 4). The combination of pLexA-mrb142/pB42AD-rga222 displayed ∼100-fold elevated levels of activity and for the reciprocal combination β-galactosidase activity elevated 20-fold (Table 4, combinations 3 and 7). This difference is likely to result from steric constraints. Among the remaining combinations, high LacZ activity levels were only measured in the mrb1/mrb1 combination (Table 4). These results provide evidence for a strong and specific interaction between Mrb1 and Rga2 as well as for Mrb1 self-interaction.
Table 4.
Interaction among Mrb1, Lga2, and Rga2
| Combination | pLexA Construct | pB42AD Construct | β-Galactosidase Activitya |
|---|---|---|---|
| 1 | pLexA-mrb142 | pB42AD-mrb141 | 295.2 ± 78.3 (6/1)b |
| 2 | pLexA-mrb142 | pB42AD-lga244 | 17.8 ± 11.2 (8/1) |
| 3 | pLexA-mrb142 | pB42AD-rga222 | 1364.4 ± 140.2 (6/2) |
| 4 | pLexA-lga243 | pB42AD-mrb141 | 8.4 ± 5.6 (8/1) |
| 5 | pLexA-lga243 | pB42AD-lga244 | 6.6 ± 3.7 (9/1) |
| 6 | pLexA-lga243 | pB42AD-rga222 | 10.8 ± 3.3 (8/1) |
| 7 | pLexA-rga222 | pB42AD-mrb141 | 252.8 ± 47.8 (11/1) |
| 8 | pLexA-rga222 | pB42AD-lga244 | 2.6 ± 2.4 (11/1) |
| 9 | pLexA-rga222 | pB42AD-rga222 | 11.4 ± 2.2 (12/1) |
| 10 | pLexA-mrb142 | –c | 0.1 ± 0.2 (4/1) |
| 11 | pLexA-lga243 | – | 1.2 ± 0.5 (6/1) |
| 12 | pLexA-rga222 | – | 0.2 ± 0.3 (4/1) |
| 13 | – | pB42AD-mrb141 | 9.3 ± 5.2 (11/1) |
| 14 | – | pB42AD-lga244 | 5.3 ± 4.4 (5/1) |
| 15 | – | pB42AD-rga222 | 7.0 ± 1.4 (4/1) |
| 16d | pGB-E1473 | pW2546-GA | 135.3 ± 21.1 (4/6) |
| 17d | pGB-E2473 | pW2546-GA | 5.1 ± 3.3 (4/1) |
| 18d | – | pW2546-GA | 4.9 ± 2.2 (4/1) |
| 19 | PLexA | pB42AD | 11.7 ± 3.7 (4/2) |
β-Galactosidase activity was assayed as described in Methods; β-galactosidase activity in Miller units ± sd.
(x/y): x is the number of independent transformants tested for each quantification; y is the number of repetitions performed with each series.
In this combination, either the pB42AD or the pLexA construct was omitted.
As positive control the combination pGB-E1473/pW2546-GA, containing coding regions for interacting domains of the bE1 and bW2 proteins fused to either the DNA binding or activation domain of GAL4 (Kämper et al., 1995), was used. As negative controls, the combination pGB-E2473/pW2546-GA, in which bE and bW gene fragments are derived from the same allele, and pW2546-GA alone were included.
Subcellular Localization of Mrb1, Lga2, and Rga2
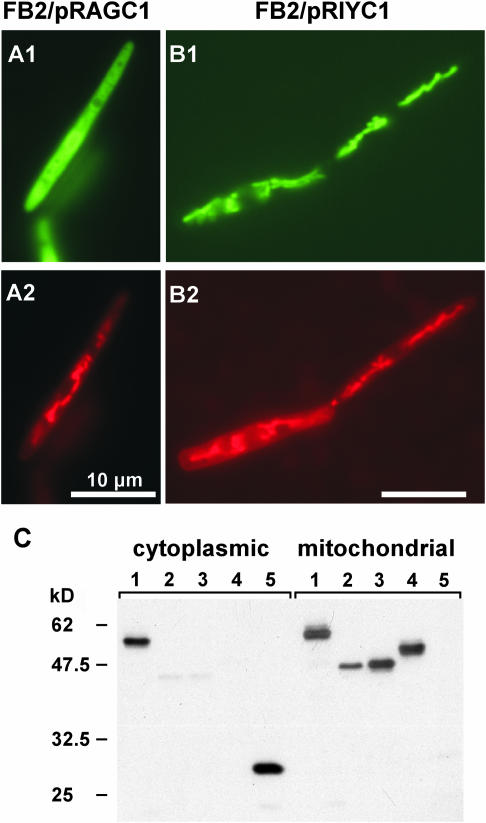
The genetic relationship among Mrb1, Lga2, and Rga2 suggests that all three proteins reside within mitochondria. To assess whether the N terminus of Rga2 specifies an import signal, the coding region for the initial 61 amino acids was translationally fused to eGFP in plasmid pRAGC1. GFP fluorescence in U. maydis pRAGC1 transformants was distributed throughout the cytoplasm (Figure 5A), revealing that the N-terminal Rga2 sequence contains no targeting information. To localize the full-length Rga2 protein, we constructed plasmid pRIYC1, which allows constitutive expression of the complete rga2 sequence fused to the YFP gene, encoding yellow fluorescent protein. Remarkably, pRIYC1 transformants exhibited a typical mitochondrial fluorescence pattern (Figure 5B), suggesting an alternative mitochondrial import mechanism for Rga2. To substantiate mitochondrial localization of Mrb1, Lga2, and Rga2, we investigated whether the respective eGFP or YFP fusion derivatives cofractionated with mitochondria. Immunoblot analysis corroborated the expected mitochondrial localization of all three fusion proteins (Figure 5C). Furthermore, mitochondrial localization of Rga2 was independent from Mrb1 as assessed from analysis of strain FB2Δmrb1/pRIYC1 (Figure 5C). For the Mrb1-eGFP fusion, a substantial amount was also detected in the cytoplasmic fraction, whereas this was not the case for the Lga2 or Rga2 fusions. Taken together, these results provide firm evidence that Mrb1, Lga2, and Rga2 are mitochondrial proteins and might point to an additional cytoplasmic function of Mrb1.
Figure 5.
Subcellular Localization of Rga2 and Immunodetection of Mrb1, Lga2, and Rga2 in Mitochondrial Fractions.
(A) and (B) Localization of Rga2. U. maydis strains FB2/pRAGC1 (A) and FB2/pRIYC1 (B) were grown in liquid CM/Glu medium and assayed for GFP or YFP fluorescence ([A1] and [B1]) and distribution of the marker dye CM-H2Xros ([A2] and [B2]). Bars in all panels = 10 μm.
(C) Proteins from U. maydis strains (1) FB2/pMGH1, (2) FB2/pRIYC1, (3) FB2Δmrb1/pRIYC1, (4) FB2/pLIGC1, and (5) CB35 were loaded. Strain CB35 expresses eGFP under the constitutive otef promoter (Basse et al., 2000). Cytoplasmic (each 50 μg) and mitochondrial protein fractions (each 5 μg) were loaded and immunostained using a monoclonal GFP IgG mouse antibody. The predicted sizes (kD) of fusion proteins are 52.0 (1), 45.8 (2,3), 46.6 (4), and 26.9 (5). Predicted mitochondrial target sequences were subtracted from the Mrb1 and Lga2 sequences for molecular weight calculation. Numbers at the left refer to positions of molecular mass standards in kD. The existence of a cytoplasmic portion of the Mrb1-eGFP fusion was verified in three independent experiments.
