Abstract
Insulin resistance represents one of the mechanisms underlying the link between type 2 diabetes (T2D) and Alzheimer's disease (AD), and we explored its in vivo neurobiology related to cognition based on a pathway-based genetic association analyses. Eighty-seven mild cognitive impairment (MCIs) subjects and 135 matched controls (HCs) were employed at baseline, and they underwent functional MRI scans, clinical evaluations and exon sequencings of 20 genes related to brain insulin resistance. A longitudinal study for an average of 35 months was performed to assess their cognitive decline over time. By using cognition as the phenotype, we detected genes that modified cognitive impairments, including AKT2, PIK3CB, IGF1R, PIK3CD, MTOR, IDE, AKT1S1 and AKT1. Based on these loci, the mass univariate modeling was utilized to construct the functional network. The MCIs showed disconnections mainly in the cerebellum-frontal-temporal regions, while compensations may occur in frontal-parietal regions to maintain the overall network efficiency. Moreover, the behavioral significance of the network was highlighted, as topological characteristics of the medial temporal lobe and the prefrontal cortex partially determine longitudinal cognitive decline. Our results suggested that the restoration of insulin activity represents a promising therapeutic target for alleviating cognitive decline associated with T2D and AD.
Keywords: Alzheimer's disease, Insulin resistance, Cognition, Genetic polymorphism, Neuroimaging
Highlights
-
•
Genetic variations of brain insulin resistance influence cognition.
-
•
Insulin pathway modifies the topological characteristics of brain networks.
-
•
Brain overall efficiency predicts the cognitive changes over time.
-
•
Relieving brain insulin resistance may alleviate cognitive declines.
1. Introduction
The interactions between type 2 diabetes (T2D) and Alzheimer's disease (AD) have received increasing attention. Epidemiologic studies have shown that AD risk is increased by T2D (Profenno et al., 2010), and an exploration of the mechanisms regarding how T2D increases the risk of AD will help to reveal the pathomechanisms of AD and find ways to relieve cognitive impairments in both diseases. A reasonable starting point concerning this field is the pathophysiological features shared by the two diseases, and insulin resistance represents a promising target. Peripheral insulin resistance represents a key causative factor for T2D, and its role in AD has also been highlighted. Subjects with peripheral insulin resistance are more likely to develop AD, and associations have been detected with peripheral insulin resistance and AD-related pathology in brains (Rasgon et al., 2011, Sims-Robinson et al., 2010).
Moreover, it is logical to assume that the dysfunctional insulin signaling within the brain has more a direct and important role in AD processes. Accumulating evidence has indicated that the brain itself develops insulin resistance, including the changed binding and sensitivity of the insulin pathway-related receptors and the expression levels of relevant molecules (Moloney et al., 2010, Talbot and Wang, 2014). Recently, disrupted insulin signaling associated with AD was further clarified, as reduced responses of the insulin receptor (IR)/insulin receptor substrate 1 (IRS1)/PI3K/AKT and IGF-1 receptor (IGF-1R)/IRS2/PI3K pathways were demonstrated in AD brains (Talbot et al., 2012). More importantly, the dysfunction took place independently of diabetic status and APOE ε4 genotype, and it gradually deteriorated as AD progressed (Talbot et al., 2012). At the cellular and molecular level, insulin signaling interferes with Aβ degradation and transportation of Aβ out of the brain to modify Aβ deposition (Carro et al., 2002, Farris et al., 2003). Further, insulin deficiency promotes the phosphorylation of tau, leading to deteriorated accumulations of neurofibrillary tangles (Schubert et al., 2003). In addition to Aβ and tau, insulin signaling disorders also promote neuro-inflammation, apoptosis, oxidative stress, impairments of energy metabolism and synaptic disconnections (Sims-Robinson et al., 2010), all of which lead to the development of AD. To conclude, brain insulin signaling plays pivotal roles in AD processes, and further studies are needed to clarify the in vivo neurobiology of brain insulin resistance underlying cognitive impairment.
Genetic association analyses allow us to solve this issue. Compared with traditional case/control designs, quantitative trait association studies massively increase the statistical power to decrease the required sample sizes (Potkin et al., 2009). Previously, cognitive performances have been successfully used as quantitative phenotypes, leading to the identification of loci that influence brain function and cognition (Almasy et al., 2008, Papassotiropoulos et al., 2006). Moreover, converging evidences have suggested that the pathophysiological processes of AD begin many years before the diagnosis of dementia, and the “preclinical” phase of AD provides critical opportunities to reveal the mechanisms underlying AD development (Sperling et al., 2011). Mild cognitive impairment (MCI) has been suggested to be a boundary area between normal aging and dementia, and MCI subjects (MCIs) are high-risk individuals for AD (Sperling et al., 2011). Therefore, exploring the interactions with insulin resistance-related genetic polymorphisms and cognitive impairments in the MCIs would support the implications of insulin resistance in AD process, identify the missing heritage for AD and lead to novel insights into relieving cognitive declines in both T2D and AD.
Moreover, genetic imaging studies allow us to assess the in vivo functional activity of targeted variations. For instance, the APOE genotype was suggested to influence the aging trajectory of the default mode network (DMN) (Shu et al., 2016), and single nucleotide polymorphisms (SNPs) of GSK3β were associated with altered neuronal activity mainly within the precuneus (Pcu) and the inferior parietal lobe (IPL) (Biffi et al., 2010), whereas the influences of insulin resistance on brain activity are undefined. More importantly, owing to the multifactorial nature of insulin resistance, the joint effects of multi-SNP should be explored to better reveal insulin-brain interactions, which could be achieved by mass univariate modeling (Bai et al., 2016a, Bai et al., 2016b, Inkster et al., 2010).
In the present study, our first aim was to investigate the associations of cognitive performances and SNPs corresponding to the brain insulin resistance pathway. Second, insulin pathway-related brain topological networks were constructed to access the integrated functional activities of SNPs with cognitive relevance. Third, to clarify the behavior significances of the established network, we explored whether the network connectivity could predict cognition changes over time.
2. Material and methods
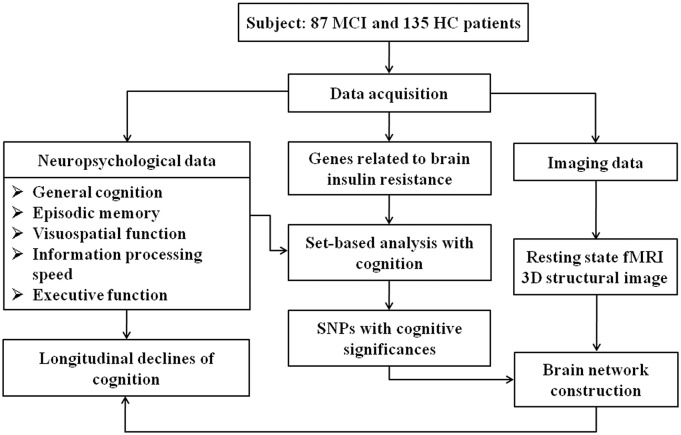
An overview of the research design and methods is shown in Fig. S1.
Fig. S1.
