ABSTRACT
Invasive Candida albicans infections are a serious health threat for immunocompromised individuals. Fluconazole is most commonly used to treat these infections, but resistance due to the overexpression of multidrug efflux pumps is of grave concern. This study evaluated the ability of five synthetic organotellurium compounds to reverse the fluconazole resistance of C. albicans clinical isolates. Compounds 1 to 4, at <10 μg/ml, ameliorated the fluconazole resistance of Saccharomyces cerevisiae strains overexpressing the major C. albicans multidrug efflux pumps Cdr1p and Mdr1p, whereas compound 5 only sensitized Mdr1p-overexpressing strains to fluconazole. Compounds 1 to 4 also inhibited efflux of the fluorescent substrate rhodamine 6G and the ATPase activity of Cdr1p, whereas all five of compounds 1 to 5 inhibited Nile red efflux by Mdr1p. Interestingly, all five compounds demonstrated synergy with fluconazole against efflux pump-overexpressing fluconazole-resistant C. albicans clinical isolates, isolate 95-142 overexpressing CDR1 and CDR2, isolate 96-25 overexpressing MDR1 and ERG11, and isolate 12-99 overexpressing CDR1, CDR2, MDR1, and ERG11. Overall, organotellurium compounds 1 and 2 were the most promising fluconazole chemosensitizers of fluconazole-resistant C. albicans isolates. Our data suggest that these novel organotellurium compounds inhibit pump efflux by two very important and distinct families of fungal multidrug efflux pumps: the ATP-binding cassette transporter Cdr1p and the major facilitator superfamily transporter Mdr1p.
KEYWORDS: multidrug resistance, efflux pumps, organotellurium, yeasts
INTRODUCTION
Opportunistic fungal infections can become life threatening for immunocompromised individuals (1, 2). The main etiological agent of invasive mycoses remains Candida albicans despite a notable increase of non-albicans Candida species in recent years (3). Azoles, especially the narrow-spectrum triazole fluconazole, are the most commonly used class of antifungal to treat Candida infections (4, 5). Their target, lanosterol 14α-demethylase (Erg 11p), is an essential enzyme of ergosterol biosynthesis, a key component of the fungal cell membrane (6). However, the often prolonged prophylactic treatment of immunocompromised patients with fluconazole has led to the inevitable rise of resistant C. albicans clinical isolates. The major azole resistance mechanisms include (i) the overexpression and/or (ii) mutation of the azole drug target Erg11p, (iii) the inactivation of ERG6 (7), but most commonly involve (iv) the overexpression of multidrug efflux pumps such as the prototype fungal multidrug efflux pumps C. albicans Cdr1p and Mdr1p (8).
ATP-binding cassette (ABC) transporters use ATP hydrolysis and major facilitator superfamily (MFS) transporters use the proton motive force to protect cells from lethal concentrations of azoles by transporting toxic compounds across their biological membranes into the cell exterior (9). The two main ABC transporters associated with resistance to fluconazole in C. albicans are Cdr1p and Cdr2p, with Cdr1p contributing most to the fluconazole resistance of clinical isolates (10). Overexpression of Mdr1p can also cause reduced fluconazole susceptibilities in C. albicans (11). Azole resistance development of clinical isolates is often a multifaceted process, and fluconazole-resistant C. albicans clinical isolates often contain more than one cause of azole resistance that usually accumulate over a prolonged period of time of fluconazole exposure (12–14).
The inhibition of multidrug efflux pumps by nonantifungal agents has been considered a promising strategy to reverse fluconazole resistance (9, 15). Many compounds, including FK506 (16), unnarmicins A and C (17), chalcone derivatives (18), disulfiram (19), curcumin (20), ibuprofen (21), and some promising milbemycins (22, 23), are capable of restoring the fluconazole susceptibility of C. albicans- and C. glabrata-resistant isolates, as well as the innately fluconazole-resistant C. krusei. However, fewer compounds have been described to reverse fluconazole resistance of Mdr1p-overexpressing clinical isolates. Examples of Mdr1p inhibitors are cerulenin analogues (24), the monoamine oxidase A inhibitor clorgyline (which inhibits a broad range of fungal ABC multidrug efflux transporters, as well as Mdr1p) (25), and compounds with a core cyclobutene-dione ring (26). A recent study by Reis de Sá et al. (27) demonstrated that synthetic organotelluride compounds inhibited the prototype Saccharomyces cerevisiae ABC multidrug efflux pump Pdr5p, a homolog of C. albicans Cdr1p. We sought here to evaluate the inhibitory potential of these organotellurides on the major C. albicans multidrug efflux pumps and to test their ability to reverse the fluconazole resistance of C. albicans clinical isolates known to overexpress variations of these efflux pumps.
RESULTS
Organotellurium compounds partially restore the fluconazole sensitivity of S. cerevisiae strains overexpressing C. albicans Cdr1p and Mdr1p.
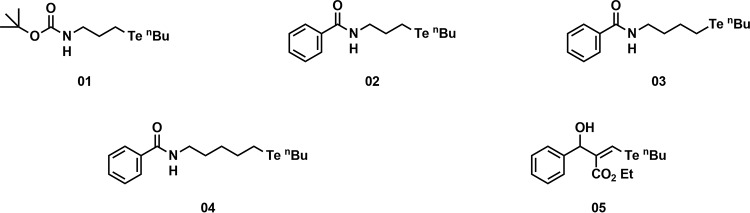
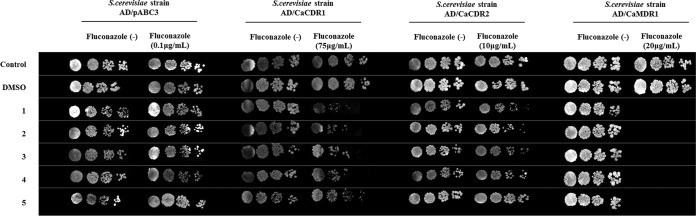
Qualitative chemosensitization assays of S. cerevisiae AD overexpressing the major C. albicans multidrug efflux pumps Cdr1p, Cdr2p, and Mdr1p showed that compounds 1, 2, 3, and 4, but not compound 5 (Fig. 1), partially restored the fluconazole sensitivity of CaCdr1p-overexpressing cells (Fig. 2). Interestingly, all five organotellurium compounds had little effect on the fluconazole susceptibility of strains overexpressing the close CaCdr1p homolog CaCdr2p, but they also reversed the fluconazole resistance of CaMdr1p-overexpressing strains (Fig. 2).
FIG 1.
Structures of the five synthetic organotellurium compounds.
FIG 2.
Organotellurium compounds chemosensitize S. cerevisiae AD strains overexpressing the C. albicans efflux pumps CaCdr1p, CaCdr2p, and CaMdr1p to fluconazole (FLC). Columns labeled “fluconazole (−)” show the growth of serial 5-fold dilutions of the indicated S. cerevisiae strains on YPD agar in the absence of FLC; columns labeled “fluconazole (+)” show the growth of the same serial dilutions of the indicated strains on YPD medium in the presence of subinhibitory concentrations of FLC (∼1/2 to ∼1/4 of their respective MICs). Cells grown on YPD with or without FLC only or on 0.5% DMSO were used as positive controls. Rows 1, 2, 3, 4, and 5 show the growth of the same cells in the presence of 100 μM compounds 1 to 5 (34, 35, 36, 38, and 40 μg/ml, respectively).
Quantification of these findings using two-dimensional checkerboard assays confirmed these results: synergy between compounds 1 to 4 and fluconazole was determined for AD/CaCDR1 and for all five compounds for AD/CaMDR1 cells (Table 1). However, all compounds could only partially restore the fluconazole sensitivity of CaCdr1p- and CaMdr1p-overexpressing cells; the fluconazole MICs of AD/CaCDR1 and AD/CaMDR1 cells in the presence of inhibitory concentrations of compounds 1 to 5 remained relatively high (75 and 3.8 μg/ml, respectively [Table 1]) compared to the sensitive control strain AD/pABC3 (0.2 μg/ml [data not shown]).