Expression of lga2 in Strains Lacking mrb1 Reduces Fitness, Affects Mitochondrial Morphology, and Triggers Degradation of Mitochondrial DNA
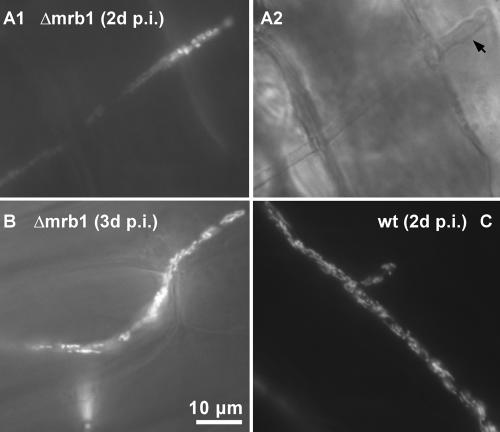
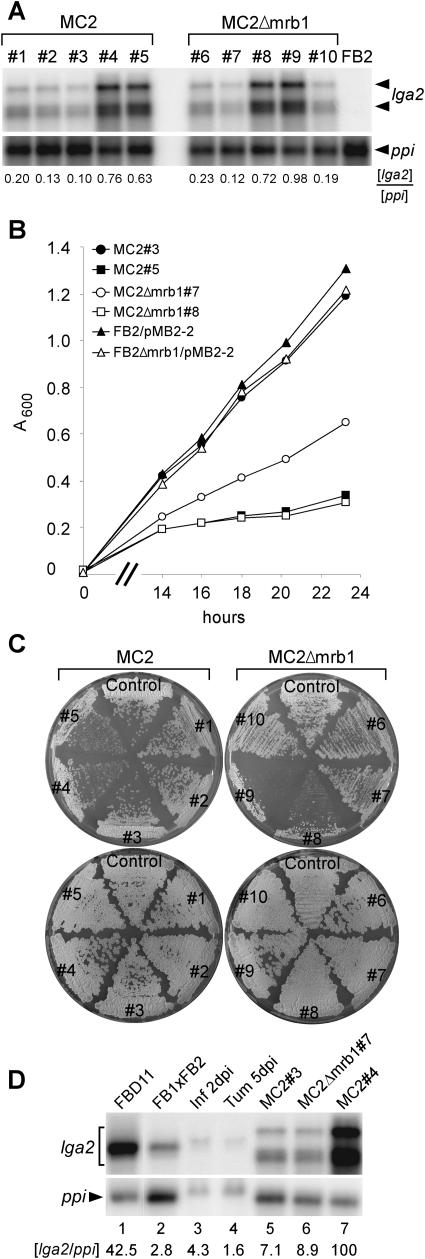
Expression of lga2 is undetectable in haploid strains but is strongly induced in response to the formation of an active bE/bW complex in the dikaryon (Urban et al., 1996b; Romeis et al., 2000). We therefore analyzed the shape of mitochondria in Δmrb1 dikaryotic hyphae during pathogenic growth. Hyphae resulting from fusion of Δmrb1/pMB2-2 strains were only rarely detected during the investigated period from 2 to 4 DAI and mainly displayed diffuse or speckled fluorescence patterns (Figures 6A and 6B). By contrast, wild-type hyphae carrying the pMB2-2 reporter contained thread-like mitochondrial structures (Figure 2C) in the majority of cells but also showed the presence of highly fragmented mitochondrial aggregates in a small percentage (Figure 6C). Prompted by this observation, we investigated the consequences of lga2 expression in haploid cells. Plasmid pCLN1 provided for conditional lga2 expression under the arabinose-inducible crg1 promoter. This plasmid was ectopically integrated into the genome of U. maydis strains FB2/pMB2-2 and FB2Δmrb1/pMB2-2 expressing eGFP targeted to mitochondria. RNA gel blot analysis showed that the resulting transformants, designated MC2 and MC2Δmrb1, respectively, expressed lga2 under inducing conditions but varied in their relative expression levels up to 10-fold (Figure 7A). This variation reflects copy number differences as demonstrated by DNA gel blot analysis (data not shown). Strain FB2 showed no lga2 expression in CM/Ara medium as expected (Figure 7A). Growth in noninducing CM/Glu medium revealed no significant differences among MC2, MC2Δmrb1, and their progenitor strains, and this was also apparent on solid CM/Glu plates (Figure 7C; data not shown). When shifted to CM/Ara medium, growth of all cultures was diminished, reflecting the carbon source preference (Figure 7C; data not shown). Remarkably, under these conditions, growth was negatively influenced by the extent of lga2 overexpression, and this effect was more pronounced in the absence of mrb1 (Figures 7A and 7B). Growth differences related to the level of lga2 expression were also seen when MC2Δmrb1 strains were grown on solid CM/Ara medium. Growth of strains MC2Δmrb1#8, 9 was markedly reduced compared with the progenitor strain and strains MC2Δmrb1#6,7,10 that express lga2 at lower levels (Figure 7C). FB2/pMB2-2 derived transformants like strains MC2#4, 5 that expressed high levels of lga2 were also affected in growth (Figure 7C). It appears that high level expression of lga2 interferes with growth; the few colonies that arise may have developed compensatory mutations.
Figure 6.
Mitochondrial Structures in Dikaryotic Hyphae during Pathogenic Growth.
(A) to (C) Maize plants were inoculated with mixtures of FB1Δmrb1/pMB2-2 and FB2Δmrb1/pMB2-2 ([A] and [B]) or FB1/pMB2-2 and FB2/pMB2-2 (C). Tissue samples collected at the time points indicated from the 3rd maize leaf blade beneath the inoculation site were assayed by epifluorescence to detect mitochondrial-localized GFP ([A1], [B], and [C]). The bar refers to all panels. d p.i., days after inoculation.
(A2) DIC microscopy from (A1). The arrow marks the hyphal tip.
Figure 7.
Influence of lga2 Expression on Cellular Growth in the Presence or Absence of mrb1 and Comparison with lga2 Expression under Natural Conditions.
(A) Expression analysis of MC2 (a2b2) and MC2Δmrb1 strains after shifting from CM/Glu to CM/Ara medium. An RNA gel blot of total RNA (7 μg per lane) was hybridized with a 32P-labeled lga2 fragment. Radioactive signals (both for lga2) were quantified and standardized by comparison with signals obtained from the ppi gene.
(B) Growth comparison of MC2 and MC2Δmrb1 strains in liquid CM/Ara medium at 28°C. The time scale (x axis) refers to the time points after shifting from CM/Glu to CM/Ara medium. MC2#3, closed circle; MC2#5, closed square; MC2Δmrb1#7, open circle; MC2Δmrb1#8, open square; FB2/pMB2-2, closed triangle; FB2Δmrb1/pMB2-2, open triangle. Very similar results were obtained with strains MC2#2, 4 and MC2Δmrb1#6, 9 (data not shown).
(C) Individual strains were streaked out from nourseothricin containing solid PD medium on solid CM/Ara (top) or CM/Glu (bottom) medium and incubated for 64 h at 28°C. Control (left), FB2/pMB2-2; control (right), FB2Δmrb1/pMB2-2.
(D) Comparison of lga2 expression in MC2 and MC2Δmrb1 strains with expression during distinct stages of development. The strains and strain combinations indicated at the top (lanes 1 and 2) were grown on CM-charcoal plates for 48 h for RNA preparation. Lanes 3 and 4, RNA was isolated from infected (FB1xFB2) leaves and leaf tumors 2 and 5 DAI, respectively. Lanes 5 to 7, see (A); 7 μg (lanes 1 and 5 to 7), 15 μg (lane 2), and 80 μg (lanes 3 and 4) RNA were loaded. Two identical RNA gel blots were hybridized with 32P-labeled lga2 and ppi fragments, respectively. Radioactive signals were quantified and the ratios of lga2/ppi hybridization signals were calculated with the most intensive signal (lane 7) set to 100. For lanes 5 to 7, both lga2 signals were used for quantification. Different transcript sizes are probably because of variations in transcription start sites.
