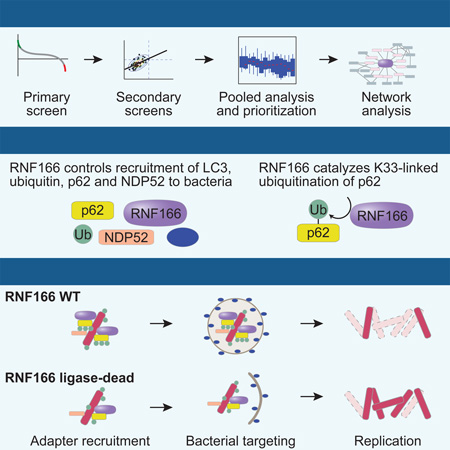
Summary
Xenophagy is a form of selective autophagy that involves the targeting and elimination of intracellular pathogens through several recognition, recruitment, and ubiquitination events. E3 ubiquitin ligases control substrate selectivity in the ubiquitination cascade; however, systematic approaches to map the role of E3 ligases in antibacterial autophagy have been lacking. Here we screened more than 600 putative human E3 ligases, identifying E3 ligases that are required for adaptor protein recruitment and LC3-bacteria colocalization, critical steps in antibacterial autophagy. An unbiased informatics approach pinpointed RNF166 as a key gene that interacts with the autophagy network and controls the recruitment of ubiquitin as well as the autophagy adaptors p62 and NDP52 to bacteria. Mechanistic studies demonstrated that RNF166 catalyzes K29- and K33-linked polyubiquitination of p62 at residues K91 and K189. Thus, our study expands the catalog of E3 ligases that mediate antibacterial autophagy and identifies a critical role for RNF166 in this process.
Graphical Abstract
Introduction
Macroautophagy (hereafter referred to as autophagy) is a homeostatic cellular process wherein constituents of the cytosol are encapsulated in a de novo-generated, double-membraned vesicle termed the autophagosome. Formation of the autophagosome is mediated by autophagy-related (ATG) proteins, and fusion of the autophagosome with lysosomes leads to degradation of autophagosomal contents (Mizushima and Komatsu, 2011). Under starvation conditions, autophagy operates non-selectively by recycling components of the cytosol for nutritional purposes. However, autophagy can also be selective, with specific substrates ranging from protein aggregates to damaged organelles (Levine et al., 2011; Randow and Youle, 2014).
Selective autophagy is also essential for cell-autonomous defense against pathogens including intracellular bacteria, viruses, and parasites (Deretic, 2011; Orvedahl et al., 2010; Selleck et al., 2015). A broad range of gram positive and gram negative bacteria, including Salmonella enterica serovar Typhimurium (Birmingham et al., 2006), Shigella flexneri (Ogawa et al., 2005), Listeria monocytogenes (Py et al., 2007), group A Streptococcus (Joubert et al., 2009; Thurston et al., 2009), Francisella tularensis (Case et al., 2014), Yersinia enterocolitica (Murthy et al., 2014), and Mycobacterium tuberculosis (Gutierrez et al., 2004), are restricted by autophagy. Despite the importance of autophagy in antibacterial defense, we currently have an incomplete understanding of how such diverse bacterial species are recognized, targeted, and eliminated by autophagy.
The ability of host cells to target a variety of pathogens that have each evolved differing invasion and niche establishment strategies indicates the existence of a synergistic defense network of target recognition molecules and adaptors that activate autophagy at distinct steps of the invasion process. Although bacteria have evolved a number of mechanisms to evade detection by the host, the host cell elicits multiple signals to target and recognize bacteria. For example, bacteria-containing vesicles that accumulate diacylglycerol subsequently become the target of autophagy, and bacteria that escape this pathway expose host glycans on their damaged vacuoles, which are targeted for autophagic degradation by galectin-8 (Shahnazari et al., 2010; Thurston et al., 2012). Another mechanism of host defense involves coating the invading bacteria or bacteria-associated proteins with polyubiquitin chains (Collins et al., 2009; Fiskin et al., 2016; Fujita et al., 2013; Katsuragi et al., 2015; Khaminets et al., 2016). Adaptor proteins then sense the bacterial ubiquitin coat and recruit autophagy adaptors to initiate engulfment of the bacteria by autophagy. There are currently four known ubiquitin-binding autophagy adaptors, namely NDP52, p62, NBR1, and optineurin, which recognize and bind to ubiquitinated substrates. These adaptors then bind to ubiquitin-like proteins of the LC3 (ATG8) family displayed on the phagophore membrane through a degenerate LC3-interacting region (Rogov et al., 2014; Sorbara and Girardin, 2015). NDP52, p62, and optineurin are each required to restrict bacterial proliferation and therefore each executes unique functions (Kang et al., 2015; Katsuragi et al., 2015; Thurston et al., 2009; Zheng et al., 2009).
Distinct interacting proteins have been identified for each of the known adaptors, potentially contributing to their non-redundant functions. Previous studies have suggested that while p62 and NDP52 are independently recruited to diverse pathogens, they exert host species-dependent effects on cell-autonomous defense (Judith et al., 2013; Selleck et al., 2015) as well as pathogen-dependent effects (Katsuragi et al., 2015; Mostowy et al., 2010; Stanley and Cox, 2013). Additionally, p62 and NDP52 each have been shown to be important at multiple steps in selective autophagy (Katsuragi et al., 2015; McEwan and Dikic, 2014; Verlhac et al., 2015). Ubiquitination plays a key role in multiple steps of bacterial recognition and targeting, and it is likely that these post-translational modifications regulate substrate specificity in the autophagy pathway (Fiskin et al., 2016; Khaminets et al., 2016). E1, E2, and E3 enzyme cascades control the linkage of ubiquitin to target proteins. The human genome contains two known E1s, several dozen E2s, and hundreds of E3 ubiquitin ligases, which play crucial roles in many cellular signaling pathways (Huttlin et al., 2015; Ordureau et al., 2015). The addition of polyubiquitin chains at lysine residues alter the target protein’s function. For example, K48-linked polyubiquitination is primarily associated with protein degradation; K27- and K29-linked ubiquitination have been linked with lysosomal degradation, and K11-linked ubiquitination is involved in cell cycle control (Kuang et al., 2013).
The diversity of E3 ligases suggests that they function to control specificity of the ubiquitination cascade and several E3 ligases have been implicated in the control of nonselective starvation-induced autophagy (Deng et al., 2015; Kuang et al., 2012; Kuang et al., 2013; Li et al., 2015; McEwan and Dikic, 2014). Recently, the E3 ligases LRSAM1 and PARK2 (also known as parkin) were demonstrated to be necessary for innate targeting of bacteria by antibacterial autophagy (Huett et al., 2012; Manzanillo et al., 2013). LRSAM1 targets Salmonella, Listeria, the autophagy susceptible strain of Shigella (Shigella ΔicsB), and adherent invasive Escherichia coli, but not M. tuberculosis, for K6- and K27-linked polyubiquitination, whereas PARK2 functions in innate resistance to M. tuberculosis, driving predominantly K63- and to a lesser extent K48-linked ubiquitination. There are likely additional E3 ligases that function during antibacterial autophagy. These E3 ligases can modulate the breadth of defense and the type of cellular response by generating different combinations of ubiquitin chain linkages on target proteins.
Systematic approaches to map the role of E3 ligases in antibacterial autophagy have been lacking. Here we screen a library of putative E3 ligases for their ability to regulate xenophagic targeting of bacteria. We identify a subset of E3 ligases functioning in antibacterial autophagy, and establish the E3 ligase RNF166 as a key gene interacting with autophagy-related proteins. Furthermore, we show that RNF166 promotes atypical K29- and K33-mediated ubiquitination of p62.