TABLE 1.
Checkerboard assay of S. cerevisiae strains overexpressing multidrug efflux pumps CaCDR1 and CaMDR1a
| Strain and compound | Compound concn (μg/ml) |
FLC concn (μg/ml) |
FICI | Outcome | ||||
|---|---|---|---|---|---|---|---|---|
| MIC1* | MIC2* | FIC | MIC1 | MIC2 | FIC | |||
| AD/CaCDR1 | ||||||||
| 1 | 69 | 0.54 | 0.008 | 300 | 75 | 0.25 | 0.26 | Synergy |
| 2 | 70 | 0.14 | 0.002 | 300 | 75 | 0.25 | 0.25 | Synergy |
| 3 | 73 | 0.07 | 0.001 | 300 | 75 | 0.25 | 0.25 | Synergy |
| 4 | 75 | 0.59 | 0.008 | 300 | 75 | 0.25 | 0.26 | Synergy |
| AD/CaMDR1 | ||||||||
| 1 | 69 | 0.13 | 0.002 | 15 | 3.8 | 0.25 | 0.25 | Synergy |
| 2 | 70 | 0.27 | 0.004 | 15 | 3.8 | 0.25 | 0.25 | Synergy |
| 3 | 73 | 1.13 | 0.016 | 15 | 3.8 | 0.25 | 0.27 | Synergy |
| 4 | 75 | 0.14 | 0.002 | 15 | 3.8 | 0.25 | 0.25 | Synergy |
| 5 | 80 | 0.63 | 0.008 | 15 | 3.8 | 0.25 | 0.26 | Synergy |
MIC values were determined by a broth microdilution assay according to CLSI protocol M27-A3 (2008) using 50% growth reduction as the cutoff for the reported MIC values. MIC1, MIC of compound or FLC alone; MIC2, MIC of compound or FLC in the presence of FLC or compound, respectively. FIC, fractional inhibitory concentration; FICI, fractional inhibitory concentration index. *, values were converted from μM to μg/ml.
Organotellurides cause intracellular accumulation of the CaCdr1p and CaMdr1p efflux pump substrates R6G and Nile red.
R6G and Nile red are substrates of CaCdr1p, but only Nile red is a substrate of CaMdr1p (28). As demonstrated in Fig. 3, the glucose-dependent rhodamine 6G (R6G) efflux of AD/CaCDR1 cells was inhibited by compounds 1, 2, 3, and 4. Nile red efflux mediated by CaMdr1p of S. cerevisiae AD/CaMDR1 cells was inhibited by compounds 1, 2, 3, 4, and 5 (Fig. 4).
FIG 3.
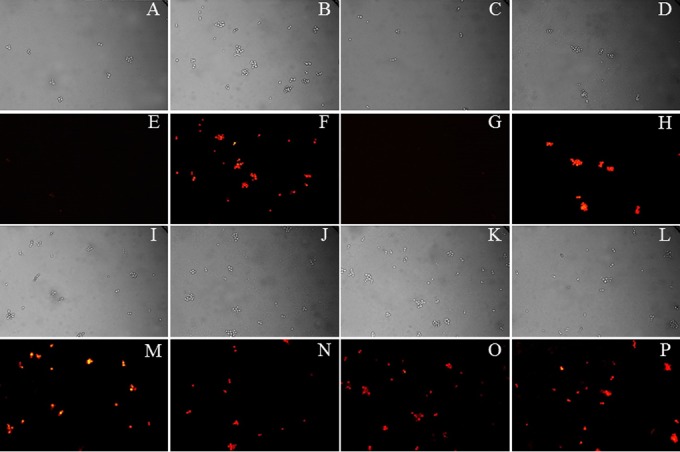
Effect of organotellurium compounds 1 to 4 on the intracellular accumulation of R6G in S. cerevisiae AD/CaCDR1. Panels A to D and panels I to L show the bright-field images of the corresponding fluorescence microscopy images shown in panels E to H and M to P. AD/CaCDR1 cells preloaded with R6G were incubated under the following conditions: A and E, 0.2% glucose; B and F, no glucose; C and G, 0.2% glucose + DMSO; D and H, 0.2% glucose + compound 1; I and M, 0.2% glucose + compound 2; J and N, 0.2% glucose + compound 3; and K and O, 0.2% glucose + compound 4. Panels L and P show cell images for the R6G-preloaded sensitive control strain S. cerevisiae AD in the presence of 0.2% glucose. The pump-inhibitory effects of compounds 1 to 5 were tested at 100 μM (34, 35, 36, 38, and 40 μg/ml, respectively).
FIG 4.
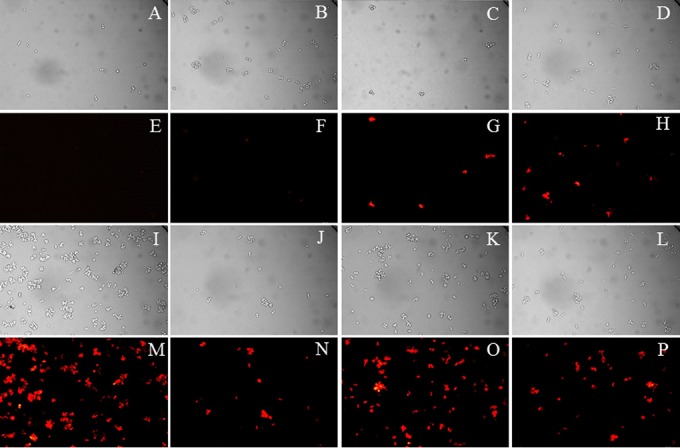
Effect of organotellurium compounds 1 to 5 on the intracellular accumulation of Nile red in S. cerevisiae AD/CaMDR1. The panels A to D and I to L show the bright-field images of the corresponding fluorescence microscopy images shown in panels E to H and M to P. AD/CaMDR1 cells preloaded with Nile red were incubated under the following conditions: A and E, PBS (pH 7.2) without any compound; B and F, + 0.5% DMSO; C and G, + compound 1; D and H, + compound 2; I and M, + compound 3; J and N, + compound 4; and K and O, + compound 5. Panels L and P show cell images for the Nile red-preloaded sensitive control strain S. cerevisiae AD in PBS (pH 7.2) without the addition of any compound. The pump-inhibitory effects of compounds 1 to 5 were tested at 100 μM (34, 35, 36, 38, and 40 μg/ml, respectively).
These results were confirmed by flow cytometry (Fig. 5 and 6). As expected, the glucose-dependent R6G efflux of S. cerevisiae AD/CaCDR1 cells was inhibited by 100 μM concentrations of compounds 1, 2, 3, and 4 (i.e., 34, 35, 36, and 38 μg/ml, respectively), leading cells to accumulate as much R6G inside their cells as had the sensitive control strain AD/pABC3 (Fig. 5). Compound 5 (40 μg/ml), on the other hand, exhibited no detectable CaCdr1p efflux pump activity (Fig. 5). When we assessed the Nile red efflux of AD/CaMDR1 cells, we found that these, too, accumulated as much Nile red inside their cells in the presence of organotellurium compounds 1 to 5 as had the sensitive control strain AD/pABC3 (Fig. 6).
FIG 5.