An overview of the research design and methods.
2.1. Participants
The Affiliated ZhongDa Hospital of Southeast University Research Ethics Committee approved the investigation, and each participant provided written informed consent. At baseline, 87 MCIs and 135 matched HCs were enrolled via newspaper advertisements and community health screenings. Every participant went through clinical evaluations, sequencings of targeted genes and MRI scans. Six HCs and seven MCIs with genotype call rates < 90% were excluded from the genetic set-based analysis. Further, four HCs were not included in the imaging genetics analysis because of excessive motion artifacts. An average 35-month follow-up study was performed, and 57 MCIs and 64 HCs returned for clinical evaluation (the follow-ups of HCs were paused when comparative numbers of MCIs and HCs participated in the follow-ups). At follow-up, four HCs developed into MCI and seven MCIs reverted to normal cognition, and all these participants were excluded in the follow-up analyses. Sixteen of the remaining 50 MCIs converted into AD (c-MCIs). Finally, 60 HCs and 50 MCIs (including 16 c-MCIs and 34 nc-MCIs) were employed in the longitudinal analysis.
2.2. Clinical evaluation
Participants underwent the same comprehensive clinical evaluations at baseline and follow-up, including the demographic information, the history of past illness and neuropsychological testing. The Mini-Mental State Examination (MMSE) and the Mattis Dementia Rating Scale-2 (MDRS-2) were used to assess general cognition. The neuropsychological battery mainly comprised the Auditory Verbal Learning Test-20-min delayed recall (AVLT-20-min DR), the Rey-Osterrieth Complex Figure Test with 20-min delayed recall, the Trail Making Test - A and B, the Stroop Color and Word Test A, B, and C, the Verbal Fluency Test, the Digital Span Test, the Semantic Similarity Test and the Clock Drawing Test, which were used to evaluate episodic memory, visuospatial function, information processing speed and executive function (detailed information given in Table S1). The presence or absence of diabetes was determined by medical history and medical records.
2.3. Inclusion and exclusion criteria
All participants were 54- to 80-year-old Han Chinese. They were all right-handers and had an education of > 8 years. MCIs were employed according to the recommendations (Albert et al., 2011, McKhann et al., 2011), as follows: (1) subjective memory impairment; (2) objective memory impairment: score of AVLT-20-min DR less than or equal to 1.5 standard deviations of age- and education-adjusted norms; (3) no or minimal impairment of general cognition: MMSE score ≥ 24 or MDRS-2 score ≥ 120; (4) a Clinical Dementia Rating of 0.5, with at least a 0.5 in the memory domain; (5) relatively intact daily activities: score of activities of daily living ≤ 25; and (6) absence of dementia or insufficient to meet the Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV) and the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease (NIA-AA). Further, HCs were required to have MMSE scores ≥ 26 and MDRS-2 scores > 120.
The exclusion criteria were as follows: (1) histories of neurological or psychiatric diseases; (2) contraindications in the MRI scans; or (3) gross brain structural abnormalities revealed by MRI scans.
2.4. ALFF analysis
The detailed information about MRI data acquisition and image preprocessing was provided in Supplementary material. The preprocessed image data were used for the ALFF analysis. Briefly, the resulting data were transformed to the frequency domain, and the power spectrum was acquired. The obtained power spectrum was square root transformed and averaged across frequencies from 0.01 to 0.08 Hz. The averaged square root was taken as the ALFF. To exclude the possibility that brain activity (i.e., the ALFF) changes were attributed to the different brain structures, voxel-wise-based gray matter volume correction was performed (detailed information given in Supplementary material).
2.5. Gene and SNP selections
Although the pathway of peripheral insulin resistance is available in the KEGG database (http://www.kegg.jp/), the insulin resistance within the brain is our focus. Therefore, we chose genes based on previous studies, and only the key components that have been demonstrated to mediate insulin resistance in AD brains were included (i.e., INS, INSR, IDE, IGF1, IGF1R, IRS1, IRS2, PIK3CA, PIK3CB, PIK3CD, PIK3CG, AKT1, AKT2, AKT1S1, AKT3, GSK3B, MTOR, ERK, JNK and PDPK1). High-throughput sequencing was carried out covering the 3′UTR, coding exon and 5′UTR of the genes mentioned above, extending to the 25 bases from 3′ end and 25 bases from 5′ end by HiSeq Sequencer (Illumina, Inc., San Diego, CA). In total, 374 SNPs of 20 genes were sequenced (details provided in Table S2). Moreover, rs7412 and rs429358 were also sequenced to determine the APOE genotype, which was introduced as a covariate.
2.6. Statistical analysis
2.6.1. Demographic and neuropsychological data
At both the baseline and follow-up, a composite Z score analysis for each cognitive domain was performed, as previously described (Shu et al., 2016, Xie et al., 2012). Briefly, the individual raw scores for each test were transformed to Z scores according to the means and standard deviations across all the participants (for tests that were measured by time, the reciprocal of the raw time was used in the subtractions), and then the composite Z scores for each cognitive domain represented the mean values of the relevant tests (details given in Table S1). To compare the demographic and neuropsychological data between HCs and MCIs, two-sample t-tests and χ2 tests (only for gender and diabetes) were utilized. The Z scores for each domain were used as phenotypes in the next genetic association analyses.
2.6.2. Genetic association analyses to select candidate SNPs with cognitive significance
Genetic association analyses were performed using the set-based tests in PLINK v1.08 (http://pngu.mgh.harvard.edu/~purcell/plink/). SNPs that passed the quality control according to the following criteria were retained for the association analysis: call rate > 0.95, minor allele frequency > 0.05 and P > 0.05 for Hardy-Weinberg disequilibrium tests. Further, participants with a genotype call rates < 0.9 were excluded from the set-based analysis (n = 13). SNPs within one gene were generated as one set, and the analysis was performed as follows. First, linear regression analysis for each single SNP was performed, and then up to five independent (r2 < 0.5) SNPs with P < 0.05 were selected for each set. Then, 1000 permutations were performed, and the empirical p-value for each set (EMP) represented the number of times the permuted set-statistic exceeded the original one for that set. To exclude the possibility that the cognitive differences were due to other factors, the effects of age, gender, education, APOE ε4, group (i.e., MCI or HC) and diabetes were introduced as covariates. It should be noted that SNPs with cognitive relevance were included in the subsequent imaging genetic analyses.
2.6.3. Mass univariate modeling to select brain regions for network construction
The analysis was performed in a similar procedure to that of previous investigations (Bai et al., 2016a, Bai et al., 2016b, Inkster et al., 2010). For each SNP related to cognitive performance, genotype-disease interactions were evaluated based on a general linear model. In detail, a 2 × 3 ANOVA for each selected SNP was performed using SPM8 (genotype status: 3-level covariates; disease: 2-level covariates, MCI and HC). It should be noted that for SNPs with MAF < 31%, rare homozygous and heterozygous groups were merged into one group (i.e., 2 × 2 ANOVAs were performed). Moreover, the influences of age, gender, education, APOE ε4 and diabetes were corrected. The Monte Carlo stimulations were applied for the imaging space corrections (voxel-wise P < 0.05, cluster sizes larger than 6165 mm3, FWHM = 6 mm; http://afni.nimh.nih.gov/pub/dist/doc/manual/AlphaSim.pdf). The regions with ALFF differences that survived the Monte Carlo stimulations were taken as regions of interest (ROIs) to construct the brain network.
