ABSTRACT
A novel erm(44) gene variant, erm(44)v, has been identified by whole-genome sequencing in a Staphylococcus saprophyticus isolate from the skin of a healthy person. It has the particularity to confer resistance to macrolides and lincosamides but not to streptogramin B when expressed in S. aureus. The erm(44)v gene resides on a 19,400-bp genomic island which contains phage-associated proteins and is integrated into the chromosome of S. saprophyticus.
KEYWORDS: MLSB, antibiotic resistance, phages, coagulase-negative staphylococci, 23S RNA methylase, Staphylococcus, macrolides-lincosamides-streptogramin B
TEXT
Staphylococcus saprophyticus is a bacterium which is widespread in the environment and in animals and may also occur on the skin of humans. It is known as a major cause of urinary tract infection and cystitis in humans (1). Although macrolides and lincosamides are not used for the treatment of urinary tract infections, they are among the antibiotics of choice for the treatment of other infectious diseases, such as pulmonary infection, and their use may contribute to the selection of resistance in bacteria of the normal human flora, including staphylococci (2). Resistance to macrolide, lincosamide, and streptogramin (MLSB) antibiotics in staphylococci has been associated with erythromycin ribosome methylase (erm) genes (Fig. 1) which methylate the 23S rRNA at position A2058, preventing binding of the MLSB antibiotics (3). The erm(44) gene, originally found in Staphylococcus xylosus from bovine mastitis milk (4), has also been recently identified in a S. saprophyticus isolate from river water (5) and has now been identified in S. saprophyticus from human skin.
FIG 1.
Relationship tree of erythromycin resistance methylases (Erm) detected in different Staphylococcus species. Amino acid (aa) identity and nucleotide (nt) identity were obtained by sequence alignment and clustering with BioNumerics 7.6 (Applied Maths). Comparison settings were as follows: standard algorithm for pairwise alignment; open gap penalty, 100%; unit gap penalty, 0%; and unweighted pair group method using average linkages (UPGMA). Methylase genes that were detected in Staphylococcus only by PCR and/or hybridization and whose sequences are not available [e.g., erm(F), erm(G), erm(Q)] were not included (http://faculty.washington.edu/marilynr/).
Three of 10 healthy human volunteers who did not receive MLSB antibiotics and who were participating to a large project aiming at determining the effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans (ANTIRESDEV project [www.ucl.ac.uk/antiresdev]; UK ethics approval number EC 10/H0806/12) were found to harbor Staphylococcus saprophyticus on the skin. The strains were isolated on sheep blood agar plates and identified using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) (microflex LT; Bruker Daltonic GmbH, Bremen, Germany). MICs of MLSB antibiotics erythromycin, clindamycin, virginiamycin S1, and pristinamycin 1A were determined by the microdilution method in Mueller-Hinton broth, and one strain (N041) showed resistance to erythromycin and clindamycin according to the EUCAST interpretation criteria (6). As this strain did not contain any known erm genes as determined using a microarray (7), whole-genome sequencing was performed at the UZH/ETH Functional Genomics Center (Zurich, Switzerland) by Life Technologies Ion Torrent semiconductor sequencing using a 400-bp library on a 314v2 chip. Comparisons of all contigs with currently annotated erm genes using BLAST identified an erm gene which showed the closest relatedness to erm(44) from S. xylosus JW4341 with 81% amino acid (aa) and 85% DNA identity and to erm(44) from S. saprophyticus A ER Ab-7 with 84% aa sequence identity and 83% DNA identity (Fig. 1). The newly detected erm gene encodes a 243-aa protein containing an rRNA adenine dimethylase signature (PS01131) as found in other erm methylases (8). It was not preceded by any intact leader peptides, neither by a complete IFVI motif nor by inverted repeat sequences, which are essential for induction and translational attenuation of erm genes (3, 9–11), likely explaining constitutive expression of this erm gene as determined by MIC analysis (Table 1). Putative −10 (TTTTAAAAT) and −35 (TTGCCT) promoter sequences were found 27 bp and 48 bp upstream of the start codon, respectively.
TABLE 1.