Results
A siRNA Screen Identifies Multiple Putative E3 Ubiquitin Ligases that Affect Antibacterial Autophagy
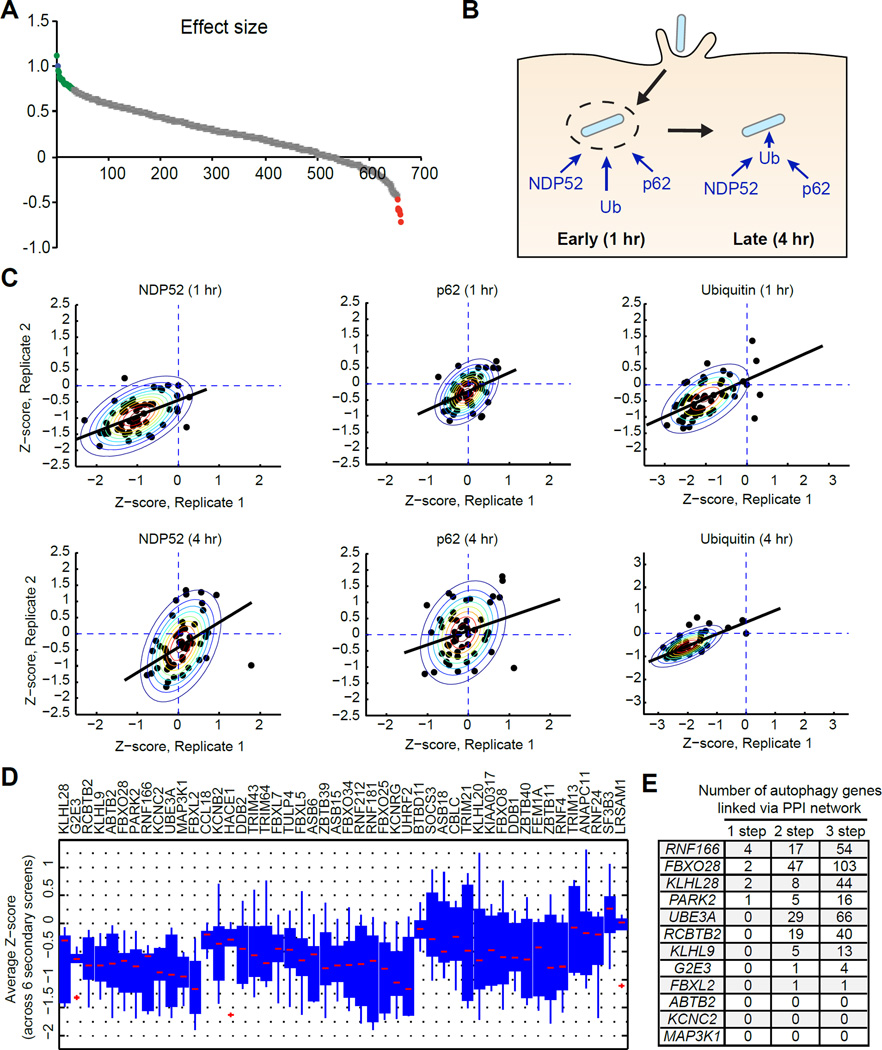
To define the scope of E3 ubiquitin ligases functioning in targeting of bacteria to autophagy, we used an siRNA library containing pools of three siRNAs for each of the 617 putative E3 ligases (Huttlin et al., 2015) in the human genome to knock down E3 ligase expression. At 1 hr postinfection, cells were monitored for changes in the colocalization of S. Typhimurium with GFP-LC3, a core autophagy gene that is critical for multiple steps in autophagy (Birmingham et al., 2006). The effect size of each siRNA was determined using a non-targeting siRNA as a negative control and an siRNA against the core autophagy gene ATG16L1 as a positive control (Figure 1A). Of the 617 genes, knockdown of 48 genes significantly reduced LC3 colocalization with Salmonella at 1 hr postinfection with an effect size of ≥ 50% of the positive control, suggesting that these genes play roles in promoting LC3-dependent targeting to bacteria. Additionally, a role for each of the 48 genes in LC3-bacteria colocalization was confirmed using single siRNAs from the deconvoluted pools. Notably, E3 ligases with known activity in cell-autonomous defense against bacteria, including LRSAM1, PARK2, and TRIM21 (Rakebrandt et al., 2014), scored as hits in this screen. Additional genes from the screen further supported the validity of our method. For example, HACE1 has been shown to directly ubiquitinate optineurin to control autophagy in lung cancer cells (Liu et al., 2014), and the E3 ligases DDB1, TRIM21, and TRIM13 have each been reported to function in selective or bulk autophagy (Antonioli et al., 2014; Kimura et al., 2015; Tomar et al., 2012).
Figure 1. A Core Contingent of E3 Ubiquitin Ligases Function Throughout Antibacterial Autophagy.
(A) Distribution of effect size for each siRNA on GFP-LC3-Salmonella colocalization at 1 hr postinfection. Effect size was calculated as a fraction relative to a non-targeting negative control siRNA and a positive control siRNA targeting ATG16L1. Genes that significantly decreased GFP-LC3-Salmonella colocalization are shown in green. Genes that significantly increased GFP-LC3-Salmonella colocalization are shown in red and genes that did not significantly change colocalization are shown in gray.
(B) Schematic of antibacterial autophagy targeting strategies. During early bacterial targeting (1 hr), autophagy adaptors can bind independently of ubiquitin. During late bacterial targeting (4 hr), autophagy adaptor proteins bind via ubiquitinated substrates.
(C) Z-normalized secondary screen colocalization data. Scatter plots show data from two independent runs for either p62, NDP52, or ubiquitin colocalization at the given time point. Raw data was standardized with z-score computation using mean and standard deviation of negative controls. Contour plots show bivariate Gaussian distribution fit to the data. Black line shows linear regression function fit to the normalized data.
(D) Box plots show pooled average Z-scores from each screen per gene. Genes are ordered based on the number of times the average Z-score was negative for all secondary screens and the magnitude of the Z-score.
(E) Table of 12 autophagy genes that can be linked to each candidate gene (column 1) via 1-, 2- or 3-step protein-protein interactions derived from Bioplex database.
Given the known function of LRSAM1 and PARK2 in antibacterial autophagy, we anticipated that individual E3 ligases would likely play discrete roles in the recruitment of adaptor proteins at specific times postinfection (Shibutani and Yoshimori, 2014). Bacterial targeting can be divided into early (1 hr postinfection) and late (4 hr postinfection) events. At the 1-hr timepoint in Salmonella infection, the membranes of a subset of Salmonella-containing vacuoles typically become compromised, exposing the bacteria to the cytoplasm, where they become associated with ubiquitinated proteins and subsequently targeted to autophagosomes (Figure 1B). This early targeting (1 hr postinfection) is characterized as being galectin-8-dependent, ubiquitin-independent targeting by NDP52 and likely involves both direct bacterial sensing as well as sensing of damaged membrane remnants by host proteins (Figure 1B). Later bacterial targeting (4 hr postinfection) is thought to be ubiquitin-dependent and likely entails direct recognition of the bacteria. In addition, the adaptor protein p62 has been shown to function in targeting ubiquitinated bacteria for autophagy (Yoshikawa et al., 2009; Zheng et al., 2009). Therefore, we tested the 48 siRNAs for effects on the colocalization of ubiquitin, p62, and NDP52 with Salmonella at both 1 hr and 4 hr postinfection (Figure 1C).
Colocalization rates were normalized to their respective non-targeting control siRNA and Z-score normalization was performed with respect to the mean and standard deviation of the negative controls. A negative Z-score implied a decrease in marker colocalization compared to the negative control. Data from duplicate runs were normalized independently (Figure 1C). Z-score normalization also allows a direct comparison of phenotypic output from different assays in a quantitative manner. We pooled average Z-scores of each gene (averaged over duplicate runs) from each of the six secondary screens and ordered them based on the number of times the average Z-score was negative across six secondary screens and the magnitude of the average Z-score across assays (Figure 1D). Using this method, we determined that each of the 48 genes decreased colocalization of bacteria with two or more markers at the time points investigated (Table S1). Eleven of the 48 genes altered NDP52-bacteria colocalization with no effect on p62 recruitment (Table S1). From this analysis, 12 genes were prioritized that consistently scored below zero in all colocalization assays at each time point, suggesting they control both p62-dependent and NPD52-dependent bacterial targeting, potentially at the earliest stages.
