Abstract
Autophagy allows cells fundamental adaptations to metabolic needs and to stress. Using autophagic bulk degradation cells can clear crosslinked macromolecules and damaged organelles that arise under redox stress. Accumulation of such debris results in cellular dysfunction and is observed in aged tissue and senescent cells. Conversely, promising anti-aging strategies aim at inhibiting the mTOR pathway and thereby activating autophagy, to counteract aging associated damage. We have inactivated autophagy related 7 (Atg7), an essential autophagy gene, in murine keratinocytes (KC) and have found in an earlier study that this resulted in increased baseline oxidative stress and reduced capacity to degrade crosslinked proteins after oxidative ultraviolet stress. To investigate whether autophagy deficiency would promote cellular aging, we studied how Atg7 deficient (KO) and Atg7 bearing cells (WT) would respond to stress induced by paraquat (PQ), an oxidant drug commonly used to induce cellular senescence.
Atg7 deficient KC displayed increased prostanoid signaling and a pro- mitotic gene expression signature as compared to the WT. After exposure to PQ, both WT and KO cells showed an inflammatory and stress-related transcriptomic response. However, the Atg7 deficient cells additionally showed drastic DNA damage- and cell cycle arrest signaling. Indeed, DNA fragmentation and –oxidation were strongly increased in the stressed Atg7 deficient cells upon PQ stress but also after oxidizing ultraviolet A irradiation. Damage associated phosphorylated histone H2AX (γH2AX) foci were increased in the nuclei, whereas expression of the nuclear lamina protein lamin B1 was strongly decreased. Similarly, in both, PQ treated mouse tail skin explants and in UVA irradiated mouse tail skin, we found a strong increase in γH2AX positive nuclei within the basal layer of Atg7 deficient epidermis.
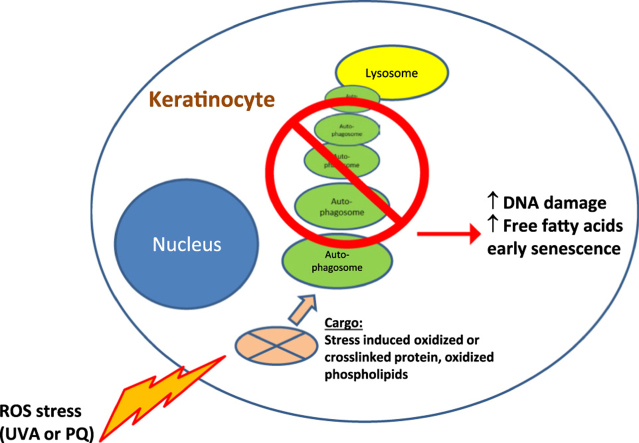
Atg7 deficiency significantly affected expression of lipid metabolic genes. Therefore we performed lipid profiling of keratinocytes which demonstrated a major dysregulation of cellular lipid metabolism. We found accumulation of autophagy agonisitic free fatty acids, whereas triglyceride levels were strongly decreased. Together, our data show that in absence of Atg7/autophagy the resistance of keratinocytes to intrinsic and environmental oxidative stress was severely impaired and resulted in DNA damage, cell cycle arrest and a disturbed lipid phenotype, all typical for premature cell aging.
Graphical abstract
Highlights
-
•
Autphagy deficiency by Atg7 deletion in keratinocytes makes them sensitive to ROS damage.
-
•
ROS stress causes exacerbated DNA damage and distorted lipid composition in Atg7 KO.
-
•
Also in vivo the autophagy deficient cells are prone to enhanced DNA damage.
-
•
Without Autphagy ROS from UV and intrinsic sources may pose higher risk of mutation.
1. Introduction
Mammalian skin is permanently exposed to intrinsic and extrinsic oxidative stressors which modify cellular macromolecules rendering them non-functional or transforming them to reactive and potentially dangerous products. Modified or oxidized macromolecules negatively affect cell function and eventually cause cell- and tissue failure, and this is often correlated to aging associated pathologies. Consequently, mechanisms to control redox damage have evolved, which comprise the inducible synthesis of cellular antioxidants and of enzymes that detoxify reactive species in the skin [1], [2]. Damaged molecules have to be repaired (e.g. DNA modifications) or have to be disposed of by degradation or secretion. A feedback on the state of damage repair determines cell fate and is exerted by the master guardian tumor protein p53 (p53), which can impose growth arrest, cellular senescence or apoptosis, depending on the sensed level of damage. If damage is repairable, p53 induces reversible growth arrest and blocks cell cycle progression during the repair of UV- induced DNA damage [3]. The disposal of damaged material other than DNA and restoration of proteome redox homeostasis is facilitated by various redox inducible cellular degradation systems [4]. These comprise an inducible conformation of the proteasome specialized to degrade oxidized proteins [5], [6] but also (macro-) autophagy [7]. Autophagy facilitates bulk degradation of oxidized protein and of damaged organelles, most importantly mitochondria [8], [9]. This clearing function is one likely cause for the anti-aging effect of autophagy, together with its metabolic control of insulin homeostasis and the inhibition of undesired pro-inflammatory signaling [10]. Conversely, autophagy deficient model systems including humans and other mammals often show typical signs of aging/senescence related pathologies [11]. We have recently generated an epidermal specific Atg7 deficient mouse to investigate the role of macroautophagy in epidermal biology [12], which surprisingly showed that autophagy is not required for the normal formation and function of the epidermis in homeostasis. What we however observed in cultured keratinocytes (KC) was a severely disturbed clearing of stress induced high molecular weight crosslinked proteins and accumulation of oxidized phospholipids in vitro.
As oxidative damage plays a major role in photoaging of the skin [13], and ROS imbalance can cause cellular senescence in the epidermal compartment [14] we investigated here the role of autophagy in ROS stress of epidermal keratinocytes. We tested how murine keratinocytes deficient in Atg7 would react to a stress protocol using the drug paraquat (PQ) which is widely applied to induce cellular senescence by ROS overproduction in various cell types [15]. We found that loss of autophagy strongly impaired the resistance of epidermal keratinocytes to extrinsic and intrinsic redox stress and resulted in severely increased DNA damage and a senescent phenotype. Atg7 deficient KC also displayed a lipid profile observed in mitochondrial damage, which was enhanced upon PQ stress. In skin organ explants treated with PQ and in murine skin exposed to UVA we could also find increased numbers of basal cells of the epidermis that were positive for γH2AX, a marker for DNA photodamage but also for cellular senescence. A role for autophagy in protecting basal epidermal cells from oxidative damage, as experienced after UVA exposure adds to the known cytoprotective functions of autophagy and underlines the potential for autophagy modulation in protecting skin from deleterious damage but also in counteracting skin aging.
2. Material and methods
2.1. Mice and primary keratinocyte culture
Atg7 f/f mice on a B6/CBA background [16] were crossed to K14::Cre mice, strain Tg(KRT14-cre)1Amc/J (also B6/CBA, Jackson Laboratories, Bar Harbor, ME) to yield Atg7 f/f K14::Cre offspring as described previously [12], [17]. These epidermal Atg7 knockout mice (hereafter: KO) were compared to Atg7 f/f littermates or age- and sex-matched Atg7 f/f mice (hereafter: WT). GFP-LC3 transgenic mice have been described previously [18]. Primary cell culture of keratinocytes was prepared from the tails of WT, KO and GFP-LC3 transgenic mice at the age of 1–2 month as described before [19]. The cells were suspended in keratinocyte growth medium-2 (KGM-2; Lonza, Basel, Switzerland) and plated in 6-well plates or 4-well chamber slides coated with bovine collagen (Vitrogen, Palo Alto, CA) and experiments were performed without further passaging. Mice were maintained according to the animal welfare guidelines of the Medical University of Vienna, Austria. The in vivo treatments had been authorized under Austrian law by the protocols TVA 66.009/0123-II/10b/2010 and 0090-WF/II/3b/2014.
2.2. Stress treatment
Cell cultures - At 50% confluence, cells were treated with 20 µM PQ (Sigma, St Louis, MO) or irradiated with UVA-1 (340–400 nm) emitted from a Sellamed 3000 device (Sellas, Ennepetal, Germany) at a distance of 20 cm as described before [20]. An irradiation time of 5 min was determined with a Waldmann (Villingen, Germany) UVmeter to yield a total fluence of 20 J/cm2. During the irradiation cells were kept in phosphate-buffered saline on a cooling plate at 25 °C.
Skin explants – Dorsal tail skin was prepared from freshly sacrificed mice and residual subcutaneous fat was removed by scraping. Tissue explants were floated in culture medium containing 20 µM PQ for 48 h before analysis.
In vivo irradiation - Mice were anaesthetized and tails were sham irradiated or irradiated with 40 J/cm2 of UVA in accordance with the above noted animal protocol. 24 h after irradiation mice were sacrificed and skin samples were taken.
