Abstract
Magnetotactic bacteria possess cellular compartments called magnetosomes that sense magnetic fields. Alignment of magnetosomes in the bacterial cell is necessary for their function, and this is achieved through anchoring of magnetosomes to filaments composed of the protein MamK. MamK is an actin homolog that polymerizes upon ATP binding. Here, we report the structure of the MamK filament at ∼6.5 Å, obtained by cryo‐Electron Microscopy. This structure confirms our previously reported double‐stranded, nonstaggered architecture, and reveals the molecular basis for filament formation. While MamK is closest in sequence to the bacterial actin MreB, the longitudinal contacts along each MamK strand most closely resemble those of eukaryotic actin. In contrast, the cross‐strand interface, with a surprisingly limited set of contacts, is novel among actin homologs and gives rise to the nonstaggered architecture.
Keywords: Cryo‐EM, actin, magnetosome, filaments
Introduction
The bacterial cytoskeleton plays essential roles in cell organization and shape. It mainly consists of filaments composed of distant homologs of the mammalian cytoskeleton protein, actin.1, 2 However, unlike mammalian cells where a single actin filament performs multiple functions through the recruitment of various binding partners, bacteria have evolved individual actin homologs for each role. Such bacterial actins include, MreB, essential for cell shape maintenance, ParM and AlfA, which play a role in plasmid segregation, and FtsA, involved in cell division.3 In all cases, the protein possesses ATPase activity, and filament formation is induced by binding to ATP, while hydrolysis to ADP leads to filament disassembly and ADP release.4
Structural analyses have revealed the common fold shared by all the actin‐like proteins, with two domains (I and II), each divided into a and b subdomains.3 ATP binds between the domains, inducing a conformational change that promotes assembly. Recent high‐resolution cryo‐Electron Microscopy (EM) and X‐ray crystallography studies have revealed the architecture of a number of actin‐like filaments, including mammalian actin (hereafter referred to as F‐actin),5, 6 the bacterial actin homologs MreB7 and ParM,8 and the archaeal filament Crenactin.9, 10 However, despite the structural similarities of the protomers and the use of the same face of the protomer for longitudinal assembly contacts, a wide range of filament arrangements arise from variation in cross‐strand as well as longitudinal contacts, including single‐ and double‐stranded, staggered and nonstaggered, right‐handed and left‐handed helices.2 For this reason, the helical architecture of other actin‐like filaments remains difficult to predict.
Magnetotactic bacteria such as Magnetospirillum magneticum AMB‐1 are capable of aligning in the earth's magnetic field, which allows them to swim towards sources of oxygen.11 This is achieved through the formation of magnetosomes, ∼50 nm membrane‐bound cellular compartments in which single magnetite or greigite crystals are biomineralized.12, 13 Efficient orientation in magnetic fields arises from alignment of multiple magnetosomes, which are positioned in a linear array that acts much like a compass needle. Alignment of magnetosomes is achieved by apparent binding to a cytoplasmic filament consisting of the magnetosome‐specific actin homolog MamK.14, 15, 16, 17, 18
In the model magnetotactic bacterium M. magneticum AMB‐1, MamK was shown to possess ATPase activity, and to form filaments with a unique architecture upon ATP binding.19, 20 MamK has been proposed to interact with the magnetosome components MamJ and MamE, as well as with a number of other proteins.21 MamJ and LimA have also been shown to promote MamK dynamics, suggesting a regulatory role in magnetosome anchoring to the MamK filament.22 A second MamK homolog, MamK‐like, was identified in the AMB‐1 genome, away from the magnetosome gene island. MamK‐like was shown to co‐localize with the MamK filament,23, 24 although the physiological relevance of this protein is poorly understood.
To further understand the structure of the MamK filament, we determined its structure at a resolution of ∼6.5 Å using cryo‐EM. We used this map to generate an atomic model of the MamK filament. These results confirm the previously proposed double‐stranded, nonstaggered organization,19 and reveals the molecular contacts in the oligomer. Intriguingly, the cross‐strand interaction responsible for the nonstaggered arrangement is limited to one bulge in domain Ia contacting the adjacent subunit, leading to a large gap between strands. Comparison with other actin‐like filaments show that while MamK's cross‐strand lateral interaction is largely unique, the longitudinal interactions along the helix are most similar to that of F‐actin, despite the fact that MamK is more similar to MreB at the sequence level.
