Abstract
PolC is one of two essential replicative DNA polymerases found in the Gram-positive bacterium Bacillus subtilis. The B. subtilis replisome is eukaryotic-like in that it relies on a two DNA polymerase system for chromosomal replication. To quantitatively image how the replicative DNA polymerase PolC functions in B. subtilis, we applied photobleaching-assisted microscopy, three-dimensional superresolution imaging, and single-particle tracking to examine the in vivo behavior of PolC at single-molecule resolution. We report the stoichiometry of PolC proteins within each cell and within each replisome, we elucidate the diffusion characteristics of individual PolC molecules, and we quantify the exchange dynamics for PolC engaged in lagging strand synthesis. We show that PolC is highly dynamic: this DNA polymerase is constantly recruited to and released from a centrally located replisome, providing, to our knowledge, new insight into the organization and dynamics of the replisome in bacterial cells.
Introduction
All cells must accurately duplicate and segregate their chromosomal DNA, and failure to do so is a critical underlying cause of >80 different human diseases and genetic disorders (1). Replisomes are multiprotein assemblies that are responsible for DNA replication in all cells (2, 3, 4). DNA polymerases are the essential replisome components that synthesize new DNA with high fidelity (5, 6). It is therefore appropriate that great efforts have been made to better understand how DNA polymerases function both in vitro and in vivo. For example, the stoichiometry and architecture of the replicative DNA polymerase holoenzyme (Pol III) have been well characterized in the model organism Escherichia coli (7, 8, 9). Although E. coli has served as a prototype for understanding DNA synthesis in vivo, and although some E. coli DNA replication features are conserved across species, the replisomes of many other bacterial species have distinct organizations and operate differently (10, 11, 12). In particular, the replisome in the Gram-positive model bacterium Bacillus subtilis does not seem to replicate DNA by actively tracking along DNA, in contrast to the E. coli replisome (7, 13, 14). Rather, the B. subtilis replisome has been shown to reside in a more restricted location through which template DNA is pulled in and newly synthesized DNA is extruded (15), although the replicative DNA polymerase in B. subtilis has not been analyzed at single-molecule resolution. In addition, B. subtilis utilizes two distinct essential DNA polymerases, PolC and DnaE, for genome replication (10, 12). Overall, the in vivo composition and architecture of the elongating replicative DNA polymerase in B. subtilis is poorly understood.
In vitro reconstitution of the B. subtilis replisome has demonstrated that PolC (163 kD) is responsible for all leading strand synthesis as well as most lagging strand synthesis, whereas the more error prone and much slower DNA replicase DnaE (25 nt/s for DnaE compared to ∼500 nt/s for PolC) plays a crucial role in initiating lagging strand synthesis (10, 16). DnaE is important for extending the lagging strand RNA primer before handing off to PolC, which then completes replication of the Okazaki fragment (10). The synergistic relationship between two polymerases in the B. subtilis replisome resembles the synergy found in eukaryotic replication. For example, in S. cerevisiae, two essential replicases (17), Pol ε and Pol δ, are, respectively, responsible for synthesizing the leading and the lagging strands (18, 19, 20). In analogy with DnaE in B. subtilis, the eukaryotic polymerase Pol α extends RNA primers for a short segment before handing off to Pol δ (21). Given these observations, the less well-understood B. subtilis replisome appears more eukaryotic-like than the E. coli replisome, and a deeper in vivo understanding of how DNA polymerase recruitment and dynamics are performed in B. subtilis will provide valuable insight into how bacteria and eukaryotic organisms use a two-DNA polymerase system for replication. Here, we apply photobleaching-assisted microscopy (22, 23), three-dimensional (3D) single-molecule superresolution microscopy (24), and single-particle tracking (25, 26) to PolC in live B. subtilis cells to measure the stoichiometry, intracellular distribution, and dynamics of the essential DNA polymerase required for the majority of leading and lagging strand synthesis.
Materials and Methods
Microscopy
B. subtilis cells from a plate were inoculated in S750 minimal medium at OD600 ∼0.1, followed by growth with shaking at 30°C for ∼3.5 h to OD600 ∼0.5–0.6. When it was used, a final concentration of 162 μM HPUra was added to the cell culture immediately before imaging. Cells were deposited on pads of 1% agarose in S750, each of which was sandwiched between two coverslips. As discussed in our prior studies, B. subtilis is highly susceptible to fluorescent impurities present on the coverslips and in the growth medium (27). The impurities produce single-molecule-like fluorescent signals that largely resemble those from PAmCherry in both brightness and photostability. We prevented such artifacts by cleaning the coverslips in an oxygen plasma (PE-50; Plasma Etch, Carson City, NV) at 200 mTorr for at least 20 min and by using freshly prepared growth medium and agarose pads each day to achieve nearly background-free imaging conditions (Fig. 1 a).
