ABSTRACT
Penicillin-binding proteins (PBPs) function as transpeptidases, carboxypeptidases, or endopeptidases during peptidoglycan synthesis in bacteria. As the well-known drug targets for β-lactam antibiotics, the physiological functions of PBPs and whether they are essential for growth are of significant interest. The pathogen Pseudomonas aeruginosa poses a particular risk to immunocompromised and cystic fibrosis patients, and infections caused by this pathogen are difficult to treat due to antibiotic resistance. To identify potential drug targets among the PBPs in P. aeruginosa, we performed gene knockouts of all the high-molecular-mass (HMM) PBPs and determined the impacts on cell growth and morphology, susceptibility to β-lactams, peptidoglycan structure, virulence, and pathogenicity. Disruptions of the transpeptidase domains of most HMM PBPs, including double disruptions, had only minimal effects on cell growth. The exception was PBP3, where cell growth occurred only when the protein was conditionally expressed on an integrated plasmid. Conditional deletion of PBP3 also caused a defect in cell division and increased susceptibility to β-lactams. Knockout of PBP1a led to impaired motility, and this observation, together with its localization at the cell poles, suggests its involvement in flagellar function. Overall, these findings reveal that PBP3 represents the most promising target for drug discovery against P. aeruginosa, whereas other HMM PBPs have less potential.
KEYWORDS: Pseudomonas aeruginosa, antimicrobial drug targets, essential genes, genetic knockout, penicillin-binding proteins, peptidoglycan
INTRODUCTION
The Gram-negative bacterium Pseudomonas aeruginosa is the fourth most commonly isolated pathogen in hospital-acquired infections, accounting for 10% of all such infections (1, 2) and 17% of cases of nosocomial pneumonia (3). P. aeruginosa infections are especially serious in immunosuppressed patients hospitalized with severe burns, cancer, and AIDS, as well as recipients of organ transplants (4). In addition, individuals with cystic fibrosis (CF) are highly susceptible to chronic pseudomonal lung infections (5, 6), and the pathogen plays a critical role in the morbidity and mortality of CF patients (7). The rate of mortality of patients infected nosocomially with P. aeruginosa is very high, with rates reported to be from ∼30 to 50% (8–10). The emergence of clinical isolates of P. aeruginosa that exhibit resistance to one or more antibiotics, including fluoroquinolones, carbapenems, and a fourth-generation cephalosporin (cefepime) that is currently the antibiotic of choice for pseudomonal infections (11), severely limits treatment options (12–17). Consequently, there is an urgent need for new therapeutic agents with activity against this pathogen (18).
For more than 60 years, β-lactam antibiotics have been highly successful in the treatment of bacterial infections. Their lethal targets are penicillin-binding proteins (PBPs), which are periplasmic enzymes that function during the final stages of peptidoglycan (PG) synthesis (19, 20). Peptidoglycan envelops the bacterial cell and is essential for cell survival, division, and maintenance of cell shape (21). PBPs are broadly divided into two groups on the basis of molecular mass (22). High-molecular-mass (HMM) PBPs function as transpeptidases (TPases) and are often essential. Class A HMM PBPs catalyze both TPase and glycosyltransferase (GTase) activities, whereas class B HMM PBPs possess only TPase activity. Low-molecular-mass (LMM) PBPs (or class C PBPs) generally function as carboxypeptidases or endopeptidases and can typically be genetically deleted without having a significant effect on cell viability or growth (19, 23).
The development of PBP inhibitors offers a potential route to the development of new antibiotics to replace β-lactams that are becoming increasingly limited in efficacy. A key step, however, is to identify which PBPs are essential or at least represent potential drug targets in a bacterial pathogen. Examination of the P. aeruginosa genome reveals 8 potential PBPs (1a, 1b, 2, 3, 3a/3x, 4, 5/6, 7), and 6 major PBPs can be detected by radiolabeling with [14C]benzylpenicillin (24, 25). PBPs 1a, 1b, 3, and 3a/3x are classified as HMM PBPs, and PBPs 4, 5/6, and 7 are classified as LMM PBPs. By homology with other PBPs, PBP1a and PBP1b are class A bifunctional TPase/GTases, and PBP2, PBP3, and PBP3x are all class B enzymes that catalyze only TPase activity. PBP3x (or PBP3a) is a homologue of PBP3 that is expressed during the stationary phase of growth (25). PBPs 4, 5, and 7 are class C LMM PBPs that would be expected to function as carboxypeptidases or endopeptidases. The impact of deleting PBPs 4, 5, and 7 has been investigated elsewhere: none are essential, and their absence does not alter the peptidoglycan structure (26).
The HMM PBPs 1a, 1b, 2, and 3 are the most likely candidates to be essential, but interestingly, no PBPs were designated essential in a library of transposon insertion mutants constructed from a pathogenic strain of P. aeruginosa, PA14 (27). In contrast, analyses of transposon insertion libraries constructed from strain PAO1 or PA14 grown in sputum from CF patients indicate that both PBPs 2 and 3 are essential (28). The physiological significance of PBPs 2 and 3 is also suggested by other studies. Although they are viable, P. aeruginosa strains lacking PBP2 exhibit a spheroidal rather than a rod-shape morphology and lowered MICs for several β-lactams, including the antipseudomonal antibiotics carbenicillin (CBC), cefotaxime, and ticarcillin (29). In addition, PBP2 appears to be a target for carbapenems (30). PBP3 is believed to be a therapeutic target because its overexpression increases the MICs of P. aeruginosa for aztreonam, cefepime, cefsulodin, and ceftazidime (25) and it appears to be targeted by fourth-generation cephalosporins (31–33) and carbapenems (30). However, its essentiality or otherwise has not been unequivocally established. Inactivation of PBP3x, which is a homologue of PBP3, has no effect on cell morphology or growth (34). At this stage, very little is known about the physiological importance of PBPs 1a and 1b.
In this study, we systematically deleted the HMM PBPs of P. aeruginosa to determine which are essential for growth and also establish how such deletions impact the pathogenicity and virulence of P. aeruginosa, susceptibility to β-lactams, cell morphology, and PG structure. This information is indispensable if any of these PBPs are to be developed as bona fide drug targets to combat P. aeruginosa infections.
RESULTS
PBP3 but not PBP 1a, 1b, or 2 is essential for growth of P. aeruginosa.
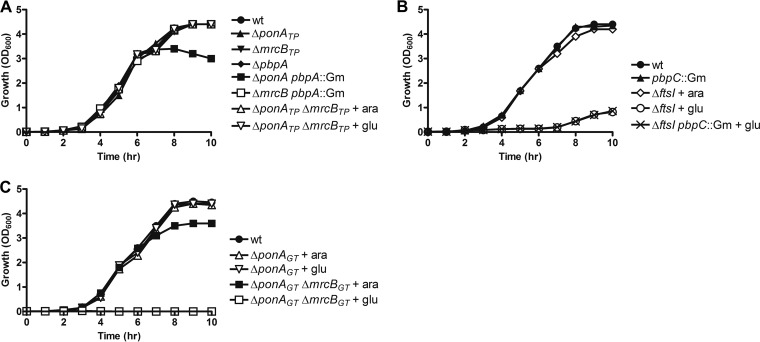
PBPs 1a, 1b, 2, and 3 were individually deleted by replacement of the respective PBP gene (or the region encoding the TPase domain for PBPs 1a and 1b) with a gentamicin (Gm) resistance cassette, followed by removal of the cassette. Single deletion of the transpeptidase (TPase) domains of PBP1a, 1b, and 2 (indicated by the subscript TP with the gene name) had no measurable impact on the growth of P. aeruginosa (Fig. 1A) (for the nomenclature used for protein and gene names, see Table 1). To assess whether the absence of a growth defect was due to a functional redundancy among these PBPs, we also made double mutants. The PBP1b/2 double mutants (ΔmrcBTP pbpA::Gm) exhibited normal growth, whereas a PBP1a/2 double mutant (ΔponATP pbpA::Gm) exhibited a slightly lower cell density after 8 h, likely due to cell lysis in stationary phase (as suggested by loose debris above the cell pellet after centrifugation). The absence of a defect when PBP2 (pbpA) was deleted in PA14 contrasts with a previous finding where a similar deletion in PAO1 resulted in a lower growth rate (29). Since a strain containing deletions of the TPase domains of PBP1a and PBP1b could not be obtained, it was necessary to construct a conditional deletion strain in which one copy of ponA under the control of the PBAD promoter was integrated into the chromosome at the attB site (ΔponATP ΔmrcBTP attB::PBAD-gfp-ponA). This promoter is induced by 0.2% arabinose but repressed with 0.2% glucose. As shown in Fig. 1A, no apparent growth defect was observed with either arabinose or glucose, indicating that both TPases of PBP1a and PBP1b are dispensable for growth. While it is possible that leaky expression was responsible for the growth of this strain, the PBAD promoter is very tightly regulated (35, 36), and even under conditions of repression, growth from the outset was the same as that for the wild type.