2.3. Immunofluorescence analysis
Mouse KC were grown on chamber slides and treated with PQ or UVA as indicated. Cells were fixed with 4% para-formaldehyde (20 min, RT), permeabilized with Triton X-100 (0.1%, 10 min, RT) and incubated overnight at 4 °C in phosphate-buffered saline (pH 7.2, 2% BSA) with the indicated primary antibodies. For in vivo experiments, the mice were sacrificed 24 h after irradiation with 40 J/cm2 UVA. The skin was separated from tails of mice and fixed with 10% formalin, paraffin embedded and microtome sections (4 µm) were immuno-stained. For tissue explant experiments, the skin was separated from sacrificed mice and cultured with paraquat in the indicated concentrations for 48 h, then immuno-stained.
As secondary antibodies goat anti-rabbit IgG (H+L), donkey anti-goat IgG (H+L), conjugated with Alexa Fluor dyes (Molecular Probes Eugene, OR, USA) were used at a dilution of 1:500. For imaging, an Olympus (Tokyo, Japan) AX 70 or when indicated a Zeiss LSM700 confocal laser microscope (Zeiss, Oberkochen, Germany) were used. All image analyses were performed under the same parameter settings. The counting analyses (50 cells per group, n=3) were performed by an observer blinded to the experimental condition.
For immunofluorescence, rabbit anti-laminB1 (ab16048; 1:1000), and anti-γH2AX (ab26350; 1:500), antibodies were obtained from Abcam Biochemicals (Cambridge, United Kingdom). Hoechst 33258 (Molecular Probes, Leiden, The Netherlands) was used to label the nuclei.
2.4. Microarrays and bioinformatic analysis
Total RNA was extracted from mouse keratinocytes after lysis with TriFast Reagent (VWR Peqlab) according to the manufacturer's instructions. The RNA cleanup and concentration was performed using the RNeasy MinElute Cleanup Kit (Qiagen) according to the manufactures's instructions. 200 ng of each sample were used for gene expression analysis with Affymetrix (Sta. Clara, CA, USA) Mouse gene 2.0 st arrays. Hybridization and scanning were performed according to manufacturer's protocol (http://www.affymetrix.com) and robust multi-array average (RMA) signal extraction and normalization were performed using custom chip description file. The experiment was performed on biological triplicate samples. Data are submitted to the GEO repository as (GSE84935). Gene sets (distinct gene symbol) with a (mean) RMA value of less than 50 in all conditions were excluded as not detectable. Moderated t-statistics (LIMMA) and a Benjamini-Hochberg multiple testing correction with a p-value <0.05 were performed using the online program Multi Experiment Viewer (MeV Version 4.9.0) which was also used to generate the expression heatmaps based on the centered log2 of the RMA values. The Euclidean Distance algorithm was used for hierarchical clustering.
2.5. Gene network and pathway analyses
Regulated genes were analyzed using of QIAGEN's Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, USA, www.qiagen.com/ingenuity). This software was used to predict which upstream regulatory events were likely to cause the observed gene expression changes between the analyzed groups and which signaling pathways were likely to be activated, both based on literature evidence. We restricted analysis to the molecule type “Genes, RNAs and Proteins”, used a z-score cutoff higher than 2 or lower than −2 which was considered to be significant [21] and sorted the upstream regulators or signaling pathways by p-value of overlap between the dataset genes and the known targets. If more than 5 regulators or pathways fulfilled the criteria, only the top 5 were depicted in the figure and only significantly changed upstream regulators and pathways were depicted. Heatmaps and activation z-scores were generated through the use of IPA (QIAGEN Redwood City, www.qiagen.com/ingenuity) and modified.
GO-Terms: We conducted Gene Ontology enrichment analyses using DAVID web server (http://david.abcc.ncifcrf.gov/) to investigate whether the regulated genes associated with certain biological functions would be overrepresented [22]. Gene Ontology analysis for the Biological Process_5 (BP_5) was performed. Enriched GO terms (p-value <0.05) were selected grouped in case of redundancy.
2.6. Comet assay and 8-OHdG assay
For DNA damage investigation, KCs were collected after 48 h PQ treatment or 24 h after UVA irradiation. For single cell gel electrophoresis (alkaline comet assay) cells were suspended at a density of 105 cells/ml in PBS (Ca2+and Mg2+ free) embedded in 1% low-melting point agarose gel on comet assay slides as described before [23]. The slides were kept for 1 h at 4 °C in lysis solution and then incubated in alkali solution (200 mM NaOH; 1 mM EDTA, pH >13) for 20 min to allow unwinding of DNA. Electrophoresis was carried out in pre-chilled alkali solution for 30 min at 20 V. The DNA was stained with DAPI. The extension of each comet was quantified using Image J software [24] and the tail moment, defined as the product of DNA in the tail and the mean distance of its migration in the tail was calculated, as well as the percentage of cells with DNA damage. Data are expressed as the mean value of 50 randomly selected cells.
To quantify oxidative DNA damage, total DNA was extracted using DNeasy Blood Mini Kit (Qiagen, Germany) on a Qiacube system (Qiagen, Germany) as described by the manufacturer and dissolved in water at 1 mg/ml. 8-OHdG levels were quantified with the OxiSelect Oxidative DNA Damage ELISA kit (Cell Biolabs, San Diego, CA) according to manufacturer's instructions. Samples were assayed in biological triplicates.
2.7. Quantitative PCR
RNA was isolated using the RNeasy 96 system (Invitrogen/Life Technologies, Grand Island, NY), and 400 ng of total RNA was reverse-transcribed with an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Quantitative PCR (qPCR) was performed using the LightCycler 480 and the LightCycler 480 SYBR Green I Master (Roche, Basel, Switzerland) with a standard protocol described before [25]. Relative quantification was performed according to the model of Pfaffl et al. [26] and the expression of the target genes was normalized to the expression of beta-2 microglobulin.
Primer sequences:
lamin B1 (Lmnb1: forward: 5‘-cagattgcccagctagaagc-3‘ reverse: 5′-ctgctccagctcttccttgt -3′); Cyclin dependent kinase1 (Cdk1: forward: 5′-ttgaaagcgaggaagaagga -3′ reverse: 5′- ccctggaggatttggtgtaa-3′); p53 (Trp53: forward: 5′-gcaactatggcttccacctg -3′ reverse: 5′-ctccgtcatgtgctgtgact -3′); cyclin-dependent kinase inhibitor 1A, p21cip1 (P21: forward: 5′-gtacttcctctgccctgctg -3′ reverse: 5‘-tctgcgcttggagtgataga -3′); arachidonate 12-lipoxygenase, 12R type (Alox12b: forward: 5′-gcctcgtggctgtagaactc -3′ reverse: 5′-atggtgctgaagcggtctac -3′); arachidonate lipoxygenase 3 (Aloxe3: forward: 5′-aggcacctgcctacaaacag -3′ reverse: 5′-atcagtgggcagaaagatgg -3′); beta-2 microglobulin (B2m forward: 5′ attcacccccactgagactg reverse: tgctatttctttctgcgtgc -3′).
2.8. Western blot
Cultured mouse KCs were washed twice with PBS and then harvested with lysis buffer (70 mM Tris-HCl, pH6.8, 1,1%SDS, 11,1% (v/v) glycerol, 0,005% bromophenol blue (BioRad Laboratories, Hercules, CA)) containing protease inhibitor cocktail (Abcam) and Pierce TM Phophatase Inhibitor Mini Tablets (Thermo Scientific) on ice and immediately sonicated. The protein content was measured using the micro BCA method (Thermo Scientific). Immunoblotting using antibodies for Lmnb1, Cdk1, p21 (ab16048, ab32384, ab109199; Abcam, all 1:1000), Active Caspase3, p53 (AF835, MAB1355; R&D systems, both 1:1000), KRT10 (PRB-159P; Covance, 1:1000) and GAPDH (5G4; HyTest, Turku, Finland; 1:2000) was performed as previously described [27].
As secondary antibody, goat anti-rabbit IgG-HRP (Biorad 170–6515) or sheep anti-mouse IgG-HRP (NA-931-V, GE Healthcare, Little Chalfont, UK; 1:10.000) were used and subsequent chemiluminescent quantification on ChemiDoc imager (Bio-Rad) was performed. The signal was measured with Image Lab 4.1 analysis software (Bio-Rad) and target bands were normalized to GAPDH.
2.9. Statistical analysis
Each experiment was performed independently at least three times. The data are expressed as the mean±standard deviation (SD). Statistical analysis was performed using ANOVA, followed by Student-Newman Keuls (SNK) post-hoc test. Statistical significance was considered at p<0.05. Statistical analysis of the microarray data is described above.
3. Results
Earlier findings suggested that autophagy conveys resistance to redox stress in epidermal cells, as autophagy deficient keratinocytes displayed increased protein crosslinking and lipid oxidation upon oxidizing ultraviolet radiation [12]. We here investigated whether the responses to paraquat (PQ; 1,1′-dimethyl-4,4′-bipyridinium), would differ between autophagy competent keratinocytes (KC) and autophagy deficient KC. PQ induces ROS generation and protein oxidation in murine keratinocytes [28] and is also widely used to induce mitochondrial dysfunction and finally cellular senescence. It was reported that PQ initiated autophagosome formation in various cell types [29].