MIC of erythromycin, clindamycin, pristinamycin Ia, and virginiamycin S1 for different Staphylococcus strains, as determined by broth microdilution
| Strain | Characteristic(s) or origin | Reference or source | Antibiotic resistance gene(s)a | MIC (μg/ml)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLI | iCLI | PIA | iPIA | VS1 | iVS1 | ||||
| S. saprophyticus | ||||||||||
| N041 | Human nose skin sample | This study | erm(44)v | 128 | >256 | >256 | 32 | 32 | 32 | 32 |
| S. aureus | ||||||||||
| RN4220 | Recipient strain for electrotransformation, plasmid free | 18 | <0.5 | <0.25 | NA | 4 | NA | 8 | NA | |
| RN4220/pBUS1-HC | RN4220 with cloning vector pBUS1-HC | 12 | tet(L) | <0.5 | <0.25 | NA | 4 | NA | 8 | NA |
| RN4220/pBUS1-Pcap-HCc | RN4220 with pBUS1-HC containing cap promoter | 12 | tet(L) | <0.5 | <0.25 | NA | 2 | NA | 8 | NA |
| RN4220/pBJW13 | RN4220 with erm(44) from S. xylosus JW4341 cloned into pBUS1-Pcap | 4 | tet(L), erm(44) | >256 | >256 | >256 | 8 | 8 | 16 | 32 |
| RN4220/pLI50-erm(44) | RN4220 with erm(44) from S. saprophyticus A ER Ab-7 cloned into pLI50 | 5 | blaTEM-1, catpC194, erm(44) | >256 | <0.25 | 256 | 4 | 64 | 32 | 128 |
| RN4220/pBCS0714 | RN4220 with erm(44)v from S. saprophyticus N041 and its regulatory region cloned into pBUS1-HC | This study | tet(L), erm(44)v | 16 | >256 | >256 | 2 | 1 | 8 | 4 |
| RN4220/pBCS0814 | RN4220 with erm(44)v from S. saprophyticus N041 cloned into pBUS1-Pcap-HC | This study | tet(L), erm(44)v | 16 | >256 | >256 | 2 | 1 | 8 | 4 |
Antibiotic resistance genes and functions: blaTEM-1, β-lactamase; catpC194, chloramphenicol acetyltransferase; tet(L), tetracycline efflux; erm(44) and erm(44)v, 23S rRNA methylase.
Abbreviations: ERY, erythromycin; CLI, clindamycin; PIA, pristinamycin IA; VS1, virginiamycin S1; iCLI, iPIA, and iVS1, 2 μg/ml erythromycin added to the broth for the detection of inducible resistance to clindamycin (iCLI), pristinamycin IA (iPIA), and virginiamycin S1 (iVS1); NA, not applicable.
Vector pBUS1-Pcap-HC is a pBUS1-HC derivate that harbors the cap promoter of the S. aureus type 1 capsular polysaccharide biosynthesis gene cluster.
The functionality of the erm gene of strain N041 was assessed after cloning into the shuttle vector pBUS1-HC was performed (12), generating plasmid pBSC0714, where the gene was expressed with its own promoter. The presence of pBCS0714 in S. aureus RN4220 led to an increase of the MIC of erythromycin to 16 μg/ml and of clindamycin to ≥256 μg/ml, while the MICs for the streptogramins pristinamycin Ia and virginiamycin S1 did not increase compared to those seen with the S. aureus RN4220 recipient strain alone and a RN4220 strain harboring pBUS1-HC or pBUS1-Pcap-HC. To verify this uncommon phenotype, the erm gene was placed under the control of a strong cap promoter in plasmid pBSC0814, confirming both the erythromycin and clindamycin phenotype and the absence of increased MICs for streptogramin B pristinamycin and virginiamycin in RN4220 (Table 1), in contrast to the results seen with the closely related erm(44) from S. xylosus JW4341 and that from S. saprophyticus A ER Ab-7 (4, 5). Due to the sequence identity being above the 80% threshold for a new erm determinant and to an altered phenotype compared to that seen with the original erm(44) from S. xylosus when expressed in S. aureus, the erm gene identified in S. saprophyticus N041 was assigned the name erm(44)v according to the nomenclature of the MLSB resistance genes (http://faculty.washington.edu/marilynr/) (13). However, the possibility cannot be excluded that erm(44)v might confer resistance to streptogramin B in S. saprophyticus due to the presence of a specific inducer which is absent in S. aureus RN4220.
