Abstract
The images of flash‐frozen biological macromolecules produced by cryo‐electron microscopy (EM) can be used to generate accurate, three‐dimensional, electric potential maps for these molecules that resemble X‐ray‐derived electron density maps. However, unlike electron density maps, electric potential maps can include negative features that might for example represent the negatively charged, backbone phosphate groups of nucleic acids or protein carboxylate side chains, which can complicate their interpretation. This study examines the images of groups that include charged atoms that appear in recently‐published, high‐resolution EM potential maps of the ribosome and β‐galactosidase. Comparisons of simulated maps of these same groups with their experimental counterparts highlight the impact that charge has on the appearance of electric potential maps.
Keywords: electric potential, electron density, electron scattering, X‐ray scattering, electron atomic scattering factor, electron microscopy
Introduction
Recent technological advances in cryo‐electron microscopy (EM) have resulted in a revolution in structural biology.1, 2, 3 The resolutions of the single‐particle EM images being obtained have improved dramatically. For example, a 2.9‐Å resolution ribosome map was reported recently, and maps have been produced for β‐galactosidase and glutamate dehydrogenase that have resolutions of 2.2‐Å resolution, and 1.80 Å, respectively.4, 5, 6, 7 EM is about to become a serious competitor of X‐ray crystallography as a method for determining the structures of macromolecules at atomic resolution.8
Prominent among the questions that will have to be answered in order for EM to realize its full potential are: (1) how best to refine atomic models obtained from EM maps, and (2) how best to assess their accuracy.9 One gets the impression that many in the EM community believe that the methods X‐ray crystallographers have developed for solving these problems will suffice. However, as we are about to see, it may not be that simple because the interpretation of the electric potential (EP) maps generated by electron microscopy is much more challenging than the interpretation of the electron density (ED) maps X‐ray crystallographers work with.
Since the 1930s, it has been clear that electron scattering patterns report on the distribution electrostatic potential within molecules, which is determined both by the positions of atomic nuclei and by the associated electron density distributions,10 but that X‐ray scattering provides information about electron density distributions only.11 As has long been realized,12, 13, 14 this difference has implications that cannot be ignored.15 For example, an ED map that represents a macromolecule should be everywhere positive, but the corresponding EP map need not be. In a high resolution EP map, atoms carrying net negative charges will be represented by features that consist of positive centers surrounded by negative halos, and at lower resolutions, the EP may be zero or negative at those locations. It follows that the density modification methods X‐ray crystallographers use to improve phases, which involve the suppression of the negative features in their maps, may produce misleading results if applied to EP maps. Nevertheless, not only have crystallographic density modification procedures been applied to EP maps,16, 17 but also it appears that X‐ray atomic scattering factors have been used to refine atomic models derived from EM maps.18, 19
Here we report some observations that have emerged from inspection of the EP distributions that correspond to each of the 5719 nucleotide residues represented in an EM‐derived atomic structure of the E. coli ribosome‐EF‐Tu complex (PDB accession number of 5AFI),5 which was determined at a resolution of 2.90 Å. That structure was solved by fitting an X‐ray crystallographic structure of the E. coli ribosome (PDB accession number of 4V9D)20 into the experimental EP map obtained by the authors; there is every reason to believe that it is fundamentally correct. Nevertheless, but not surprisingly, the EP distributions that represent base pairs in this map do not look like the corresponding electron density distributions in the ED map from which the parent structure was obtained. In an effort to better understand why this is so, density maps were computed for base pairs at several different resolutions using: (1) X‐ray atomic scattering factors, (2) neutral, single‐atom electron scattering factors, and (3) a similar set of electron scattering factors that made it possible to represent the fact that the two non‐bridging oxygen atoms of backbone phosphodiester groups carry half a negative charge each.
It is clear that the differences between maps computed for base pairs using X‐ray structure factors and isolated, neutral‐atom electron structure factors are modest, but that neither corresponds well to what it is seen in experimental EP maps of nucleic acids. Atomic charges must be taken into account if a reasonable level of agreement between experimental EP maps and model maps is to be achieved. None of this should come as a surprise.12, 13 The reason for presenting these observations here is to stimulate the biological EM community to pay more attention to these issues going forward.