FIG 1.
Impact of pbp gene deletions on the growth of PA14. (A) Single and double deletions of the HMM PBPs PBP1a (ponA), PBP1b (mrcB), and PBP2 (pbpA) (the subscript TP indicates deletion of the transpeptidase domain only). (B) Deletion of PBP3x (pbpC) and conditional deletion of PBP3 (ftsI). (C) Deletion of the GTase domains of PBP1a and PBP1b (the subscript GT indicates deletion of the glycosyltransferase domain only). The P. aeruginosa strains were cultivated in LB broth (with arabinose or glucose, where indicated), and cell growth was monitored by measuring the optical density at 600 nm. wt, wild type.
TABLE 1.
Protein and gene names, loci in PA14 and PAO1, PBP class, and length of protein (in P. aeruginosa) for P. aeruginosa PBPs
| PBP | Gene | PA14 locus | PAO1 locus | PBP class | No. of amino acids |
|---|---|---|---|---|---|
| 1a | ponA | 66670 | 5045 | A | 822 |
| 1b | mrcB | 62200 | 4700 | A | 774 |
| 2 | pbpA | 12060 | 4003 | B | 646 |
| 3 | ftsI | 57425 | 4418 | B | 579 |
| 3a/3x | pbpC | 35190 | 2272 | B | 565 |
| 4 | dacB | 24690 | 3047 | C | 476 |
| 5/6 | dacC | 12100 | 3999 | C | 386 |
| 7 | pbpG | 53020 | 0869 | C | 310 |
The only strain that failed to grow directly was the one lacking PBP3 (ftsI). To ensure that this was due to the essentiality of PBP3 and was not due to insufficient colony screening, we made a conditional ftsI mutant with an integrated PBAD-ftsI at the attB site in a manner similar to that used for ponA. In the presence of glucose, the ΔftsI strain displayed an extended lag phase and a longer doubling time, but in the presence of arabinose, growth was restored to the wild-type level (Fig. 1B). Overall, these data reveal that under laboratory culture conditions, only PBP3 is essential for growth and there is significant redundancy in the TPase function among the remaining HMM PBPs.
Late growth of the ΔftsI strain.
During the growth experiments, we observed slight growth of the ΔftsI strain in the presence of glucose after about 7 h (Fig. 1B). To examine whether this was caused by derepression of the PBAD promoter after depletion of glucose, we resupplemented glucose at 6 h postinoculation, but the same late growth was still observed (data not shown). In addition, when an overnight culture of the ΔftsI strain that had been grown in the presence of glucose was diluted into fresh LB with glucose, no growth was again observed, indicating that the slow growth was not due to a mutation that upregulated the expression of PBP3. Since PBP3x exhibits 48% sequence identity with PBP3 and is mainly expressed in the stationary phase (34), we also considered whether the late growth was due to the expression of PBP3x. To test this, we constructed a ΔftsI pbpC::Gm double mutant strain (CW81), but in the presence of glucose, the same late-growth phenotype was observed (Fig. 1B). As expected, deletion of pbpC alone did not affect cell growth, consistent with previous findings that PBP3x is not essential (34). Hence, we conclude that the most likely reason for the late growth of the ΔftsI strain is the gradual accumulation of transcripts due to leaky expression from the promoter.
GTase domains of PBPs 1a and 1b are not individually essential.
Although the TPase domains of PBP1a and PBP1b appear not to be essential, these bifunctional PBPs also contain a glycosyltransferase (GTase) domain that might be required for growth. To address this, we constructed strains with conditional deletion of the GTase domain (indicated by the subscript GT with the gene name), the ΔponAGT and ΔponAGT ΔmrcBGT strains (Table 2), in which a copy of PBAD-ponA was integrated at the attB site. Sequencing confirmed that the TPase domain remained in frame in these GTase deletion mutants. In the presence of glucose, deletion of the GTase domain of PBP1a did not affect cell growth (Fig. 1C). Simultaneous mutation of both the PBP1a and PBP1b GTase domains, however, completely repressed growth, and this was reversed by adding arabinose to induce the expression of PBP1a (Fig. 1C). These data indicate that at least one GTase domain is required for cell viability in P. aeruginosa. Escherichia coli similarly fails to grow after deletion of both PBP1a and PBP1b, though, in that case, a distinction between the TPase and GTase domains was not made (37).
TABLE 2.
Bacterial strains and plasmids used or constructed in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| Stellar | E. coli HST08 strain for cloning of plasmids | Clontech |
| JM109 | E. coli strain for cloning of plasmids | Promega |
| P. aeruginosa strains | ||
| PA14 | Wild type | 68 |
| CW26 | ponATP::Gm | This study |
| CW28 | mrcBTP::Gm | This study |
| CW29 | ΔponATP | This study |
| CW30 | ΔponATP pbpA::Gm | This study |
| CW31 | ΔmrcBTP | This study |
| CW34 | ΔmrcBTP pbpA::Gm | This study |
| CW36 | pbpA::Gm | This study |
| CW37 | ΔpbpA | This study |
| CW46 | ΔftsI attB::PBAD-gfp-ftsI | This study |
| CW47 | ΔponATP ΔmrcBTP attB::PBAD-gfp-ponA | This study |
| CW49 | ΔponAGT attB::PBAD-gfp-ponA | This study |
| CW50 | ΔponAGT ΔmrcBGT attB::PBAD-gfp-ponA | This study |
| CW52 | ΔponATP attB::PBAD-gfp-ponA | This study |
| CW57 | PA14 containing pMP7605-PBP1a | This study |
| CW58 | PA14 containing pMP7605-PBP1b | This study |
| CW59 | PA14 containing pMP7605-PBP2 | This study |
| CW60 | PA14 containing pMP7605-PBP3 | This study |
| CW61 | PA14 containing pJN-flhF-mCherry | This study |
| CW80 | pbpC::Gm | This study |
| CW81 | ΔftsI pbpC::Gm attB::PBAD-gfp-ftsI | This study |
| Plasmids | ||
| pEX18ApGw | Suicide vector for gene knockout in P. aeruginosa, Gmr CBCr | 62 |
| pEX18ApGW-pbp1aTP | Suicide vector for deletion of the PBP1a TPase domain, Gmr CBCr | This study |
| pEX18ApGW-pbp1bTP | Suicide vector for deletion of the PBP1b TPase domain, Gmr CBCr | This study |
| pEX18ApGW-pbp2 | Suicide vector for deletion of pbp2, Gmr CBCr | This study |
| pEX18ApGW-pbp3 | Suicide vector for deletion of pbp3, Gmr CBCr | This study |
| pEX18ApGW-pbp3x | Suicide vector for deletion of pbp3x, Gmr CBCr | This study |
| pEX18ApGW-pbp1aGT | Suicide vector for deletion of PBP1a GTase domain, Gmr CBCr | This study |
| pEX18ApGW-pbp1bGT | Suicide vector for deletion of PBP1b GTase domain, Gmr CBCr | This study |
| pMini-CTX1 | Integration vector in P. aeruginosa, Tcr | 69 |
| pBAD24 | Plasmid with PBAD promoter induced by l-arabinose, CBCr | 35 |
| pCAB | Derivative of pMini-CTX1 containing PBAD promoter, Tcr | This study |
| pCAB-gfp-pbp1a | Derivative of pCAB containing PBAD-gfp-ponA, Tcr | This study |
| pCAB-gfp-pbp3 | Derivative of pCAB containing PBAD-gfp-ftsI, Tcr | This study |
| pMP7605 | E. coli-P. aeruginosa shuttle vector expressing mCherry, Gmr | 61 |
| pMP7605-PBP1a | Derivative of pMP7605 expressing mCherry-PBP1a, Gmr | This study |
| pMP7605-PBP1b | Derivative of pMP7605 expressing mCherry-PBP1b, Gmr | This study |
| pMP7605-PBP2 | Derivative of pMP7605 expressing mCherry-PBP2, Gmr | This study |
| pMP7605-PBP3 | Derivative of pMP7605 expressing mCherry-PBP3, Gmr | This study |
| pJN-flhF-mCherry | Derivative of pJN105 expressing FlhF-mCherry, Gmr | 46 |
CBCr, carbenicillin resistant; Gmr, gentamicin resistant; Tcr, tetracycline resistant.
