ABSTRACT
Polymyxin B-based combinations have emerged as a mainstay treatment against carbapenem-resistant Escherichia coli (CREC). We investigated the activity of polymyxin B-based two-antibiotic combinations against CREC using time-kill studies (TKS) and validated the findings in a hollow-fiber infection model (HFIM). TKS were conducted using 5 clinical CREC strains at 5 log10 CFU/ml against 10 polymyxin B-based two-antibiotic combinations at maximum clinically achievable concentrations. HFIMs simulating dosing regimens with polymyxin B (30,000U/kg/day) and tigecycline (100 mg every 12 h) alone and in combination were conducted against two CREC strains at 5 log10 CFU/ml over 120 h. Emergence of resistance was quantified using antibiotic-containing media. Phenotypic characterization (growth rate and stability of resistant phenotypes) of the resistant isolates was performed. All five CREC strains harbored carbapenemases. Polymyxin B and tigecycline MICs ranged from 0.5 mg/liter to 2 mg/liter and from 0.25 mg/liter to 8 mg/liter, respectively. All antibiotics alone did not have bactericidal activity at 24 h in the TKS, except for polymyxin B against two strains. In combination TKS, only polymyxin B plus tigecycline demonstrated both bactericidal activity and synergy in two out of five strains. In the HFIM, polymyxin B alone was bactericidal against both CREC strains before regrowth was observed at 8 h. Phenotypically stable polymyxin B-resistant mutants were observed for both strains, with a reduced growth rate observed in one strain. Tigecycline alone resulted in a slow reduction in bacterial counts. Polymyxin B plus tigecycline resulted in rapid and sustained bactericidal killing up to 120 h. Polymyxin B plus tigecycline is a promising combination against CREC. The clinical relevance of our results warrants further investigations.
KEYWORDS: hollow-fiber infection model, antibiotic combination testing, polymyxin B, carbapenem-resistant Enterobacteriaceae
INTRODUCTION
Over the past decade, the emergence of carbapenem-resistant Enterobacteriaceae (CRE) has challenged the utility of carbapenems worldwide (1, 2). Infections caused by carbapenem-resistant Escherichia coli (CREC) are particularly problematic for several reasons. First, E. coli is a major etiologic agent for a range of life-threatening infections, such as ventilator-associated pneumonia and complicated intra-abdominal infections (2, 3). Second, genes encoding carbapenem resistance in E. coli (e.g., those encoding K. pneumoniae carbapenemases [KPCs] and New Delhi metallo-beta-lactamases [NDM]) are located on mobile genetic elements, facilitating the propensity for rapid spread in both the hospital and community settings (1–3). Third, CREC often exhibits broad-spectrum resistance to other classes of antibiotics, including beta-lactams, aminoglycosides, and fluoroquinolones (1–3). This has resulted in a paucity of effective treatment options against CREC, leaving clinicians in a major dilemma when faced with such infections.
In light of the futility of carbapenems against CREC, clinicians are now turning to polymyxin B, an antibiotic once sidelined due to concerns about unacceptable nephrotoxicity risks, for the treatment of infections caused by CREC (4). Unfortunately, increasing reports of polymyxin heteroresistance have suggested that rapid resistance to polymyxins can develop upon treatment with polymyxin B monotherapy, especially upon exposure to subtherapeutic polymyxin concentrations (5, 6). To circumvent this phenomenon, experts have advocated that polymyxins should be used in combination with one or more antibiotics for the treatment of CREC (4, 7, 8). When selecting a polymyxin B-based combination against CREC, one must consider the pharmacokinetic/pharmacodynamic (PK/PD) properties of the antibiotics and optimize the PK/PD target attainment of both polymyxin B and the adjuvant antibiotic employed (4, 7, 8).
In this study, we evaluated the activities of multiple antibiotics in combination with polymyxin B in vitro against CREC, using 24-h time-kill studies (TKS) to identify the most promising polymyxin B-based two-antibiotic combination against CREC. We then validated the activity of the most active combination using a hollow-fiber infection model (HFIM), subjecting the CREC strains to clinically relevant fluctuating concentrations of both polymyxin B and the adjuvant antibiotic. Additionally, we described the phenotypic properties of any resistant isolates that emerged when CREC was subjected to the fluctuating antibiotic concentrations in the HFIM.
(The results of this study were presented in part at the 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 5 to 9 September 2014.)
RESULTS
Phenotypic and genotypic characterization.
The MICs of the five CREC isolates to various antibiotics are shown in Table 1. All five isolates were resistant to amikacin, levofloxacin, rifampin, cefepime, meropenem, doripenem, imipenem, and aztreonam. No interpretative standards are provided by the CLSI for Enterobacteriaceae against polymyxin B or tigecycline (9). Polymyxin B MICs ranged from 0.5 mg/liter to 2 mg/liter, while tigecycline MICs ranged from 0.25 mg/liter to 8 mg/liter. The type of beta-lactamase gene, presence of changes in porin gene expression, and presence of efflux pumps in the five CREC isolates are shown in Table 2. All five CREC isolates harbored genes encoding carbapenemases; three isolates (CREC01, CREC03, and CREC05) harbored blaNDM, one (CREC02) harbored blaOXA-48, and one (CREC04) harbored blaKPC-2. The presence of efflux pumps was exhibited in all five CREC isolates based on phenotypic testing; in addition, three isolates (CREC01, CREC02, and CREC03) had downregulation of porin gene expression.
TABLE 1.