Figure 1.
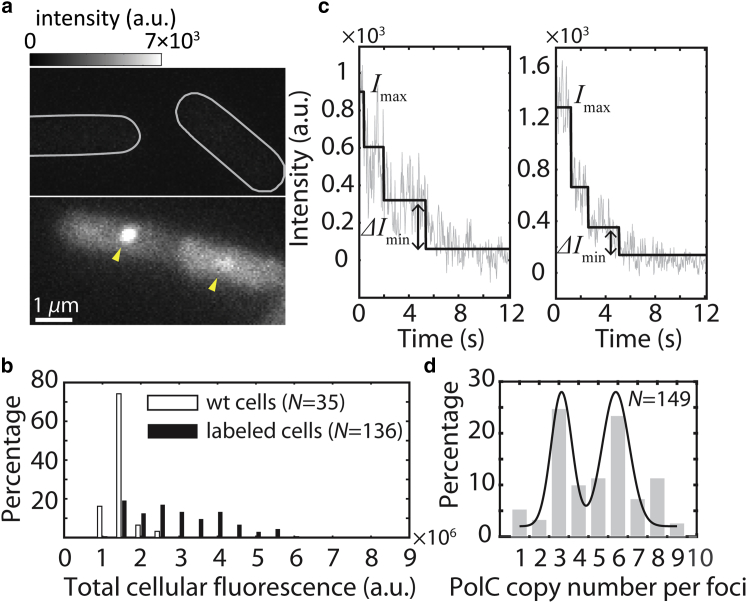
PolC stoichiometry within each cell and at each replisome. (a) Representative fluorescence image of wt B. subtilis cells (upper) and cells with PolC-mCherry (lower). PolC-mCherry foci are highlighted by yellow arrows. (b) Distribution of initial total cellular fluorescence intensity for wt cells (median = 1.43 × 106) and cells labeled with PolC-mCherry (median = 2.77 × 106). The difference in fluorescence indicates the total intensity of mCherry in labeled cells. (c) Representative background-subtracted intensity time traces (gray lines) of two different PolC-mCherry foci undergoing photobleaching, where different intensity states (black lines) are identified by maximum likelihood estimation. The left panel shows typical, fairly uniform steps, whereas the right panel shows a scenario where multiple photobleaching events occurred within a single frame. (d) Distribution of PolC copy numbers at each PolC-mCherry focus. The black line is a guide to the eye. To see this figure in color, go online.
After preparation, the sample was mounted on a wide-field inverted microscope (Olympus, Melville, NY) for imaging, with appropriate excitation, dichroic, and emission filters (Semrock, Rochester, NY) placed along the light path to achieve optimal signal/noise. Fluorescence emission was collected by a 1.40 NA 100× oil-immersion phase-contrast objective and detected on a 512 × 512 pixel electron-multiplying charge-coupled device detector Evolve camera (Photometrics, Princeton Instruments, Acton, MA).
In our photobleaching-assisted microscopy, the cells were illuminated by a 561-nm laser (Sapphire 561-50; Coherent, Bloomfield, CT) with a power density of 50 W/cm2, and images were recorded at 30 ms/frame. In our 3D superresolution imaging, a weak cylindrical lens in the emission pathway between the microscope and the camera induced astigmatism (24), such that the ellipticity of the microscope point spread function changed based on the axial (z) position of the emitting molecule (Fig. S3 in the Supporting Material). To visualize only 1–3 copies of PolC-PAmCherry at a time, a 200-ms 405-nm laser (model No. 405-100; Coherent) pulse with power density ∼100 W/cm2 was used to photoactivate PAmCherry, and PolC-PAmCherry molecules were subsequently imaged by a 561-nm laser with a power density of 400 W/cm2 at 50 ms/frame.
Image processing and data analysis
Phase-contrast images of cells were segmented using a custom MATLAB script (The MathWorks, Natick, MA), and subsequent analyses were all performed within cell boundaries. We purposely discarded all chain-forming cells when taking and analyzing images to avoid ambiguities associated with cell segmentation.
In both photobleaching-assisted microscopy and 3D superresolution microscopy, initial estimates for signal locations were made based on band-pass filtered images. For photobleaching-assisted microscopy (Fig. 1), the center pixel of each mCherry focus was determined from the filtered image. In each raw data image, the average intensity of all pixels within 3 pixels of this center pixel was measured. Background fluorescence intensity for each cell in each imaging frame was determined from the average intensity value of all other pixels within the cell not belonging to any mCherry focus. This average background intensity of each cell was subtracted from the intensity of each mCherry focus. The resulting background-corrected focus intensity was recorded over time to monitor the photobleaching process for that mCherry focus and to generate the intensity time traces as in Fig. 1 c.