A PBP1a and PBP2 double mutant shows reduced fitness.
To determine whether disruption of PBP genes alters the fitness of strains, we performed in vitro competition assays, in which each mutant strain was cultured with the wild-type strain PA14, starting at a 1:1 ratio. As shown in Fig. S1 in the supplemental material, deletion of ponATP, mrcBTP, pbpA, or ponA and mrcBTP did not significantly impact the fitness of P. aeruginosa. Only the simultaneous deletion of ponA and pbpA reduced the fitness by approximately 3-fold compared with that of the wild type. These experiments indicate that P. aeruginosa can tolerate a single deletion of nonessential PBPs without a significant reduction of fitness in vitro.
Susceptibility of pbp mutants to β-lactams.
As PBPs are targets of β-lactams, we examined whether deletion of HMM PBPs alters the susceptibility of P. aeruginosa to these antibiotics. Lowering of the MICs in a specific deletion strain would indicate the functional importance of that PBP (29) and increases in the MIC may indicate a role for the deleted PBP in resistance to β-lactams. All the β-lactams selected, with the exception of penicillin G, are used to treat pseudomonal infections. MICs were determined by the broth dilution method and are shown in Table 3. Compared to the MICs for the wild-type strain PA14, deletion of ponATP or mrcBTP had a minimal impact on the MICs of all antibiotics tested. Only the ΔponATP ΔmrcBTP double deletion strain was more susceptible and showed a 2-fold decrease in the MICs of cefsulodin and cefepime. The MICs of penicillin G, however, were decreased by 4-fold in the ponATP mutant and 8-fold in the ponATP and mrcBTP double deletion mutant. The relatively high MIC of penicillin observed for wild-type strain PA14 is consistent with the previously reported MICs of ampicillin and amoxicillin for PAO1 (29). Deletion of pbpA caused an 8-fold reduction in the MIC of penicillin G and a 4-fold decrease in the MIC of imipenem but, surprisingly, did not change the MIC of doripenem, which is also a carbapenem. These reductions in MICs are significantly less than those reported for wild-type strain PAO1 with a pbpA deletion, where the MIC was reduced 100-fold for ampicillin and 18-fold for carbenicillin (29).
TABLE 3.
Antibiotic susceptibilities of pbp mutant strains in PA14
| Strain | Genotype | MICa (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| IMP | DOP | CEP | CESD | CAZ | PIP | PenG | ATM | ||
| PA14 | Wild type | 1 | 0.25 | 2 | 4 | 4 | 4 | 2,048 | 8 |
| CW29 | ΔponATP | 1 | 0.25 | 2 | 2 | 4 | 4 | 2,048 | 8 |
| CW31 | ΔmrcBTP | 1 | 0.25 | 2 | 2 | 4 | 4 | 512 | 8 |
| CW37 | ΔpbpA | 0.25 | 0.25 | 2 | 4 | 4 | 4 | 256 | 8 |
| CW46b | ΔftsI | 0.25 | 0.125 | 0.25 | 0.5 | 0.5 | 1 | 64 | 1 |
| CW30 | ΔponATP pbpA::Gm | 0.25 | 0.25 | 2 | 16 | 4 | 64 | 1,024 | 8 |
| CW34 | ΔmrcBTP pbpA::Gm | 0.25 | 0.25 | 2 | 4 | 4 | 4 | 512 | 8 |
| CW47b | ΔponATP rmrcBTP | 1 | 0.25 | 1 | 2 | 2 | 4 | 256 | 8 |
IMP, imipenem; DOP, doripenem; CEP, cefepime; CESD, cefsulodin; CAZ, ceftazidime; PIP, piperacillin; PenG, penicillin G; ATM, aztreonam.
Strains CW46 and CW47 were grown in LB supplemented with 0.2% glucose, which was used to repress ectopic expression of PBP3 and PBP1a, respectively.
Simultaneous deletion of mrcBTP and pbpA in PA14 had an effect similar to that of deletion of pbpA alone, but interestingly, a double deletion of ponATP and pbpA significantly increased the MICs of cefsulodin and piperacillin by 4-fold and 16-fold, respectively. Finally, conditional deletion of ftsI decreased the MICs of all β-lactams tested: 4-fold for imipenem, 2-fold for doripenem, 8-fold for cefepime, cefsulodin, and ceftazidime, 4-fold for piperacillin, 8-fold for aztreonam, and 32-fold for penicillin G. These data are consistent with an essential function for PBP3 because in its absence P. aeruginosa was more susceptible to the lethal action of antibiotics.
Altered cell morphology of pbp mutants.
We also examined the morphology of PBP mutant strains using scanning electron microscopy (SEM). As shown in Fig. 2A, the ΔponATP, ΔmrcBTP, and ΔponATP ΔmrcBTP deletion strains appeared very similar to PA14, although the mean cell lengths tended to be slightly shorter (Fig. 2B). Deletion of pbpA or ftsI, though, did affect the cell morphology. The absence of pbpA reduced the mean cell length by approximately 30% (Fig. 2B), and while they still had a rod shape, the cells appeared somewhat wider. This differs from the spherical morphology observed in a pbpA deletion strain of PAO1 (29). Concurrent deletion of ponATP or mrcBTP with pbpA produced a similar effect. In sharp contrast, deletion of ftsI had a significant effect on cell morphology. Cells appeared as tangles of long filaments (Fig. 2A), indicative of impaired cell separation. These data show that P. aeruginosa PBP3 is required for cell division, similar to its E. coli counterpart (37).
FIG 2.
Cell morphology and cell length of pbp mutants. (A) The effect of HMM PBP deletion on cell morphology was determined by scanning electron microscopy. Cultures from early log phase were harvested and prepared as described in Materials and Methods. (B) Cell length of pbp mutants. More than 100 cells were counted and measured by the use of ImageJ software. Student's t test was performed for determination of the significance of the differences between the wild type and the pbp mutants. ***, P < 0.0001. The median average length is shown in parentheses.
Since deletion of ftsI causes impairment of cell separation, we reasoned that treatment of P. aeruginosa with a β-lactam at sub-MIC levels should generate a morphology similar to that of the ftsI deletion strain. In keeping with this hypothesis, treatment of PA14 with a sub-MIC of cefsulodin (0.5 μg/ml, 1/8 MIC) resulted in the formation of long filaments, in contrast to the rod morphology of untreated cells (Fig. 3). This is in keeping with PBP3 inhibition causing filamentation of P. aeruginosa, as is the case in E. coli (38).
