Abstract
DNA replication in Eukaryotes is a highly dynamic process that involves several dozens of proteins. Some of these proteins form stable complexes that are amenable to high‐resolution structure determination by cryo‐EM, thanks to the recent advent of the direct electron detector and powerful image analysis algorithm. But many of these proteins associate only transiently and flexibly, precluding traditional biochemical purification. We found that direct mixing of the component proteins followed by 2D and 3D image sorting can capture some very weakly interacting complexes. Even at 2D average level and at low resolution, EM images of these flexible complexes can provide important biological insights. It is often necessary to positively identify the feature‐of‐interest in a low resolution EM structure. We found that systematically fusing or inserting maltose binding protein (MBP) to selected proteins is highly effective in these situations. In this chapter, we describe the EM studies of several protein complexes involved in the eukaryotic DNA replication over the past decade or so. We suggest that some of the approaches used in these studies may be applicable to structural analysis of other biological systems.
Keywords: Cryo‐EM, DNA replication, eukaryotic DNA replication, origin recognition complex, replicative helicase
Introduction
Thanks to the recent advance in direct electron detection technology and image processing methods, cryo‐EM is now a high resolution structural tool that is complementary to X‐ray crystallography and NMR.1, 2, 3 In the cases of membrane proteins and large protein complexes, cryo‐EM is particularly advantageous over other methods because the method requires smaller amount of samples and can tolerate some level of sample heterogeneity.4
The DNA double helix structure and its implication for replication were discovered over half a century ago.5, 6 Since then, a wealth of knowledge has accumulated about how a genome is duplicated at the cellular level.7, 8 But less is known at the molecular and mechanistic level, particularly in eukaryotes.9, 10, 11, 12 Unlike the well‐conserved molecular machinery for protein and RNA synthesis, the DNA replication machinery is less conserved and appears to have evolved twice independently in Bacteria and Archaea/Eukaryota.13 Indeed, many key proteins involved in DNA replication, such as the replicative helicase and DNA polymerases are unrelated between Bacteria and Archaea/Eukaryota. The bacterial helicases move on the lagging strand DNA template in a 5′‐to‐3′ direction whereas the eukaryotic helicases move on the leading strand DNA template in a 3′‐to‐5′ direction.14 Therefore, it is important to understand how DNA replication is executed in eukaryotes, in order to discern DNA replication defects to human medical conditions such as cancers.15, 16 DNA replication in bacteria and archaea has been reviewed recently.17, 18, 19 We focus this review on the most recent structural studies of eukaryotic DNA replication.
Eukaryotic DNA replication starts at many discrete sites in the genome, referred to as the origins of replication.20 There are several hundred replication origins in lower eukaryotes such as yeast and tens of thousands of origins in mammals.21, 22 To initiate DNA replication, a eukaryotic cell assembles a six‐protein complex called Origin Recognition Complex (ORC) at the replication origins in the G1 phase of the cell cycle.23 ORC, with the help of additional initiation factors Cdc6 and Cdt1, recruit the replication helicase core Mcm2‐7 hexamer.24, 25, 26, 27, 28, 29 The Mcm2‐7 is recruited onto double stranded DNA (dsDNA) as an inactive double hexamer.30, 31 At the G1‐to‐S transition, the double hexamers are converted to two active helicases comprised of Cdc45‐Mcm2‐7‐GINS (CMG) that each encircles single stranded DNA (ssDNA) to carry out bi‐directional duplex DNA separation.32 The eukaryotic CMG helicase does not work alone; it physically interacts and coordinates with DNA primase and polymerases to synthesize DNA on both leading and lagging strands.9, 33, 34 In the following sections, we review recent structural studies of these DNA replication complexes, focusing primarily on EM‐based studies. Some of the studies are at low resolution by negative stain EM but nevertheless capture key dynamic complexes and transient states, while others are at near atomic resolution by using the state‐of‐the‐art cryo‐EM facilities. Together, these studies have begun to shed light on the eukaryotic DNA replication mechanism. We conclude the review by providing a brief outlook on what's to come in the near future.
ORC Structure and Mapping ORC Subunits by MBP Fusion
ORC is a six‐protein complex, comprised of Orc1 through Orc6, named in the descending order of the protein's molecular weight.23 ORC is highly conserved among eukaryotes.35 ORC is an ATPase and binds DNA in an ATP‐dependent manner. The Saccharomyces cerevisiae ORC (ScORC) recognizes over 300 DNA replication origin sequences, the most well‐characterized of which is the autonomously replicating sequence 1 (ARS1).36 However, there are no defined origin sequences in Schizosaccharomyces pombe and higher organisms. The S. pombe and the Drosophila melanogaster ORC (SpORC and DmORC) preferentially bind to supercoiled DNA.37, 38 Therefore, ORC binding may be defined by the local DNA and chromatin topological structure and environment.39, 40, 41 Negative stain EM study has revealed that the ScORC is a crescent‐like bi‐lobed structure that is 16 nm long and 12 nm wide with a main gap [Fig. 1(A)]. The DmORC has a similar shape, and the shape is not markedly changed upon hyper phosphorylation.42, 43 Binding of the S. cerevisiae Cdc6 to ORC induces a change in its DNA footprint, and the binding requires the Orc1 ATPase.44 EM study has revealed that Cdc6 fills the gap between Orc1 and Orc2 in ORC and reorients an N‐terminal domain of Orc1 44, 45 [Fig. 1(B)].
Figure 1.