To further filter E3 ligases of interest, we next performed a network analysis to evaluate which of the 12 candidate genes might functionally connect with the known autophagy protein-protein interaction network. Using the Bioplex protein interactome database, we mapped first-, second-, and third-degree interactors of each of the 12 candidate E3 ligases with a curated list of known autophagy-associated genes derived from published databases (Huttlin et al., 2015; Lipinski et al., 2010; McKnight et al., 2012; Orvedahl et al., 2011; Sorbara and Girardin, 2015; Szyniarowski et al., 2011) (Figure 1E and Table S2). We found four genes that directly interacted with one or more autophagy-associated genes, including RNF166, FBXO28, KLHL28, and PARK2. Among the four genes with direct interactions to one or more autophagy genes, RNF166 formed a network with 24 other proteins including four direct interactors (Figure S1). Taken together, these data suggest that at least 48 E3 ligases are involved in antibacterial autophagy, pointing to a substantial requirement for E3 ubiquitin ligases in this process. Additionally, we identify a high-confidence set of 12 E3 ligases that are likely functioning in the recruitment of ubiquitin, NDP52, and/or p62 to bacteria.
RNF166 Is Required for the Early Recruitment of Autophagy Adaptors to Salmonella
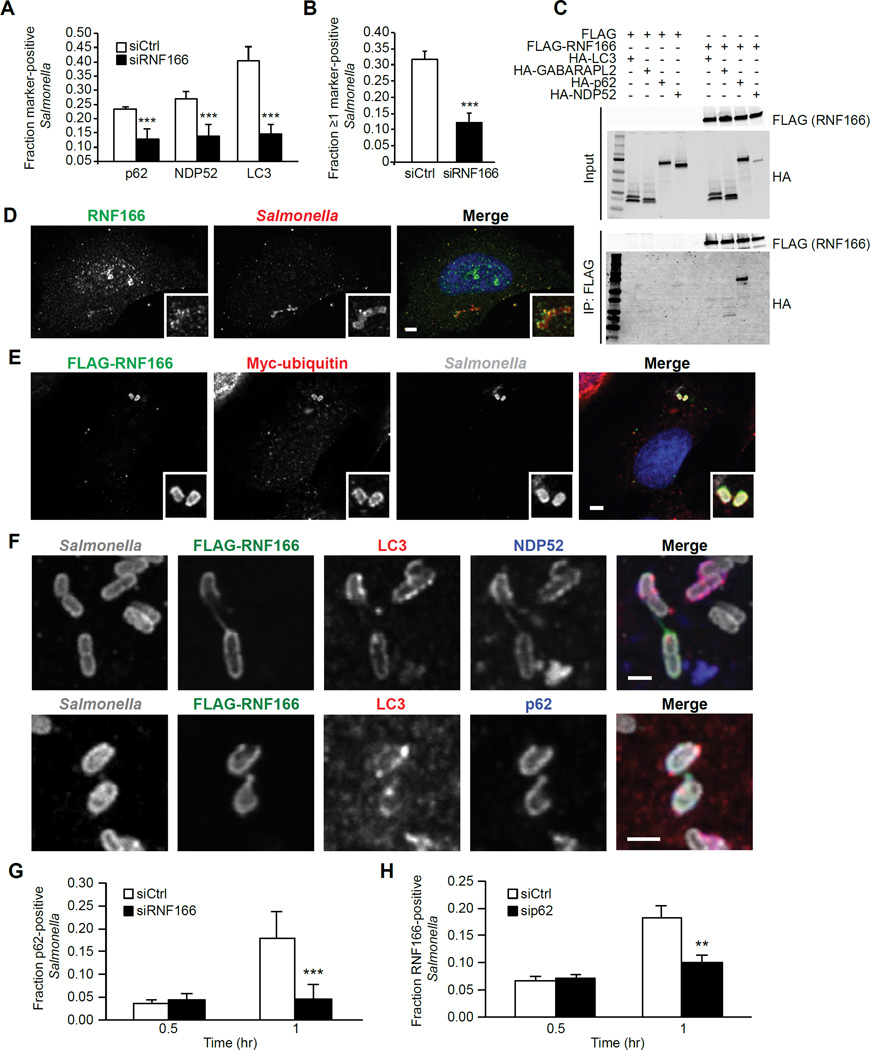
Little is known about the proteins involved in the early recruitment of p62. Given our finding that RNF166 was required for LC3-Salmonella colocalization as well as p62 recruitment to Salmonella, we selected RNF166 for further analysis. First, we used confocal microscopy to confirm the results of our high-throughput screen, which suggested that RNF166 is required for bacterial targeting by autophagy adaptors. Knockdown of RNF166 resulted in decreased proportions of bacteria that colocalized with p62, NDP52, or LC3 (Figures 2A and S2A). The majority of studies analyzing adaptor recruitment to bacteria evaluate the colocalization of a single marker with bacteria at a given time point. Although useful, this method does not allow for the enumeration of bacteria that are marked with more than one adaptor. We therefore developed a four-color imaging approach using confocal microscopy to analyze bacteria colocalization with relevant proteins. Using this method, we next assessed the fraction of all intracellular bacteria that simultaneously localized with one or more canonical antibacterial autophagy markers (LC3, NDP52, or p62) (Figures S2B and S2C). We found that the fraction of Salmonella that colocalized with 3 markers simultaneously was unchanged in RNF166-deficient cells (Figure S2C). In contrast, when we evaluated colocalization of one or more markers with Salmonella, we found that in cells expressing non-targeting siRNA, 32% of all bacteria colocalized with one or more markers simultaneously at 1 hr postinfection. An siRNA targeting RNF166 reduced this number to 12% (Figure 2B). These data suggest that in the absence of RNF166 the recruitment of single adapters to Salmonella is decreased, but that the population that colocalizes with 3 markers at the same time is unaffected. To determine whether RNF166 binds the autophagy apparatus, we expressed FLAG-tagged RNF166 in HEK293T cells along with HA-tagged p62, NDP52, LC3B, or GABARAPL2 and immunoprecipitated with an anti-FLAG antibody. Of the four target proteins, only p62 co-immunoprecipitated with RNF166, suggesting this interaction is specific to p62 (Figure 2C).
Figure 2. RNF166 Is Required to Recruit the Autophagy Apparatus to Salmonella and Interacts with p62.
(A) HeLa cells were treated with a non-targeting siRNA or siRNA targeting RNF166 for 48 hr. Cells were then infected with Salmonella for 1 hr and the fraction of Salmonella colocalizing with either p62, NDP52, or LC3 was enumerated. Data represent means ± SEM, n = 100 infected cells per group, data pooled from three independent experiments.
(B) RNF166-depleted HeLa cells were infected with Salmonella for 1 hr and co-stained for endogenous LC3, p62, and NDP52. The fraction of all intracellular bacteria that colocalized with ≥ 1 marker (LC3, p62, and/or NDP52) was determined. Data represent means ± SEM, n = 100 infected cells per group, data pooled from three independent experiments.
(C) HEK293T cells were transfected for 24 hr with constructs expressing FLAG alone or FLAGRNF166 and HA-tagged autophagy proteins as indicated. Proteins were immunoprecipitated with anti-FLAG antibodies. Data are representative of four independent experiments.
(D) Confocal images of HeLa cells infected with Salmonella for 1 hr and stained for endogenous RNF166. Insets indicate areas of bacterial colocalization. Data are representative of three independent experiments.