FIG 3.
Cell morphology of PA14 treated with a sub-MIC level of cefsulodin. PA14 was inoculated into LB broth at 1/8 MIC (0.5 μg/ml) of cefsulodin and harvested after overnight growth. The cell morphology was examined by transmission electron microscopy.
Peptidoglycan composition in pbp mutants.
To test whether the absence of HMM PBPs alters the cell wall structure in P. aeruginosa, the peptidoglycan composition profile from mid-log-phase cultures of PBP deletion strains was analyzed using reverse-phase high-performance liquid chromatography (HPLC). Individual muropeptides were assigned on the basis of the profile of E. coli strain MC6RFl, as it appears similar to P. aeruginosa (39, 40). Compared to the findings for wild-type strain PA14, all mutant P. aeruginosa strains had the same HPLC profile and all showed a similar degree of cross-linking (Table 4). The only exception was a slight decrease in the proportion of peptidoglycan monomers and a correspondingly slight increase in peptidoglycan trimers in the pbpA deletion strain. The peptidoglycan composition of the ftsI conditional mutant could not be investigated because insufficient material for HPLC analysis was obtained due to poor growth in the presence of glucose. Overall, these data show that deletion of HMM PBPs 1a, 1b, and 2 does not significantly impact the peptidoglycan structure.
TABLE 4.
Peptidoglycan composition of pbp mutants in PA14
| Genotype | Relative abundancea (%) |
|||
|---|---|---|---|---|
| Cross-links | Monomers | Dimers | Trimers | |
| Wild type | 26.03 ± 0.88 | 49.66 ± 2.27 | 45.16 ± 1.13 | 5.18 ± 0.19 |
| ΔponATP | 24.14 ± 1.32 | 53.27 ± 3.46 | 41.97 ± 1.65 | 4.75 ± 0.96 |
| ΔmrcBTP | 25.46 ± 0.18 | 51 ± 1.65 | 43.2 ± 2.60 | 5.79 ± 1.68 |
| ΔpbpA | 27.88 ± 0.60 | 46 ± 1.51 | 46.2 ± 2.90 | 7.17 ± 1.43 |
| ΔponATP pbpA::Gm | 24.415 ± 0.58 | 52.71 ± 2.78 | 42.63 ± 0.71 | 4.65 ± 0.45 |
| ΔmrcBTP pbpA::Gm | 27.21 ± 0.81 | 47.5 ± 0.92 | 46.7 ± 1.37 | 5.79 ± 0.31 |
Data are means ± standard errors from three independent experiments.
Motility of pbp mutant strains.
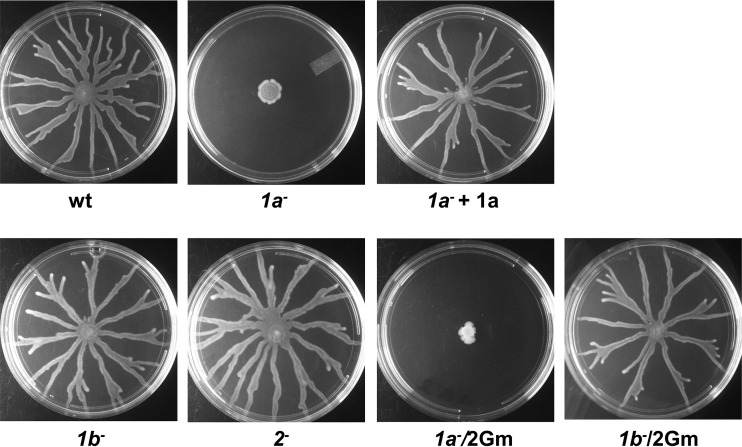
The importance of motility for P. aeruginosa pathogenicity (41) led us to determine the swarming capability of PBP mutants. Deletion of mrcBTP or pbpA had no effect on swarming motility. However, deletion of ponATP severely impaired swarming (Fig. 4), and this defect was reversed by complementation of the gene on a plasmid. Interestingly, introduction of mrcB could partially restore the motility of the ponATP mutant (data not shown). Since swarming also requires rhamnolipid surfactants (42), we also measured rhamnolipid levels in the PBP mutant strains by the anthrone method (43) but found no differences in rhamnolipid levels between the mutant strains and the wild-type strain (data not shown). These data suggest an involvement of PBP1a in cell motility. It remains to be established whether this is direct involvement or, for instance, a consequence of the bacterial stress response being triggered due to alterations of PG (44).
FIG 4.
Swarming motility of pbp mutants of PA14. Five microliters of an overnight culture was spotted onto an M9 plate (0.5% agar) and incubated for 18 h at 37°C. Swarming profiles were recorded by a digital camera.
Subcellular location of HMM PBPs in PA14.
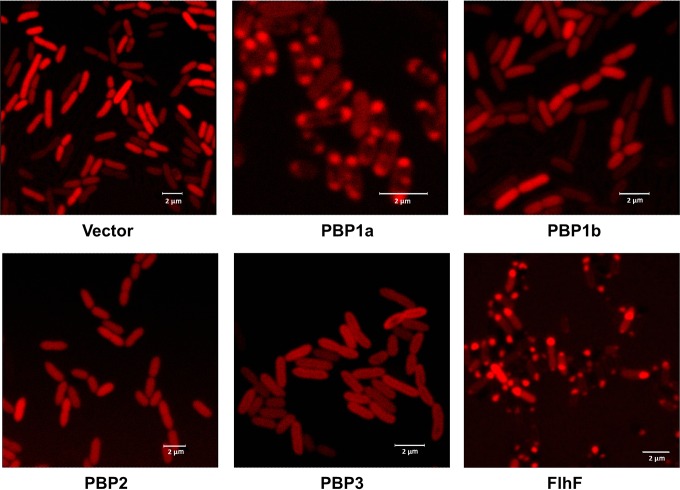
Some HMM PBPs, such as PBP2x and PBP2b of Streptococcus pneumoniae (45), exhibit a specific cellular localization. To observe the cellular localization of HMM PBPs in P. aeruginosa, we constructed N-terminal mCherry fusions of each of the four HMM PBPs (PBPs 1a, 1b, 2, and 3). We first verified that known phenotypes for each deletion were restored to the wild type by the respective mCherry-PBP fusion. Thus, expression of mCherry-PBP1a and mCherry-PBP1b completely (PBP1a) or partially (PBP1b) restored cell motility in the ponATP deletion strain, mCherry-PBP2 prevented cell lysis of the ΔponATP pbpA::Gm double mutant at stationary phase, and mCherry-PBP3 restored the growth rate of the ftsI deletion strain in the presence of glucose (data not shown). Examination of overnight cultures of the strains by confocal microscopy revealed that PBPs 1b and 2 were diffusely distributed throughout the cell (Fig. 5). In contrast, PBP1a mostly localized at both cell poles. Surprisingly, PBP3 did not localize in the middle of cells but seemed to anchor around the cell envelope. As expected, the empty vector expressing mCherry alone was diffusely distributed in the cytoplasm, and the known polar protein FlhF, which regulates the localization of the flagellum in P. aeruginosa (46), was primarily located at one pole. We also determined PBP localization at the mid-log phase, when cell division is more active, but similar patterns were observed. The exception was PBP1a, which was found to be distributed around the cell (data not shown).
FIG 5.
Subcellular location of HMM PBPs in PA14. P. aeruginosa PBPs were fused with mCherry at the N terminus of each protein and transformed into PA14. One microliter of an overnight culture was spotted on a slide and covered by a thin layer of 1.2% agarose. A known polar protein, FlhF, was used as a polar control. The empty vector expressing mCherry was used as a negative control. The location of fusion proteins was observed using a Leica SP2 confocal microscope.