We isolated primary KC from the tails of mice with cre-recombinase-targeted deletion of Atg7 [16] driven by the keratin K14 promoter (Atg7 f/f K14::cre, hereafter called “WT”) and their autophagy competent, cre negative siblings (Atg7 f/f, hereafter called knockout “KO”) which lack epidermal Atg7, as we described earlier [12]. First, we determined that 20 µM PQ would not affect cell viability or induce significant apoptosis within 48 h of treatment (Supplementary Fig. 1A-C). Similar to what we had observed earlier after UVA exposure, PQ treatment resulted in lipidation of LC3 to form LC3-II (Supplementary Fig. 1D) and also to autophagosome formation (Supplementary Fig. 1E), visualized in LC3-GFP transgenic KC, showing that PQ initiates autophagy [30]. PQ treatment also resulted in accumulation of crosslinked, high molecular weight (HMW) modifications of the autophagy adaptor protein Sequestosome 1 (Sqstm1, thereafter p62) in autophagy deficient KC (Supplementary Fig. 1E), in line with what we had observed with other oxidative stressors.
We extracted total RNA from WT and KO cells 48 h after exposure to 20 µM PQ and performed transcriptional profiling with gene level microarrays. Next, we determined the genes that were significantly regulated by Atg7 deficiency and/or PQ treatment, and then systematically characterized the cellular processes affected by the knockout with two bioinformatic approaches. First, we used “Ingenuity Pathways Analysis” software (IPA) to investigate whether the composition of the regulated gene groups suggested activation or inhibition of canonical signaling pathways or upstream transcriptional regulators. As a second method we used Gene Ontology (GO) analysis to characterize cellular processes affected by the knockout.
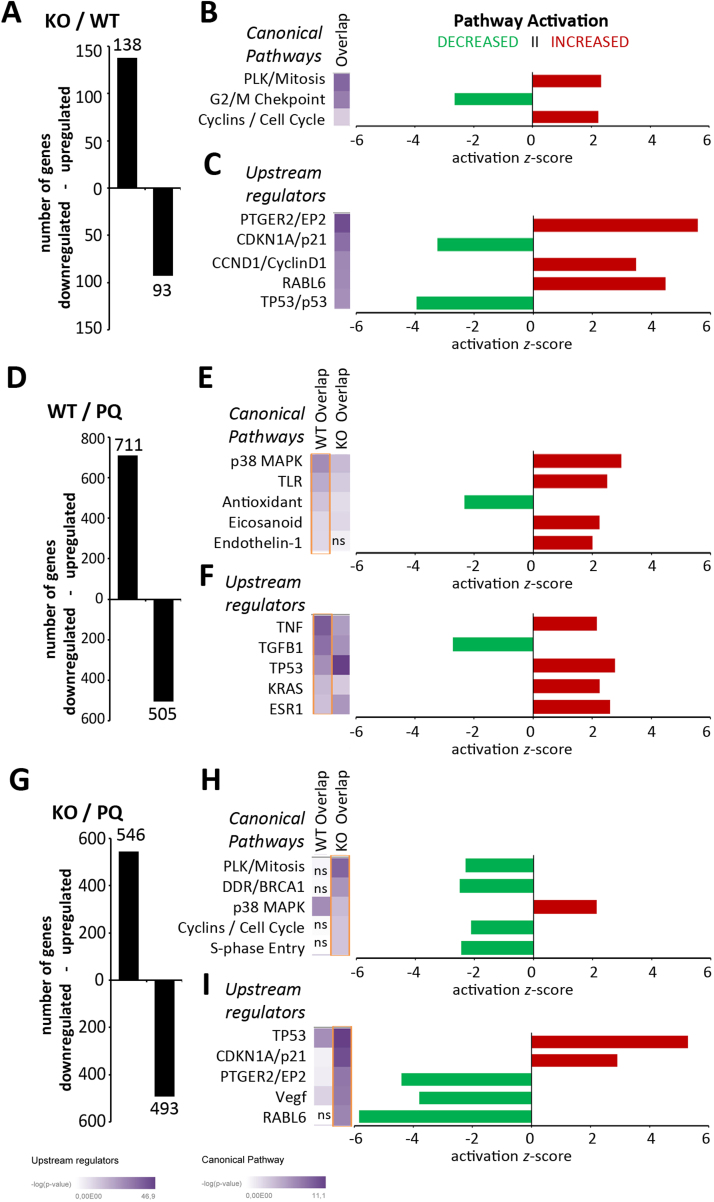
At baseline conditions the Atg7 knockout resulted in 138 genes being upregulated and 93 genes being downregulated more than two-fold compared to WT cells (Fig. 1A). We analyzed with IPA whether composition and regulation pattern of these gene groups indicated activation of canonical signaling pathways. In the Atg7 KO cells pro-mitotic and positive cell cycle regulatory pathways were found activated whereas DNA damage checkpoint regulation was decreased (Fig. 1B). The analysis of upstream regulatory elements suggested activation of prostaglandin E2 receptor (EP2), Cyclin D1 and RAS Oncogene Family-Like 6 (RABL6) signaling, and inhibition of p53 and cyclin-dependent kinase inhibitor 1 A (p21) signaling (Fig. 1C). The GO analysis of this dataset yielded similar results, as terms grouped as “cell cycle regulation”, “nucleosomal organization” and “DNA repair” were significantly regulated in the KO cells (Supplementary Fig. 2A). Atg7 can, independently of its function in autophagy, regulate cell cycle arrest via the p53/p21 axis in fibroblasts [31], and our findings indicate that Atg7 may likewise negatively regulate cell cycle signaling in cultured KC. A cell cycle analysis indeed showed a higher number of cells in S phase, we did however not observe increased proliferation of the KO cells within 48 h after seeding (Supplementary Fig. 2J and Supplementary Fig. 1). Of note, the increased EP2 signaling supports our previous finding that lipid mediators with EP2 agonistic activity were elevated in lipid extracts isolated from Atg7 deficient KC [12].
Fig. 1.
Gene regulation by paraquat in Atg7 bearing (WT) and Atg7 deficient (KO) mouse keratinocytes. Primary tail skin keratinocytes were treated with paraquat (20 µM) or left untreated for 48 h. Then RNA was extracted and global gene expression was assayed with microarray technology (Affymetrix mouse gene 2.0 ST). Numbers of up- and downregulated genes in untreated KO compared to WT KC (A), in PQ treated WT cells (D), and in PQ treated KO cells (G). Activation of canonical pathways and upstream regulators was analyzed using IPA software. Heatmaps depicting the p-value for activation of the pathways (B, E, H) and upstream regulators (C, F, I), respectively of both genotypes are shown. Heatmaps showing regulation by paraquat are shown for both genotypes next to each other, sorted for WT in E and F (yellow boxed) and for KO in H and I (yellow boxed). Next to the heatmaps the bar graphs indicate positive (red) or negative (green) activation z-scores for the listed pathways or upstream activators. Experiments were perfomed in triplicates; for A, D, G a p<0.05 (moderated t-statistics (LIMMA), Benjamini-Hochberg corrected) was regarded as significant. For B, C,E,F,H,I the top 5 pathways or regulators with a significant activation score (above 2 or below −2), are shown; pathways not significantly regulated in the other genotype are marked with “ns” within the heatmap. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we investigated the transcriptional responses to paraquat, which resulted in the upregulation of 711 and downregulation of 505 genes in WT cells, whereas in KO cells 546 genes were up- and 493 were downregulated (Fig. 1D, Supplementary Table 1). In Atg7 bearing KC, the PQ treatment activated the p38 stress signaling-, the TLR signaling-, the eicosanoid- and endothelin signaling pathways (Fig. 1E). Activation of p53, Tnfa, Kras and Esr1 and inhibition of Tgfb1 signaling were predicted as upstream regulatory events (Fig. 1F). The GO analysis likewise indicated reduced expression of cell cycle and proliferation genes, and revealed increased expression of lipid metabolism genes (Supplementary Fig. 2B).
Genes in the p38 Map kinase, eicosanoid, and Toll-like receptor (TLR) networks were regulated in a similar way by PQ in the knockouts (Fig. 1E,H) indicating that these signaling cascades were not affected by the knockouts. A major difference to the responses in the wildtype cells was however observed, as PQ treatment induced a transcriptional signature of strong cell cycle arrest and DNA damage signaling in the Atg7 deficient cells (Fig. 1H,I). This response included inhibition of cyclin and pro-mitotic gene expression and activation of p21 as upstream regulator. The GO analysis confirmed that cell cycle regulatory genes were decreased in the knockouts after PQ stress, and identified lipid metabolic genes to be enriched within the PQ regulated group in Atg7 KO cells (Supplementary Fig. 2C). Expression heatmaps showing mean induction by PQ compared to sham treated in both genotypes of genes within the p38, cyclin/cell cycle, DNA damage response, eicosanoid, Plk/mitotic and TLR networks, respectively, are shown in Supplementary Fig. 2D-I. Cell cycle analysis revealed a strong, but not complete reduction of cells in S-phase in both genotypes upon PQ treatment (Supplementary Fig. 2J).