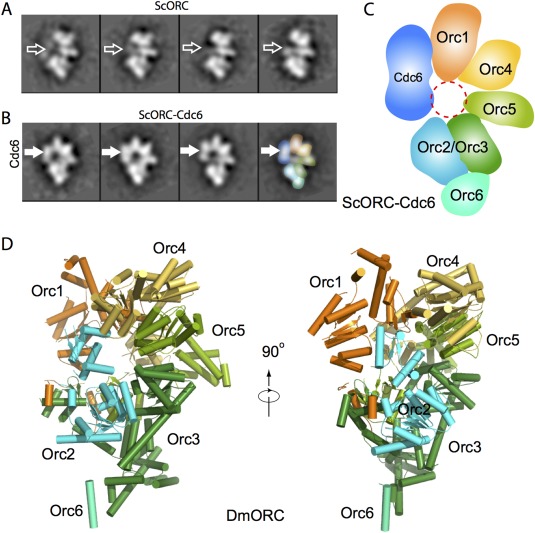
Structures of ScORC and DmORC as determined by EM and X‐ray crystallography, respectively. A: 2D class averages of the purified ORC in the presence of the 66 bp ARS1‐containing dsDNA. B: Class averages of ORC‐Cdc6‐DNA. The thicker horizontal arrows mark the absence (A) or presence (B) of the Cdc6 density. C: A sketch of the subunit arrangement of ORC‐Cdc6 as determined by the MBP fusion approach. D: Crystal structure of the fly ORC in a side and an edge view (PDB ID: 4XGC).
In order to localize Orc1‐6 subunits, a maltose binding protein (MBP) was systematically fused to the N‐ or C‐termini of the five largest ScORC subunits, one subunit at a time, to generate ten MBP‐fused ScORC variants.46 The extra MBP density was visualized in 2D class averages. By comparing with non‐tagged ScORC, the order of ScORC subunits was shown to be Orc1:Orc4:Orc5:Orc2/Orc3 with the smallest subunit Orc6 binding to Orc2/Orc3 [Fig. 1(C)]. A recent crystal structure of DmORC at 3.5 Å resolution further confirmed the basic subunit arrangement,47 and showed that DmORC is a two‐layered and notched ring structure, with a collar of the winged‐helix domains sitting on top of the AAA+ ring [Fig. 1(D)]. However, the crystal structure is in an auto‐inhibited conformation, because Orc1 AAA+ domain is rotated more than 90°, which disrupts the interaction with the neighboring Orc4. A lesson we learned from this work is that sometimes going bigger is better in order to obtain optimal samples for EM studies. ORC alone is fairly flexible. Therefore, a crystallographer would identify the flexible sequences and then trim them in order to derive a stable core for crystallization. However, we found that ORC can be stabilized by ATPγS and DNA, and the structure is further rigidified when Cdc6 bridges the gap between Orc1 and Orc2, and of course this contributes to the larger size of the ORC‐Cdc6‐DNA, which is better for image alignment.
EM study of ScORC was the first systematic application of MBP for subunit mapping.46 The method has since been applied to other complexes such as the proteasome lid.48 MBP can also be inserted into an internal location to map domains and subunits.49, 50 A modified approach was recently reported in which MBP is crosslinked to a target protein complex that contains an unnatural amino acid.51 Internal insertion with GFP is an alternative approach.52 Internal insertion of AviTag sequence followed by biotinylation and tagging with a monovalent variant of streptavidin was also shown to be a power approach for subunit mapping.53
Direct‐Mixing EM Catches ORC‐Cdc6 in the Act of Loading a Cdt1‐Bound Mcm2‐7 Hexamer
The function of the ORC‐Cdc6 is to load the replicative helicase core, the hetero‐hexameric Mcm2‐7 complex, onto the duplex DNA. In vitro experiments have shown that ORC‐Cdc6 loads a double hexamer on DNA.30, 31 How ORC‐Cdc6 accomplishes this feat was unknown because when the Mcm2‐7 double hexamer assembles, ORC‐Cdc6 is already dissociated. A series of direct mixing experiments were designed to capture the loading intermediate in which ORC‐Cdc6 is still bound to Mcm2‐7.49 Assuming ATP hydrolysis was needed to start the loading reaction, ATP was added to the ORC, Cdc6, ARS‐1‐containing DNA, Cdt1, and Mcm2‐7 mixture. The authors attempted to block the reaction completion by varying molar ratio and concentration of the components, by using ATPase mutants, including Orc1, Orc4, Orc5, and Cdc6 mutants, and by varying the length of DNA ‐ knowing that longer DNA was required to form the Mcm2‐7 double hexamer. All those efforts were unsuccessful. The authors eventually succeeded in capturing ORC‐Cdc6 in the act of loading the first Cdt1‐bound Mcm2‐7 when they replaced ATP with ATPγS. Not surprisingly, the sample was heterogeneous. But after 2D classification, a novel 3‐tiered structure was observed: the tilted top tier was ORC‐Cdc6 in side view, and the lower two parallel tiers were the Mcm2‐7 hexamer in side view with an attached Cdt1 [Fig. 2(A)].
Figure 2.
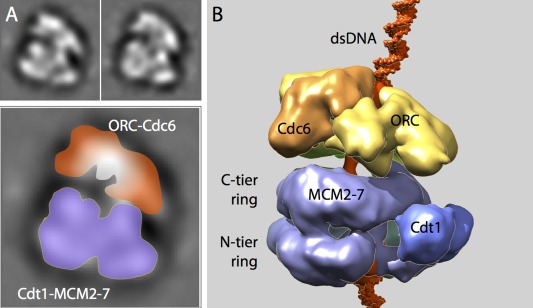
The loading mechanism of Cdt1‐Mcm2‐7 onto dsDNA by ORC‐Cdc6 ATPase as revealed by the OCCM architecture. A: Two class averages of cryo‐EM images of OCCM formed by direct mixing ORC, Cdc6, DNA, Cdt1, and Mcm2‐7 in the presence of ATPγS. For scale, the box size is 27 nm. An enlarged view is shown at the bottom with the top‐tier ORC‐Cdc6 and the lower two‐tier Cdt1‐MCM2‐7 labeled. B: Cryo‐EM 3D map of the OCCM complex purified by a magnetic beads pulldown method (EMDB ID: EMD‐5625). ORC is displayed in yellow, Cdc6 in orange, Mcm2‐7 in purple, Cdt1 in blue, and DNA in red. Atomic structures of short DNA segments are modeled on the top and bottom of the structure. At this stage, the first Mcm2‐7 hexamer already fully encircles the dsDNA.