MICs for the five Escherichia coli strains employed in this study
| Antibiotic | MIC (mg/liter) for strain: |
||||
|---|---|---|---|---|---|
| CREC01 | CREC02 | CREC03 | CREC04 | CREC05 | |
| Amikacin | ≥128 | ≥128 | ≥128 | 64 | 32 |
| Levofloxacin | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 |
| Rifampin | 16 | 32 | ≥64 | ≥64 | ≥64 |
| Polymyxin B | 2 | 1 | 1 | 0.5 | 0.5 |
| Tigecycline | 0.5 | 0.5 | 0.25 | 1 | 8 |
| Cefepime | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 |
| Meropenem | ≥64 | 32 | ≥64 | 16 | ≥64 |
| Doripenem | ≥16 | 8 | ≥16 | 4 | ≥16 |
| Imipenem | ≥32 | ≥32 | ≥32 | ≥32 | ≥32 |
| Aztreonam | ≥128 | ≥128 | ≥128 | ≥128 | ≥128 |
| Piperacillin-tazobactam | ≥256/4 | ≥256/4 | ≥256/4 | ≥256/4 | ≥256/4 |
TABLE 2.
Molecular mechanisms of resistance of the five Escherichia coli strains employed in this study
| Mechanism of resistance | Presence in strain: |
||||
|---|---|---|---|---|---|
| CREC01 | CREC02 | CREC03 | CREC04 | CREC05 | |
| Genetic | blaCTXM-1, blaCMY, blaNDM | blaTEM, blaCTXM-1, blaOXA-48 | blaCTXM-1, blaCMY, blaNDM | blaTEM, blaKPC-2 | blaCTXM-1, blaCMY, blaNDM |
| Downregulation of porin gene expression | OmpF | OmpC, OmpF | OmpC, OmpF | None | None |
| Presence of efflux pumps | Yes | Yes | Yes | Yes | Yes |
TKS.
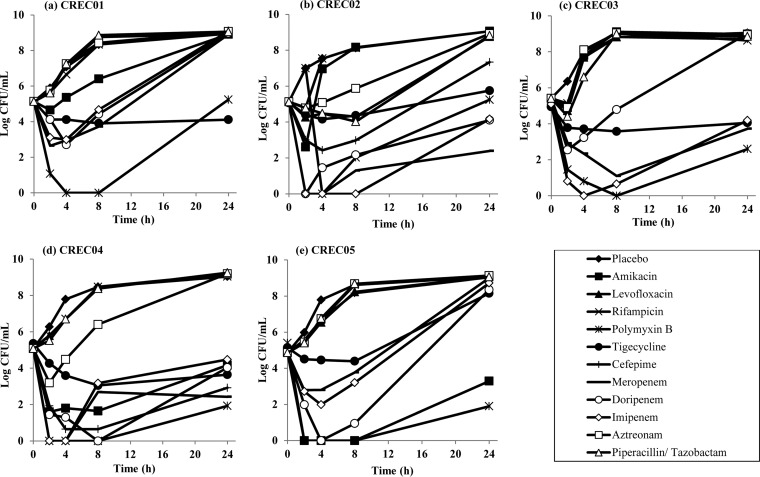
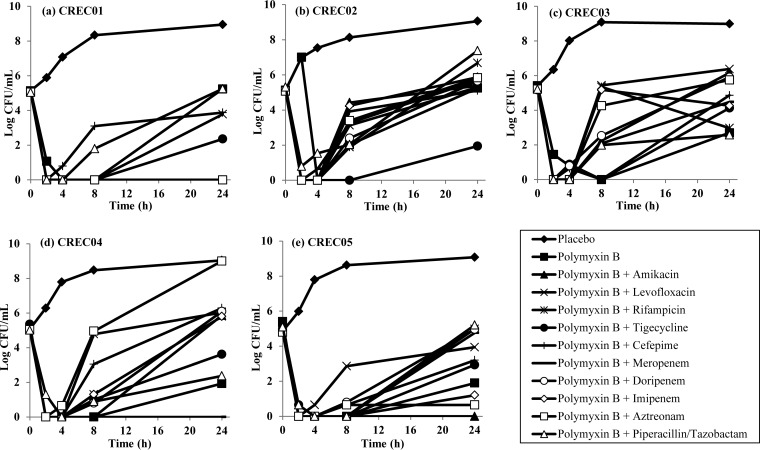
The time-kill profiles of the single and combination antibiotics over 24 h are shown in Fig. 1 and 2, respectively. As shown, all antibiotics alone did not have bactericidal activity at 24 h, except for polymyxin B against CREC04 and CREC05. In combination time-kill studies (TKS), a reduction in bacterial counts was observed for all polymyxin B-based combinations against all five CREC strains at 2 to 4 h, followed by various degrees of regrowth (an initial reduction from the starting inoculum followed by an increase in bacterial counts) for most combinations. Effective combinations appeared to be highly strain specific, and all tested combinations except polymyxin B plus tigecycline demonstrated antagonism against one or more CREC strains (Table 3). Polymyxin B plus amikacin, polymyxin B plus tigecycline, polymyxin B plus meropenem, polymyxin B plus imipenem, and polymyxin B plus aztreonam were bactericidal against 2/5 strains each; among these, only polymyxin B plus tigecycline also demonstrated synergy against both strains.
FIG 1.
Microbiological response of CREC01 (a), CREC02 (b), CREC03 (c), CREC04 (d), and CREC05 (e) over 24 h upon exposure to various single-antibiotic regimens in TKS.
FIG 2.
Microbiological response of CREC01 (a), CREC02 (b), CREC03 (c), CREC04 (d), and CREC05 (e) over 24 h upon exposure to polymyxin B alone and to various polymyxin B-based 2-antibiotic combinations in TKS.
TABLE 3.
Summary of the pharmacodynamic endpoints achieved by the polymyxin B-based two-antibiotic combinations in the in vitro TKS at 24 h against the five CREC isolates
| Combination | Resulta for strain: |
||||
|---|---|---|---|---|---|
| CREC01 | CREC02 | CREC03 | CREC04 | CREC05 | |
| Polymyxin B + amikacin | B/S | I | A | A | B |
| Polymyxin B + levofloxacin | I | I | A | A | A |
| Polymyxin B + rifampin | B/S | I | I | A | A |
| Polymyxin B + tigecycline | B/S | B/S | I | I | I |
| Polymyxin B + cefepime | I | I | A | A | I |
| Polymyxin B + meropenem | B/S | I | A | B | A |
| Polymyxin B + doripenem | B/S | I | A | A | A |
| Polymyxin B + imipenem | B/S | I | I | A | B |
| Polymyxin B + aztreonam | B/S | I | A | A | B |
| Polymyxin B + piperacillin-tazobactam | I | I | I | I | A |
A, antagonism; B, bactericidal; I, indifference; S, synergism.