In the case of 3D superresolution microscopy (Fig. 2), unfiltered images were fit at the positions of the initial guesses with an asymmetric Gaussian function,
| (1) |
where Ibg, A, x0, and y0, respectively, denote the background intensity, the amplitude of emission, and the x- and y-center positions of the molecule. The widths of the signal in the x direction and the y direction characterize the elliptical emission signal, and we precalibrated the relationship between and z position using 0.1 μm TetraSpeck beads (Thermo Fisher Scientific, Waltham, MA) and a PIFOC piezo objective scanner (Physik Instrumente, Karlsruhe, Germany) (Fig. S3). For each localization, we then obtained the z position of the molecule by finding the z value that minimizes the difference between the measured and values.
Figure 2.
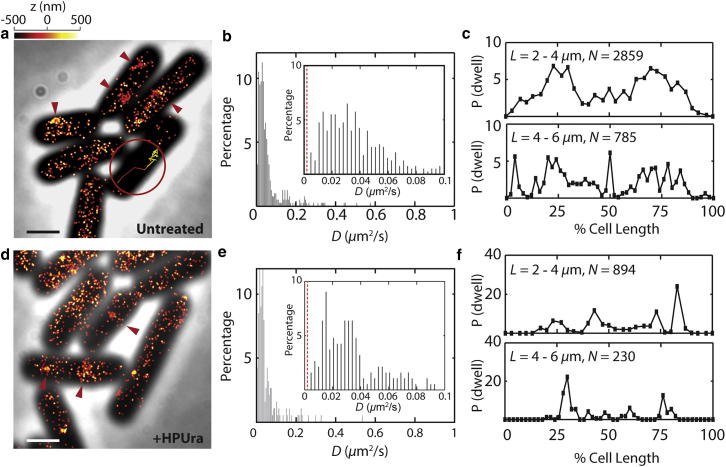
Diffusion and dwelling behavior of PolC-PAmCherry in live B. subtilis cells. (a) 3D superresolution reconstruction image of PolC-PAmCherry in untreated B. subtilis, overlaid on a phase contrast image of the cells. The position of each localization is indicated by a single dot with width corresponding to the average localization precision in the lateral plane (25 nm). The axial (z) position is color-coded according to the color bar above. (Red arrows) Regions of PolC enrichment in the cell. (Inset) A representative, color-coded 3D single-molecule trajectory illustrates a PolC-PAmCherry molecule making a transition from diffusing (bottom) to dwelling (top). (b) Distribution of PolC-PAmCherry diffusion coefficients, D, in untreated cells. (Inset) Zoom-in on the 0–0.1 μm2/s region of the original histogram. (Red dashed line) Average apparent diffusion coefficient (0.003 μm2/s) for stationary PolC-PAmCherry molecules measured in fixed cells. (c) Localization probabilities of dwelling events along the longitudinal cellular axis in untreated cells. L: cell length, N: total number of dwelling events analyzed. (d) 3D superresolution reconstruction image of PolC-PAmCherry in HPUra-treated cells. (e) Distribution of PolC-PAmCherry diffusion coefficients, D, in HPUra-treated cells. (f) Localization probabilities of dwelling events in HPUra-treated cells. Scale bars = 1 μm. To see this figure in color, go online.
The diffusion coefficients of single-molecule trajectories were calculated from the mean squared displacement versus time lag. We analyzed all trajectories at least 10 frames long, and in our calculations we used the squared displacements associated with the first one-third of the time lags to minimize errors associated with higher time lags.
Results and Discussion
Copy number of PolC within a cell
To investigate the copy number of PolC in live B. subtilis cells, we constructed B. subtilis strains natively expressing PolC fused to the red fluorescent protein mCherry (28) as the sole source of PolC. Western blotting of PolC fusions demonstrated that each PolC fusion used in this study was intact (Fig. S1). We measured growth curves and found that cells bearing each fusion showed no growth defects at 37°C in rich medium: the wild-type control strain PY79 doubled in 24.4 min, cells with PolC-mCherry doubled in 22.3 min, and cells with PolC-PAmCherry doubled in 21.1 min (data not shown). Under identical imaging conditions, the fluorescence intensity level in cells harboring PolC-mCherry was elevated compared to the intensity in unlabeled wild-type (wt) PY79 cells (Fig. 1, a and b). The elevated fluorescence emission comes from all the PolC-mCherry molecules in the cytosol. The average number of PolC molecules per cell can be approximated by subtracting the median background fluorescence of wt cells from the median fluorescence intensity of labeled cells and then dividing the background-subtracted labeled cell fluorescence intensity by the intensity of a single PolC-mCherry molecule (Fig. S2). Our estimation from 136 cells indicates that on average, 61 ± 6 (SE) copies of PolC are present within each cell; this copy number is slightly higher than the 40 copies of Pol III core enzymes quantified in E. coli (29); our estimation of PolC copies likely represents a lower bound, considering the possible presence of nonfluorescent or immature PolC fusion copies likely to be present in vivo.