To assess whether lga2 expression under natural conditions reached comparable levels as during conditional expression, RNA gel blot analysis was performed. RNA was isolated from a mixture of FB1 and FB2 strains cultivated for 48 h on solid CM-charcoal medium to allow mating and formation of dikaryotic hyphae, from the diploid strain FBD11 growing filamentously under the same conditions, from infected leaf and tumor tissues 2 and 5 DAI, respectively, and from strains MC2#3, 4, and MC2Δmrb1#7 expressing lga2 to different extents. For standardization, the ratio of the lga2 to the constitutive ppi signal was determined. This revealed that lga2 levels in strain FBD11 reached almost the same high levels as in the strongly lga2 overexpressing strain MC2#4 (Figure 7D). lga2 levels after mating of strains FB1 and FB2 were ∼15-fold reduced compared with FBD11, reflecting that in a cross of compatible sporidia not all cells had formed a dikaryon. During biotrophic growth, lga2 expression was ∼10-fold reduced 2 DAI, reaching levels similar to those from strains MC2#3 and MC2Δmrb1#7, and then further decreased by 5 DAI (Figure 7D). Together, this could indicate that under natural conditions high extensive lga2 transcript levels are confined to a short period after cell fusion.
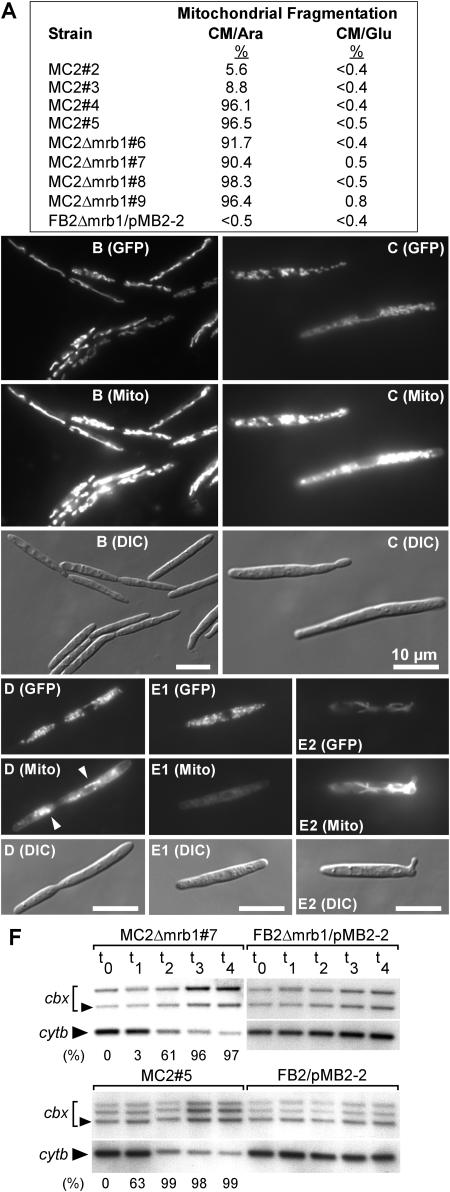
During growth in glucose-containing medium, all progenitor strains as well as all transformants displayed thread-like mitochondrial networks (data not shown). Strikingly, after shifting to arabinose, all transformants highly overexpressing lga2 predominantly (96 to 98%) displayed excessive accumulation of dot-like fluorescent fragments throughout the cytoplasm (Figures 8A, 8D, and 8E1), indicative of mitochondrial fragmentation. Among the transformants that expressed lga2 to a lower extent (Figure 7A), a high proportion (90 to 92%) of MC2Δmrb1 cells displayed fragmented mitochondria (Figures 8A and 8C), whereas mitochondrial networks were seen in the majority of MC2 cells (Figures 8A and 8B). We monitored the time course of mitochondrial fragmentation in strains MC2#4 and MC2Δmrb1#8, which strongly overexpress lga2. Starting with intact mitochondria 2.5 h after shifting to inducing conditions, fragmented mitochondria were observed in 10 and 15% of MC2#4 and MC2Δmrb1#8 cells, respectively. The percentage further increased to 74 and 83%, respectively, after an additional 6 h. This indicates that lga2 expression directly affects mitochondrial morphology.
Figure 8.
Overexpression of lga2 Influences Mitochondrial Morphology, Activity, and Integrity of mtDNA.
(A) Mitochondrial fragmentation (%) in the indicated strains during logarithmic growth 17 to 19 h after transfer to liquid CM/Ara medium. Mitochondrial fragmentation was assessed from distribution of mitochondrial GFP fluorescence (see below). At least 200 cells were included for each calculation.
(B) to (E) Strains MC2 (a2b2) and MC2Δmrb1 were assayed 20 h after transfer to liquid CM/Ara medium for GFP fluorescence (GFP) and distribution of the marker dye CM-H2Xros (Mito) by fluorescence and DIC light microscopy. Representative examples of mitochondrial patterns are shown. MC2#3 (B), MC2Δmrb1#7 (C), MC2#5 (D), and MC2Δmrb1#9 ([E1] and [E2]). Arrowheads in (D) point to spot-like fluorescence. The strong mitotracker fluorescence in thread-like mitochondria (E2) is compared with the hardly detectable fluorescence of fragmented mitochondria (E1) in the same strain. Exposure time for fluorescence photography: 600 ms ([B] and [C]); 2 s ([D] and [E]). All pictures ([B] to [E]) were prepared using Adobe Photoshop, applying identical processing functions and only those that could be applied equally to all pixels of the image being used. Bars in all panels = 10 μm.
(F) Strains indicated on top were precultured in liquid CM/Glu medium, transferred to liquid CM/Ara, and incubated for additional 4 (t1), 8 (t2), 12 (t3), and 14 h (t4). Samples were removed at these time points for analysis of mtDNA degradation. An additional sample was collected from cells immediately before transfer in CM/Ara (t0). Total DNA was digested with BamHI/StuI and simultaneously hybridized with digoxigenin-labeled cytb and cbx (isolated from pSL-Cbx) fragments. The lower band represents the 1823-bp StuI/BamHI fragment from mitochondrial cytb. The 4330-bp band (arrowhead) represents the genomic StuI/BamHI cbx fragment. Additional bands stem from plasmid pMB2-2 (cbx) integrated in one or two copies. The percentage of fragmented mitochondria (determined from at least 100 cells) at the various time points is indicated at the bottom.
To assess respiratory activity in MC2 and MC2Δmrb1 strains expressing lga2, cells were incubated with the mitochondrial marker dye CM-H2Xros, whose fluorescence and sequestration into mitochondria is limited to actively respiring cells. In contrast with MC2#3 and MC2Δmrb1#7 strains expressing lower levels of lga2 (Figures 8B and 8C), mitochondrial fluorescence was markedly reduced in highly expressing lga2 transformants as assessed from rapid fluorescence fading of mitochondrial fragments. Upon excitation, fluorescence either diminished to a few spots (Figure 8D) or was barely detectable (Figure 8E1). Reduction in mitochondrial fluorescence was always associated with the highly fragmented mitochondrial phenotype and was not observed in the minority of cells exhibiting a thread-like mitochondrial pattern (Figure 8E2). Collectively, these results reflect the interaction between Mrb1 and Lga2 and show that overexpression of lga2 interferes with metabolic activity and leads to mitochondrial fragmentation.