Results and Discussion
Cryo‐EM structure of the MamK filament
We had previously reported a structure of the M. magneticum AMB‐1 MamK filament at ∼12 Å, revealing a double‐stranded architecture with a unique nonstaggered arrangement.19 However, the limited resolution did not allow us to build an atomic model with sufficient accuracy to investigate the molecular details of filament assembly. We therefore collected a new dataset of MamK filaments [Fig. 1(A)], using direct electron detector technology. This allowed us to obtain a 3D reconstruction of the filament to a resolution of ∼6.5 Å [Fig. 1(C) and Table 1]. The filaments were obtained in the presence of ATP, however previous biochemical studies have shown that MamK rapidly hydrolyzes ATP to ADP and releases Pi (approximately 0.2 μm min−1/μm protein).19 Since we assembled the MamK filaments by incubating the protein in the presence of ATP for ∼5 min prior to grid preparation, it is likely that the imaged filaments have ADP bound.
Figure 1.

Cryo‐EM reconstruction of the MamK filament at 6.5 Å. (A) Representative electron micrograph of frozen‐hydrated MamK. The scale bar is in black at the bottom. (B) Representative class average obtained from a 250‐segment subset of the dataset. The 8‐subunit repeat is indicated. (C) Fourier Shell Correlation curve for the 3D reconstruction. The gold‐standard resolution definition of 0.143% is indicated with a blue dotted line. (D) Final 3D reconstruction of the MamK filament, shown at a contour level of 5.0. The complete map is shown in grey on the left, and the two strands are colored in dark blue and khaki, respectively, on the right.
Table 1.
Structure Determination Parameters and Model Statistics
| Data Acquisition Parameters | |
|---|---|
| Electron energy (kV) | 200 |
| Electron dose (e‐/Å2) | 40 |
| Pixel size | 1.26 |
| Number of movies collected | 240 |
| frames/movie | 24 |
| Defocus range (nm) | −1 to −2 |
| Number of segments picked | 8.129 |
| Box sixe (pixels) | 320 |
| Helical step (pixels) | 120 |
| Model Parameters | |
| Resolution (Å) | 6.5 |
| Symmetry | |
| Rotation (°) | 53.8 |
| Rise (Å) | 24 |
| Dihedral | C2 |
| Number of Atoms (Monomer) | |
| Protein | 2506 |
| Other | 28 |
| RMS deviation | |
| Bond length (Å) | 0.012 |
| Bond angle (°) | 1.4 |
| Ramachandran Plot Values (%) | |
| Most favored | 89.9 |
| Allowed | 7.4 |
| Disallowed | 2.8 |
The reconstruction [Fig. 1(D)] possesses clear secondary structure element features, with notably an extended lateral loop in domain Ia corresponding to the hydrophobic loop found in MreB. A large gap between subunits, visible in the class averages [Fig. 1(B)] and in the reconstruction [Fig. 1(D)], forms a cavity at the center of the structure.
Modeling of the MamK filament structure
We next sought to obtain an atomic model of the MamK filament based on the cryo‐EM map. To that end, we generated a structural model of the MamK monomer, using the MreB crystal structure (PDB ID: 4CZG)7 as a template. As mentioned above, the MamK filaments likely are in the ADP‐bound conformation. We therefore placed an ADP molecule, and the coordinating Mg2+ ion, in the active site, by superposing the ADP‐bound structure of MreB (PDB ID: 4CZG).
This protomer homology model, including an ADP and Mg2+ bound, was refined in the cryo‐EM map by successive rounds of manual building, and refinement using Rosetta (See Materials and Methods, Supplementary Methods and supplementary information Fig. S1 for details). The procedure converged to a local minimum with low energy, excellent fit to EM density [Fig. 2(A)], and good overall geometry (Table 1), with a Procheck score25 of −0.16 and a MolProbity clashscore26 of 10.49.
Figure 2.
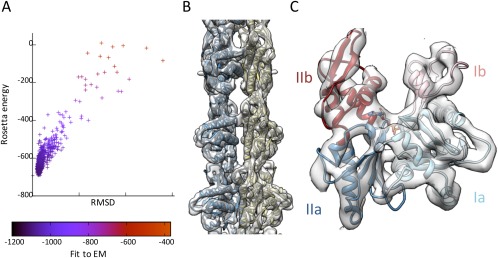
EM‐guided modeling of the MamK filament. (A) Results of the Rosetta Relax refinement step. The energy of each decoy model is plotted versus the RMSD to backbone of the lowest‐energy model, and color‐coded for the fit to the EM map density. (B) Complete atomic model of the MamK filament fitted into the EM map density, shown at a contour level of 5.0. Individual protomers from each strand are colored in dark blue and khaki respectively. (C) Close‐up view of a single MamK molecule, with the EM map shown at a contour level of 5.0. The four actin domains, Ia, Ib, IIa, and IIb are indicated and colored differentially.