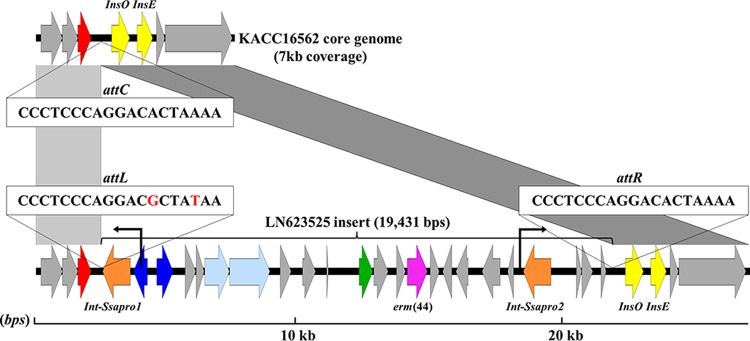
The erm(44)v gene was located on a putative 19,400-bp genomic island (GenBank accession no. LN623525) which is absent in the MLSB-susceptible strain S. saprophyticus KACC16562 (GenBank accession no. AHKB01; Fig. 2). In contrast to erm(44) from S. xylosus JW4341, which is situated on a prophage ΦJW4341-pro (4), the genomic composition of the island described here shows a rather heterogeneous composition of open reading frames (ORFs) remotely resembling that of a temperate siphoviral bacteriophage, SaPImw2, with the common presence of one terminase, two primases, two transcriptional regulators, and an integrase belonging to the tyrosine type of bacterial phage integrases (Int-Ssapro1; NCBI conserved domain number cd01189) (Fig. 2) (14). The genomic island contains an additional integrase of the same type (Int-Ssapro2; NCBI conserved domain number cd01189) at its distal end which potentially played a role in the integration and recombination of the genomic island into the S. saprophyticus genome. However, no conjugal transfer of macrolide resistance into S. aureus 80Cr5 (rifampin resistant [Rifr]) (15) and S. saprophyticus 7108R (a rifampin-resistant mutant of 7108) (16) was observed by filter mating (17) using different donor-recipient ratios (106:108, 108:108, and 108:106 cells/ml) and 10 μg/ml erythromycin and 100 μg/ml rifampin in the brain heart infusion (BHI) agar selective plates. No circular form could be observed by PCR using GoTaq polymerase (Promega) and plasmid DNA (NucleoBond PC 100; Macherey-Nagel) as the template and using primer1 (5′-CCCGTTGTTACGGGGTTTCT) and primer2 (5′-GCGATAAAGAGCATTTTGATTTTCC) (annealing temperature, 55°C; extension time, 2 min), reading outward of the genomic island (Fig. 2).
FIG 2.
Insertion site of genomic island in S. saprophyticus N041 (GenBank accession no. LN623525) and core genome of S. saprophyticus KACC16562 (GenBank accession no. NZ_AHKB00000000.1). Gray areas represent high similarity at the nucleotide level (>98%). Arrows represent positions and orientations of open reading frames (ORFs). New ML resistance gene erm(44)v is shown in pink. The 19-bp putative insertion site attC and the duplicated sites attL and attR in the N041 genome are shown. Two transposases of the core genome (InsO_Ssapro and InsE_Ssapro [abbreviated as InsO and InsE]) are indicated in yellow, the metal-dependent phosphodiesterase in red, and the two flanking integrases of the genomic island (Int-Ssapro1 and Int-Ssapro2) in orange. Additional genes are colored according to sequence and function: transcription regulators are dark blue; replication genes (including the primase gene) are light blue; the terminase gene is green; genes encoding hypothetical proteins are gray. Primers for the circular form test are indicated with a black arrow.
Analysis of Staphylococcus whole-genome sequences using a MaGe microscope platform (https://www.genoscope.cns.fr/agc/microscope/home/) revealed that the genetic island containing erm(44)v inserted into a chromosomal hot spot, as most strains annotated in MaGe show large sequence variation at this specific locus. The genomic island integrated at a specific 19-bp integration site (attC [CCCTCCCAGGACACTAAAA]) situated between a metal-dependent phosphodiesterase and two tandem transposases (InsO_Ssapro and InsE_Ssapro; NCBI conserved protein family numbers COG2801 and COG2963) (Fig. 2). Attachment site attC was duplicated in the N041 strain with one perfect copy downstream (attR) and one imperfect copy upstream (attL) of the genomic island (Fig. 2).
This report describes an erm(44) gene variant, erm(44)v, in a human isolate of S. saprophyticus which does not confer decreased susceptibility to streptogramin B in S. aureus, in contrast to the erm(44) gene from S. xylosus from milk and from S. saprophyticus from river water. However, besides this uncommon phenotype, the erm(44)v was found, like erm(44) from S. xylosus (4), on an element containing genes associated with phages, indicating that phage-associated elements may play a role in the spread of MLSB resistance.