2.6.4. Construction of network and explorations of the characteristics
(a) The ROIs mentioned above were defined as nodes of the network, and the Fisher's Z-transformed Pearson's correlation coefficient (CC) between every two ROIs represented the weighted edges between the two nodes. During the CC calculations, the effects of motion parameters, cerebrospinal fluid and white matter were regressed. As 13 ROIs were selected (details given in Results), a 13 × 13 matrix was obtained for each participant. Thus, a network with 13 nodes and 78 undirected edges was established for each participant.
(b) In addition to the undirected edges (ROI-to-ROI connectivity), the topological characteristics for each node were calculated to better illustrate the overall efficiency of the network. The degree Si described the extent to which nodei is relevant to the constructed network according to the following formula:
| (1) |
where Wij represents the weighted edge connecting node i and node j, which is the Fisher's Z-transformed CC value between brain region i and brain region j.
(c) To explore the disease-related differences of the constructed network, every undirected weighted edge and the topological characteristics of each node were compared between the HCs and MCIs. Two-sample t-tests were utilized, and P < 0.05 was considered statistically significant.
(d) The multivariate linear regression analyses were utilized to explore whether the network connectivity was related to the longitudinal declines for each cognitive domain, which were described as M scores. Briefly, the longitudinal changes (follow-up raw values minus baseline raw values) for each individual test were transformed to Z scores, and the M scores represented the mean values of the relevant tests for each domain. In order to exclude the possibility that the cognitive declines were attributed to other factors, the effects of age, gender, education, APOE and diabetes status were corrected. The analyses were performed for MCIs and HCs respectively. P < 0.05 was considered statistically significant. For the ROI-to-ROI connectivity (i.e., edges), the analyses were performed in HCs as follows:
| (2) |
We also investigated whether the topological characteristics of the network influenced the cognitive changes using the same formula:
| (3) |
Furthermore, as mentioned above, there were two subgroups of the MCIs (i.e., c-MCIs and nc-MCIs). Thus, for the analyses of MCIs, the subgroup status was also introduced as covariates to correct the effects of subgroup-related differences. The formula was listed as follows:
| (4) |
where β0 is the intercept of the fitting line, and β1 is the effect of the network connectivity (i.e., weighted edges and topological characteristics). β2, β3, β4, β5, β6 and β7 are the effects of age, gender, education, APOE ε4, diabetes and subgroup (i.e., c-MCIs and nc-MCIs, only for the MCIs), respectively, which are covariates of no interest.
3. Results
3.1. Demographic and cognitive data
No significant differences in the demographic data were detected between the MCIs and HCs (all P > 0.05). At both baseline and follow-up, the MCIs showed significant deficits in general cognition, episodic memory, visuospatial function, information processing speed and executive function (all P < 0.001, as shown in Table 1 and detailed information given in Table S1).
Table 1.
Demographic and neuropsychological data.
| Baseline |
Follow-up |
|||||
|---|---|---|---|---|---|---|
| MCI (n = 80) | HC (n = 127) | P | MCI (n = 50) | HC (n = 60) | P | |
| Age (years) | 69.53 ± 7.43 | 68.88 ± 6.67 | 0.085 | 71.14 ± 7.21 | 71.27 ± 5.86 | 0.93 |
| Gender (M/F) | 47/40 | 65/70 | 0.354 | 28/22 | 28/32 | 0.33 |
| Education (years) | 11.83 ± 3.18 | 12.28 ± 2.99 | 0.178 | 11.79 ± 3.18 | 12.66 ± 2.95 | 0.10 |
| Diabetic (Y/N) | 12/75 | 20/115 | 0.832 | 7/43 | 12/48 | 0.43 |
| Composite Z scores of each cognitive domain | ||||||
| General cognition | − 0.62 ± 1.05 | 0.39 ± 0.52 | < 0.001 | − 0.48 ± 1.37 | 0.35 ± 0.16 | < 0.001 |
| Episodic memory | − 0.75 ± 0.70 | 0.48 ± 0.50 | < 0.001 | − 0.73 ± 0.73 | 0.56 ± 0.46 | < 0.001 |
| Visuospatial function | − 0.40 ± 1.12 | 0.26 ± 0.82 | < 0.001 | − 0.42 ± 1.32 | 0.32 ± 0.50 | < 0.001 |
| Information processing speed | − 0.45 ± 0.75 | 0.29 ± 0.74 | < 0.001 | − 0.63 ± 0.92 | 0.33 ± 0.77 | < 0.001 |
| Executive function | − 0.45 ± 0.81 | 0.29 ± 0.81 | < 0.001 | − 0.55 ± 0.96 | 0.31 ± 0.57 | < 0.001 |
Data was represented as mean ± SD. Details are shown in Table S1.
3.2. Interaction with brain insulin resistance and cognition
The genetic set-based analyses revealed that eight of the 20 candidate genes were associated with cognitive performances at the threshold of EMP < 0.05. ATK2 (rs41275750, rs33933140) and AKT1 (rs3803304, rs2494735) were related to general cognition and information processing speed, respectively. IGF1R (rs1815009) and PIK3CB (rs2305268) influenced episodic memory. Further, associations were detected between executive function and PIK3CD (rs72633865), IDE (rs1887922), AKT1S1 (rs3810268) and MTOR (rs4845988, rs3737611, rs17235612 and rs7524202) (13 SNPs in total, details given in Table 2). Furthermore, it should be noted that none set influenced the disease status (i.e., the HC or MCI; for all the 20 sets, P > 0.05 in logistic case/control analyses, data not shown).
Table 2.
Set-based analysis reveals the cognitive significances of SNPs related to the pathway of brain insulin resistance.
| GENE | NSNP | NSIG | ISIG | EMP | SNPs | Cognition |
|---|---|---|---|---|---|---|
| AKT2 | 8 | 2 | 2 | 0.029 | rs41275750, rs33933140 | General cognition |
| PIK3CB | 1 | 1 | 1 | 0.003 | rs2305268 | Episodic memory |
| IGF1R | 23 | 6 | 1 | 0.02 | rs1815009 | Episodic memory |
| PIK3CD | 8 | 1 | 1 | 0.001 | rs72633865 | Execution |
| MTOR | 47 | 30 | 4 | 0.002 | rs4845988,rs3737611, rs17235612,rs7524202 | Execution |
| IDE | 7 | 1 | 1 | 0.026 | rs1887922 | Execution |
| AKT1S1 | 5 | 2 | 1 | 0.038 | rs3810268 | Execution |
| AKT1 | 15 | 6 | 2 | 0.01 | rs3803304, rs2494735 | Information processing speed |
NSNP: number of SNPs in set; NSIG: total number of SNPs below p-value 0.05 in the linear regression analysis; ISIG: number of significant SNPs also passing linkage disequilibrium-criterion; EMP, empirical set-based p-value; SNPs, list of single nucleotide polymorphisms.