(E) Confocal images of HeLa cells transfected for 24 hr with FLAG-tagged RNF166 and Myctagged ubiquitin infected with Salmonella for 1 hr. Insets indicate areas of bacterial colocalization with FLAG-RNF166 and Myc-ubiquitin. Data are representative of three independent experiments.
(F) Confocal images of HeLa cells transfected for 24 hr with FLAG-tagged RNF166 and infected with Salmonella for 1 hr. Cells expressing FLAG-RNF166 were co-stained for LC3 and NDP52 (top panels) or LC3 and p62 (bottom panels). Data are representative of three independent experiments.
(G) HeLa cells treated with a non-targeting siRNA or siRNA targeting RNF166 for 48 hr were infected with Salmonella for the indicated time periods. Cells were stained for endogenous p62. Shown is the fraction of Salmonella colocalizing with p62 enumerated for at least 100 cells. Data represent means ± SEM, n = 100 infected cells per group, data pooled from three independent experiments.
(H) HeLa cells were treated with a non-targeting siRNA or siRNA targeting p62 for 48 hr, infected with Salmonella for the indicated time periods, and stained for endogenous RNF166. Shown is the fraction of Salmonella colocalizing with RNF166 at the indicated time points. Data represent means ± SEM, n = 150 infected cells per group, data pooled from three independent experiments.
For all panels, **p < 0.01, ***p < 0.001, Student’s t test.
See also Figure S2.
RNF166 is a p62-Interacting Protein that Colocalizes with S. Typhimurium
We next evaluated whether the RNF166-p62 interaction occurs in the context of antibacterial autophagy. To test this possibility, we examined whether RNF166 colocalized with bacteria. HeLa cells expressing RNF166 were infected with Salmonella and stained for RNF166 at 1 hr postinfection. We observed that both endogenous RNF166 and overexpressed RNF166 localized to Salmonella (Figures 2D and 2E). Additionally, we confirmed colocalization of RNF166 with ubiquitin, p62, NDP52, and LC3 around Salmonella (Figures 2E, 2F, and S2D), suggesting that RNF166 colocalizes with the autophagy apparatus during bacterial targeting. Our results suggest that RNF166 functions in the early intracellular recognition of bacteria. To confirm that RNF166 is indeed acting to recruit p62 during the initial targeting stages, we quantified early p62 localization to Salmonella. We observed peak p62 colocalization 1 hr postinfection, with 18 ± 2.7% of Salmonella colocalized with p62, an effect that was abolished by depletion of RNF166 (Figure 2G). No differential recruitment was observed at 30 minutes post-infection (Figure 2G). Interestingly, the fraction of Salmonella that colocalized with RNF166 was similar to that of Salmonella colocalization with p62; 18 ± 2% (Figure 2H). This frequency is also similar to that of Salmonella colocalization with LC3 during antibacterial autophagy (Birmingham et al., 2006). Depletion of p62 resulted in a significant reduction in RNF166 colocalization with Salmonella at 1 hr postinfection, suggesting that the localization of p62 and RNF166 to bacteria is co-dependent (Figures 2G and 2H).
RNF166 Mediates Atypical Ubiquitination of p62
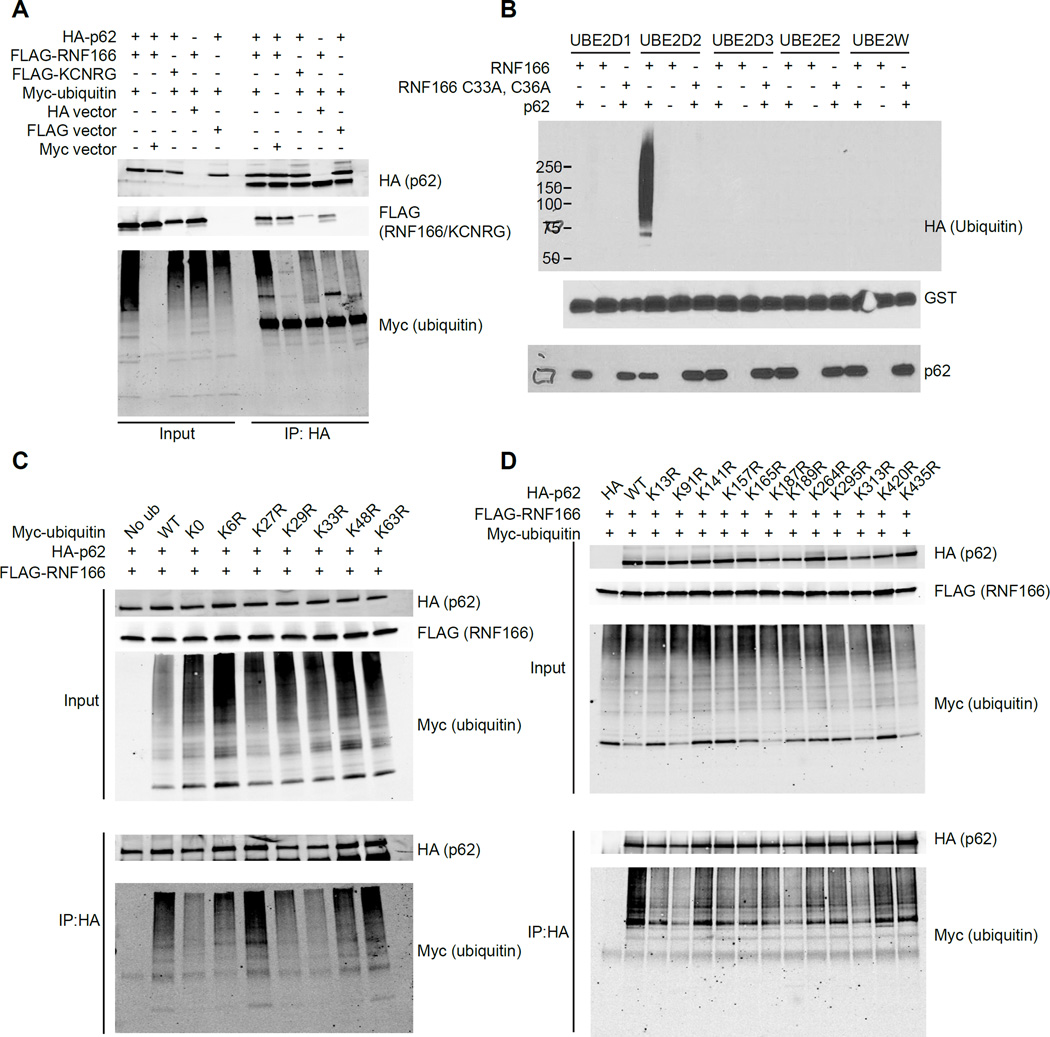
Given that RNF166 is an E3 ligase that likely functions through ubiquitination of target proteins, we hypothesized that RNF166 directly ubiquitinates p62 and that this ubiquitination regulates its function. To test this hypothesis, we co-transfected HEK293T cells with RNF166, p62, and ubiquitin and infected them for 1 hr with Salmonella. Immunoprecipitation of p62 under these conditions demonstrated that p62 was significantly ubiquitinated in the presence of RNF166 (Figure 3A). To confirm the specificity of this interaction, we replaced RNF166 with another E3 ligase that also disrupted p62-bacteria colocalization in our screen, KCNRG, and observed no ubiquitination of p62 (Figure 3A). To further validate this finding and show that RNF166 can directly ubiquitinate p62, we used an in vitro ubiquitination assay with recombinant UBA1 (E1), E2 enzymes, HA-ubiquitin, GST-RNF166, and SUMO-p62. To determine the specific E2 enzyme required for the reaction, we tested a panel of the five most likely E2 enzymes identified by proteomics (Markson et al., 2009). Of the five E2s tested, ubiquitination of p62 was observed only in the presence of all defined proteins and the E2 enzyme UBE2D2 (Figure 3B). Furthermore, a ligase-dead RNF166 mutant (RNF166 C33A, C36A), was unable to drive p62 ubiquitination under the same conditions (Figure 3B). Thus, these data confirm that RNF166 directly ubiquitinates p62.