Results and Discussion
Backbone phosphate groups have smaller potentials than corresponding bases
The most striking differences between the EP map for the E. coli ribosome structure5 reported for 5AFI and the corresponding ED map have to do with the appearance of phosphate groups. In X‐ray derived ED maps of structures that include nucleic acid, phosphate groups are invariably the most conspicuous component of every nucleotide. In the 5AFI EP map, on the other hand, the amplitudes of the peaks that correspond to the backbone phosphate groups of over 99.5% the nucleotides represented are much smaller than the amplitudes of the features that represent their bases (Fig. 1, Supporting Information Fig. S1). The backbone phosphate groups of ∼ 0.5% have EP peaks comparable in magnitude to those their bases, but it is invariably the case that these phosphate groups have divalent Mg(II) ions associated with them.
Figure 1.
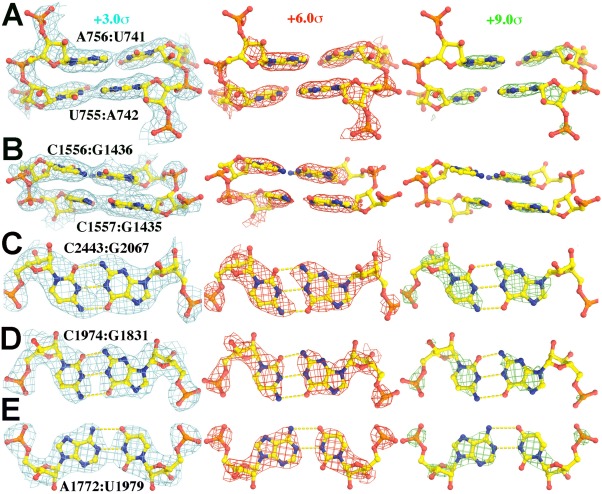
Experimental EP map of portions of the nucleic acid part of the E. coli ribosome‐EF‐Tu complex (5AFI5), contoured at +3.0σ (cyan, left), +6.0σ (red, middle), and +9.0σ (green, right). (A) and (B) show a side‐on view of two base pairs in a helix found in a highly ordered part of the structure (A), and a less ordered part (B) of the structure. (C) and (D) are face on views of selected Watson‐Crick G:C base pairs) and (E) is a face‐on view of an A:U base pair. Original EM‐derived atomic models (PDB accession number in parenthesis along with citation reference) were used in all figures in this paper with the exception of simulations in which H atoms were added. An expanded view of duplexes for (A) and (B) can be found in Supporting Information Figure S1.
A more detailed examination of individual nucleotides reveals another feature of nucleic acid EP maps that distinguishes them from the ED maps of nucleic acids [Fig. 1(C,D)]. The exocyclic amine groups of bases have higher EP values than their exocyclic carbonyl oxygen groups, independent of what kinds of base‐base interaction they are involved in.21 In ED maps, the exocyclic groups of nucleic acid bases look alike.
Simulated EP maps for RNA base pairs
To understand why EP maps of RNA nucleotides look the way they do, three different kinds of EP and ED maps were computed for base pairs (see Methods): (i) ED maps based on independent‐atom, spherically‐averaged X‐ray scattering factors, (ii) EP maps in which atoms are represented using independent, neutral‐atom, spherically‐averaged electron scattering factors, and (iii) EP maps based on a similar set of electron atomic scattering factors that use the electron scattering factor22, 23 for O‐ to represent the negative charges on non‐bridging phosphate oxygen atoms (Fig. 2).
Figure 2.
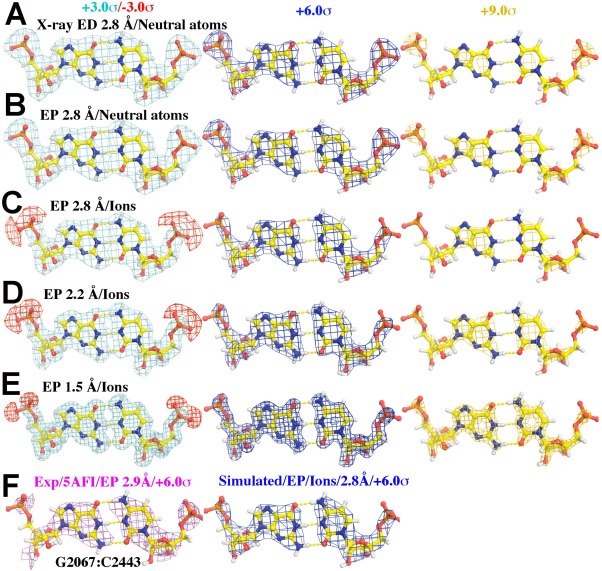
Simulated and experimental maps for GC base pairs contoured at +3.0σ (cyan, left)/‐3.0σ (red, left), +6.0σ (blue, middle), and +9.0σ (gold, right) for (A) through (E). (A) X‐ray ED maps simulated using independent, neutral atoms X‐ray scattering factors at 2.80‐Å resolution. (B) EP maps simulated using neutral atom electron scattering factors at 2.80‐Å resolution. (C‐E) EP maps using partially ionized electron scattering factors for OP1 and OP2 at 2.80‐Å (C), 2.20‐Å (D), and 1.50‐Å (E) resolution. (F) An experimental EP map of a G:C pair determined at 2.90‐Å resolution and contoured at +6.0σ (cyan, left) compared to a simulated EP map at 2.80‐Å resolution similar to (C) contoured at +6.0σ (blue, right).