Stoichiometry of PolC at replisome
As expected for a spatially restricted replisome, and consistent with previous studies (2), PolC-mCherry forms distinct foci at the midcell position or near the quarter-cell positions (Fig. 1 a). The PolC-mCherry foci likely represent the site(s) of active DNA replication (2). Photobleaching experiments and in vitro measurements of E. coli have found that three copies of DnaE are positioned at each replication fork (7, 30), while other in vitro studies have found two copies (9). It is therefore important to determine the stoichiometry of the essential DNA polymerase PolC in B. subtilis. Performing photobleaching-assisted microscopy on live cells with PolC-mCherry, we observe PolC-mCherry foci photobleaching in real time (Fig. 1 c). To determine the stoichiometry of PolC at the site of DNA replication, we divided the intensity of the detected state (31) corresponding to the highest copy number of PolC (Imax) by the intensity change corresponding to the smallest jump between two states (ΔImin). Compared to simply counting the number of photobleaching steps, this method gives accurate stoichiometry information even when multiple copies of molecules are photobleached within a single frame (Fig. 1 c, right panel). The distribution of PolC-mCherry copy numbers (Fig. 1 d) indicates that three—or in the case where two sister replication forks spatially overlap, six—copies of PolC are present at each replication fork. If PolC were responsible in vivo for just leading strand replication, we would have expected approximately one PolC molecule at each fork and two at forks that spatially overlap. Our observation that three copies of PolC are present at each fork is most supportive of the model that PolC is responsible for leading strand replication and the majority of lagging strand replication in vivo (10). Taken together with our estimation that ∼61 total copies of PolC are present within each cell, these findings pose an intriguing question: what is the benefit or purpose for cells to express so many copies of PolC that do not appear to be replisome-associated?
In vivo localization and diffusion of PolC
To visualize both replisome-associated and free cytosolic PolC, we replaced the mCherry label on PolC with the photoactivatable red fluorescent protein PAmCherry (32). PAmCherry allows us to visualize the dynamics of individual PolC proteins at the single-molecule level by stochastically switching a small subset (1–3 molecules per cell) of the many PolC-PAmCherry molecules into a fluorescent state at a time. To follow the motion of single PolC molecules in ∼1 μm thick cells, we tracked each photoactivated molecule with astigmatism-based 3D superresolution microscopy to simultaneously resolve the lateral (x, y) as well as the axial (z) position of single PolC molecules (Fig. S3) (24). 3D tracking allowed us to unambiguously resolve PolC motion and dwelling within the 3D confines of a live bacterial cell.
Consistent with what was observed for PolC-mCherry (Fig. 1 a), we detect a population of PolC-PAmCherry specifically enriched near the mid-cell or quarter-cell positions (red arrows in Fig. 2 a), whereas the rest of the PolC-PAmCherry copies were distributed throughout the cell (Fig. 2 a). Furthermore, using single-particle tracking, we unambiguously detected the PolC-PAmCherry molecules in 3D, including occasional direct observations of a molecule changing from a fast-diffusing mode to a slow dwelling motion (Fig. 2 a, inset). Many PolC-PAmCherry molecules diffuse at a rate slower than 0.1 μm2/s (Fig. 2 b), but none were strictly stationary, as all molecules have diffusion coefficients larger than the apparent diffusion coefficient of truly stationary PolC-PAmCherry molecules (0.003 μm2/s) measured in fixed cells (Fig. S4). Referring to other DNA-binding proteins of comparable sizes (33, 34), the range of PolC-PAmCherry diffusion coefficients indicates that the slower moving PolC proteins are engaged in confined motion, and we interpret the slow moving PolC-PAmCherry as a subpopulation actively engaged in DNA replication.