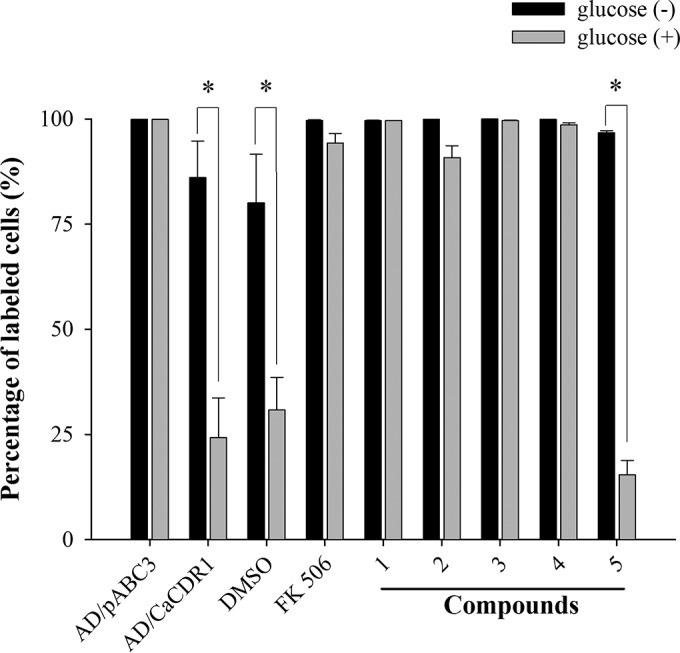
Quantification by flow cytometry of the CaCdr1p pump-inhibitory effects of organotellurium compounds 1 to 5. The bars indicate the percent R6G accumulation relative to the sensitive control strain AD/pABC3, which was set to 100%, in the absence (■) or presence () of 0.2% glucose of R6G-preloaded AD/CaCDR1 cells incubated in the presence of DMSO, 10 μM FK506 (8.0 μg/ml), or 100 μM compounds 1 to 5 (34, 35, 36, 38, and 40 μg/ml, respectively). The data represent the means ± the standard errors from three independent experiments (*, P < 0.05).
FIG 6.
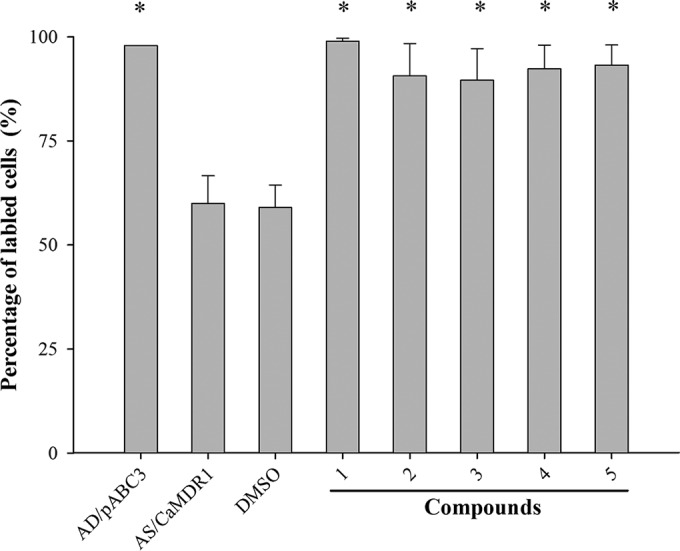
Quantification by flow cytometry of the CaMdr1p pump-inhibitory effects of organotellurium compounds 1 to 5. Gray bars indicate the percent Nile red accumulation relative to the sensitive control strain AD/pABC3 in the presence of PBS (pH 7.2), which was set to 100% of Nile red-preloaded AD/CaMDR1 cells incubated in the presence of buffer only (PBS, pH 7.2), buffer including the vehicle control of 0.5% DMSO, or buffer plus 100 μM concentrations of compounds 1 to 5 (34, 35, 37, 38, and 40 μg/ml, respectively), dissolved in DMSO, as indicated. The data represent the means ± the standard errors from three independent experiments (*, P < 0.05 compared to AD/CaMDR1).
Organotellurium compounds inhibit the CaCdr1p ATPase activity.
The CaCdr1p ATPase activity of AD/CaCDR1 plasma membrane preparations was strongly inhibited by organotellurium compounds 1, 2, 3, and 4, with 50% inhibitory concentrations (IC50s) ranging from 0.17 μg/ml (compound 3) to 0.38 μg/ml (compound 1; Table 2), but compound 5 at 40 μg/ml inhibited the Cdr1p ATPase activity by <30% (data not shown).
TABLE 2.
IC50 values of organotellurides against the CaCdr1p ATPase activity
| Compound | Mean IC50 (μg/ml) ± SDa |
|---|---|
| 1 | 0.38 ± 0.07 |
| 2 | 0.20 ± 0.03 |
| 3 | 0.17 ± 0.02 |
| 4 | 0.18 ± 0.03 |
The data represent the means from three independent experiments.
Determining the CC50s of organotellurides against mammalian cells.
We used a spectrophotometric cell viability assay to test the cytotoxicities of the five organotellurium compounds against three different mammalian cell lines (i.e., J774, HaCat, and HFF cells). As shown in Fig. 7, all five compounds had 50% cytotoxicity concentrations (CC50s) that were significantly larger than 100 μM (i.e., 34 to 40 μg/ml), the concentration required to completely inhibit CaCdr1p and CaMdr1p efflux pump functions (see Fig. 3 to 6).
FIG 7.
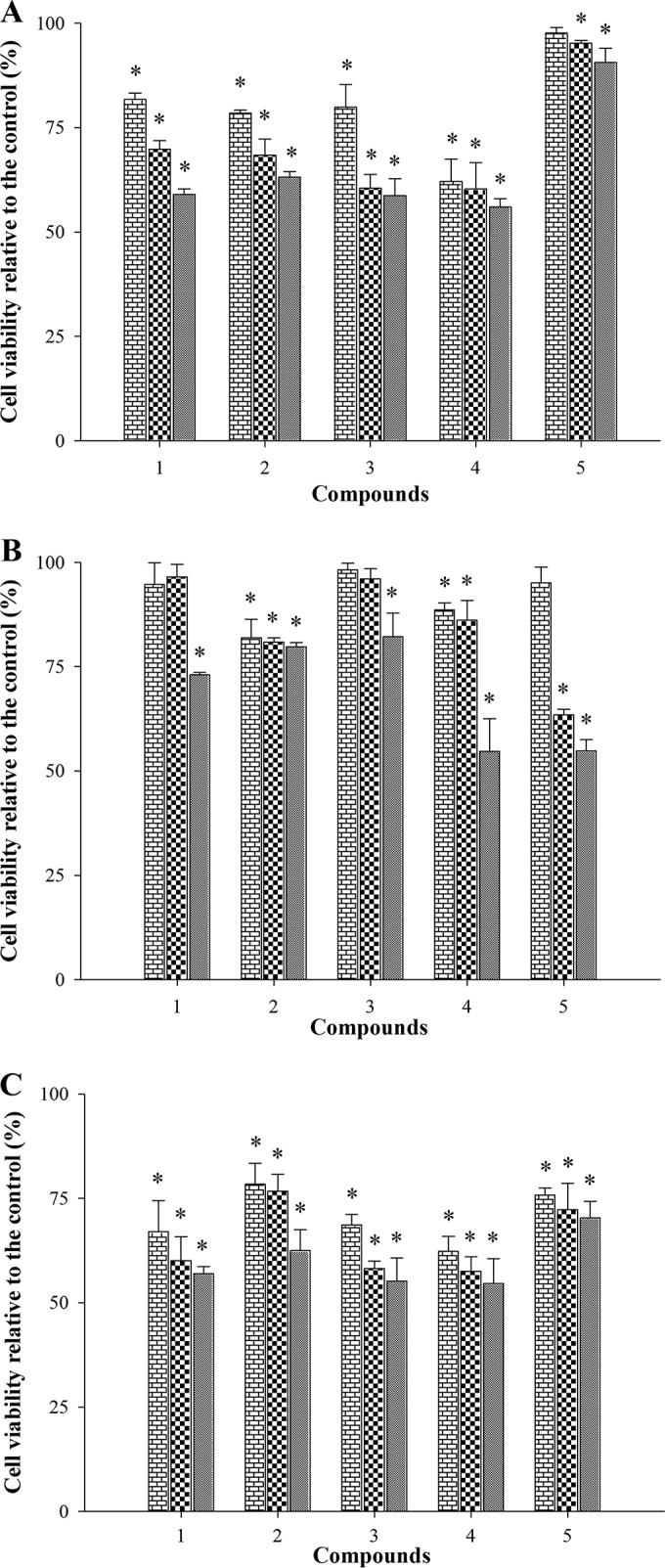
Test of the cytotoxicity of organotellurides. Three different mammalian cell lines were used to test the cytotoxicity (see Materials and Methods for further details) of the organotellurium compounds 1 to 5; J774 (A [macrophages]), HFF (B [fibroblasts]), and HaCaT (C [keratinocytes]). Each compound was tested at three 25, 50, and 100 μM concentrations (i.e., compound 1 was tested at 8, 17, and 34 μg/ml, compound 2 at 9, 18, and 35 μg/ml, compound 3 at 9, 18, and 36 μg/ml, compound 4 at 9, 19, and 38 μg/ml, and compound 5 at 10, 20, and 40 μg/ml, respectively). The data are the means ± the standard errors from three independent experiments.