HFIM.
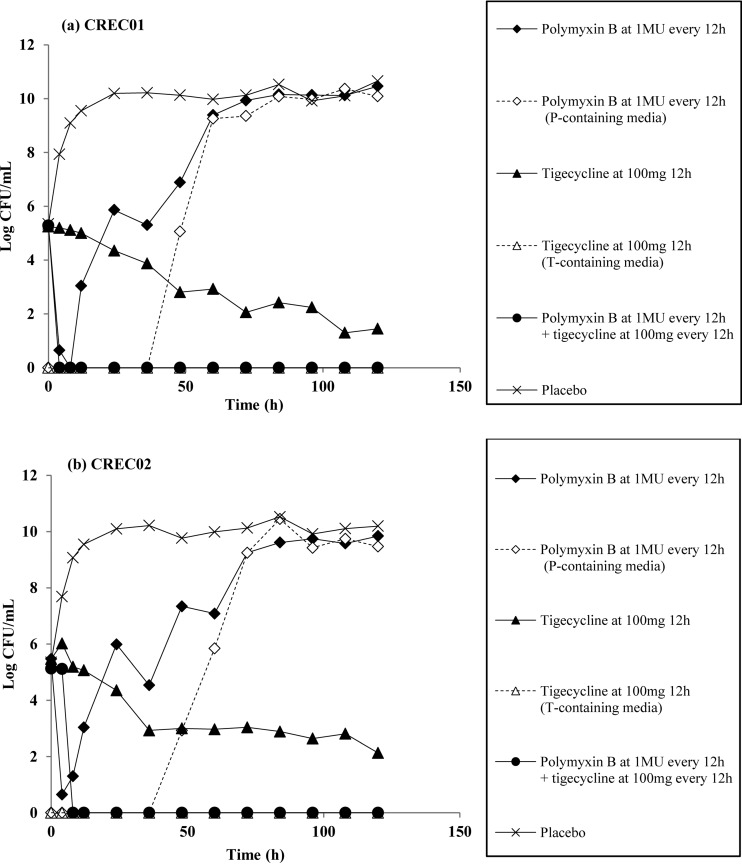
We validated polymyxin B plus tigecycline in our hollow-fiber infection model (HFIM) against CREC01 and CREC02 based on our TKS results. The time courses of the bacterial burden over 120 h against polymyxin B plus tigecycline are shown in Fig. 3. Against both strains, polymyxin B alone exhibited bactericidal activity up to 8 h. This was followed by rapid regrowth, reaching approximately 9 log10 CFU/ml at 60 h to 72 h. This increase in bacterial burden was accompanied by an increase in the polymyxin B-resistant subpopulation in the polymyxin B-supplemented medium. Exposure to tigecycline alone in the HFIM resulted in a slow reduction in bacterial counts, with a decrease of 2 log10 CFU/ml from the baseline inoculum observed only after 48 h. No tigecycline-resistant subpopulation was observed upon exposure to tigecycline alone. Rapid bactericidal killing was observed within 4 h for both CREC01 and CREC02 when exposed to clinically fluctuating concentrations of polymyxin B plus tigecycline; this suppression was sustained for both isolates up to 120 h.
FIG 3.
Microbiological response of CREC01 (a) and CREC02 (b) over 120 h upon exposure to polymyxin B and tigecycline singly and in combination in an HFIM.
PK validation.
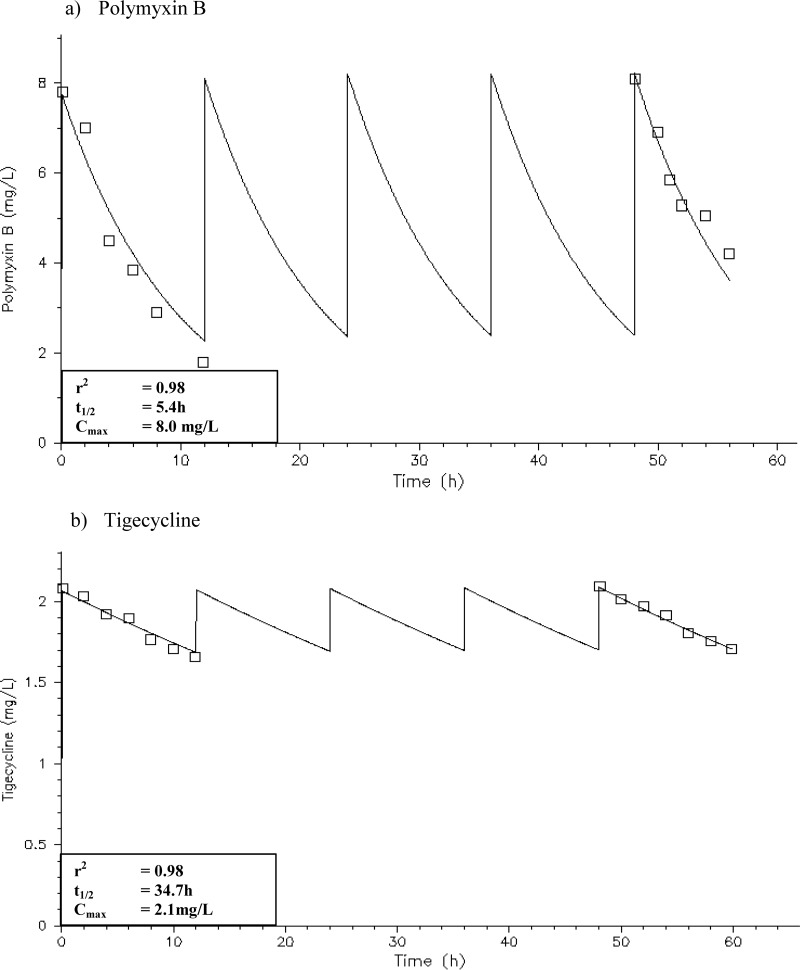
The simulated polymyxin B and tigecycline exposures in the HFIM are shown in Fig. 4. All simulated polymyxin B and tigecycline exposures were satisfactory; the simulated antibiotic exposures had an r2 value of 0.98.