Previously, we observed with fluorescence microscopy that two B. subtilis replisome subunits, the processivity clamp protein DnaN and the clamp loader protein DnaX, both engage in subtle motions confined within a small domain of ∼100 nm (35); these movements likely represent the dynamics of the B. subtilis replisome in general. Here, defining consecutive steps with 3D displacements <100 nm as dwelling, we next probed where such dwelling events occur for PolC, and whether the PolC dwelling positions were consistent with the location of other replisomal subunits reported earlier, and how the dwelling positions depend on cell growth rate. We found that in most cells of typical lengths of 2–4 μm, PolC predominantly dwelled at the quarter-cell positions (Fig. 2 c), consistent with diffraction-limited images of PolC, DnaX, and DnaN (2, 36). Thus, the slower moving PolC molecules appear to be interacting with the replication fork. In fast-growing B. subtilis cells, new rounds of genome replication can start before cell division occurs, giving rise to three or even four replication forks per cell (15). In our experiment, we did indeed observe that the spatial distribution of PolC dwelling events is more dispersed in longer cells with lengths of 4–6 μm (Fig. 2 c). For example, in longer cells, PolC tends to dwell at the cell center in addition to at the quarter positions, indicating that PolC molecules are actively engaged in DNA replication at multiple sites in fast growing cells.
Notably, the change from fast to slow motion for PolC at the replisome is similar to the behavior we previously observed for the DNA mismatch repair protein MutS (35). It is possible that both proteins exhibit similar dynamics at the replisome simply because DNA binding on the chromosome occurs more readily at the replication fork where the DNA is unwound and more accessible. Alternatively, the fast-to-slow transition quantified at the replisome could be a general behavior of the many replisomal and repair proteins that have affinity for DnaN or single-stranded DNA binding (SSB) protein; these two replication proteins have many known binding partners and therefore might directly recruit proteins, causing the fast-to-slow transition we observed for PolC and MutS in vivo. The latter scenario would suggest that DnaN and SSB protein function like a hub through which DNA polymerases, repair proteins, and replisome components are recruited to and released from active sites of DNA replication. Such an exchange represents a convenient mechanism by which the cell could confine proteins to efficiently coordinate the recruitment of more dynamic proteins that are important for genome replication, maintenance, and repair.
We asked if PolC would persist through interactions with other replication proteins in the absence of ongoing DNA replication. We hypothesized that rapid replication fork arrest would not disperse PolC, and that PolC would continue to be actively recruited to existing replisome regions, while new replisome complexes would not be assembled in the absence of ongoing DNA synthesis. To test the effect of rapid replication fork arrest on PolC localization and dynamics, we treated cells with the DNA replication inhibitor HPUra (10, 37), which rapidly arrests DNA synthesis through a DNA damage-independent process by specifically inhibiting PolC activity (10, 37). HPUra immediately blocks DNA synthesis without collapsing the replisome complex, as previously determined through bulk fluorescence imaging (38). 3D superresolution images of HPUra-treated cells show that PolC still forms foci at the quarter-cell and midcell positions (Fig. 2 d), and the overall distribution of PolC diffusion coefficients remains similar to that of untreated cells (Fig. 2 e). However, the spatial distribution of dwelling events was affected by HPUra treatment: whereas PolC still preferentially slows down at quarter-cell positions, the spatial distribution is much more localized compared to untreated cells, reflecting a less diffuse and more distinct localization probability within a cell (Fig. 2 f). These HPUra experiments demonstrate that pausing DNA replication will stop formation of new replisome complexes, but will not prevent PolC from engaging in the fast-to-slow diffusion transitions which we suggest occur through protein-protein interactions that take place at the replication fork. Our results further suggest that PolC is recruited only after the assembly of the replisome has begun.
Dwell time of PolC at the replisome
Under normal conditions, in addition to interacting with various replisomal subunits, PolC also interacts with DNA extensively during replication. Thus, although PolC undergoes dynamic exchange at the replisome regardless of ongoing DNA replication, the extent to which PolC dwells at the replisome likely depends on whether it is engaged in DNA synthesis. To test the relationship between PolC dwell times and DNA synthesis, we quantified the dwell times for PolC during normal growth and after HPUra treatment.