3.3. Gene-based brain network construction
Of the 13 SNPs with cognitive significance, 9 SNPs were identified to influence neuronal activity (ALFF), as revealed by the MCI-genotype interactions, including rs1815009 (IGF1R), rs1887922 (IDE), rs2305268 (PIK3CB), rs3803304 (AKT1), rs72633865 (PI3KCD), rs17235612 and rs4845988 (MTOR), rs33933140 and rs41275750 (AKT2). Furthermore, the regions with ALFF differences that survived the minimum non-stationary AlphaSim-correction were widely detected in cerebellar-cortical regions (13 regions in total and detailed information given in Table 3).
Table 3.
Brain regions extracted from the genotype-by-MCI interactions.
| GENE | SNP | Allele | Peak MNI | Cluster size (mm3) | Peak F value | Brain region |
|---|---|---|---|---|---|---|
| IGF1R | rs1815009 | CT | − 51 − 57 48 | 7425 | 9.1 | LIPL |
| IDE | rs1887922 | CT | 42 33 39 | 6210 | 10.5 | RMFG |
| PIK3CB | rs2305268 | CT | 9 –60 − 45 | 10,017 | 9.0 | RcpCbm |
| − 36 − 12 − 33 | 13,689 | 9.2 | LHip/PHG | |||
| − 12 − 12 72 | 6750 | 7.1 | BPCL | |||
| AKT1 | rs3803304 | CG | 69 –36 27 | 6291 | 14.9 | RSTG |
| 21 51 18 | 6156 | 11.2 | RSFG | |||
| rs2494735 | TC | NA | ||||
| PI3KCD | rs72633865 | CG | 18–100 0 | 8667 | 14.5 | RIOG |
| MTOR | rs17235612 | CT | 9 –48 − 54 | 11,907 | 19.6 | RcpCbm |
| 69 –30 21 | 14,067 | 28.4 | RINS | |||
| rs4845988 | AG | − 57 − 6 − 18 | 15,417 | 21.4 | LMTG | |
| rs3737611 | AG | NA | ||||
| rs7524202 | TC | NA | ||||
| AKT1S1 | rs3810268 | CT | NA | |||
| AKT2 | rs33933140 | AG | − 15 6 –30 | 8883 | 8.5 | LSTG |
| rs41275750 | CG | − 6 57 18 | 6237 | 9.6 | LMFG/SFG |
All of these regions survived the Monte Carlo correction. LIPL, left inferior parietal lobule; R/LMFG, right/left middle frontal gyrus; RcpCbm, right cerebellum posterior lobe; LHip/PHG, left Hippocampus/ParaHippocampal; BPCL, bilateral Paracentral Lobule; R/LSTG, right/left superior temporal gyrus; R/LSFG, right/left superior frontal gyrus; RIOG, right inferior occipital gyrus; RINS, right insula; LMTG, left middle temporal gyrus.
3.4. Characteristics and behavior significances of the network
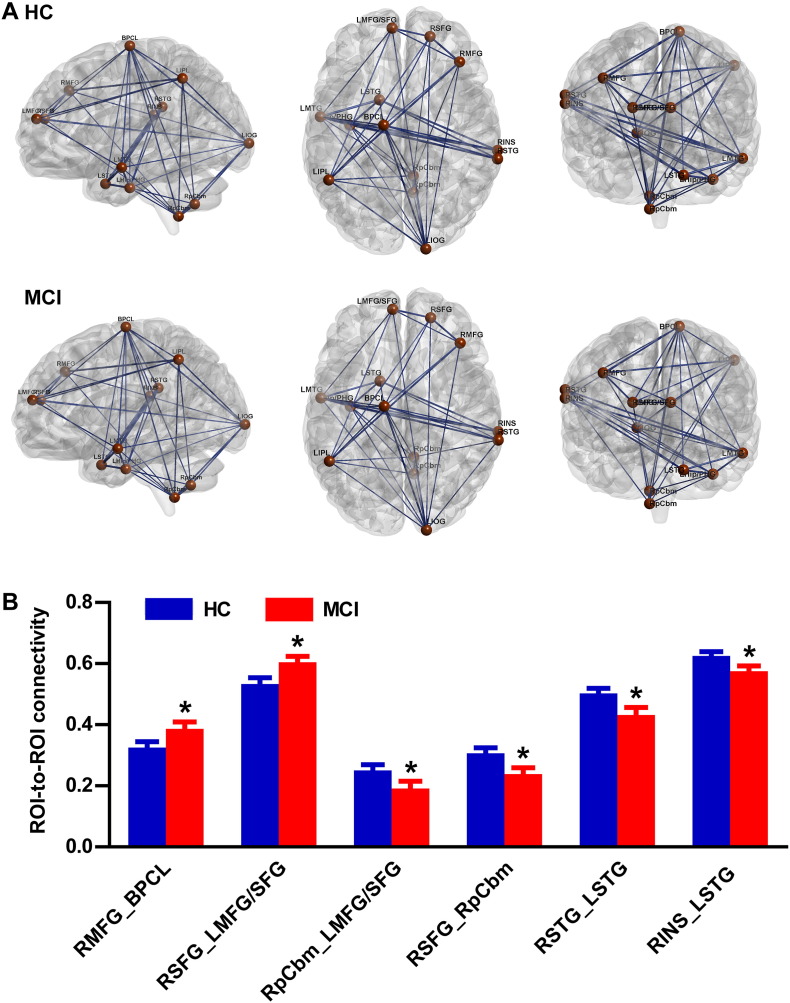
After AlphaSim-correction, the 13 brain regions remained significant for the imaging space, which were taken as ROIs to construct the brain network, and a unidirectional weighted network with 13 nodes and 78 edges was obtained for each participant. The average connectivity pattern for the HCs and MCIs is illustrated in Fig. 1A, and only the edges with weights (CC values between two ROIs) larger than 0.3 are shown.
Fig. 1.
A. Insulin resistance pathway-based unidirectional weighted networks with 13 nodes and 78 edges for MCIs and HCs. The connectivity about the thresholds (r = 0.3) was shown. The figure was created using BrainNet Viewer (http://www.nitrc.org/projects/bnv/). B. Changed ROI-to-ROI connectivity between HCs and MCIs. *Indicates significant differences for MCI compared with HC, P < 0.05.
3.4.1. Weights of edges in the network
As shown in Fig. 1B, of the 78 unidirectional edges, two edges with significantly increased connectivity were detected in the MCIs compared with the HCs, including the RMFG-to-BPCL (T = 2.04, P = 0.04) and RSFG-to-LMFG/SFG (T = 2.41, P = 0.017) connectivities. The MCIs also showed significantly decreased connectivity in the four edges, including RpCbm-to-LMFG/SFG (T = − 2.0, P = 0.047), RSFG-to-RpCbm (T = − 2.423, P = 0.016), RSTG-to-LSTG (T = − 2.475, P = 0.014) and RINS-to-LSTG (T = − 2.27, P = 0.024) connectivities. However, no corrections were found with the cognitive changes and edge weights in neither HCs nor MCIs (all P > 0.05).