A recent study had found that mitochondrial ROS overproduction in keratinocytes affected epidermal stem cell marker gene expression [32]. The KO keratinocytes displayed increased expression of Kif14, Kif11, Ccnb1, Cempe and Cdc20 in basal state, which was reduced to WT levels upon PQ stress, whereas in WT cells the expression of these genes was not affected by PQ (Supplementary Fig. 3A). The same was observed for the anti-senescence checkpoint kinases Bub1 and Bub1b (Supplementary Fig. 3B). At the same time, genes related to keratinocyte differentiation (Keratin 10, Klf9, CD36) of genes specific for basal epidermal KC (Dll1 and integrin alpha 6), and the KC hyperproliferation marker Krt6b were not significantly changed, with the exception of loricrin, which was slightly reduced in the KO (Supplementary Fig. 3C).
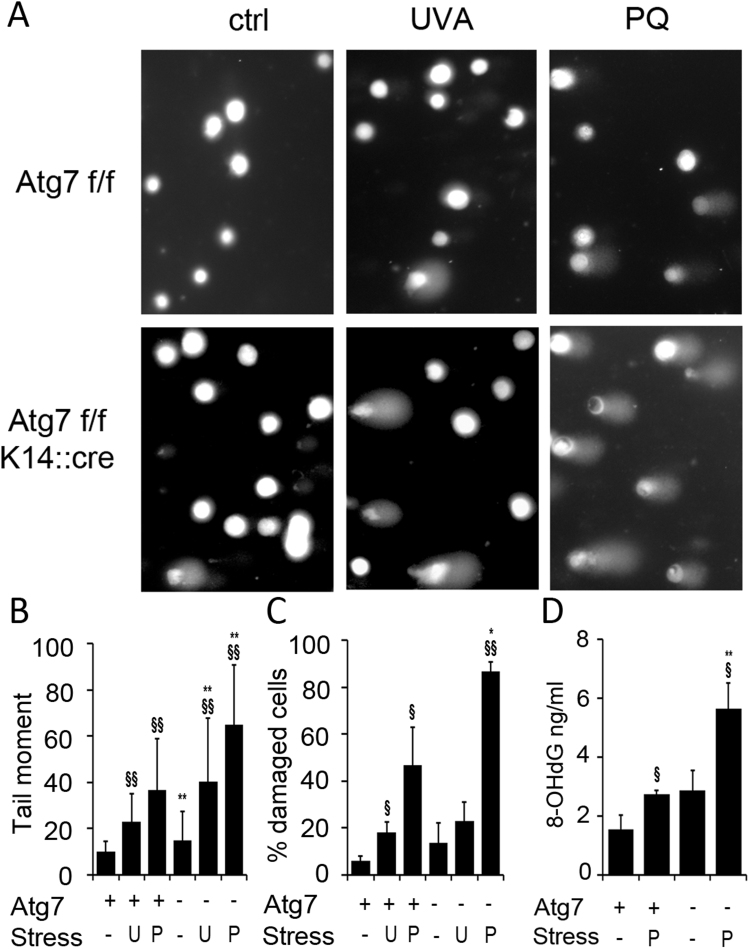
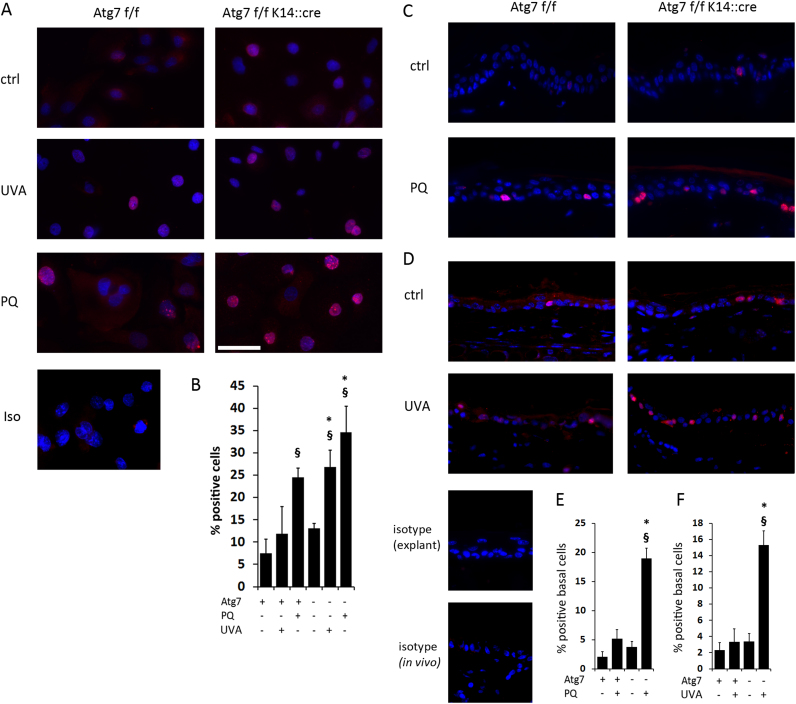
We next investigated whether biological effects of Atg7 deficiency suggested by bioinformatics and by previous work could be confirmed with complementary, biochemical and cell biological methods. First, we investigated whether the KO cells would show exacerbated DNA damage after ROS stress. We performed single cell electrophoresis (comet assay) to observe the total DNA damage in cells that had either been treated with PQ or had been exposed to 20 J/cm2 of UVA as alternative, relevant oxidative stressor (Fig. 2A), and calculated the mean tail moment as well as the percentage of cells showing DNA damage. PQ treatment and, to a lesser extent, UVA exposure led to significantly increased tail moment (Fig. 2B) and increase in percentage of comets in WT cells (Fig. 2C). Both tail moment and percentage of DNA damaged cells was significantly higher in Atg7- deficient KCs after PQ treatment compared with wild type KCs, while the increase in UVA induced damage was not significant in the KO cells (Fig. 2 B,C). UVA and UVA-induced singlet oxygen cause oxidative DNA damage, which is a major mutagenic factor [33], and 8-OHdG is one major DNA modification induced by UVA in basal layers of the skin [34]. As our earlier studies have suggested a redox imbalance in Atg7 KO we investigated whether the 8-OHdG levels would be increased in Atg7 KO and after PQ stress. PQ treatment induced significant increase in 8-OHdG levels in WT cells, and this was further significantly increased in autophagy deficient KC (Fig. 2D).
Fig. 2.
Autophagy deficiency increases oxidative DNA damage. Keratinocytes were either sham treated or exposed to PQ (20 µM) or UVA (20 J/cm2) and DNA damage assayed 24 h (UVA) or 48 h (PQ) after stress with comet assay and 8-OhdG immunoassay. (A) Representative images of the comet assay perfomed on Atg7 bearing and Atg7 deficient cells. (B) Each bar represents the mean average of the tail moment (product of DNA in the tail and the mean distance of its migration) of 50 randomly selected cells. (C) Percentage of cells displaying DNA damage (comets). (D) 8-OHdG levels in were quantified by immunoassay. Samples were assayed in biological triplicates. Error bars in B-D indicate +/− SD (n=3), Significant differences upon treatment are indicated by §§ (p<0.01) and § (p<0.05), differences between WT and KO are indicated by **(p<0.01) and *(p<0.05) and were determined by ANOVA, followed by Student-Newman Keuls (SNK) post-hoc test.