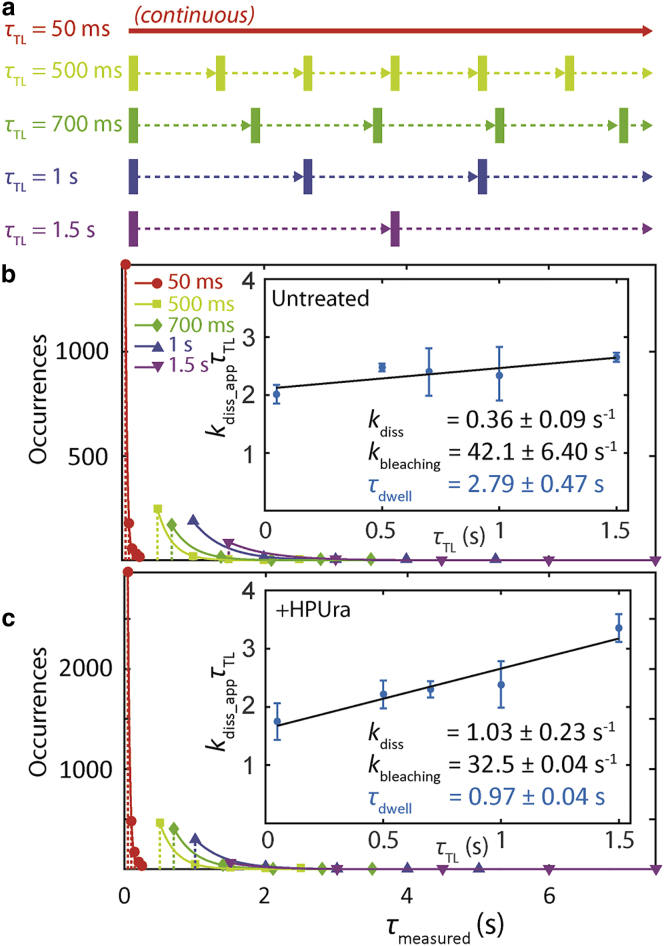
Due to the limited photostability of fluorescent proteins, continuous illumination yields single-molecule PolC-PAmCherry trajectories that last on average for only ∼15 imaging frames (∼750 ms) before PAmCherry undergoes irreversible photobleaching. We therefore performed time-lapse imaging to capture dwelling behavior of PolC-PAmCherry molecules at multiple timescales (39). In our time-lapse imaging mode, every frame is still captured with a 50-ms image integration time (τint), but a time delay (τdelay) of 0–1.45 s is introduced between each pair of consecutive frames. The time-lapse period (τTL) is defined as the sum of τint and τdelay (Fig. 3 a).
Figure 3.
Dwell time analysis for PolC-PAmCherry. (a) Scheme for time-lapse imaging. Every imaging frame is captured with a 50-ms integration time (solid rectangles), and a time delay (dashed arrows) is introduced between each pair of consecutive frames. The time-lapse period (τTL) is the sum of the integration time and the time delay. (b) Dwell time distributions for PolC-PAmCherry in untreated cells. For clarity, the distributions are shown as stem plots, and the color corresponds to τTL as described in (a). (Solid lines) Fits to the exponential decay in Eq. 3. (Inset) Linear fit (black line) of (kdiss_app × τTL) versus τTL, from which the dissociation rate constant kdiss, the photobleaching rate constant kbleaching, and the dwell time constant τdwell are obtained according to Eq. 4. Errors bars are from four rounds of bootstrapping. (c) Dwell time distributions and analysis for PolC-PAmCherry in HPUra-treated cells. To see this figure in color, go online.
To measure the timescale of PolC dwelling events, we model the interaction between PolC and various replisomal subunits as a simple association/dissociation reaction:
| (2) |
This reaction has a forward reaction rate constant of kdiss_app. This apparent dissociation rate of PolC is caused by PAmCherry photobleaching as well as PolC dissociating from the replisome. The kdiss_app can be obtained by plotting the distribution of PolC dwell times, τmeasured, for each of five time-lapse imaging experiments performed with τTL = 50, 500, and 700 ms, and 1 and 1.5 s, respectively (Fig. 3 b). As above, two sequential localizations separated by < 100 nm in 3D are counted as one dwelling event (Fig. S5). In each case, τmeasured = (n – 1) × τTL, where n is the number of sequential frames where dwelling events occurred, and the distribution of τmeasured follows an exponential decay function:
| (3) |
During imaging, two Poisson processes contribute to the observed apparent rate of dissociation of PolC-PAmCherry: (1) photobleaching of PAmCherry with a rate constant of kbleaching, and (2) PolC dissociating from the replisome with a rate constant of kdiss. Photobleaching of PAmCherry occurs only when the laser illumination is on (i.e., only during τint), while the physical dissociation of PolC from the replisome can occur any time during the time-lapse interval τTL. These two processes contribute independently to the apparent dissociation, and respectively produce each of the two terms on the right-hand side of (39):
| (4) |
Equation 4 indicates that the relation between kdiss_app × τTL and τTL is linear. The average dwell time, τdwell, of PolC can thus be extracted by plotting kdiss_app × τTL versus τTL (Fig. 3 b, inset): dividing the y intercept of this equation by τint yields the photobleaching rate constant kbleaching = 42.1 ± 6.4 s−1, and the slope corresponds to the real dissociation rate constant kdiss = 0.36 ± 0.09 s−1, which is the reciprocal of the PolC dwell time constant τdwell = 2.79 ± 0.47 s. It should be noted that τdwell does not depend on whether or how long PolC has been dwelling before PAmCherry was photoactivated, as exponential distributions are by definition memoryless: the dwell time distribution remains the same even if PolC-PAmCherry molecules have been bound to the replisome for some amount of time before being photoactivated and imaged.