Organotellurium compounds significantly increased the fluconazole susceptibilities of clinically resistant C. albicans isolates.
Encouraged by these findings, we tested whether the five organotellurium compounds could also sensitize fluconazole-resistant C. albicans clinical isolates. We chose three clinical C. albicans isolates because they had previously been shown (14) to be fluconazole resistant due to the overexpression of the ABC efflux pumps CDR1 and CDR2 (isolate 95-142), the MFS efflux pump MDR1, and the drug target ERG11 (isolate 96-25) or due to the overexpression of all four genes (isolate 12-99).
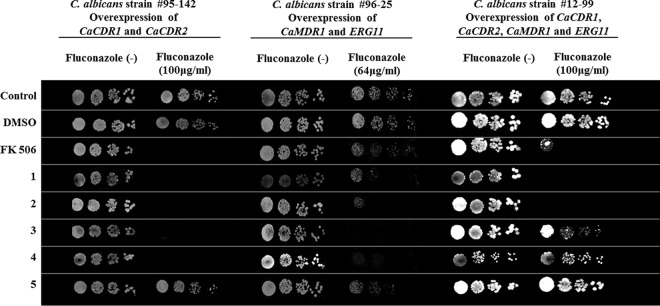
The chemosensitization assays indicated that compounds 1, 2, 3, and 4, but not compound 5, could sensitize isolate 95-142 to fluconazole (Fig. 8). This was expected for a CDR1-overexpressing isolate because compounds 1 to 4, but not compound 5, also chemosensitized AD/CaCDR1 cells to fluconazole (Fig. 2 and Table 1). Compounds 1 to 5 had very similar effects on isolate 96-25 (Fig. 8), which was a bit surprising because compound 5 did chemosensitize AD/CaMDR1 cells to fluconazole (Fig. 2). Even less predictable were the results observed for C. albicans 12-99, with only compounds 1 and 2 chemosensitizing the cells to fluconazole (Fig. 8). In contrast, the results for the known CaCdr1p, but not CaMdr1p, efflux pump inhibitor FK506 (8.0 μg/ml) were as expected (i.e., it was able to chemosensitize isolates 95-142 and 12-99, but not isolate 96-25, to fluconazole [Fig. 8]). The quantitative checkerboard assays validated these findings (Table 3). It thus appears that organotellurium compounds 1 and 2 are promising lead inhibitors of CaCdr1p and CaMdr1p and are capable of either partially (strains 95-142 and 12-99) or completely (strain 96-25) sensitizing resistant C. albicans clinical isolates to fluconazole.
FIG 8.
Organotellurium compounds 1 to 5 chemosensitize C. albicans clinical FLC-resistant isolates. “Fluconazole (−)” columns show the growth of serial 5-fold dilutions of the indicated C. albicans isolates on Sabouraud agar in the absence of FLC; the fluconazole-positive columns show the growth of the same serial dilutions of the indicated isolates on Sabouraud medium in the presence of subinhibitory concentrations of FLC (i.e., ∼1/5 of the MICFLC for C. albicans isolates 95-142 and 12-99 and almost as much FLC as its MICFLC for C. albicans 96-25). Cells grown on Sabouraud agar with or without FLC only or 0.5% DMSO were used as positive controls, and cells grown on 10 μM FK506 (i.e., 8.0 μg/ml) were used as negative controls. Rows 1, 2, 3, 4, and 5 show the growth of the same cells in the presence of 100 μM concentrations of compounds 1 to 5 (i.e., 34, 35, 37, 38, and 40 μg/ml, respectively).
TABLE 3.
Checkerboard assays of fluconazole-resistant C. albicans clinical isolates overexpressing the multidrug efflux pumps CDR1, CDR2, and MDR1 and/or overexpressing the drug target ERG11 as indicateda
| Strain and compound | Compound concn (μg/ml) |
FLC concn (μg/ml) |
FICI | Outcome | ||||
|---|---|---|---|---|---|---|---|---|
| MIC1* | MIC2* | FIC | MIC1 | MIC2 | FIC | |||
| 95-142 (CDR1/CDR2) | ||||||||
| 1 | 69 | 0.13 | 0.002 | 64 | 16 | 0.25 | 0.25 | Synergy |
| 2 | 70 | 0.55 | 0.008 | 64 | 16 | 0.25 | 0.26 | Synergy |
| 3 | 73 | 0.14 | 0.002 | 64 | 16 | 0.25 | 0.25 | Synergy |
| 4 | 76 | 0.29 | 0.004 | 64 | 16 | 0.25 | 0.25 | Synergy |
| 96-25 (MDR1/ERG11) | ||||||||
| 1 | 69 | 0.13 | 0.002 | 32 | 2 | 0.06 | 0.06 | Synergy |
| 2 | 70 | 0.14 | 0.002 | 32 | 8 | 0.25 | 0.25 | Synergy |
| 3 | 36 | 0.14 | 0.004 | 32 | 2 | 0.06 | 0.06 | Synergy |
| 4 | 38 | 0.15 | 0.004 | 32 | 2 | 0.06 | 0.06 | Synergy |
| 5 | 80 | 80 | 1 | 32 | 32 | 1 | 2 | Indifferent |
| 12-99 (all four genes) | ||||||||
| 1 | 69 | 0.13 | 0.002 | 128 | 32 | 0.25 | 0.25 | Synergy |
| 2 | 70 | 0.14 | 0.002 | 128 | 32 | 0.25 | 0.25 | Synergy |
MIC values were determined by a broth microdilution assay according to CLSI protocol M27-A3 (2008) using 50% growth reduction as the cutoff for the reported MIC values. MIC1, the MIC of the compound or FLC alone; MIC2, the MIC of the compound or FLC in the presence of FLC or compound, respectively. FIC, fractional inhibitory concentration; FICI, fractional inhibitory concentration index. *, values were converted from μM to μg/ml.
Determination of C. albicans CDR1, CDR2, MDR1, and ERG11 mRNA expression levels.