The effect of lga2 overexpression on mitochondrial morphology as well as metabolic activity led us to consider mitochondrial DNA (mtDNA) degradation as cause for this phenotype. To assess DNA integrity, total DNA was isolated at various time points after transfer of strains MC2#3, MC2#5, MC2Δmrb1#7, and MC2Δmrb1#8 and control strains FB2/pMB2-2 and FB2Δmrb1/pMB2-2 to CM/Ara (Figure 8A). DNA was subjected to DNA gel blot analysis using probes from the cytochrome b (cytb) and the cbx genes to discriminate between mtDNA and genomic DNA, respectively. In strains MC2#5 and MC2Δmrb1#8 strongly overexpressing lga2 as well as in strain MC2Δmrb1#7, which moderately overexpressed lga2 (Figure 7A), the mtDNA signal started to disappear between 4 and 8 h after transfer to inducing conditions compared with the nuclear cbx signal (Figure 8F; data not shown). At the 4 h time point (t1), fragmentation was mainly confined to mitochondria of the strongly overexpressing strain, whereas at the 8 h time point (t2), this was also apparent in strain MC2Δmrb1#7 weakly overexpressing lga2. In these strains, the percentage of highly fragmented mitochondria further increased to ≥96% until 12 h after transfer (Figure 8F). By contrast, ratios of genomic to mtDNA appeared similar in respective control strains lacking pCLN1 as well as in strain MC2#3 moderately overexpressing lga2 (Figure 8F; data not shown). These results demonstrate a relationship between mitochondrial fragmentation and loss of mtDNA.
Role of lga2 and rga2 in Uniparental Mitochondrial Inheritance?
The observation that overexpression of lga2 induced loss of mtDNA prompted us to investigate whether lga2 plays a role in mitochondrial inheritance as previously suggested (Urban et al., 1996a). The recent release of the mtDNA sequence from U. maydis strain 521 (Whitehead Institute) served as a basis for a comparison of the mitochondrial genomes of strains FB2 (a2b2) and BUB7 (a1b3) and led to the identification of a HindIII restriction fragment length polymorphism (2.3 kb in strain FB2 and 4.5 kb in strain BUB7). To investigate mitochondrial inheritance, we infected plants with mixtures of strain BUB7 and strain FB2 as well as mixtures of BUB7 and two independent FB2Δlga2Δrga2 strains. For strain combinations including Δlga2Δrga2 mutants, six tumors were collected each, and for the wild-type combination, 12 tumors were collected from independently infected plants 7 DAI. DNA gel blot analysis with a probe detecting the polymorphic fragment revealed in all 24 DNA preparations that the band specific for strain BUB7 predominated; in only three probes from wild-type infected tumors an additional faint band specific for FB2 was detected (data not shown). This provides evidence for uniparental mitochondrial inheritance in U. maydis and suggests that the presence of the lga2 and rga2 genes basically have no influence on the outcome.
DISCUSSION
In this study, we have shown that Mrb1 is an important mitochondrial virulence determinant in U. maydis. Most intriguingly, our data have revealed that Mrb1 is only required for pathogenicity in the presence of the a locus genes lga2 and rga2 and to a lesser extent when rga2 is absent. Lga2 and Rga2 proteins were also shown to reside in mitochondria, and Mrb1 and Rga2 were able to interact in the two-hybrid system of yeast. Expression of lga2 from a regulated promoter resulted in fragmentation of tubular structures of mitochondria, a reduction of mitochondrial metabolism, and loss of mtDNA, suggesting a mitochondrial maintenance function for Mrb1.
Mrb1 Is a Member of p32 Family Proteins
Based on its primary sequence and localization, Mrb1 is a member of the p32 protein family that localize predominantly to mitochondria. Despite common structural motifs (Jiang et al., 1999; Hayman et al., 2001), p32 family proteins seem to function in different processes, of which many are attributed to extramitochondrial locations (Krainer et al., 1991; Yu et al., 1995; Petersen-Mahrt et al., 1999; Braun et al., 2000; Van Scoy et al., 2000 and references therein). Only few studies address mitochondrial functions. Among these, a role of p32 in calcium homeostasis was proposed based on structural similarity to calsequestrin (Jiang et al., 1999). Additionally, p32 was found to be associated with mitochondrial protein kinase Cμ and implicated in its activity (Storz et al., 2000). p22 of T. brucei is involved in mitochondrial RNA editing and was shown to stimulate the binding properties of the mitochondrial guide RNA binding protein RBP16 (Hayman et al., 2001). Human p32 and Mam33p of yeast are primarily localized in mitochondria and form oligomeric complexes (Muta et al., 1997; Seytter et al., 1998; Jiang et al., 1999). The demonstration of Mrb1 self-interaction suggests that Mrb1 also adopts an oligomeric structure. However, a mitochondrial function of p32 and Mam33p has not been established conclusively. For Mam33p, it has been demonstrated that it is functionally important for maintaining oxidative phosphorylation in yeast (Muta et al., 1997), but such a role was not detected in an independent study (Seytter et al., 1998). Comparable analysis of Δmrb1 mutants did not reveal growth differences on either fermentable or nonfermentable carbon sources, ruling out a physiologically important role of Mrb1 in mitochondrial functions of haploid cells.
The Interplay among Mrb1, Lga2, and Rga2
We observed that mrb1 was dispensable for pathogenicity of haploid, solopathogenic strains derived from FB1 (a1b1), whereas pathogenicity was abolished in solopathogenic strains derived from FB2 (a2b2). This effect is reminiscent of a recent investigation, which showed that a ste20 homolog in U. maydis was particularly required in an a2 background for pheromone-induced mfa2 gene expression and mating (Smith et al., 2004). Most intriguingly, our data have revealed that Mrb1 is required for pathogenicity in normal crosses of compatible strains, but this requirement ceases when the a2 mating partner is mutant in the a2 locus genes lga2 and rga2. The contribution of lga2 and rga2 to this effect is not equal, and Lga2 acts as major player in preventing pathogenic development of Δmrb1 mutants. Whereas Rga2 alone is unable to interfere with pathogenic development of Δmrb1 strains, a concerted action between Lga2 and Rga2 is suggested by the finding that the Δmrb1Δrga2 mutants are less severely affected in pathogenicity than Δmrb1 mutants, in which lga2 and rga2 are intact. Lga2 and Rga2 were also shown to reside in mitochondria, and Mrb1 and Rga2 interacted in the yeast two-hybrid system. Rga2 represents a small, hydrophilic protein of 18.9 kD with a high content of basic amino acids (20.3%) that are predicted to adopt α-helical structures. In line with the basic nature of Rga2, many of the described interaction partners of p32 family proteins are characterized by basic regions (T. brucei RBP16, human ASF/SF2, and Epstein-Barr nuclear antigen-1), suggesting the participation of electrostatic interactions (Hayman et al., 2001 and references therein). The fact that Rga2 localizes to mitochondria in the absence of an N-terminal targeting sequence implicates the existence of an alternative targeting mechanism for which there is precedence (Biswas and Getz, 2002; Stan et al., 2003). At present we have no data concerning functional domains for the mitochondrial localization of Rga2, but its presence in mitochondria does not require Mrb1.
BLAST and PFAM database searches provided no clues to the function of either lga2 or rga2. The tight linkage of pheromone precursor and receptor genes in mating type (MAT) loci is common in basidiomycetes and has also been described in Coprinus cinereus and Schizophyllum commune (Halsall et al., 2000; Kothe et al., 2003). However, these species do not contain genes related to lga2 and rga2 in their MAT loci. BLAST searches (TBLASTN) of the first release of the C. cinereus genome from the Broad Institute revealed no sequences homologous to either lga2 or rga2 in other parts of the genome but detected a protein sequence similar to Mrb1 (E value, 3e-19). It is thus an interesting possibility that this system for control of mitochondrial function is specific to the plant pathogenic basidiomycetes.