As expected [Fig. 2(A)], the electron density associated with the phosphate groups in base pairs in these simulated ED maps is higher than that corresponding to their bases, and the same is true for the EP maps computed using neutral‐atom electron scattering factors [Fig. 2(B)]. The correspondence between model EP features and experimental EP features improves substantially if OP1 and OP2 atoms are both assigned half negative charges, the reason being that the resulting negative potential cancels some of the positive potential associated with P atoms [Fig. 2(C)]. As contour levels rise, phosphate groups disappear from the simulated EP maps before bases, just the way they do in experimental EM maps (Fig. 1). When the ionized atomic model simulation is examined as a function of resolution from 2.80‐Å, 2.20‐Å, and then to 1.50‐Å resolution, similar features are observed [Fig. 2(C–E)].
What these simulations do not explain is the low visibility of exocyclic oxygens relative to exocyclic amines. This observation is consistent with the fact that at resolutions below ∼ 2.2 Å, the electric potential peaks of individual atoms obtained by Fourier inversion of isolated‐atom electron structure factors (see International Tables for Crystallography22) vary in amplitude in the order C > N > O. Only at resolutions higher than 2.2 Å, does the amplitude order become O > N > C, as is in ED maps. However, in the resolution range relevant here, these amplitude distinctions are small, and not all that different from what is expected for X‐rays. Instead, it is again highly likely that this difference in visibility is a manifestation of the distribution of partial charges in nucleotide bases. These partial charge distributions have been examined many times over the years using many different approaches, and while the outcomes of these studies have varied, there is qualitative agreement about the charges associated with exocyclic groups.21 All exocyclic oxygens are strongly electronegative. Their charges are of the order of ‐ 0.5e. The nitrogen atoms of their exocyclic amines are also electronegative, but because the hydrogen atoms bonded to them are electropositive, the net charge of these groups is close to zero or slightly positive. These differences in charge should be enough to explain the differences in visibility between the two groups.
It is important to realize that the independent, spherical atom calculations done to obtain the ED and EP maps in Figure 2 ignore the redistributions of valence electrons associated with covalent bond formation.24 Experience shows that the impact of these redistributions on ED maps is so small that it is hard to visualize them at all in maps having resolutions worse than 1 Å. This is not the case for EP maps. The magnitude of electron scattering factors is proportional to the difference between the nuclear charge of an atom and its X‐ray structure factor, which at low resolutions is a small difference between large numbers that are comparable in magnitude. For that reason, the redistributions of valence electrons associated with bond formation, which are likely to alter the spherically averaged X‐ray structure factor of an atom somewhat, but more important, make the non‐spherically averaged X‐ray structure factor of that atom far more anisotropic than it otherwise would be, have a comparatively large impact on electron structure factors at low‐resolution. Thus the effects of bonding will be larger in EP maps that include low resolution data than they are in EP maps computed using only data collected at high‐resolution, where nuclear charges dominate scattering factors.12, 13
Effects of charge on base pairing geometry and atomic positions
In most of the interpretations of biological molecular structures based on experimental EP maps published to date, no account has been taken of charges, even those associated with species like the Mg(II) cation, which are usually modeled as Mg(0) atoms.5 As far as we know, only a handful of protein crystallographers have ever done otherwise. However, the effects are modest in the X‐ray case because the difference in X‐ray scattering factor between Mg(II) and Mg(0), for example, is only about 20% at zero scattering angle, and diminishes rapidly as resolution increases. The magnitude of the errors made by neglecting the charge on magnesium ions is certain to be much larger in the EM case. The amplitudes of the electron scattering factor of the two species differ by orders of magnitude at low resolution,22 Mg(II) ≫ Mg(0), and their differences do not die out until the resolution reaches ∼ 3 Å.