Obtaining the first view of the loading intermediate, later called OCCM referring to ORC‐Cdc6‐Cdt1‐Mcm2‐7, was an important first step, because it immediately showed that the two Mcm2‐7 hexamers in the Mcm2‐7 double hexamer have to be loaded by ORC‐Cdc6 one at a time. And the image provided a method for preparing the elusive loading intermediate in larger quantity and better quality. A magnetic bead pull‐down method was subsequently developed to isolate the intermediate in the presence of ATPγS.49 In this method, the biotinylated ARS1 origin DNA‐containing plasmid is first attached to streptavidin‐labeled magnetic beads, and then protein complexes assembled on DNA can then be pulled by a magnet. With the purified OCCM complex, a 14‐Å 3D EM map was determined by cryo‐EM images recorded in a 4K x 4K CCD camera [Fig. 2(B)]. The MBP‐fused or MBP‐inserted OCCM variants were used to determine which subunit in ORC‐Cdc6 interacts with which subunit in Cdt1‐Mcm2‐7. The cryo‐EM structure revealed that the ORC‐Cdc6 attaches onto the C‐terminal AAA+ domains of the Mcm2‐7 hexamer, leaving the N‐terminal surface of the Mcm2‐7 exposed, thus available for interaction with the second Mcm2‐7 hexamer. Such organization makes sense considering the final loading product, the Mcm2‐7 double hexamer, is a head‐to‐head dimer.
Direct Mixing Coupled with Cross‐Linking Captures Transient OCM and OCMM Loading Intermediates
Capturing OCCM revealed how the first Mcm2‐7 hexamer was loaded on the DNA, but raises the question of how the second Mcm2‐7 hexamer is loaded. There were several hypotheses, chief among them was that two ORC‐Cdc6 load the two Mcm2‐7 hexamers separately, and the two Mcm2‐7 hexamers on DNA are able to interact head‐to‐head to form the double hexamer.54 Such hypothesis makes sense in that the origin DNA sequence is a lot longer than what can be covered by a single ORC‐Cdc6. A major problem with the hypothesis is that eukaryotic replication origins are not palindromic, so it is unclear how two ORC‐Cdc6 complexes can bind to the origin DNA in opposing directions. We sought to address this question by directly observing the loading intermediates by EM immediately following the OCCM formation. But this is an inherently difficult experiment, because the reaction is locked in the stage of OCCM formation if ATPγS is used [Fig. 3(A)], and using ATP assembles the double hexamer and allows the dissociation of ORC‐Cdc6 from the double hexamer. We, therefore, utilized chemical crosslinking via a time course study by stopping the loading reaction of the directly mixed samples in the presence of ATP at 1 min, 7 min, or 30 min with the addition of glutaraldehyde to a final concentration of 0.1%. In the 1 min crosslinked sample, we observed a structure termed OCM that is very similar to OCCM, except that Cdt1 was missing [Fig. 3(B)]. In sample crosslinked at 7 min reaction time, the number of OCM decreased but a new structure called ORC‐Cdc6‐Mcm2‐7‐Mcm2‐7 (OCMM) appeared [Fig. 3(C)]. The number of OCMM virtually diminished in reaction mixtures crosslinked at 30 min, leaving almost exclusively the double hexamer [Fig. 3(D)]. It is clear that upon ATP hydrolysis, Cdt1 is immediately released from DNA leading to the formation of ORC‐Cdc6‐Mcm2‐7 (OCM). And OCM would then go on to recruit the second Mcm2‐7 hexamer.55 We can derive from these series of experiments that OCMM is not an in vitro impassable binding artifact, because the quantity of OCMM drops accompanying the rise of the number of the double hexamers.50 Therefore, this work demonstrates that one ORC‐Cdc6, not two, load the Mcm2‐7 double hexamer [Fig. 3(E)]. Two extensive single molecule FRET studies further substantiated this conclusion.56, 57 Therefore, the EM work with only 2D class average and at a low resolution provides an example for how direct mixing, coupled with crosslinking and image analysis, can provide critical biological insights.
Figure 3.
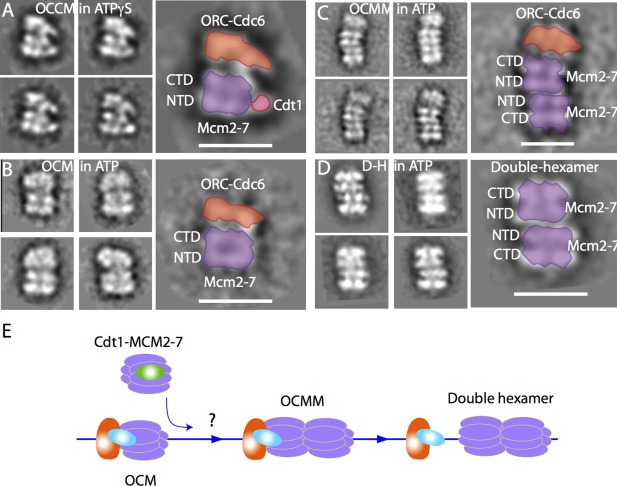
Capturing the loading intermediates OCM and OCMM by direct mixing and cross‐linking. A: Cryo‐EM 2D class averages of OCCM in 1 mM ATPγS. B−D: Four class averages of three loading intermediates observed by negative stain EM in 1 mM ATP: OCM (B), OCMM (C), and the Mcm2‐7 double hexamer (D). Note that panel (C) for OCMM is on a smaller scale. Scale bar is 20 nm. E: A sketch showing that OCM is the loader of the second Cdt1 bound Mcm2‐7 hexamer, and OCMM is an on‐pathway intermediate for the double hexamer assembly. ORC is in orange, Cdc6 in cyan, Cdt1 in green, and Mcm2‐7 in purple. The dsDNA is shown as a blue line.