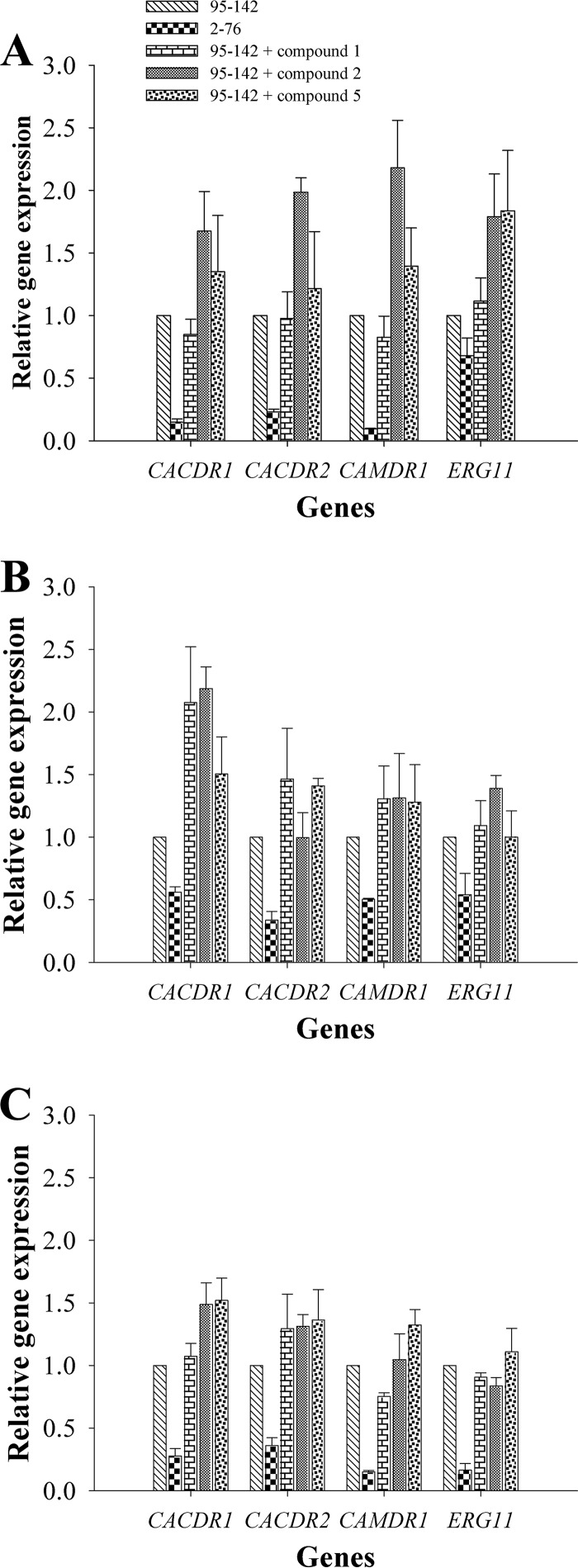
To ascertain whether the observed fluconazole-sensitizing effect of clinical C. albicans isolates was a direct effect of the organotellurium compounds on Cadr1p and CaMdr1p efflux pump function and not an indirect effect caused by reduced expression levels, we quantified the effects of the organotellurides on the mRNA expression levels of CDR1, CDR2, and MDR1 and the azole drug target ERG11. We only tested the effects of compounds 1 and 2, the most promising organotelluride chemosensitizers, and compound 5 was used as a “negative” control because it did not chemosensitize CDR1-overexpressing cells to fluconazole.
The reverse transcription-quantitative PCR (RT-qPCR) results showed that all three resistant isolates (95-142, 96-25, and 12-99) showed significantly higher CDR1, CDR2, MDR1, and ERG11 mRNA expression levels than did the fluconazole-sensitive control strain C. albicans 2-76 (the only exception was ERG11 of strain 95-142; Fig. 9), confirming previously published results (14). However, compounds 1, 2, and 5 did not downregulate CDR1, CDR2, MDR1, or ERG11 mRNA levels; instead, some of these compounds caused a slight (i.e., no more than 2-fold) increase in mRNA levels for some of the four genes in some of the strains tested (Fig. 9). These data indicated that the observed chemosensitizing effects of compounds 1 to 5 were most likely caused by direct interactions of these compounds with C. albicans CaCdr1p and CaMdr1p and/or other unknown factors that indirectly affected CaCdr1p and/or CaMdr1p efflux pump function.
FIG 9.
Effect of organotellurides on CaCDR1, CaCDR2, CaMDR1, and CaERG11 mRNA expression levels of C. albicans clinical FLC-resistant isolates. (A) mRNA expression levels of C. albicans 95-142 in the presence of compounds 1, 2, and 3. (B) mRNA expression levels of C. albicans 96-25. (C) mRNA expression levels of C. albicans 12-99. All mRNA levels were normalized relative to the housekeeping gene ACT1. The data represent the means ± the standard errors from three independent experiments, and each value was then compared to its ACT1 control (*, P < 0.05). The mRNA expression levels of a wild-type, FLC-sensitive C. albicans clinical isolate (2-76) are shown for comparison.
DISCUSSION
Previous studies demonstrated the inhibitory effect of synthetic organotellurium compounds on ABC transporters associated with multidrug resistance of human cancer cells (29–31). Reis de Sá et al. recently demonstrated that synthetic organotellurium compounds also inhibit the fungal multidrug ABC efflux pump S. cerevisiae Pdr5p, a homolog of C. albicans Cdr1p (27). The present study is an extension of that study intended to assess the chemosensitization potential of the same compounds against fluconazole-resistant C. albicans clinical isolates.
Our data showed that compounds 1 to 4, but not compound 5, partially chemosensitized AD/CaCDR1 cells, but not AD/CaCDR2 cells, to fluconazole, suggesting that compounds 1 to 4 specifically inhibit the fluconazole efflux of CaCdr1p, but not CaCdr2p, like the Cdr1p specific efflux pump inhibitors FK506 and enniatin (32), FK520 (33), and unnarmicins A and C (17). It would appear that CaCdr1p and CaCdr2p have very similar substrate transport profiles but that their inhibitor sensitivities differ quite significantly (34). Interestingly, all five of organotellurium compounds 1 to 5 partially chemosensitized AD/CaMDR1 cells to fluconazole, a finding similar to the results observed for clorgyline (25), a small antidepressant molecule that could reverse the fluconazole resistance of AD/CaCDR1 and AD/CaMDR1 strains. Although compound 5 shares the tellurium-butyl side chain at one end with compounds 1 to 4 and has a phenyl group like compounds 2 to 4 on the other end of the molecule, compound 5 is otherwise quite dissimilar to compounds 1 to 4 (Fig. 1), which may explain its altered specificity (i.e., compound 5 specifically inhibited CaMdr1p). Specific CaMdr1p inhibitors are rare. Examples include cerulenin analogues (24) or synthetic heterocyclic compounds containing a cyclobutene-dione core (26). Consistent with the fluconazole chemosensitization results, compounds 1 to 4 also inhibited R6G efflux of CaCdr1p, and all five of compounds 1 to 5 inhibited Nile red efflux of Mdr1p-overexpressing cells. However, in this case, CaCdr1p and CaMdr1p were fully inhibited by compounds 1 to 5, causing CaCdr1p- or CaMdr1p-overexpressing cells to accumulate as much R6G or Nile red as the sensitive control strain AD/pABC3.
We have previously shown that compounds 1 to 4 are potent ATPase inhibitors of S. cerevisiae Pdr5p (IC50 ∼ 0.52 μg/ml) (27). Here, we have demonstrated that the same compounds demonstrated similar ATPase inhibitory activities against C. albicans CaCdr1p, with IC50s ranging from 0.17 to 0.38 μg/ml. Disulfiram (19) and some d-octapeptides (35) are other examples of CaCdr1p efflux pump inhibitors affecting pump ATPase activity. Inhibition of the pump ATPase activity is, however, not the only mechanism blocking CaCdr1p-mediated fluconazole transport. There are molecules that reverse fluconazole resistance mediated by CaCdr1p without affecting its pump ATPase activity (e.g., chalcone derivatives [18], farnesol [36], and curcumin [20]). Although we cannot say how these compounds inhibit C. albicans Cdr1p and Mdr1p, it is certain that some of the organotellurium compounds 1 to 5 could chemosensitize C. albicans-resistant isolates to fluconazole. The set of clinically resistant isolates tested included strains expressing the following fluconazole-resistant mechanisms: (i) overexpression of CDR1 and CDR2 (isolate 95-142); (ii) overexpression of MDR1 and ERG11 (isolate 96-25); and (iii) overexpression of all four fluconazole drug resistance-associated genes CDR1, CDR2, MDR1, and ERG11 (isolate 12-99). As expected from the S. cerevisiae studies described above, compounds 1 and 2 could chemosensitize all three resistant isolates to fluconazole. However, the results for compounds 3, 4, and 5 were not quite as expected for CaCdr1p (compounds 3 and 4) and CaMdr1p (compounds 3, 4, and 5) efflux pump inhibitors. As expected, compounds 3 and 4 fluconazole-chemosensitized isolates 95-142 and 96-25; however, they could not sensitize 12-99 cells to fluconazole. Similarly, compound 5 was expected to fluconazole-chemosensitize CaMdr1p overexpressing isolate 96-25, but it could not chemosensitize any of the three isolates. It is quite likely that additional unknown factors contributed to these unexpected chemosensitization patterns.
