Abstract
Background
Methicillin-resistant Coagulase-negative Staphylococci (MR-CoNS) have emerged as an important cause of nosocomial infections especially in patients with prosthetic devices and implants. This study was conducted with an aim to determine the prevalence of methicillin resistance among CoNS isolates at a tertiary care center by both phenotypic and genotypic methods.
Methods
This cross sectional study was carried out from September 2011 to February 2014 in which 150 non-repetitive clinical isolates of CoNS were identified at the species level by conventional phenotypic methods. Cefoxitin disk (30 μg) diffusion testing was used to determine methicillin resistance and confirmed by detection of mecA gene by polymerase chain reaction (PCR).
Results
Out of 150 CoNS isolates, 51 were methicillin resistant by cefoxitin disk diffusion method. Out of these 51 isolates, mecA gene was detected only in 45 isolates. Moreover, mecA gene was also detected in 4 isolates, which were cefoxitin sensitive. Thus, the prevalence of methicillin resistance among CoNS was found to be 32.7% by PCR.
Conclusion
The prevalence of methicillin resistance among Coagulase-negative Staphylococci (CoNS) was 32.7% by PCR detection of mecA gene. The sensitivity and specificity of cefoxitin disk diffusion method against mecA gene detection by PCR were found to be more than 90%. It can be concluded from this study that cefoxitin disk diffusion test can be used as a useful screening method to detect methicillin resistance among CoNS isolates. However, detection of mecA gene by PCR remains a more accurate method of detecting methicillin resistance among CoNS.
Keywords: Methicillin-resistant Coagulase-negative Staphylococci (MR-CoNS), Cefoxitin disk, mecA gene, Polymerase chain reaction (PCR)
Introduction
Coagulase-negative staphylococci (CoNS) are one of the most commonly isolated organisms in the clinical microbiology laboratory. They were earlier considered to be clinically insignificant contaminants, when isolated from clinical specimens due to their ubiquitous nature.1 The frequent isolation of CoNS from intravenous catheters, blood and other normally sterile body fluids presents a persistent interpretive challenge to clinical microbiologists. However, their role as significant pathogens following surgery and in patients with prosthetic devices has now been well established. The possible explanation for their current clinical importance and increased prevalence includes being commensals on the skin, selection pressure due to widespread use of broad-spectrum antibiotics in the hospital and their ability of adherence and formation of biofilms on the surfaces of vascular catheters and other medical devices.1
Emergence and increasing prevalence of methicillin-resistant Coagulase-negative Staphylococci (MR-CoNS) are a cause for concern. National Nosocomial Infection Survey data of USA revealed that from 1980 to 1989, the proportion of nosocomial CoNS resistant to methicillin, oxacillin, or nafcillin increased from 20% to 60%. Most of these MR-CoNS were also resistant to multiple antimicrobial agents.2
Staphylococcal chromosome cassette (SCC) mec is a resistance island present in the genome of methicillin-resistant isolates, where mec is the genetic element, that is responsible for resistance to methicillin.2 mecA gene encodes for a particular penicillin-binding protein (PBP) called PBP2A, which has a low affinity for methicillin and most of the other β-lactam drugs and is thus responsible for the intrinsic resistance of these isolates to almost all β-lactams. Such multidrug-resistant strains are becoming a significant threat to public health.2
This study was conducted with an aim to determine the prevalence of methicillin resistance among CoNS isolates by both phenotypic and genotypic methods.
Material and methods
This cross sectional study was carried out from September 2011 to February 2014. A total of 150 non-repetitive clinical isolates of Coagulase-negative Staphylococci were isolated from various clinical specimens received in microbiology laboratory of a tertiary care centre. Isolates from blood samples and intravascular catheter tips were interpreted as being pathogenic, and not merely contaminants, by collecting paired blood cultures and simultaneous collection of blood sample from peripheral vein respectively. Same CoNS species isolated from urine more than once was interpreted as being pathogenic and not merely contaminant.
Identification of CoNS at species level
All CoNS isolates were identified at the species level by conventional phenotypic methods such as colony morphology, Gram's stain, catalase test, slide and tube coagulase test, susceptibility to novobiocin and polymyxin B, PYR test, acetoin production, ornithine decarboxylase test and fermentation of glucose, maltose, sucrose, mannitol, trehalose and mannose.
Identification of MR-CoNS by phenotypic method
Cefoxitin disc (30 μg) is used as a surrogate marker for prediction of mecA gene mediated resistance to oxacillin and is the preferred method of testing of methicillin resistant CoNS. All the isolates were subjected to Cefoxitin disc diffusion testing using a 30 μg cefoxitin disc. A 0.5 Mc Farland standard suspension of the isolate was made and lawn culture done on Muller–Hinton agar (MHA) plate. Plates were incubated at 37 °C for 24 h and zone diameters were measured. An inhibition zone diameter of ≤24 mm was reported as Methicillin resistant and ≥25 mm was reported as Methicillin sensitive.