Because our measurements do not discriminate between PolC engaged in leading or lagging strand synthesis, and because many more copies of PolC are required to synthesize the lagging strands due to the shorter length of the Okazaki fragments, we hypothesize that the majority of our measurements of PolC-PAmCherry correspond to PolC synthesizing the lagging strands. In B. subtilis, the speed of the replication fork is ∼500 nt/s (10, 37). With our measured average dwell time close to 3 s, these data correspond to a PolC molecule synthesizing ∼1500 nucleotides each time it is recruited to the replisome, which is consistent with the typical length of ∼1–2 kbp for a single Okazaki fragment on the lagging strand for B. subtilis (10). Unlike lagging strands that are synthesized in short segments, the leading strands are synthesized more continuously by a single PolC protein for an extended period of time. Therefore, the dwell time for PolC engaged in leading strand synthesis is likely to be significantly longer than that of PolC synthesizing the lagging strand, and we do indeed occasionally detect PolC copies dwelling for much longer; we attribute these longer (up to ∼5–10 s) dwell times to PolC synthesizing the leading strand. If these measurements do in fact represent PolC engaged in leading strand synthesis, we would expect that 2500–5000 nucleotides are incorporated during the longer dwell times that we observe. However, when we attempted to fit our measured dwell time distributions with two timescales, the percentage contribution of the second population at all τTL values was always smaller than 2%, and the fit uncertainty associated with the dwell time of the second population was too large to make meaningful conclusions. Besides technical challenges related to the mismatch in timescales of fast and slow dwell times, the small number of observations of long-dwelling PolC molecules might also be a realistic reflection of the relatively small percentage of PolC engaged in leading strand synthesis compared to PolC synthesizing Okazaki fragments on the lagging strand.
To determine whether the PolC dwell time is affected by arresting DNA replication, we calculated the PolC dwell time in the presence of HPUra, and we found that τdwell decreased to 0.97 ± 0.04 s (Fig. 3 c). Because no DNA replication takes place after HPUra is added, the residual dwelling behavior of PolC can be explained in two ways: (1) the dwell times of all PolC molecules at the replisome are reduced, and the residual dwell time is solely due to interactions between PolC and certain replisome proteins, such as the replication sliding clamp DnaN, as well as from transient, nonspecific interactions between PolC and DNA (in this case, the decreased τdwell measured here establishes a baseline for discriminating between replicating PolC and these transient interactions); or (2) among the three PolC copies at the replisome, two have had their dwell time reduced to ∼0 s, but the dwell time of the third copy is not affected by HPUra. In the latter case, the third PolC copy is not involved in DNA replication concurrently with the other two, but rather waits until one of the other two copies is released from either leading or lagging strand replication. Further studies are required to discriminate between these two possibilities.
Conclusions
Here, we have applied photobleaching-assisted microscopy, 3D superresolution microscopy, and single-particle tracking to quantify the copy number, localization, and dynamics of the essential DNA polymerase PolC in live B. subtilis cells. At the single-molecule level, PolC is highly diffusive, but dwells at specific subcellular positions on a timescale consistent with that required to synthesize one Okazaki fragment. Furthermore, our results show that PolC constantly undergoes dynamic exchange centered on a replisome as it is recruited to and released from an active site of DNA replication. Our results support the model that during genome replication, the relatively confined replisome is assembled and then functions as a central hub to coordinate the recruitment of PolC and other more dynamic proteins to the site of active DNA synthesis. Our stoichiometric analysis of PolC showing three and six copies in a focus supports in vitro models that PolC is responsible for leading strand and the majority of lagging strand replication (10). Furthermore, our imaging data and dwell-time analysis suggest that PolC exchanges on a rapid timescale, leading to the possibility that the seemingly large number of PolC molecules in the cell is necessary to ensure a sufficiently high probability of PolC recruitment to the replisome via PolC-DnaN (β-clamp) interactions. Therefore, a large number of PolC molecules per cell would ensure that PolC is always positioned at the assembled replisome to engage in leading and lagging strand synthesis. Such a mechanism would allow for efficient synthesis of each Okazaki fragment as PolC rapidly exchanges; this mechanism would also ensure rapid exchange on the leading strand in the event that PolC encounters a block, lesion, or other impediment.
Author Contributions
Yi L., J.W.S., L.A.S., and J.S.B. designed research; Yi L., Yilai L., and J.W.S. performed research; Yi L., L.A.S., and J.S.B. analyzed data; and Yi L., Yilai L., J.W.S., L.A.S., and J.S.B. wrote the article.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant No. R01-GM107312 to L.A.S.; and J.W.S. was supported in part by the National Institutes of Health (NIH) National Research Service award No. T32-GM007544.