In vitro virulence and in vivo pathogenicity of pbp mutants.
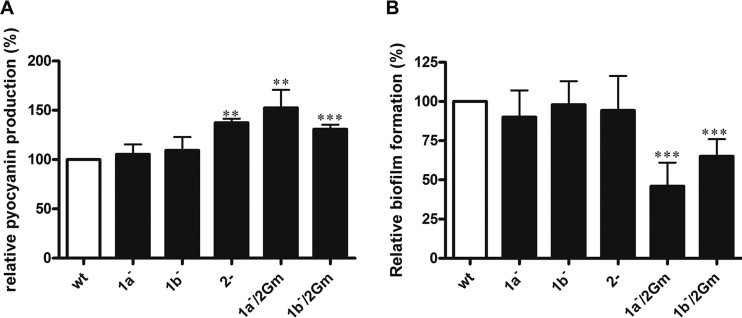
As assessments of virulence, we measured the pyocyanin levels and biofilm formation of the mutant strains. Deletion of ponATP or mrcBTP did not significantly affect pyocyanin levels compared to those in the wild type (Fig. 6A). Deletion of pbpA, however, increased pyocyanin levels by approximately 30%, and this increased to 50% when pbpA and ponATP were simultaneously disrupted. Given that the double deletion strain started autolysis when entering stationary phase (Fig. 1A), this finding suggests that conditions of stress can lead to increased pyocyanin production. A concurrent pbpA and mrcBTP deletion produced a lesser increase in the level of pyocyanin production. The ΔponATP pbpA::Gm and ΔmrcBTP pbpA::Gm double PBP mutants formed 50% and 30% less biofilm than the wild type, respectively, but single deletions of ponATP, mrcBTP, and pbpA had no effect (Fig. 6B). Overall, these data suggest that the loss of PBP2 can alter virulence phenotypes in vitro, especially when coupled with the deletion of PBP1a or PBP1b.
FIG 6.
Virulence-associated phenotypes of pbp mutants in vitro. (A) Pyocyanin production of pbp mutants. (B) Static biofilm formation on pegged lids. Data are presented as the mean ± SE from three independent experiments. Student's t test was performed for determination of the significance of the differences between the wild type and the pbp mutants. **, P < 0.001; ***, P < 0.0001.
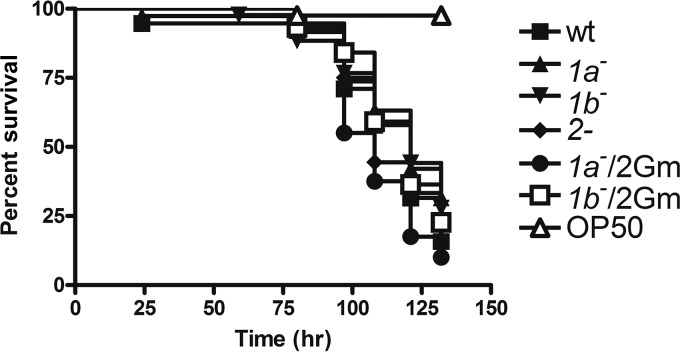
As a more direct measure of pathogenicity, we determined the toxicity of PBP mutants in Caenorhabditis elegans, a validated model for assessment of the virulence of P. aeruginosa (47, 48). Overnight cultures of PBP mutants were fed to synchronized worms, whose survival was then monitored every 12 h. The pathogenicity of single PBP deletion strains was essentially unchanged compared to that of the wild type (Fig. 7) and is consistent with the minimal effects on the virulence phenotypes observed in vitro.
FIG 7.
Survival of C. elegans infected by PA14 or the pbp mutants. Overnight cultures of PA14 or the pbp mutants were used to feed synchronized worms at the L4 stage. Worm survival was monitored every 12 h. The results of a representative experiment from a total of four are shown.
DISCUSSION
P. aeruginosa is a major nosocomial pathogen for which new treatment options are urgently needed due to high levels of resistance to current antibiotics (18). One approach for drug discovery is to identify antimicrobials with activity against novel molecular targets. These efforts often founder, however, due to the inaccessibility of the target, difficulty in developing an assay suitable for high-throughput screening, toxicity against human homologues, or other reasons. Another approach is to develop new antimicrobials with activity against existing targets. Such targets have the advantage of being proven in terms of accessibility and lethality, and there is a lower likelihood of adverse events arising due to toxicity. An outstanding example of this is penicillin-binding proteins (PBPs), the lethal targets for β-lactams (19, 20). For many years, β-lactams have been highly successful as antibiotics, but their efficacy is waning due to resistance mechanisms, such as β-lactamases. By developing new non-β-lactam inhibitors of PBPs that circumvent the action of β-lactamases, PBPs could be resurrected as clinical targets. Examples of this approach are the development of boronic acids and diazabicyclooctanes as PBP inhibitors (38, 49). Diazabicyclooctanes are known to inhibit E. coli PBP2 and exhibit antimicrobial activity against P. aeruginosa (strain PAO1) (38). They also increase the antimicrobial potency of β-lactams, including ceftazidime (50). Another approach is to develop new β-lactams, where an antimicrobial advantage could be gained by covalent inhibition of more than one PBP, rather than just one (51). P. aeruginosa has 8 known PBPs, and key to the present study was the identification of those that are essential for survival or at least are important with respect to inhibition by β-lactams or pathogenicity. The overall goal is to exploit PBPs thus identified as drug targets.
We found that only PBP3 is required for growth (see below), whereas single deletions of PBPs 1a, 1b, and 2 had little or no impact on growth. The nonessentiality of pbpA contrasts with the findings of a transposon insertion mutagenesis study, where this gene appeared to be essential in both PA14 and PAO1 (26), but agrees with the findings of a previous study where it was also deleted but found to be nonessential (29). Why PBP2 appears to be essential when it is disrupted by transposon insertion but nonessential when the entire open reading frame (ORF) is deleted is unclear but serves as a caution that different approaches to the investigation of essentiality can yield very different outcomes. Strains with double deletions of the TPase domains of HMM PBPs (PBPs 1a/1b, 1a/2, and 1b/2) were also viable. Redundancy between the respective GTase domains of PBPs 1a and 1b was also observed, with one domain being able to substitute for the other in the deletion strains (although in this case, both could not be deleted simultaneously). Taken together, this is evidence of functional overlap between the HMM PBPs in P. aeruginosa for both TPase and GTase activities. The apparent dispensability of HMM PBPs has also been observed in other bacteria, especially E. coli, where none are essential individually and only the simultaneous deletion of PBPs 1a and 1b is lethal (37). Similarly, PBPs 1a, 1b, and 2a (class A) and PBP2b (class B) can be deleted in Streptococcus gordonii, and only PBP2x is essential (52). In contrast, both of the HMM PBPs (PBPs 1 and 2) in Neisseria gonorrhoeae are considered essential (53), and this may reflect the relatively small complement of PBPs in this organism. It is, however, interesting to note how the essential PBPs of P. aeruginosa and E. coli are different, which cautions against making assumptions about the essentiality of PBP homologs across species.
The underlying reason for the apparent redundancy of PBPs in bacteria is an open question (54). One reason is that individual PBPs may have specific physiological roles that are not detected under cell culture conditions where there is an ample supply of nutrients and oxygen. A given PBP may be required at a specific stage in the cell cycle or subcellular localization, and only if this condition is known and can be reproduced in a laboratory setting would the essentiality of that PBP then become apparent. It is also worth noting that in bacteria like P. aeruginosa, which have comparatively large genomes, genetic redundancy is more likely due to the presence of paralogs of the genes under study (55).