Aside from drug efflux pump inhibition, there are other ways to chemosensitize C. albicans isolates resistant to fluconazole (9). Thymol, for instance, not only inhibits fluconazole efflux but also inhibits CDR1 and MDR1 transcription (37). However, qPCR assays revealed that compounds 1 and 2, the most promising lead inhibitors, did not modulate the transcription of CDR1, CDR2, MDR1, and ERG11 to a relevant extent in any of the three resistant isolates. Tests also confirmed that all five organotellurium compounds had low cytotoxicity (CC50, 34 to 40 μg/ml), an important property for any potential drug used in fluconazole combination therapy.
We conclude that organotellurium compounds 1 to 5, especially compounds 1 and 2, are useful lead compounds for novel efflux pump inhibitors of C. albicans Cdr1p and Mdr1p that may be used as adjuvants in combination therapy with fluconazole for the successful treatment of C. albicans infections.
MATERIALS AND METHODS
Chemicals, drugs, and stock solutions.
Rhodamine 6G (R6G), Nile red, ATP disodium (ATP) salt, HEPES, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich, St. Louis, MO. FK506, also known as tacrolimus, was purchased from Tecoland (Irvine, CA), and fluconazole was obtained from the university pharmacy (UFJF, Juiz de Fora-MG, Brazil). Nile red and FK506 stocks were dissolved in DMSO, R6G stocks were dissolved in 50 mM HEPES-NaOH buffer (pH 7.0), ATP stocks were dissolved in Tris-HCl buffer (pH 7.0), and fluconazole stocks were dissolved in distilled water.
Synthetic organotelluride compounds.
The organotelluride compounds (Fig. 1) were synthesized as previously described (38–41). All compounds were stored in a desiccator at 4°C, and stock solutions were prepared in DMSO.
Yeast strains and culture conditions.
The yeast strains used in this study are listed in Table 4. Typically, yeast cultures were incubated in YPD medium (1% yeast extract, 2% peptone and 2% glucose) by shaking at 150 rpm for ∼17 h at 30°C (S. cerevisiae) or 37°C (C. albicans), and the cells were harvested at logarithmic growth phase.
TABLE 4.
Yeast strains used in this study
| Species and strain | Description | Reference |
|---|---|---|
| Saccharomyces cerevisiae | ||
| AD/pABC3 | Strain containing empty transformation cassette | 32 |
| AD/CaCDR1 | Strain overexpressing CaCDR1 | 32 |
| AD/CaCDR2 | Strain overexpressing CaCDR2 | 32 |
| AD/CaMDR1 | Strain overexpressing CaMDR1 | 32 |
| Candida albicans | ||
| 2-76 | FLC-susceptible clinical isolate | 14 |
| 95-142 | FLC-resistant clinical isolate overexpressing CDR1 and CDR2 | 14 |
| 96-25 | FLC-resistant clinical isolate overexpressing MDR1 and ERG11 | 14 |
| 12-99 | FLC-resistant clinical isolate overexpressing CDR1, CDR2, MDR1, and ERG11 | 14 |
Fluconazole chemosensitization assays.
The chemosensitization assays were performed as described previously (20, 27). In brief, 5-μl aliquots of undiluted and three 5-fold serial dilutions of logarithmic (optical density at 600 nm [OD600] = 0.1 × 106 or ∼1 × 106 cells/ml) S. cerevisiae cell suspensions were placed onto sterile polystyrene six-well YPD agar plates that were supplemented with synthetic organotellurium compounds in the presence or absence of subinhibitory (∼1/2 to ∼1/4 of the MIC) fluconazole concentrations. The plates were then incubated at 30°C for 48 h to visualize cell growth.
The chemosensitization assays for the clinical C. albicans isolates were performed with slight modifications. Fivefold serial dilutions were prepared using a starting cell suspension of 6 × 105 cells/ml, and cells were incubated on Sabouraud instead of YPD agar at 37°C rather than at 30°C.
Checkerboard assays.
In order to evaluate whether individual organotellurium compounds exhibit synergy or indifference in combination with fluconazole against S. cerevisiae and C. albicans strains, checkerboard assays were performed as described previously (42) according to Clinical and Laboratory Standards Institute 2008 protocol M27-A3, but with slight modifications to the protocol (i.e., S. cerevisiae cells were grown in YPD at 30°C because they cannot grow in the recommended RPMI medium, and C. albicans growth was monitored at 37°C rather than at 35°C). Individual wells of a 96-well microtiter plate containing YPD liquid medium with various concentrations of fluconazole in combination with various concentrations of the organotelluride test compounds were inoculated with 104 logarithmically grown S. cerevisiae cells, followed by incubation at 30°C for 48 h. For the C. albicans isolates, 2.5 × 103 logarithmically grown cells/ml were used as an inoculum, and the cells were incubated in RPMI 1640 (Sigma-Aldrich, USA) medium at 37°C for 48 h, after which cell growth (i.e., the OD600) was monitored with a Fluostar Optima (BMG Labtech, Germany) plate reader.
The MIC of a compound was defined as the lowest concentration that caused a ≥50% reduction of cell growth (MIC50). Synergy between fluconazole and organotellurides was determined by calculating the fractional inhibitory concentration index (FICI). FICI values of ≤0.5, >0.5 to ≤4.0 and >4.0 indicated synergy, indifference, or antagonistic interactions for different drug combinations (6).
R6G and Nile red accumulation assays.
The accumulation of the fluorescent multidrug efflux pump substrates R6G (substrate of CaCdr1p) and Nile red (substrate of both efflux pumps CaCdr1p and CaMdr1p) were used as another measure to evaluate the pump-inhibitory activity of the five organotellurides. The accumulation of fluorescence inside cells was assessed by fluorescence microscopy and flow cytometry.
The microscopic assessment of the accumulation of fluorescent efflux pump substrates was conducted as previously described (43), including minor modifications. The yeast cells were incubated by shaking at 150 rpm in 200 ml of YPD medium at 30°C for 17 h and harvested at mid-log phase by centrifugation at 5,000 × g and 4°C for 5 min. The cells were then washed twice with 10 ml of 10 mM phosphate-buffered saline (PBS; pH 7.2), and the cell pellets were resuspended in 10 ml of PBS (pH 7.2) and starved for 2 h at 4°C. The starved cells were harvested at 5,000 × g for 5 min, resuspended in 10 ml of PBS (pH 7.2), and 2 × 106 cells/ml were then incubated in the presence of R6G (15 μM) or Nile red (7 μM) in 500 μl at 30°C for 30 min to preload the cells with the fluorescent dyes. Preloaded cells were harvested and incubated at 30°C for 30 min in 500 μl of PBS in the presence or absence of the organotelluride test compounds. The R6G accumulation assay also contained 0.2% glucose because the ABC transporter Cdr1p requires ATP as an energy source for R6G efflux. However, the addition of glucose was not required for the Nile red accumulation assay because the MFS transporter CaMdr1p uses the electrochemical gradient across the plasma membrane rather than ATP as an energy source for Nile red efflux. The cells were harvested by centrifugation at 5,000 × g for 5 min, washed twice with PBS (pH 7.2), resuspended in ∼200 μl of PBS (pH 7.2), and finally analyzed by confocal microscopy (Eclipse E400; Nikon, Tokyo, Japan).
Cells for flow cytometry of R6G and Nile red accumulation were prepared in the same way. However, in that case the cells were analyzed with an Acouri C6 flow cytometry analyzer (Becton Dickinson, USA) using the accompanying Acouri C6 software package.
Plasma membrane preparation.
S. cerevisiae plasma membranes were isolated as previously described (44). Isolated plasma membranes were stored in liquid nitrogen until further use.
CaCdr1p ATPase assays.