FIG 4.
Pharmacokinetic profiles in the HFIM with polymyxin B at 30,000 U/kg/day or at least 1 MU every 12 h (a) and tigecycline at 100 mg every 12 h (b).
Phenotypic characterization of the resistant isolates.
The MICs of the resistant isolates recovered from polymyxin-B supplemented plates at 120 h (the end of the HFIM experiments) are shown in Table 4. An increase in MICs to polymyxin B was observed for both the CREC01 (polymyxin B MIC = 8 mg/liter) and CREC02 (polymyxin B MIC = 16 mg/liter) colonies recovered from the polymyxin B-supplemented plates. No changes in the susceptibilities to other antibiotics were observed. After 10 and 20 days of serial passages on antibiotic-free medium, no reversal of polymyxin B MICs was observed, which suggested the stability of the resistance phenotype in the postexposure isolates. A slight reduction in growth rate was observed in the post-polymyxin B exposure isolates (mean best-fit growth rate constant [Kg] = 1.451/h) for CREC01 compared to the parent strain (Kg = 1.477/h). In contrast, a large reduction in growth rate was observed for the CREC02 post-polymyxin B exposure isolates (Kg of CREC02 parent strain = 1.706/h; Kg of CREC02 postexposure subpopulation = 1.120/h), suggesting a large reduction in biofitness upon polymyxin B exposure for CREC02.
TABLE 4.
Comparison of susceptibilities of resistant isolates CREC01 and CREC02 before and after exposure to polymyxin B in an HFIM
| Antibiotic | MIC (mg/liter) for strain: |
|||
|---|---|---|---|---|
| CREC01 |
CREC02 |
|||
| Before polymyxin B exposure | After polymyxin B exposure | Before polymyxin B exposure | After polymyxin B exposure | |
| Amikacin | ≥128 | ≥128 | ≥128 | ≥128 |
| Levofloxacin | ≥64 | 32 | ≥64 | ≥64 |
| Rifampin | ≥64 | ≥64 | ≥64 | ≥64 |
| Polymyxin B | 2 | 8 | 1 | 16 |
| Tigecycline | 0.5 | ≤0.25 | 0.5 | 0.5 |
| Cefepime | ≥64 | ≥64 | ≥64 | ≥64 |
| Meropenem | ≥64 | ≥64 | 32 | 32 |
| Doripenem | ≥16 | ≥16 | 8 | ≥16 |
| Imipenem | ≥32 | ≥32 | ≥32 | ≥32 |
| Aztreonam | ≥128 | ≥128 | ≥128 | ≥128 |
| Piperacillin-tazobactam | ≥256/4 | ≥256/4 | ≥256/4 | ≥256/4 |
DISCUSSION
Our study investigated the activity of multiple antibiotics in combination with polymyxin B in vitro against CREC in TKS and validated the most promising polymyxin B-based combination in an HFIM. The results from our HFIM experiments showed that the combination of polymyxin B and tigecycline was rapidly bactericidal within 4 h of antibiotic exposure and limited the emergence of polymyxin B resistance up to 120 h.
The antibiotic concentrations employed in the TKS and the dosage regimens simulated in the HFIM experiments were based on careful consideration of the concentration-time profiles of the unbound antibiotics that can be achieved in patients when administered at maximum doses (10, 11). The polymyxin B concentrations achieved in our HFIM were similar to those in the most recent pharmacokinetic studies on polymyxin B; in a population pharmacokinetic study by Sandri et al., exposure to a polymyxin B dose of 30,000 U/kg/day (administered every 12 hours) resulted in a median steady-state peak unbound concentration of 8.51 mg/liter and a median steady-state unbound trough concentration of 2.25 mg/liter (12). The tigecycline concentrations employed in our HFIM experiments were based on achievable unbound tissue concentrations instead of serum concentrations (13). This is because tigecycline distributes widely into tissues and has no utility in the treatment of bacteremia in clinical practice but is instead commonly employed for the treatment of skin/soft tissue and intra-abdominal infections (14).
We employed five clinical CREC strains harboring an array of carbapenem resistance mechanisms, including carbapenemases, enhanced efflux pump expression, and/or altered membrane permeability due to porin loss, in our study to reflect the variability of carbapenem resistance mechanisms encountered in our local setting (15). An NDM-producing strain and an OXA-producing strain were eventually selected for the HFIM experiments, as these carbapenemases were reported to be the most common among Enterobacteriaceae in Singapore (15, 16). In the TKS experiments, all five strains were susceptible to polymyxin B (MIC range, 0.5 to 2 mg/liter); however, regrowth was demonstrated in three out of the five strains exposed to polymyxin B alone, suggesting potential heteroresistance among these strains. No single polymyxin B-based combination was effective against all five strains, which may be attributable to the differences in the mechanisms of molecular resistance. Interestingly, despite similar molecular resistance mechanisms mediating carbapenem resistance in CREC01 and CREC03, the effective polymyxin B-based combinations against each strain appeared to be highly strain specific. Of note, all polymyxin B-carbapenem combinations were bactericidal against CREC01 at 24 h, while none were bactericidal against CREC03. We postulate that the observed difference may be attributed to the differences in the type and/or degree of porin loss; a more detailed work-up on the resistance mechanisms in these strains will be required (17). Antagonism was observed in all polymyxin B-based combinations except polymyxin B plus tigecycline against one or more CREC strains. This finding suggested that the empirical selection of polymyxin B-based combinations can be counterproductive and emphasized the importance of in vitro testing in guiding the selection of effective combinations (18, 19).