Mcm2‐7 Double Hexamer: Subunit Mapping by MBP Insertion and Cryo‐EM Atomic Model
The architecture of the Mcm2‐7 double hexamer, i.e. which subunit in one hexamer interacts with which subunit in the other hexamer, is important for understanding how the double hexamer is subsequently activated. In the absence of a high‐resolution structure, an MBP labeling approach was used to address this question.50 The challenge of mapping out the double hexamer was the pseudo symmetry. Without a distinctive 2D view, identifying a MBP in a 2D average is not useful. So, 3D mapping was required [Fig. 4(A–C)]. The N‐ or C‐terminal fusion approach did not work in this case, because the termini of a protein are usually flexible. Therefore, MBP was inserted into a loop region between two predicted domains in several Mcm proteins. This way, MBP was constrained at both sides and less flexible. And indeed the MBP density was clearly detected in 3D reconstructions, leading to the determination of the double hexamer architecture50 [Fig. 4(C)]. The structure showed that the DNA gate between Mcm2‐Mcm5 in each hexamer is not aligned between the two hexamers, the two hexamers are tilted and twisted against each other, and the head‐to‐head juxtaposition of double hexamer generates a multi‐subunit binding site for activating kinase DDK.
Figure 4.
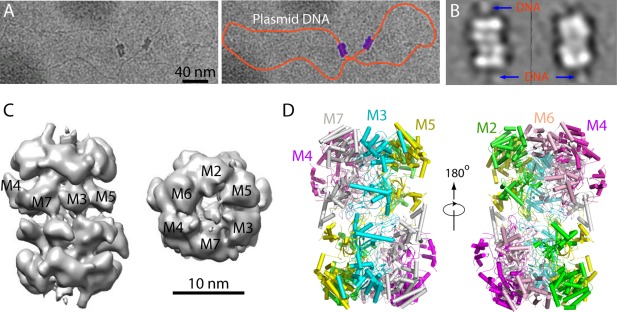
Cryo‐EM structure of the inactive Mcm2‐7 double hexamer. A: Cryo‐EM micrograph of two Mcm2‐7 double hexamers loaded in vitro onto a plasmid DNA. Left panel is the raw image, and the right panel shows the trace of the plasmid by an orange curve and the 2 double hexamers in purple. B: 2D class averages of the double hexamers on plasmid showing that DNA passes through the length of the double hexamer cylinder. C: A negative stain EM 3D map. Subunit identity is labeled based on subunit mapping by MBP insertion approach. D: Atomic model of the double hexamer based on a 3.8 Å resolution cryo‐EM map (PDB ID: 3JA8).
The atomic model of the double hexamer was recently derived by the Tye and Gao labs from a 3.8 Å resolution cryo‐EM 3D map58 [Fig. 4(D)]. Notably, the sample used for this study was purified from the G1 chromatin of the budding yeast rather than assembled in vitro. The structure revealed that the six available nucleotide‐binding sites are mostly occupied yet in different conformations, the central DNA channel is narrow and kinked, and the helix 2‐insertion loops (H2I) of Mcm6, Mcm2, and Mcm5 are arranged spirally matching the DNA pitch while the H2I of Mcm3, Mcm7, and Mcm4 are nearly planar. These structural features were suggested to be linked to how the origin duplex DNA is initially separated.
EM Structures of the Active Eukaryotic CMG Helicases
There is a significant gap in our understanding of double hexamer activation and separation.59 What we do know is that Mcm2‐7 is only the core of a fully active helicase, which is Cdc45‐Mcm2‐7‐GINS (CMG) containing two addition units, Cdc45 and the four‐protein complex GINS.60, 61, 62 The eukaryotic helicase is shown to encircle the leading‐strand DNA and moves in 3′‐5′ direction while excluding the lagging strand.63, 64 Negative stain EM of the fly CMG showed that the two‐tiered Mcm2‐7 ring form the main channel and Cdc45 and GINS bind from the side and apparently form a smaller side channel.65, 66 EM image analysis further revealed that streptavidin bound to a biotin that is labeled at the duplex DNA end is located above the motor ring in the fly CMG, consistent with the idea that the motor ring pushes against the duplex.67 Most recently, higher resolution cryo‐EM maps have been reported for both the fly and yeast CMG helicases by using images recorded on direct electron detectors.68, 69 The fly CMG structure in the presence of a forked DNA was determined to 7–10 Å resolution. The DNA fork contained a 50‐bp duplex region with a 16‐nt overhang on the 5′‐end and 40‐nt poly‐T on the 3′‐end. 3D reconstruction of the vitrified helicase complex revealed a conformation change between a compact and a relax state, depending on the nucleotide substrate engagement status.68 Only a short stretch of ssDNA of six nucleotides was cleared resolved between the C‐tier motor ring and the N‐tier ring. The bound ssDNA inside the central channel appears to make contact with the pre‐sensor I helix and helix‐2 insertion loop of Mcm7, Mcm4, and Mcm6. Observation of single strand DNA but not double strand DNA inside the helicase channel is in agreement with the idea that helicase tracks along single stranded DNA.63, 64
The yeast CMG helicase was determined to a resolution of 3.7 to 4.8 Å, leading to an atomic model of the 11‐protein complex69 [Fig. 5(A–D)]. The most rigid part of the structure, including Cdc45, GINS, and the N‐tier ring of Mcm2‐7, was resolved to 3.8 Å resolution. This leads to an atomic model of the full‐length yeast GINS and an atomic model of the yeast Cdc4569 [Fig. 5(A,B)]. Previously reported human GINS structures all lacked the B‐domain of the Psf1 subunit.70, 71, 72 This domain is found in the EM structure to be responsible for interaction with Cdc45. The yeast Cdc45 is highly similar to the human Cdc45 structure, containing two α/β domains that are linked by two linker loops and a middle insertion domain.73 Focused 3D classification of the flexible C‐terminal motor ring of Mcm2‐7 resulted in two different conformations, a compact and an extended structure at 4.8 and 4.7 Å resolution, respectively [Fig. 5(C,D)]. In these two conformations, the N‐terminal regions of Mcm2–7, braced by Cdc45–GINS, form a rigid platform, while the C‐tier AAA+ motor ring appears to be capable of moving up and down longitudinally. These structures led to the proposal that the eukaryotic helicase functions like an oil‐rig pumpjack attached to a stable platform, nodding up and down. Such DNA translocation mechanism is analogous to the inchworm‐like mechanism found in many nonreplicative monomeric helicases,74, 75, 76 but deviates significantly from the rotational mechanism proposed for other hexameric helicases.77, 78, 79 Interestingly, the Mcm5 winged‐helix domain is inserted into the central channel, apparently blocking the entry of double‐stranded DNA and supporting a steric‐exclusion DNA‐unwinding model.64
Figure 5.