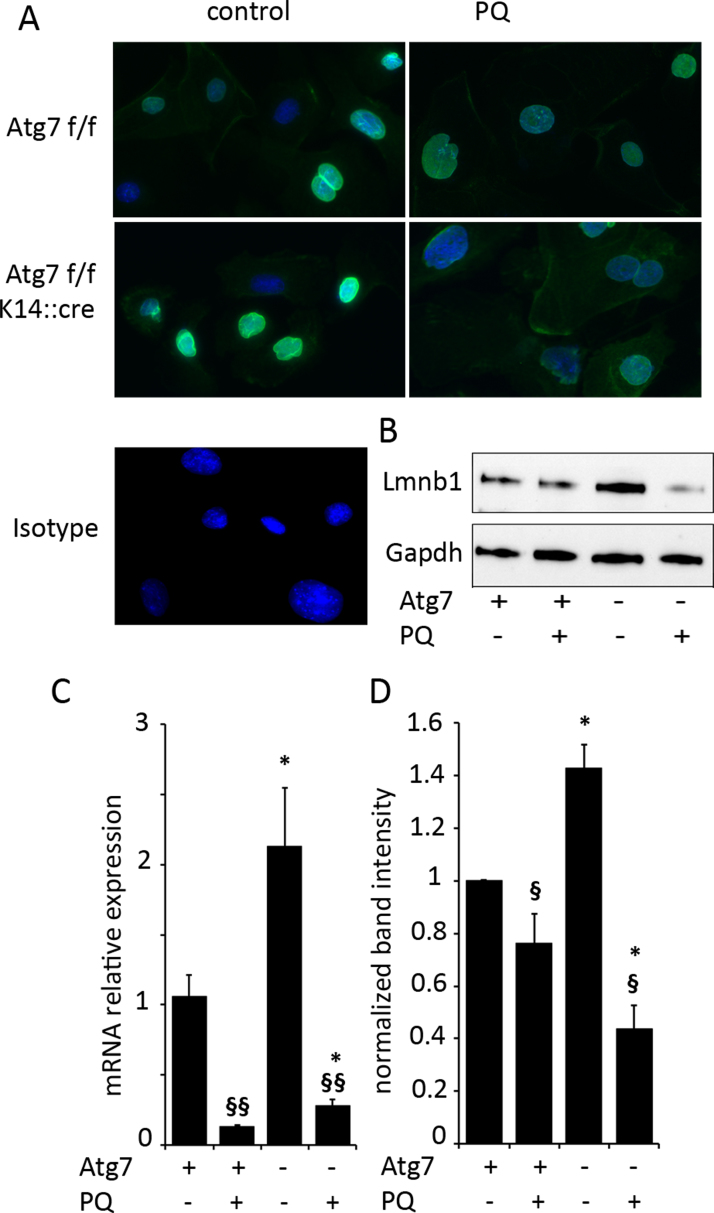
We next investigated how Atg7 deficiency would influence the degradation of the nuclear lamina protein LaminB1, which is decreased in early stages of DNA damage induced cellular senescence [35]. When we compared LaminB1 immunofluorescence (IF) staining in untreated WT and KO cells, the signal was stronger in the Atg7 deficient cells. Upon PQ exposure however, the LaminB1 staining was strongly decreased, and more markedly so in the KO than in the WT cells (Fig. 3A). To quantify this observation, we performed western blot (WB) and quantitative PCR (qPCR) analyses of LaminB1 expression. WB analysis confirmed the IF results on protein level (Fig. 3B, D), and qPCR showed that PQ strongly inhibits LaminB1 expression in both WT and KO (Fig. 3C), however the relative mRNA expression levels were not lower in treated KO than in WT. Atg7 may contribute directly to LaminB1 protein degradation, as has been described recently in an oncogenic stress model [36] and this may explain the increase in LaminB1 staining in untreated knockouts. Our data show that Atg7 is dispensable for degradation of LaminB1 upon PQ induced ROS stress and that LaminB1 protein is even stronger decreased in the knockouts.
Fig. 3.
Lamin B1 in Atg7 deficient KC – accumulation in absence of stress, increased degradation upon PQ exposure. (A) Keratinocytes were exposed to PQ (20 µM) and LaminB1 protein was assayed with immunofluorescence microscopy after 48 h. Representative images of three independent experiments are shown. Lamin B1 protein was assayed by immunoblotting (B) and mRNA relative expression, normalized to beta-2-microglobulin expression was quantified by qPCR (C). The protein band intensities, normalized to Gapdh, of three independent experiments are shown in (D). Significant differences upon treatment are indicated by §§ (p<0.01) and § (p<0.05), differences between WT and KO are indicated by **(p<0.01) and *(p<0.05) and were determined by ANOVA, followed by Student-Newman Keuls (SNK) post-hoc test.
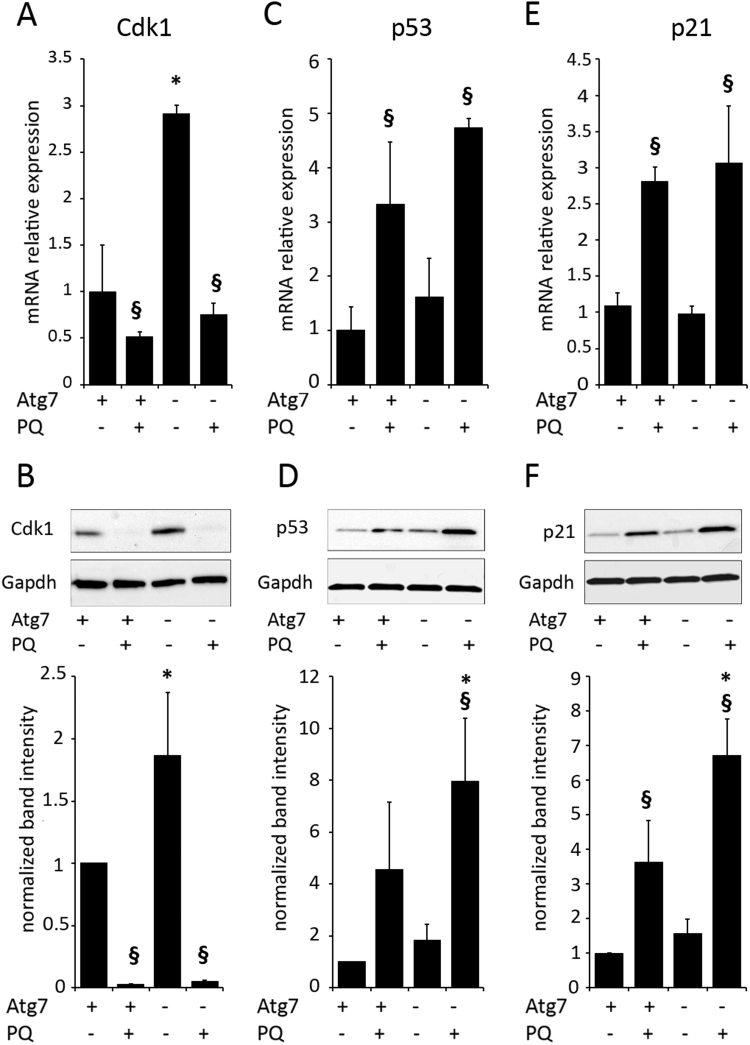
Next, we investigated whether Atg7 deficiency in PQ stressed cells would affect the expression of key growth arrest mediators that are active in promotion of cellular senescence. The microarray data had shown that p53, p21 and Cdk1 were regulated by PQ and the knockout, whereas p16 expression was below detection level. Using qPCR we could verify that PQ significantly decreased expression of Cdk1 in WT and KO cells, whereas the knockout cells showed higher baseline expression of Cdk1 (Fig. 4A). Using WB we could show that this was reflected on protein level, with a stronger Cdk1 signal in untreated KO and undetectable Cdk1 upon PQ treatment (Fig. 4B). p53 and the downstream mediator p21 were induced by PQ on mRNA and protein level, and the induction was increased in the knockouts on protein level for both proteins (Fig. 4C-F). In order to verify that the cell cycle arrest was not induced by keratinocyte differentiation, we performed a Keratin 10 immunoblot which showed that this protein was not expressed as consequence of the stress protocol (Supplementary Fig. 4). Interestingly, while expression levels of most differentiation genes were not affected by PQ treatment, several late cornified envelope (Lce) and small proline rich proteins (Sprr) gene class members of the epidermal differentiation complex (EDC) were highly induced by paraquat (not shown), in line with their recently identified redox dependent regulation via Nrf2 [37].
Fig. 4.
Cdk1, p53 and p21 regulation in autophagy deficient KC. Keratinocytes were exposed to PQ (20 µM) for 48 h and the relative mRNA expression of Cdk1 (A), p53 (C) and p21 (E) was quantified by pPCR (normalized to expression of beta-2-microglobulin). Protein levels of Cdk1 (B), p53 (D) and p21 (F) were assayed by immunoblotting and the protein band intensities, normalized to Gapdh, of three independent experiments are shown in the bar graphs below the blots (+/− SD of three independent experiments). Significant differences upon treatment are indicated by §§ (p<0.01) and § (p<0.05), differences between WT and KO are indicated by **(p<0.01) and *(p<0.05) and were determined by ANOVA, followed by Student-Newman Keuls (SNK) post-hoc test.
The central mediator that conveys DNA damage associated senescence - signaling is the histone H2AX, which exerts growth arrest via p53 [38] when phosphorylated on Serine 139 (called γH2AX). This modification is detectable at sites of (UV induced-) DNA damage, including strand breaks, stalled replication forks or oxidative DNA damage [39] and is required to maintain cellular senescence [40]. Thus, we investigated whether autophagy deficient KC would display differences in phosphorylation of H2AX compared to WT cells at baseline and upon PQ stress.
We did not observe a significant difference in the number of cells displaying γH2AX foci between non-stressed cultured WT and KO cells. Upon PQ exposure, there was a significant increase in positive nuclei in WT cells, which was again significantly higher in KO cells. In parallel, we exposed the cells to UVA, which did not cause a significant rise in positive nuclei in WT cells, but did so in KO cells (Fig. 5A, B).
Fig. 5.