Figure 3. RNF166 Mediates K29- and K33-Linked Ubiquitination of p62.
(A) HEK293T cells were transfected with indicated constructs followed by immunoprecipitation of HA-p62. Representative blot is shown from four independent experiments.
(B) GST-RNF166 and SUMO-p62 were incubated together or separately in the presence of recombinant UBE1 (E1), various E2 ubiquitin-conjugating enzymes as indicated, and HAubiquitin. Representative blot is shown from three independent experiments.
(C) HEK293T cells were transfected with FLAG-RNF166, HA-p62 and one of the indicated Mycubiquitin constructs with single point mutations at the indicated lysine residues. Proteins were immunoprecipitated with anti-HA antibodies and immunoblots were performed with antibodies against HA and Myc to detect ubiquitinated proteins. Representative blot is shown from four independent experiments.
(D) HEK293T cells were co-transfected with Myc-ubiquitin, FLAG-RNF166, and HA-p62 with the indicated mutations. Proteins were immunoprecipitated with anti-HA antibodies and immunoblots were performed with antibodies against HA and Myc to detect ubiquitinated proteins. Representative blot is shown from three independent experiments.
Ubiquitin contains seven lysines through which polyubiquitin chains can be assembled, with specific linkages determining the fate of the substrate. Several of these linkages, including K48- and K63-based linkages, have been associated with the bacterial ubiquitin coat during antibacterial autophagy (Collins et al., 2009; Manzanillo et al., 2013; van Wijk et al., 2012). To determine which ubiquitin linkage is critical for RNF166-mediated ubiquitination of p62 we used site-directed mutagenesis to generate lysine to arginine substitutions at each of the seven lysines in ubiquitin. HEK293T cells were co-transfected with RNF166, p62, and either wildtype (WT) ubiquitin, a ubiquitin containing no lysines (K0), or ubiquitin containing the indicated arginine substitutions. Transfected cells were infected for 1 hr with Salmonella and p62 was immunoprecipitated. Loss of ubiquitinated p62 was observed with the K0 ubiquitin and K33R ubiquitin as well as with K29R to a lesser extent (Figure 3C). However, no change in ubiquitination of p62 was observed using K48R or K63R mutants, suggesting that these linkages are not critical for RNF166-mediated ubiquitination of p62 (Figure 3C). Finally, we sought to identify the ubiquitinated lysine residues in p62. We individually mutated each of the 12 ubiquitinated lysine residues in p62 to arginine (Hornbeck et al., 2015) and tested the ability of RNF166 to ubiquitinate each of these p62 mutants. Arginine substitutions at K91 and K189, and to a lesser extent K313, resulted in significant decreases in co-immunoprecipitated ubiquitin compared to wild-type p62 (Figure 3D). Taken together, these data suggest that RNF166 drives K29- and K33-linked ubiquitination of p62.
RNF166 Is Necessary to Limit Bacterial Replication
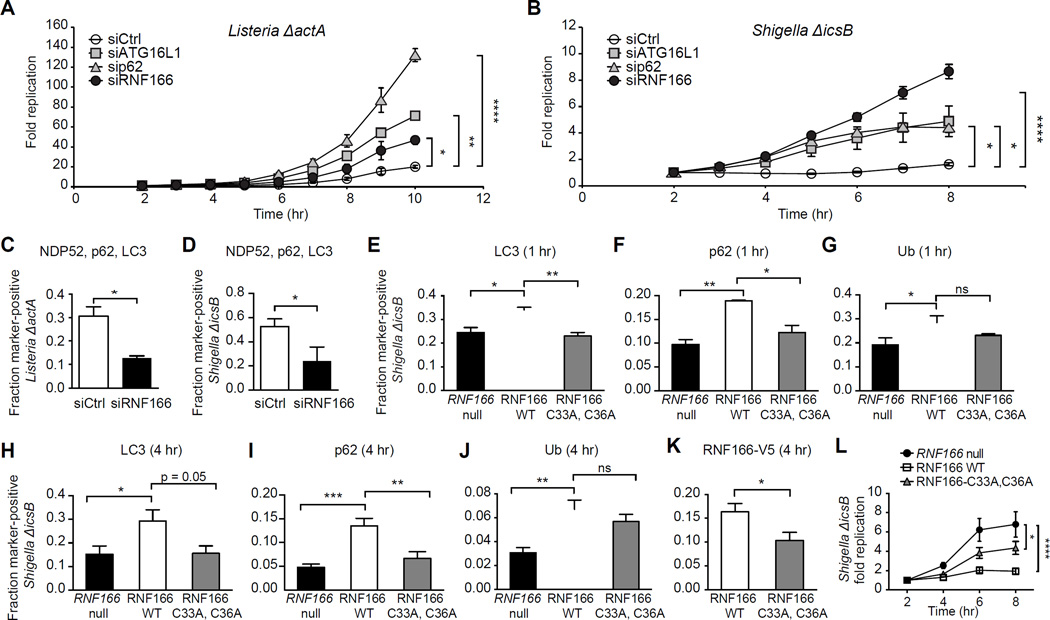
We next determined the role of RNF166 in limiting bacterial replication. NDP52 and p62 have differing roles in the autophagic recognition of bacteria and subsequent bacterial clearance. Specifically, NDP52 has been previously shown to restrain Salmonella replication (Thurston et al., 2009); conversely, p62 is predominantly required to control Listeria and Shigella replication, but is less important in the control of Salmonella replication (Yoshikawa et al., 2009; Zheng et al., 2009). Consistent with these reports, we observed no significant effects on intracellular replication of Salmonella in cells treated with siRNF166 compared to a non-targeting control (Figure S3A). Therefore, we next tested the requirement of RNF166 in restricting the intracellular replication of autophagy-susceptible strains of Shigella and Listeria. HeLa cells treated with a non-targeting siRNA or siRNF166 were infected with autophagy-susceptible strains Listeria ΔactA or Shigella ΔicsB expressing luciferase, and monitored for intracellular replication following gentamycin treatment. In the absence of RNF166, Listeria ΔactA replication increased more than 2-fold over the time course analyzed, with levels comparable to those observed in the absence of ATG16L1 (Figure 4A). Replication of Shigella ΔicsB also increased more than 4-fold in the absence of RNF166, with levels higher than those observed in the absence of either ATG16L1 or p62 (Figure 4B). Immunofluorescence studies confirmed that endogenous RNF166 is recruited to both Shigella ΔicsB (Figure S3B) and Listeria ΔactA (Figure S3C). These data suggest that RNF166 functions as an important component of autophagic targeting of cytosol-adapted pathogenic bacteria.
Figure 4. RNF166 Is Required to Inhibit the Intracellular Replication of Listeria and Shigella.
(A) HeLa cells treated with a non-targeting siRNA or siRNA targeting RNF166, p62, or ATG16L1 for 48 hr were infected with Listeria ΔactA expressing luciferase. Cells were treated with gentamicin to remove extracellular bacteria and relative light units were monitored over the indicated time course. Fold replication represents light units over time compared to 2 hr postinfection. Data represent means ± SEM, n = 8.
(B) Cells were treated as in (A) and infected with Shigella ΔicsB expressing luciferase. Data represent means ± SEM, n = 8.
(C, D) HeLa cells were treated with a non-targeting siRNA or siRNA targeting RNF166 for 48 hr, then infected with Listeria ΔactA (C) or Shigella ΔicsB (D) for 1 hr and co-stained for endogenous NDP52, p62, and LC3. The fraction of colocalization of each intracellular bacterium simultaneously with NDP52, p62, and LC3 was scored. > 50 bacteria from three independent experiments were analyzed. Data represent means + SEM.