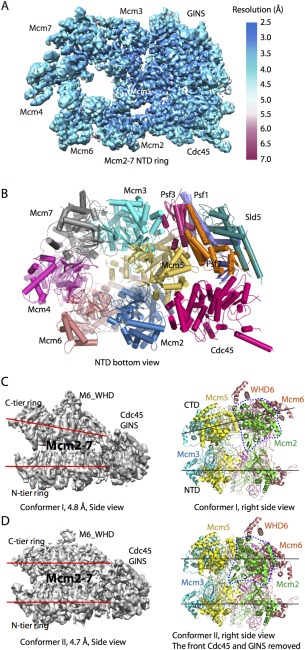
Cryo‐EM structure of the yeast CMG helicase. A: 3D cryo‐EM map of CMG in the bottom N‐terminal view color coded by local resolution (EMD ID: 6534). B: CMG atomic model viewed from the N‐tier face (PDB ID: 3JC6). C: CMG conformer I at 4.8 Å resolution (EMD ID: 6536, PDB ID: 3JC6). D: CMG conformer II at 4.7 Å resolution (EMD ID: 6535; PDB ID: 3JC5). The two red or black lines in C, D: highlight the distinct configuration of the two conformers. The dashed blue line encircles Mcm2 AAA+ domain. This domain is rotated by 30° in conformer II. Note that the right panels in (C, D) are rotated by 90° with respect to the left panels.
Direct Mixing EM Captures Several Yeast Replisome Assemblies
At the core of the eukaryotic replisome are replicative helicase CMG, the leading strand polymerase ε, the lagging strand polymerase δ, and the primase‐polymerase α.14, 80, 81, 82 Knowing how these key machines coordinate with each other during parental DNA unwinding and daughter DNA synthesis is important to understand the replication mechanism. Pol ε is a four‐protein complex comprised of catalytic subunit Pol2, and Dpb2, Dpb3, and Dpb4, and has a bi‐lobed overall shape as revealed by EM analysis.83 Only the structure of catalytic domain of the catalytic subunit Pol2 has been solved to atomic resolution.84 A physical interaction was reported between CMG and Pol ε.85 Using direct mixing of CMG and pol ε and negative stain EM, it was later shown that the two protein complexes indeed formed a super binary complex called CMG‐E86 [Fig. 6(A–C)]. Interestingly, in the 3D EM map, Pol ε is positioned on the C‐terminal side of CMG helicase, making direct contacts with the motor ring of the Mcm2‐7 hexamer, as well as with Cdc45 and GINS [Fig. 6(C)]. The overall architecture of the CMG‐E complex was further confirmed by crosslinking mass spectrometry.86 It was shown that Pol ε lies on the C‐terminal side of the Mcm2‐7 ring and that Pol ε subunits crosslink with Mcm2, Mcm5, and Mcm6.
Figure 6.
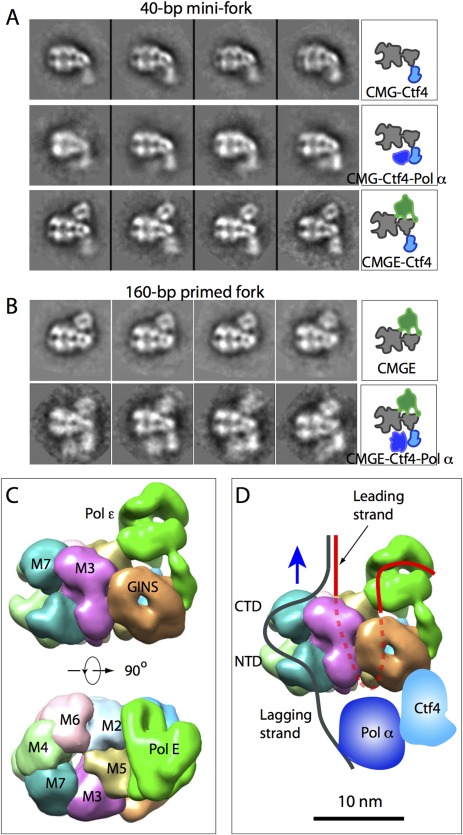
Direct mixing EM captures several replisome complexes. A: Mixing purified proteins in the presence of 40‐bp mini fork DNA, CGM was found in complex with Ctf4 (top row), Ctf4 and Pol α (middle row), and Pol ε and Ctf4. B: Mixing in the presence of 160‐bp primed forked DNA, CMG was found in complex with Pol ε (top row) and with Pol ε, Ctf4, and Pol α (bottom row). C: 3D EM map of CMG‐Pol ε in a side and top motor ring view (EMD ID: 6465). D: A cartoon showing the replisome architecture with a possible DNA pathway sketched. This architecture is derived with the assumption that the C‐tier motor ring pushes against the dsDNA.
The polymerase α–primase is a complex of four‐proteins: Pol1, Pol12, Pri1, and Pri2. Crystal structures of the catalytic domain of Pol1 and the human primase PriS‐PriL heterodimer, a homolog of the yeast Pri1‐Pri2, have been reported.87, 88 Negative stain EM has shown that polymerase α‐primase is a dumbbell‐like particle with the polymerase and primase located in separate lobes.89, 90 Study combining X‐ray crystallography and negative stain EM showed that the Ctf4 is a trimer that physically associates Pol α with the CMG helicase.91 Ctf4 contains a flexible N‐terminal WD40 domain and a C‐terminal β‐propeller domain that mediates Ctf4 trimerization. Such architecture is consistent with the scaffolding role of Ctf4 in the replisome. Negative stain EM and image analysis of the direct mixed sample of Pol α and Ctf4 showed that Ctf4 trimer alone binds to Cdc45 and GINS at the N‐tier side of Mcm2‐7 in the CMG complex [Fig. 6(A)]. In the direct mixed sample of CMG, Ctf4 and Pol α, a super‐ternary complex of CMG‐Ctf4‐Pol α was observed, showing that Pol α binds next to Ctf4 right below the Mcm2‐7 N‐tier ring [Fig. 6(A)].