The final model, shown in Fig. 2(B,C) in the EM map density, possesses the characteristics of other actin family members, with the four canonical domains clearly identifiable. Clear density for helices is visible, as well as for most loops and strands. The location of the nucleotide corresponds to very strong density (still visible at contour level 8.5 in Chimera), with the ADP molecule located in the cavity between domains Ib and IIb. ADP binding likely induces the two domains to rotate, as described previously for MreB and F‐actin, thus allowing oligomerization. Due to the limited resolution, the exact orientation of the nucleotide cannot be determined unambiguously here, but was modeled nonetheless to prevent the refinement procedure from fitting surrounding loops into the region of the density corresponding to ADP. We note nonetheless that in our model, the ADP molecule and the Mg2+ ion are both coordinated in a chemically sensible manner (supplementary information Fig. S2), with a number of residues conserved across actin homologs involved in the expected interactions. In particular, we note that Glu 143, shown previously to be essential for ATPase activity in MamK, is in a suitable position to act as a proton donor to the catalytic reaction, as proposed for MreB.6
Molecular details of the interactions in the MamK filament
While a few actin‐like filaments have been reported to be single stranded (crenactin9, 10) or four‐stranded (Alp12,27 BtParM28), most of them possess a staggered double‐stranded arrangement, with intimate contacts between strands. In contrast, the MamK filament is nonstaggered, with a large gap between adjacent strands [Fig. 1(D) and supplementary information Fig. S3), and the only contacts are formed between a loop in domain Ia and a helix spanning domains IIa and IIb [Fig. 3(A)]. Notably, this loop is largely absent in MreB [Fig. 6(A)], which adopts an anti‐parallel filament architecture.
Figure 3.

Lateral contacts involved in strands assembly. (A) Two adjacent molecules in opposing strands of the MamK filament model are shown in cartoon representation, in dark blue and khaki, respectively, with the EM map density at a contour level of 5.0 shown in grey. The top panel is represented from the side of the filament, and the bottom panel corresponds to a view from the top of the filament. (B) Close‐up view of the lateral contacts across strands in the MamK filament model shown with a black box in (A), with interacting residues in sticks. Two salt‐bridges, between Glu 81 and Arg 195, and between Arg 85 and Glu 202, form the interface, stabilized by additional hydrogen bonds.
Figure 6.
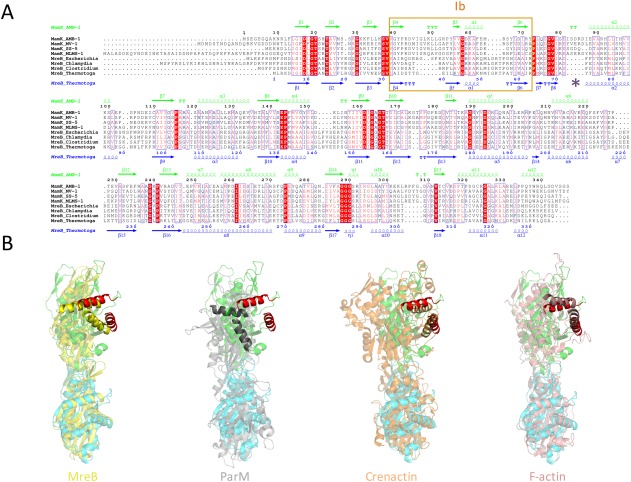
Comparison with other actin‐like filaments. (A) Multiple sequence alignment of MamK and MreB orthologs from a number of bacterial strains. Identical residues are in a red box, similar residues are in red character. The secondary structure elements for MamK and MreB are shown at the top in green, and at the bottom in blue, respectively. Domain Ib is indicated with a pink box, and the loop involved in cross‐strand contacts in MamK is shown with a purple star. (B) Comparison of the filament arrangement between MamK (green and cyan) and MreB (crystal structure‐derived filament, yellow), ParM (gray), Crenactin (orange), and F‐actin (pink). For each of them, two adjacent molecules along the helical axis are shown, where the bottom subunit is aligned to the bottom (green) subunit of MamK. For clarity, two helices from domain Ia are in red in the MamK structures, and highlighted in the other bacterial actins. Despite the closer sequence and structural similarity of the MamK protomer to MreB, the top (green) subunit of MamK aligns more closely to that of F‐actin.