Since essentiality is not the only factor governing the legitimacy of a PBP drug target, we also investigated the impact of PBP deletions on a number of phenotypes, including susceptibility to β-lactams, cell morphology, peptidoglycan structure, swarming motility, virulence factor production, and pathogenicity, in a C. elegans model of infection. As with the cell growth experiments, we found that deletions of most HMM PBPs (PBPs 1a, 1b, and 2) had little or no impact. Only deletion of PBP2 elicited a slight change in cell morphology by making cells shorter and wider, although the rod morphology was maintained. In contrast, the deletion of PBP 1a or 3 had major but different effects. First, the loss of PBP1a severely impaired swarming motility (Fig. 4) and PBP1a localized to the cellular poles of P. aeruginosa with a unipolar bias (Fig. 5). Although more investigation is needed, this at least implies an involvement of PBP1a in flagellum activity. If so, this may be an example of a specific physiological function of a PBP that underlies the apparent redundancy among PBPs and one that manifested only when swarming motility was measured.
The lesser effects on cell growth, morphology, and MICs when PBP2 was deleted in PA14 (this study) than when it was deleted in PAO1 (29) are of significant interest, as this PBP is considered a potential drug target (30, 38). The reason for these differences is not clear but suggests that, at least for PBP2, the background strain affects the severity of the phenotypes. More investigation of the importance of PBP2 in other strains of P. aeruginosa, including clinical strains, is therefore warranted.
The major finding of this study is that PBP3 is apparently the only PBP essential in P. aeruginosa. This was initially suggested by the growth failure of a PBP3 deletion strain and subsequently confirmed by the occurrence of growth when the protein was endogenously expressed under the control of the PBAD promoter (Fig. 1). In contrast to the situation with PBP2, our finding is in agreement with the findings obtained with a transposon insertion library, where PBP3 (ftsI) was found to be essential in both PAO1 and PA14 (28). It is also consistent with active-site mutations of PBP3 arising in P. aeruginosa (PAO1) exposed to increasing concentrations of meropenem (56). As demonstrated by the filamentous nature of the PBP3 deletion strain (Fig. 2), we also found that PBP3 is essential for cell division in P. aeruginosa, consistent with its established role in E. coli (57–60). Finally, the importance of PBP3 was also demonstrated by the significant decrease in the MICs of several antibiotics for the PBP3 deletion strain, and, indeed, this was the only deletion strain for which MICs were lower (Table 3). This is congruent with the overexpression of PBP3 increasing the MICs of aztreonam, cefepime, cefsulodin, and ceftazidime (25). Overall, these data suggest that, among the PBPs in P. aeruginosa, PBP3 has the highest potential for development as a drug target. Importantly, we also show that the remaining HMM PBPs (PBPs 1a and 1b and possibly also PBP2) are not likely to be fruitful targets.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 2. The P. aeruginosa and E. coli strains were routinely cultured at 37°C in LB medium (Miller) unless otherwise specified. To maintain plasmids, the medium was supplemented with 50 μg/ml carbenicillin (CBC), 30 μg/ml gentamicin (Gm), or 20 μg/ml tetracycline (TC) for E. coli and 200 μg/ml CBC, 30 μg/ml Gm, or 200 μg/ml TC for P. aeruginosa.
Manipulation of DNA and generation of pbp mutants.
The names of the PBPs and their respective genes are given in Table 1, and the plasmids used in this study are listed in Table 2. All plasmids were constructed using an In-Fusion kit (Clontech) except where indicated. The primers are listed in Table S1 in the supplemental material. To generate an arabinose-inducible integration vector (pCAB), an araC-PBAD fragment of 1.3 kb was excised from pBAD24 (35) with ClaI (converted to a blunt end) and KpnI and inserted into pMini-CTX1 restricted with SacI (converted to a blunt end) and KpnI. For complementation assays, pbp genes were inserted into pCAB linearized by NcoI. To observe the subcellular localization of PBPs, pbp genes were inserted into pMP7605 (61) linearized by BamHI. To construct gene knockout plasmids, two homologous arms of each pbp gene, as well as a fragment encoding Gm resistance (Gmr), were amplified, purified, and inserted into pEX18ApGw (62) linearized by EcoRI and HindIII. All plasmid constructs were confirmed by sequencing.
To generate each pbp deletion mutant, a gene deletion plasmid was transformed into strain PA14 by electroporation as described previously (63). The antibiotic resistance of the transformants was determined on LB-Gm and LB-CBC plates. Gmr CBC-susceptible (CBCs) colonies were putative replacement mutants, which were then confirmed by PCR with flanking screening primers and sequencing. To generate marker-free deletion strains, the Gmr cassette of the replacement mutants was removed by Flp recombinase, and the Flp plasmid was subsequently removed by sucrose counterselection. The deletion of the target gene was confirmed by PCR and sequencing.
To conditionally delete PBP3, one copy of ftsI was integrated into the chromosome of PA14 (wild type) at the attB site using the integration vector pCAB. In this vector, the target gene is under the control of the promoter PBAD, which is induced by 0.2% l-arabinose but repressed by 0.2% d-glucose. Genomic ftsI was deleted in the presence of arabinose. Similarly, for conditional knockouts of the TPase or the GTase domain of both PBP1a and PBP1b, one copy of ponA was integrated into the chromosome. For deletion of the GTase and TPase domains of PBP1a, amino acids 60 to 230 and 419 to 743, respectively, were deleted. For equivalent deletions in PBP1b, amino acids 160 to 343 and 424 to 694, respectively, were deleted. For PBP2, which is a monofunctional TPase, 87% of its ORF (amino acids 54 to 612) was deleted. Similarly, for PBP3 and PBP3x, 88% (amino acids 34 to 542) and 97% (amino acids 1 to 549) of the respective ORFs were deleted. Double mutants had either double deletions or single deletions combined with replacement of the second gene with a Gmr cassette.
In vitro competition assay.
PA14 and pbp replacement mutant strains were cultivated to early log phase (optical density at 600 nm [OD600] = 0.5). Cells were harvested, washed, and suspended in fresh medium. Cultures of the wild type and mutant were then mixed together in a 1:1 ratio in 10 ml LB broth, with the initial OD600 of each strain being 0.01. Since the ΔponATP pbpA::Gm double mutant (Table 2) exhibited autolysis in early stationary phase, cultures were incubated with shaking at 37°C for 7 h before serial dilution and spreading on LB plates (for total viable cell counts) and LB-Gm plates (for mutant cell counts only) after overnight incubation. The competition index is defined as the number of colonies with Gm resistance divided by the number of colonies grown on LB alone. A total of three independent experiments were performed.
MIC determination.
The susceptibilities of the pbp mutants to β-lactams were determined as MICs using the broth dilution method (64). One hundred microliters of an early-log-phase culture (prepared as described above) was diluted to an OD600 of 0.001 (corresponding to 104 to 105 CFU/ml) and dispensed into each well of a 96-well plate containing a serially diluted β-lactam. After 20 h of incubation at 37°C, the OD600 was measured using a BioTek Synergy HT plate reader. For each pbp mutant, three independent MIC assays were performed for each β-lactam.
SEM.
Cultures of the pbp mutants were prepared as described above, and cells were harvested at early log phase (OD600 = 0.5). For the ΔftsI and ΔponATP ΔmrcBTP mutants, the pellets were harvested at 4 h postinoculation because at this time the expression of PBP3 or PBP1a was expected to be fully repressed by glucose (Fig. 1). Samples were fixed in 2% phosphate-buffered glutaraldehyde for 1 h, rinsed in 0.1 M phosphate buffer, and fixed in 2% osmium tetroxide for 1 h. Samples were then dehydrated in 50% ethanol for 15 min, 100% ethanol for 15 min, and 100% hexamethyldisilazane for 5 min. Hexamethyldisilazane was replaced once, and samples were air dried, mounted on scanning electron microscopy (SEM) stubs, and sputter coated with gold-palladium (35-nm coating). The samples were examined in a JEOL 5410 SEM, and representative digital images were recorded. The lengths of P. aeruginosa cells were measured using ImageJ software (65).
