Abstract
Four Aeromonas strains from clinical and environmental samples differed from known species on the basis of rpoD gene sequence. Multilocus phylogenetic analysis and in silico DNA-DNA hybridization confirmed them as four new species even though their 16S rRNA gene sequence similarity with their closest relatives was >98.7%, as occurred for other Aeromonas spp.
Keywords: Aeromonas, faeces, new species, oysters, taxonomy, water
The genus Aeromonas belongs to the Family Aeromonadaceae and includes oxidase-positive, facultatively anaerobic, Gram-negative bacilli [1], [2]. To date, 32 species are recognized, and 19 so far have been implicated in human diarrhoea, bacteraemia or wound infections, and are considered human opportunistic pathogens [2], [3], [4], [5]. New data have demonstrated that Aeromonas spp. are true enteropathogens [5].
Recently a group of four Aeromonas strains could not be assigned to any species. Two of them, 1178CT and 113634T, were recovered from patients with diarrhoea from two Spanish hospitals; stool samples were collected in plastic containers. Strain 1178CT was isolated, in the absence of other microbes, from the diarrhoeic faeces of a 45-year-old man with vomiting and fever over 2 months. Strain 113634T was recovered from a 32-year-old woman with abdominal pain lasting several days. One (AOSE3-14AT) of the other two strains was isolated from Crassostrea gigas harvested from Alfacs Bay (River Ebro Delta, Spain), and the other (AE207T) was isolated from Lake Pyhäjärvi water (Finland).
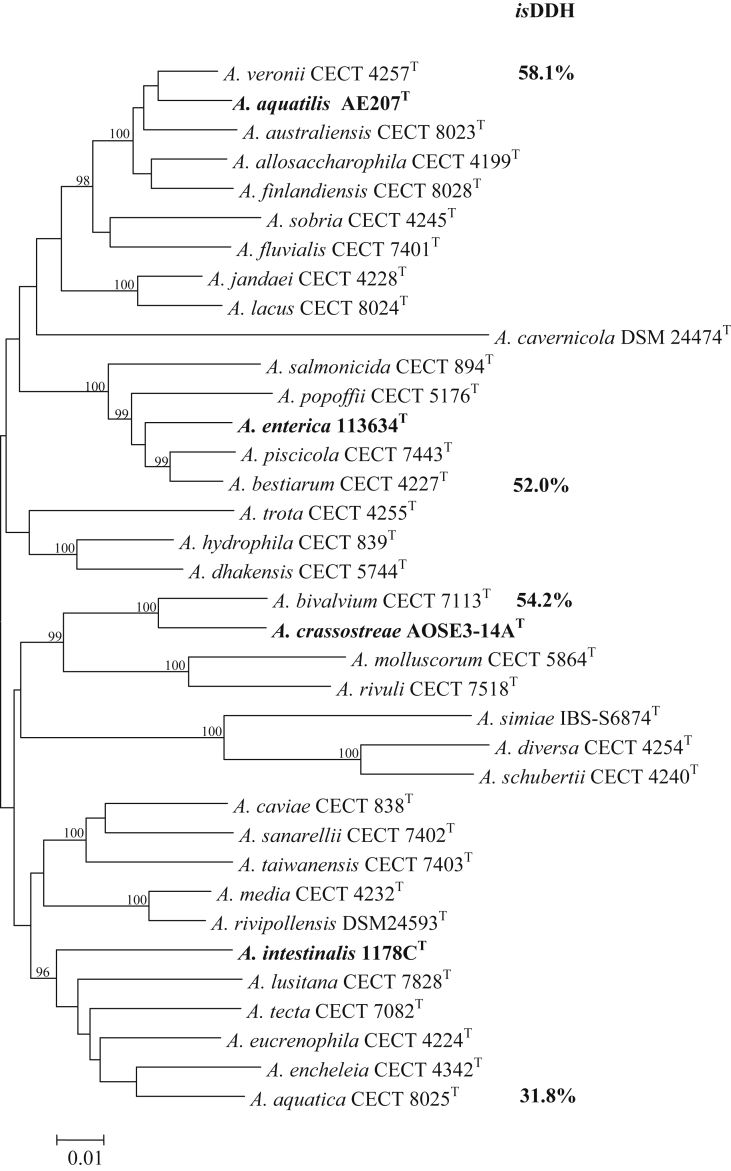
The phylogenetic tree constructed with the rpoD gene, which is a gene we have described earlier as a first-line tool for the identification of Aeromonas spp. [2], [6], indicated that the four strains formed independent branches from the rest of species, and this was confirmed with the tree constructed with the concatenated sequences of six housekeeping genes (rpoD, gyrB, gyrA, recA, dnaJ and dnaX, 3558 bp) (Fig. 1), all performed as previously described [3]. The interspecies 16S rRNA gene sequence (1367 bp) similarity of the four strains with the types of their closest Aeromonas species was >98.7% (Supplementary Fig. S1). This is in agreement with the high values of >98.7 to 100% observed among several accepted Aeromonas species [2], [3], [6], [7]. The closest species to clinical strain 1178CT was A. jandaei (98.85% similarity), while for strain 113634T they were A. salmonicida (99.86%), A. bestiarum (99.71%) and A. piscicola (99.71%). Strain AOSE3-14AT grouped with A. encheleia (99.93% similarity) and AE207T with A. tecta (99.50% similarity) (Supplementary Fig. S1). The genomes of the four new species were sequenced, and the in silico DNA-DNA hybridization (isDDH) values with the genomes of the type strains of the closest species was calculated as described elsewhere [3]. The four strains showed isDDH values of <70%, which confirmed them as four new species (Fig. 1 and Supplementary Fig. S1).