Many EM investigators have employed tools from X‐ray crystallography for interpreting EP maps, and have even used X‐ray atomic scattering factors instead of electron atomic scattering factors.5, 6, 7 The consequences of so doing are unlikely to be large when it comes to the initial fitting of RNA sequences into EP maps because once the positions of the base and ribosyl moieties of a nucleotide and its neighbors on both sides have been defined, the positions of the phosphate groups that link them together are largely determined. However, these practices are likely to produce problems during model refinement, the symptoms of which will be small mis‐positionings of nucleotides, and atom‐to‐atom variations in B‐factors that have nothing to do with structural disorder.
Charge and the visibility of protein carboxylate groups
The observations just described motivated us to examine the appearance of charged residues in the protein regions of the ribosome EP map of concern here, as well as in the EP map now available for β‐galactosidase,6 It has been realized for a long time that charge affects the appearance of side chains in the EP maps of proteins.24, 25 Thus we were not surprised to find that the carboxylate groups of aspartic and glutamic acids are virtually invisible in the ribosome EP map, but that there is density for many of the amide groups of asparagines and glutamines (Fig. 3, Supporting Information Fig. S2). The former carry charges of −1 e, of course, while the latter are uncharged. In some cases, in the EP map for β‐galactosidase, which has higher resolution, the oxygens of carboxylate groups that are hydrogen bond acceptors can be distinguished from those that are not (Fig. 4). By contrast, there is density for the positively charged epsilon amino groups of many lysine resides (Supporting Information Fig. S2). Other model calculations we have done indicate that these differences in visibility may again be manifestations of the effect of charge on EP maps (data not shown). In this connection it is important to point out that the side chains of Asp and Glu are liable to decarboxylate when exposed to ionizing radiation, particularly X‐ray radiation.26, 27 Thus in some instances, radiation damage may contribute to the absence of density for these groups in the EP maps.28
Figure 3.
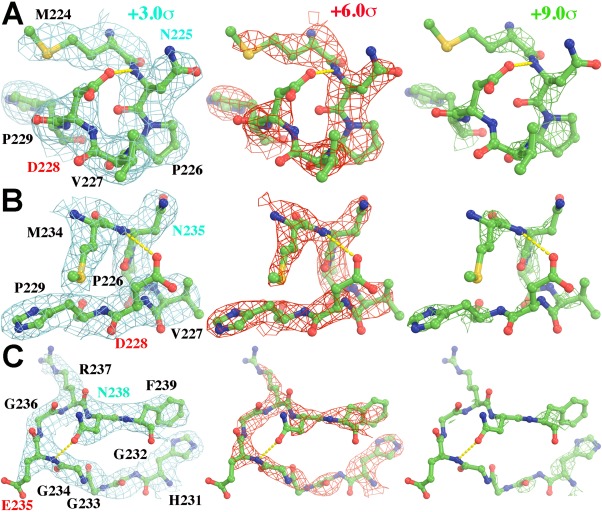
Portions of the experimental EP map reported for segments of protein in the ribosome (5AFI5) are shown that include Asp, Glu, and Asn side chains (Asn residues, cyan labels; and carboxylate residues, red labels), contoured at +3.0σ (cyan, left), +6.0σ (red, middle), and +9.0σ (green, right). (A, B) Two views of residues M224‐P229 in chain C. (C) Residues H231‐F239 in chain C.
Figure 4.
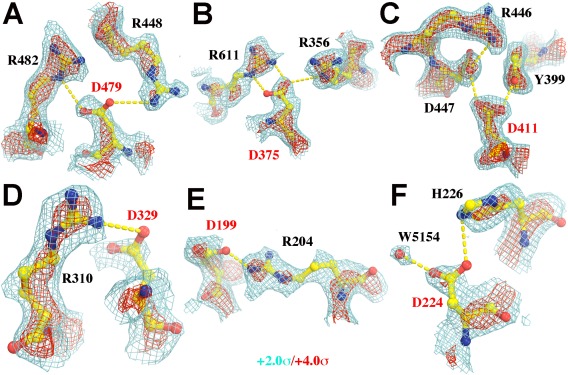
Portions of the experimental EP map reported for the segments in β‐galactosidase (5A1A6) contoured at +2.0σ (cyan) and +4.0σ (red). Asp residues are labeled in red. (A) D479. (B) D375, (C) D411. It is noted that both Oδ1 and Oδ2 atoms of this Asp residue are inside the iso‐potential envelope at the +4.0σ level. (D) D329. (E) D199. (F) D224. Additional views of Gln and Glu residues can be found in Supporting Information Figure S2.