Detection of mecA gene by polymerase chain reaction
All the CoNS isolates were subjected to polymerase chain reaction (PCR) for detection of mecA gene, which encodes the low-affinity penicillin-binding protein PBP 2A, the main factor responsible for the methicillin resistance.
DNA extraction
DNA extraction was done from all isolates of CoNS using the QIAamp DNA mini kits from QIAGEN, Germany. Manufacturer's instructions were followed for extracting DNA from the fresh cultures.
Primers
Primers were procured from Sigma–Aldrich Chemicals Pvt. Ltd. and are shown in Table 1.
Table 1.
Sequences of forward and reverse primers used for detection of mecA gene in MR-CoNS isolates.
| Gene | Primer sequence | No of bases | nmol | Amplicon size |
|---|---|---|---|---|
| mec A | (F): 5′-GTAGAAATGACTGAACGTCCGATAA-3′ | 25 | 38.9 | 310 bp |
| (R): 5′-CCAATTCCACATTGTTTCGGTCTAA-3′ | 25 | 28.9 |
PCR reaction mixtures
PCR reaction mixtures were prepared under laminar flow under strict precautions to prevent cross contamination. Amplification was carried out with the following thermal cycling profile: initial denaturation for 4 min at 94 °C, 35 cycles of amplification consisting of 1 min at 94 °C, 1 min at 50 °C, and 1 min at 72 °C, with 7 min at 72 °C for the final extension.
Gel electrophoresis
DNA fragments were analyzed by electrophoresis in 0.5× Tris-borate-EDTA on a 1% agarose gel stained with ethidium bromide. The gel was viewed under UV transilluminator and was documented with the help of digital camera attached with the transilluminator and to the computer. Specific bands of 310 bp for mecA gene were considered as positive PCR reaction, with no band in negative control.
Statistical analysis
Comparison of cefoxitin disc diffusion testing for methicillin resistance with detection of mecA gene by PCR was done for parameters like sensitivity, specificity, positive predictive value and negative predictive value.
Antibiotic susceptibility testing
All 150 isolates of CoNS were tested for their susceptibility to various antibiotics by Kirby Bauer disc diffusion method. Since Kirby Bauer disc diffusion method is not recommended for susceptibility testing of CoNS to vancomycin, determination of minimum inhibitory concentration (MIC) has to be done either by E-test or agar dilution method. In this study, we used vancomycin E-strips (Bio-Merieux) to determine the MIC of vancomycin. MIC ≤4 μg/ml was interpreted as sensitive according to CLSI guidelines.
Results
The most common clinical sample from which CoNS were isolated was blood (52/150) followed by intravascular catheter tip (43/150), urine (41/150) and pus (14/150). The isolation of CoNS from blood samples was interpreted with the help of paired blood samples from two peripheral veins. Isolation from intravascular catheter tips was also correlated by simultaneously collecting blood sample from a peripheral vein and isolating the same organism from both the samples. Same CoNS species isolated from urine sample more than once was interpreted as being pathogenic and not merely contaminant.
The age of the patients from whom CoNS were obtained ranged from 3 months to 77 years of age. It was noticed that among the 150 isolates, 99 were from male patients and 51 were from female patients. Male to female ratio was approximately 2:1. Maximum numbers of isolates were from age group 21 to 30 years comprising 42.7% of the total followed by age group 31–40 years (26.7%).
The samples from which CoNS were isolated were collected from various wards including ICU, acute wards, other wards, and OPD. Out of 150 isolates, 73 were from ICU (48.7%), 18 were from acute wards (12%), 9 were from other wards (6%), and 50 isolates were from OPD (33.3%) samples.
Out of 150 isolates of CoNS, the most common species isolated was Staphylococcus epidermidis (90/150) followed by Staphylococcus saprophyticus (41/150) and Staphylococcus hemolyticus (19/150).