Expression of lga2 Affects Mitochondrial Morphology
Experiments in which lga2 was expressed from the inducible crg1 promoter in wild-type and Δmrb1 mutant cells demonstrated an interference with growth and mitochondrial function. Ectopic expression of lga2 changed mitochondrial morphology from a tubular structure to a dotted appearance, and the extent of mitochondrial fragmentation was determined by the level of lga2 expression and appeared exaggerated in Δmrb1 mutants, corroborating the interaction between Mrb1 and Lga2. This points to the existence of a mitochondrial target of Lga2. The mitochondrial network is a highly dynamic structure and is maintained by balanced membrane fusion and fission events. In S. cerevisiae, mitochondrial fusion and tubular morphology are essential for correct mitochondrial segregation during vegetative growth and sexual development (Nunnari et al., 1997; Yaffe, 1999; Berger and Yaffe, 2000; Westermann, 2002). A component of the yeast mitochondrial fusion machinery is FzoI, encoding a putative GTPase located in the mitochondrial outer membrane (Rapaport et al., 1998). Yeast fzo1 mutants are deficient in mitochondrial fusion and contain fragmented mitochondria that lose their DNA (Hermann et al., 1998; Rapaport et al., 1998). Recently, it has been determined that Fzo1 levels are regulated by MDM30 (Fritz et al., 2003). MDM30 encodes a mitochondrial protein that lacks significant homologies to other proteins in the database; however, it contains at its N terminus an F-box domain. The F-box is a motif of ∼45 amino acids that serves as an interaction site for the Skp1 protein, a component of the Skp1-Cdc53 (cullin)-F-box complex involved in ubiquitin-dependent protein degradation (Patton et al., 1998). Intriguingly, the Lga2 sequence from amino acids 103 to 142 matches the F-box consensus in 17 positions and shows significant similarity to the Mdm30 F-box sequence (Figure 9C). This similarity is consistent with a role of Lga2 in controlling mitochondrial fusion. The existence of an Fzo1 homolog in U. maydis (E value, 2e-77) opens a number of testable predictions to address the function of Lga2 in U. maydis more specifically.
Figure 9.
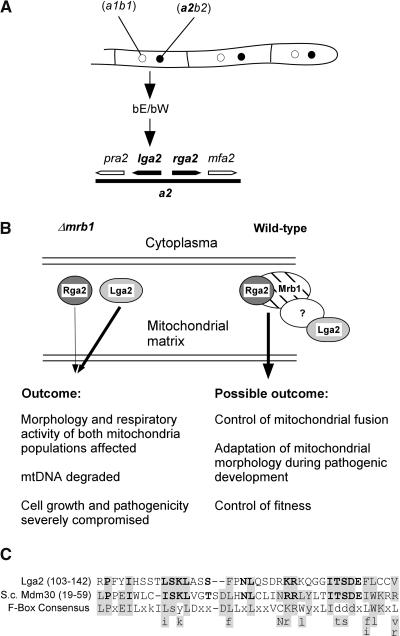
Model for the Interplay among Mrb1, Rga2, and Lga2.
(A) Arrangement of the lga2 and rga2 genes on the a2 mating-type locus and direct control of lga2 expression by an active bE/bW complex formed in the dikaryon.
(B) In Δmrb1 mutants, Lga2 and Rga2 interfere with mitochondrial fusion in an uncontrolled manner and thus affect cellular fitness and fungal virulence. The extent of negative contribution relies on the observed genetic relationship and is reflected by the different thickness of the arrows. Although the presence of Lga2 alone is already sufficient to compromise pathogenicity, Rga2 can only enhance this effect in an Lga2-dependent manner. In wild-type cells, Mrb1 directly controls Rga2, whereas Lga2 is indirectly controlled, possibly by complex formation with a yet unidentified protein. The complex is presumed to control mitochondrial fusion and to be needed for adaptation of mitochondrial morphology and function in response to special demands during the host interaction. It may thus provide for control of fungal fitness in planta.
(C) The putative F-Box motif of Lga2 is aligned with the respective sequence of Mdm30 (S. cerevisiae) and an F-box consensus sequence (Patton et al., 1998). Residues that match F-box sequences are shaded. Identical or highly similar amino acids between Lga2 and Mdm30 are in boldface. Gaps have been introduced at a variable position of the F-box consensus sequence to increase the alignment. Small letters refer to amino acids that are not predominant in a position. Letters beneath the consensus refer to amino acids that fit the Lga2 or Mdm30 motifs and are found in at least two other F-boxes (Bai et al., 1996; Patton et al., 1998).
The features uncovered for Lga2 may also relate to apoptosis (Bossy-Wetzel et al., 2003; Priault et al., 2003), and it is conceivable that the fragmentation of mitochondria observed after induction of lga2, as well as its cytotoxicity, are secondary effects and result from permeabilization of mitochondrial membranes. Furthermore, based on mtDNA degradation in response to lga2 overexpression, we expect a reduction in cytochrome c oxidase activity, which could result in increased reactive oxygen species and a decrease in membrane potential as suggested for S. cerevisiae during apoptosis (Ludovico et al., 2002).
A Model for the Role of mrb1, lga2, and rga2 during the Sexual Life Cycle
Although it is clear that Lga2, Rga2, and Mrb1 localize to mitochondria and are likely to be interconnected in their functions, the expression of the respective genes is quite distinct: mrb1 is constitutively expressed (except in teliospores) and rga2 shows a low basal level that is induced after pheromone stimulation and remains at this level in the dikaryon, whereas lga2 expression is undetectable in haploid cells, is induced by pheromone, and is strongly upregulated by the bE/bW heterodimer (Urban et al., 1996b; Romeis et al., 2000).
The reduction in tumor formation seen in combinations of FB1 and FB2Δmrb1 strains, but not in reciprocal crosses, implicates that the lga2 levels attained before fusion might be sufficiently high to interfere with mitochondrial integrity when mrb1 is absent. During subsequent pathogenic development, lga2 levels are still high enough (Figure 7D) to cause mitochondrial fragmentation and mtDNA degradation in the absence of mrb1, consequently reducing metabolic activity, whereas mrb1 would counteract these detrimental effects in infections with wild-type strains. Based on microscopic investigation of pathogenic development of mrb1 mutants, we suggest that the interference of lga2 expression with mitochondrial function specifically affects hyphal proliferation in planta. Because pathogenicity was compromised in dikaryotic hyphae resulting from a mixture of FB1Δmrb1 and FB2Δmrb1 strains, both mitochondria types should suffer from the absence of mrb1 (Figure 9).
Why would a fungus develop such a system in which expression of the mitochondrial protein Lga2 can become cytotoxic and interfere with subsequent pathogenic development? The lga2 and rga2 genes map to the a2 mating-type locus, an 8.5-kb region that is dissimilar from the a1 locus and is thus inherited as one unit (Urban et al., 1996a). This assures their presence in all mating events. Based on this linkage and a presumed mitochondrial function of Lga2, a role in mitochondrial inheritance was suggested (Urban et al., 1996a). Previous cytoduction assays, which relied on mitochondrial segregation in the progeny of dikaryotic cells that emerged from haploid U. maydis cells differing in their a loci, provided no evidence for preferred mitochondrial inheritance under axenic culture conditions (Trueheart and Herskowitz, 1992). Data provided in this communication suggest that uniparental inheritance of mitochondria does occur during the biotrophic growth phase, and it will be interesting to elucidate when the decision is made. However, lga2 and rga2 did not basically affect the outcome of this segregation process. Although we cannot formally exclude the possibility of Lga2/Rga2 effects on mitochondrial inheritance in other strain combinations, we consider this possibility unlikely.