Increased γH2AX positive nuclei in stressed, Atg7 deficient KC in vitro (PQ, UVA), in tissue explants (PQ) and in vivo (UVA). (A) Keratinocytes were either sham treated or exposed to PQ (20 µM) or UVA (20 J/cm2) and γH2AX immunofluorescence was assayed 24 h (UVA) or 48 h (PQ) after stress. (B) Quantification of the percentage of γH2AX positive cells from three independent experiments (Bars +/− SD). (C) Tail skin explants were exposed to PQ (20 µM, 48 h), and sections were analyzed with immunohistochemistry for γH2AX. (D) Tails were irradiated in vivo with UVA (40 J/cm2), sections were prepared after 24 h and analyzed for γH2AX. For C, D representative pictures are show from one of two experiments with each three animals per group. (E, F) Quantification of positive nuclei in one representative experiment (n=3 per group; error bars: +/- SD). Significant differences upon treatment are indicated by §§ (p<0.01) and § (p<0.05), differences between WT and KO are indicated by **(p<0.01) and *(p<0.05) and were determined by ANOVA, followed by Student-Newman Keuls (SNK) post-hoc test.
Next, we studied the formation of γH2AX foci in keratinocytes within the epidermis. We used an organ explant model to investigate the effect of PQ exposure (Fig. 5C, E), and we in vivo irradiated mouse tail skin with UVA (Fig. 5D, F) and then assessed γH2AX formation. In both model systems, the autophagy deficient keratinocytes within the basal layer of the epidermis displayed enhanced numbers of nuclei with γH2AX foci.
We had shown earlier that phospholipid oxidation is increased in Atg7 KO cells, and the bioinformatic analyses (Fig. 1, Suppl. Fig. 2) had indicated that lipid- and eicosanoid metabolism could be affected by the knockout and under PQ stress. We thus performed a screening of the major neutral lipid classes in WT and KO cells with or without PQ stress. The knockout alone resulted in a strong decrease in the percentage of triglycerides (TG) of total neutral lipids from 47% to 22%, and a concomitant increase in free fatty acids (FFA) from 17% to 33% and an increase in sterols from 36% to 45%. Treatment of WT cells with PQ had a similar effect, in decreasing the percentage of TG from 47 to 31 and increasing FFA from 17% to 35%, while the relative amount of sterol remained virtually unchanged. Treating the KO with PQ resulted in an additive effect and shifted the balance between TG and FFA further towards FFA with 38% and the Triglycerides down to 17%, while sterol levels also here remained unaffected by PQ treatment at 45% (Fig. 6A).
Fig. 6.
GC/MS analysis of neutral lipid distribution and of fatty acid species, Total lipids were extracted from cultured WT and KO kerationcytes treated for 48 h with PQ (20 µM) or sham treated. A semiquantitative analysis of neutral lipids (free fatty acids, sterols and triglycerides) was performed by GC/MS. Relative amount of each lipid species was calculated from integrated area ratios. (A) Distribution of TG, FFA and Sterols within each group. (B) Distribution of the major FFA species within each group (n=3). Significant differences upon treatment are indicated by § (p<0.05), differences between WT and KO are indicated by *(p<0.05) and were determined by ANOVA, followed by Student-Newman Keuls (SNK) post-hoc test.
We next investigated the composition of the FFA, and calculated the percentage of the Myristic acid (14:0), Palmitic acid (16:0), Palmitoleic acid (16:1), Stearic acid (18:0) and Oleic Acid (18:1) in the samples. The relative amounts of 14:0, 16:0 and 18:1 were increased, whereas 18:0 was strongly and 16:1 was slightly decreased in the KO cells. PQ treatment of the WT cells resulted in a very similar change in FFA pattern than did the knockout compared to untreated WT cells. PQ treatment of KO cells led to a further increase in 18:1, the 14:0 and 18:0 shares did not change, 16:1 levels were increased and 16:0 levels decreased (Fig. 6B).
4. Discussion
The declining capacity of aged cells and tissues to keep up redox homeostasis or to restore it after stress is a major cause for age-associated accumulation of damage and pathological changes. Cell and tissue aging is, in addition to sun exposure a risk factor to develop actinic keratoses and skin cancers [41], the incidence of which rises in the aging, sun exposed population. UV light can directly cause DNA photoproducts or elicit DNA modifications indirectly, as consequence of UV induced redox events, both causative for mutation and malignancy.
Aged cells frequently display defects in the mitochondrial electron transport chain which contributes to higher ROS levels, together with a decline in proteasomal function, impaired redox responses and reduced DNA damage repair capacity. Enhanced ROS contribute to chronologic aging [42] and ultraviolet light can additionally generate ROS, accelerate aging [43] and further impair the DNA repair [44]. Thus aged, UV exposed cells are especially prone to acquire deleterious DNA damage. Cells can respond to DNA- and protein damage by immediate repair, immediate cell death or forms cellular senescence. Cellular senescence withdraws the cells from the cell cycle and halts proliferation, and that allows repair of the damage over a longer timespan or the alerting of the immune system for clearance of the cell [45], [46].
Autophagy is one fallback repair mechanism for cells that have suffered major ROS damage which cannot be restored by proteasomal degradation or detoxification pathways alone [47]. ROS can activate autophagy via oxidation of cysteine residues in the autophagy core component Atg4 [48]. Under redox stress p53, Stat3, Nrf2 and NF-kB are regulators which coordinate the fine tuning of inflammation, antioxidant responses, DNA repair or cell death with autophagy [49].
4.1. Atg7 deficiency causes dysregulation of cell cycle and stem cell markers
When comparing global gene expression of WT to KO cells in absence of ROS stress, we observed a striking activation of pro-mitotic pathways and upstream regulators in the KOs, pointing to decreased p53/p21 signaling and increased cell cycle progression signaling. This finding was also reflected on protein level, with an increase in CDK1 and LaminB1 protein expression. Atg7 can, independently of its function in autophagy, regulate cell cycle arrest via the p53/p21 axis in fibroblasts [31]. Our findings indicate that Atg7 may likewise contribute to negatively regulating cell cycle gene transcription in cultured KC, which we found to be in S phase more frequently in the Atg7 knockouts. Further, autophagy can degrade the nuclear lamin B1 including associated chromatin fragments, by a nuclear to cytoplasmic transport mechanism in fibroblasts [36]. In cultured, unstressed keratinocytes we indeed observed accumulation of LaminB1, so this mechanism may be active to control the turnover of LaminB1 in homeostasis. Further, stem cell markers genes (Ccnb1, Kif11, Cenpe, Cdc20, Cdca5, Kif14) were higher expressed in the autophagy deficient cells in homeostasis. This phenotype had no consequence on proliferation of the cultured cells and, as discussed below, was more than reverted under redox stress.
Under ROS stress elicited by paraquat, pathway analysis of the transcription pattern identified p38 MAPK, TLR and eicosanoid signaling as activated in the WT keratinocytes. In the autophagy deficient cells PQ treatment additionally led to an increased p53 response and strong downregulation of mitosis- and cell cycle progression- associated genes (Cyclins A, -B, -E families, Plk1 and –dependent genes, Cdk1 and Cdc25 B, -C among others). The expression signature was typical for cell cycle arrest at G2/M and G1/S checkpoints, indicative of severe DNA damage signaling in the KOs under ROS stress. This was confirmed also on protein level, as p53 and p21 were increased compared to stressed WT cells, and CDK1 was not detectable in the stressed KO, which usually leads to deep cell cycle arrest. Further, LaminB1 protein was strongly decreased in the autophagy deficient and stressed cells as compared to stressed WT cells. Thus autophagy is neither required for stress induced degradation of LaminB1 nor for activation of cell cycle arrest or cellular senescence via the p53/p21 axis and via Cdk1 in primary murine keratinocytes.
In autophagy deficient murine fibroblasts ROS, DNA damage and γH2AX signal were found increased [31] similar to our findings in KC. In contrast to KC there was an increase in proliferation in FB that suggests context- and cell type specificity for interaction of autophagy with p53 signaling. Indeed, epithelial cells display unique features in the context of ROS induced cellular senescence, including DNA single strand break accumulation in nuclear foci, p38 MAPK activation, lack of p53 Ser15 phosphorylation and an inactive ATM/ATR dependent damage repair [50].
4.2. Increased DNA damage and senescence markers in stressed autophagy deficient KC
Both PQ and UVA can elicit oxidative DNA damage and strand breaks [51], [52], and both types of damage were strongly exacerbated in Atg7 deficient, stressed KC.
In the response to UV radiation of various wavelengths γH2AX can appear in association with nucleotide excision repair, at stalled replication forks, in S – phase apoptosis [53], and at sites of repair of oxidative DNA damage [39] in addition to its traditionally ascribed role in DSB repair. Autophagy can clear nuclear remnants positive for γH2AX that arise upon severe cell cycle disturbance [54]. Furthermore, autophagy directs the nucleotide excision repair complex to sites of UV induced DNA damage [55], a process involving p62 [56] thus, disturbed repair or clearance of damaged DNA may contribute to the increase in actual DNA damage we observed.