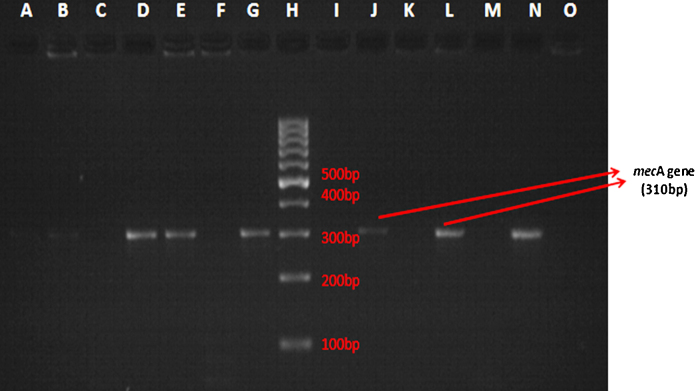
A total of 51 isolates of CoNS were found to be resistant to cefoxitin by Kirby–Bauer disk diffusion method. All the 150 isolates were subjected to PCR for detection of mecA gene. Out of these isolates, mecA gene (bp size 310) was found to be present only in 49 isolates (Fig. 1). Therefore, prevalence of mecA gene mediated methicillin resistance among CoNS isolates was found to be 32.7%. PCR detected mecA gene only in 45 isolates out of 51, which were cefoxitin resistant. Interestingly, there were 4 isolates which were classified as methicillin susceptible by cefoxitin disk diffusion test, in which mecA gene was detected by PCR. The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of cefoxitin disk diffusion testing for methicillin resistance was found to be 91.8%, 94%, 88.2% and 96% respectively.
Fig. 1.
Gel electrophoresis showing presence of mecA gene (band at 310 bp). H – 100 bp molecular marker. I – negative control for mecA gene (Staphylococcus aureus ATCC 25923). G – positive control for mecA gene (Staphylococcus aureus ATCC 43300). D, E, J, L, N – band at 310 bp denoting mecA gene in clinical isolates of CoNS.
Out of 90 isolates of S. epidermidis, 36 isolates (40%) were found to methicillin resistant by PCR, whereas 8 out of 41 isolates (19.5%) of S. saprophyticus and 5 out of 19 isolates (26.3%) of S. hemolyticus were found methicillin resistant by PCR.
Antimicrobial susceptibility testing of all the MR-CoNS isolates was done by Kirby–Bauer disk diffusion method. Susceptibility to vancomycin was determined by E-test method. All the isolates were found to be resistant to penicillin. In contrast, all the isolates were found to be sensitive to vancomycin, teicoplanin and linezolid. The resistance to gentamicin, amikacin, erythromycin, ciprofloxacin and levofloxacin of MR-CoNS isolates was found to be 42%, 36%, 60%, 59%, and 52%, respectively, as against 36%, 30%, 57%, 52% and 50% for methicillin-sensitive CoNS isolates respectively.
Discussion
Coagulase negative staphylococci (CoNS) were previously overlooked as contaminants. However, in the last few decades, they have emerged as important potential pathogens due to the increasing use of implants and increasing number of severely debilitated patients in hospitals.3
Historically, resistance to the antistaphylococcal, penicillinase-stable penicillins has been referred to as “methicillin resistance,” and the acronym MR-CoNS (methicillin-resistant Coagulase-negative Staphylococci) is still commonly used, even though methicillin is currently not the agent of choice for testing or treatment. SCC mec is a resistance island present in the genome of MR-CoNS isolates, where mec is the genetic element that is responsible for resistance to methicillin.2 mecA gene encodes for a particular PBP called PBP2A which has a low affinity for methicillin and most of the other β-lactam drugs and is consequently responsible for the intrinsic resistance to most of the β-lactams.5 Since its initial detection in the 1960s, the incidence of infections caused by MR-CoNS is on the rise.4
Kirby–Bauer disk diffusion test, agar or broth dilution and agar screen methods are the only standardized means of identifying methicillin resistance in the clinical microbiology laboratory. These tests have many shortcomings, because there are numerous factors, which can alter the interpretation of the results. These factors include inoculum size, incubation time and temperature, pH of the medium, salt concentration of the medium and exposure to β-lactam antibiotics. Speedy and precise identification of methicillin resistance among CoNS isolates has vital implications for the treatment of infected patients.4 Errors in detection of methicillin resistance can lead to increased health care costs, treatment failures and unfavorable clinical consequences (Table 1).
In this study, we compared cefoxitin disk diffusion test for detecting methicillin resistance against detection of mecA gene by polymerase chain reaction (PCR).
Out of 150 isolates of CoNS, 51 were found to be methicillin resistant by cefoxitin disk diffusion method. Out of these 51 isolates, mecA gene was detected only in 45 isolates. This may be because of resistance to methicillin by mechanisms other than mecA gene and indicates that mecA gene detection may not be accurate enough to guide therapy for CoNS infections. It has been reported earlier that mecA gene assay performed less optimally with the CoNS as compared to Staphylococcus aureus.5
Moreover, PCR detected mecA gene in 4 isolates which were found to be methicillin sensitive by cefoxitin disk diffusion method. All these isolates, which were labeled as methicillin sensitive by the phenotypic method, should be regarded as potentially methicillin-resistant isolates bearing the mecA gene6 and should not be classified as methicillin susceptible in spite of their susceptibility to beta lactam antibiotics. The sensitivity, specificity, PPV, and NPV of cefoxitin disk diffusion test against PCR for detection of MR-CoNS was 91.8%, 94%, 88.2%, and 96% respectively.