When CMG, Pol ε, Pol α, and Ctf4 were mixed together and analyzed by negative stain EM, a super‐quaternary complex of CMG‐Pol ε‐Pol α‐Ctf4 was observed in 2D class averages. It was clear from the images that Pol ε rides on top (C‐tier motor ring) of the helicase, while the Pol α polymerase‐primase trails behind the helicase [Fig. 6(B)]. This architecture was quite unexpected because in any textbook drawing of the DNA replisome, eukaryotic or prokaryotic, polymerases are always placed behind the replicative helicase to synthesize nascent DNA on the leading and lagging single stranded DNA templates. It is well established that the leading strand threads the central channel of the helicase while the lagging strand is excluded outside.63, 64 If the motor domain indeed pushes against the duplex, a concept widely accepted and experimentally supported by a DNA labeling study on the fly CMG,67 then the leading‐strand DNA template would have to travel a long distance before reaching Pol ε: it has to first thread through the central channel of the Mcm2–7 hexamer and then make a U‐turn at the bottom to reach Pol ε at the top of CMG [Fig. 6(D)].
Summary
The past several years have witnessed breathtaking advance in biochemical analyses of DNA replication in the eukaryotic systems.30, 57, 62, 63, 64, 92, 93, 94, 95 Accompanying these rapid biological advances are several insightful structural works done by electron microscopy and X‐ray crystallography. We now know the fly ORC structure at atomic level detail.47 We understand how Cdc6 fills the gap between Orc1 and Orc2 in the crescent‐shaped ORC structure, converting the ORC from an origin DNA binder to an active helicase‐loading platform.45 We know the Mcm2‐7 double hexamer is loaded onto double stranded DNA by ORC‐Cdc6 one at a time.49 The first Cdt1 bound Mcm2‐7 hexamer is loaded by ORC‐Cdc6 on DNA, and the second Cdt1‐bound Mcm2‐7 is not loaded by a second ORC‐Cdc6, is loaded instead by the same ORC‐Cdc6 that is in complex with the first Mcm2‐7 hexamer.50, 94 In other words, the first recruited Mcm2‐7 hexamer plays an active role in recruiting the second Mcm2‐7 hexamer. We also have the atomic model of the inactive Mcm2‐7 double hexamer as well as the active helicase the CMG complex.58, 69 Even a crude yet surprising architecture of the eukaryotic replisome has been worked out.86
Many key questions remain. We don't yet know at the atomic level how the first and then the second Cdt1‐bound Mcm2‐7 hexamers are recruited to DNA by ORC‐Cdc6, and we know almost nothing at the structural level how the inactive Mcm2‐7 double hexamer is activated by a handful of enzymes and co‐factors including DDK, CDK, Sld2, Sld3, Sld4, Dpb11, Cdc45, and GINS.59 There is a paucity of structural information on how CMG helicase interacts with the DNA at the replication fork.68 We don't yet have high‐resolution information on how the helicase interacts with the polymerases and primase, let alone with many other replication factors. We can anticipate that some of these structures will be forthcoming in the next several years. But others may be much harder due to the highly flexible nature of the replisome.
Cryo‐EM has played an important role in our current understanding of the DNA replication mechanism. With further improvement in electron detector quantum efficiency and better correction of beam‐induced specimen movement,96 cryo‐EM will be able to elucidate the replication mechanism at even greater precision. However, given the complexity and the dynamic nature of the replisome, X‐ray crystallography and NMR will certainly continue to play an essential role in this quest. Crosslinking mass spectrometry has been brought into the fold for many protein complexes.86, 97, 98 Recent years have seen an increased use of fluorescence‐based single molecule experiments to complement cryo‐EM study of the molecule machines.99, 100 All these tools are highly complementary and essential if we are to gain a full understanding of any biological system, but particularly for a system as complex and critical as the DNA replication.
Acknowledgments
The EM work described here on yeast DNA replication initiation was in collaboration with Bruce Stillman Lab at Cold Spring Harbor Laboratory and Christian Speck Lab at Imperial College London. The EM study of the yeast replisome was a collaboration with Michael O'Donnell lab at the Rockefeller University.
References
- 1. Henderson R (2013) Structural biology: ion channel seen by electron microscopy. Nature 504:93–94. [DOI] [PubMed] [Google Scholar]
- 2. Bai XC, McMullan G, Scheres SH (2015) How cryo‐EM is revolutionizing structural biology. Trends Biochem Sci 40:49–57. [DOI] [PubMed] [Google Scholar]
- 3. Cheng Y (2015) Single‐particle cryo‐EM at crystallographic resolution. Cell 161:450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng Y, Grigorieff N, Penczek PA, Walz T (2015) A primer to single‐particle cryo‐electron microscopy. Cell 161:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watson JD, Crick FH (1953) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171:737–738. [DOI] [PubMed] [Google Scholar]
- 6. Watson JD, Crick FH (1953) Genetical implications of the structure of deoxyribonucleic acid. Nature 171:964–967. [DOI] [PubMed] [Google Scholar]
- 7. Kornberg A, Baker TA (1992) DNA replication, W.H. Freeman and Co, New York. [Google Scholar]
- 8. Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Ann Rev Biochem 71:333–374. [DOI] [PubMed] [Google Scholar]
- 9. MacNeill S (2012) The eukaryotic replisome a guide to protein structure and function Subcellular biochemistry. New York: Springer, Dordrecht, pp. 1. [Google Scholar]
- 10. Costa A, Hood IV, Berger JM (2013) Mechanisms for initiating cellular DNA replication. Ann Rev Biochem 82:25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Araki H (2013) Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication. Genes Cells 18:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaplan DL, Springer International Publishing AG. The Initiation of DNA Replication in Eukaryotes (2016) Springer International Publishing, Cham, pp. Online‐Resource.