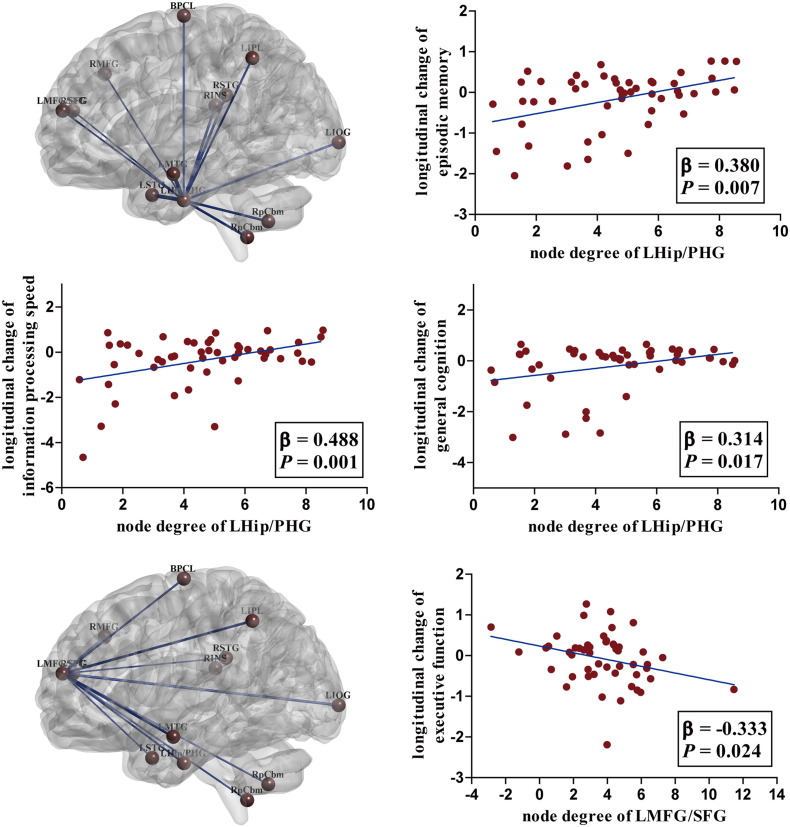
3.4.2. Degrees of nodes in the network
There were not significant disease-related differences concerning the S values of any of the 13 nodes between the HCs and MCIs (all P > 0.05), while the topological characteristics showed behavior relevance. As illustrated in Fig.2, for MCIs, the higher node degrees of the LHip/PHG predicted less decline in episodic memory (β = 0.380, P = 0.007), information processing speed (β = 0.488, P = 0.001) and general cognition (β = 0.314, P = 0.017). However, the oppose associations were detected between the topological characteristics of LMFG/SFG and changes of execution (β = − 0.333, P = 0.024). It should be noted that, in addition to the S values of the network, the cognitive declines were also attributed to other factors, including age, education and APOE genotype. Moreover, the subgroup-related differences (c-MCIs and nc-MCIs) regarding the cognitive changes were also detected (detailed information was provided in Table S3). No associations between the node degrees and cognitive changes were detected for the HCs.
Fig. 2.
The topological pattern and behavior significances. For MCIs, the node degree of LHip/PHG was positively related with the longitudinal changes of episodic memory, information processing speed and general cognition. However, the oppose associations were detected between the node degree of LMFG/SFG and executive function.
4. Discussion
Using the genetic association analyses, we provided in vivo evidence that the pathway of brain insulin resistance modifies cognitive performance and further showed that the influences occurred in the absence of diabetes. The functional MRI led to the identification of brain networks related to the pathway, which partially reveal the mechanisms underlying cognitive injury related to brain insulin disorder. Compared to the matched HCs, the MCIs showed regional deficits in connectivity, while compensations may take place to maintain the overall efficiency of the network. However, the node degree may correspond with disease progression because it predicts cognitive changes over time, especially the degree of medial temporal lobe and prefrontal cortex. These findings are compatible with the key role of insulin signaling in maintaining hippocampal function underlying AD development, as shown in postmortem analyses (Talbot et al., 2012).
4.1. The association with brain insulin resistance and AD-related intermediate phenotypes
IDE is a very promising gene that influences cognitive performance because of its biological function and location near LOAD linkage peaks, i.e., the chromosome 10q (Bertram et al., 2000). Various variants of IDE have been analyzed for their associations with AD risk in different races (Bertram et al., 2007), and it has been debated whether IDE represents a potential AD susceptibility gene because inconsistent results have been obtained (Bertram et al., 2007). However, it has been replicated in several independent samples that rs1887922 exhibits interactions with AD-related quantitative measures, including performances of MMSE, loads of Aβ plaques and densities of neurofibrillary tangles (Blomqvist et al., 2005, Prince et al., 2003). Our present result substantiated the previous findings by firstly showing that the influences of rs1887922 on cognition can also be detected in the Han nationality. In particular, we further identified that the influences were mainly embodied in executive domain. Thereby, this result might suggest that brain regions that regulate execution are targets for IDE, such as the prefrontal cortex (PFC). Regarding the IGF1R, rs2229765 has gained attention as a likely AD candidate locus because of its association with plasma levels of IGF-1 (Bonafe et al., 2003). Subsequent studies have suggested this loci is related to the risk of vascular dementia (Garcia et al., 2006), while negative results were obtained regarding the associations of rs2229765 and AD risk in investigations performed in Caucasians (Garcia et al., 2006). The current study, by using the intermediate phenotypes, reported that rs1815009 of IGF1R may have a role in determining AD severity, further supporting the important roles of IGF1R in AD development.
In terms of the down-stream molecules, we also detected their associations with cognition. Highly phosphorylated IRS-1 has been consistently discovered in AD brains (Ma et al., 2009, Moloney et al., 2010, Talbot et al., 2012), and such phosphorylation suppresses the ability of IRS-1 to transmit signals to downstream molecules (Boura-Halfon and Zick, 2009). Feedback inhibition primarily by AKT, MTOR, GSK-3 and PKC and feedforward inhibition by JNK and IKK regulate the phosphorylation process (Talbot and Wang, 2014), and the expression levels of these molecules predict memory performances (Talbot et al., 2012), indicating their key roles in AD. Our results are in accordance with the idea that SNPs of the molecules that control the phosphorylation of IRS-1 also determine the performances of multiple cognitive domains. However, it must be noted that the genome-wide association studies (GWAS) did not highlight the SNPs discussed presently in AD process (i.e. except for rs2305268, rs72633865 and rs4845998, all the other SNPs were included in the GWAS analysis, but none of them reached the genome-wide significance) (Lambert et al., 2013). Furthermore, our present case/control analyses also obtained negative associations between MCI status and brain insulin genes (data not shown). Regarding the inconsistence, we could assume that the effect size of each individual SNP were too small, and the pathway-based quantitative trait association studies may better reveal the interactions with AD and brain insulin resistance. In addition, these results could imply that the brain insulin disorder may not directly lead to the occurrence of AD, but play a role in determining the AD severity (i.e., the cognitive impairments). The validations of the functional activities of these loci should be given great priority. Previous investigations have highlighted the biological significance of these SNPs. For instance, the genotype of AKT1 in rs3803304 predicted longevity (Pawlikowska et al., 2009), and rs33933140 of AKT2 was related to radiation pneumonitis in cancer patients who received radiation therapy (Tang et al., 2016). However, the data on their functional activities in AD process are very limited. Moreover, none of the SNPs are missense mutations, meaning that they do not result in amino acid changes of the relevant proteins. We might assume that these SNPs function by modifying the expression levels of corresponding molecules, and this idea must be further verified. By taking advantages of functional MRI, we attempted to explore the topological alterations of brain function associated with insulin resistance, which helps to reveal gene-brain-behavior interactions.