γH2AX is involved in mitotic checkpoint control, the maintenance of stem cells and in cellular senescence [40], where it exerts growth arrest via p53. It also promotes secretion of senescence associated cyto/chemokines termed the SASP [38], which also we have found increased in stressed KO cells.
The combination of γH2AX and p53/p21 signals, downregulation of Cdc2/Cdk1 and the enhanced breakdown of LaminB1 in the absence of cell death we found in the stressed, autophagy deficient keratinocytes points to enhanced cellular senescence in vitro. In vivo, transit- amplifying (TA) epidermal keratinocytes are replenished from epidermal stem cells, and it has been proposed that increased TA cell senescence can drive reduction of stem cells and contribute to epidermal thinning [32], [57]. Further research will identify whether autophagy deficiency or – impairment would affect the biology of epidermal stem cells and contributes to skin aging in vivo as our data suggest. Autophagy is a critical factor that acts tumor suppressive in nontransformed cells, but promotes tumors by allowing alternative nutrient utilization [58]. This also was observed in epidermal tumors [59], and as cellular ROS increase, autophagic activity declines in aging and we found increased DNA damage in absence of autophagy, epidermal cell transformation in (photo) aging requires further research.
4.3. Atg7 deficiency affects neutral lipid composition in cultured KC
Autophagy as a metabolic master regulator can also affect lipid metabolism, as it can facilitate the lipolysis but also lipogenesis, thus controlling the levels of FFA and triglycerides [60], [61]. Lipid metabolic genes (Cyp51, Idi1, Hsd17b7) were among the top regulated genes in Atg7 deficient stressed KC, and interestingly these genes were also found strongly induced in mitochondrial dysfunction models [62]. Further ELOVL6, which converts C16 to C18 FA and may regulate mitochondrial function by stearylation of the transferrin receptor [63] was induced in the knockouts and also by PQ.
Both palmitic acid (16:0) and oleic acid (18:1) can induce autophagy [64] and interestingly, we have found these two FFA to accumulate in autophagy deficient cells, and 18:1 to increase even more under redox stress. Intracellular accumulation of oleic acid induces p53, and which may feed into the increased p53 activity in the stressed KO cells [65]. The beneficial effects of dietary oleic acid supplementation have recently been proposed to be dependent on their autophagy agonistic effect [66]. Conversely, when FFA are neither stored in TG nor degraded by autophagy in aging cells, their lipotoxic effects may become dominant [67], [68], [69] and contribute to inflammation in senescent cells [70].
Prostaglandin E2 receptor (EP2) signaling was activated in KO cells. This observation is in line with our previous finding, that the Atg7 deficient cells accumulated oxidized lipid mediators like 1-palmitoyl-2-epoxyisoprostane E2-sn-glycero-3-phosphorylcholine (PEIPC) [12] which are endogenous agonists of EP2 [71]. As EP2 signaling contributes to senescence in fibroblasts [72], and EP2 deletion reduces oxidative damage and severity of Alzheimer's disease [73], which suggests EP2 signaling as a potential link between defective autophagy and senescence/aging. Recently, it was shown, that in aged dermal fibroblasts and brain tissue the autophagic activity was declined [74], underlining the potential impact of autophagy and lipid mediators in age associated diseases.
Taken together, our data show that multiple manifestations of ROS stress and senescence in keratinocytes are affected by autophagy, adding evidence that functional autophagy protects cells from damage triggered by stress that causes - or is associated with - aging. In the absence of Atg7/autophagy cells display a lipid composition and lipid signaling that may not only correlate to cellular redox stress but also promote cellular aging. This adds to our previous finding that autophagy is induced by- and degrades oxidized phospholipids, which as danger associated molecular patterns (DAMPS) affect responses to aging promoting stress.
Autophagy deficient KC are highly susceptible to redox stress - induced p53- and DNA damage signaling. Thus UVA, the most ubiquitous redox stressor for the skin and elevated ROS in aged cells may be much more mutagenic when autophagy is deficient or impaired.
Conflict of interest
JG is co-founder of Evercyte GmbH and TAmiRNA GmbH.
Acknowledgements
We are grateful to Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) and Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) for providing ATG7-floxed and GFP-LC3 transgenic mice, respectively. The financial support of the Federal Ministry of Science, Research, and Economy (BMWFW) of Austria and the National Foundation for Research, Technology, and Development of Austria is gratefully acknowledged.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2016.12.015.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Schafer M., Werner S. Nrf2--A regulator of keratinocyte redox signaling. Free Radic. Biol. Med. 2015;88:243–252. doi: 10.1016/j.freeradbiomed.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 2.Kammeyer A., Luiten R.M. Oxidation events and skin aging. Ageing Res. Rev. 2015;21:16–29. doi: 10.1016/j.arr.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Latonen L., Laiho M. Cellular UV damage responses--functions of tumor suppressor p53. Biochim. Biophys. Acta. 2005;1755:71–89. doi: 10.1016/j.bbcan.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Reeg S., Grune T. Protein oxidation in aging: does it play a role in aging progression? Antioxid. Redox Signal. 2015;23:239–255. doi: 10.1089/ars.2014.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickering A.M., Linder R.A., Zhang H., Forman H.J., Davies K.J. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J. Biol. Chem. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohn A., Konig J., Grune T. Protein oxidation in aging and the removal of oxidized proteins. J. Proteom. 2013;92:132–159. doi: 10.1016/j.jprot.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Pajares M., Jimenez-Moreno N., Dias I.H., Debelec B., Vucetic M., Fladmark K.E. Redox control of protein degradation. Redox Biol. 2015;6:409–420. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic. Biol. Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baraibar M.A., Friguet B. Oxidative proteome modifications target specific cellular pathways during oxidative stress, cellular senescence and aging. Exp. Gerontol. 2013;48:620–625. doi: 10.1016/j.exger.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Rubinsztein D.C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Kroemer G. Autophagy: a druggable process that is deregulated in aging and human disease. J. Clin. Investig. 2015;125:1–4. doi: 10.1172/JCI78652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y., Zhang C.F., Rossiter H., Eckhart L., Konig U., Karner S. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J. Investig. Dermatol. 2013;133:1629–1637. doi: 10.1038/jid.2013.26. [DOI] [PubMed] [Google Scholar]
- 13.Hudson L., Bowman A., Rashdan E., Birch-Machin M.A. Mitochondrial damage and ageing using skin as a model organ. Maturitas. 2016 doi: 10.1016/j.maturitas.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Velarde M.C., Flynn J.M., Day N.U., Melov S., Campisi J. Mitochondrial oxidative stress caused by Sod2 deficiency promotes cellular senescence and aging phenotypes in the skin. Aging. 2012;4:3–12. doi: 10.18632/aging.100423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung T., Hohn A., Catalgol B., Grune T. Age-related differences in oxidative protein-damage in young and senescent fibroblasts. Arch. Biochem. Biophys. 2009;483:127–135. doi: 10.1016/j.abb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossiter H., Konig U., Barresi C., Buchberger M., Ghannadan M., Zhang C.F. Epidermal keratinocytes form a functional skin barrier in the absence of Atg7 dependent autophagy. J. Dermatol. Sci. 2013;71:67–75. doi: 10.1016/j.jdermsci.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossiter H., Barresi C., Pammer J., Rendl M., Haigh J., Wagner E.F. Loss of vascular endothelial growth factor a activity in murine epidermal keratinocytes delays wound healing and inhibits tumor formation. Cancer Res. 2004;64:3508–3516. doi: 10.1158/0008-5472.CAN-03-2581. [DOI] [PubMed] [Google Scholar]
- 20.Gruber F., Bicker W., Oskolkova O.V., Tschachler E., Bochkov V.N. A simplified procedure for semi-targeted lipidomic analysis of oxidized phosphatidylcholines induced by UVA irradiation. J. Lipid Res. 2012;53:1232–1242. doi: 10.1194/jlr.D025270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang d.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Song X., Mosby N., Yang J., Xu A., Abdel-Malek Z., Kadekaro A.L. Alpha-MSH activates immediate defense responses to UV-induced oxidative stress in human melanocytes. Pigment Cell Melanoma Res. 2009;22:809–818. doi: 10.1111/j.1755-148X.2009.00615.x. [DOI] [PubMed] [Google Scholar]
- 24.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruber F., Mayer H., Lengauer B., Mlitz V., Sanders J.M., Kadl A. NF-E2-related factor 2 regulates the stress response to UVA-1-oxidized phospholipids in skin cells. FASEB J. 2010;24:39–48. doi: 10.1096/fj.09-133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2003–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rendl M., Ban J., Mrass P., Mayer C., Lengauer B., Eckhart L. Caspase-14 expression by epidermal keratinocytes is regulated by retinoids in a differentiation-associated manner. J. Investig. Dermatol. 2002;119:1150–1155. doi: 10.1046/j.1523-1747.2002.19532.x. [DOI] [PubMed] [Google Scholar]