Among the simulations done were a series in which H atoms were either included or excluded, as they often are in X‐ray crystallographic calculations. It was found that contribution of H scattering power to the atoms to which those H atoms are bonded is relatively small in simulated EP maps at medium resolution (data not shown), contrary to an earlier suggestion.24 In X‐ray crystallography, the H+ cation is invisible because it has no electron. In EM, on the other hand, it is highly visible because it has a positive charge. Thus while deprotonated and protonated carboxylate groups look almost the same in ED maps, they are easily distinguished in EP maps, and EP maps may provide unique information about protonation state of catalytically important residues inside enzymes that X‐ray crystallography cannot. For example, D411 in β‐galactosidase6 must be protonated because there is positive density for both its Oδ1 and Oδ2 atoms in the β‐galactosidase EP map (Fig. 4). As already noted, this is not the case for most other Asp residues (Fig. 4).
It should be emphasized that the simulations described above were done to obtain qualitative insights into the reasons why EP maps do not look like ED maps. Anyone intent using the structure of a macromolecule to calculate an EP map that accurately corresponds to that molecule's experimental EP map must address a host of issues24 that have been ignored here, e.g.,: (1) the effects of bonding on atomic structure factors, which were mentioned earlier, (2) the distributions of partial and full charges within residues, (3) the impact of counter‐ions, and solvent structure on electric potential distributions, (4) the variations in dielectric constant within and around the molecule of interest, and (5) the effects of polarizability on charge distributions. These are all formidable problems, but comparisons between the EP and ED maps of macromolecules may offer experimental insights into them that are of great interest.
In conclusion, recent advances in cryo‐EM technology are certain to result in a flood of new atomic‐resolution structures for macromolecules. The purpose of this report is to emphasize the importance of taking charge into account in the interpretation of the high‐resolution EP maps that are now being produced in abundance.
Methods
The G2607:C2443 Watson‐Crick base pair was taken from the 5AFI atomic model,5 placed in P1 unit cell with a = b = c = 25 Å, and α = β = γ = 90°, and atomic B‐factors were set at 20.00 Å2. The structure factors were calculated by Fourier transformation of X‐ray or electron atomic scattering factors22, 23 for all atoms j = 1, …, N,
| (Eq. 1) |
where X is fractional coordinate vectors, H is miller index vector of the reciprocal P1 lattice, f j is atomic scattering factor for j‐th atoms either for electron or X‐ray radiation, and B j is the atomic B‐factor. In some instances the non‐bridging oxygen atoms OP1 and OP2 of the backbone phosphate groups of RNA molecules were treated as carrying half a negative charge each. In this case, the structure factor for each oxygen was a 50:50 blend of the electron scattering factors for O− 1 and neutral O.
EP maps or ED maps were calculated using the CCP package29 and examined using both Coot30 and Pymol,31 and figures were made using Pymol. The root‐mean‐square deviations used for contouring maps were those provided by the program Pymol, which is known to differ slightly with some other programs in this regard under certain conditions. The absolute values of root‐mean‐square deviations may vary significantly with resolution in both EP and ED maps. One should focus on relative contour levels.
Conflict of Interest Statement
The authors declare no conflict of interest in publishing results of this study.
Supporting information
Supporting Information.
Acknowledgment
The authors thank Drs. R. Henderson, F. Sigworth, and Y. Xiong for insightful discussion during this study, and Mr. G. Banerjee and Ms. I. Ghosh for bringing this subject to our attention. This work was in part supported by National Institutes of Health Grant P01 GM022778.