4.2. The joint actions of the insulin resistance pathway on brain function
Peripheral insulin resistance has been suggested to influence AD-related MRI markers. For instance, the homeostasis model assessment of insulin resistance 2, a measure used to quantify insulin resistance, showed negative associations with volumes of the hippocampus (Rasgon et al., 2011), the medial prefrontal cortex (mPFC) and medial temporal regions (Morris et al., 2014). Moreover, positive relations were detected with the peripheral levels of IGF-1 and total brain volume (Westwood et al., 2014). Regarding the functional network, higher insulin levels resulted in DMN disconnections for participants without diabetes, especially the connectivity between mPFC and Hip/PHG (Kenna et al., 2013). Further, deficits of the DMN could also be detected in patients with T2D, and higher insulin resistance predicted more serious injury of the DMN (Chen et al., 2014). Using genetic association analyses, Silver et al. identified genetics variations linked to insulin resistance that determined brain atrophy over time, including HK2, PIK3R3, PIK3CG, ACACA and G6PC (Silver et al., 2012). Our data extend our understanding as to how brain insulin resistance influences brain function.
Within the functional network based on the pathway of brain insulin resistance, the MCIs showed disconnections mainly in the cerebellum-frontal-temporal cortex, in which reduced response to insulin signaling were found by molecular analyses (Moloney et al., 2010, Talbot et al., 2012). These disturbances could represent one of the mechanisms underlying cognitive impairment induced by insulin resistance. Additionally, increased functional activities within the bilateral frontal-parietal lobes were also found in MCIs. According to the scaffolding theory of aging and cognition (Reuter-Lorenz and Park, 2014), aging is associated with improved functional recruitments of the frontal-parietal brain to compensate for the neural losses of other regions. Therefore, we assumed the increased connectivity in MCIs corresponded to accelerated compensation to maintain relatively intact cognition. Furthermore, it may also suggest that the insulin function in frontal-parietal regions was relatively preserved compared to other regions, which also fits the findings from the postmortem analyses (Moloney et al., 2010, Talbot et al., 2012). More importantly, we found behavioral significance of the functional network. None of the edge connectivities were related to cognition, but the node degree of the LHip/PHG predicted longitudinal changes of multiple cognitive domains. Higher node degrees corresponded to better cognition that was sustained over time. The associations provided convincing evidence to support that the Hip/PHG were targeted regions of insulin disorder and the pivotal role of Hip/PHG in AD processes (Biessels and Reagan, 2015, Talbot et al., 2012). Inversely, the node degrees of the LMFG/SFG were negatively related to the changes in execution, which is in line with the key role of the frontal lobes in controlling execution and is supportive of the compensatory nature for the increased involvements of frontal brains. The higher node degree identified a greater need for compensation, suggesting more severe disease. As the disease progressed, the abilities of the brain to provide effective compensation would eventually diminish, at which time patients show severe cognitive deficits (Rao et al., 2015).
4.3. Limitations to be declared
There were several methodological issues in this study. First, as the pathway of brain insulin resistance has not been completely clarified, some genes related to the disorder may have been missed. Further studies are needed to better reveal the pathway of brain insulin resistance. Second, the absent evaluations of AD-related pathology could lead to the heterogeneity of MCIs, meaning that some MCIs employed may not meet the criteria of “MCI due to AD” (Dubois et al., 2010). Third, the intervals between baseline and follow-up investigations appeared to be long. We cannot better understand the trajectories of cognitive declines for the participants. Especially for the c-MCIs, we did not know that when they converted into AD, thus the duration of the MCI was not considered in the analysis. Furthermore, we tried to control the influences of other demographic and genetic factors in the analysis, but the age range of the participants was fairly wide. More importantly, the ages of c-MCIs were significantly larger than the nc-MCIs (detailed information regarding the demographic and neuropsychological data was provided in Table S4). Last, owing to the relatively small number of participants, we did not perform multiple comparison corrections in the gene-based analyses. We look forward to further studies with larger sample sizes and better experimental designs to verify our present results.
To conclude, our results identified that neurobiological alterations related to brain insulin resistance modify AD progress by influencing the overall efficiency of brain functional networks, providing evidence that supports that treating insulin resistance represents a promising therapeutic target for alleviating cognitive declines associated with T2D and AD.
The following are the supplementary data related to this article.
87 MCIs and 135 HCs were employed at baseline, and all of them went through clinical evaluations, sequencings of targeted genes and MRI scans. Set-based gene association analyses were performed to selected genes with cognitive relevance, which were used to construct functional network. The association analyses were performed with the topological characteristics of the constructed network and cognitive declines over time.
Conflict of interest
None.
Acknowledgements
This research was partly supported by the National Natural Science Foundation of China (No. 91332104, 81500919, 81671665); Natural Science Foundation of Jiangsu Province (No. BK20160071); Six Talent Peaks Project in Jiangsu Province (No. 2015-WSN-003); Program for New Century Excellent Talents in University (No. NCET-13-0117); Key Program for Clinical Medicine and Science and Technology: Jiangsu Province Clinical Medical Research Center (No·BL2013025); National High-tech R.D Program (863 Program) (No. 2015AA020508); Fundamental Research Funds for the Central Universities and Graduate Candidate Research Innovation Program of Jiangsu Province (No·KYLX15_0188).