In the MamK atomic model reported here, two salt bridges between Glu 81 and Arg 195, and Arg 85 and Glu 202 respectively, form the interface, further stabilized by the adjacent Asn 198 [Fig. 3(B)]. The overall cross‐strand buried interface area is 516 Å2, compared to 901 for MreB, 733.7 Å2 for ParM and 839.3 Å2 for F‐actin (for ParM and F‐actin, due to the staggered filament architecture the buried area of one subunit to two adjacent molecules were summed). In addition to the smaller cross‐strand buried surface area in MamK, a large cavity (6786 Å3, supplementary information Fig. S3) is present between the strands, which is not observed in other actin filaments.
The key aspect defining the twist and rise of the helix is the longitudinal interaction between subunits along the helical axis. In the MamK filament, this interaction primarily consists in a hairpin loop in domain IIb forming contacts with domain IIa of the next molecule (Fig. 4), similar to the longitudinal interface observed in other actin‐like filaments. While the cleft between domains Ib and IIb is largely negatively charged due to the ADP molecule, the top edge of the molecule possesses a net positive charge, that accommodates the overall positively charged bottom edge (Fig. 4(A)]. More specifically, two salt bridges are formed in our MamK model: between Lys 240 (domain IIb of subunit i) and Glu 266 (domain IIa of subunit i + 1), and between R65 (domain Ib of subunit i) and E275 (domain IIa of subunit i + 1), as illustrated in Figure 4(B). Additional polar residues including Asn 210 and Asn 212 contribute to stabilizing the interaction through hydrogen bonds.
Figure 4.
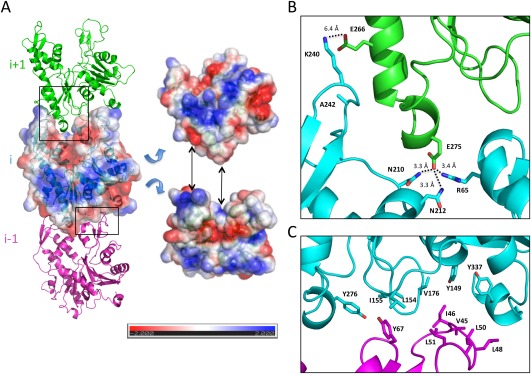
Longitudinal contacts in filament assembly. (A) Three adjacent molecules (i − 1, i, and i + 1) along the helical axis are shown in cartoon representation, in magenta, cyan, and green, respectively. For the central i molecule, a surface representation is also shown, colored according to surface charge. Tilted views of the protomer with charge representation is on the right, illustrating the charge complementarity between a positively charged top edge and a negatively charged bottom edge. (B) Close‐up view of the interface between domain IIb from molecule i and domain IIa of molecule i + 1 (indicated by a black box in (A)), with interacting residues in sticks. Two sets of interactions, each including a salt bridge and additional bonds, can be identified. We note that the distance between Glu 266 and Lys 240 is too large for a bona fide salt bridge, which could be due to errors in our structural model in this region, or could indicate long‐range electrostatics interactions. (C) Close‐up view of the interface between domain Ib from molecule i − 1 and domain Ia of molecule i (indicated by a black box in (A)), with interacting residues in sticks. Several hydrophobic residues, particularly in the loop between residues 48 and 51 (molecule i − 1) and between residues 149 and 155 (molecule i) are exposed.
In addition, we note that domain Ib is located in close proximity to domain Ia of the adjacent subunits. However, analysis of our structural model did not reveal any direct interaction in this region, consistent with the lack of continuous density in the cryo‐EM map. Nonetheless, several surface‐exposed hydrophobic residues are present in this region (Fig. 4(C)) that may help stabilize the oligomer.