(E-G) Quantification of LC3 (E), p62 (F), and ubiquitin (G) recruitment to Shigella ΔicsB at 1 hr postinfection in RNF166-null HeLa cells expressing the indicated constructs. Data represent means ± SEM, n = 125 infected cells per group, data pooled from three independent experiments.
(H-K) Quantification of LC3 (H), p62 (I), ubiquitin (J), and RNF166-V5 (K) recruitment to Shigella ΔicsB in RNF166-null HeLa expressing the indicated constructs. Data represent means ± SEM, n = 125 infected cells per group, data pooled from three independent experiments.
(L) Intracellular replication of Shigella ΔicsB expressing luciferase in RNF166-null HeLa cells expressing the indicated constructs. Cells were treated with gentamicin to remove extracellular bacteria and relative light units were monitored over the indicated time course. Fold replication represents light units over time compared to 2 hr postinfection. Data represent means ± SEM, n = 8.
For all panels, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. Student’s t test for (C, D, K), one-way ANOVA with multiple comparisons for (A, B, E-J, and L).
See also Figures S3 and S4.
We next evaluated whether loss of RNF166 altered recruitment of autophagy adaptors to Listeria ΔactA and Shigella ΔicsB. Consistent with our replication data, Listeria ΔactA and Shigella ΔicsB exhibited a 2–3-fold reduction in bacteria that simultaneously colocalized with p62, NDP52, and LC3 in RNF166-deficient cells (Figures 4C and 4D). This result is in contrast to results obtained with Salmonella in which this p62/NDP52/LC3-positive population was largely unchanged in the absence of RNF166 (Figure S2C). These data suggest an inverse correlation between intracellular bacterial replication and the simultaneous accumulation of p62, NDP52, and LC3 around bacteria. This may explain the observed phenotypic differences in intracellular replication between Salmonella, Listeria ΔactA, and Shigella ΔicsB in the absence of RNF166, with Listeria ΔactA and Shigella ΔicsB exhibiting a more significant block in downstream accumulation of multiple adaptors.
Finally, to validate the requirement for RNF166 and RNF166 ligase activity in antibacterial autophagy we generated an RNF166-null HeLa cell line using CRISPR/Cas9 (Figures S3D and S3E). It is known that p62 plays a role in role during non-selective or bulk autophagy therefore we employed an LC3 flux assay to determine if loss of RNF166 alters bulk autophagy. In this immunoblot assay, levels of lipidated LC3-II are compared to LC3-I and an increase in the ratio of LC3-II to LC3-I corresponds to an increase in autophagy (Kang et al., 2015). To determine whether RNF166 functions broadly in starvation-induced autophagy, cells were treated with Torin 1, an mTOR inhibitor and inducer of bulk autophagy, or Torin 1 and lysosomal protease inhibitors E64d/pepstatin A to evaluate autophagic flux. No differences in autophagic flux were observed in RNF166-null cells, suggesting that RNF166 functions specifically in antibacterial autophagy (Figure S3F) (Thoreen et al., 2009).
We focused on evaluating RNF166 in the context of Shigella ΔicsB infection, as this is where the loss of RNF166 had the most significant effect. Consistent with our siRNA data, recruitment of LC3, p62, and ubiquitin to Shigella ΔicsB at 1 hr postinfection was significantly reduced in RNF166-null cells compared to the same cells rescued with WT RNF166 (Figures 4E-4G). The E3 ligase-dead mutant (RNF166 C33A, C36A) was sufficient to rescue recruitment of ubiquitin, but not LC3 or p62, suggesting that ubiquitin modification of bacteria is unchanged in the absence of RNF166 ligase activity (Figures 4E-4G). Both WT RNF166 and the ligase-dead mutant localized to Shigella ΔicsB equally at this time point (Figure S3G and S3H). We also investigated recruitment of these proteins to Shigella ΔicsB at 4 hr postinfection and found LC3, p62, and ubiquitin recruitment was significantly decreased in RNF166-null cells; furthermore, LC3 and p62 recruitment were also decreased in cells expressing the ligase-dead mutant RNF166 (Figures 4H-4J). Importantly, a significant decrease in the recruitment of the ligase-dead RNF166 to Shigella ΔicsB was observed at 4 hr post-infection, suggesting differences in bacteria-host protein complex stability over time in the absence of RNF166 ligase activity (Figure 4K). No difference in the recruitment of NDP52 to Shigella ΔicsB was observed at either 1 hr or 4 hr postinfection (Figure S3I and S3J). Additionally, the ligase-dead mutant RNF166 was sufficient to completely rescue defects in LC3, p62, and ubiquitin colocalization with Salmonella in RNF166-null cells (Figure S4A-C). These data are consistent with bacterial species-specific requirements for RNF166 and the E3 ligase activity of RNF166 in antibacterial autophagy.
Finally, intracellular replication of Shigella ΔicsB was significantly increased in RNF166-null cells compared to the same cells rescued with wild-type RNF166 (Figure 4L). The ligase-dead RNF166 mutant was able to partially rescue the observed replication phenotype, suggesting that RNF166 may have important roles beyond ubiquitination. Taken together, these data demonstrate that RNF166 limits intracellular replication of Shigella and that the E3 ligase activity of RNF166 is important for this function.
Discussion
Autophagy represents a fundamental host cell response to invasion by a variety of bacteria. The accumulation of a ubiquitin coat is a central component in the autophagic targeting of intracellular pathogens (Collins et al., 2009; Katsuragi et al., 2015; Khaminets et al., 2016). Recent studies have provided a global analysis of the ubiquitinome of Salmonella-infected cells identifying numerous host and bacterial proteins that are ubiquitinated or deubiquitinated upon infection (Fiskin et al., 2016). Systematic approaches have also revealed key contributions of TRIM proteins to autophagy regulation and substrate recognition (Mandell et al., 2014). Other E3 ligases have also been shown to regulate non-selective autophagy (Liu et al., 2016; Xu et al., 2014; Zhang et al., 2015). Our LC3 screen as well as our secondary screening of adaptor recruitment to Salmonella revealed that multiple E3 ligases are involved at various steps in antibacterial autophagy. This broad use of E3 ligases across multiple steps likely adds redundancy to the system, allowing for diverse pathogen targeting.
We identified 48 E3 ligases that control recruitment of LC3 to bacteria from a total of 617 putative E3 ligases. From this group, we identified a core group of 12 E3 ligases that alter the recruitment of NDP52, p62, and ubiquitin to bacteria, suggesting that they function at upstream steps in pathogen targeting. From this core contingent, we focused on RNF166, which has multiple protein-protein interactions in known autophagy networks (Figure 1) (Orvedahl et al., 2011; Sorbara and Girardin, 2015). Of the 12 genes, Parkin (PARK2) and KLHL9 have been previously implicated in the earliest steps in targeting Mycobacteria and Salmonella, respectively, during antibacterial autophagy, thus validating the experimental and computational framework used in the analysis (Begun et al., 2015; Manzanillo et al., 2013). Another gene within this core group, UBE3A, has been shown to disrupt protein aggregate clearance in Huntington’s disease models, a process known to involve selective autophagy (Maheshwari et al., 2014). Additionally, TRIM13 and TRIM21, which scored in our LC3 colocalization screen, have each been shown to function in selective autophagy (Kimura et al., 2015; Tomar et al., 2012). It is therefore likely that some of the identified E3 ligases function broadly in ubiquitination events involved in different forms of selective autophagy including xenophagy, aggrephagy, and mitophagy. Additionally, analysis from the STRING database suggests that UBE2D2 interacts with 9 of the 48 E3 ligases that scored in our screen, suggesting that UBE2D2 may be an important E2 ubiquitin-conjugating enzyme regulating autophagic processes (Szklarczyk et al., 2015). Further studies will be required to understand the contribution of the various identified ligases to antibacterial autophagy.