References
- 1. Henderson R (2015) Overview and future of single particle electron cryomicroscopy. Arch Biochem Biophys 581:19–24. [DOI] [PubMed] [Google Scholar]
- 2. Subramaniam S, Kuhlbrandt W, Henderson R (2016) CryoEM at IUCrJ: a new era. IUCr J 3:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Subramaniam S, Earl LA, Falconieri V, Milne JL, Egelman EH (2016) Resolution advances in cryo‐EM enable application to drug discovery. Curr Opin Struct Biol 41:194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shalev‐Benami M, Zhang Y, Matzov D, Halfon Y, Zackay A, Rozenberg H, Zimmerman E, Bashan A, Jaffe CL, Yonath A, Skiniotis G (2016) 2.8‐A cryo‐EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania. Cell Rep 16:288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H (2015) Structure of the E. coli ribosome‐EF‐Tu complex at <3 A resolution by Cs‐corrected cryo‐EM. Nature 520:567–570. [DOI] [PubMed] [Google Scholar]
- 6. Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JL, Subramaniam S (2015) 2.2 A resolution cryo‐EM structure of beta‐galactosidase in complex with a cell‐permeant inhibitor. Science 348:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JL, Subramaniam S (2016) Breaking cryo‐EM resolution barriers to facilitate drug discovery. Cell 165:1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nogales E (2016) The development of cryo‐EM into a mainstream structural biology technique. Nat Meth 13:24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glaeser RM (2016) How good can cryo‐EM become? Nat Meth 13:28–32. [DOI] [PubMed] [Google Scholar]
- 10. Mott NF (1930) The scattering of electrons by atoms. Proc Royal Soc Lond A127:658–665. [Google Scholar]
- 11. James RW (1930) X‐ray crystallography, the fifth edition, Methuen's Monographs on Physical Subjects. Bell & Son's, London. [Google Scholar]
- 12. Chang S, Head‐Gordon T, Glaeser RM, Downing KH (1999) Chemical bonding effects in the determination of protein structures by electron crystallography. Acta Cryst A55:305–313. [DOI] [PubMed] [Google Scholar]
- 13. Koritsanszky T, Flaig R, Zobel D, Krane H, Morgenroth W, Luger P (1998) Accurate experimental electronic properties of dl‐proline monohydrate obtained within 1 day. Science 279:356–358. [DOI] [PubMed] [Google Scholar]
- 14. Yonekura K, Kato K, Ogasawara M, Tomita M, Toyoshima C (2015) Electron crystallography of ultrathin 3D protein crystals: atomic model with charges. Proc Natl Acad Sci USA 112:3368–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moore P (2012) Visualizing the invisible: imaging techniques for the structural biologist. Oxford; New York: Oxford University Press. [Google Scholar]
- 16. Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y (1997) Surface of bacteriorhodopsin revealed by high‐resolution electron crystallography. Nature 389:206–211. [DOI] [PubMed] [Google Scholar]
- 17. Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y (1999) The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. J Mol Biol 286:861–882. [DOI] [PubMed] [Google Scholar]
- 18. Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T (2005) Lipid‐protein interactions in double‐layered two‐dimensional AQP0 crystals. Nature 438:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonen T, Sliz P, Kistler J, Cheng Y, Walz T (2004) Aquaporin‐0 membrane junctions reveal the structure of a closed water pore. Nature 429:193–197. [DOI] [PubMed] [Google Scholar]
- 20. Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH (2011) Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Srinivasan AR, Sauers RR, Fenley MO, Boschitsch AH, Matsumoto A, Colasanti AV, Olson WK (2009) Properties of the nucleic‐acid bases in free and Watson‐Crick hydrogen‐bonded states: Computational insights into the sequence‐dependent features of double‐helical DNA. Biophys Rev 1:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prince E (2004) International Tables for Crystallography. Volume C., London, U.K: Kluwer Academic Publishers. [Google Scholar]
- 23. Peng LM (1999) Electron atomic scattering factors and scattering potentials of crystals. Micron 30:625–648. [Google Scholar]
- 24. Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R (1996) Electron‐crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol 259:393–421. [DOI] [PubMed] [Google Scholar]
- 25. Kuhlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light‐harvesting complex by electron crystallography. Nature 367:614–621. [DOI] [PubMed] [Google Scholar]
- 26. Ravelli RB, McSweeney SM (2000) The 'fingerprint' that X‐rays can leave on structures. Structure 8:315–328. [DOI] [PubMed] [Google Scholar]
- 27. Weik M, Ravelli RB, Kryger G, McSweeney S, Raves ML, Harel M, Gros P, Silman I, Kroon J, Sussman JL (2000) Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci USA 97:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayward SB, Glaeser RM (1979) Radiation damage of purple membrane at low temperature. Ultramicroscopy 04:201–210. [DOI] [PubMed] [Google Scholar]
- 29. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Cryst D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Cryst D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 31. DeLano WL (2002) The PyMOL Molecular Graphics System. http://www.pymol.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.