The longitudinal interaction is essential for filament assembly in vitro and in vivo
To verify the role of the molecular contacts in filament assembly, we mutated residues at the MamK longitudinal interface. Specifically, we engineered a double mutant where the salt bridge‐forming residue Lys 240 was mutated to an Ala, and the nearby Ala 242 was mutated to Glu, so as to reverse the surface charge on this region of the protein [Fig. 4(A,B)]. The resulting mutant protein (hereafter referred to as MamKMut) was soluble and could be purified to similar yield as the wild‐type protein (MamKWT). We next used light scattering to monitor assembly at various concentrations. As shown in Figure 5(A), MamKWT readily assembles into filaments (in ∼1 min) in the presence of ATP and Mg2+ at 10 µM. In contrast, MamKMut assembly is severely impaired; it did not form filaments at 10 µM, and only slow filament assembly (in ∼5 min) was observed at 35 µM. This indicates that the mutation increases critical concentration, or slows down nucleation such that we can no longer observe it in the course of this experiment. In addition, we spiked MamKMut at 9 µM with 1 µM MamKWT, which induced limited filament formation (MamKWT did not show any assembly at 1 µM under the conditions used in this assay, which is slightly above the critical concentration reported previously, likely due to the difference in KCl concentration). This suggests that the mutation hinders filament nucleation, but retains the capacity to assemble into filaments once suitable nuclei are formed. Negative‐stain electron microscopy was used to confirm that MamKWT forms filaments at 20 µM, but not at 2 µM, while MamKMut can form filaments at 20 µM only in the presence of low amount of MamKWT [Fig. 5(B)].
Figure 5.
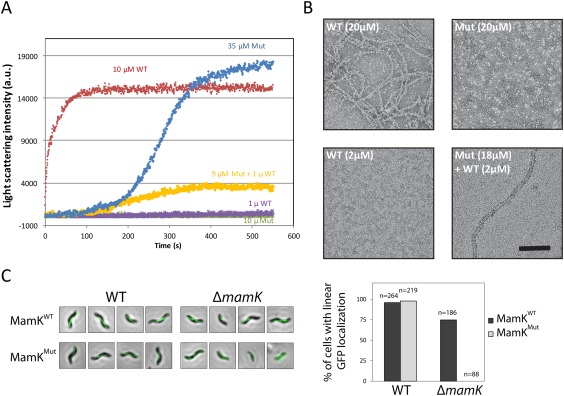
Mutation of the longitudinal interface leads to assembly deficiency. (A) Light scattering was used to monitor polymerization of MamKWT and MamKMut, at various protein concentrations. For each sample, the light scattering intensity (in arbitrary units) is plotted over time. MamKMut has a higher critical assembly concentration, due to its reduced nucleation capacity. (B) Negative‐stain electron micrographs of MamKWT and MamKMut, at various protein concentration, confirming that the light scattering signal in (A) corresponds to filament assembly. (C) GFP‐tagged MamKWT or MamKMut was expressed in AMB‐1 (left) or a mutant strain lacking the mamK gene (right), and the resulting bacteria were observed by fluorescence microscopy. A green fluorescent filament is observed when MamKWT is expressed, regardless of the strain. When MamKMut is expressed, it is incorporated in the MamK filament in the WT strain, but cannot assemble on its own in the ΔmamK strain. The quantification of the corresponding images is shown in the right panel.
Finally, we sought to investigate the interface mutant on MamK assembly in vivo. We had shown previously that a GFP‐fused MamK forms fluorescent filaments, both in the wild‐type AMB‐1 strain, that includes a chromosomal copy of the mamK gene (WT), as well as in a mamK‐deletion strain (ΔmamK) [Fig. 5(C), top panels]. MamKMut‐GFP also formed fluorescent filaments in the WT strain, confirming that the mutation did not affect protein integration into the MamK filament. However, in the ΔmamK strain, MamKMut‐GFP led to diffuse fluorescence, demonstrating that the mutation prevents filament formation in vivo.
Comparison with other actin‐like filaments
MamK shows significant sequence similarity (24%) to the bacterial actin MreB, with a number of identifiable conserved motifs [Fig. 6(A)]. Indeed, the MreB structure was used as a template to generate the initial MamK model, and unsurprisingly monomers of MreB and MamK have a significant structure similarity (backbone RMSD of 9.4 Å for the whole molecule, 3.3 Å for domains Ia and IIa only). In contrast, the sequence identity with other actin‐like molecules is limited to the active‐site residues involved in ATP binding and hydrolysis.2 However, the helical arrangement of each MamK strand differs from that observed in the MreB crystal structure [Fig. 6(B) and supplementary information Fig. S4]. Domain Ib adopts a different architecture in the two proteins, with several inserted or deleted loops [Fig. 6(A)], which correlates with a different filament interface. The ParM filament also adopts a dramatically different arrangement to that of MamK [Fig. 6(B)]. In contrast, while more dissimilar at the sequence level, the helical arrangement of F‐actin resembles that of MamK [Fig. 6(B), right panel], with notably a similar helical rotation (27° for F‐actin, 24° for MamK). Notably, domain Ib in the F‐actin structure is most similar to that of MamK, with similarly positioned loops pointing toward the adjacent subunit. Interestingly, domain Ia of the Crenactin filament structure overlap well to that of MamK, but domain II is largely rotated [Fig. 6(B)] in the former.