Owing to the fact that all phenotypic consequences of lga2 expression are counteracted by Mrb1, we suggest the existence of a complex containing Mrb1, Lga2, and Rga2. In light of the finding of Mrb1 self-interaction and interactions between Rga2 and Mrb1, the complex is expected to be of higher order and may contain additional proteins not yet identified (Figure 9). This complex is predicted to control mitochondrial fusion. Mitochondrial morphology has been shown to be important in response to new energy demands and during toxin exposure (Bereiter-Hahn and Vöth, 1994; Bossy-Wetzel et al., 2003 and references therein). In addition, it has been shown that intracellular mitochondrial networks allow the dissipation of energy between remote parts of the cell and can compensate for uneven oxygen supply (Skulachev, 2001). Increasing evidence suggests that mitochondrial structure is regulated during development, and this can involve transcriptional regulation of genes encoding factors of the fission/fusion machinery (Yaffe, 1999; Honda and Hirose, 2003 and references therein). The existence of such a system in U. maydis and its tight control through the mating type loci suggests that mitochondrial morphology needs adaptation during the switch from saprophytic to biotrophic growth. It will be a challenge to find out what special demands the fungus has to cope with in the apoplast of the host plant and how this is linked to fungal fitness via the control of mitochondrial morphology and function.
Recently, the mitochondrial Fow1 protein of Fusarium oxysporum was identified as a first mitochondrial virulence determinant of a plant pathogenic fungus (Inoue et al., 2002). Fow1 displays high similarity to mitochondrial carrier proteins of yeast. Reminiscent to mrb1 mutant strains, fow1 mutants were specifically affected during colonization of plant tissue, and it was suggested that Fow1 might have functions uncoupled from primary mitochondrial functions in response to the host environment.
This study is also reminiscent of a recent investigation, which showed that the virulence of hybrid species of the basidiomycete fungus Heterobasidion annosum, a pathogen of conifers, is controlled by the mitochondrial genome (Olson and Stenlid, 2001). However, in this system, the nature of the mitochondrial factor involved remains to be determined. Our observation that Mrb1 is specifically required for pathogenicity in the presence of the a2 mating-type locus emphasizes the importance of mitochondrial proteins in controlling virulence in a given nuclear background. In addition, this study has uncovered that products of mating-type genes affect mitochondrial morphology and has documented a critical connection between mitochondrial morphology and fungal development during the pathogenic growth phase.
METHODS
Strains and Growth Conditions
Escherichia coli strain TOP10 (Invitrogen, Carlsbad, CA) was used as host for plasmid amplification. Ustilago maydis strains, growth conditions, and mating tests were described previously (Basse et al., 2000, 2002). For arabinose induction, precultures grown in CM/Glu (1% glucose [w/v]) were washed twice in water and resuspended at equal densities before inoculation in CM/Ara (1% arabinose). Cultures were shaken at 200 rpm at 28°C. Solid CM medium also contained 2% Bacto-agar. Solid YPG medium consisted of 2% Bacto-agar, 1% yeast extract, 2% Bacto-peptone, and 3% glycerol. Plant infections were done with the variety Early Golden Bantam (Olds Seed, Madison, WI) as described (Gillissen et al., 1992). Yeast strain EGY48[p8op-LacZ], which carries LEU2 and LacZ under the control of LexA operators, was obtained from Clontech (Palo Alto, CA) and cultivated according to the manufacturer's instructions.
DNA and RNA Procedures
DNA and RNA procedures were performed as described (Basse et al., 2000). Labeling and hybridization with digoxigenin followed the manufacturer's procedure (Roche, Indianapolis, IN). The correctness of all plasmid constructs was verified by sequencing (Automatic DNA Isolation and Sequencing; Max-Planck-Institut, Köln, Germany). Integration of all constructs into genomic U. maydis DNA was either confirmed by PCR or by DNA gel blot analysis. Nucleotide sequences were compared using BLAST, PSI-BLAST (Altschul et al., 1997), and PFAM (http://www.sanger.ac.uk/Software/Pfam). Predictions of mitochondrial targeting sequences, membrane spanning domains, and secondary structures were made with MITOPROT, TMPRED, TMHMM, and SOPM (http://www.expasy.ch). Genomic and mtDNA sequences of U. maydis were retrieved from the first release from the Whitehead Institute (http://www.broad.mit.edu/annotation/fungi/ustilago_maydis/index.html).
DNA Fragments and Investigation of Mitochondrial Inheritance
As selectable markers for U. maydis, hygromycin (hph) and carboxin resistance (ip) cassettes were isolated as NotI fragments as described (Basse et al., 2002). A nourseothricin (nat) resistance cassette was described previously (Müller et al., 1999). For RNA gel blot analysis, a fragment from the constitutively expressed ppi gene (Bohlmann, 1996) and an lga2 cDNA fragment spanning the complete ORF were used.
A 394-bp fragment was amplified from the mitochondrial U. maydis cytb gene using primers Mit1 (5′-CCTTCTAAGTGCTATTCCATGG-3′)/Mit2 (5′-AGCATAGAACGGTAGTAGGTACCAC-3′). An mtDNA fragment of 554 bp was amplified from total U. maydis DNA using primers M18 (5′-CACGTTGTAGAAGCTTGGCTAATCC-3′)/M29 (5′-GCCTACTTCATGGTTGTATTAGGAG-3′) to detect a HindIII restriction fragment length polymorphism between U. maydis strains FB2 (a2b2) and BUB7 (a1b3).
Deletion of mrb1
The NheI-NotI fragment, comprising mrb1 isolated from pS (Basse et al., 2002), was introduced into the EcoRI and HindIII sites of pUC18 to yield pXM. The 608-bp genomic BssHII-KpnI fragment was replaced in pXM by the ip and hph cassettes, respectively. The deletion comprised the complete mrb1 ORF except 85 bp at the 5′end and 123 bp at the 3′end. To replace the resident mrb1 gene with the Δmrb1 allele, pXMDmrb1-ip and pXMDmrb1-hph linearized at the NdeI and Eco47III sites, respectively, were introduced into U. maydis strains FB1, FB2, and SG200.
Deletion of lga2 and rga2
The generation of deletions spanning the lga2 and rga2 genes was performed by a PCR-based approach (Kämper, 2004). The lga2 and rga2 double deletion comprised positions 5780 to 7908 of the mfa2 locus (GenBank accession number U37796). The individual lga2 and rga2 deletions comprised positions 5780 to 6678 and 7385 to 7908, respectively, of the mfa2 locus.
Fusion Constructs
For construction of eGFP reporter constructs pMB2-1 and pMB2-2, pXM was amplified with the primers xs1 5′-ATAGTCATGATCGCACGCAACTTGTGC-3′/xs2 5′-GAGCTCATGACCGAAAACGTGCGCATACC-3′ and xs1/xs3 5′-GCCACCATGGCGTCGTATTCGCCTTGACC-3′, respectively. PCR products were cleaved with BspHI and/or NcoI and introduced into the NcoI site of pHD3 (Basse et al., 2002). pMB2-1 and pMB2-2 were linearized with AatII and integrated ectopically in the genome of U. maydis. For construction of the mrb1:eGFP fusion construct pMGH1, pXM was amplified using the mrb1-specific primers 5′-CAATGATATCGAATCCGCTCTCGAGCG-3′/5′-GATTCCATGGCATTGATGAAATCGCGC-3′. The PCR product was cleaved with EcoRV/NcoI and ligated to eGFP isolated as NcoI-EcoRV fragment (Basse et al., 2000). pMGH1 was derived from this construct by ligating the hph cassette into the EcoRV site. This plasmid was linearized with Eco47III for introduction into U. maydis. Homologous integration of pMGH1 was confirmed in all wild-type and FB2Δmrb1 transformants. For construction of pLIGC1, genomic FB2 DNA was amplified with primers 5′-GAGCTCATGATGCTACTGTCCTATATCC-3′/5′-GAGCTCATGACGTCCAGGAAAACGCACATC-3′. The PCR product was cleaved with BspHI and inserted into the NcoI site of pHD3. For construction of pRIYC1, genomic FB2 DNA was amplified with primers RG1/RG2 (5′-CATGCCATGGAAAAGATTCGCCCCACATTG-3′/5′-GAGCTCATGAAACGCTTTCGTCTCCACCTTC-3′). PCR products were digested with NcoI/BspHI and inserted into the NcoI site of plasmid p123-YFP, which enables YFP expression under the otef promoter (Weber et al., 2003). For construction of pRAGC1, genomic FB2 DNA was amplified with primers RG1/RG3 (5′-GAGCTCATGAGACGAGGTGCGTCAGC-3′). The resulting PCR product was cleaved with NcoI/BspHI and introduced into the NcoI site of pHD3. All plasmids were linearized at the SspI site for homologous integration into the ip locus (Basse et al., 2000).