Accession number(s).
The erm(44)v-containing element and its insertion region in S. saprophyticus N041 have been deposited in the DDBJ/ENA/GenBank database under accession number LN623525.
ACKNOWLEDGMENTS
This study was carried out with financial support from the Commission of the European Communities, specifically, the Infectious Diseases research domain of the Health theme of the 7th Framework Programme, contract 241446, “The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiotas at various body sites,” and research grant 35-539 from the Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland.
We thank Sandra Neumann and Sören Gatermann (Ruhr-Universität Bochum) for providing S. saprophyticus 7108, Stefan Schwarz and Andrea T. Feßler (Friedrich-Loeffler-Institut, Neustadt-Mariensee, Germany) for providing vector pLI50 containing erm(44) from S. saprophyticus A ER Ab-7, and Alexandra Collaud (Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland) for technical assistance.
REFERENCES
- 1.Raz R, Colodner R, Kunin CM. 2005. Who are you–Staphylococcus saprophyticus? Clin Infect Dis 40:1–5. doi: 10.1086/428353. [DOI] [PubMed] [Google Scholar]
- 2.Le Bouter A, Leclercq R, Cattoir V. 2011. Molecular basis of resistance to macrolides, lincosamides and streptogramins in Staphylococcus saprophyticus clinical isolates. Int J Antimicrob Agents 37:118–123. doi: 10.1016/j.ijantimicag.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Wipf JR, Perreten V. 2016. Discovery of novel MLSB resistance methylase genes and their associated genetic elements in staphylococci. Curr Clin Microbiol Rep 3:42–52. doi: 10.1007/s40588-016-0030-x. [DOI] [Google Scholar]
- 4.Wipf JRK, Schwendener S, Perreten V. 2014. The novel MLSB resistance gene erm(44) is associated with a prophage in Staphylococcus xylosus. Antimicrob Agents Chemother 58:6133–6138. doi: 10.1128/AAC.02949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendlandt S, Hess S, Li J, Fessler AT, Wang Y, Kadlec K, Gallert C, Schwarz S. 2015. Detection of the macrolide-lincosamide-streptogramin B resistance gene erm(44) and a novel erm(44) variant in staphylococci from aquatic environments. FEMS Microbiol Ecol 91:fiv090. doi: 10.1093/femsec/fiv090. [DOI] [PubMed] [Google Scholar]
- 6.EUCAST. 2016. Breakpoint tables for interpretation of MICs and zone diameters, version 6.0, 2016. The European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org. [Google Scholar]
- 7.Strauss C, Endimiani A, Perreten V. 2015. A novel universal DNA labeling and amplification system for rapid microarray-based detection of 117 antibiotic resistance genes in Gram-positive bacteria. J Microbiol Methods 108:25–30. doi: 10.1016/j.mimet.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Schwendener S, Perreten V. 2012. New MLSB resistance gene erm(43) in Staphylococcus lentus. Antimicrob Agents Chemother 56:4746–4752. doi: 10.1128/AAC.00627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisblum B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother 39:797–805. doi: 10.1128/AAC.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisblum B. 1998. Macrolide resistance. Drug Resist Updat 1:29–41. doi: 10.1016/S1368-7646(98)80212-4. [DOI] [PubMed] [Google Scholar]
- 11.Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. 2007. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev 20:79–114. doi: 10.1128/CMR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwendener S, Perreten V. 2015. New shuttle vector-based expression system to generate polyhistidine-tagged fusion proteins in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol 81:3243–3254. doi: 10.1128/AEM.03803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppälä H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novick RP, Christie GE, Penadés JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel HW, Soedirman N, Rost JA, van Leeuwen WJ, van Embden JD. 1980. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J Bacteriol 142:407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatermann S, Marre R, Heesemann J, Henkel W. 1988. Hemagglutinating and adherence properties of Staphylococcus saprophyticus: epidemiology and virulence in experimental urinary tract infection of rats. FEMS Microbiol Immunol 1:179–185. [DOI] [PubMed] [Google Scholar]
- 17.Perreten V, Giampà N, Schuler-Schmid U, Teuber M. 1998. Antibiotic resistance genes in coagulase-negative staphylococci isolated from food. Syst Appl Microbiol 21:113–120. doi: 10.1016/S0723-2020(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 18.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]