Editor: Antoine van Oijen.
Footnotes
Five figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)31033-5.
Contributor Information
Lyle A. Simmons, Email: lasimm@umich.edu.
Julie S. Biteen, Email: jsbiteen@umich.edu.
Supporting Material
References
- 1.DePamphilis M.L., editor. DNA Replication and Human Disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. [Google Scholar]
- 2.Lemon K.P., Grossman A.D. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 3.Migocki M.D., Lewis P.J., Harry E.J. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol. Microbiol. 2004;54:452–463. doi: 10.1111/j.1365-2958.2004.04267.x. [DOI] [PubMed] [Google Scholar]
- 4.Berkmen M.B., Grossman A.D. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol. Microbiol. 2006;62:57–71. doi: 10.1111/j.1365-2958.2006.05356.x. [DOI] [PubMed] [Google Scholar]
- 5.Johnson A., O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 6.McHenry C.S. DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 7.Reyes-Lamothe R., Sherratt D.J., Leake M.C. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgescu R.E., Kurth I., O’Donnell M.E. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat. Struct. Mol. Biol. 2011;19:113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohrmann P.R., Correa R., McHenry C.S. The DNA polymerase III holoenzyme contains γ and is not a trimeric polymerase. Nucleic Acids Res. 2016;44:1285–1297. doi: 10.1093/nar/gkv1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders G.M., Dallmann H.G., McHenry C.S. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Sun J., Shi Y., O’Donnell M.E. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dervyn E., Suski C., Ehrlich S.D. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. doi: 10.1126/science.1066351. [DOI] [PubMed] [Google Scholar]
- 13.Joshi M.C., Bourniquel A., Bates D. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc. Natl. Acad. Sci. USA. 2011;108:2765–2770. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates D., Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemon K.P., Grossman A.D. Movement of replicating DNA through a stationary replisome. Mol. Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 16.Le Chatelier E., Bécherel O.J., Jannière L. Involvement of DnaE, the second replicative DNA polymerase from Bacillus subtilis, in DNA mutagenesis. J. Biol. Chem. 2004;279:1757–1767. doi: 10.1074/jbc.M310719200. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T.A. Balancing eukaryotic replication asymmetry with replication fidelity. Curr. Opin. Chem. Biol. 2011;15:620–626. doi: 10.1016/j.cbpa.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgescu R.E., Schauer G.D., O’Donnell M.E. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife. 2015;4:e04988. doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nick McElhinny S.A., Gordenin D.A., Kunkel T.A. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pursell Z.F., Isoz I., Kunkel T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel T.A., Burgers P.M. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burnette D.T., Sengupta P., Kachar B. Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc. Natl. Acad. Sci. USA. 2011;108:21081–21086. doi: 10.1073/pnas.1117430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonson P.D., Rothenberg E., Selvin P.R. Single-molecule-based super-resolution images in the presence of multiple fluorophores. Nano Lett. 2011;11:5090–5096. doi: 10.1021/nl203560r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang B., Wang W., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 26.Elmore S., Müller M., Woldringh C.L. Single-particle tracking of oriC-GFP fluorescent spots during chromosome segregation in Escherichia coli. J. Struct. Biol. 2005;151:275–287. doi: 10.1016/j.jsb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Tuson H.H., Aliaj A., Biteen J.S. Addressing the requirements of high-sensitivity single-molecule imaging of low-copy-number proteins in bacteria. ChemPhysChem. 2016;17:1435–1440. doi: 10.1002/cphc.201600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaner N.C., Campbell R.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 29.Maki H., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the α subunit, devoid of nuclease activities. J. Biol. Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- 30.McInerney P., Johnson A., O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol. Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Watkins L.P., Yang H. Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 32.Subach F.V., Patterson G.H., Verkhusha V.V. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elf J., Li G.W., Xie X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uphoff S., Reyes-Lamothe R., Kapanidis A.N. Single-molecule DNA repair in live bacteria. Proc. Natl. Acad. Sci. USA. 2013;110:8063–8068. doi: 10.1073/pnas.1301804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y., Schroeder J.W., Biteen J.S. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc. Natl. Acad. Sci. USA. 2015;112:E6898–E6906. doi: 10.1073/pnas.1507386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmons L.A., Davies B.W., Walker G.C. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol. Cell. 2008;29:291–301. doi: 10.1016/j.molcel.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J.D., Sanders G.M., Grossman A.D. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goranov A.I., Katz L., Grossman A.D. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. USA. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gebhardt J.C., Suter D.M., Xie X.S. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.