Construction of pCLN1
A genomic lga2 fragment extending the complete ORF sequence at the 3′ site was amplified from FB2 DNA using primers 5′- GGAATTCCATATGATGCTACTGTCCTATATCC-3′/5′-GCAGGATATCGTTAGCAAGAAGATACACGACC-3′. The PCR product was cleaved with NdeI/EcoRV and inserted into the HindIII/NdeI sites of plasmid pRU11, which contains the crg1 promoter (Bottin et al., 1996; Brachmann et al., 2001). pCLN1 was obtained from this plasmid by inserting a nat cassette into the PmlI site. pCLN1 was linearized with ScaI for transformation into U. maydis.
Transformation of b1 and b2 Alleles
Plasmids pUb2-2 and pUb1 contain 8-kb BamHI fragments encompassing the complete bE2 and bE1 genes, respectively (Schulz et al., 1990). Both plasmids carry the hph gene. pUb2-2 linearized with NotI was introduced into U. maydis strains FB1 and FB1Δmrb1. pUb1 was linearized with MluI and introduced into strains FB2 and FB2Δmrb1. Integration of plasmids was confirmed by PCR using primers that bind in the variable portions of bW2 and bW1/bE1, respectively. The presence of an active bE1/bW2 complex in pUB1 transformants was verified by pheromone stimulation with the compatible donor strain FB6b (a1b2), and the presence of an active bE2/bW1 complex was verified by pheromone stimulation assays with strain FB6a (a2b1).
Interaction among Mrb1, Lga2, and Rga2
For construction of pLexA-mrb142, a mrb1 fragment was amplified from positions 124 to 825 of the mrb1 ORF and introduced into the BamHI and NcoI sites of pLexA (Gyuris et al., 1993). For construction of pLexA-lga243 and pLexA-rga222, lga2 and rga2 fragments were amplified from a FBD11 cDNA preparation (Basse et al., 2000) from positions 127 to 648 (lga2) and 64 to 477 (rga2) and introduced into the BamHI/NcoI and EcoRI/NcoI sites of pLexA. For construction of pB42AD-mrb141, pB42AD-lga244, and pB42AD-rga222, fragments were amplified from positions 121 to 825, 130 to 648, and 64 to 477 of the ORF regions of mrb1, lga2, and rga2, respectively, and introduced into the XhoI site of pB42AD (Gyuris et al., 1993). Plasmids pGB-E1473, pGB-E2473, and pW2546-GA have been described (Kämper et al., 1995). Transformation of yeast strain EGY48[p8op-LacZ] followed Clontech's instructions. For quantification of β-galactosidase activity according to Miller (1972), cells were grown in galactose/raffinose-containing selective medium to an OD600 of 0.8 to 1.4.
Isolation of Mitochondria from U. maydis
U. maydis strains were grown in CM/Glu (1%) medium. Protoplasts were prepared as described (Schulz et al., 1990) using an STC solution lacking CaCl2 (20 mM Tris-HCl, pH 7.5, and 1 M sorbitol). Mitochondria were prepared from protoplasts according to the yeast protocol (Yaffe, 1991). The cytoplasmic fraction was collected from the 9500g supernatant. The determination of mitochondrial protein concentrations followed the same protocol.
Immunodetection
Immunoblot analysis was performed as described (Basse et al., 2000) using a monoclonal GFP IgG mouse antibody (Roche).
Microscopy
Microscopy and processing of images was performed as described (Basse et al., 2000). Cell wall components were stained with 2 μg/mL of Calcofluor-white (Sigma, St. Louis, MO) in PBS. The existence of dikaryotic hyphae was verified by 4′,6-diamidino-2-phenylindole (DAPI) staining: U. maydis cells grown on solid PD charcoal medium were fixed with 4% (w/v) formaldehyde and stained with 0.5 μg/mL of DAPI (Sigma) in PBS, pH 7.2, at 60°C. Samples were observed with DIC optics or under fluorescence microscopy (excitation/emission for DAPI or Calcofluor-white, eGFP, YFP: 365/>397 nm; 450 to 490/515 to 565 nm; 480 to 520/505 to 565 nm). To visualize mitochondria, U. maydis cells were incubated with 0.5 μM Mito-Tracker CM-H2Xros (Molecular Probes, Eugene, OR) in the same medium as used for cell culture for 20 min at 25°C and observed under fluorescence microscopy (excitation/emission: 546/>590 nm). Staining with CBE followed the method of Brundrett et al. (1996).
The nucleotide sequence data referred to the mig2 locus from position 8423 to 10,542 (Basse et al., 2002), comprising mrb1 coding and 5′/3′ noncoding sequences, have been submitted to the GenBank database under accession number AY574042. GenBank accession numbers for the sequences of the U. maydis mfa2 locus and the cytb gene are U37796 and AB040663, respectively.
Acknowledgments
We thank Christine Kerschbamer for help in generating Δmrb1 strains and initial pathogenicity experiments and Kathrin Stelter and Susanne Berger for assistance in mapping mtDNA sequences. In addition, we would like to thank two anonymous reviewers for their suggestions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christoph W. Basse (basse@staff.uni-marburg.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022657.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustlago maydis. Development 122, 2965–2976. [DOI] [PubMed] [Google Scholar]
- Basse, C.W., Kolb, S., and Kahmann, R. (2002). A maize-specifically expressed gene cluster in Ustilago maydis. Mol. Microbiol. 43, 75–93. [DOI] [PubMed] [Google Scholar]
- Basse, C.W., and Steinberg, G. (2004). Ustilago maydis, model system for analysis of the molecular basis of fungal pathogenicity. Mol. Plant. Pathol. 5, 83–92. [DOI] [PubMed] [Google Scholar]
- Basse, C.W., Stumpferl, S., and Kahmann, R. (2000). Characterization of a Ustilago maydis gene specifically induced during the biotrophic phase: Evidence for negative as well as positive regulation. Mol. Cell. Biol. 20, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn, J., and Vöth, M. (1994). Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27, 198–219. [DOI] [PubMed] [Google Scholar]
- Berger, K.H., and Yaffe, M.P. (2000). Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8, 508–513. [DOI] [PubMed] [Google Scholar]
- Biswas, T.K., and Getz, G.S. (2002). Import of yeast mitochondrial transcription factor (Mtf1p) via a nonconventional pathway. J. Biol. Chem. 277, 45704–45714. [DOI] [PubMed] [Google Scholar]
- Bohlmann, R. (1996). Isolierung und Charakterisierung von filamentspezifisch exprimierten Genen aus Ustilago maydis. PhD dissertation (Munich, Germany: LMU München).
- Bölker, M., Genin, S., Lehmler, C., and Kahmann, R. (1995). Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73, 320–325. [Google Scholar]
- Bölker, M., Urban, M., and Kahmann, R. (1992). The a mating type locus of Ustilago maydis specifies cell signaling components. Cell 68, 441–450. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel, E., Barsoum, M.J., Godzik, A., Schwarzenbacher, R., and Lipton, S.A. (2003). Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr. Opin. Cell Biol. 15, 706–716. [DOI] [PubMed] [Google Scholar]
- Bottin, A., Kämper, J., and Kahmann, R. (1996). Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253, 342–352. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Weinzierl, G., Kämper, J., and Kahmann, R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42, 1047–1063. [DOI] [PubMed] [Google Scholar]
- Braun, L., Ghebrehiwet, B., and Cossart, P. (2000). gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 19, 1458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett, M., Bougher, N., Dell, B., Grove, T., and Malajczuk, N. (1996). Working with Mycorrhizas in Forestry and Agriculture, Monograph 32. (Canberra, Australia: Australian Centre for International Agricultural Research).