Our findings in the HFIM experiments corroborated our TKS results. Exposure to polymyxin B alone resulted in an initial bactericidal killing followed by regrowth, which is possibly due to the selective amplification of the preexisting polymyxin B-resistant subpopulations when exposed to polymyxin B alone. Upon exposure to fluctuating concentrations of tigecycline alone, a slow reduction in bacterial burden was observed for both CREC isolates, with less than 1 log10 CFU/ml reduction after 24 h. Our findings were similar to those in previous reports, which showed that tigecycline demonstrated a slow but sustained bacteriostatic effect against Enterobacteriaceae (20). There was no emergence of tigecycline-resistant subpopulations despite prolonged treatment with tigecycline monotherapy, which is contrast to a previous HFIM study by Lim et al., who demonstrated that tigecycline-resistant Klebsiella pneumoniae mutants were isolated from an initially tigecycline-susceptible strain after 240 h of tigecycline exposure at clinically relevant concentrations (11).
To estimate the biological costs associated with resistance development, we conducted a pairwise time-growth study between the resistant isolates recovered from the antibiotic-supplemented plates in the HFIM and their corresponding ancestral strains (21). Interestingly, a significant reduction in the growth rate of the polymyxin B-resistant isolates recovered from the antibiotic-supplemented plates was seen in CREC02, while only a slight reduction in growth rate was observed in the CREC01 post-polymyxin B exposure isolates. This suggested that the magnitude of biological costs that is imposed by the development of resistance mutations can vary among strains and may be dependent on a multitude of factors, including the initial biofitness of the ancestral strain and the type and degree of resistance acquired upon antibiotic exposure. It should be noted that the lack of measureable cost based on growth experiments under in vitro conditions does not necessarily mean that the costs of resistance are nonexistent, as an in vitro biofitness deficit in bacteria does not equate to in vivo biofitness (21). Further in vivo work-up will be required to fully investigate the cost of resistance development on in vivo virulence of these postexposure isolates.
To date, a limited number of previous studies have investigated the activity of polymyxin (polymyxin B or colistin)-based combinations against CREC; however, most of these studies only conducted TKS or checkerboard assays over 24 h (20, 22, 23). Betts et al. further explored the efficacy of tigecycline plus colistin against three CREC strains in a Galleria mellonella model, but the relevance of an invertebrate model in predicting human therapeutics remains questionable (23). In contrast, our present study is the first to investigate polymyxin B-based combinations against CREC using an HFIM, subjecting the CREC strains to clinically relevant concentrations of antibiotics for a prolonged duration. The main limitation of our study was that we were able to validate only a single polymyxin B-based combination in the HFIM experiments due to their labor-intensive nature. Given the strain-specific activity of various polymyxin B-based combinations in our TKS, future studies will be required to further validate the activity of other polymyxin B-based combinations in an HFIM system.
Conclusion.
We have shown in our study that the combination of polymyxin B and tigecycline at clinically relevant fluctuating concentrations was rapidly bactericidal against CREC and suppressed the emergence of polymyxin B resistance. Our future studies will include validating other combinations found effective against CREC in an HFIM system, as well as investigating the in vivo and clinical relevance of these combinations.
MATERIALS AND METHODS
Bacterial isolates.
Five nonclonal, clinical strains of CREC (CREC01 to CREC05), previously collected as part of a nationwide surveillance study from 2011 to 2012, were obtained from the largest tertiary hospital in Singapore (1,700 beds). Genus identity was ascertained using Vitek 2 ID-GN cards (bioMérieux, Inc., Hazelwood, MO). The CREC strains were stored at −80°C in Cryobank (Thermo Scientific, Singapore) storage vials, and fresh isolates were subcultured twice on 5% sheep blood agar plates (Thermo Scientific, Singapore) for 24 h at 35°C before each experiment.
Phenotypic and genotypic characterization.
MICs to multiple antibiotics were determined in cation-adjusted Mueller-Hinton II broth (Ca-MHB) (BBL, Sparks, MD) using commercial broth microdilution panels (Trek Diagnostics, East Grinstead, UK) and interpreted in accordance to the CLSI guidelines (9, 10). All experiments involving tigecycline were performed using freshly prepared Ca-MHB. PCR was employed to determine the presence of genes encoding extended-spectrum β-lactamases (ESBLs) (blaTEM and blaSHV), plasmid-mediated AmpCs, metallo-β-lactamases (MBLs) (blaVIM, blaIMP, blaSIM, blaGIM, blaSPM, and blaNDM), and KPCs (24). Expression of the ompC and ompF porin genes was examined by quantitative reverse transcription-PCR (RT-PCR), with expression levels normalized to the housekeeping rpoB gene (25). Relative quantities of mRNA of each gene of interest were determined using the comparative threshold cycle method, calibrating against standard E. coli ATCC 25922. A reduction in porin gene expression of ≥10-fold was considered indicative of reduced porin gene expression (26). To elucidate the presence of efflux pumps, levofloxacin MICs were determined in the presence and absence of the efflux pump inhibitor phenylalanine-arginine β-naphthylamide (50 μg/ml), in accordance to a previously published protocol (27). Levofloxacin MICs that were ≥4-fold lower in the presence of an efflux pump inhibitor were considered to be indicative of an elevated efflux mechanism (27).
Antimicrobial agents.