The inhibitory effects of individual test compounds on the CaCdr1p ATPase activity were assessed as previously described (27). To determine the CaCdr1p-specific ATPase activities of plasma membranes isolated from CaCdr1p-overexpressing AD/CaCDR1 cells, the background ATPase activities of plasma membranes isolated from the negative-control strain AD/pABC3 were subtracted.
Cytotoxicity tests against mammalian cell lines.
Cytotoxicity assays against macrophages (J774), keratinocytes (HaCaT), and fibroblasts (HFFs) were performed as previously described (45). Briefly, after incubating 104 cells in individual wells of a 96-well plate containing 100 μl of Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum for 24 h at 37°C, the cells were washed with the same medium and resuspended in 100 μl of fresh medium containing increasing concentrations of organotellurides. After 48 h of incubation at 37°C, 20 μl of a thiazolyl blue tetrazolium blue–MTT (Sigma-Aldrich) solution (5 mg/ml) was added to each well, and the plate incubated in the dark at 37°C for an additional 4 h. After removal of the supernatant, formazan crystals were solubilized with 100 μl of DMSO, and the cell metabolic activity was determined by measuring the formazan absorbance at 595 nm with a Fluostar Optima (BMG Labtech) plate reader.
RT-qPCR.
qPCR quantification of mRNA expression levels was also performed as previously described (21). Briefly, 10-ml portions of mid-log-phase cells (106 cells/ml) were incubated in YPD broth in the presence or absence of organotelluride compounds at 37°C with shaking for 120 min. After the cells were harvested, the total RNA was extracted using an RNeasy minikit (Qiagen Sciences, Germantown, MD) and quantified with a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA). First-strand cDNA was synthesized with a high-capacity cDNA reverse transcription kit from Applied Biosystems (Life Technologies) using a Veriti 96-well thermal cycler (Applied Biosystems) under the following conditions: 10 min at 25°C, 120 min at 37°C, and 5 min at 85°C. For the real-time qPCRs, the cDNAs of interest were CaCDR1, CaCDR2, CaMDR1, and CaERG11, and the housekeeping gene ACT1 was used as an internal control. The DNA oligomer primers used in this study are listed in Table 5. qPCRs (20 μl) consisted of 2 μl of first-strand cDNA templates, 10 μl of SYBR Green master mix (Applied Biosystems/Life Technologies), and DNA oligomer primers at 0.5 μM (CaCDR1, CaCDR2, and CaMDR1) or 0.7 μM (CaACT1 and CaERG11). All qPCRs were performed in a StepOne real-time PCR system (Applied Biosystems). To confirm PCR product specificity, melting curves (from 65 to 95°C) for each PCR amplification were established, and the results were analyzed with the StepOne software.
TABLE 5.
DNA oligomer primers used in this study
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| CaCDR1 | Forward | TGCCAAACAATCCAACAA |
| Reverse | CGACGGATCACCTTTCATACGA | |
| CaCDR2 | Forward | AAGGTTTTGATGCTACTGC |
| Reverse | GTCGGACATGTGGCTCAAA | |
| CaMDR1 | Forward | GTGTTGGCCCATTGGTTTTCAGTC |
| Reverse | CCAAAGCAGTGGGGATTTGTAG | |
| ERG11 | Forward | GGTGGTCAACATACTTCTGCTTC |
| Reverse | GTCAAATCATTCAAATCACCACCT | |
| ACT1 | Forward | AAGAATTGATTTGGCTGGTAGAGA |
| Reverse | TGGCAGAAGATTGAGAAGAAGTTT |
Statistical analysis.
All experiments were performed in triplicate, and the data are presented as means ± the standard errors. Values with a probability level of 5% (P < 0.05) in Student t tests were considered significant.
ACKNOWLEDGMENTS
We thank our lab assistant, Geralda Rodrigues Almeida, for her great support. We also thank Richard Cannon and Brian Monk (University of Otago—New Zealand) and Theodore White (University of Missouri) for donating the yeast mutants and C. albicans-resistant isolates used in this study.
This study was funded by FAPERJ, FAPESP, CNPq, and CAPES for financial support and scholarships. This study was also supported in part by the University of São Paulo through the NAP-CatSinQ (Research Core in Catalysis and Chemical Synthesis).
REFERENCES
- 1.Maeda T, Babazono A, Nishi T, Yasui M, Matsuda S, Fushimi K, Fujimori K. 2015. The impact of opportunistic infections on clinical outcome and healthcare resource uses for adult T cell leukaemia. PLoS One 10:1. doi: 10.1371/journal.pone.0135042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang JL, Kung HC, Lei WC, Yao M, Wu UI, Hsu SC, Lin CT, Li CC, Wu SJ, Hou HA, Chou WC, Huang SY, Tsay W, Chen YC, Chang SC, Ko BS, Tien HF. 2015. High incidences of invasive fungal infections in acute myeloid leukemia patients receiving induction chemotherapy without systemic antifungal prophylaxis: a prospective observational study in Taiwan. PLoS One 10:e0128410. doi: 10.1371/journal.pone.0128410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quindos G. 2014. Epidemiology of candidaemia and invasive candidiasis: a changing face. Rev Iberoam Micol 31:42–14. doi: 10.1016/j.riam.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Autmizguine J, Guptill JT, Cohen-Wolkowiez M, Benjamin DK Jr, Capparelli EV. 2014. Pharmacokinetics and pharmacodynamics of antifungals in children: clinical implications. Drugs 74:891–909. doi: 10.1007/s40265-014-0227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthaiou DK, Christodoulopoulou T, Dimopoulos G. 2015. How to treat fungal infections in ICU patients. BMC Infect Dis 15:205. doi: 10.1186/s12879-015-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odds FC, Brown AJ, Gow NA. 2003. Antifungal agents: mechanisms of action. Trends Microbiol 11:272–279. doi: 10.1016/S0966-842X(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 7.Xie JL, Polvi EJ, Shekhar-Guturja T, Cowen LE. 2014. Elucidating drug resistance in human fungal pathogens. Future Microbiol 9:523–542. doi: 10.2217/fmb.14.18. [DOI] [PubMed] [Google Scholar]
- 8.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. 2015. Mechanisms of antifungal drug resistance. Cold Spring Harbor Perspect Med 5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes AR, Lin YH, Niimi K, Lamping E, Keniya M, Niimi M, Tanabe K, Monk BC, Cannon RD. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother 52:3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa C, Dias PJ, Sa-Correia I, Teixeira MC. 2014. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front Physiol 5:197. doi: 10.3389/fphys.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombo AL, Janini M, Salomao R, Medeiros EA, Wey SB, Pignatari AC. 2009. Surveillance programs for detection and characterization of emergent pathogens and antimicrobial resistance: results from the Division of Infectious Diseases, UNIFESP. An Acad Bras Cienc 81:571–587. doi: 10.1590/S0001-37652009000300020. [DOI] [PubMed] [Google Scholar]
- 13.Kanafani ZA, Perfect JR. 2008. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis 46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 14.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother 46:1704–1713. doi: 10.1128/AAC.46.6.1704-1713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Hou Y, Chen X, Gao Y, Li H, Sun S. 2014. Combination of fluconazole with non-antifungal agents: a promising approach to cope with resistant Candida albicans infections and insight into new antifungal agent discovery. Int J Antimicrob Agents 43:395–402. doi: 10.1016/j.ijantimicag.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Egner R, Bauer BE, Kuchler K. 2000. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol Microbiol 35:1255–1263. doi: 10.1046/j.1365-2958.2000.01798.x. [DOI] [PubMed] [Google Scholar]