In contrast to the longitudinal contact, which is formed by similar protomer surfaces in all actin homologs, the cross‐strand interface is unique to each filament. In MamK, the presence of an insertion loop in domain Ia would not permit the interface found in MreB and other actin homologs. Instead, a different type of filament is formed, and the loop causes each strand to be significantly distanced from each other, with a large cavity in‐between molecules. The very limited buried interface within cross‐strand contacts suggests that it would not permit dimerization without filament assembly. It is likely that the double‐stranded architecture of actins helps stabilize and rigidify the filamentous structure, while the specific arrangement may form specific interfaces for the recruitment of adapters.
Based on these observations, we propose that the structure of domain Ib is the key element that dictates filament twist angle and hand, while domain IIb is mainly responsible for the recruitment of the adjacent subunit upon nucleotide binding. Additional loops and insert regulate the cross‐strand interface. If that hypothesis is correct, it should then be possible to alter the helical arrangement of a given actin‐like homolog, by engineering chimeric proteins that possess domains from other proteins. This could prove useful for the design of new protein‐based biomaterials consisting of filaments with any desired twist angle.
Materials and Methods
Protein expression, purification, and filament assembly
Purification of MamK, and filament assembly, has been described previously.19 Briefly, a plasmid containing the MamK gene under T7 promoter was transformed into E. coli BL21 pLysS. Transformants were grown to mid‐log phase, and protein expression was induced with 1 mM IPTG at 20°C for 16 h. After cell harvesting and lysis, the protein was purified using two successive Ammonium Sulfate cuts in MamK buffer (50 mM HEPES pH 7.0, 50 mM KCl). The obtained protein was further purified by gel filtration, using a Superdex200 column.
The MamKMut‐encoding plasmid for protein expression and purification was created by site‐directed mutagenesis using the primer set below:
Forw: gagttccgcgccgagggcgcgcccgtgcgcgct
Rev: agcgcgcacgggcgcgccctcggcgcggaactc
In the case of MamKMut, AmSO4 cuts were taken at 22.5% saturation, instead of 20% for MamKWT.
For electron microscopy, MamK filaments were formed by incubating with 5 mM ATP and 5 mM MgCl2 for 5 min at 4°C, in MamK buffer.
Electron microscopy
For negative‐stain experiments, protein samples were applied to glow‐discharged carbon‐coated grids, and stained with 1% uranyl formate. Images were collected on a FEI Tecnai G2 Spirit 120 kV TEM equipped with a Gatan Ultrascan 4000 CCD Camera.
For cryo‐experiments, samples were applied to glow‐discharged holey grids, blotted for 6 s, and plunged in liquid ethane using a Vitrobot (FEI). Images were acquired on a FEI Tecnai G2 F20 200 kV Cryo‐S/TEM equipped with a Gatan K‐2 Summit Direct Detect camera with a pixel size of 1.26 Å/pixel.
Image processing and 3D reconstruction
Two hundred and forty images were collected using Leginon,29 movie frames were aligned with Appion30 and the defocus parameters were estimated with CTFFIND.31 Filament were picked using the filament option of the Particle Picker program of Appion, and particle stacks were generated in Appion, using a box size of 320 pixels and a helical step of 120 pixels, leading to 8129 individual overlapping segments, corresponding to ∼32,500 unique MamK particles. Class averages were generated with Xmipp.32
3D reconstruction was obtained by iterative helical real space reconstruction33 in Spider,34 using a featureless cylinder as the initial model and 25 rounds of gold standard refinement.35 The reconstruction converged to a map with an estimated resolution of 6.5 Å. The final helical parameters were 24° azimuthal rotation and 53.8 Å axial rise per helical repeat.
The map was deposited to the EMDB (accession number EMD‐8180).
Model building and refinement in rosetta
Because of the low sequence identity between MamK and MreB (24%), we used the secondary structure‐based threading server Phyre36 to generate an initial MamK model, using the MreB crystal structure (PDB ID: 4CZJ) as a template. The initial model showed reasonable fit to the EM map density, with the exception of domain Ib that was largely positioned outside of the map density. In addition, a C‐terminal helix was predicted in MamK, which is present in the Thermotoga maritima MreB homolog but not in the Caulobacter crescentus one, that was used for the initial model. This helix was therefore not initially modeled, although clear density was visible in the EM map. We then used Coot37 to manually position domain Ib in the map density, and to build the C‐terminal helix.