NDP52 and p62 are non-redundant adaptor proteins that recruit LC3 to ubiquitin-associated cargos (Selleck et al., 2015). NDP52 has been relatively well described in relation to both early (1 hr postinfection, ubiquitin-independent) and late (4-hr post infection, ubiquitin-dependent) bacterial targeting (Cemma et al., 2011; Kang et al., 2015; Thurston et al., 2009; Thurston et al., 2012); however, the mechanisms of p62-mediated targeting of bacteria have been less clearly defined. Additionally, the proteins mediating ubiquitination of p62 are currently unknown. Here we show that RNF166 is necessary for targeting LC3, p62, and ubiquitin to Salmonella, Listeria ΔactA, and Shigella ΔicsB, but limits the intracellular replication of only Listeria ΔactA and Shigella ΔicsB. This finding suggests that multiple E3 ligases can function in the innate targeting of the autophagy machinery to invading bacteria without necessarily inhibiting the pathogens’ ability to replicate. The most likely explanation for this observation is that different E3 ligases promote differing ubiquitin chain linkages; for example, PARK2 decorates M. tuberculosis with K63-linked chains, whereas LRSAM1 modifies Salmonella with K6- and K27-linked ubiquitin chains, and these linkages promote specific downstream signaling (or degradation) pathways (Guo et al., 2016). Depending on the bacterial life cycle, specific ubiquitin linkages and downstream signaling events may be more relevant for elimination of some pathogens compared to others.
Our results demonstrate that RNF166 can bind p62, but not NDP52 or the related ATG8 family proteins LC3B and GABARAPL2. p62 is a multi-domain protein that includes an N-terminal PB1 domain, an LC3-interacting region (LIR) motif mediating the interaction with ATG8 family proteins, and a C-terminal UBA domain that binds ubiquitin with low affinity (Johansen and Lamark, 2011; Long et al., 2008; Long et al., 2010; Vadlamudi et al., 1996). The N-terminal PB1 domain mediates interaction with several other proteins as well as homo-oligomerization, which is important for autophagosome formation (Lamark et al., 2003). Furthermore, deletion of the PB1 domain or oligomerization-inhibiting mutations decrease the interaction with both LC3B and ubiquitin in pull-down assays, suggesting that oligomerization may increase the interaction with these binding partners (Itakura and Mizushima, 2011). Notably, one of the lysines in p62 (K91) that is ubiquitinated by RNF166 lies within the PB1 domain, suggesting that ubiquitination of K91 may facilitate p62 oligomerization.
Intriguingly, we found that RNF166-mediated ubiquitination of p62 is composed of atypical K29- and K33-based polyubiquitin linkages. Little is known about these linkage types, and proteins generating and recognizing these chains in eukaryotic cells have remained elusive (Kimura et al., 2015; Michel et al., 2015). The E3 ligase KLHL20, which also scored in our primary screen for LC3 colocalization to bacteria, has been found to drive K33-mediated ubiquitination of coronin 7, which is necessary for post-Golgi trafficking (Antonioli et al., 2014). In both endocytic and secretory pathways, ubiquitin modification of membrane proteins serves as a sorting signal for their delivery to specific destinations through interaction with a number of ubiquitin-binding adaptor proteins (Begun et al., 2015), suggesting that K29- and K33-based linkages may be broadly involved in intracellular cargo trafficking. Thus, we have identified K29- and K33-linked ubiquitination as a signal for selective autophagy. Further studies will help elucidate the role of RNF166 and these atypical ubiquitin linkages in p62-mediated antibacterial autophagy.
We found that loss of RNF166 was associated with decreases in the levels of p62, LC3, and ubiquitin around Salmonella, Listeria, and Shigella. Additionally, RNF166 ligase activity was required for RNF166 colocalization with Shigella at later time points but not for Salmonella colocalization. These findings are consistent with a model in which ubiquitinated p62 acts as a scaffold to recruit downstream adaptors to bacteria at early time points and then helps maintain a stable complex. However, the contribution of RNF166 to intracellular bacterial restriction is highly pathogen dependent. Taken together, our data reveal that ubiquitination of p62 is a critical event in the targeting of multiple bacterial species to autophagy and pinpoint RNF166 as a previously uncharacterized mediator of these ubiquitination events.
Experimental Procedures
Infection Assays
Bacterial infection assays were performed as previously described (Huett et al., 2009).
Immunofluorescence
Cells were washed once in PBS before being fixed and permeabilized with methanol at −20°C for 3 min followed by PBS wash. Coverslips were stained with appropriate primary antibodies in PBS + 10% donkey serum for 1 hr at room temperature, washed three times with PBS, then stained with appropriate secondary antibodies plus Hoechst 33342 in PBS with 10% serum for 1 hr. For screening purposes, 96-well glass-bottomed plates were imaged using an ImagXpress Micro XLS (Molecular Devices) at 40x magnification. For all other purposes, coverslips were imaged using a Leica SP5 confocal microscope.
In vitro Ubiquitination
RNF166 (and RNF166 C33A-C36A) and p62 were expressed as His6-GST or His6-SUMO fusions, respectively, from a pET28-derived vector in Codon Plus RIPL. Bacteria were grown in terrific broth amended with 0.5% glucose, 2 mM MgSO2, 0.375% aspartic acid, 100 μg/mL kanamycin and 34 μg/ml Cm at 37°C to OD 600 1.0. The cultures were cooled on ice then induced with 0.5 mM IPTG at 16°C for ~18 hr. Cell p ellets were stored at −80°C until purification. Cell pellets were thawed on ice, then lysed in buffer A (50 mM HEPES pH 8, 500 mM NaCl, 10% glycerol, 1 mM TCEP) with 1x bug buster reagent, 1x lysonase, 25 mM Im, and protease inhibitors (Roche). After removal of insoluble material by centrifugation, the supernatant was applied to a 5 mL His-Trap HP that was then washed with buffer A containing 50 mM Im and eluted with 250 mM Im. Eluates were further purified by SEC in 50 mM HEPES pH 8, 150 mM NaCl, 10% glycerol, and 1 mM TCEP. Desired fractions were concentrated and the protein was aliquoted and frozen in lN2.
In vitro ubiquitination reactions contained 50 nM GST-RNF166, 500 nM SUMO-p62, 50 mM HEPES, 5 mM MgCl2, 1 mM DTT, 2.5 mM ATP, 100 mM NaCl, 0.1% Triton X-100, 2.5 nM E1 (UBE1, Lifesensors), 200 nM E2 (UBE2D1, UBE2D2, UBE2D3, UBE2E2, UBE2W from Lifesensors), and 5 μM HA-ubiquitin (Boston Biochem) at pH 7.5. Reactions had a final volume of 25 μL and were initiated by the addition of HA-ubiquitin. After 90 min at 37°C, reactions were quenched by the addition of an equal amount of 2x SDS loading buffer.
Generation of RNF166 Knockout Cells
The first and fifth exons of RNF166 were targeted in HeLa cells using the px330 plasmid CRISPR system as previously described (Ran et al., 2013). Briefly, 20-nucleotide guide sequences complementary to exons 1 and 5 of RNF166 were cloned into px330 as described. The Cas9 vector containing RNF166-specific sgRNA sequence was then used to transfect HeLa cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. 48 hr post transfection, cells were plated in limiting dilution in 96-well plates to isolate single clones. Knockout was verified by Western blot.