- 13. Leipe DD, Aravind L, Koonin EV (1999) Did DNA replication evolve twice independently? Nucleic Acids Res 27:3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Donnell M, Langston L, Stillman B (2013) Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 5:a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH (2001) DNA replication licensing and human cell proliferation. J Cell Sci 114:2027–2041. [DOI] [PubMed] [Google Scholar]
- 16. Jackson AP, Laskey RA, Coleman N (2014) Replication proteins and human disease. Cold Spring Harb Perspect Biol 6:a013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mott ML, Berger JM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5:343–354. [DOI] [PubMed] [Google Scholar]
- 18. Duderstadt KE, Berger JM (2008) AAA+ ATPases in the initiation of DNA replication. Crit Rev Biochem Mol Biol 43:163–187. [DOI] [PubMed] [Google Scholar]
- 19. Ishino S, Kelman LM, Kelman Z, Ishino Y (2013) The archaeal DNA replication machinery: past, present and future. Genes Genet Syst 88:315–319. [DOI] [PubMed] [Google Scholar]
- 20. Marks AB, Smith OK, Aladjem MI (2016) Replication origins: determinants or consequences of nuclear organization? Curr Opin Genet Dev 37:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyrien O (2015) Peaks cloaked in the mist: the landscape of mammalian replication origins. J Cell Biol 208:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng C, Luo H, Zhang X, Gao F (2015) Recent advances in the genome‐wide study of DNA replication origins in yeast. Front Microbiol 6:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bell SP, Stillman B (1992) ATP‐dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128–134. [DOI] [PubMed] [Google Scholar]
- 24. Liang C, Weinreich M, Stillman B (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81:667–676. [DOI] [PubMed] [Google Scholar]
- 25. Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF (1996) An essential role for the Cdc6 protein in forming the pre‐replicative complexes of budding yeast. Nature 379:180–182. [DOI] [PubMed] [Google Scholar]
- 26. Santocanale C, Diffley JF (1996) ORC‐ and Cdc6‐dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. embo J 15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 27. Nishitani H, Lygerou Z, Nishimoto T, Nurse P (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404:625–628. [DOI] [PubMed] [Google Scholar]
- 28. Tanaka S, Diffley JF (2002) Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2‐7 during G1 phase. Nat Cell Biol 4:198–207. [DOI] [PubMed] [Google Scholar]
- 29. Mimura S, Seki T, Tanaka S, Diffley JF (2004) Phosphorylation‐dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 431:1118–1123. [DOI] [PubMed] [Google Scholar]
- 30. Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C (2009) A double‐hexameric MCM2‐7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA 106:20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF (2009) Concerted loading of Mcm2‐7 double hexamers around DNA during DNA replication origin licensing. Cell 139:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bell SD, Botchan MR (2013) The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 5:a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan DL (2016). The initiation of DNA replication in eukaryotes. Cham: Springer, pp 1. [Google Scholar]
- 34. O'Donnell M, Li H (2016) The eukaryotic replisome goes under the microscope. Curr Biol 26:R247–R256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Stillman B (2012) The origin recognition complex: a biochemical and structural view. Subcell Biochem 62:37–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817–823. [DOI] [PubMed] [Google Scholar]
- 37. Remus D, Beall EL, Botchan MR (2004) DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC‐DNA binding. embo J 23:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Houchens CR, Lu W, Chuang RY, Frattini MG, Fuller A, Simancek P, Kelly TJ (2008) Multiple mechanisms contribute to Schizosaccharomyces pombe origin recognition complex‐DNA interactions. J Biol Chem 283:30216–30224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372–376. [DOI] [PubMed] [Google Scholar]
- 40. Calvi BR, Byrnes BA, Kolpakas AJ (2007) Conservation of epigenetic regulation, ORC binding and developmental timing of DNA replication origins in the genus Drosophila. Genetics 177:1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM (2011) Chromatin signatures of the Drosophila replication program. Genome Res 21:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M (2006) Nucleotide‐dependent conformational changes in the DnaA‐like core of the origin recognition complex. Nat Struct Mol Biol 13:684–690. [DOI] [PubMed] [Google Scholar]
- 43. Clarey MG, Botchan M, Nogales E (2008) Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol 164:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Speck C, Chen Z, Li H, Stillman B (2005) ATPase‐dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 12:965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H (2012) Cdc6‐induced conformational changes in ORC bound to origin DNA revealed by cryo‐electron microscopy. Structure 20:534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H (2008) The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 105:10326–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bleichert F, Botchan MR, Berger JM (2015) Crystal structure of the eukaryotic origin recognition complex. Nature 519:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun J, Evrin C, Samel SA, Fernandez‐Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H (2013) Cryo‐EM structure of a helicase loading intermediate containing ORC‐Cdc6‐Cdt1‐MCM2‐7 bound to DNA. Nat Struct Mol Biol 20:944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun J, Fernandez‐Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H (2014) Structural and mechanistic insights into Mcm2‐7 double‐hexamer assembly and function. Genes Dev 28:2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dambacher CM, Lander GC (2015) Site‐specific labeling of proteins for electron microscopy. J Struct Biol 192:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ciferri C, Lander GC, Nogales E (2015) Protein domain mapping by internal labeling and single particle electron microscopy. J Struct Biol 192:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ (2012) The molecular architecture of human Dicer. Nat Struct Mol Biol 19:436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yardimci H, Walter JC (2014) Prereplication‐complex formation: a molecular double take?. Nat Struct Mol Biol 21:20–25. [DOI] [PubMed] [Google Scholar]
- 55. Evrin C, Fernandez‐Cid A, Riera A, Zech J, Clarke P, Herrera MC, Tognetti S, Lurz R, Speck C (2014) The ORC/Cdc6/MCM2‐7 complex facilitates MCM2‐7 dimerization during prereplicative complex formation. Nucleic Acids Res 42:2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chistol G, Walter JC (2015) Single‐molecule visualization of MCM2‐7 DNA loading: seeing is believing. Cell 161:429–430. [DOI] [PubMed] [Google Scholar]