- 28.Black A.T., Gray J.P., Shakarjian M.P., Laskin D.L., Heck D.E., Laskin J.D. Increased oxidative stress and antioxidant expression in mouse keratinocytes following exposure to paraquat. Toxicol. Appl. Pharm. 2008;231:384–392. doi: 10.1016/j.taap.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Polo R.A., Niso-Santano M., Ortiz-Ortiz M.A., Gomez-Martin A., Moran J.M., Garcia-Rubio L. Inhibition of paraquat-induced autophagy accelerates the apoptotic cell death in neuroblastoma SH-SY5Y cells. Toxicol. Sci. 2007;97:448–458. doi: 10.1093/toxsci/kfm040. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo A.A. Vol. 12. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) pp. 1–222. (Autophagy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee I.H., Kawai Y., Fergusson M.M., Rovira I.I., Bishop A.J., Motoyama N. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velarde M.C., Demaria M., Melov S., Campisi J. Pleiotropic age-dependent effects of mitochondrial dysfunction on epidermal stem cells. Proc. Natl. Acad. Sci. USA. 2015;112:10407–10412. doi: 10.1073/pnas.1505675112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kielbassa C., Roza L., Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 34.Agar N.S., Halliday G.M., Barnetson R.S., Ananthaswamy H.N., Wheeler M., Jones A.M. The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations: a role for UVA in human skin carcinogenesis. Proc Natl. Acad. Sci. USA. 2004;101:4954–4959. doi: 10.1073/pnas.0401141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freund A., Laberge R.M., Demaria M., Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou Z., Xu C., Donahue G., Shimi T., Pan J.A., Zhu J. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafer M., Farwanah H., Willrodt A.H., Huebner A.J., Sandhoff K., Roop D. Nrf2 links epidermal barrier function with antioxidant defense. EMBO Mol. Med. 2012;4:364–379. doi: 10.1002/emmm.201200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodier F., Coppe J.P., Patil C.K., Hoeijmakers W.A., Munoz D.P., Raza S.R. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greinert R., Volkmer B., Henning S., Breitbart E.W., Greulich K.O., Cardoso M.C. UVA-induced DNA double-strand breaks result from the repair of clustered oxidative DNA damages. Nucleic Acids Res. 2012;40:10263–10273. doi: 10.1093/nar/gks824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turinetto V., Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam M., Ratner D. Cutaneous squamous-cell carcinoma. N. Engl. J. Med. 2001;344:975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 42.Balaban R.S., Nemoto S., Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Tulah A.S., Birch-Machin M.A. Stressed out mitochondria: the role of mitochondria in ageing and cancer focussing on strategies and opportunities in human skin. Mitochondrion. 2013;13:444–453. doi: 10.1016/j.mito.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Courdavault S., Baudouin C., Charveron M., Canguilhem B., Favier A., Cadet J. Repair of the three main types of bipyrimidine DNA photoproducts in human keratinocytes exposed to UVB and UVA radiations. DNA Repair. 2005;4:836–844. doi: 10.1016/j.dnarep.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serrano M., Blasco M.A. Cancer and ageing: convergent and divergent mechanisms. Nat. Rev. Mol. Cell Biol. 2007;8:715–722. doi: 10.1038/nrm2242. [DOI] [PubMed] [Google Scholar]
- 47.Hohn A., Jung T., Grune T. Pathophysiological importance of aggregated damaged proteins. Free Radic. Biol. Med. 2014;71:70–89. doi: 10.1016/j.freeradbiomed.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 48.Scherz-Shouval R., Shvets E., Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3:371–373. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 49.Pietrocola F., Izzo V., Niso-Santano M., Vacchelli E., Galluzzi L., Maiuri M.C. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Nassour J., Martien S., Martin N., Deruy E., Tomellini E., Malaquin N. Defective DNA single-strand break repair is responsible for senescence and neoplastic escape of epithelial cells. Nat. Commun. 2016;7:10399. doi: 10.1038/ncomms10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribas G., Frenzilli G., Barale R., Marcos R. Herbicide-induced DNA damage in human lymphocytes evaluated by the single-cell gel electrophoresis (SCGE) assay. Mutat. Res. 1995;344:41–54. doi: 10.1016/0165-1218(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 52.Chen Q., Niu Y., Zhang R., Guo H., Gao Y., Li Y. The toxic influence of paraquat on hippocampus of mice: involvement of oxidative stress. Neurotoxicology. 2010;31:310–316. doi: 10.1016/j.neuro.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Cleaver J.E. gammaH2Ax: biomarker of damage or functional participant in DNA repair "all that glitters is not gold!". Photochem. Photobiol. 2011;87:1230–1239. doi: 10.1111/j.1751-1097.2011.00995.x. [DOI] [PubMed] [Google Scholar]
- 54.Rello-Varona S., Lissa D., Shen S., Niso-Santano M., Senovilla L., Marino G. Autophagic removal of micronuclei. Cell Cycle. 2012;11:170–176. doi: 10.4161/cc.11.1.18564. [DOI] [PubMed] [Google Scholar]
- 55.Qiang L., Zhao B., Shah P., Sample A., Yang S., He Y.Y. Autophagy positively regulates DNA damage recognition by nucleotide excision repair. Autophagy. 2016;12:357–368. doi: 10.1080/15548627.2015.1110667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hewitt G., Carroll B., Sarallah R., Correia-Melo C., Ogrodnik M., Nelson G. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy. 2016:1–14. doi: 10.1080/15548627.2016.1210368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castilho R.M., Squarize C.H., Chodosh L.A., Williams B.O., Gutkind J.S. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galluzzi L., Pietrocola F., Bravo-San Pedro J.M., Amaravadi R.K., Baehrecke E.H., Cecconi F. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cosway B., Lovat P. The role of autophagy in squamous cell carcinoma of the head and neck. Oral Oncol. 2016;54:1–6. doi: 10.1016/j.oraloncology.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 61.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroder T., Kucharczyk D., Bar F., Pagel R., Derer S., Jendrek S.T. Mitochondrial gene polymorphisms alter hepatic cellular energy metabolism and aggravate diet-induced non-alcoholic steatohepatitis. Mol. Metab. 2016;5:283–295. doi: 10.1016/j.molmet.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan C.Y., Virtue S., Bidault G., Dale M., Hagen R., Griffin J.L. Brown adipose tissue thermogenic capacity is regulated by Elovl6. Cell Rep. 2015;13:2039–2047. doi: 10.1016/j.celrep.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niso-Santano M., Malik S.A., Pietrocola F., Bravo-San Pedro J.M., Marino G., Cianfanelli V. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park E.J., Lee A.Y., Chang S.H., Yu K.N., Kim J.H., Cho M.H. Role of p53 in the cellular response following oleic acid accumulation in chang liver cells. Toxicol. Lett. 2014;224:114–120. doi: 10.1016/j.toxlet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 66.Enot D.P., Niso-Santano M., Durand S., Chery A., Pietrocola F., Vacchelli E. Metabolomic analyses reveal that anti-aging metabolites are depleted by palmitate but increased by oleate in vivo. Cell Cycle. 2015;14:2399–2407. doi: 10.1080/15384101.2015.1064206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic control of autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tchkonia T., Morbeck D.E., von Z.T., van D.J., Lustgarten J., Scrable H. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo W., Pirtskhalava T., Tchkonia T., Xie W., Thomou T., Han J. Aging results in paradoxical susceptibility of fat cell progenitors to lipotoxicity. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1041–E1051. doi: 10.1152/ajpendo.00557.2006. [DOI] [PubMed] [Google Scholar]
- 70.Suganami T., Nishida J., Ogawa Y. A paracrine loop between adipocytes and macrophages aggravates inflammatory changes: role of free fatty acids and tumor necrosis factor alpha. Arterioscler. Thromb. Vasc. Biol. 2005;25:2062–2068. doi: 10.1161/01.ATV.0000183883.72263.13. [DOI] [PubMed] [Google Scholar]
- 71.Li R., Mouillesseaux K.P., Montoya D., Cruz D., Gharavi N., Dun M. Identification of prostaglandin E2 receptor subtype 2 as a receptor activated by OxPAPC. Circ. Res. 2006;98:642–650. doi: 10.1161/01.RES.0000207394.39249.fc. [DOI] [PubMed] [Google Scholar]
- 72.Martien S., Pluquet O., Vercamer C., Malaquin N., Martin N., Gosselin K. Cellular senescence involves an intracrine prostaglandin E2 pathway in human fibroblasts. Biochim. Biophys. Acta. 1831;1217–27:2013. doi: 10.1016/j.bbalip.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 73.Liang X., Wang Q., Hand T., Wu L., Breyer R.M., Montine T.J. Deletion of the prostaglandin E2 EP2 receptor reduces oxidative damage and amyloid burden in a model of Alzheimer's disease. J. Neurosci. 2005;25:10180–10187. doi: 10.1523/JNEUROSCI.3591-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ott C., Konig J., Hohn A., Jung T., Grune T. Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 2016;10:266–273. doi: 10.1016/j.redox.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material