To further refine the obtained model, we applied the Relax application in Rosetta 3.5, using the EM map as a restraint.38 Energy and fit to EM map were calculated using the Scoring application in Rosetta. We then used the lowest‐energy model to generate a MamK tetramer encompassing all four interfaces in the MamK filament, and repeated the relax procedure so as to refine the interface side‐chains. The detailed sets of Rosetta commands and flags used are shown in the Supplementary methods section. Geometry parameters for the final model, obtained with MolProbity,26 are presented in Table 1.
The resulting model was deposited to the PDB (accession ID 5JYG).
Sequence and structure analysis
The multiple sequence alignment was obtained with ClustalW,39 and the corresponding figure was generated using ESPript.40
All structure figures were generated with either PyMOL41 or Chimera.42
Electrostatic charges were calculated using APBS.43 Buried surface areas were measured with PISA.44 RMSD between structures were obtained in PyMol. Cavity volumes were determined with the 3V server,45 using default parameters.
MamK bulk polymerization assays
MamK polymerization was assessed by right angle light scattering at ambient temperature as previously described.19 Briefly, protein samples were dialyzed into polymerization buffer (10 mM Tris‐HCl, pH 7.4, 75 mM KCl, 10% glycerol, 1 mM DTT). Polymerization was initiated by the addition of 5 mM ATP‐MgCl2 at t0 and light scattering intensities were measured over time. Background signals, prior to the addition of the nucleotide, were subtracted from the data for visualization. All measurements were performed in triplicate.
MamK in vivo polymerization
The in vivo assembly of MamK was assessed through use of a GFP fusion as previously described.14 To investigate the effect of mutations A238E/K240A a derivative of plasmid was created through site‐directed mutagenesis using the primer pair described above. Both plasmids were conjugated into M. magneticum AMB‐1 using Escherichia coli WM3064 as donor strain. Cells were cultured and visualized by fluorescence microscopy.
Supporting information
Supporting Information 1
Supplementary Methods: Detailed flags and procedures employed for the EM‐guided modeling of MamK.
Acknowledgments
We are grateful to Frank DiMaio for help with Rosetta. E. Ozyamak and A. Komeili were supported by a grant from the National Institute of General Medical Sciences (R01GM084122). JMK acknowledges the University of Washington for funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Olga Draper's current address is Office of Environment, Health and Safety, University of California, Berkeley, California.
Meghan E. Byrne's current address is Public Library of Science, San Francisco.
References
- 1. Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC (2015) The evolution of compositionally and functionally distinct actin filaments. J Cell Sci 128:2009–2019. [DOI] [PubMed] [Google Scholar]
- 2. Bork P, Sander C, Valencia A (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 89:7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ozyamak E, Kollman JM, Komeili A (2013) Bacterial actins and their diversity. Biochemistry 52:6928–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dominguez R, Holmes KC (2011) Actin structure and function. Ann Rev Biophys 40:169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S (2015) Structure of the F‐actin‐tropomyosin complex. Nature 519:114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galkin VE, Orlova A, Vos MR, Schroder GF, Egelman EH (2015) Near‐atomic resolution for one state of F‐actin. Structure 23:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van den Ent F, Izore T, Bharat TA, Johnson CM, Lowe J (2014) Bacterial actin MreB forms antiparallel double filaments. eLife 3:e02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bharat TA, Murshudov GN, Sachse C, Lowe J (2015) Structures of actin‐like ParM filaments show architecture of plasmid‐segregating spindles. Nature 523:106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Izore T, Duman R, Kureisaite‐Ciziene D, Lowe J (2014) Crenactin from Pyrobaculum calidifontis is closely related to actin in structure and forms steep helical filaments. FEBS Lett 588:776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Braun T, Orlova A, Valegard K, Lindas A‐C, Schroder GF, Egelman EH (2015) Archaeal actin from a hyperthermophile forms a single‐stranded filament. Proc Natl Acad Sci USA 112:9340–9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lefevre CT, Bazylinski DA (2013) Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev 77:497–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rahn‐Lee L, Komeili A (2013) The magnetosome model: insights into the mechanisms of bacterial biomineralization. Front Microbiol 4:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barber‐Zucker S, Keren‐Khadmy N, Zarivach R (2016) From invagination to navigation: the story of magnetosome‐associated proteins in magnetotactic bacteria. Protein Sci 25:338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komeili A, Li Z, Newman DK, Jensen GJ (2006) Magnetosomes are cell membrane invaginations organized by the actin‐like protein MamK. Science 311:242–245. [DOI] [PubMed] [Google Scholar]