Antibodies
The following antibodies were used: mouse anti-FLAG antibody clone M2 (Sigma-Aldrich); rabbit anti-LC3 clone APG8C (Sigma-Aldrich); mouse anti-actin clone AC-15 (Sigma-Aldrich); rabbit anti-HA (Sigma-Aldrich); mouse anti-HA clone HA-7(Sigma-Aldrich); mouse anti-myc (Covance); mouse anti-NDP52 (Abcam); rabbit anti-NDP52; rabbit anti-RNF166 (Abgent); guinea pig anti-p62 (American Research Products); Alexa Fluor 488-conjugated anti-rabbit; Alexa Fluor 488-conjugated anti-mouse; Alexa Fluor 594-conjugated anti-rabbit; Alexa Fluor 594-conjugated anti-mouse; Alexa Fluor 594-conjugated anti-goat; Alexa Fluor 647-conjugated anti-rabbit; (Jackson Immunology) and FK2 anti-ubiquitin (Enzo Life Sciences).
Statistical analysis
Statistical analyses were performed using Graphpad Prism software. For comparisons between two groups, an unpaired Students t test was used. For comparison of groups of three or more, a one-way ANOVA with multiple comparisons was used.
Supplementary Material
Acknowledgments
We thank Natalia Nedelsky for editorial and graphics assistance. This work was supported by funding from The Leona M. and Harry B. Helmsley Charitable Trust and the National Institutes of Health grant AI109725 to R.J.X.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
R.J.H., L.A.B., J.S.R., V.M., G.L.P., and V.J. performed experiments. R.J.H, K.G.L., and G.G. analyzed data. R.J.H., K.G.L., and R.J.X. designed research. R.J.H., K.G.L. and R.J.X. wrote the paper. The authors report no conflicts of interest.
References
- Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C, Dengjel J, et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev. Cell. 2014;31:734–746. doi: 10.1016/j.devcel.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Begun J, Lassen KG, Jijon HB, Baxt LA, Goel G, Heath RJ, Ng A, Tam JM, Kuo SY, Villablanca EJ, et al. Integrated Genomics of Crohn's Disease Risk Variant Identifies a Role for CLEC12A in Antibacterial Autophagy. Cell Rep. 2015;11:1905–1918. doi: 10.1016/j.celrep.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Case ED, Chong A, Wehrly TD, Hansen B, Child R, Hwang S, Virgin HW, Celli J. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell. Microbiol. 2014;16:862–877. doi: 10.1111/cmi.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:341–345. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, De Maziere A, van Dijk S, Carlsson F, Klumperman J, Brown EJ. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Jiang C, Chen L, Jin J, Wei J, Zhao L, Chen M, Pan W, Xu Y, Chu H, et al. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell. 2015;58:804–818. doi: 10.1016/j.molcel.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskin E, Bionda T, Dikic I, Behrends C. Global Analysis of Host and Bacterial Ubiquitinome in Response to Salmonella Typhimurium Infection. Mol. Cell. 2016;62:967–981. doi: 10.1016/j.molcel.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Bian W, Zhang Y, Li H. Expression in Escherichia coli, purification and characterization of LRSAM1, a LRR and RING domain E3 ubiquitin ligase. Protein Expr. Purif. 2016 doi: 10.1016/j.pep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huett A, Ng A, Cao Z, Kuballa P, Komatsu M, Daly MJ, Podolsky DK, Xavier RJ. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J. Immunol. 2009;182:4917–4930. doi: 10.4049/jimmunol.0803050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Judith D, Mostowy S, Bourai M, Gangneux N, Lelek M, Lucas-Hourani M, Cayet N, Jacob Y, Prevost MC, Pierre P, et al. Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep. 2013;14:534–544. doi: 10.1038/embor.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, Lu T, Yankner BA, Campisi J, Elledge SJ. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, Deretic V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 2015;210:973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang E, Okumura CY, Sheffy-Levin S, Varsano T, Shu VC, Qi J, Niesman IR, Yang HJ, Lopez-Otin C, Yang WY, et al. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet. 2012;8:e1003007. doi: 10.1371/journal.pgen.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang E, Qi J, Ronai Z. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem. Sci. 2013;38:453–460. doi: 10.1016/j.tibs.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang L, Zhou J, Luo S, Huang R, Zhao C, Diao A. Nedd4 E3 ubiquitin ligase promotes cell proliferation and autophagy. Cell Prolif. 2015;48:338–347. doi: 10.1111/cpr.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, Wu PR, Lin MY, Jiang ST, Tsai TF, et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol. Cell. 2016;61:84–97. doi: 10.1016/j.molcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J. Biol. Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B, Williamson MP, Layfield R, Searle MS. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J. Mol. Biol. 2010;396:178–194. doi: 10.1016/j.jmb.2009.11.032. [DOI] [PubMed] [Google Scholar]
- Maheshwari M, Shekhar S, Singh BK, Jamal I, Vatsa N, Kumar V, Sharma A, Jana NR. Deficiency of Ube3a in Huntington's disease mice brain increases aggregate load and accelerates disease pathology. Hum. Mol. Genet. 2014;23:6235–6245. doi: 10.1093/hmg/ddu343. [DOI] [PubMed] [Google Scholar]
- Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Munch J, Kirchhoff F, Simonsen A, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19:1905–1911. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Dikic I. Cullins keep autophagy under control. Dev. Cell. 2014;31:675–676. doi: 10.1016/j.devcel.2014.12.010. [DOI] [PubMed] [Google Scholar]
- McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 2012;31:1931–1946. doi: 10.1038/emboj.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel MA, Elliott PR, Swatek KN, Simicek M, Pruneda JN, Wagstaff JL, Freund SM, Komander D. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol. Cell. 2015;58:95–109. doi: 10.1016/j.molcel.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, Gouin E, Demangel C, Brosch R, Zimmer C, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Ordureau A, Munch C, Harper JW. Quantifying ubiquitin signaling. Mol. Cell. 2015;58:660–676. doi: 10.1016/j.molcel.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- Rakebrandt N, Lentes S, Neumann H, James LC, Neumann-Staubitz P. Antibody- and TRIM21-dependent intracellular restriction of Salmonella enterica. Pathog Dis. 2014;72:131–137. doi: 10.1111/2049-632X.12192. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Selleck EM, Orchard RC, Lassen KG, Beatty WL, Xavier RJ, Levine B, Virgin HW, Sibley LD. A Noncanonical Autophagy Pathway Restricts Toxoplasma gondii Growth in a Strain-Specific Manner in IFN-gamma-Activated Human Cells. MBio. 2015;6:e01157-01115. doi: 10.1128/mBio.01157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, Yoshimori T. Autophagosome formation in response to intracellular bacterial invasion. Cell. Microbiol. 2014;16:1619–1626. doi: 10.1111/cmi.12357. [DOI] [PubMed] [Google Scholar]
- Sorbara MT, Girardin SE. Emerging themes in bacterial autophagy. Curr. Opin. Microbiol. 2015;23:163–170. doi: 10.1016/j.mib.2014.11.020. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Cox JS. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr. Top. Microbiol. Immunol. 2013;374:211–241. doi: 10.1007/82_2013_332. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyniarowski P, Corcelle-Termeau E, Farkas T, Hoyer-Hansen M, Nylandsted J, Kallunki T, Jaattela M. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy. 2011;7:892–903. doi: 10.4161/auto.7.8.15770. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar D, Singh R, Singh AK, Pandya CD, Singh R. TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim. Biophys. Acta. 2012;1823:316–326. doi: 10.1016/j.bbamcr.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosineindependent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J. Biol. Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell. 2012;47:797–809. doi: 10.1016/j.molcel.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlhac P, Gregoire IP, Azocar O, Petkova DS, Baguet J, Viret C, Faure M. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host Microbe. 2015;17:515–525. doi: 10.1016/j.chom.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Xu C, Feng K, Zhao X, Huang S, Cheng Y, Qian L, Wang Y, Sun H, Jin M, Chuang TH, et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy. 2014;10:2239–2250. doi: 10.4161/15548627.2014.981792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Zhang T, Dong K, Liang W, Xu D, Xia H, Geng J, Najafov A, Liu M, Li Y, Han X, et al. G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. Elife. 2015;4:e06734. doi: 10.7554/eLife.06734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.