In case of methicillin-sensitive CoNS (MS-CoNS) isolates, PCR results showing lesser consistency with those from Cefoxitin susceptibility tests have been reported earlier.7 This may be due to the fact that heterogeneous expression of resistance varies more for MS-CoNS as compared to methicillin-sensitive Staphylococcus aureus (MSSA) and the subpopulation of resistant cells is smaller for MS-CoNS than for MSSA.2 These reports confirm that expression of methicillin resistance is more variable for MS-CoNS and the detection of the mecA gene is important in interpreting methicillin susceptible staphylococci.
In the present study, the prevalence of methicillin resistance among CoNS was found to be 32.7%. The prevalence varies among different countries of the world.8 The prevalence of MR-CoNS in our study is slightly lower than other studies done by James et al.9 from Mexico and Singhal et al.10 from India, who reported a prevalence of 41.6%, 53.4% and 62.7% respectively.
Antimicrobial resistance pattern of CoNS was done by Kirby–Bauer disk diffusion method and the results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. As, vancomycin disk is not recommended for susceptibility testing for CoNS isolates, determination of vancomycin MIC by E-test method was carried out in this study.
All the 49 isolates of MR-CoNS were found to be resistant to penicillin. The resistance to gentamicin, amikacin, erythromycin, ciprofloxacin, and levofloxacin of MR-CoNS isolates was found to be 42%, 36%, 60%, 59%, and 52% respectively as against 36%, 30%, 57%, 52% and 50% for methicillin-sensitive CoNS isolates respectively. This finding is of particular concern, since high prevalence of such drug resistant MR-CoNS isolates leaves the clinician with very few alternative treatment options. MR-CoNS isolates with reduced susceptibility to glycopeptides have been reported. However, fortunately, in the present study, all the CoNS isolates were found to be sensitive to vancomycin, teicoplanin, and linezolid.
Conclusion
The prevalence of methicillin resistance among CoNS was 32.7% by PCR detection of mecA gene. The sensitivity and specificity of cefoxitin disk diffusion method against mecA gene detection by PCR were found to be 91.8% and 94% respectively. It can be concluded from this study that cefoxitin disk diffusion test can be used as a useful screening method to detect methicillin resistance among CoNS isolates. However, detection of mecA gene by PCR remains a faster and more accurate method of detecting methicillin resistance among CoNS.
Conflicts of interest
The authors have none to declare.
References
- 1.Pfaller M.A., Herwaldt L.A. Laboratory, clinical and epidemiological aspects of coagulase-negative Staphylococci. Clin Microbiol Rev. 1988;1(3):281–299. doi: 10.1128/cmr.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katayama Y., Ito T., Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohan U., Jindal N., Aggarwal P. Species distribution and antibiotic sensitivity pattern of coagulase negative Staphylococci isolated from various clinical specimens. Indian J Med Microbiol. 2002;20(1):45–46. [PubMed] [Google Scholar]
- 4.Gold H.S., Moellering R.C., Jr. Antimicrobial drug resistance. N Engl J Med. 1996;335(19):1445–1453. doi: 10.1056/NEJM199611073351907. [DOI] [PubMed] [Google Scholar]
- 5.Shreshtha N.K., Tuohy M.J., Hall G.S., Isada C.M., Procop G.W. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J Clin Microbiol. 2002;40(7):2659–2661. doi: 10.1128/JCM.40.7.2659-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ralllapalli S., Verghese S., Verma R.S. Validation of multiplex PCR strategy for simultaneous detection and identification of methicillin-resistant Staphylococcus aureus. Indian J Med Microbiol. 2008;26(4):361–364. doi: 10.4103/0255-0857.43580. [DOI] [PubMed] [Google Scholar]
- 7.Lan M.O., Qi-nan W. Rapid detection of methicillin-resistant Staphylococci using polymerase chain reaction. Int J Infect Dis. 1997;2:15–20. [Google Scholar]
- 8.Schmitz F.J., Fluit A.C., Gondolf M. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of Staphylococci from 19 European hospitals. J Antimicrob Chemother. 1999;43:253–259. [PubMed] [Google Scholar]
- 9.James P.J., Butcher I.A., Gardner E.R., Hamblin D.L. Methicillin resistant Staphylococcus epidermidis in infection of HIP arthroplasties. J Bone Joint Surg (Br) 1994;76:725–727. [PubMed] [Google Scholar]
- 10.Singhal R., Mohanty S.D., Seema S.B., Das B., Kapil A. Species distribution and antimicrobial susceptibility of coagulase negative Staphylococci in a tertiary care hospital. Indian J Med Res. 2006;123:569–570. [PubMed] [Google Scholar]