Eleven antibiotics were used in the experiments. Aztreonam and rifampin were purchased from Toronto Research Chemicals. Cefepime was purchased from Kemimac(s) Pte Ltd. Amikacin, levofloxacin, and polymyxin B were purchased from Sigma-Aldrich. Doripenem was obtained from Shionogi & Co. Imipenem was provided by Merck & Co. Meropenem was provided by Astra Zeneca Inc. Tigecycline and piperacillin-tazobactam were provided by Pfizer Inc. Aliquots of stock solutions of all antibiotics except rifampin were prepared in sterile water and stored at −80°C. Prior to each experiment, the aliquot was thawed and diluted to the desired concentrations with Ca-MHB. Rifampin was dissolved in dimethyl sulfoxide (DMSO) before being serially diluted in sterile water to the desired concentration. The final DMSO concentration (<1%, vol/vol) did not affect bacterial growth (9).
TKS.
Time-kill studies (TKS) were performed on all five CREC strains with amikacin, aztreonam, cefepime, doripenem, imipenem, levofloxacin, meropenem, piperacillin-tazobactam, rifampin, and tigecycline singly and in two-antibiotic combinations with polymyxin B. The antibiotic concentrations used were based on maximal clinically relevant unbound concentrations when maximum antibiotic doses were administered and are shown in Table 5 (13, 28–37). The procedures for the TKS have been described in detail in our previous study (10). Briefly, 15 ml of log-phase bacterial suspensions in Ca-MHB was transferred to sterile flasks containing 1 ml of antibiotics and placed into a shaker water bath at 35°C (final bacterial concentration, approximately 5 log10 CFU/ml [1 × 105 CFU/ml to 5 × 105 CFU/ml]). At 0, 2, 4, 8, and 24 h, samples were obtained in duplicate from each flask and washed with sterile normal saline to minimize drug carryover. Viable counts were obtained by dropping serial 10-fold dilutions of the reconstituted samples onto Mueller-Hinton agar (MHA) plates (Thermo Scientific, Singapore) and enumerating at 24 h. The lower limit of detection for the colony counts was 2.6 log10 CFU/ml.
TABLE 5.
Simulated antibiotic dosing regimens and corresponding drug concentrations
| Drug | Simulated dosing regimen | Concn (mg/liter) | Reference |
|---|---|---|---|
| Amikacin | 15–20 mg/kg every 24 h | 80 | 28 |
| Aztreonam | 6 g every 24 h (infused over 24 h) | 18 | 29 |
| Cefepime | 2 g every 8 h | 200 | 30 |
| Doripenem | 1 g every 8 h (infused over 4 h) | 13 | 31 |
| Imipenem | 1 g every 6 h (infused over 0.5 h) | 32 | 32 |
| Levofloxacin | 750 mg every 24 h | 8 | 33 |
| Meropenem | 2 g every 8 h (infused over 3 h) | 64 | 34 |
| Piperacillin-tazobactam | 4.5 g every 6 h (infused over 4 h) | 75/15 | 35 |
| Polymyxin B | 30,000 IU/kg/day or at least 1 MU every 12 h | 2 | 36 |
| Rifampin | 600 mg every 12 h | 2 | 37 |
| Tigecycline | 100 mg every 12 h | 2 | 13 |
Experimental setup for the HFIM.
A hollow-fiber infection model (HFIM) (FiberCell Systems, Frederick, MD), first described by Blaser, was used to determine the killing activity of the antibiotic combinations when bacteria were exposed to clinically relevant fluctuating antibiotic concentrations (38). A schematic diagram of the HFIM has been previously published (11, 38, 39). Two bacterial isolates (CREC01 and CREC02) were tested against polymyxin B plus tigecycline (the most active antibiotic combination in the TKS). The single-antibiotic regimens and a placebo control were also tested. Twenty milliliters of log-phase bacteria (∼5 log10 CFU/ml), prepared as described above, was injected into the extracapillary compartment of the hollow-fiber cartridge and subjected to the different antibiotic exposures for up to 120 h in a humidified incubator set at 35°C. The antibiotic exposures simulated the steady-state pharmacokinetic profiles when polymyxin B at 30,000 IU/kg/day, or least 1 MU every 12 h, or tigecycline at 100 mg every 12 h was administered to patients with multidrug resistant Gram-negative bacterial infections (10, 12). Maintenance doses were given in accordance with clinical dosing frequencies for the individual drugs to reattain the target concentrations in the HFIM.
Microbiological response.
Serial samples were collected aseptically from the HFIM at 0 h (baseline), 4 h, 8 h, 24 h, 36 h, 48 h, 60 h, 72 h, 84 h, 96 h, 108 h, and 120 h in duplicates to determine the total and resistant bacterial subpopulation viable cell count. For the total bacterial population, the samples were cultured on MHA plates as described above; for resistant bacterial subpopulations, the samples were cultured on MHA supplemented with the exposed antibiotic at 3× the MIC of the exposed agent. The lower limit of detection for the colony counts was 2.6 log10 CFU/ml.
Pharmacokinetic validation.
Serial 1-ml samples were obtained every two hours from the central reservoir of the HFIM from 0 h to 12 h and 48 h to 60 h in triplicates and kept at −80°C until analysis. The antibiotic concentrations in the samples were assayed using a validated liquid chromatography-mass spectrometry (LC-MS) method as described previously (10). The pharmacokinetic models of the antibiotics were obtained by fitting a one-compartment linear model to the observations using ADAPT II (40). All samples were analyzed within 1 month from the completion of the HFIM studies.
Phenotypic characterization of the resistant isolates.