- 57. Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP (2015) Single‐molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 161:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li N, Zhai Y, Zhang Y, Li W, Yang M, Lei J, Tye BK, Gao N (2015) Structure of the eukaryotic MCM complex at 3.8 A. Nature 524:186–191. [DOI] [PubMed] [Google Scholar]
- 59. Tognetti S, Riera A, Speck C (2015) Switch on the engine: how the eukaryotic replicative helicase MCM2‐7 becomes activated. Chromosoma 124:13–26. [DOI] [PubMed] [Google Scholar]
- 60. Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/Mcm2‐7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA 103:10236–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ilves I, Petojevic T, Pesavento JJ, Botchan MR (2010) Activation of the MCM2‐7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37:247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF (2015) Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC (2011) Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yardimci H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, Walter JC (2012) Bypass of a protein barrier by a replicative DNA helicase. Nature 492:205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM (2011) The structural basis for MCM2‐7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Petojevic T, Pesavento JJ, Costa A, Liang J, Wang Z, Berger JM, Botchan MR (2015) Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc Nat Acad Sci USA 112:E249–E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan‐Gibson K, Fleck RA, Botchan MR, Berger JM (2014) DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife 3:e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abid Ali F, Renault L, Gannon J, Gahlon HL, Kotecha A, Zhou JC, Rueda D, Costa A (2016) Cryo‐EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun 7:10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yuan Z, Bai L, Sun J, Georgescu R, Liu J, O'Donnell ME, Li H (2016) Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol 23:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS (2007) Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci USA 104:12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choi JM, Lim HS, Kim JJ, Song OK, Cho Y (2007) Crystal structure of the human GINS complex. Genes Dev 21:1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F (2007) Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol 14:388–396. [DOI] [PubMed] [Google Scholar]
- 73. Simon AC, Sannino V, Costanzo V, Pellegrini L (2016) Structure of human Cdc45 and implications for CMG helicase function. Nat Commun 7:11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75–84. [DOI] [PubMed] [Google Scholar]
- 75. Fischer CJ, Maluf NK, Lohman TM (2004) Mechanism of ATP‐dependent translocation of E.coli UvrD monomers along single‐stranded DNA. J Mol Biol 344:1287–1309. [DOI] [PubMed] [Google Scholar]
- 76. Lee JY, Yang W (2006) UvrD helicase unwinds DNA one base pair at a time by a two‐part power stroke. Cell 127:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Singleton MR, Sawaya MR, Ellenberger T, Wigley DB (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101:589–600. [DOI] [PubMed] [Google Scholar]
- 78. Enemark EJ, Joshua‐Tor L (2006) Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275. [DOI] [PubMed] [Google Scholar]
- 79. Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA (2012) The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA (2008) Division of labor at the eukaryotic replication fork. Mol Cell 30:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Johansson E, Dixon N (2013) Replicative DNA polymerases. Cold Spring Harb Perspect Biol 5:a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Plosky BS (2014) More division of labor at the eukaryotic replication fork. Molecular Cell 56:467–468. [DOI] [PubMed] [Google Scholar]
- 83. Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E (2006) Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo‐electron microscopy. Nat Struct Mol Biol 13:35–43. [DOI] [PubMed] [Google Scholar]
- 84. Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer‐Eriksson AE, Johansson E (2014) Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat Struct Mol Biol 21:49–55. [DOI] [PubMed] [Google Scholar]
- 85. Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME (2014) CMG helicase and DNA polymerase epsilon form a functional 15‐subunit holoenzyme for eukaryotic leading‐strand DNA replication. Proc Natl Acad Sci USA 111:15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O'Donnell ME (2015) The architecture of a eukaryotic replisome. Nat Struct Mol Biol 22:976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kilkenny ML, Longo MA, Perera RL, Pellegrini L (2013) Structures of human primase reveal design of nucleotide elongation site and mode of Pol alpha tethering. Proc Natl Acad Sci USA 110:15961–15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Perera RL, Torella R, Klinge S, Kilkenny ML, Maman JD, Pellegrini L (2013) Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. Elife 2:e00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Klinge S, Nunez‐Ramirez R, Llorca O, Pellegrini L (2009) 3D architecture of DNA Pol alpha reveals the functional core of multi‐subunit replicative polymerases. embo J 28:1978–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nunez‐Ramirez R, Klinge S, Sauguet L, Melero R, Recuero‐Checa MA, Kilkenny M, Perera RL, Garcia‐Alvarez B, Hall RJ, Nogales E, Pellegrini L, Llorca O (2011) Flexible tethering of primase and DNA Pol alpha in the eukaryotic primosome. Nucleic Acids Res 39:8187–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak‐Vinkovic D, Labib K, Costa A, Pellegrini L (2014) A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 510:293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME (2014) Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol 21:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Samel SA, Fernandez‐Cid A, Sun J, Riera A, Tognetti S, Herrera MC, Li H, Speck C (2014) A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2‐7 onto DNA. Genes Dev 28:1653–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Duzdevich D, Warner MD, Ticau S, Ivica NA, Bell SP, Greene EC (2015) The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single‐molecule level. Mol Cell 58:483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Masai H (2015) Building up the machinery for DNA replication. Cell Cycle 14:3011–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Henderson R (2015) Overview and future of single particle electron cryomicroscopy. Arch Biochem Biophys 581:19–24. [DOI] [PubMed] [Google Scholar]
- 97. Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. [DOI] [PubMed] [Google Scholar]
- 98. Chait BT, Cadene M, Olinares PD, Rout MP, Shi Y (2016) Revealing higher order protein structure using mass spectrometry. J Am Soc Mass Spectrom 27:952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chen B, Frank J (2016) Two promising future developments of cryo‐EM: capturing short‐lived states and mapping a continuum of states of a macromolecule. Microscopy 65:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Frank J (2016) Whither ribosome structure and dynamics research? J Mol Biol 428:3565–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]