- Fritz, S., Weinbach, N., and Westermann, B. (2003). Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell 14, 2303–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen, B., Bergemann, J., Sandmann, C., Schroer, B., Bölker, M., and Kahmann, R. (1992). A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68, 647–657. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Halsall, J.R., Milner, M.J., and Casselton, L.A. (2000). Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics 154, 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman, M.L., Miller, M.M., Chandler, D.M., Goulah, C.C., and Read, L.K. (2001). The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA binding activity. Nucleic Acids Res. 29, 5216–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G.J., Thatcher, J.W., Mills, J.P., Hales, K.G., Fuller, M.T., Nunnari, J., and Shaw, J.M. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell. Biol. 143, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, S., and Hirose, S. (2003). Stage-specific enhanced expression of mitochondrial fusion and fission factors during spermatogenesis in rat testis. Biochem. Biophys. Res. Commun. 311, 424–432. [DOI] [PubMed] [Google Scholar]
- Inoue, I., Namiki, F., and Tsuge, T. (2002). Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14, 1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Zhang, Y., Krainer, A.R., and Xu, R.M. (1999). Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl. Acad. Sci. USA 96, 3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann, R., Steinberg, G., Basse, C., Feldbrügge, M., and Kämper, J. (2000). Ustilago maydis, the causative agent of corn smut disease. In Fungal Pathology, J.W. Kronstad, ed (Dordrecht, The Netherlands: Kluwer), pp. 347–371.
- Kämper, J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271, 103–110. [DOI] [PubMed] [Google Scholar]
- Kämper, J., Reichmann, M., Romeis, T., Bölker, M., and Kahmann, R. (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81, 73–83. [DOI] [PubMed] [Google Scholar]
- Kothe, E., Gola, S., and Wendland, J. (2003). Evolution of multispecific mating-type alleles for pheromone perception in the homobasidiomycete fungi. Curr. Genet. 42, 268–275. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Mayeda, A., Kozak, D., and Binns, G. (1991). Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66, 383–394. [DOI] [PubMed] [Google Scholar]
- Kronstad, J.W., and Leong, S.A. (1989). Isolation of two alleles of the b locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA 86, 978–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludovico, P., Rodrigues, F., Almeida, A., Silva, M.T., Barrientos, A., and Corte-Real, M. (2002). Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Espinoza, A.D., Garcia-Pedrajas, M.D., and Gold, S.E. (2002). The Ustilaginales as plant pests and model systems. Fungal Genet. Biol. 35, 1–20. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 352–355.
- Müller, P., Aichinger, C., Feldbrügge, M., and Kahmann, R. (1999). The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34, 1007–1017. [DOI] [PubMed] [Google Scholar]
- Muta, T., Kang, D., Kitajima, S., Fujiwara, T., and Hamasaki, N. (1997). p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272, 24363–24370. [DOI] [PubMed] [Google Scholar]
- Nunnari, J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, Å., and Stenlid, J. (2001). Mitochondrial control of fungal hybrid virulence. Nature 411, 438. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt, S.K., Estmer, C., Ohrmalm, C., Matthews, D.A., Russell, W.C., and Akusjarvi, G. (1999). The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18, 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault, M., Camougrand, N., Kinnally, K.W., Vallette, F.M., and Manon, S. (2003). Yeast as a tool to study Bax/mitochondrial interactions in cell death. FEMS Yeast Res. 4, 15–27. [DOI] [PubMed] [Google Scholar]
- Rapaport, D., Brunner, M., Neupert, W., and Westermann, B. (1998). Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150–20155. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Brachmann, A., Kahmann, R., and Kämper, J. (2000). Identification of a target gene for the bE-bW homeodomain protein complex in Ustilago maydis. Mol. Microbiol. 37, 54–66. [DOI] [PubMed] [Google Scholar]
- Schulz, B., Banuett, F., Dahl, M., Schlesinger, R., Schäfer, W., Martin, T., Herskowitz, I., and Kahmann, R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60, 295–306. [DOI] [PubMed] [Google Scholar]
- Seytter, T., Lottspeich, F., Neupert, W., and Schwarz, E. (1998). Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae is related to the human complement receptor gC1q-R. Yeast 15, 303–310. [DOI] [PubMed] [Google Scholar]
- Skulachev, V.P. (2001). Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 26, 23–29. [DOI] [PubMed] [Google Scholar]
- Smith, D.G., Garcia-Pedrajas, M.D., Hong, W., Yu, Z., Gold, S.E., and Perlin, M.H. (2004). An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot. Cell 3, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1994). Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol. Res. 98, 347–355. [Google Scholar]
- Spellig, T., Bölker, M., Lottspeich, F., Frank, R.W., and Kahmann, R. (1994). Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13, 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan, T., Brix, J., Schneider-Mergener, J., Pfanner, N., Neupert, W., and Rapaport, D. (2003). Mitochondrial protein import: Recognition of internal import signals of BCS1 by the TOM complex. Mol. Cell. Biol. 23, 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, P., Hausser, A., Link, G., Dedio, J., Ghebrehiwet, B., Pfizenmaier, K., and Johannes, F.J. (2000). Protein kinase C [micro] is regulated by the multifunctional chaperon protein p32. J. Biol. Chem. 275, 24601–24607. [DOI] [PubMed] [Google Scholar]
- Trueheart, J., and Herskowitz, I. (1992). The a locus governs cytoduction in Ustilago maydis. J. Bacteriol. 174, 7831–7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, M. (1995). Pheromonantwort beim Maisbrandpilz Ustilago maydis. PhD dissertation (Munich, Germany: LMU München).
- Urban, M., Kahmann, R., and Bölker, M. (1996. a). The biallelic a mating type locus of Ustilago maydis: Remnants of an additional pheromone gene indicate evolution from a multiallelic ancestor. Mol. Gen. Genet. 250, 414–420. [DOI] [PubMed] [Google Scholar]
- Urban, M., Kahmann, R., and Bölker, M. (1996. b). Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251, 31–37. [DOI] [PubMed] [Google Scholar]
- Van den Brulle, J., Steidl, S., and Brakhage, A.A. (1999). Cloning and characterization of an Aspergillus nidulans gene involved in the regulation of penicillin biosynthesis. Appl. Environ. Microbiol. 65, 5222–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Scoy, S., Watakabe, I., Krainer, A.R., and Hearing, J. (2000). Human p32: A coactivator for Epstein-Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology 275, 145–157. [DOI] [PubMed] [Google Scholar]
- Weber, I., Gruber, C., and Steinberg, G. (2003). A class V myosin required for mating, hyphal growth and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15, 2826–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, B. (2002). Merging mitochondria matters. Cellular role and molecular machinery of mitochondrial fusion. EMBO Rep. 3, 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P. (1991). Analysis of mitochondrial function and assembly. Methods Enzymol. 194, 631–634. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.P. (1999). The machinery of mitochondrial inheritance behavior. Science 283, 1493–1497. [DOI] [PubMed] [Google Scholar]
- Yu, L., Shang, Z., Loewenstein, P.M., Desai, K., Tang, Q., Mao, D., Symington, J.S., and Green, M. (1995). Molecular cloning and characterization of a cellular protein that interacts with the human immunodeficiency virus type 1 tat transactivator and encodes a strong transcriptional activation domain. J. Virol. 69, 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]