LETTER
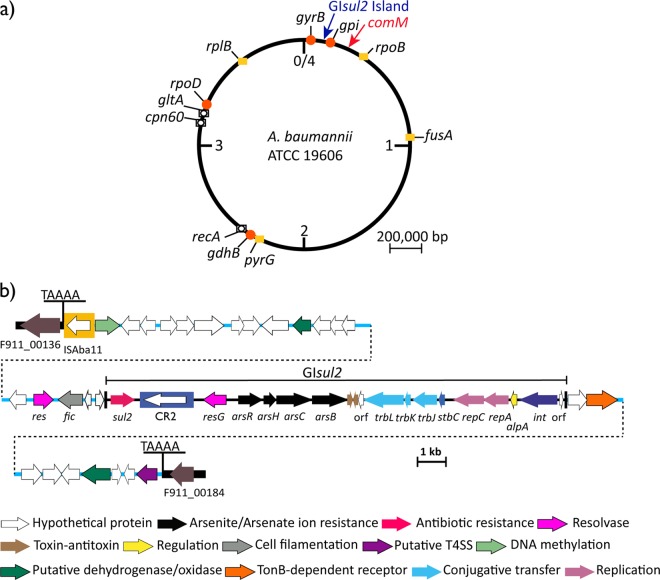
During the course of routine antibiotic resistance testing, we noticed that Acinetobacter baumannii ATCC 19606, which was isolated prior to 1948 (1) and is often used in genetic studies, is resistant to the sulfonamide compound sulfamethoxazole (MIC, >128 mg/liter). Three groups have sequenced the genome of ATCC 19606 (GenBank accession numbers JMRY01000000 [2], ACQB01000000, and APRG01000000). The draft genome in GenBank (accession number APRG01000000) was used to search for a gene (sul1, sul2, or sul3) accounting for the sulfonamide resistance phenotype as it includes longer contigs than the others. The sul2 sulfonamide resistance gene was found in a 210,382-bp contig (supercont1.1.C1; GenBank accession number APRG01000001). The context of the sul2 gene was analyzed, and it is located in GIsul2 (genomic island sul2) (Fig. 1b), a 15.5-kbp integrative element (3).
FIG 1.
Schematic representation of the GIsul2 island found in the genome of A. baumannii ATCC 19606. (a) Chromosomal location of the GIsul2 island relative to the multilocus sequence type (MLST) markers used in Oxford schemes (red dots) or in Institut Pasteur schemes (yellow boxes) or both types of schemes (open circles on boxes). (b) Structure of the GIsul2 island in the chromosome of A. baumannii ATCC 19606. Arrows indicate the extent and orientation of genes. The central blue line indicates the backbone of the genomic island, and the thick horizontal black line represents the chromosome, with locus identifiers indicated at the bottom of the panel. The extent of GIsul2 is also shown. Functions of the genes/orfs are color coded and indicated at the bottom of the panel. Scale bars are also shown. T4SS, type IV secretion system.
GIsul2 was reannotated and found to include several genes encoding putative arsenate and arsenite resistance proteins and a toxin-antitoxin, in addition to the resolvase and an integrase noted previously (3) (Fig. 1). However, ATCC 19606 was not significantly resistant to arsenite and arsenate ions. GIsul2 in ATCC 19606 lacks ISAba1, which is found upstream of sul2 in A. baumannii ATCC 17978 (3). In A. baumannii ATCC 17978, GIsul2 is now known to be located in a large conjugative plasmid, pAB3 (GenBank accession number CP012005). As the MIC for ATCC 17978 is also >128 mg/liter of sulfamethoxazole, the promoter in ISAba1 is not essential for high-level sulfonamide resistance.
Inspection of the sequences on either side of GIsul2 in ATCC 19606 revealed that it is located within a larger genomic island that has been inserted into the chromosome of ATCC 19606 (Fig. 1b). This genomic island, including GIsul2, is 36,157 bp long and is located between two open reading frames with locus identifiers F911_00136 and F911_00184 in the data located at GenBank accession number APRG01000001 (Fig. 1). These two reading frames are adjacent to one another in the genome of many other A. baumannii strains, e.g., ATCC 17978-mf and A1 (GenBank accession numbers CP012004 and CP010781, respectively). This genomic island is flanked by 5-bp target site duplication (Fig. 1b) indicative of transposition, and there is a copy of ISAba11 at one end (Fig. 1). Seven fragments (ranging in size from 155 to 3,467 bp) of the 13-kb and 6.5-kb regions flanking GIsul2 match parts of pXBB1-9 (GenBank accession number CP010351), which is a very large (398,857-bp) plasmid in Acinetobacter johnsonii strain XBB1, with DNA identities ranging from 96.4% to 98.7%. Hence, it appears that ISAba11, a 1,101-bp insertion sequence belonging to the IS701 family that creates 5-bp target site duplications (4), has mobilized a large DNA segment from a plasmid related to pXBB1-9 and inserted it into the chromosome of ATCC 19606. However, it is not possible to infer when GIsul2 became part of this segment. To date, several IS elements, e.g., ISEcp1, ISKpn23, and ISAba1, have also been shown to mobilize adjacent DNA segments (5–7).
GIsul2 and segments derived from it have been found in various resistance regions, and it has been suggested that GIsul2 is the main element responsible for the mobilization of the sul2 gene (3, 8–10). Here, we described the occurrence of GIsul2 in a new location leading to sulfonamide resistance in ATCC 19606.
ACKNOWLEDGMENT
This study and M.H. were supported by NHMRC Project Grant 1079616.
REFERENCES
- 1.Hugh R, Reese R. 1968. A comparison of 120 strains of Bacterium anitratum Schaub and Hauber with the type strain of this species. Int J Syst Evol Microbiol 18:1–3. [Google Scholar]
- 2.Davenport KW, Daligault HE, Minogue TD, Bruce DC, Chain PS, Coyne SR, Jaissle JG, Koroleva GI, Ladner JT, Li PE, Palacios GF, Scholz MB, Teshima H, Johnson SL. 2014. Draft genome assembly of Acinetobacter baumannii ATCC 19606. Genome Announc 2:e00832-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigro SJ, Hall RM. 2011. GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J Antimicrob Chemother 66:2175–2176. doi: 10.1093/jac/dkr230. [DOI] [PubMed] [Google Scholar]
- 4.Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother 66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 5.Partridge SR. 2016. Mobilization of blaBKC-1 by ISKpn23? Antimicrob Agents Chemother 60:5102–5104. doi: 10.1128/AAC.00785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob Agents Chemother 47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigro SJ, Hall RM. 2016. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother 71:1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 8.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Harmer CJ, Hall RM. 30 August 2016. PCR-based typing of IncC plasmids. Plasmid doi: 10.1016/j.plasmid.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Anantham S, Harmer CJ, Hall RM. 2015. p39R861-4, a type 2 A/C2 plasmid carrying a segment from the A/C1 plasmid RA1. Microb Drug Resist 21:571–576. doi: 10.1089/mdr.2015.0133. [DOI] [PubMed] [Google Scholar]