- 15. Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D (2006) An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114. [DOI] [PubMed] [Google Scholar]
- 16. Katzmann E, Muller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schuler D (2011) Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol 82:1316–1329. [DOI] [PubMed] [Google Scholar]
- 17. Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schuler D (2010) Loss of the actin‐like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense . Mol Microbiol 77:208–224. [DOI] [PubMed] [Google Scholar]
- 18. Cornejo E, Subramanian P, Li Z, Jensen GJ, Komeili A (2016) Dynamic remodeling of the magnetosome membrane is triggered by the initiation of biomineralization. mBio 7:e01898–e01815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ozyamak E, Kollman J, Agard DA, Komeili A (2013) The bacterial actin MamK: in vitro assembly behavior and filament architecture. J Biol Chem 288:4265–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sonkaria S, Fuentes G, Verma C, Narang R, Khare V, Fischer A, Faivre D (2012) Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin‐like homolog, MamK. PLoS One 7:e34189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan W, Xie C, Lv J (2012) Screening for the interacting partners of the proteins MamK & MamJ by two‐hybrid genomic DNA library of Magnetospirillum magneticum AMB‐1. Curr Microbiol 64:515–523. [DOI] [PubMed] [Google Scholar]
- 22. Draper O, Byrne ME, Li Z, Keyhani S, Cueto Barrozo J, Jensen G, Komeili A (2011) MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol 82:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abreu N, Mannoubi S, Ozyamak E, Pignol D, Ginet N, Komeili A (2014) Interplay between two bacterial actin homologs, MamK and MamK‐Like, is required for the alignment of magnetosome organelles in Magnetospirillum magneticum AMB‐1. J Bacteriol 196:3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rioux J‐B, Philippe N, Pereira S, Pignol D, Wu L‐F, Ginet N (2010) A second actin‐like MamK protein in Magnetospirillum magneticum AMB‐1 encoded outside the genomic magnetosome island. PLoS One 5:e9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laskowski RA, MacArthur MW, Thornton JM (1998) Validation of protein models derived from experiment. Curr Opin Struct Biol 8:631–639. [DOI] [PubMed] [Google Scholar]
- 26. Chen VB, Arendall IIIWB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popp D, Narita A, Lee LJ, Ghoshdastider U, Xue B, Srinivasan R, Balasubramanian MK, Tanaka T, Robinson RC (2012) Novel actin‐like filament structure from Clostridium tetani. J Biol Chem 287:21121–21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang S, Narita A, Popp D, Ghoshdastider U, Lee LJ, Srinivasan R, Balasubramanian MK, Oda T, Koh F, Larsson M, Robinson RC (2016) Novel actin filaments from Bacillus thuringiensis form nanotubules for plasmid DNA segregation. Proc Natl Acad Sci USA 113:E1200–E1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. [DOI] [PubMed] [Google Scholar]
- 30. Lander GC, Staff SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P‐W, Lyumkis D, Potter CS, Carragher B (2009) Appion: an integrated, database‐driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rohou A, Grigorieff N (2015) CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de la Rosa‐Trevin JM, Oton J, Marabini R, Zaldivar A, Vargas J, Carazo JM, Sorzano COS (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol 184:321–328. [DOI] [PubMed] [Google Scholar]
- 33. Egelman EH (2010) Reconstruction of helical filaments and tubes. Methods Enzymol 482:167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J (2008) SPIDER image processing for single‐particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3:1941–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scheres SH, Chen S (2012) Prevention of overfitting in cryo‐EM structure determination. Nat Methods 9:853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y, Baker D (2015) Atomic‐accuracy models from 4.5‐A cryo‐electron microscopy data with density‐guided iterative local refinement. Nat Methods 12:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinform Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 40. Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. The PyMOL Molecular Graphics System, Schrödinger, LLC, 2006. [Google Scholar]
- 42. Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157:281–287. [DOI] [PubMed] [Google Scholar]
- 43. Unni S, Huang Y, Hanson RM, Tobias M, Krishnan S, Li WW, Nielsen JE, Baker NA (2011) Web servers and services for electrostatics calculations with APBS and PDB2PQR. J Comput Chem 32:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. [DOI] [PubMed] [Google Scholar]
- 45. Voss NR, Gerstein M (2010) 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res 38:W555–W562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1
Supplementary Methods: Detailed flags and procedures employed for the EM‐guided modeling of MamK.