References
- Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C., Gamst A., Holtzman D.M., Jagust W.J., Petersen R.C., Snyder P.J., Carrillo M.C., Thies B., Phelps C.H. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L., Gur R.C., Haack K., Cole S.A., Calkins M.E., Peralta J.M., Hare E., Prasad K., Pogue-Geile M.F., Nimgaonkar V., Gur R.E. A genome screen for quantitative trait loci influencing schizophrenia and neurocognitive phenotypes. Am. J. Psychiatry. 2008;165:1185–1192. doi: 10.1176/appi.ajp.2008.07121869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., Liao W., Yue C., Pu M., Shi Y., Yu H., Yuan Y., Geng L., Zhang Z. Genetics pathway-based imaging approaches in Chinese Han population with Alzheimer's disease risk. Brain Struct. Funct. 2016;221:433–446. doi: 10.1007/s00429-014-0916-4. [DOI] [PubMed] [Google Scholar]
- Bai F., Yuan Y., Shi Y., Zhang Z. Multiple genetic imaging study of the association between cholesterol metabolism and brain functional alterations in individuals with risk factors for Alzheimer's disease. Oncotarget. 2016;7:15315–15328. doi: 10.18632/oncotarget.8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L., Blacker D., Mullin K., Keeney D., Jones J., Basu S., Yhu S., McInnis M.G., Go R.C., Vekrellis K., Selkoe D.J., Saunders A.J., Tanzi R.E. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Biessels G.J., Reagan L.P. Hippocampal insulin resistance and cognitive dysfunction. Nat. Rev. Neurosci. 2015;16:660–671. doi: 10.1038/nrn4019. [DOI] [PubMed] [Google Scholar]
- Biffi A., Anderson C.D., Desikan R.S., Sabuncu M., Cortellini L., Schmansky N., Salat D., Rosand J., Alzheimer's Disease Neuroimaging, I Genetic variation and neuroimaging measures in Alzheimer disease. Arch. Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist M.E., Chalmers K., Andreasen N., Bogdanovic N., Wilcock G.K., Cairns N.J., Feuk L., Brookes A.J., Love S., Blennow K., Kehoe P.G., Prince J.A. Sequence variants of IDE are associated with the extent of beta-amyloid deposition in the Alzheimer's disease brain. Neurobiol. Aging. 2005;26:795–802. doi: 10.1016/j.neurobiolaging.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Bonafe M., Barbieri M., Marchegiani F., Olivieri F., Ragno E., Giampieri C., Mugianesi E., Centurelli M., Franceschi C., Paolisso G. Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J. Clin. Endocrinol. Metab. 2003;88:3299–3304. doi: 10.1210/jc.2002-021810. [DOI] [PubMed] [Google Scholar]
- Boura-Halfon S., Zick Y. Phosphorylation of IRS proteins, insulin action, and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2009;296:E581–E591. doi: 10.1152/ajpendo.90437.2008. [DOI] [PubMed] [Google Scholar]
- Carro E., Trejo J.L., Gomez-Isla T., LeRoith D., Torres-Aleman I. Serum insulin-like growth factor I regulates brain amyloid-beta levels. Nat. Med. 2002;8:1390–1397. doi: 10.1038/nm1202-793. [DOI] [PubMed] [Google Scholar]
- Chen Y.C., Jiao Y., Cui Y., Shang S.A., Ding J., Feng Y., Song W., Ju S.H., Teng G.J. Aberrant brain functional connectivity related to insulin resistance in type 2 diabetes: a resting-state fMRI study. Diabetes Care. 2014;37:1689–1696. doi: 10.2337/dc13-2127. [DOI] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., Cummings J.L., Dekosky S.T., Barberger-Gateau P., Delacourte A., Frisoni G., Fox N.C., Galasko D., Gauthier S., Hampel H., Jicha G.A., Meguro K., O'Brien J., Pasquier F., Robert P., Rossor M., Salloway S., Sarazin M., de Souza L.C., Stern Y., Visser P.J., Scheltens P. Revising the definition of Alzheimer's disease: a new lexicon. Lancet Neurol. 2010;9:1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J., Ahmadi A., Wonnacott A., Sutcliffe W., Nagga K., Soderkvist P., Marcusson J. Association of insulin-like growth factor-1 receptor polymorphism in dementia. Dement. Geriatr. Cogn. Disord. 2006;22:439–444. doi: 10.1159/000095803. [DOI] [PubMed] [Google Scholar]
- Inkster B., Nichols T.E., Saemann P.G., Auer D.P., Holsboer F., Muglia P., Matthews P.M. Pathway-based approaches to imaging genetics association studies: Wnt signaling, GSK3beta substrates and major depression. NeuroImage. 2010;53:908–917. doi: 10.1016/j.neuroimage.2010.02.065. [DOI] [PubMed] [Google Scholar]
- Kenna H., Hoeft F., Kelley R., Wroolie T., DeMuth B., Reiss A., Rasgon N. Fasting plasma insulin and the default mode network in women at risk for Alzheimer's disease. Neurobiol. Aging. 2013;34:641–649. doi: 10.1016/j.neurobiolaging.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., Russo G., Thorton-Wells T.A., Jones N., Smith A.V., Chouraki V., Thomas C., Ikram M.A., Zelenika D., Vardarajan B.N., Kamatani Y., Lin C.F., Gerrish A., Schmidt H., Kunkle B., Dunstan M.L., Ruiz A., Bihoreau M.T., Choi S.H., Reitz C., Pasquier F., Cruchaga C., Craig D., Amin N., Berr C., Lopez O.L., De Jager P.L., Deramecourt V., Johnston J.A., Evans D., Lovestone S., Letenneur L., Moron F.J., Rubinsztein D.C., Eiriksdottir G., Sleegers K., Goate A.M., Fievet N., Huentelman M.W., Gill M., Brown K., Kamboh M.I., Keller L., Barberger-Gateau P., McGuiness B., Larson E.B., Green R., Myers A.J., Dufouil C., Todd S., Wallon D., Love S., Rogaeva E., Gallacher J., St George-Hyslop P., Clarimon J., Lleo A., Bayer A., Tsuang D.W., Yu L., Tsolaki M., Bossu P., Spalletta G., Proitsi P., Collinge J., Sorbi S., Sanchez-Garcia F., Fox N.C., Hardy J., Deniz Naranjo M.C., Bosco P., Clarke R., Brayne C., Galimberti D., Mancuso M., Matthews F., European Alzheimer's Disease, I, Genetic, Environmental Risk in Alzheimer's, D, Alzheimer's Disease Genetic, C, Cohorts for, H, Aging Research in Genomic, E, Moebus S., Mecocci P., Del Zompo M., Maier W., Hampel H., Pilotto A., Bullido M., Panza F., Caffarra P., Nacmias B., Gilbert J.R., Mayhaus M., Lannefelt L., Hakonarson H., Pichler S., Carrasquillo M.M., Ingelsson M., Beekly D., Alvarez V., Zou F., Valladares O., Younkin S.G., Coto E., Hamilton-Nelson K.L., Gu W., Razquin C., Pastor P., Mateo I., Owen M.J., Faber K.M., Jonsson P.V., Combarros O., O'Donovan M.C., Cantwell L.B., Soininen H., Blacker D., Mead S., Mosley T.H., Jr., Bennett D.A., Harris T.B., Fratiglioni L., Holmes C., de Bruijn R.F., Passmore P., Montine T.J., Bettens K., Rotter J.I., Brice A., Morgan K., Foroud T.M., Kukull W.A., Hannequin D., Powell J.F., Nalls M.A., Ritchie K., Lunetta K.L., Kauwe J.S., Boerwinkle E., Riemenschneider M., Boada M., Hiltuenen M., Martin E.R., Schmidt R., Rujescu D., Wang L.S., Dartigues J.F., Mayeux R., Tzourio C., Hofman A., Nothen M.M., Graff C., Psaty B.M., Jones L., Haines J.L., Holmans P.A., Lathrop M., Pericak-Vance M.A., Launer L.J., Farrer L.A., van Duijn C.M., Van Broeckhoven C., Moskvina V., Seshadri S., Williams J., Schellenberg G.D., Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q.L., Yang F., Rosario E.R., Ubeda O.J., Beech W., Gant D.J., Chen P.P., Hudspeth B., Chen C., Zhao Y., Vinters H.V., Frautschy S.A., Cole G.M. Beta-amyloid oligomers induce phosphorylation of tau and inactivation of insulin receptor substrate via c-Jun N-terminal kinase signaling: suppression by omega-3 fatty acids and curcumin. J. Neurosci. 