- 17.Tanabe K, Lamping E, Adachi K, Takano Y, Kawabata K, Shizuri Y, Niimi M, Uehara Y. 2007. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochem Biophys Res Commun 364:990–995. doi: 10.1016/j.bbrc.2007.10.110. [DOI] [PubMed] [Google Scholar]
- 18.Lacka I, Konieczny MT, Bulakowska A, Kodedova M, Gaskova D, Maurya IK, Prasad R, Milewski S. 2015. Chemosensitization of multidrug resistance by the oxathiolone-fused chalcone derivatives. Front Microbiol 6:783. doi: 10.3389/fmicb.2015.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla S, Sauna ZE, Prasad R, Ambudkar SV. 2004. Disulfiram is a potent modulator of multidrug transporter Cdr1p of Candida albicans. Biochem Biophys Res Commun 322:520–525. doi: 10.1016/j.bbrc.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 20.Sharma M, Manoharlal R, Shukla S, Puri N, Prasad T, Ambudkar SV, Prasad R. 2009. Curcumin modulates efflux mediated by yeast ABC multidrug transporters and is synergistic with antifungals. Antimicrob Agents Chemother 53:3256–3265. doi: 10.1128/AAC.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ricardo E, Costa-de-Oliveira S, Dias AS, Guerra J, Rodrigues AG, Pina-Vaz C. 2009. Ibuprofen reverts antifungal resistance on Candida albicans showing overexpression of CDR genes. FEMS Yeast Res 9:618–625. doi: 10.1111/j.1567-1364.2009.00504.x. [DOI] [PubMed] [Google Scholar]
- 22.Lamping E, Ranchod A, Nakamura K, Tyndall JD, Niimi K, Holmes AR, Niimi M, Cannon RD. 2009. Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrob Agents Chemother 53:354–369. doi: 10.1128/AAC.01095-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva LV, Sanguinetti M, Vandeputte P, Torelli R, Rochat B, Sanglard D. 2013. Milbemycins: more than efflux inhibitors for fungal pathogens. Antimicrob Agents Chemother 57:873–886. doi: 10.1128/AAC.02040-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diwischek F, Morschhauser J, Holzgrabe U. 2009. Cerulenin analogues as inhibitors of efflux pumps in drug-resistant Candida albicans. Archiv Pharmazie 342:150–164. doi: 10.1002/ardp.200800160. [DOI] [PubMed] [Google Scholar]
- 25.Holmes AR, Keniya MV, Ivnitski-Steele I, Monk BC, Lamping E, Sklar LA, Cannon RD. 2012. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob Agents Chemother 56:1508–1515. doi: 10.1128/AAC.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keniya MV, Fleischer E, Klinger A, Cannon RD, Monk BC. 2015. Inhibitors of the Candida albicans major facilitator superfamily transporter Mdr1p responsible for fluconazole resistance. PLoS One 10:e0126350. doi: 10.1371/journal.pone.0126350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reis de Sá LF, Toledo FT, de Sousa BA, Goncalves AC, Tessis AC, Wendler EP, Comasseto JV, Dos Santos AA, Ferreira-Pereira A. 2014. Synthetic organotelluride compounds induce the reversal of Pdr5p-mediated fluconazole resistance in Saccharomyces cerevisiae. BMC Microbiol 14:201. doi: 10.1186/s12866-014-0201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, Sklar LA. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394:87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebert SP, Wetzel B, Myette RL, Conseil G, Cole SP, Sawada GA, Loo TW, Bartlett MC, Clarke DM, Detty MR. 2012. Chalcogenopyrylium compounds as modulators of the ATP-binding cassette transporters P-glycoprotein (P-gp/ABCB1) and multidrug resistance protein 1 (MRP1/ABCC1). J Med Chem 55:4683–4699. doi: 10.1021/jm3004398. [DOI] [PubMed] [Google Scholar]
- 30.Myette RL, Conseil G, Ebert SP, Wetzel B, Detty MR, Cole SP. 2013. Chalcogenopyrylium dyes as differential modulators of organic anion transport by multidrug resistance protein 1 (MRP1), MRP2, and MRP4. Drug Metab Dispos 41:1231–1239. doi: 10.1124/dmd.112.050831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawada GA, Raub TJ, William Higgins J, Brennan NK, Moore TM, Tombline G, Detty MR. 2008. Chalcogenopyrylium dyes as inhibitors/modulators of P-glycoprotein in multidrug-resistant cells. Bioorg Med Chem 16:9745–9756. doi: 10.1016/j.bmc.2008.09.065. [DOI] [PubMed] [Google Scholar]
- 32.Lamping E, Monk BC, Niimi K, Holmes AR, Tsao S, Tanabe K, Niimi M, Uehara Y, Cannon RD. 2007. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot Cell 6:1150–1165. doi: 10.1128/EC.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gauthier C, Weber S, Alarco AM, Alqawi O, Daoud R, Georges E, Raymond M. 2003. Functional similarities and differences between Candida albicans Cdr1p and Cdr2p transporters. Antimicrob Agents Chemother 47:1543–1554. doi: 10.1128/AAC.47.5.1543-1554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanabe K, Lamping E, Nagi M, Okawada A, Holmes AR, Miyazaki Y, Cannon RD, Monk BC, Niimi M. 2011. Chimeras of Candida albicans Cdr1p and Cdr2p reveal features of pleiotropic drug resistance transporter structure and function. Mol Microbiol 82:416–433. doi: 10.1111/j.1365-2958.2011.07820.x. [DOI] [PubMed] [Google Scholar]
- 35.Niimi K, Harding DR, Parshot R, King A, Lun DJ, Decottignies A, Niimi M, Lin S, Cannon RD, Goffeau A, Monk BC. 2004. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a d-octapeptide derivative. Antimicrob Agents Chemother 48:1256–1271. doi: 10.1128/AAC.48.4.1256-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma M, Prasad R. 2011. The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans. Antimicrob Agents Chemother 55:4834–4843. doi: 10.1128/AAC.00344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad A, Khan A, Manzoor N. 2013. Reversal of efflux mediated antifungal resistance underlies synergistic activity of two monoterpenes with fluconazole. Eur J Pharm Sci 48:80–86. doi: 10.1016/j.ejps.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Keppler AF, Gariani RA, Lopes DG, Comasseto JV. 2009. Lithium butylchalcogenolate induced Michael-aldol tandem sequence: easy and rapid access to highly functionalized organochalcogenides and unsaturated compounds. Tetrahedron Lett 50:2181–2184. doi: 10.1016/j.tetlet.2009.02.158. [DOI] [Google Scholar]
- 39.Sousa BA, Dos Santos AA. 2012. A facile, versatile, and mild Morita-Baylis-Hillman-type reaction for the modular one-pot synthesis of highly functionalized MBH adducts. Eur J Organic Chem 2012:3431–3436. doi: 10.1002/ejoc.201200371. [DOI] [Google Scholar]
- 40.Sousa BA, Gariani KAFRA, Comasseto JV, Dos Santos AA. 2012. Metallic chalcogenolates mediated modular Michael-aldol cascade reaction: an easy route to multi-functionalized chalcogenides and Morita-Baylis-Hillman adducts. Tetrahedron 68:1046–1053. [Google Scholar]
- 41.Vargas F, Toledo FT, Comasseto JV. 2010. N-Functionalized organolithium compounds via tellurium/lithium exchange reaction. J Brazil Chem Soc 21:2072–2078. doi: 10.1590/S0103-50532010001100007. [DOI] [Google Scholar]
- 42.Barchiesi F, Falconi Di Francesco L, Scalise G. 1997. In vitro activities of terbinafine in combination with fluconazole and itraconazole against isolates of Candida albicans with reduced susceptibility to azoles. Antimicrob Agents Chemother 41:1812–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Gomes AS, Curvelo JA, Soares RM, Ferreira-Pereira A. 2012. Curcumin acts synergistically with fluconazole to sensitize a clinical isolate of Candida albicans showing a MDR phenotype. Med Mycol 50:26–32. doi: 10.3109/13693786.2011.578156. [DOI] [PubMed] [Google Scholar]
- 44.Ferreira-Pereira A, Marco S, Decottignies A, Nader J, Goffeau A, Rigaud JL. 2003. Three-dimensional reconstruction of the Saccharomyces cerevisiae multidrug resistance protein Pdr5p. J Biol Chem 278:11995–11999. doi: 10.1074/jbc.M212198200. [DOI] [PubMed] [Google Scholar]
- 45.Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]