FIG. 1.
Neighbour-joining phylogenetic tree obtained with concatenated sequences of six housekeeping genes (rpoD, gyrB, gyrA, recA, dnaJ and dnaX, 3558 bp) showing position of four new Aeromonas spp. (bold) relative to 32 Aeromonas spp. Numbers at nodes represent bootstrap percentages obtained by repeating analysis 1000 times. Scale bar = 0.01 estimated substitution per site. In silico DNA-DNA hybridization (isDDH) represents genetic similarity obtained between genome of each of four new species and genomes of their closest neighbour species.
The clinical strains 1178CT and 113634T were initially identified with MicroScan W/A and Vitek II, respectively. In both cases, the identifications obtained were erroneous and masked these new species under the species A. hydrophila and A. sobria, respectively.
More than one phenotypical characteristic distinguished the four new species from their closest relatives. The most important were the nonacid production from d-sucrose but production from lactose by strain 1178CT, the production of acid from l-fucose by 113634T, the use dl-lactate by AOSE3-14AT and the mannitol-negative reaction of strain AE207T that also showed to be susceptible to the vibriostatic agent O/129.
The names proposed are as follows: ‘Aeromonas intestinalis’ (in.tes.ti.na′lis, N.L. fem. adj. intestinalis, ‘pertaining to the intestine’), ‘Aeromonas enterica’ (en.te′ri.ca, Gr. n. enteron, ‘gut, bowel, intestine’; L. fem. suff. -ica, suffix used with the sense ‘pertaining to’; N.L. fem. adj. enterica, ‘pertaining to intestine’), ‘Aeromonas crassostreae’ (crass.os′tre.ae, N.L. gen. n. crassostreae, ‘of the oyster genus Crassostrea’) and ‘Aeromonas aquatilis’ (a.qua′ti.lis, L. fem. adj. aquatilis ‘aquatic, growing in water’).
Nucleotide sequence accession number
The GenBank/European Molecular Biology Laboratory/DNA Data Bank of Japan accession numbers of the 16S rRNA gene sequences of strains 1178CT (= CECT 8980T = LMG 29048T), 113634T (= CECT 8981T = LMG 29049T), AOSE3-14AT (= CECT 8982T = LMG 29050T) and AE207T (= CECT 8026T = LMG 26714T) are LT630759, LT630760, LT630761 and LT630765, respectively. The rpoD, gyrB, gyrA, recA, dnaJ and dnaX of the other strains of the four novel species have also been deposited under the accession numbers LT630710–LT630712 and LT630716, LT630717–LT630719 and LY630723, LT630724–LT630726 and LT630730, LT630731–LT630733 and LT630737, LT630738–LT630740 and LT630744, LT630745–LT630747 and LT630751, respectively.
Deposit in a culture collection
All the type strains have been deposited in the Culture Collections of Spain (CECT) and Belgium (LMG): ‘A. intestinalis’ 1178CT (= CECT 8980T = LMG 29048T), ‘A. enterica’ 113634T (= CECT 8981T = LMG 29048T), ‘A. aquatilis’ AE207T (= CECT 8026T = LMG 26714T) and ‘A. crassostreae’ AOSE3-14AT (= CECT 8982T = LMG 29050T).
Acknowledgements
This study was supported by the projects JPIW2013-095-C03-03 of MINECO (Spain) and AQUAVALENS of the Seventh Framework Program (FP7/2007-2013) grant agreement 311846 from the European Union. We thank A. Oren, Hebrew University of Jerusalem, for supervising and correcting etymology of the species names.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.nmni.2016.11.019.
Conflict of Interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Janda J.M., Abbott S.L. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueras M.J., Beaz-Hidalgo R. Aeromonas infections in humans. In: Graf J., editor. Aeromonas. Caister Academic Press; Norfolk: 2015. pp. 65–108. [Google Scholar]
- 3.Beaz-Hidalgo R., Latif-Eugenín F., Hossain M.J., Berg K., Niemi R.M., Rapala J. Aeromonas aquatica sp. nov., Aeromonas finlandiensis sp. nov. and Aeromonas lacus sp. nov. isolated from Finnish waters associated with cyanobacterial blooms. Syst Appl Microbiol. 2015;38:161–168. doi: 10.1016/j.syapm.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Marti E., Balcázar J.L. Aeromonas rivipollensis sp. nov., a novel species isolated from aquatic samples. J Basic Microbiol. 2015;55:1435–1439. doi: 10.1002/jobm.201500264. [DOI] [PubMed] [Google Scholar]
- 5.Teunis P., Figueras M.J. Reassessment of the enteropathogenicity of mesophilic Aeromonas species. Front Microbiol. 2016;7:1395. doi: 10.3389/fmicb.2016.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueras M.J., Beaz-Hidalgo R., Collado L., Martínez-Murcia A.J. Recommendations for a new bacterial species description based on analyses of the unrelated genera Aeromonas and Arcobacter. Bull Bergeys Int Soc Microb Syst. 2011;2:1–16. [Google Scholar]
- 7.Rossi-Tamisier M., Benamar S., Raoult D., Fournier P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int J Syst Evol Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.