The following phenotypic tests were conducted to characterize the resistant isolates (if any) that emerged in the HFIM experiments: (i) susceptibility testing using commercial broth microdilution panels to screen for potential phenotypic changes in resistance patterns, (ii) serial passages on drug-containing and drug-free media to evaluate stability of the resistant phenotypes, and (iii) time-growth studies to evaluate the biofitness of the resistant subpopulation. MIC testing for the exposed antibiotic(s) was performed on at least two colonies of the resistant isolates recovered from antibiotic-supplemented plates throughout the HFIM experiment to confirm the presence of resistance. MICs to other antibiotics were also determined with at least two colonies of the resistant isolates obtained at 120 h. For the serial passages, the resistant isolates were subjected to 20 days of passages on drug-free MHA and MHA supplemented with the exposed antibiotic at 3× the MIC, and MIC testing for all the antimicrobial agents was performed at the 10-day and 20-day time points. To carry out the time-growth studies, 24 ml of log-phase bacteria (∼5 log10 CFU/ml), prepared as described above, was transferred to 50-ml sterile conical flasks and incubated in a shaker water bath at 35°C. At 0 (baseline), 1, 2, 3, 4, 5, 6, and 24 h, serial 1-ml samples were obtained (in triplicate) and quantified as described above. The exponential growth of the bacterial population over 24 h was analyzed using an adapted mathematical model (41).
Pharmacodynamic endpoints.
Bactericidal activity was defined as a ≥3 log10 CFU/ml decrease in the colony count from the initial inoculum at 24 h (11). Synergy was defined as a ≥2 log10 CFU/ml decrease in the colony count by the drug combination compared with its most active constituent and a ≥2 log10 CFU/ml decrease from the initial inoculum at 24 h (11). Indifference was defined as a <2 log10 CFU/ml change at 24 h by the combination compared with that by the most active single agent (11). Antagonism was defined as defined as an increase of ≥2 log10 CFU/ml with the combination in comparison with the viable bacterial count obtained with the most active single agent (11).
ACKNOWLEDGMENTS
We thank the clinical microbiology staff of Singapore General Hospital and Changi General Hospital for assisting in the collection of the bacterial isolates.
Andrea L. Kwa, Tze-Peng Lim, and Winnie Lee have received funding for research from Janssen-Cilag and Merck Sharp and Dohme (I.A.) Corp. None of the companies provided any funding for this study.
This work was supported in part by Singapore General Hospital Research Grant SRG/C3/02/2014, National Medical Research Council Centre Grant NMRC/CG/016/2013, and unrestricted grant funding from Pfizer Inc. (WS 832447 and WS 776979). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:1–11. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Doi Y, Paterson DL. 2015. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Y, Lee W, Kwa AL. 2015. Polymyxin B versus colistin: an update. Expert Rev Anti Infect Ther 13:1481–1497. doi: 10.1586/14787210.2015.1093933. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. 2011. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother 66:946–947. doi: 10.1093/jac/dkr007. [DOI] [PubMed] [Google Scholar]
- 7.Zavascki AP, Bulitta JB, Landersdorfer CB. 2013. Combination therapy for carbapenem-resistant Gram-negative bacteria. Expert Rev Anti Infect Ther 11:1333–1353. doi: 10.1586/14787210.2013.845523. [DOI] [PubMed] [Google Scholar]
- 8.Perez F, El Chakhtoura NG, Papp-Wallace KM, Wilson BM, Bonomo RA. 2016. Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: can we apply “precision medicine” to antimicrobial chemotherapy? Expert Opin Pharmacother 17:761–781. doi: 10.1517/14656566.2016.1145658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing: 25th informational supplement. M100-S25. CLSI, Wayne, PA, USA. [Google Scholar]
- 10.Cai Y, Lim TP, Teo JQ, Suranthran S, Lee W, Hong Y, Chan EC, Tan TY, Tan TT, Koh TH, Hsu LY, Kwa AL. 2016. In vitro activity of polymyxin B in combination with various antibiotics against extensively drug-resistant Enterobacter cloacae with decreased susceptibility to polymyxin B. Antimicrob Agents Chemother 60:5238–5246. doi: 10.1128/AAC.00270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim TP, Cai Y, Hong Y, Chan EC, Suranthran S, Teo JQ, Lee WH, Tan TY, Hsu LY, Koh TH, Tan TT, Kwa AL. 2015. In vitro pharmacodynamics of various antibiotics in combination against extensively drug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 59:2515–2524. doi: 10.1128/AAC.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 13.Rodvold KA, Gotfried MH, Cwik M, Korth-Bradley JM, Dukart G, Ellis-Grosse EJ. 2006. Serum, tissue and body fluid concentrations of tigecycline after a single 100 mg dose. J Antimicrob Chemother 58:1221–1229. doi: 10.1093/jac/dkl403. [DOI] [PubMed] [Google Scholar]
- 14.De Rosa FG, Corcione S, Di Perri G, Scaglione F. 2015. Re-defining tigecycline therapy. New Microbiol 38:121–136. [PubMed] [Google Scholar]
- 15.Teo J, Cai Y, Lim TP, Tan TT, Kwa AL. 2016. Carbapenem resistance in Gram-negative bacteria: the not-so-little problem in the little red dot. Microorganisms 4:13. doi: 10.3390/microorganisms4010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balm MN, Ngan G, Jureen R, Lin RT, Teo JW. 2013. OXA-181-producing Klebsiella pneumoniae establishing in Singapore. BMC Infect Dis 13:58. doi: 10.1186/1471-2334-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong JH, Clancy CJ, Cheng S, Shields RK, Chen L, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Characterization of porin expression in Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob Agents Chemother 57:2147–2153. doi: 10.1128/AAC.02411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Y, Chua NG, Lim TP, Teo JQ, Lee W, Kurup A, Koh TH, Tan TT, Kwa AL. 2016. From bench-top to bedside: a prospective in vitro antibiotic combination testing (iACT) service to guide the selection of rationally optimized antimicrobial combinations against extensively drug resistant (XDR) Gram negative bacteria (GNB). PLoS One 11:e0158740. doi: 10.1371/journal.pone.0158740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai B, Cai Y, Liew YX, Chua NG, Teo JQ, Lim TP, Kurup A, Ee PL, Tan TT, Lee W, Kwa AL. 2016. Clinical efficacy of polymyxin monotherapy versus nonvalidated polymyxin combination therapy versus validated polymyxin combination therapy in extensively drug-resistant Gram-negative Bacillus infections. Antimicrob Agents Chemother 60:4013–4022. doi: 10.1128/AAC.03064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pournaras S, Vrioni G, Neou E, Dendrinos J, Dimitroulia E, Poulou A, Tsakris A. 2011. Activity of tigecycline alone and in combination with colistin and meropenem against Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae strains by time-kill assay. Int J Antimicrob Agents 37:244–247. doi: 10.1016/j.ijantimicag.