2009;29:9078–9089. doi: 10.1523/JNEUROSCI.1071-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney A.M., Griffin R.J., Timmons S., O'Connor R., Ravid R., O'Neill C. Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer's disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol. Aging. 2010;31:224–243. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Morris J.K., Vidoni E.D., Perea R.D., Rada R., Johnson D.K., Lyons K., Pahwa R., Burns J.M., Honea R.A. Insulin resistance and gray matter volume in neurodegenerative disease. Neuroscience. 2014;270:139–147. doi: 10.1016/j.neuroscience.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papassotiropoulos A., Stephan D.A., Huentelman M.J., Hoerndli F.J., Craig D.W., Pearson J.V., Huynh K.D., Brunner F., Corneveaux J., Osborne D., Wollmer M.A., Aerni A., Coluccia D., Hanggi J., Mondadori C.R., Buchmann A., Reiman E.M., Caselli R.J., Henke K., de Quervain D.J. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Pawlikowska L., Hu D., Huntsman S., Sung A., Chu C., Chen J., Joyner A.H., Schork N.J., Hsueh W.C., Reiner A.P., Psaty B.M., Atzmon G., Barzilai N., Cummings S.R., Browner W.S., Kwok P.Y., Ziv E., Study of Osteoporotic, F Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin S.G., Turner J.A., Guffanti G., Lakatos A., Torri F., Keator D.B., Macciardi F. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cogn. Neuropsychiatry. 2009;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince J.A., Feuk L., Gu H.F., Johansson B., Gatz M., Blennow K., Brookes A.J. Genetic variation in a haplotype block spanning IDE influences Alzheimer disease. Hum. Mutat. 2003;22:363–371. doi: 10.1002/humu.10282. [DOI] [PubMed] [Google Scholar]
- Profenno L.A., Porsteinsson A.P., Faraone S.V. Meta-analysis of Alzheimer's disease risk with obesity, diabetes, and related disorders. Biol. Psychiatry. 2010;67:505–512. doi: 10.1016/j.biopsych.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Rao S.M., Bonner-Jackson A., Nielson K.A., Seidenberg M., Smith J.C., Woodard J.L., Durgerian S. Genetic risk for Alzheimer's disease alters the five-year trajectory of semantic memory activation in cognitively intact elders. NeuroImage. 2015;111:136–146. doi: 10.1016/j.neuroimage.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon N.L., Kenna H.A., Wroolie T.E., Kelley R., Silverman D., Brooks J., Williams K.E., Powers B.N., Hallmayer J., Reiss A. Insulin resistance and hippocampal volume in women at risk for Alzheimer's disease. Neurobiol. Aging. 2011;32:1942–1948. doi: 10.1016/j.neurobiolaging.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter-Lorenz P.A., Park D.C. How does it STAC up? Revisiting the scaffolding theory of aging and cognition. Neuropsychol. Rev. 2014;24:355–370. doi: 10.1007/s11065-014-9270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M., Brazil D.P., Burks D.J., Kushner J.A., Ye J., Flint C.L., Farhang-Fallah J., Dikkes P., Warot X.M., Rio C., Corfas G., White M.F. Insulin receptor substrate-2 deficiency impairs brain growth and promotes tau phosphorylation. J. Neurosci. 2003;23:7084–7092. doi: 10.1523/JNEUROSCI.23-18-07084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H., Shi Y., Chen G., Wang Z., Liu D., Yue C., Ward B.D., Li W., Xu Z., Chen G., Guo Q., Xu J., Li S.J., Zhang Z. Opposite neural trajectories of apolipoprotein E 4 and 2 alleles with aging associated with different risks of Alzheimer's disease. Cereb. Cortex. 2016;26:1421–1429. doi: 10.1093/cercor/bhu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M., Janousova E., Hua X., Thompson P.M., Montana G., Alzheimer's Disease Neuroimaging, I Identification of gene pathways implicated in Alzheimer's disease using longitudinal imaging phenotypes with sparse regression. NeuroImage. 2012;63:1681–1694. doi: 10.1016/j.neuroimage.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims-Robinson C., Kim B., Rosko A., Feldman E.L. How does diabetes accelerate Alzheimer disease pathology? Nat. Rev. Neurol. 2010;6:551–559. doi: 10.1038/nrneurol.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R.A., Aisen P.S., Beckett L.A., Bennett D.A., Craft S., Fagan A.M., Iwatsubo T., Jack C.R., Jr., Kaye J., Montine T.J., Park D.C., Reiman E.M., Rowe C.C., Siemers E., Stern Y., Yaffe K., Carrillo M.C., Thies B., Morrison-Bogorad M., Wagster M.V., Phelps C.H. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & dementia. J. Alzheimer's Assoc. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K., Wang H.Y. The nature, significance, and glucagon-like peptide-1 analog treatment of brain insulin resistance in Alzheimer's disease. Alzheimers Dement. 2014;10:S12–S25. doi: 10.1016/j.jalz.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K., Wang H.Y., Kazi H., Han L.Y., Bakshi K.P., Stucky A., Fuino R.L., Kawaguchi K.R., Samoyedny A.J., Wilson R.S., Arvanitakis Z., Schneider J.A., Wolf B.A., Bennett D.A., Trojanowski J.Q., Arnold S.E. Demonstrated brain insulin resistance in Alzheimer's disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Invest. 2012;122:1316–1338. doi: 10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu B., Li J., Wu H., Yang J., Zhou X., Yi M., Li Q., Yu S., Yuan X. Genetic variants in PI3K/AKT pathway are associated with severe radiation pneumonitis in lung cancer patients treated with radiation therapy. Cancer medicine. 2016;5:24–32. doi: 10.1002/cam4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood A.J., Beiser A., Decarli C., Harris T.B., Chen T.C., He X.M., Roubenoff R., Pikula A., Au R., Braverman L.E., Wolf P.A., Vasan R.S., Seshadri S. Insulin-like growth factor-1 and risk of Alzheimer dementia and brain atrophy. Neurology. 2014;82:1613–1619. doi: 10.1212/WNL.0000000000000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Bai F., Yu H., Shi Y., Yuan Y., Chen G., Li W., Chen G., Zhang Z., Li S.J. Abnormal insula functional network is associated with episodic memory decline in amnestic mild cognitive impairment. NeuroImage. 2012;63:320–327. doi: 10.1016/j.neuroimage.2012.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
87 MCIs and 135 HCs were employed at baseline, and all of them went through clinical evaluations, sequencings of targeted genes and MRI scans. Set-based gene association analyses were performed to selected genes with cognitive relevance, which were used to construct functional network. The association analyses were performed with the topological characteristics of the constructed network and cognitive declines over time.