TEM.
For transmission electron microscopy (TEM), an overnight culture of wild-type strain PA14 was pelleted and rinsed in phosphate buffer. The pellet was suspended in 2% osmium tetroxide for 5 min and rinsed with distilled water twice. The suspension was applied to a carbon-coated Formvar grid and allowed to dry. Cell shape was recorded with a JEOL 1011 electron microscope.
Analysis of peptidoglycan cross-links.
Peptidoglycan was isolated from the P. aeruginosa strains as described previously (39, 66). Briefly, cells were lysed in boiling SDS, and the peptidoglycan was harvested by ultracentrifuge. The proteins in the suspension were then removed by protease E, and peptidoglycan was digested by mutanolysin. The sample was reduced by sodium borohydride and adjusted to pH 3 to 4 with 20% (vol/vol) orthophosphoric acid. All steps were performed at room temperature.
The peptidoglycan fragments were analyzed by reverse-phase HPLC (Waters) using a HyperClone octyldecyl silane (C18) column (5 μm, 250 by 4.6 mm) equilibrated in 50 mM sodium phosphate (pH 4.35) with sodium azide (8 mg/liter); 75 mM sodium phosphate (pH 4.95) and 50% (vol/vol) methanol were used as buffer B. After sample (40 μl) injection, muropeptides were eluted with a 0 to 60% linear gradient of buffer B over 30 min at a flow rate of 1 ml/min. The UV absorbance of the eluates was monitored at 205 nm. The degree of cross-linking was calculated as described previously (40, 66).
Subcellular location of HMM PBPs.
PBP genes were cloned into pMP7605 to generate N-terminal mCherry fusion proteins. pJN-flhF-mCherry was used as a control. Each plasmid was then transformed into the wild-type strain PA14. The culture was taken in the mid-log phase and stationary phase for all the P. aeruginosa strains. One microliter of culture was spotted on a clean slide coated with a thin layer of 1.2% agarose and allowed to dry for 5 min. The subcellular localization of HMM PBPs (PBPs 1a, 1b, 2, and 3) was observed with a Leica SP2 confocal microscope with excitation at 587 nm and emission at 610 nm.
Determination of pyocyanin, biofilm formation, and motility of pbp deletion strains.
The quantification of pyocyanin in pbp mutant strains was performed as described previously (67). Briefly, supernatants of overnight cultures were harvested by centrifugation, and pyocyanin was extracted from the supernatants with equal volumes of chloroform and acidified with 0.2 N HCl. The absorbance of the samples at 520 nm was measured. A total of three independent experiments were performed, and the pyocyanin levels of the pbp mutants were normalized to those of the wild type.
In order to test swarming motility, overnight cultures of the pbp mutant strains (5 μl) were spotted on M9 medium (22 mM KH2PO4, 12 mM Na2HPO4, 20 mM NH4Cl, 8.6 mM NaCl, and 0.5% granulated agar supplemented with 1 mM CaCl2, 1 mM MgSO4, 0.2% glucose, and 0.5% Casamino Acids). The plates were incubated at 37°C for 18 h, after which the swarming pattern was recorded by digital photography.
To measure static biofilm formation, early-log-phase cultures of the pbp mutant strains were diluted into fresh LB to a final OD600 of 0.01 and dispensed into a 96-well plate (100 μl). Each sample was assayed in 16 replicate wells. The plate was covered with a pegged lid (Nunc) and incubated at 37°C for 18 h. The biofilms on the pegged lid were washed twice with sterile water and stained with 0.1% crystal violet (CV) for 15 min in a new 96-well plate with gentle shaking. The excess CV was washed off with sterile water, and the CV retained on the pegged lid was eluted with 150 μl of 95% ethanol. Solubilized CV was quantified by measuring the absorbance at 590 nm using a BioTek Synergy HT plate reader. In total, three independent experiments were performed, and the data were normalized to those for the wild type.
Pathogenicity of pbp deletion strains in C. elegans.
To test the pathogenicity of the P. aeruginosa strains, overnight cultures (100 μl) were spread onto nematode growth medium (NGM) containing 2.5% agar and 100 μg/ml 5-fluorodeoxyuridine and incubated overnight at 37°C. A culture of E. coli OP50 was used as a negative control. L4 worms of the C. elegans N2 strain (n = 30) were transferred from the synchronization plate to the inoculated NGM plates and incubated at 23°C. Three plates were used for each sample, for a total of 90 worms for each tested strain per experiment. The survival of C. elegans was monitored every 12 h using a dissection microscope. Worms that were unresponsive to touch were recorded as dead and removed from the plate. Each P. aeruginosa strain was tested in 4 independent experiments. Kaplan-Meier survival curves were plotted using GraphPad Prism (version 4.0) software, and statistical analysis was performed using the log-rank test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erika Büllesbach for guidance with the HPLC experiments and Nancy M. Smythe of MUSC's research electron microscopy service laboratory for help with electron microscopy. We also thank Zemer Gitai of Princeton University for providing the pJN-flhF-mCherry plasmid used in this study.
We declare no conflicts of interest.
This study used the MUSC Morphology, Imaging and Instrumentation Core, supported by NIGMS, NIH, grant P30 GM103342 to the South Carolina COBRE for Developmentally Based Cardiovascular Diseases. This work was supported by National Institutes of Health award AI109385 to C.D.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01651-16.
REFERENCES
- 1.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol 29:1–15. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 2.Blanc DS, Petignat C, Janin B, Bille J, Francioli P. 1998. Frequency and molecular diversity of Pseudomonas aeruginosa upon admission and during hospitalization: a prospective epidemiologic study. Clin Microbiol Infect 4:242–247. doi: 10.1111/j.1469-0691.1998.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 3.Talbot GH, Bradley J, Edwards JE Jr, Gilbert D, Scheld M, Bartlett JG. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin Infect Dis 42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 4.Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 5.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 6.Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, Ramsey BW. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis 183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 7.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 8.Zavascki AP, Barth AL, Fernandes JF, Moro AL, Goncalves AL, Goldani LZ. 2006. Reappraisal of Pseudomonas aeruginosa hospital-acquired pneumonia mortality in the era of metallo-beta-lactamase-mediated multidrug resistance: a prospective observational study. Crit Care 10:R114. doi: 10.1186/cc5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.American Thoracic Society, Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 10.Pena C, Gomez-Zorrilla S, Oriol I, Tubau F, Dominguez MA, Pujol M, Ariza J. 2013. Impact of multidrug resistance on Pseudomonas aeruginosa ventilator-associated pneumonia outcome: predictors of early and crude mortality. Eur J Clin Microbiol Infect Dis 32:413–420. doi: 10.1007/s10096-012-1758-8. [DOI] [PubMed] [Google Scholar]
- 11.Chapman TM, Perry CM. 2003. Cefepime: a review of its use in the management of hospitalized patients with pneumonia. Am J Respir Med 2:75–107. doi: 10.1007/BF03256641. [DOI] [PubMed] [Google Scholar]
- 12.Neuhauser MM, Weinstein RA, Rydman R, Danziger LH, Karam G, Quinn JP. 2003. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA 289:885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, Perez JL, Oliver A. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother 51:4329–4335. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Souli M, Galani I, Giamarellou H. 2008. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill 13(47):pii=19045 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19045. [PubMed] [Google Scholar]
- 15.Lopez-Dupla M, Martinez JA, Vidal F, Almela M, Soriano A, Marco F, Lopez J, Olona M, Mensa J. 2009. Previous ciprofloxacin exposure is associated with resistance to beta-lactam antibiotics in subsequent Pseudomonas aeruginosa bacteremic isolates. Am J Infect Control 37:753–758. doi: 10.1016/j.ajic.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Akhabue E, Synnestvedt M, Weiner MG, Bilker WB, Lautenbach E. 2011. Cefepime-resistant Pseudomonas aeruginosa. Emerg Infect Dis 17:1037–1043. doi: 10.3201/eid/1706.100358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa—a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 18.Rice LB. 2006. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin Infect Dis 43:S100–S105. doi: 10.1086/504487. [DOI] [PubMed] [Google Scholar]