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. doi: 10.1016/S1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhang R, Cai JC, Zhou HW, Nasu M, Chen GX. 2011. Genotypic characterization and in vitro activities of tigecycline and polymyxin B for members of the Enterobacteriaceae with decreased susceptibility to carbapenems. J Med Microbiol 60:1813–1819. doi: 10.1099/jmm.0.025668-0. [DOI] [PubMed] [Google Scholar]
- 23.Betts JW, Phee LM, Hornsey M, Woodford N, Wareham DW. 2014. In vitro and in vivo activities of tigecycline-colistin combination therapies against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 58:3541–3546. doi: 10.1128/AAC.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinson HM, Gautam A, Olet S, Gibbs PS, Barigye R. 2010. Molecular analysis of porin gene transcription in heterogenotypic multidrug-resistant Escherichia coli isolates from scouring calves. J Antimicrob Chemother 65:1926–1935. doi: 10.1093/jac/dkq246. [DOI] [PubMed] [Google Scholar]
- 25.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 26.Kallman O, Motakefi A, Wretlind B, Kalin M, Olsson-Liljequist B, Giske CG. 2008. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J Antimicrob Chemother 62:986–990. doi: 10.1093/jac/dkn296. [DOI] [PubMed] [Google Scholar]
- 27.Davies TA, Marie Queenan A, Morrow BJ, Shang W, Amsler K, He W, Lynch AS, Pillar C, Flamm RK. 2011. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007-09. J Antimicrob Chemother 66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 28.Tod M, Lortholary O, Seytre D, Semaoun R, Uzzan B, Guillevin L, Casassus P, Petitjean O. 1998. Population pharmacokinetic study of amikacin administered once or twice daily to febrile, severely neutropenic adults. Antimicrob Agents Chemother 42:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaPlante KL, Sakoulas G. 2009. Evaluating aztreonam and ceftazidime pharmacodynamics with Escherichia coli in combination with daptomycin, linezolid, or vancomycin in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 53:4549–4555. doi: 10.1128/AAC.00180-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tam VH, McKinnon PS, Akins RL, Drusano GL, Rybak MJ. 2003. Pharmacokinetics and pharmacodynamics of cefepime in patients with various degrees of renal function. Antimicrob Agents Chemother 47:1853–1861. doi: 10.1128/AAC.47.6.1853-1861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaruratanasirikul S, Wongpoowarak W, Kositpantawong N, Aeinlang N, Jullangkoon M. 2012. Pharmacodynamics of doripenem in critically ill patients with ventilator-associated Gram-negative bacilli pneumonia. Int J Antimicrob Agents 40:434–439. doi: 10.1016/j.ijantimicag.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Sakka SG, Glauner AK, Bulitta JB, Kinzig-Schippers M, Pfister W, Drusano GL, Sorgel F. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob Agents Chemother 51:3304–3310. doi: 10.1128/AAC.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez Navarro A, Colino Gandarillas CI, Alvarez Lerma F, Menacho YA, Dominguez-Gil A. 2005. Pharmacokinetics and pharmacodynamics of levofloxacin in intensive care patients. Clin Pharmacokinet 44:627–635. doi: 10.2165/00003088-200544060-00004. [DOI] [PubMed] [Google Scholar]
- 34.Jaruratanasirikul S, Sriwiriyajan S, Punyo J. 2005. Comparison of the pharmacodynamics of meropenem in patients with ventilator-associated pneumonia following administration by 3-hour infusion or bolus injection. Antimicrob Agents Chemother 49:1337–1339. doi: 10.1128/AAC.49.4.1337-1339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shea KM, Cheatham SC, Wack MF, Smith DW, Sowinski KM, Kays MB. 2009. Steady-state pharmacokinetics and pharmacodynamics of piperacillin/tazobactam administered by prolonged infusion in hospitalised patients. Int J Antimicrob Agents 34:429–433. doi: 10.1016/j.ijantimicag.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Kwa AL, Lim TP, Low JG, Hou J, Kurup A, Prince RA, Tam VH. 2008. Pharmacokinetics of polymyxin B1 in patients with multidrug-resistant Gram-negative bacterial infections. Diagn Microbiol Infect Dis 60:163–167. doi: 10.1016/j.diagmicrobio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Gumbo T, Louie A, Deziel MR, Liu W, Parsons LM, Salfinger M, Drusano GL. 2007. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob Agents Chemother 51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaser J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J Antimicrob Chemother 15(Suppl A):125–130. doi: 10.1093/jac/15.suppl_A.125. [DOI] [PubMed] [Google Scholar]
- 39.Bilello JA, Bauer G, Dudley MN, Cole GA, Drusano GL. 1994. Effect of 2′,3′-didehydro-3′-deoxythymidine in an in vitro hollow-fiber pharmacodynamic model system correlates with results of dose-ranging clinical studies. Antimicrob Agents Chemother 38:1386–1391. doi: 10.1128/AAC.38.6.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Argenio DZ, Schumitzky A. 1997. Biomedical simulations resource ADAPT II user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical simulations resource, University of Southern California, Los Angeles, CA, USA. [Google Scholar]
- 41.Tam VH, Schilling AN, Nikolaou M. 2005. Modelling time-kill studies to discern the pharmacodynamics of meropenem. J Antimicrob Chemother 55:699–706. doi: 10.1093/jac/dki086. [DOI] [PubMed] [Google Scholar]