- 19.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev 30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 20.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 21.Holtje JV. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goffin C, Ghuysen JM. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev 62:1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh AS, Chowdhury C, Nelson DE. 2008. Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol 16:309–317. doi: 10.1016/j.tim.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi H, Matsuhashi M, Mitsuhashi S. 1979. Comparative studies of penicillin-binding proteins in Pseudomonas aeruginosa and Escherichia coli. Eur J Biochem 100:41–49. doi: 10.1111/j.1432-1033.1979.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 25.Liao X, Hancock RE. 1997. Susceptibility to beta-lactam antibiotics of Pseudomonas aeruginosa overproducing penicillin-binding protein 3. Antimicrob Agents Chemother 41:1158–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ropy A, Cabot G, Sanchez-Diener I, Aguilera C, Moya B, Ayala JA, Oliver A. 2015. Role of Pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, beta-lactam resistance, and peptidoglycan structure. Antimicrob Agents Chemother 59:3925–3934. doi: 10.1128/AAC.05150-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legaree BA, Daniels K, Weadge JT, Cockburn D, Clarke AJ. 2007. Function of penicillin-binding protein 2 in viability and morphology of Pseudomonas aeruginosa. J Antimicrob Chemother 59:411–424. doi: 10.1093/jac/dkl536. [DOI] [PubMed] [Google Scholar]
- 30.Davies TA, Shang W, Bush K, Flamm RK. 2008. Affinity of doripenem and comparators to penicillin-binding proteins in Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:1510–1512. doi: 10.1128/AAC.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maejima T, Inoue M, Mitsuhashi S. 1991. In vitro antibacterial activity of KP-736, a new cephem antibiotic. Antimicrob Agents Chemother 35:104–110. doi: 10.1128/AAC.35.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe N, Katsu K. 1992. Bactericidal activity of cefclidin (E1040) against Pseudomonas aeruginosa under conditions simulating plasma pharmacokinetics: lack of development of chromosomally-mediated resistance to beta-lactams. J Antimicrob Chemother 30:475–487. doi: 10.1093/jac/30.4.475. [DOI] [PubMed] [Google Scholar]
- 33.Pucci MJ, Boice-Sowek J, Kessler RE, Dougherty TJ. 1991. Comparison of cefepime, cefpirome, and cefaclidine binding affinities for penicillin-binding proteins in Escherichia coli K-12 and Pseudomonas aeruginosa SC8329. Antimicrob Agents Chemother 35:2312–2317. doi: 10.1128/AAC.35.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao X, Hancock RE. 1997. Identification of a penicillin-binding protein 3 homolog, PBP3x, in Pseudomonas aeruginosa: gene cloning and growth phase-dependent expression. J Bacteriol 179:1490–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181:3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King AM, King DT, French S, Brouillette E, Asli A, Alexander JA, Vuckovic M, Maiti SN, Parr TR Jr, Brown ED, Malouin F, Strynadka NC, Wright GD. 2016. Structural and kinetic characterization of diazabicyclooctanes as dual inhibitors of both serine-beta-lactamases and penicillin-binding proteins. ACS Chem Biol 11:864–868. doi: 10.1021/acschembio.5b00944. [DOI] [PubMed] [Google Scholar]
- 39.Desmarais SM, Cava F, de Pedro MA, Huang KC. 2014. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp 83:e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quintela JC, Caparrós M, de Pedro MA. 1995. Variability of peptidoglycan structural parameters in Gram-negative bacteria. FEMS Microbiol Lett 125:95–100. doi: 10.1111/j.1574-6968.1995.tb07341.x. [DOI] [PubMed] [Google Scholar]
- 41.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Candrasekaran EV, Bemiller JN. 1980. Constituent analyses of glycosamino-glycans, p 89–96. In Whistler RL. (ed), Methods in carbohydrate chemistry. Academic Press Inc, New York, NY. [Google Scholar]
- 44.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsui HC, Boersma MJ, Vella SA, Kocaoglu O, Kuru E, Peceny JK, Carlson EE, VanNieuwenhze MS, Brun YV, Shaw SL, Winkler ME. 2014. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol 94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowles KN, Moser TS, Siryaporn A, Nyakudarika N, Dixon W, Turner JJ, Gitai Z. 2013. The putative Poc complex controls two distinct Pseudomonas aeruginosa polar motility mechanisms. Mol Microbiol 90:923–938. doi: 10.1111/mmi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan M-W, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan M-W, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inglis SR, Zervosen A, Woon EC, Gerards T, Teller N, Fischer DS, Luxen A, Schofield CJ. 2009. Synthesis and evaluation of 3-(dihydroxyboryl)benzoic acids as d,d-carboxypeptidase R39 inhibitors. J Med Chem 52:6097–6106. doi: 10.1021/jm9009718. [DOI] [PubMed] [Google Scholar]
- 50.Mendes RE, Rhomberg PR, Becker HK, Jones RN. 2013. β-Lactam activity tested in combination with β-lactamase inhibitor candidates against Enterobacteriaceae producing class A, B and D carbapenemases, abstr F-1189 Abstr 53rd Intersci Conf Antimicrob Agents Chemother, Denver, CO. American Society for Microbiology, Washington, DC: http://gm.asm.org/. [Google Scholar]
- 51.Josephine HR, Kumar I, Pratt RF. 2004. The perfect penicillin? Inhibition of a bacterial dd-peptidase by peptidoglycan-mimetic beta-lactams. J Am Chem Soc 126:8122–8123. [DOI] [PubMed] [Google Scholar]
- 52.Haenni M, Majcherczyk PA, Barblan JL, Moreillon P. 2006. Mutational analysis of class A and class B penicillin-binding proteins in Streptococcus gordonii. Antimicrob Agents Chemother 50:4062–4069. doi: 10.1128/AAC.00677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbour AG. 1981. Properties of penicillin-binding proteins in Neisseria gonorrhoeae. Antimicrob Agents Chemother 19:316–322. doi: 10.1128/AAC.19.2.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young KD. 2001. Approaching the physiological functions of penicillin-binding proteins in Escherichia coli. Biochimie 83:99–102. doi: 10.1016/S0300-9084(00)01205-0. [DOI] [PubMed] [Google Scholar]
- 55.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabot G, Zamorano L, Moya B, Juan C, Navas A, Blazquez J, Oliver A. 2016. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob Agents Chemother 60:1767–1778. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Botta GA, Park JT. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol 145:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen-Disteche M, Fraipont C, Buddelmeijer N, Nanninga N. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol Life Sci 54:309–316. doi: 10.1007/s000180050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buddelmeijer N, Beckwith J. 2002. Assembly of cell division proteins at the E. coli cell center. Curr Opin Microbiol 5:553–557. doi: 10.1016/S1369-5274(02)00374-0. [DOI] [PubMed] [Google Scholar]
- 61.Lagendijk EL, Validov S, Lamers GE, de Weert S, Bloemberg GV. 2010. Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol Lett 305:81–90. doi: 10.1111/j.1574-6968.2010.01916.x. [DOI] [PubMed] [Google Scholar]
- 62.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 65.Rismondo J, Möller L, Aldridge C, Gray J, Vollmer W, Halbedel S. 2015. Discrete and overlapping functions of peptidoglycan synthases in growth, cell division and virulence of Listeria monocytogenes. Mol Microbiol 95:332–351. doi: 10.1111/mmi.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glauner B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172:451–464. doi: 10.1016/0003-2697(88)90468-X. [DOI] [PubMed] [Google Scholar]
- 67.Farrow JM III, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. 2008. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol 190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 69.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.